Summary
Electrocardiogram (ECG) is a widely used reliable, non-invasive approach for cardiovascular disease diagnosis. With the rapid growth of ECG examinations and the insufficiency of cardiologists, accurate and automatic diagnosis of ECG signals has become a hot research topic. In this paper, we developed a deep neural network for automatic classification of cardiac arrhythmias from 12-lead ECG recordings. Experiments on a public 12-lead ECG dataset showed the effectiveness of our method. The proposed model achieved an average F1 score of 0.813. The deep model showed superior performance than 4 machine learning methods learned from extracted expert features. Besides, the deep models trained on single-lead ECGs produce lower performance than using all 12 leads simultaneously. The best-performing leads are lead I, aVR, and V5 among 12 leads. Finally, we employed the SHapley Additive exPlanations method to interpret the model's behavior at both the patient level and population level.
Subject areas: Medicine, Clinical Finding, Artificial Intelligence
Graphical abstract
Highlights
-
•
We develop a deep learning model for the automatic diagnosis of ECG
-
•
We present benchmark results of 12-lead ECG classification
-
•
We find out the top performance single lead in diagnosing ECGs
-
•
We employ the SHAP method to enhance clinical interpretability
Medicine; Clinical Finding; Artificial Intelligence
Introduction
Cardiovascular diseases (CVDs) are the leading cause of death and produce immense health and economic burdens in the United States and globally (Virani et al., 2020). The electrocardiogram (ECG) is a simple, reliable, and non-invasive approach for monitoring patients' heart activity and diagnosing cardiac arrhythmias. A standard ECG has 12 leads including 6 limb leads (I, II, III, aVR, aVL, aVF) and 6 chest leads (V1, V2, V3, V4, V5, V6) recorded from electrodes on the body surface. Accurately interpreting the ECG for a patient with concurrent cardiac arrhythmias is challenging even for an experienced cardiologist, and incorrectly interpreted ECGs might result in inappropriate clinical decisions or lead to adverse outcomes (Bogun et al., 2004).
An estimated 300 million ECGs are recorded worldwide annually (Holst et al., 1999) and keep growing. Computer-aided interpretation of ECGs has become more important, especially in low-income and middle-income countries where experienced cardiologists are scarce (World Health Organization, 2014). Therefore, accurate and automatic diagnosis of ECG signals has become a hot research interest. In past decades, automatic diagnosis of ECGs has been widely investigated with the availability of large open-source ECG datasets such as MIT-BIH Arrhythmia Database (Moody and Mark, 2001), 2017 Physionet Challenge/CinC dataset (Clifford et al., 2017), 2018 China Physiological Signal Challenge dataset (CPSC2018) (Liu et al., 2018a), PTB-XL database (Wagner et al., 2020).
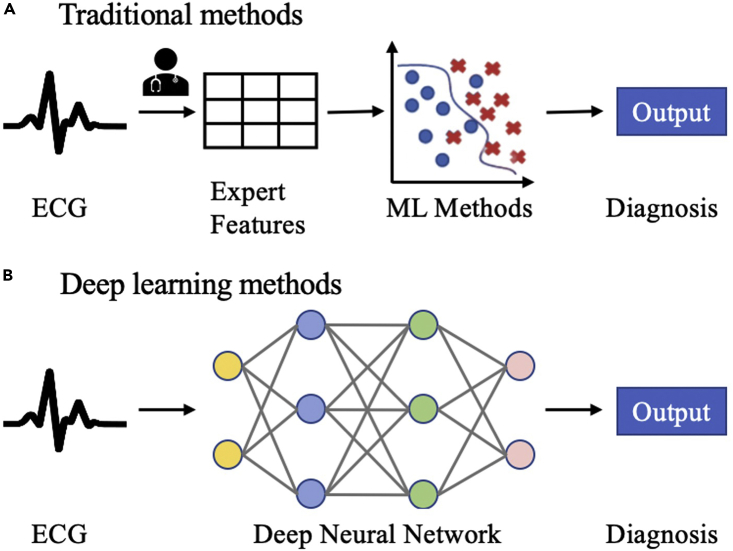
Existing models for automatic diagnosis of ECG abnormalities can be classified into two categories: traditional methods and deep learning methods. The comparison between traditional methods and deep learning methods is demonstrated in Figure 1. Traditional methods based on machine learning (ML) algorithms are of two stages; these methods require experts to engineer useful features or extract features using signal processing techniques first and then use these features to build ML classifiers (Jambukia et al., 2015, Macfarlane et al., 2005). The University of Glasgow ECG analysis program applied rule-based criteria on signal processing features and medical features for the diagnosis of ECGs (Macfarlane et al., 2005). The use of wavelet coefficients for the classification of ECGs has been investigated in (De Chazal et al., 2000). Detta et al. developed a feature-oriented method with a two-layer cascaded binary classifier and achieved the best performance in the 2017 Physionet/CinC Challenge for atrial fibrillation classification from single-lead ECGs (Datta et al., 2017).
Figure 1.
Comparison of existing models for automatic diagnosis of ECG abnormalities
(A) Two-stage traditional methods using feature engineering; (B) end-to-end deep learning methods.
However, traditional methods are limited by data quality and domain knowledge. Additional effort is required to extract expert features. The second approach is using end-to-end deep learning techniques that do not require an explicit feature extraction. Deep learning methods have made great progress in many areas (LeCun et al., 2015) such as computer vision, speech recognition, and natural language processing since 2012. Many studies have also demonstrated promising results of deep learning in the healthcare domain such as complex diagnostics spanning dermatology, radiology, ophthalmology, and pathology (Esteva et al., 2019). Recently, deep learning models have been applied to ECG data for various tasks including disease detection, annotation or localization, sleep staging, biometric human identification, denoising, and so on (Hong et al., 2020). Deep neural networks have shown initial success in cardiac diagnosis from single-lead or multi-lead ECGs (Chen et al., 2020, Datta et al., 2017, Hannun et al., 2019, He et al., 2019, Strodthoff et al., 2020, Zhu et al., 2020). A deep learning model trained on a large single-lead ECG dataset with 91,232 ECG recordings shows superior performance than cardiologists for diagnosing 12 rhythm classes (Hannun et al., 2019). Ullah et al. transformed the 1D ECG time series into a 2D spectral image through short-time Fourier transform and trained a deep learning model to classify cardiac arrhythmias (Ullah et al., 2020). Twelve-lead ECGs are the standard techniques in realistic clinical settings and can provide more valuable information compared to single-lead ECGs. Chen et al. proposed an artificial neural network that combined convolutional neural networks (CNNs), recurrent neural networks (RNNs), and attention mechanism for cardiac arrhythmias detection and won first place in the 2018 China Physiological Signal Challenge (Chen et al., 2020). Zhu et al. applied a deep learning algorithm to 12-lead ECGs to diagnosis 20 types of cardiac abnormalities, and the model performance exceeded physicians trained in ECG interpretation (Zhu et al., 2020). Besides, some studies (Hong et al., 2017, Liu et al., 2018b) showed that the performance of neural networks can be significantly improved by incorporating expert features. Despite the promising performance of deep learning models on cardiac arrhythmias diagnosis, deep learning models usually operate as black boxes, and understanding the model's behavior on making decisions is important and challenging.
In this study, we developed a deep neural network based on 1D CNNs for automatic multi-label classification of cardiac arrhythmias in 12-lead ECG recordings, and the model achieved comparable state-of-the-art performance (average F1 score is 0.813) on the CPSC2018 dataset. We also conducted experiments on single-lead ECGs and showed the performance of every single lead. In addition, we applied the SHapley Additive exPlanations (SHAP) method (Lundberg and Lee, 2017) to interpret the model's predictions at both the patient level and population level. SHAP is a game-theoretic approach to explain the model predictions and has been applied to tree-based algorithms to enhance clinical interpretability (Lundberg et al., 2020; Li et al., 2020).
To summarize, the contributions of our work are as follows:
-
•
We developed a deep neural network for automatic diagnosis of cardiac arrhythmias and the model achieved comparable state-of-the-art performance (Chen et al., 2020) on the CPSC2018 dataset.
-
•
We compared the performance of the proposed model with 4 machine learning classifiers and 3 deep learning classifiers. The result showed that the proposed model outperformed all baseline classifiers.
-
•
We conducted experiments on single-lead ECGs and the results suggested the F1 score, averaged across diagnostic classes, of the deep model trained on single-lead ECGs is 4.4%–11.8% lower than using all 12 leads, and the top-performing single leads are lead I, aVR, and V5.
-
•
To better understand the model's behavior, we employed the SHAP method to enhance clinical interpretability at both the patient level and population level.
Results and discussion
Experiment setup
Study design
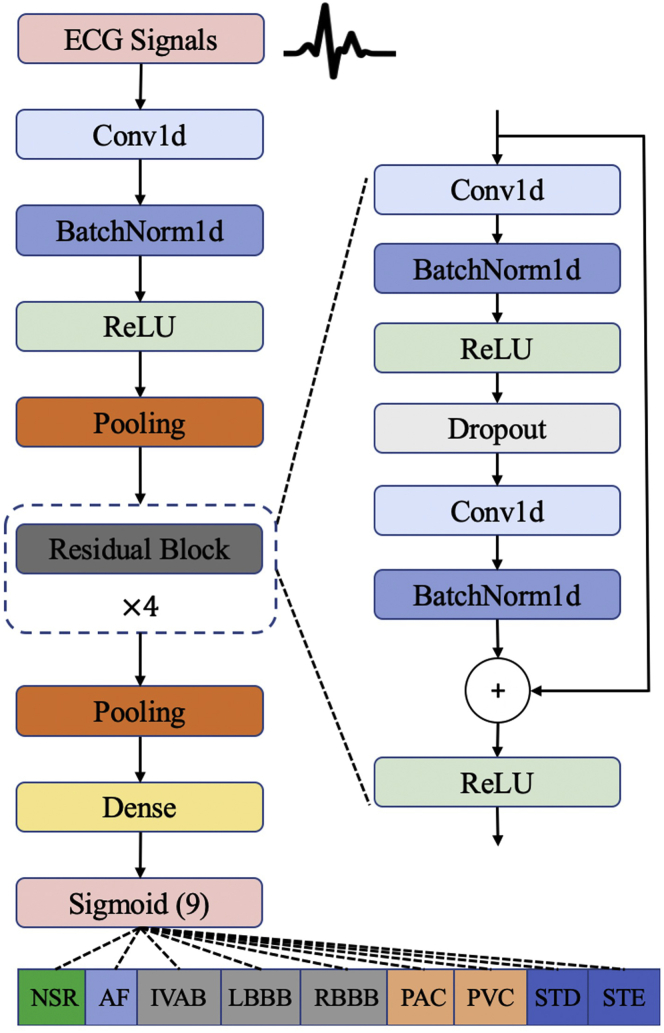
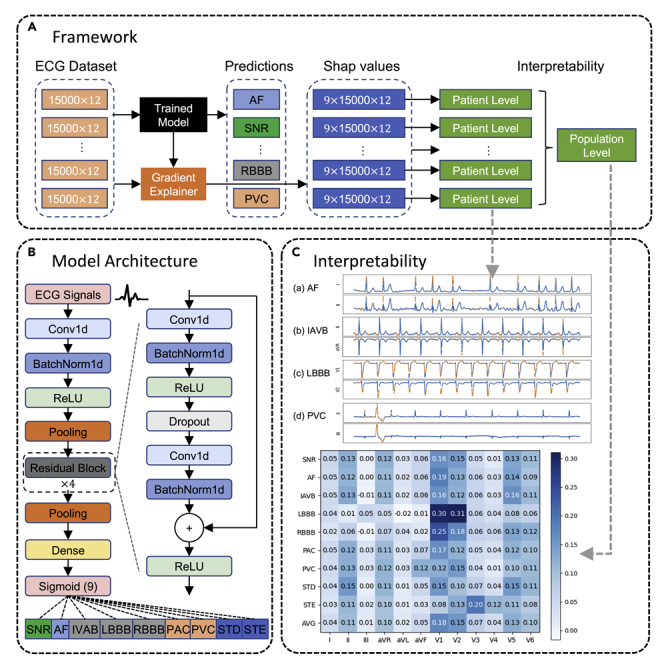
In this study, we aim to develop a deep learning model for automatic diagnosis of 12-lead ECG with 9 cardiac arrhythmias (CA) types: normal sinus rhythm (SNR), atrial fibrillation (AF), first-degree atrioventricular block (IAVB), left bundle branch block (LBBB), right bundle branch block (RBBB), premature atrial contraction (PAC), premature ventricular contraction (PVC), ST-segment depression (STD), and ST-segment elevation (STE). An example of 12-lead ECG for a patient with AF from the CPSC2018 dataset is shown in Figure S1. Patient characteristics and diagnostic class prevalence on the CPSC2018 dataset are reported in Table S1. The overview of the proposed network architecture is illustrated in Figure 2. Our proposed deep neural network accepts raw ECG inputs (12 leads, duration of 30 s, sampling rate of 500 Hz), utilizes 1D CNNs to extract deep features, and outputs the prediction results for 9 diagnostic classes.
Figure 2.
Deep neural network architecture for cardiac arrhythima diagnosis
Our deep neural network accepts raw ECG inputs (12 leads, duration of 30 s, sampling rate of 500 Hz), utilizes 1D CNNs to extract deep features, and outputs the prediction results for 9 diagnostic classes.
Twelve-lead model performance
Precision, recall, F1 score, AUC, and accuracy of the model's prediction on each cardiac arrhythmia on the test data set of 10 rounds are averaged and reported in Table 1. Overall, average AUC and accuracy of the deep learning model both exceeded 0.95, and the average F1 score was 0.813 with an average precision of 0.821 and an average recall of 0.812. Among all cardiac arrhythmias, the deep model performed best on AF and RBBB classification with an F1 score of over 0.9. However, we also observed the F1 score of STE is low as 0.535 which may be due to the significant physician disagreement in diagnosing STE from ECGs (McCabe et al., 2013).
Table 1.
Twelve-lead model performance averaged on 10-fold tests
| CA type | Precision | Recall | F1 | AUC | Accuracy |
|---|---|---|---|---|---|
| SNR | 0.814 | 0.800 | 0.805 | 0.974 | 0.948 |
| AF | 0.920 | 0.918 | 0.919 | 0.988 | 0.971 |
| IAVB | 0.868 | 0.865 | 0.864 | 0.987 | 0.974 |
| LBBB | 0.844 | 0.894 | 0.866 | 0.980 | 0.991 |
| RBBB | 0.911 | 0.942 | 0.926 | 0.987 | 0.959 |
| PAC | 0.756 | 0.720 | 0.735 | 0.949 | 0.952 |
| PVC | 0.869 | 0.839 | 0.851 | 0.976 | 0.971 |
| STD | 0.808 | 0.826 | 0.814 | 0.971 | 0.953 |
| STE | 0.603 | 0.504 | 0.535 | 0.923 | 0.974 |
| AVG | 0.821 | 0.812 | 0.813 | 0.970 | 0.966 |
To illustrate why the model is working or not working on specific examples of cardiac arrhythmias, we selected the best validation model of 10 rounds and used the confusion matrices calculated on the test data set. The confusion matrices are shown in Figure S2. Low false-negative rate and high true-negative rate were observed for all 9 classes as shown in Figure S2. For the diagnosis of AF, RBBB, and PVC, low false-positive rate and false-negative rate were observed. However, the confusion matrices showed that the model had trouble in classifying PAC, STD, and STE with a high false-negative rate. Besides, we adopted an ablation study to measure the effectiveness of data augmentation. By applying scaling and shifting during the training phase, performance on the test data set improved 1.9% and the average F1 score increases from 0.794 to 0.813. In order to estimate the statistical significance of the differences, we also applied statistical t test and observed a significant p value (i.e., ).
Comparison with baseline models
Inspired by (De Chazal et al., 2000) and (Liu et al., 2018b), we built several machine learning models with extracted expert features. To be specific, we extracted 2 types of expert features: (1) statistical features (e.g., mean, standard deviation, variance, and percentile) of raw ECG input and (2) statistics and Shannon entropy of signal processing features extracted by applying discrete wavelet decomposition. Statistical features and signal processing features are concatenated and input to machine learning classifiers. For machine learning classifiers, we considered logistic regression, random forest (RF), gradient boosting trees (GBT), and multi-layer perceptron. Besides, we considered the following 3 neural networks for time series classification as deep learning baselines:
-
•
Long short-term memory (Hochreiter and Schmidhuber, 1997) is a variant of RNN which is designed for time series processing.
-
•
Time-incremental CNN (Yao et al., 2018) combines the feature extraction ability of CNN and RNN's ability to effectively learn from time series.
-
•
InceptionTime (Inception) (Fawaz et al., 2020) is an ensemble model based on the CNN applied to time series classification.
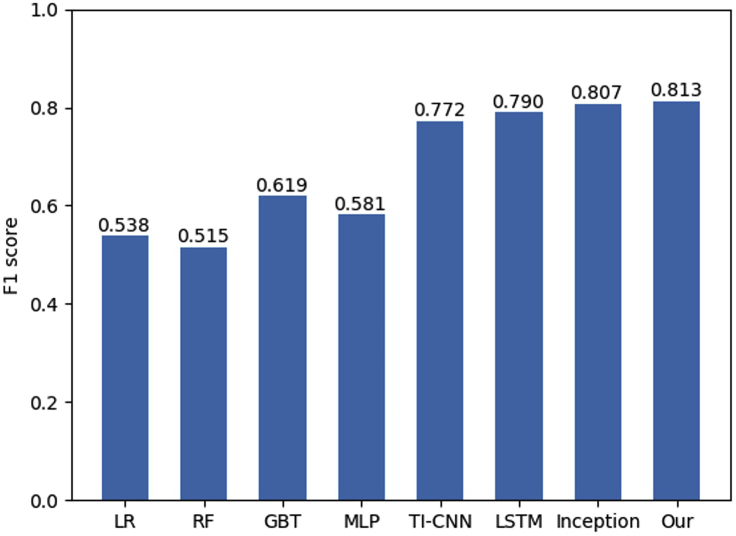
The comparison of model performance (F1 score) is shown in Figure 3. Our proposed model achieved the best performance compared to other methods. Inception showed slightly poorer performance compared to our model. Among 4 machine learning models, GBT achieved the best average F1 score of 0.619, while RF performed worst with an average F1 score of 0.515. As shown in Figure 3, it is apparent that the end-to-end deep learning model with deep features showed significant accuracy improvement compared to machine learning models. Among 8 methods, our deep learning model achieved the best performance with an average F1 score of 0.813.
Figure 3.
F1 score comparison of machine learning models and end-to-end deep learning models
Single-lead model performance
We modified the input layer of the deep neural network and trained the model on single-lead ECG inputs . Comparison of single-lead model performance measured by F1 score is summarized in Table 2. From Table 2, we observed the following: (1) in summary, the single-lead model showed inferior performance compared to using all 12 leads simultaneously. On average, the performance of the deep learning model trained on single-lead ECGs dropped by 4.4%–11.8% compared to using all 12 leads. (2) Among 12 leads, lead I, aVR, and V5 are the top-performing single leads with an F1 score of more than 0.765, and lead aVL is shown to perform worst with an average F1 score of 0.695. (3) All single leads achieved good performance on AF classification with an F1 score of over 0.9. Lead II, aVR showed the comparable best performance in the diagnosis of AF. (4) The F1 score (0.94) on RBBB classification obtained using lead V1 is significantly higher than that using any other leads which means V1 plays an important role in diagnosing RBBB. (5) The best predictive single lead for LBBB is lead I. (6) Lead I used by Apple Watch and lead II favored by cardiologists for quick review also showed very good performance on average. (7) Interestingly, although the 12-lead model achieved comparable or better performance than single-lead models for most diagnostic classes, lead I for LBBB and lead V1 for RBBB showed superior performance. We speculate that unexpected feature interactions may hurt the performance of the 12-lead model. (8) The results identified lead aVR as a useful lead in ECG interpretation while ECG interpretation mostly ignores this lead historically (Gorgels et al., 2001).
Table 2.
Comparison of single-lead model performance measured by F1 score
| CA type | I | II | III | aVR | aVL | aVF | V1 | V2 | V3 | V4 | V5 | V6 | All |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNR | 0.705 | 0.682 | 0.602 | 0.712 | 0.604 | 0.663 | 0.657 | 0.694 | 0.710 | 0.717 | 0.731 | 0.721 | 0.805 |
| AF | 0.914 | 0.927 | 0.911 | 0.929 | 0.913 | 0.908 | 0.924 | 0.913 | 0.915 | 0.922 | 0.910 | 0.905 | 0.919 |
| IAVB | 0.843 | 0.853 | 0.818 | 0.842 | 0.808 | 0.830 | 0.860 | 0.866 | 0.866 | 0.816 | 0.842 | 0.840 | 0.864 |
| LBBB | 0.897 | 0.778 | 0.783 | 0.825 | 0.802 | 0.737 | 0.860 | 0.860 | 0.804 | 0.759 | 0.813 | 0.789 | 0.866 |
| RBBB | 0.859 | 0.802 | 0.804 | 0.845 | 0.815 | 0.796 | 0.940 | 0.886 | 0.852 | 0.828 | 0.827 | 0.840 | 0.926 |
| PAC | 0.723 | 0.737 | 0.709 | 0.688 | 0.698 | 0.719 | 0.730 | 0.689 | 0.692 | 0.680 | 0.715 | 0.702 | 0.735 |
| PVC | 0.813 | 0.821 | 0.846 | 0.818 | 0.792 | 0.836 | 0.788 | 0.842 | 0.835 | 0.838 | 0.818 | 0.809 | 0.851 |
| STD | 0.695 | 0.790 | 0.627 | 0.793 | 0.573 | 0.711 | 0.615 | 0.652 | 0.702 | 0.753 | 0.781 | 0.757 | 0.814 |
| STE | 0.433 | 0.406 | 0.312 | 0.435 | 0.251 | 0.338 | 0.293 | 0.417 | 0.477 | 0.552 | 0.485 | 0.497 | 0.535 |
| AVG | 0.765 | 0.755 | 0.712 | 0.765 | 0.695 | 0.726 | 0.741 | 0.758 | 0.762 | 0.763 | 0.769 | 0.762 | 0.813 |
Model interpretability
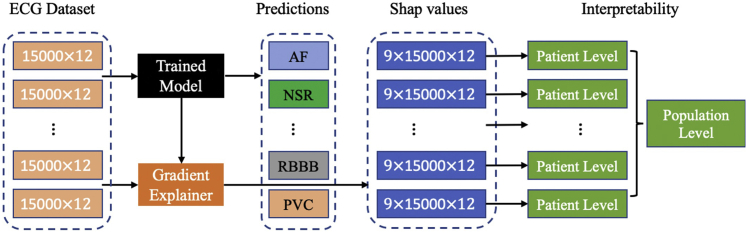
Model interpretability of deep neural networks has been a common challenge and limiting factor toward real-world applications. In addition to the promising performance achieved by our deep model in diagnosing cardiac arrhythmias, the SHAP method was used to explain model predictions. As shown in Figure 4, we demonstrated the model interpretability at both the patient level and population level through visualizations.
Figure 4.
Interpretability of the deep learning model at both the patient level and population level using SHAP values
Patient-level interpretation
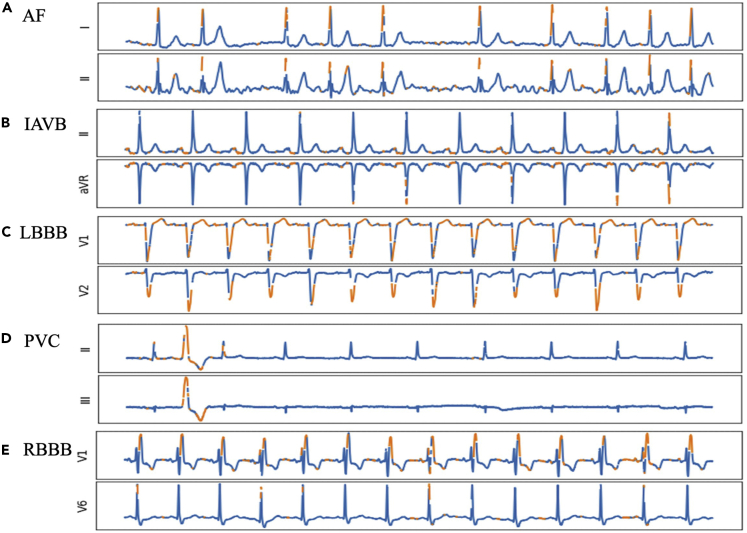
For each ECG input with the top-predicted cardiac arrhythmia class , we visualized the SHAP value matrix along with the raw ECG input matrix . The explanations of the model's prediction results for several ECG instances from different patients are shown in Figure 5. Figure 5A shows the model's identification of irregular QRS complexes (combinations of Q, R, S waves seen on a typical ECG) with the lack of P waves as a classic example of AF. This observation is consistent with the diagnostic criteria of AF (Gutierrez and Blanchard, 2011). In Figure 5B with IAVB, highlighted features show increased PR intervals (periods that extends from the beginning of the P waves until the beginning of the QRS complexes) which are used for the diagnosis of IAVB (Barold et al., 2006). Figure 5D shows a typical example of PVC. PVC happens in some sporadic periods in the ECGs, and only the period where PVC occurs is highlighted in Figure 5D which is reasonable. Figure 5C shows the model's identification of deep S waves in lead V1 for LBBB. Typically, RBBB is detected with an RSR′ QRS complex in lead V1 as shown in Figure 5E. Observations from Figures 5C and 5E are compatible with the corresponding diagnostic criteria for LBBB and RBBB (Alventosa-Zaidin et al., 2019; Goldberger et al., 2017). More interpretation results can be found in Figure S3. After reviewing the model's predicted findings with a clinician (S.Y. in the authorship), the characteristics of the ECG associated with the diagnoses were consistent with standard ECG interpretation.
Figure 5.
Explanation of the model's prediction results for several ECG instances from different patients
The features with high contribution (i.e., SHAP values) are highlighted in orange. Only the last 10 s of top 2 influential leads are displayed due to the limited space.
However, the deep learning model could make wrong predictions, and the SHAP method could learn wrong interpretations. To show this, we picked some failed cases, and the discussions are shown in Figure S4.
Population-level interpretation
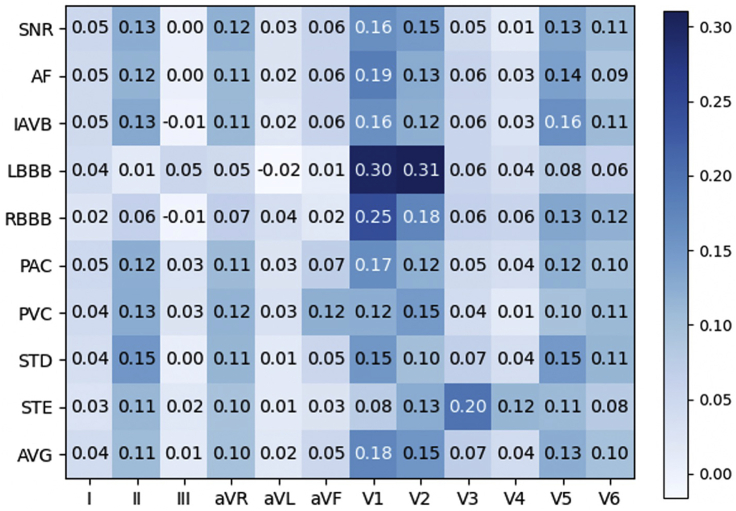
Because SHAP values are directly additive, we calculated the contribution rate of ECG leads toward each diagnosis class, which is utilized for population-level interpretation of the deep learning model as shown in Figure 4. Figure 6 demonstrates the contribution rate of ECG leads toward diagnostic classes in the 12-lead deep model. Diagnosing AF and IAVB requires visualizing P waves and the PR intervals. These findings can be seen on many leads but are best seen in leads II and V1. This is confirmed by the model's ranking of these leads of importance for the identification of these rhythms. The model's ranking of V5's importance raises the question about whether or not clinicians should look at this lead to improve ECG interpretation. LBBB's and RBBB's hallmark feature is the deep S waves in V1 and RSR′ complexes in V1, respectively. The model's identification of the importance of this lead in LBBB and RBBB is consistent with standard ECG interpretations. STD and STE are seen in an acute coronary syndrome where a region of the heart is suffering from poor oxygenation. Depending on the affected areas, STE and STD can occur in a variety of leads as seen in the distribution of the model rankings. From the average perspective, lead II, aVR, V1, V2, V5, and V6 are the most important leads in the 12-lead model. We also observe some leads (III, aVL) are associated with a low contribution rate which means these leads are possibly neglected in the 12-lead ECG model. This may be because of feature interactions among ECG leads (e.g., lead III is the difference between lead II and lead I).
Figure 6.
Population-level interpretation by calculating the contribution rate of ECG leads toward diagnostic classes in the 12-lead deep model
Limitations of the study
In this paper, we developed a deep neural network for automatic diagnosis of cardiac arrhythmias from 12-lead ECG recordings. The proposed model achieved state-of-the-art performance on the CPSC2018 dataset and employed the SHAP method to enhance clinical interpretability. However, model generalization to patients of different races should be further validated since the CPSC2018 dataset is entirely collected from China hospitals. Secondly, adversarial samples can lead to misbehaviors of deep learning models. It is crucial to test the model's robustness, protect from adversarial attacks, and avoid overoptimistic of the model. Besides, there is no objective gold standard for ECG interpretation. What combination of ECG leads could achieve better performance remains unexplored.
Resource availability
Lead contact
Ping Zhang, PhD, zhang.10631@osu.edu.
Materials availability
This study did not generate any new materials.
Data and code availability
The 12-lead ECG data set used in this study is the CPSC2018 training dataset which is released by the first China Physiological Signal Challenge (CPSC) 2018 during the seventh International Conference on Biomedical Engineering and Biotechnology. Details of the CPSC2018 dataset can be found at http://2018.icbeb.org/Challenge.html. The source code is provided and is available at https://github.com/onlyzdd/ecg-diagnosis.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
This work was funded in part by the National Science Foundation (CBET-2037398 for P.Z.).
Author contributions
Conceptualization, P.Z.; methodology, D.Z. and P.Z.; software and investigation, D.Z.; domain knowledge and validation, S.Y.; formal analysis, D.Z., X.Y., and P.Z.; writing – original draft, D.Z. and P.Z.; writing – review & editing, D.Z., S.Y., X.Y., and P.Z.; supervision, P.Z.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: April 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102373.
Supplemental information
References
- Alventosa-Zaidin M., Guix Font L., Benitez Camps M., Roca Saumell C., Pera G., Alzamora Sas M.T., Forés Raurell R., Rebagliato Nadal O., Dalfó-Baqué A., Brugada Terradellas J. Right bundle branch block: prevalence, incidence, and cardiovascular morbidity and mortality in the general population. Eur. J. Gen. Pract. 2019;25:109–115. doi: 10.1080/13814788.2019.1639667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barold S.S., Ilercil A., Leonelli F., Herweg B. First-degree atrioventricular block. J. Interv. Card. Electrophysiol. 2006;17:139–152. doi: 10.1007/s10840-006-9065-x. [DOI] [PubMed] [Google Scholar]
- Bogun F., Anh D., Kalahasty G., Wissner E., Serhal C.B., Bazzi R., Weaver W.D., Schuger C. Misdiagnosis of atrial fibrillation and its clinical consequences. Am. J. Med. 2004;117:636–642. doi: 10.1016/j.amjmed.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Chen T.M., Huang C.H., Shih E.S., Hu Y.F., Hwang M.J. Detection and classification of cardiac arrhythmias by a challenge-best deep learning neural network model. Iscience. 2020;23:100886. doi: 10.1016/j.isci.2020.100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford G.D., Liu C., Moody B., Li-wei H.L., Silva I., Li Q., Johnson A., Mark R.G. AF classification from a short single lead ECG recording: The PhysioNet/computing in cardiology challenge 2017. Comput Cardiol. 2017 doi: 10.22489/CinC.2017.065-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Puri C., Mukherjee A., Banerjee R., Choudhury A.D., Singh R., Ukil A., Bandyopadhyay S., Pal A., Khandelwal S. Identifying normal, AF and other abnormal ECG rhythms using a cascaded binary classifier. Comput Cardiol. 2017 doi: 10.22489/CinC.2017.173-154. [DOI] [Google Scholar]
- De Chazal P., Celler B., Reilly R. Using wavelet coefficients for the classification of the electrocardiogram. Proceedings of the 22nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (Cat. No. 00CH37143) 2000 doi: 10.1109/IEMBS.2000.900669. [DOI] [Google Scholar]
- Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- Fawaz H.I., Lucas B., Forestier G., Pelletier C., Schmidt D.F., Weber J., Webb G.I., Idoumghar L., Muller P.-A., Petitjean F. Inceptiontime: finding alexnet for time series classification. Data Mining Knowledge Discov. 2020;34:1936–1962. doi: 10.1007/s10618-020-00710-y. [DOI] [Google Scholar]
- Goldberger A.L., Goldberger Z.D., Shvilkin A. Elsevier Health Sciences; 2017. Clinical Electrocardiography: A Simplified Approach E-Book. [Google Scholar]
- Gorgels A.P., Engelen D., Wellens H.J. Lead aVR, a mostly ignored but very valuable lead in clinical electrocardiography∗. J. Am. Coll. Cardiol. 2001;38:1355–1356. doi: 10.1016/S0735-1097(01)01564-9. [DOI] [PubMed] [Google Scholar]
- Gutierrez C., Blanchard D.G. Atrial fibrillation: diagnosis and treatment. Am. Fam. Phys. 2011;83:61–68. [PubMed] [Google Scholar]
- Hannun A.Y., Rajpurkar P., Haghpanahi M., Tison G.H., Bourn C., Turakhia M.P., Ng A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019;25:65. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Liu Y., Wang K., Zhao N., Yuan Y., Li Q., Zhang H. Automatic cardiac arrhythmia classification using combination of deep residual network and bidirectional LSTM. IEEE Access. 2019;7:102119–102135. doi: 10.1109/ACCESS.2019.2931500. [DOI] [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Holst H., Ohlsson M., Peterson C., Edenbrandt L. A confident decision support system for interpreting electrocardiograms. Clin. Physiol. 1999;19:410–418. doi: 10.1046/j.1365-2281.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- Hong S., Wu M., Zhou Y., Wang Q., Shang J., Li H., Xie J. ENCASE: An ENsemble ClASsifiEr for ECG classification using expert features and deep neural networks. Comput Cardiol. 2017 doi: 10.22489/CinC.2017.178-245. [DOI] [Google Scholar]
- Hong S., Zhou Y., Shang J., Xiao C., Sun J. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput. Biol. Med. 2020:103801. doi: 10.1016/j.compbiomed.2020.103801. [DOI] [PubMed] [Google Scholar]
- Jambukia S.H., Dabhi V.K., Prajapati H.B. Classification of ECG signals using machine learning techniques: a survey. 015 International Conference on Advances in Computer Engineering and Applications. 2015 doi: 10.1109/ICACEA.2015.7164783. [DOI] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li X., Xu X., Xie F., Xu X., Sun Y., Liu X., Jia X., Kang Y., Xie L., Wang F. A time-phased machine learning model for real-time prediction of sepsis in critical care. Crit. Care Med. 2020 doi: 10.1097/CCM.0000000000004494. [DOI] [PubMed] [Google Scholar]
- Liu F., Liu C., Zhao L., Zhang X., Wu X., Xu X., Liu Y., Ma C., Wei S., He Z. An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J. Med. Imaging Health Inform. 2018;8:1368–1373. doi: 10.1166/jmihi.2018.2442. [DOI] [Google Scholar]
- Liu Z., Meng X., Cui J., Huang Z., Wu J. IEEE; 2018. Automatic identification of abnormalities in 12-lead ecgs using expert features and convolutional neural networks; pp. 163–167. (2018 International Conference on Sensor Networks and Signal Processing (SNSP)). [DOI] [Google Scholar]
- Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intelligence. 2020;2:2522–5839. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg S.M., Lee S.I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017:4765–4774. [Google Scholar]
- Macfarlane P., Devine B., Clark E. The university of Glasgow (Uni-G) ECG analysis program. Comput Cardiol. 2005:451–454. doi: 10.1109/CIC.2005.1588134. [DOI] [Google Scholar]
- McCabe J.M., Armstrong E.J., Ku I., Kulkarni A., Hoffmayer K.S., Bhave P.D., Waldo S.W., Hsue P., Stein J.C., Marcus G.M. Physician accuracy in interpreting potential ST-segment elevation myocardial infarction electrocardiograms. J. Am. Heart Assoc. 2013;2:e000268. doi: 10.1161/JAHA.113.000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody G.B., Mark R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001;20:45–50. doi: 10.1109/51.932724. [DOI] [PubMed] [Google Scholar]
- Strodthoff N., Wagner P., Schaeffter T., Samek W. Deep learning for ECG analysis: benchmarks and insights from PTB-XL. arXiv. 2020 doi: 10.1109/jbhi.2020.3022989. [DOI] [PubMed] [Google Scholar]
- Ullah A., Anwar S.M., Bilal M., Mehmood R.M. Classification of arrhythmia by using deep learning with 2-D ECG spectral image representation. Remote Sens. 2020;12:1685. doi: 10.3390/rs12101685. [DOI] [Google Scholar]
- Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N. Heart disease and stroke statistics—2020 update: a report from the American Heart Association. Circulation. 2020:E139–E596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Wagner P., Strodthoff N., Bousseljot R.D., Kreiseler D., Lunze F.I., Samek W., Schaeffter T. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data. 2020;7:1–15. doi: 10.1038/s41597-020-0495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Global Status Report on Noncommunicable Diseases 2014. Number WHO/NMH/NVI/15.1. World Health Organization; 2014. [Google Scholar]
- Yao Q., Fan X., Cai Y., Wang R., Yin L., Li Y. Time-incremental convolutional neural network for arrhythmia detection in varied-length electrocardiogram. 2018 IEEE 16th Intl Conf on Dependable, Autonomic and Secure Computing, 16th Intl Conf on Pervasive Intelligence and Computing, 4th Intl Conf on Big Data Intelligence and Computing and Cyber Science and Technology Congress(DASC/PiCom/DataCom/CyberSciTech) 2018 doi: 10.1109/DASC/PiCom/DataCom/CyberSciTec.2018.00131. [DOI] [Google Scholar]
- Zhu H., Cheng C., Yin H., Li X., Zuo P., Ding J., Lin F., Wang J., Zhou B., Li Y. Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: a cohort study. Lancet Digital Health. 2020 doi: 10.1016/S2589-7500(20)30107-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 12-lead ECG data set used in this study is the CPSC2018 training dataset which is released by the first China Physiological Signal Challenge (CPSC) 2018 during the seventh International Conference on Biomedical Engineering and Biotechnology. Details of the CPSC2018 dataset can be found at http://2018.icbeb.org/Challenge.html. The source code is provided and is available at https://github.com/onlyzdd/ecg-diagnosis.