Abstract
Purpose
To assess the contribution of a generative adversarial network (GAN) to improve intermanufacturer reproducibility of radiomic features (RFs).
Materials and Methods
The authors retrospectively developed a cycle-GAN to translate texture information from chest radiographs acquired using one manufacturer (Siemens) to chest radiographs acquired using another (Philips), producing fake chest radiographs with different textures. The authors prospectively evaluated the ability of this texture-translation cycle-GAN to reduce the intermanufacturer variability of RFs extracted from the lung parenchyma. This study assessed the cycle-GAN’s ability to fool several machine learning (ML) classifiers tasked with recognizing the manufacturer on the basis of chest radiography inputs. The authors also evaluated the cycle-GAN’s ability to mislead radiologists who were asked to perform the same recognition task. Finally, the authors tested whether the cycle-GAN had an impact on radiomic diagnostic accuracy for chest radiography in patients with congestive heart failure (CHF).
Results
RFs, extracted from chest radiographs after the cycle-GAN’s texture translation (fake chest radiographs), showed decreased intermanufacturer RF variability. Using cycle-GAN–generated chest radiographs as inputs, ML classifiers categorized the fake chest radiographs as belonging to the target manufacturer rather than to a native one. Moreover, cycle-GAN fooled two experienced radiologists who identified fake chest radiographs as belonging to a target manufacturer class. Finally, reducing intermanufacturer RF variability with cycle-GAN improved the discriminative power of RFs for patients without CHF versus patients with CHF (from 55% to 73.5%, P < .001).
Conclusion
Both ML classifiers and radiologists had difficulty recognizing the chest radiographs’ manufacturer. The cycle-GAN improved RF intermanufacturer reproducibility and discriminative power for identifying patients with CHF. This deep learning approach may help counteract the sensitivity of RFs to differences in acquisition.
Supplemental material is available for this article.
© RSNA, 2020
See also the commentary by Alderson in this issue.
Summary
A generative adversarial network accurately translates texture between manufacturers at image level on chest radiographs, reduces the intermanufacturer variability of radiomic features, and improves radiomic diagnostic accuracy, allowing for improving retrospective and multicenter radiomic studies.
Key Points
■ Image texture translation using a generative adversarial network reduces the intermanufacturer variability of radiomic features (RFs) extracted from chest radiographs and has the potential to improve radiomic diagnostic accuracy.
■ This texture translation, applied before RF extraction, may dampen the risk of systematic biases and improve the statistical power of retrospective and multicenter radiomic studies.
Introduction
The recent development of radiomics has raised hope for improving the diagnostic, prognostic, and predictive accuracy of radiologic examinations (1,2). Although several studies have reported improvement in diagnostic procedures and patient treatment using radiomics and statistical methods, such as machine learning (ML) (3), concerns have arisen over the reproducibility of quantitative radiomic features (RFs) extracted from radiologic images (4). Numerous studies have revealed RFs to vary depending on the manufacturer, reconstruction algorithm, or even image characteristics, potentially resulting in the nonreproducibility of RFs (5–8).
RF reproducibility is an essential aspect of radiomic studies because the nonreproducibility of RFs may add noise to the data and thus dampen the studies’ statistical power, possibly resulting in false-negative findings (ie, type II statistical errors). Another worrisome consequence of RF nonreproducibility may consist in introducing confounding factors in statistical models (eg, if all patients undergo chest radiography using a machine from a given manufacturer, and all controls without disease undergo radiography performed using a machine from another manufacturer), potentially resulting in false-positive findings (ie, type I statistical errors). However, the sources of the RF nonreproducibility are likely to be difficult to control in clinical studies, especially when using a large multicenter design or retrospective cohorts, as is often the case in radiomic studies. Moreover, although concern about the potential lack of reproducibility due to intermanufacturer variability arose with respect to radiomic studies, this may similarly apply to deep learning research, as these algorithms are sensitive to subvisual patterns in images (9,10). Thus, a method that counteracts changes in image texture at the image level due to differences in acquisition using platforms developed by different manufacturers could improve the quality of both radiomic and deep learning studies, thereby dampening the risks of false-positive and false-negative findings.
Recently, generative adversarial networks (GANs) have emerged with the ability to learn to mimic any kind of data distribution (11,12). They have been employed to transform an image from one source domain to a target domain, thereby generating, on the basis of photography, counterfeit images from a renowned painter (13,14). A specific kind of GAN called a cycle-GAN has been developed to translate texture at the image level (15). Here, we aim to leverage the recent development of GANs to perform texture translation at the image level on radiologic images acquired with units from different manufacturers with the objective to improve RF intermanufacturer reproducibility. We first developed a cycle-GAN model to transfer the texture of chest radiographs among manufacturers. Then, we assessed the cycle-GAN’s ability to improve RF reproducibility, mislead both ML algorithms and radiologists into misclassifying the manufacturer of counterfeit (or fake) images, and modify the diagnostic performance of RFs in classifying disease on a heterogeneous dataset.
Materials and Methods
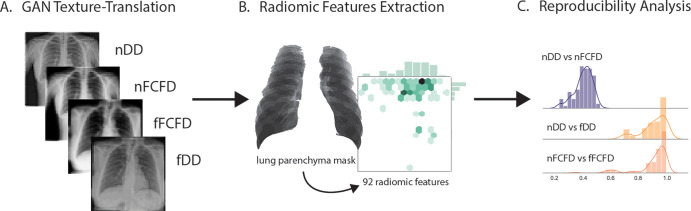
The study protocol was approved by the ethics committee of the Geneva State, with the reproducibility analysis pipeline displayed in Figure 1.
Figure 1:
Radiomic feature (RF) reproducibility analysis pipeline. A, A cycle-GAN was first trained to translate textures between manufacturers: Philips DigitalDiagnost (DD) and Siemens Fluorospot Compact FD (FCFD). B, Following texture translation, 92 RFs were extracted from lung parenchyma for each native and fake chest radiograph of an independent testing dataset. C, The intermanufacturer RF variability was compared between pairs of native and translated chest radiographs in this independent dataset computing the concordance correlation coefficient for each RF. fDD = fake DD image, fFCFD = fake FCFD image, GAN = generative adversarial network, nDD = native DD image, nFCFD = native FCFD image.
Training and Testing Datasets
A training dataset was retrospectively built by retrieving and preprocessing 6528 consecutive upright frontal chest radiographs, both with normal and abnormal findings, obtained in 2017 at our institution using two different radiographic unit manufacturers: the Philips DigitalDiagnost (DD) (Best, the Netherlands) and the Siemens Fluorospot Compact FD (FCFD) (Erlangen, Germany). After developing our GAN model, an initial independent testing dataset of 914 consecutive chest radiographs (including those with normal and pathologic findings, with 457 chest radiographs from each manufacturer) was retrospectively collected for the same two manufacturers (see Appendix E1 [supplement]). A second testing dataset of 200 chest radiographs was also retrospectively collected after GAN model development (see Appendix E1 [supplement]) to evaluate the hypothesis that cycle-GAN improves the classification of patients with or without congestive heart failure (CHF) using RFs.
GAN Model
To perform texture translation between manufacturers, we used a cycle-GAN model adapted from Zhu et al (15). After training the cycle-GAN with architecture similar to that of Zhu et al (15) (Fig 2 and Appendix E1 [supplement] for details), we obtained two generator networks that could translate chest radiograph texture from a source manufacturer to a target one. The first generator translated a texture from the DD to the FCFD manufacturer by transforming an original chest radiograph from the DD set (native DD images [nDDs]) to match the FCFD type, producing a fake FCFD image (fFCFD) (Fig 2, A); the other generator performed the inverse texture translation, from FCFD to DD, producing fake DD images (fDDs) based on native FCFD images (nFCFDs) (Fig 2, B). We used these two generator networks to produce the fDDs and fFCFDs from chest radiographs of the testing set and to assess the quality of chest radiograph texture translation. Thus, we obtained two sets of 457 fake chest radiographs using each manufacturer (fDDs and fFCFDs), paired with their respective sets of 457 images from the original dataset (nFCFDs and nDDs, respectively). We computed a structural similarity index measure with 95% confidence intervals for all images of the testing set as a general indicator of the GAN cycle’s consistency (see Appendix E1 [supplement]). This similarity measure was calculated between each input and its reconstructed version (ie, the same image after passing sequentially through the two generators).
Figure 2:
GAN model. A, An nDD is fed into a generator (GDDtoFCFD), which translates its texture to match the FCFD type, producing an fFCFD image, based on the discriminator feedback (DFCFD). B, The inverse translation is similarly performed on the basis of a second generator and discriminator pair (GFCFDtoDD and DDD, respectively). The two discriminators (DFCFD and DDD) are trained to identify native and fake images produced by their corresponding generators (GDDtoFCFD and GFCFDtoDD, respectively), providing quality feedback concerning the counterfeit images to their corresponding generator. The cycle-GAN network architecture is similar to the one used by Zhu et al (15), except for image input and output shape (here, 512 × 512 pixels). DD = Philips DigitalDiagonost, DDD = DD discriminator, DFCFD = FCFD discriminator, FCFD = Siemens Fluorospot Compact FD, fDD = fake DD image, fFCFD = fake FCFD image, GAN = generative adversarial network, GDDtoFCFD = generator translating DD to FCFD, GFCFDtoDD = generator translating FCFD to DD, nDD = native DD image, nFCFD = native FCFD image.
Reproducibility of RFs
We used original and fake chest radiographs from the first independent testing set to compare RF reproducibility before and after the GAN texture translation. We computed RFs from the entire lung parenchyma of each chest radiograph using pyradiomics, extracting 92 default features (ie, without image filtering or shape features; see Appendix E1 [supplement]) (16) and tested for a reduction in RF intermanufacturer variability before and after the GAN texture translation using the concordance correlation coefficient (CCC), as defined by Lin (17) (see Appendix E1 [supplement]). An RF with a CCC greater than or equal to 0.85 was considered a reproducible feature, which was similar to the approach of Choe et al (18). As our GAN would translate the texture from one manufacturer to the other, we hypothesized that CCC would be improved when comparing one type of native chest radiograph with its paired fake cycle-GAN–generated chest radiograph (ie, nDD vs fDD; nFCFD vs fFCFD), as compared with the CCC between pairs of native or fake images (ie, nDD vs nFCFD or fDD vs fFCFD, respectively).
ML classification of the manufacturer.—Given that GAN is likely to reduce the intermanufacturer difference between RFs, we hypothesized that ML classifiers trained to recognize the manufacturer of native chest radiographs would be misled when trying to identify the manufacturer of fake chest radiographs. Thus, we trained five common ML classifiers to enable them to identify the manufacturer using native chest radiographs based on the previously extracted 92 RFs. We then assessed the performance of these five ML classifiers in distinguishing the manufacturers of native and fake chest radiographs, using 10-fold cross validation (see Appendix E1 [supplement]). Correct manufacturer recognition was defined as the original manufacturer for native chest radiographs and target manufacturer for fake chest radiographs (eg, FCFD class for original DD image translated to FCFD by the GAN). Thus, if ML classifiers identified fake chest radiographs as belonging to the target manufacturer class instead of the original one, they would be considered to have been misled by the GAN texture translation.
Radiologic classification of the manufacturer.—Given that chest radiograph characteristics are likely to depend on image features specific to each manufacturer, we hypothesized that experienced radiologists would accurately distinguish the manufacturer of native chest radiographs, yet be misled by GAN texture translation in recognizing the manufacturer of fake chest radiographs. To test this hypothesis, we asked two radiologists (S.P.M. and X.M., 12 and 19 years of experience) to review native and fake chest radiographs and to identify their manufacturer. The two radiologists were not involved in GAN model development and read chest radiographs together to reach a consensus on manufacturer. During this task, radiologists were asked to identify the manufacturer, regardless of whether the image was processed by the GAN or not (see Appendix E1 [supplement]). As for ML classifiers, the correct manufacturer recognition was defined as the original manufacturer for native chest radiographs and the target manufacturer for fake chest radiographs. If radiologists identified fake chest radiographs as belonging to the target manufacturer type, they would be considered to have been misled by the GAN.
Improvement of Diagnostic Performance of RFs
We used native and fake chest radiographs from the second independent testing set to compare the discriminative power of RFs before and after the GAN texture translation. We computed the same default 92 RFs from the first testing set, from a region of interest manually placed in the apex of the right lung (30-mm radius, not covering mediastinal or parietal structures) by a senior resident in radiology (J.H., 4 years of experience). We then assessed the performance of a support-vector-machine classifier in distinguishing chest radiographs from patients with and those without CHF based on these 92 RFs, before (native RFs) and after (translated RFs) GAN texture translation, also using 10-fold cross validation. We also assessed the performance of the same classifier on the same 92 RFs after feature harmonization with ComBat (publicly available at https://github.com/Jfortin1/ComBatHarmonization/tree/master/R), following methods described in the study by Orlhac et al (19), with manufacturer as a batch variable. Finally, we compared the classification performance between native, translated, and ComBat RFs using the McNemar test. The study’s aim was to assess changes in CHF classification depending on the nature of the RFs (native RFs, translated RFs, or ComBat RFs) and was not a comparison between radiologist versus RF classification performance.
Results
GAN Cycle Consistency
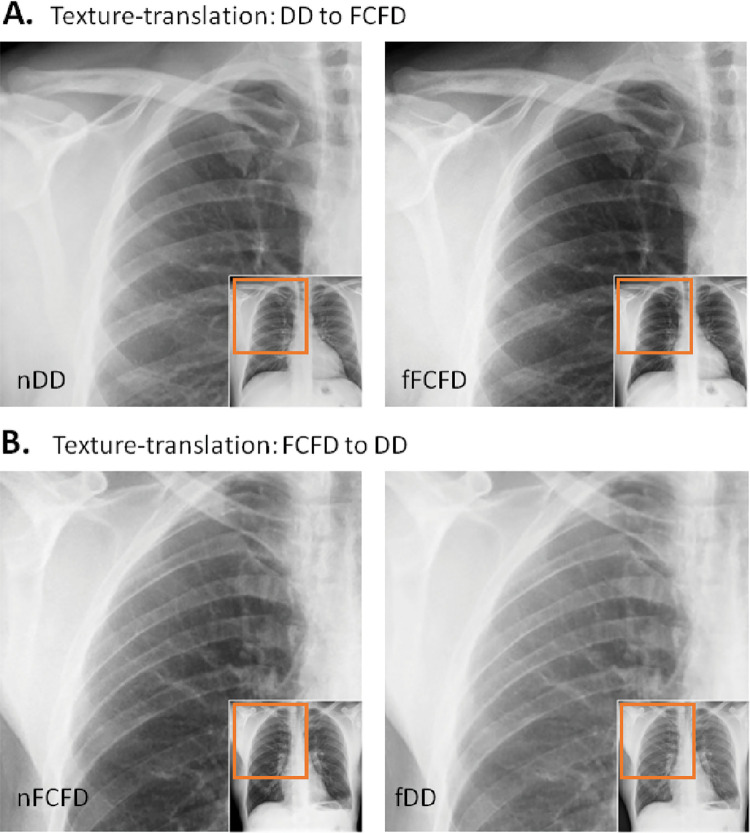
After GAN training, we observed a very high structural similarity index measure for the independent testing set, supporting accurate intermanufacturer texture translation by the GAN (95% confidence intervals: DD manufacturer: 0.9943, 0.9947; FCFD manufacturer: 0.9947, 0.9950). As GAN focuses on texture translation, changes between the manufacturers mostly occurred at high spatial frequencies, whereas the objects’ global structures (eg, thorax shape) were hardly altered (see illustrative cases in Fig 3).
Figure 3:
Texture translation between radiographs from different manufacturers. A, Texture translation from the Philips DigitalDiagonost (DD) to the Siemens Fluorospot Compact FD (FCFD) with the original (left) and its corresponding fake (right) chest radiograph, shows between-manufacturer changes occurring at high spatial frequencies, with global thoracic structures hardly altered. B, Texture translation from the FCFD to the DD. fDD = fake DD image, fFCFD = fake FCFD image, nDD = native DD image, nFCFD = native FCFD image.
Reproducibility of RFs
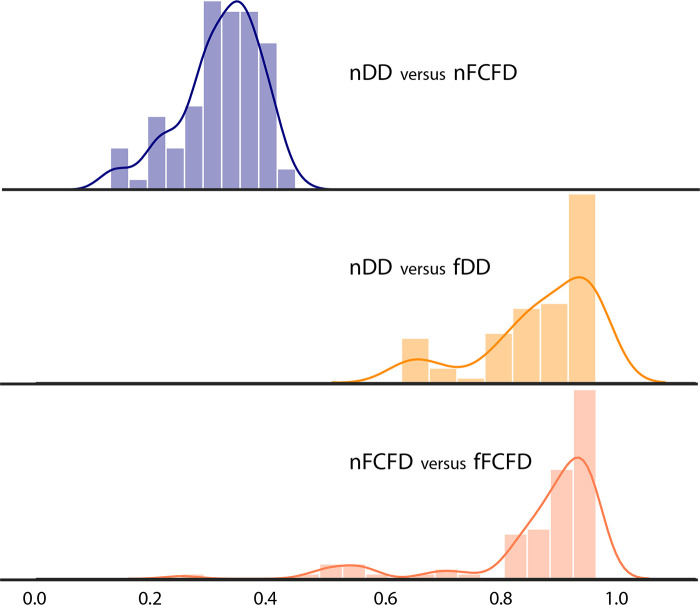
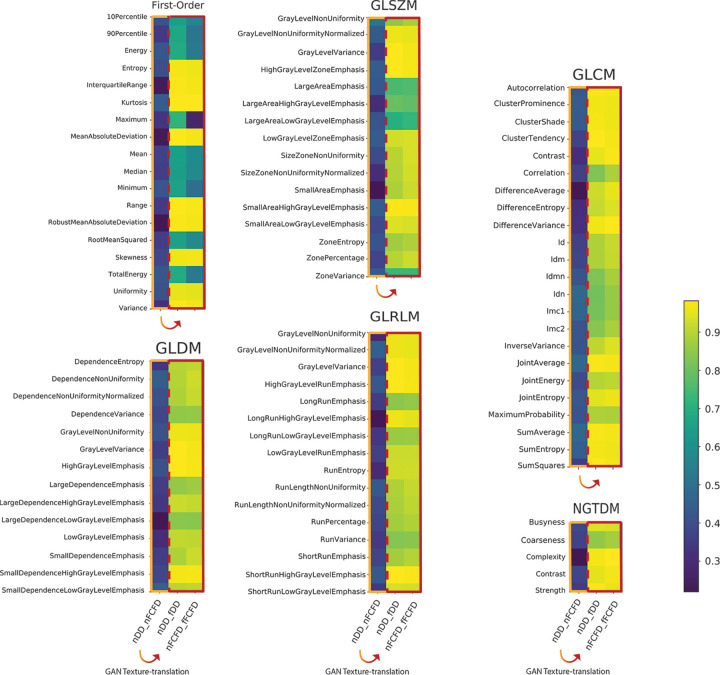
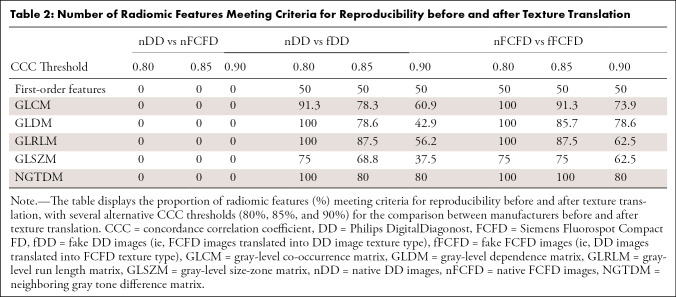
RF reproducibility was assessed by measuring the CCC between RFs before and after GAN texture translation. A summary of the result groups by class of RF is available in Table 1, and a graphical representation of CCC for all RFs is available in Figures 4 and 5. Detailed results for all RFs are available in Table E1 (supplement). Among the 92 RFs, none were considered reproducible before texture translation (native chest radiographs), whereas 72.8% were considered reproducible after texture translation from nFCFDs to fDDs (alternative CCC threshold: 0.80: 83.7%, 0.85: 72.8%, and 0.90: 52.2%), and 79.3% of RFs were considered reproducible after texture translation from nDDs to fFCFDs (alternative CCC threshold: 0.80: 85.9%, 0.85: 79.3%, and 0.90: 66.3%) (see Table 2 for details).
Table 1:
Concordance Correlation Coefficient between Manufacturers before and after Texture Translation
Figure 4:
Distribution of the concordance correlation coefficient between the two manufacturers before and after texture translation. fDD = fake Philips DigitalDiagonost image, fFCFD = fake Siemens Fluorospot Compact FD image, nDD = native Philips DigitalDiagonost image, nFCFD = native Siemens Fluorospot Compact FD image.
Figure 5:
Concordance correlation coefficient (CCC) heatmap for all radiomic features (RFs) before and after texture translation. The heatmaps display the CCC of all RF groups by classes of RF. In each RF class, the left column (framed in orange) compared native RFs (ie, nDDs to nFCFDs). The two other columns (framed in red) compared native to translated RFs (ie, nDDs to fDDs and nFCFDs to fFCFDs). Numerical values of CCC for all RFs are available in Table E1 (supplement). DD = Philips DigitalDiagonost, FCFD = Siemens Fluorospot Compact FD, fDD = fake DD image, fFCFD = fake FCFD image, GAN = generative adversarial network, GLCM = gray-level co-occurrence matrix, GLDM = gray-level dependence matrix, GLRLM = gray-level run length matrix, GLSZM = gray-level size zone matrix, nDD = native DD image, nFCFD = native FCFD image, NGTDM = neighboring gray tone difference matrix.
Table 2:
Number of Radiomic Features Meeting Criteria for Reproducibility before and after Texture Translation
Classification Using RFs
Given that the GAN model reduced the intermanufacturer difference of RFs, we tested the hypothesis that ML classifiers, trained to recognize the manufacturer based on RFs extracted from native chest radiographs, would be misled when trying to identify the manufacturer of fake chest radiographs, despite their having never been explicitly exposed to fake chest radiographs during training. All ML classifiers showed good accuracy for identifying the manufacturer of native chest radiographs, with accuracies exceeding 95% (Table E2A [supplement]) and performed above chance level (P < .001; Wilcoxon signed rank tests). Interestingly, these ML classifiers were similarly accurate at distinguishing the manufacturer for fake chest radiographs but recognized the target manufacturers instead of the original ones. Thus, ML classifiers were misled by GAN texture translation as hypothesized, providing further evidence that GAN accurately translates texture between manufacturers.
Radiologic Evaluation
As our GAN accurately translated texture between manufacturers, we hypothesized that radiologists would be misled by GAN texture translation when trying to identify the manufacturer of fake chest radiographs but would accurately identify the manufacturer of native chest radiographs. As hypothesized, we found that although radiologists correctly identified the manufacturer of native chest radiographs with 85.0% accuracy, they identified the target manufacturer for fake chest radiographs 88.3% of the time for fDDs (vs nFCFDs) and 74.6% of the time for fFCFDs (vs nDDs) (Table E2B [supplement]). All of these classifications were performed above chance level (P < .001; permutation testing).
Improving Disease Classification
The discriminative power of RFs was compared before and after the GAN texture translation, as well as with the ComBat harmonization method. Thus, a support-vector-machine classifier trained on native RFs showed an accuracy of 55% in discriminating CHF from non-CHF chest radiographs (sensitivity: 54%, specificity: 56%). This accuracy rose to 64.5% (sensitivity: 64%, specificity: 65%) for RFs after ComBat harmonization and to 73.5% (sensitivity: 77%, specificity: 70%) for RFs after texture translation with GAN. Radiomic discriminative performance was significantly better when comparing native to translated RFs (χ2: 18.78, P < .001) and native RFs to ComBat RFs (χ2: 7.90, P = .005). Interestingly, diagnostic accuracy was significantly better after GAN texture translation (translated RFs) as compared with ComBat RFs (χ2: 4.01, P = .045).
Discussion
As RFs are sensitive to acquisition settings and protocols, they may be nonreproducible between different manufacturers. To counteract this issue, we developed a texture-translation deep learning algorithm that improves the intermanufacturer reproducibility of RFs. By using three independent cohorts of patients for model development and evaluation, we have shown that GAN texture translation can reduce the intermanufacturer variability of RFs and improve the discriminative power of RFs for specific diseases as compared with other methods for improving RF reproducibility.
The nonreproducibility of RFs, due to intermanufacturer variability, is an essential concern in radiomic studies and, possibly, in deep learning research (20,21). Large multicenter studies are often performed using different radiologic materials or in a clinical setting, restricting the use of standardized imaging materials or protocols. This nonreproducibility of RFs may result in studies with lower statistical power and potentially false-negative findings or, on the contrary, to systematic biases in statistical models and potentially false-positive findings. Herein, we have provided evidence that GANs can translate texture between the manufacturers of chest radiographs, that this texture translation significantly improves the intermanufacturer reproducibility of RFs, and that it improves disease classification accuracy based on RFs, as compared with other methods for correcting RF variability. This texture translation even managed to mislead both radiologists and ML classifiers trying to identify the manufacturer, as based on the visual image inspection or RFs, respectively. The latter is of particular interest because many radiomic studies use ML methods based on RFs to make predictions from radiologic images. Finally, we showed that RFs after texture translation have higher discriminative power for identifying patients with CHF, as compared with both native RFs and RFs harmonized with compensation methods. Altogether, texture translation, using GANs, can reduce the intermanufacturer RF variability and improves diagnostic accuracy, which may improve retrospective or multicenter radiomic studies by dampening their risk of systematic biases and improving their statistical power.
An essential advantage of texture translation using GANs lies in its ability to work at an image level; it is thus agnostic to the kind of processing performed at a later time point on radiologic images. This contrasts with other methods developed to improve RF reproducibility by acting directly on individual RFs, such as compensation methods (19,22,23). This way, we anticipate that texture translation using GANs may be valuable for image analysis methods beyond radiomics, such as deep learning, because it directly corrects potential biases at an image level. Indeed, deep learning algorithms are susceptible to subvisual image features, which can bias the classification process (9,10).
Several important limitations should be noted. One of the major GAN drawbacks lies in the technical requirement needed to develop a model and in its potential failure cases (15). These failure cases can make the model incapable of generating a wide variety of images after training (model collapse) or they may prevent the generator and discriminator from reaching equilibrium during training (convergence failure), thus preventing proper texture translation. Because of the growing interest in radiomics, deep learning, and other qualitative imaging techniques, manufacturers could readily implement the development of texture-translation GAN models (eg, with translation toward a common texture type). We also did not observe failure cases in our two testing datasets, which might be due to the fact that chest radiographs are less prone to failure, thanks to their better homogeneity compared with images typically used to train GANs (eg, the natural images used in Zhu et al’s study [15]). Second, chest radiograph resolution was downsampled to 512 × 512 pixels because of the computational resources of GAN development on large images. This is likely to result in loss of information and performance degradation during the GAN evaluation we performed. Third, our study focused on standard radiography, whereas most radiomic studies employ either CT or MRI. This constraint was due to the current difficulty in developing GAN models on large three-dimensional images, notably because of computational resources. Fourth, reproducibility of the RFs was assessed over the entire lung parenchyma, whereas these features are usually extracted from well-defined lesions. In a future study, we wish to apply our method on a cohort with benign and malignant lung lesions to evaluate the contribution of texture translation in a clinical diagnostic task. Fifth, the evaluation of our GAN model to improve diagnosis of CHF is preliminary, and the reported performance is suboptimal in a clinical context. Further studies are therefore needed to evaluate how texture translation leads to diagnostic improvement. Given the interest in and rapid development of deep learning and GANs in particular, we anticipate that sufficient computational resources will be made rapidly available to perform texture translation on CT and MR images, based on our work on two-dimensional radiologic images.
In conclusion, a GAN is capable of translating textures between manufacturers and improving intermanufacturer RF reproducibility. By working directly at the image level, this technique improves the intermanufacturer concordance of RFs extracted from chest radiographs and has the potential to improve radiomic diagnostic accuracy. Our work on two-dimensional radiologic images could serve as a basis for developing three-dimensional texture translation GANs aimed at improving statistical models in retrospective or multicenter quantitative radiologic studies.
APPENDIX
Acknowledgments
We would like to thank Michel Kocher, PhD, and Simon Burgermeister, MSc, for their helpful comments about this work.
S.M. and J.H. contributed equally to this work.
Supported by a Research and Development Grant of the Geneva University Hospital (PRD 11-2017-2).
Disclosures of Conflicts of Interest: S.M. disclosed no relevant relationships. J.H. disclosed no relevant relationships. M.G.P. work supported in part by the Center for Biomedical Imaging (CIBM) of the Geneva-Lausanne Universities and the EPFL. S.P.M. disclosed no relevant relationships. D.V.D.V. disclosed no relevant relationships. X.M. disclosed no relevant relationships.
Abbreviations:
- CCC
- concordance correlation coefficient
- CHF
- congestive heart failure
- DD
- Philips DigitalDiagnost
- FCFD
- Siemens Fluorospot Compact FD
- fDD
- fake DD image
- fFCFD
- fake FCFD image
- GAN
- generative adversarial network
- ML
- machine learning
- nDD
- native DD image
- nFCFD
- native FCFD image
- RF
- radiomic feature
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 2017;37(5):1483–1503. [DOI] [PubMed] [Google Scholar]
- 3.Thrall JH, Li X, Li Q, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol 2018;15(3, pt B):504–508. [DOI] [PubMed] [Google Scholar]
- 4.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 2018;102(4):1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol 2015;50(11):757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H, Park CM, Lee M, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One 2016;11(10):e0164924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 2017;44(3):1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 2018;288(2):407–415. [DOI] [PubMed] [Google Scholar]
- 9.Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks. ArXiv 1312.6199 [preprint] https://arxiv.org/abs/1312.6199. Posted December 21, 2013.
- 10.Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples. ArXiv 1412.6572 [preprint] https://arxiv.org/abs/1412.6572. Posted December 20, 2014.
- 11.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, eds. Advances in neural information processing systems 27. Red Hook, NY: Curran Associates, 2014; 2672–2680. [Google Scholar]
- 12.Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Process Mag 2018;35(1):53–65. [Google Scholar]
- 13.Taigman Y, Polyak A, Wolf L. Unsupervised cross-domain image generation. ArXiv 1611.02200 [preprint] https://arxiv.org/abs/1611.02200. Posted November 7, 2016.
- 14.Yi Z, Zhang H, Tan P, Gong M. Dual GAN: unsupervised dual learning for image-to-image translation. In: Proceedings of 2017 IEEE International Conference on Computer Vision (ICCV). Venice, Italy: Institute of Electrical and Electronics Engineers, 2017; 2868–2876. [Google Scholar]
- 15.Zhu J, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. ArXiv 1703.10593 [preprint] https://arxiv.org/abs/1703.10593. Posted March 30, 2017.
- 16.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989;45(1):255–268. [PubMed] [Google Scholar]
- 18.Choe J, Lee SM, Do KH, et al. Deep learning-based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology 2019;292(2):365–373. [DOI] [PubMed] [Google Scholar]
- 19.Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology 2019;291(1):53–59. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan DC, Obuchowski NA, Kessler LG, et al. ; RSNA-QIBA Metrology Working Group . Metrology standards for quantitative imaging biomarkers. Radiology 2015;277(3):813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14(3):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. Springer series in statistics. New York, NY: Springer, 2001. [Google Scholar]
- 23.Orlhac F, Boughdad S, Philippe C, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med 2018;59(8):1321–1328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.