Abstract
Bacteria are constantly adapting to their environment by sensing extracellular factors that trigger production of intracellular signaling molecules, known as second messengers. Recently, 2’3,’-cyclic nucleotide monophosphates (2’,3’-cNMPs) were identified in Escherichia coli and have emerged as possible novel signaling molecules. 2’,3’-cNMPs are produced through endonucleolytic cleavage of short RNAs by the T2 endoribonuclease, RNase I; however, the physiological roles of RNase I remain unclear. Our transcriptomic analysis suggests that RNase I is involved in regulating numerous cellular processes, including nucleotide metabolism, motility, acid sensitivity, metal homeostasis and outer membrane morphology. Through a combination of deletion strain and inhibitor studies, we demonstrate that RNase I plays a previously unknown role in E. coli stress resistance by affecting pathways that are part of the defense mechanisms employed by bacteria when introduced to external threats, including antibiotics. Thus, this work provides insight into the emerging roles of RNase I in bacterial signaling and physiology and highlights the potential of RNase I as a target for antibacterial adjuvants.
Graphical Abstract
INTRODUCTION
While bacteria play essential roles in many aspects of human health, as evidenced by the growing body of work on the human microbiome,1, 2 bacterial infections also can wreak havoc, particularly if the infectious microbes are antibiotic resistant. Currently, over 2 million people develop hospital-acquired infections each year, which costs the United States an additional $20 billion in direct healthcare costs annually.3–7 Increasing levels of resistance are found among Gram-negative strains, which are particularly hard to treat due to the presence of the outer membrane. Therefore, identifying proteins and pathways involved in bacterial resistance mechanisms is essential to the development of new antibacterial agents. A complementary method to traditional antibiotics is the identification of adjuvants that modulate proteins/pathways involved in inherent mechanisms that provide resistance to current antibacterial treatments, which has the potential to extend the utility of these agents.8 The adjuvant/antibiotic strategy can be very effective, as evidenced by the co-dosing of amoxicillin and clavulanic acid, a β-lactam antibiotic and a β-lactamase inhibitor, respectively.9
Bacterial cells constantly respond and adapt to their environment, allowing them to respond to external stressors, such as antibiotics, through a number of mechanisms, including modifications of antimicrobial targets, decreased drug uptake, activation of efflux mechanisms, and changes to important metabolic pathways.10 While the main source of defense for Gram-positive bacteria is the dense cell wall, Gram-negative bacteria have a selectively permeable outer membrane, composed of lipopolysaccharides and lipoproteins, such as porins, which serve as a channel for small molecules and protect the cell against antibacterial compounds by providing structural integrity, protection against envelope stress, and transport of the antibiotics across the membrane.11 Additionally, bacterial cell membranes contain transporters that pump various classes of antimicrobials out of the cell before they reach their target.12
Given the importance of the bacterial cell wall in protecting bacteria from external stressors, it follows that proteins in the periplasm and outer membrane of Gram-negative bacteria may be involved in regulating bacterial resistance mechanisms. Recently, RNase I, a RNase T2 superfamily member that is widely distributed within bacteria and is found in both the cytoplasm and periplasm of E. coli,13 was demonstrated to regulate biofilm formation.14 The E. coli RNase I deletion strain (Δrna) exhibited a hyper-biofilm phenotype (~10-fold increase in biofilm formation, as compared to WT) that was linked to an upregulation of curli structural proteins, which are a major component of the E. coli biofilm matrix.14
To elucidate the range of cellular pathways regulated by RNase I and possible roles in resistance to external stressors, the present work describes transcriptome-wide changes in E. coli Δrna, enabling identification of cellular processes governed by RNase I. Our studies have linked RNase I to the regulation of diverse cellular processes, including biofilm formation, motility, acid resistance, β-lactam tolerance, and nucleotide metabolism. To probe the potential of RNase I as a target to downregulate Gram-negative bacterial resistance pathways, inhibitors based on non-hydrolyzable oligonucleotide scaffolds were generated and tested for their ability to modulate outer membrane morphology. Taken together, this work suggests that RNase I regulates many bacterial defense mechanisms and may serve as a target for a generation of novel antibacterial adjuvants.
RESULTS & DISCUSSION
RNase I Transcriptomics
Given the relative dearth of information on the physiological effects of RNase I, transcriptomic analysis was used to develop an understanding of the breadth of affected processes. These gene expression studies revealed ~800 genes dysregulated in Δrna relative to WT (Figure 1A). RNase I modulates genes encoding diverse protein classes, including transcription factors, transporters, and hydrolases (Figure 1B), linking RNase I to numerous cellular functions, including biofilm and motility.14 A similar case exists for transcripts encoding components of nucleotide metabolism.
Figure 1.
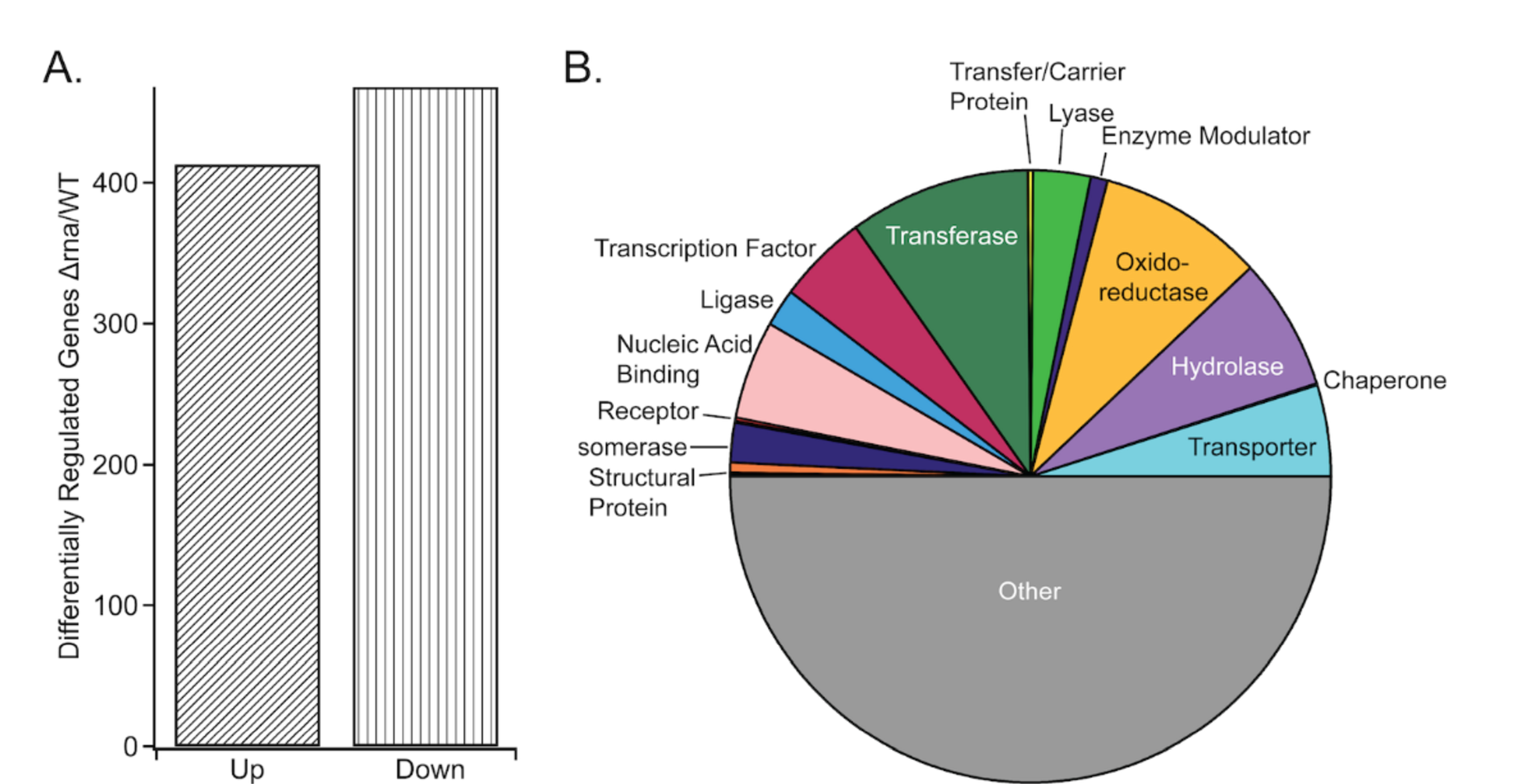
A) Numbers of genes differentially regulated in Δrna vs WT E. coli. B) Enzyme classes for the differentially regulated genes.
The rna gene produces both a cytoplasmic and periplasmic variant of RNase I; the cytoplasmic version displays a proclivity for the hydrolysis of small (~2–12-residue) oligoribonucleotides (oligoRNAs) in vitro, while the periplasmic form is non-specific.15 The different variants of RNase I potentially modulate gene expression through distinct mechanisms. For example, the dysregulation of genes involved in peptidoglycan biosynthesis suggests a potential function for periplasmic RNase I in cell wall assembly. Importantly, the rate of mRNA decay is not altered in E. coli lacking the rna gene,16 demonstrating that RNase I, unlike oligoribonuclease,17 is not directly modulating transcript abundance through mRNA degradation. Alternatively, the in vitro preference of cytoplasmic RNase I for the degradation of small RNA substrates suggests that small oligoRNAs could potentially accumulate in the Δrna strain and alter transcription. Interestingly, prior studies in Pseudomonas aeruginosa deficient for oligoribonuclease (which degrades short RNA substrates to 5’-NMPs) demonstrated that high levels of oligoRNAs shift transcriptional start sites across the genome.18 These findings suggest that a similar mechanism may elicit the transcriptional changes in Δrna E. coli relative to WT. In addition, accumulation of oligoRNAs could modulate transcript stability through an anti-sense mechanism. Due to the fact that E. coli encodes both RNase I and oligoribonuclease19, 20 (whereas P. aeruginosa lacks a close homolog of RNase I), additional work is necessary to probe the effect of RNase I depletion on oligoRNA levels. It also remains possible that RNase I influences transcription in a catalytically independent fashion, as catalytically inactivate T2 family RNases modulate certain cellular processes in eukaryotes.21
Due to the expansive role of 3’,5’-cAMP in regulating E. coli transcription through interaction with the 3’,5’-cAMP receptor protein (Crp),22 the intracellular concentration of 3’,5’-cAMP was quantified in WT and Δrna to probe the potential function of 3’,5’-cAMP-Crp in regulating the altered transcriptional profile in the absence of RNase I. LC-MS/MS analysis revealed barely detectable levels of 3’,5’-cAMP in both strains (limit of detection is ~150 fmol),23 which is not surprising due to the attenuation of adenylate cyclase activity in the presence of glucose24 (the carbon source in all experiments) (Supporting Information Figure S4). These data indicate that altered 3’,5’-cAMP levels are not modulating the transcriptional changes in E. coli lacking RNase I.
RNase I broadly impacts nucleotide metabolism and affects adenine sensitivity
The transcriptomic data indicated that expression of nrdAB, which encode subunits of a type I aerobic ribonucleotide reductase (RNR),25 were down-regulated ~1.9- and 2.6-fold, respectively, in Δrna relative to WT (Figure 2A). Due to the role of RNR in reducing 5’-NDPs to the corresponding 2’-deoxy 5’-NDPs, 25, 26 5’-NDP pools were quantified to assess the effect of altered RNase I on 5’-NDP metabolism. In agreement with the gene expression data, 5’-NDPs accumulated in Δrna relative to WT (Figure 2B).
Figure 2.
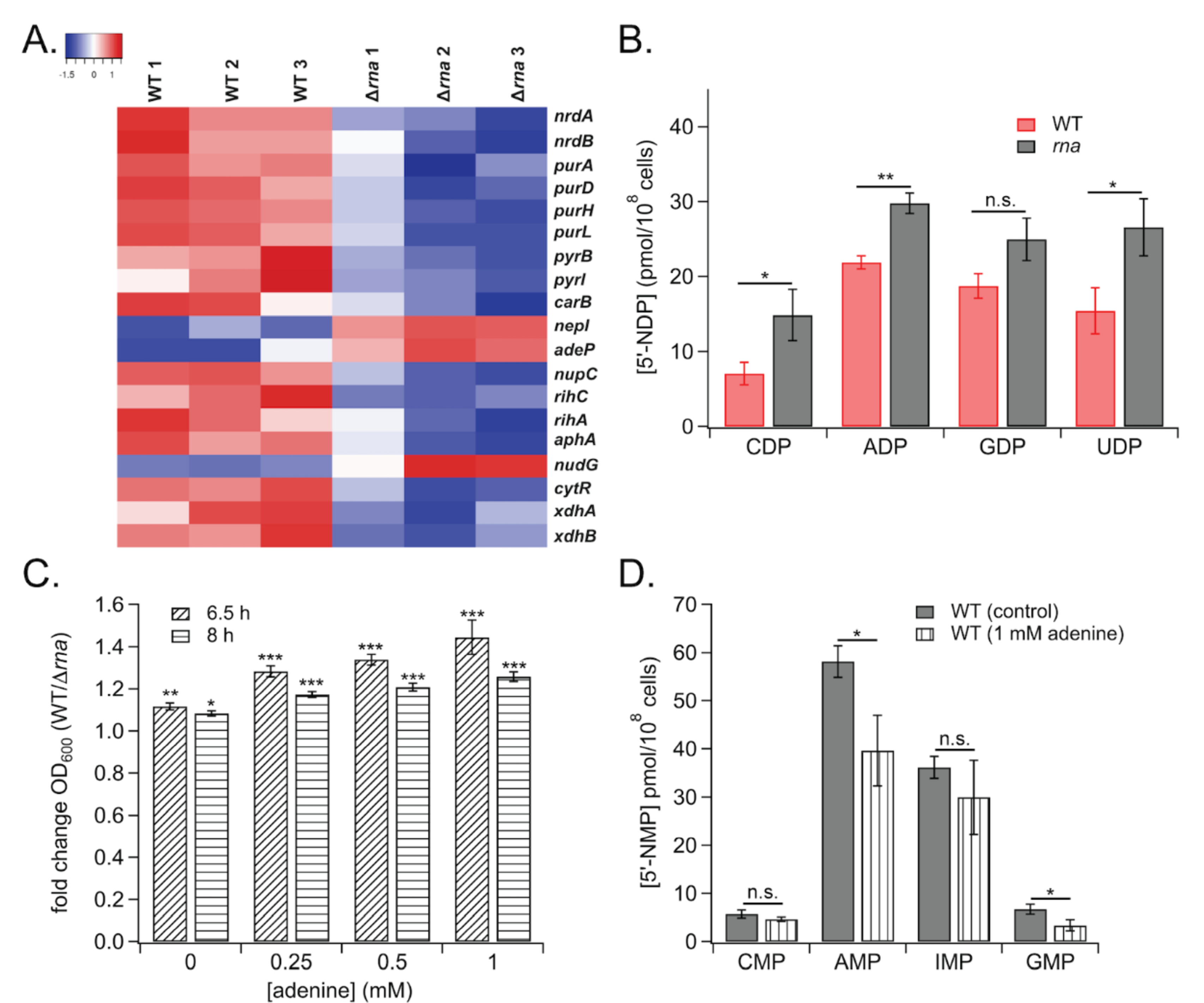
A) Heatmap of nucleotide metabolism gene expression data. B) Concentrations of NDPs in WT vs. Δrna strains. C) Relative effects of adenine on growth of WT and Δrna strains. D) Effects of adenine on NMP levels in WT. (*, ρ < 0.05; **, ρ < 0.01; ***, ρ < 0.001; n.s., not significant)
Expression of the nrdAB locus is subject to complex regulation by several transcriptional activators and repressors, including Crp,22 HNS,27 Fis,28 NrdR,29 and DnaA.28, 30 Although DNA microarray analysis indicated that mRNA levels of these transcription factors were not altered in the Δrna strain, transcription of nrdAB is governed by ATP binding to the DnaA transcriptional regulator,30 suggesting that altered nucleotide levels in Δrna could decrease RNR expression by modulating DnaA binding to the nrdAB promoter. The altered expression profile of nucleotide metabolic genes upon perturbation of RNase I levels potentially could modulate the enzymatic activity of nucleotide metabolic enzymes, as many proteins in this class are regulated allosterically or orthosterically by nucleotide binding.31–34 One such example is RNR, which is allosterically regulated by (d)NTP concentrations, both in terms of substrate specificity and reductase activity.26 Proper RNR function is vital to cell survival; Saccharomyces cerevisiae expressing a non-natural RNR1 allele with a mutated allosteric specificity binding site accumulates nearly 20-fold higher levels of dTTP and dCTP relative to wild-type concentrations,35 resulting in an elevated mutation rate across the genome.36 Due to its essentiality, RNR has been targeted in anti-cancer and antibacterial chemotherapy.37, 38 Consequently, these emerging regulatory links between RNase I and RNR suggest that T2 family RNases could be modulated to interfere with RNR function in bacteria.
Depletion of RNase I also down-regulated transcription of numerous genes involved in purine and pyrimidine nucleotide metabolism, including purDH and purL, as well as pyrBI and carB (Figure 2A), all of which function in de novo nucleotide synthesis. The Δrna strain also exhibited increased expression of nepI, encoding a purine nucleoside efflux pump, and attenuated transcription of nucleoside symporter genes nupC and adeP (Figure 2A). Other genes involved in nucleotide salvage also were down-regulated in Δrna, namely those encoding nucleoside hydrolases rihC, rihA, and nudG (Figure 2A).
Due to the impact of aberrant RNase I levels on nucleotide-related transcripts, the effect of rna deletion on adenine sensitivity was interrogated. Adenine-induced growth inhibition often is exacerbated in E. coli mutants lacking components of de novo purine biosynthesis or catabolism,39 leading to the hypothesis that RNase I may influence adenine toxicity. In support of the hypothesis, adenine more strongly inhibited the growth of RNase I-deficient E. coli relative to WT (Figure 2C). Prior studies determined that adenine toxicity in E. coli occurs primarily due to guanine nucleotide starvation39 and concentrations of 5’-GMP and -AMP were found to be lower in adenine-treated WT cultures relative to untreated control (Figure 2D). This result further suggests that altered purine metabolism mediates the differential sensitivity of WT and Δrna to adenine toxicity, and corroborates the dysregulated expression of nucleotide metabolic genes in cells lacking RNase I.
Modulation of nucleotide homeostasis could mediate some of the transcriptional and phenotypic changes observed in Δrna, as mRNA levels associated with de novo nucleotide biosynthesis and salvage genes were dysregulated (Figure 2A). Moreover, primary nucleotide metabolism influences processes such as biofilm formation in E. coli40–42 (perhaps through modulation of c-di-GMP pools) and cell wall rigidity in Lactococcus lactis.43 A prior study established that NTP concentrations impact the efficacy of transcription initiation from rRNA promoters,44 suggesting a potential mechanism through which aberrant nucleotide metabolism could drive transcriptional changes upon depletion of RNase I.
RNase I regulates β-lactam and acid sensitivity
With the rise of antibiotic resistant bacteria, it is vital to find pathways that can be targeted to prevent resistance. Transcriptomic data identified decreased expression of several genes involved in peptidoglycan maturation, such as ampH, mrcB, and murEF, and acid tolerance in Δrna vs. WT cells. Notably, the blr, lpoA, and yfeW transcripts implicated in penicillin binding and β-lactam resistance also were down-regulated in Δrna (Figure 3A), suggesting that E. coli lacking RNase I would exhibit decreased resistance to β-lactam challenge.
Figure 3.
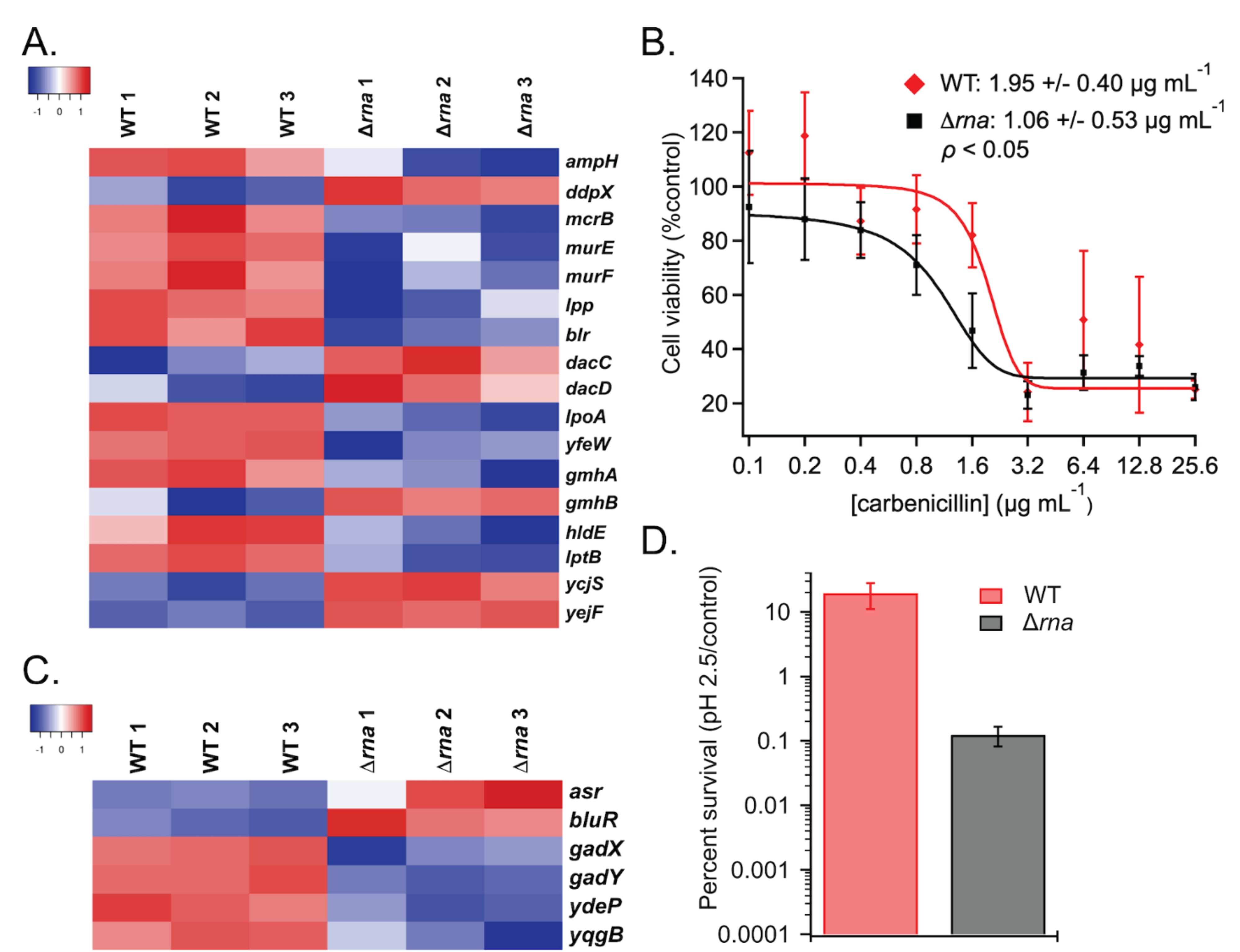
A) Heatmap of β-lactam related gene expression. B) Cell viability of WT and βrna strains grown in the presence of carbenicillin. Values for minimum concentration needed to cause 50% loss of cell viability are listed. C) Expression levels of genes related to acid resistance. D) Survival of WT and Δrna following acid challenge (ρ < 0.01).
In agreement with the gene expression data, dose-response assays revealed that Δrna was hypersensitive to carbenicillin-induced toxicity relative to WT (Figure 3B). In addition, down-regulated transcript levels of gadX and gadY, which are involved in glutamic acid-dependent acid resistance (Figure 3C), resulted in Δrna exhibiting ~100-fold decrease in acid tolerance relative to WT E. coli in a phenotypic assay (Figure 3D).
The altered sensitivity to acidic conditions and β-lactam treatment in cells with aberrant RNase I expression (Figure 3) suggests potential therapeutic relevance of RNase I. Acid tolerance is critical for colonization of the mammalian gastrointestinal tract by both pathogenic and probiotic bacterial species.45 Additionally, β-lactam antibiotics are among the most widely prescribed drugs to treat bacterial infections,46 emphasizing the potential significance of RNase I in microbial pathogenesis. Perturbation of basal RNase I levels could influence resistance to low pH and β-lactam treatment through modulation of amino acid (AA) homeostasis, as bacterial survival under these stressors is influenced by AA levels due to the role of proton-dependent AA decarboxylases and DD-transpeptidases in acid tolerance and β-lactam resistance, respectively.45, 46 AA homeostasis intersects with de novo nucleotide biosynthesis, as certain AAs are substrates of nucleotide anabolic enzymes such as PyrB and CarA.47, 48 Indeed, pyrB expression was attenuated by RNase I depletion (Figure 2A), further suggesting that dysregulation of de novo nucleotide biosynthesis alters amino acid levels and impacts resistance to acid and β-lactams. In fact, prior studies demonstrated that down-regulated expression of the pyrimidine biosynthetic gene pyrB (encoding aspartate carbamoyl transferase) increases L-Asp concentration and thus modulates peptidoglycan cross-linking in Lactococcus lactis,43 alluding to similar links between de novo nucleotide synthesis, amino acid homeostasis, and cell wall assembly in E. coli.
Copper Sensitivity and Metal Homeostasis
Microbes require copper as a cofactor for various cellular processes. Because of its redox chemistry it is used in enzymes involved in electron transport and oxidative respiration such as superoxide dismutase, cytochrome oxidase, methane mono-oxidase.49, 50 However free Cu(I) is toxic to the cells and different industries are exploiting the biocidal properties of Cu(I) as seen by its increasing use in public health protection, agriculture, and in hospital settings.51 Bacteria have evolved to evade toxicity caused by Cu(I) by tightly regulating Cu(I) levels in the cell52 through the tripartite CusABC efflux pump. This system has high specificity for removing toxic ions, Cu(I) and Ag(I) ions, out of the periplasm and cytoplasm to the outer membrane space.53, 54
Transcriptomic data in cells lacking RNase I indicated down-regulation of the cusB, cusC, and cueO genes (Figure 4A). The two former genes are involved in the CusABC pump and the latter gene oxidizes Cu(I) to Cu(II) under aerobic conditions.55 Additionally, deletion of RNase I causes increased expression ycfQ, which decreases outer membrane permeability to copper, and zitB which is a zinc/copper exporter.56, 57 To determine if RNase I affects copper sensitivity, a dose response assay was performed under both aerobic and anaerobic conditions. Under aerobic conditions Δrna and WT cells show similar dose dependence to CuSO4. However, under anaerobic conditions, Δrna cells exhibited significantly reduced copper tolerance (Figure 4B, 4C). It is important to note that the Δrna strain shows a slight growth defect even at 0 μM CuSO4 under anaerobic conditions. This finding indicates that the sensitivity to copper may not only be due to the dysregulation in the copper homeostasis systems, but also due to defects in anaerobic respiration (Figure 4D).
Figure 4.
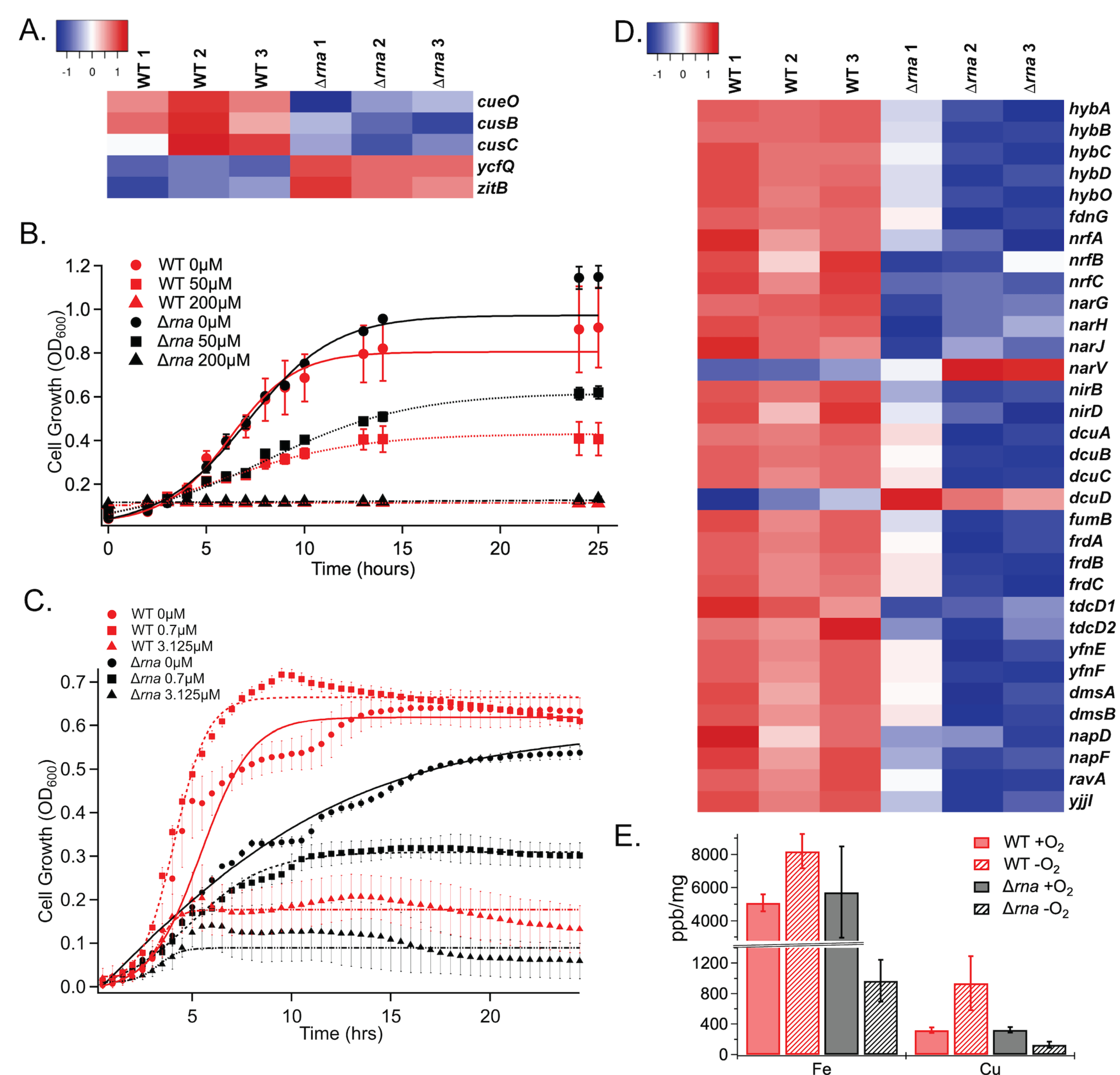
A) Copper and zinc homeostasis gene expression. Growth of WT and Δrna strains in the presence of various concentrations of CuSO4 under aerobic (B) and anaerobic (C) conditions. Differences between WT and Δrna at each concentration under aerobic conditions are not significant. Anaerobic data significance calculated by ANOVA for WT vs. Δrna strains; 0 μM and 0.7 μM, P < 0.0004; 3.125 μM, P < 0.017. D) Differential expression of genes related to metabolism. E) Levels of intracellular metals in WT and Δrna cells grown +/- O2.
To further understand how RNase I affects intracellular metal concentration, inductively coupled plasma-mass spectrometry (ICP-MS) was performed on WT and Δrna cells under both aerobic and anaerobic conditions. Cells were supplemented with a mix of metals during growth (Supporting Table S3) prior to ICP-MS analysis. The data demonstrate that under aerobic conditions, E. coli WT and Δrna cells have similar metal concentrations. However, under anaerobic conditions, Δrna exhibits 8-fold decrease intracellular concentration of 56 Fe, as compared to WT cells, and 65 Cu accumulated almost 10-fold less effectively in Δrna cells, as compared to WT cells. Further analysis of metals concentrations demonstrated that, compared to WT, Δrna struggles to maintain its intracellular metal concentrations under anaerobic conditions (Figure 4E). Like eukaryotes, bacteria require metals for growth and for cellular functions. Cellular processes such as nitrogen fixation, respiration, and metabolism are highly dependent on metals with redox properties such as Fe, Cu, Mn, Zn, Co, Ni, Mo, and Mg.58, 59 Furthermore, the transcriptomic data reveals that several anaerobic and aerobic respiration genes are dysregulated in WT vs Δrna such as hydrogenases (hybABCDO), nitrite reductases (nrfABC), and nitrate reductases (narGHJV). Dysregulation of these genes could lead to a reduced growth rate because the bacteria need to allocate energy to compensate for the lack of metals present that would normally assist in these respiratory activities.
RNase I regulates motility and outer membrane morphology
Analysis of the transcriptome in WT and Δrna revealed dysregulation of >10 genes involved in c-di-GMP signaling (Figure 5A), which regulates biofilm formation and motility, and substantial up-regulation of >30 genes involved in chemotaxis and flagellar motility in E. coli lacking RNase I (Figure 5B). Notably, these motility-associated genes include transcripts encoding methyl-accepting chemotaxis receptor proteins (MCPs; tap, tar, tsr, trg, aer),60 intracellular Che effectors (cheAW, cheRBYZ),60 transcriptional activators (flgM, fliAZ),61 flagellar biosynthesis/export proteins (flgN, fliST, flhBA, fliR),62 and components of the flagellar motor (motAB, flgEFGHI, flgKL, fliC, fliE)62 (Figure 5B). To investigate the phenotypic consequences of these altered gene expression profiles, the effect of RNase I on flagellar-dependent swimming motility was assayed. In agreement with the increased expression of chemotaxis and motility genes in Δrna relative to WT, the RNase I-deficient mutant was hypermotile (Figure 5C), and complementation of Δrna with plasmid pBAD33-rna restored WT swimming behavior (Figure 5C), demonstrating that RNase I regulates swimming motility. In addition, positive control experiments using uracil and ribose, which are known E. coli chemoattractants 63, 64, demonstrated that both the WT and Δrna strains respond normally to these established chemo-modulators (Supplementary Figure S5).
Figure 5.
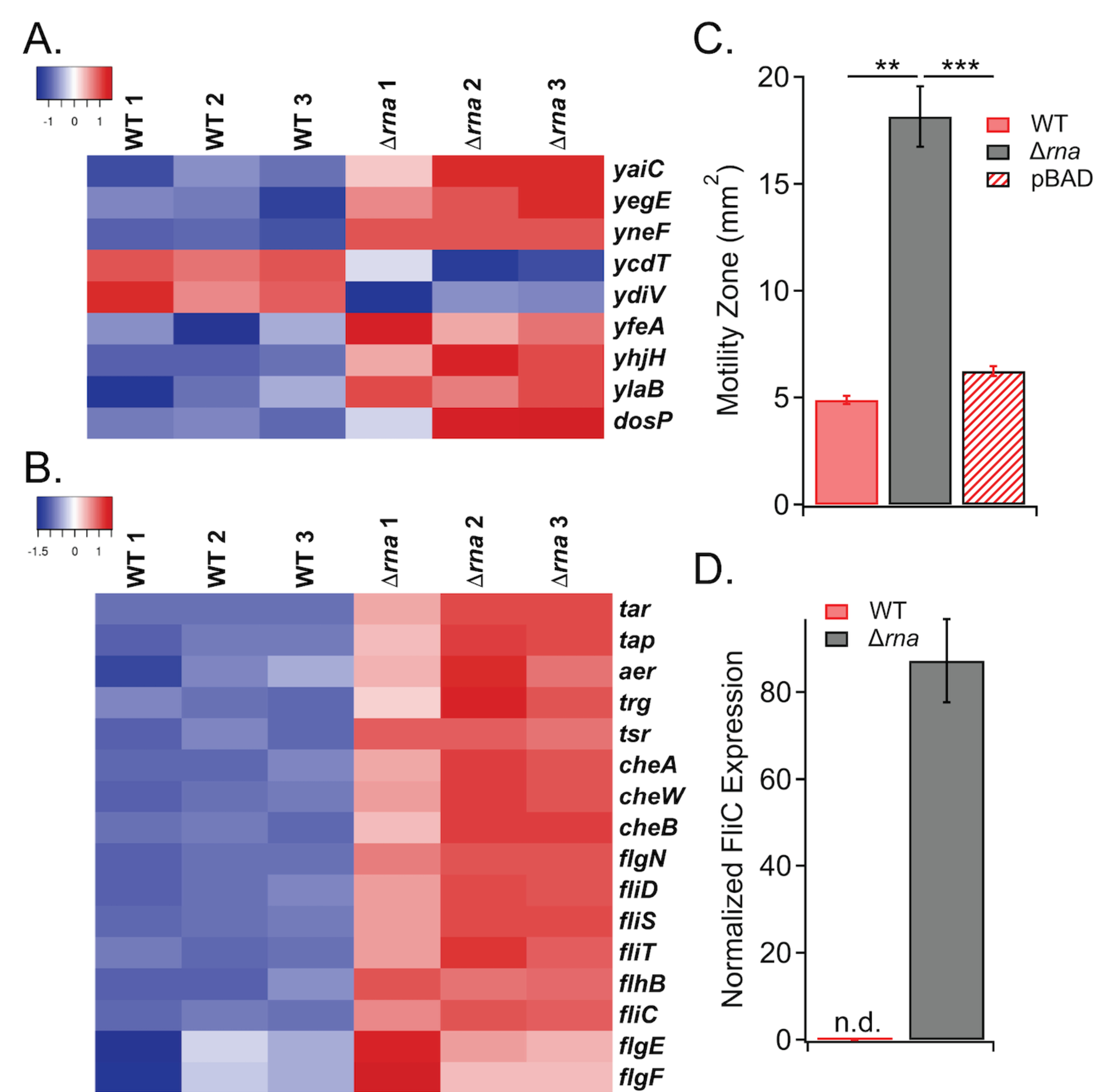
A) Differential transcript levels of c-di-GMP-related genes. B) Heat map of motility genes with differences in expression between WT and Δrna. C) Difference in motility for WT vs. Δrna. Complementation of E. coli Δrna with RNase I expressed on a plasmid (pBAD) results in WT-like motility (**, ρ < 0.01; ***, ρ < 0.001). D) The Δrna strain exhibits increased FliC protein levels.
To further probe the molecular effects of increased expression of flagellar export and assembly genes upon deletion of rna, the abundance of the FliC flagellar filament was quantified by western blot in WT and Δrna strains. In agreement with the ~8-fold up-regulation of the fliC transcript in Δrna compared to WT, the FliC protein level was elevated in the RNase I-deficient mutant (Figure 5D, Supplementary Figure S1).
Due to the increased production of flagella and sensitivity to antibiotics in the Δrna strain, changes in the outer membrane were further investigated. Outer membrane proteins are an integral component of the defense mechanisms that bacteria employ against antimicrobial compounds. The outer membrane is biosynthesized primarily through three distinct protein complexes: Bam, Lol, and Lpt which, respectively, assemble β- barrel proteins, lipoproteins, and lipopolysaccharide components.65 Transcriptomic data identified slight downregulation in proteins involved in the Bam and Lol complexes, which are involved in β- barrel and lipopolysaccharide assembly respectively (Figure 6A), and suggested possible effects on the outer membrane. To probe if the outer membrane is disturbed by deletion of RNase I and the possible dysregulation of outer membrane biogenesis machinery, electron microscopy was used to visualize morphological changes between WT and Δrna cells at exponential growth phase (Figure 6B-E).
Figure 6.
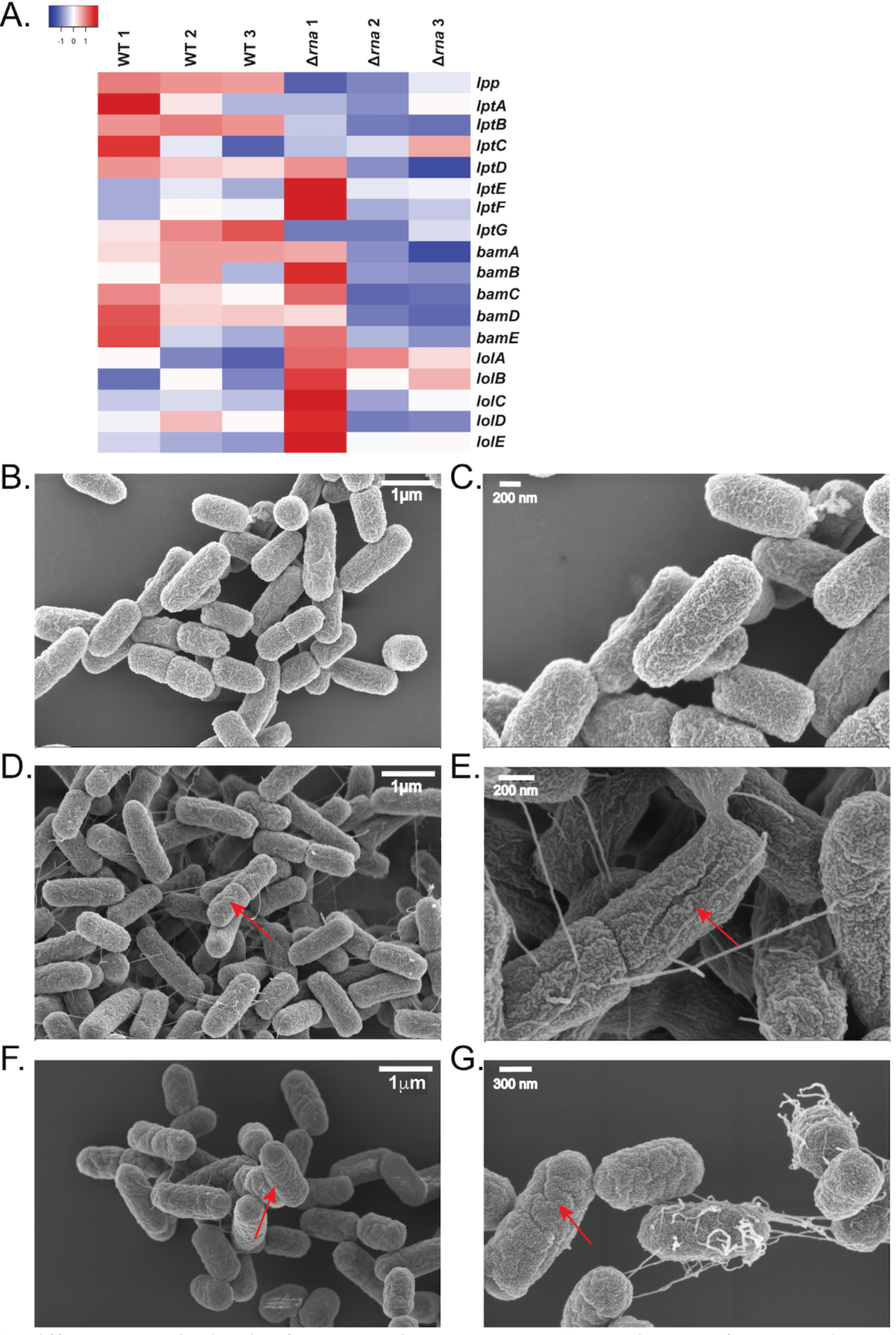
A) Different transcript levels of outer-membrane genes. B, C) TEM images of WT E. coli. D, E) TEM images of Δrna E. coli. Select membrane “wrinkles” are highlighted by arrows. F, G) WT E. coli treated with inhibitor also exhibit membrane wrinkles.
The SEM images show the WT strain cells are more homogenous shape with an approximate size of 1.29 μm ± 0.27 μm by 0.59 μm ± 0.04 μm, while Δrna strain cells are more heterogenous and have an approximate size of 1.46 μm ± 0.35 μm by 0.55 μm ± 0.04 μm (Figure 7A). At a higher magnification (100,000x), the micrographs show that outer membrane of the Δrna strain is partially disrupted, resulting in the appearance of wrinkling morphology. Additionally, at lower magnification (15,000x), increased flagellar production is observed and confirmed by immunogold staining for the flagellar structural protein FliC (Supplementary Figure S6). Taken together, these data demonstrate that RNase I modulates outer-membrane structure.
Figure 7.
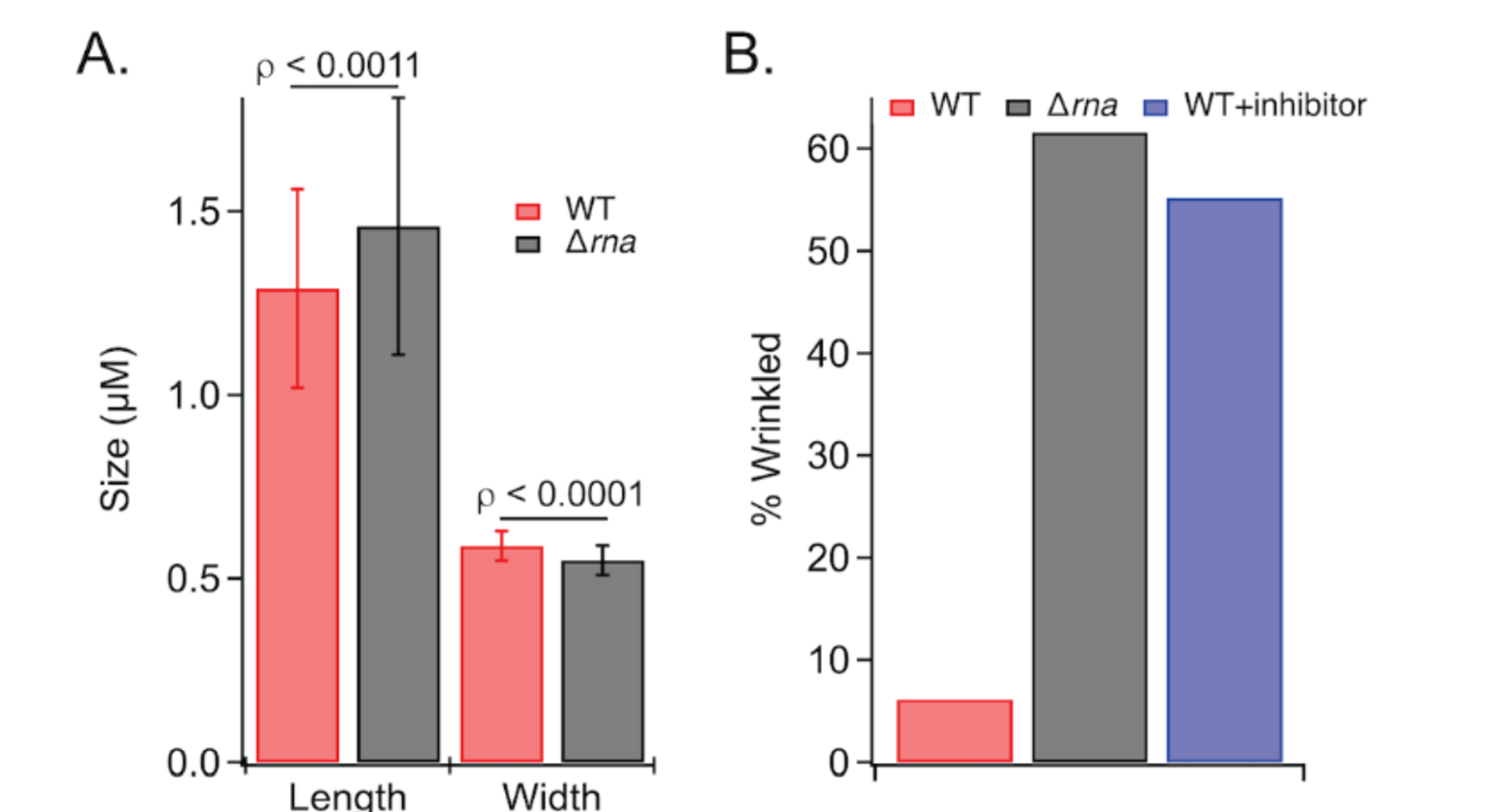
A) Comparison of WT and Δrna E. coli length and widths. B) Wrinkling morphology is observed for Δrna E. coli and WT cells incubated with RNase I inhibitor.
Inhibition of periplasmic RNase I
In order to determine if the transcriptomic and phenotypic effects are due to the absence of RNase I catalytic activity or the physical absence of RNase I, an oligonucleotide inhibitor was designed to inhibit periplasmic RNase I.66 Because periplasmic RNase I naturally targets short oligonucleotides, a substrate, based on previous work with RNase A and RNase H,67, 68 was designed as a short oligonucleotide FRET sensor to allow for monitoring RNase I activity in vitro and in vivo. In its uncleaved state, the substrate fluorescence is quenched; however, RNase I cleavage of the RNA substrate strand results in TAMRA fluorescence, which can be monitored.
Using purified RNase I, in vitro data indicate that the nuclease-resistant G-oligonucleotide Inhib1 (see Supporting Table S4 for sequence) inhibited RNase I activity; addition of 10 μM Inhib1 reduced fluorescence emission down to levels comparable to the reaction with no RNase I (Supplementary Figure S3). The inhibitor also was tested with live E. coli cells suspended in PBS buffer. Prior to addition of substrate, cells were incubated with oligo-G Inhib1 to allow binding to RNase I. The data demonstrate that 25 μM Inhib1 reduced fluorescence levels of WT cells down to levels comparable to that of Δrna cells (Supplementary Figure S3). While RNase I was reported as the periplasmic RNase in E. coli,69 it is possible that there are additional uncharacterized periplasmic RNases that are inhibited by Inhib2, as the nuclease-resistant oligonucleotide scaffold does not impart specificity to RNase I.
After validating RNase I inhibition in vitro and in cells, the inhibitor was used to interrogate the in vivo phenotypic effects of inhibiting periplasmic RNase I. Previous work has demonstrated that E. coli do not transport intact oligonucleotides through the cell membrane; therefore, the inhibitor should only act on periplasmic RNase I.70E. coli WT cells were grown in the presence of 100 μM Inhib2 (see Supporting Table S4 for sequence) and the effects on outer-membrane structure analysed by SEM. The results indicate that approximately 55% of the WT+inhibitor cells develop a wrinkled morphology, similar to that of the Δrna strain, while only 6% of WT cells grown without inhibitor exhibit the wrinkly phenotype (Figure 6F, 6G, 7B). These data demonstrate that inhibition of RNase I is sufficient to cause outer-membrane defects and highlight RNase I as a possible target for development of antibacterial adjuvants.
CONCLUSIONS
In summary, we have uncovered novel physiological roles of the T2 endoribonuclease, RNase I, in E. coli. Transcriptomic data demonstrates that deleting the rna gene encoding for RNase I results in dysregulation of approximately 800 genes involved in various metabolic and cellular processes. Through comparisons of E. coli WT and Δrna strains and development of an RNase I inhibitor, a variety of pathways modulated by RNase I have been investigated. RNase I was shown to regulate nucleotide metabolism, sensitivity to a variety of antimicrobial compounds, metal homeostasis, and outer membrane morphology. Electron microscopy demonstrated that either deletion or inhibition of RNase I results in changes to outer membrane morphology, highlighting a novel role for periplasmic RNase I. As inhibition of RNase I is sufficient to perturb the outer membrane, it may be possible to develop novel RNase I inhibitors for use as adjuvant therapy to increase efficacy of existing antibiotics.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Dr. Fred Strobel of the Emory Mass Spectrometry Core for assistance with LC-MS/MS quantitation experiments and to the Huck Institutes of the Life Sciences Microscopy Core at the Pennsylvania State University for assistance with TEM and SEM images.
FUNDING
Funding was provided by NIH (1R01GM125842–01A1) and Pennsylvania State University to EEW.
Footnotes
ACCESSION NUMBERS
Gene expression data for WT and Δrna have been submitted to ArrayExpress at EMBL-EBI (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-6095.
SUPPORTING INFORMATION
Supporting Information Available: This material is available free of charge via the Internet. Materials and methods, supporting figures, and tables can be found in the Supporting Information.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Sommer F, and Backhed F (2013) The gut microbiota-masters of host development and physiology, Nat Rev. Microbiol 11, 227–238. [DOI] [PubMed] [Google Scholar]
- 2.Kostic AD, Howitt MR, and Garrett WS (2013) Exploring host-microbiota interactions in animal models and humans, Genes Dev 27, 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. (2019) Antibiotic Resistance Threats in the United States, 2019, U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, and Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America, Clin. Infect. Dis 48, 1–12. [DOI] [PubMed] [Google Scholar]
- 5.Infectious Diseases Society of, A., Spellberg B., Blaser M., Guidos RJ., Boucher HW., Bradley JS., Eisenstein BI., Gerding D., Lynfield R., Reller LB., Rex J., Schwartz D., Septimus E., Tenover FC., and Gilbert DN. (2011) Combating antimicrobial resistance: policy recommendations to save lives, Clin. Infect. Dis 52 Suppl 5, S397–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts RR, Hota B, Ahmad I, Scott RD, Foster SD 2nd., Abbasi F., Schabowski S., Kampe LM., Ciavarella GG., Supino M., Naples J., Cordell R., Levy SB and Weinstein RA. (2009) Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship, Clin. Infect. Dis 49, 1175–1184. [DOI] [PubMed] [Google Scholar]
- 7.Dadgostar P (2019) Antimicrobial Resistance: Implications and Costs, Infect Drug Resist 12, 3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douafer H, Andrieu V, Phanstiel O, and Brunel JM (2019) Antibiotic Adjuvants: Make Antibiotics Great Again!, J. Med. Chem 62, 8665–8681. [DOI] [PubMed] [Google Scholar]
- 9.Finlay J, Miller L, and Poupard JA (2003) A review of the antimicrobial activity of clavulanate, J. Antimicrob. Chemother 52, 18–23. [DOI] [PubMed] [Google Scholar]
- 10.Munita JM, and Arias CA (2016) Mechanisms of Antibiotic Resistance, Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi U, and Lee C-R (2019) Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli, Front. Microbiol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor G, Saigal S, and Elongavan A (2017) Action and resistance mechanisms of antibiotics: A guide for clinicians, In J. Anaesthesiol. Clin. Pharmacol, pp 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannistraro VJ, and Kennell D (1991) RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli, J Bacteriol 173, 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaine BM, Martin KS, Garcia-Rodriguez JM, Jung C, Briggs L, Southwell JE, Jia X, and Weinert EE (2018) RNase I regulates Escherichia coli 2',3'-cyclic nucleotide monophosphate levels and biofilm formation, Biochem. J 475, 1491–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannistraro V, and Kennell D (1991) RNase I*, a form of RNase I, and messenger RNA degradation in Escherichia coli, J. Bacteriol 173, 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kivity-Vogel TE, D. (1967) On the metabolic inactivation of messenger RNA in Escherichia coli: Ribonuclease I and polynucleotide phosphorylase, Biochim. Biophys. Acta 138, 66–75. [DOI] [PubMed] [Google Scholar]
- 17.Andrade JM, Pobre V, Silva IJ, Domingues S, and Arraiano CM (2009) The role of 3'-5' exoribonucleases in RNA degradation, Prog Mol Biol Transl Sci 85, 187–229. [DOI] [PubMed] [Google Scholar]
- 18.Goldman S, Sharp J, Vvedenskaya I, Livny J, Dove S, and Nickels B (2011) NanoRNAs prime transcription initiation in vivo, Mol. Cell 42, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S, and Deutscher M (1999) Oligoribonuclease is an essential component of the mRNA decay pathway, Proc. Natl. Acad. Sci. U. S. A 96, 4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niyogi S, and Datta A (1975) Novel oligoribonuclease of Escherichia coli: 1) Isolation and properties, J. Biol. Chem 250, 7307–7312. [PubMed] [Google Scholar]
- 21.Luhtala N, and Parker R (2010) T2 Family ribonucleases: ancient enzymes with diverse roles, Trends Biochem. Sci 35, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng D, Constantinidou C, Hobman J, and Minchin S (2004) Identification of the CRP regulon using in vitro and in vivo transcriptional profiling, Nucleic Acids Res 32, 5874–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia X, Fontaine BM, Strobel F, and Weinert EE (2014) A facile and sensitive method for quantification of cyclic nucleotide monophosphates in mammalian organs: Basal levels of eight cNMPs and identification of 2',3'-cIMP, Biomolecules 4, 1070–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorke B, and Stulke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients, Nat. Rev. Microbiol 6, 613–624. [DOI] [PubMed] [Google Scholar]
- 25.Brignole E, Ando N, Zimanyi C, and Drennan C (2012) The prototypic class Ia ribonucleotide reductase from Escherichia coli: still surprising after all these years, Biochem. Soc. Trans 40, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torrents E (2014) Ribonucleotide reductases: essential enzymes for bacterial life, Front. Cell. Infect. Microbiol 4, doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cendra M, Juarez A, Madrid C, and Torrents E (2013) H-NS Is a novel transcriptional modulator of the ribonucleotide reductase genes in Escherichia coli, J. Bacteriol 195, 4255–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustin L, Jacobson B, and Fuchs J (1994) Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an Nrd-Lac fusion, J. Bacteriol 176, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, Aharonowitz Y, Sjoberg B, and Cohen G (2007) NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes, J. Bacteriol 189, 5012–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olliver A, Saggioro C, Herrick J, and Sclavi B (2010) DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli, Mol. Microbiol 76, 1555–1571. [DOI] [PubMed] [Google Scholar]
- 31.Cho B, Federowicz S, Embree M, Park Y, Kim D, and Palsson B (2011) The PurR regulon in Escherichia coli K-12 MG1655, Nucleic Acids Res 39, 6456–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long C, and Pardee A (1967) Cytidine triphosphate synthetase of Escherichia coli B. I. Purification and kinetics, J. Biol. Chem 242, 4715–4721. [PubMed] [Google Scholar]
- 33.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles A, and Barzu O (1995) Escherichia coli UMP kinase, a member of the aspartokinase fimaly, is a hexamer regulated by guanine nucleotides and UTP, Biochemistry 34, 5066–5074. [DOI] [PubMed] [Google Scholar]
- 34.Zhou G, Smith J, and Zalkin H (1994) Binding of purine nucleotides to 2 regulatory sites results in synergistic feedback inhibition of glutamine 5'-phosphoribosylpyrophosphate amidotransferase, J. Biol. Chem 269, 6784–6789. [PubMed] [Google Scholar]
- 35.Kumar D, Abdulovic A, Viberg J, Nilsson A, Kunkel T, and Chabes A (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools, Nucleic Acids Res 39, 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt DL, Buckland RJ, Lujan SA, Kunkel TA, and Chabes A (2016) Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools, Nucleic Acids Res 44, 1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao J, Liu X, Zhu L, and Yen Y (2013) Targeting ribonucleotide reductase for cancer therapy, Expert Opin. Ther.Targets 17, 1423–1437. [DOI] [PubMed] [Google Scholar]
- 38.Tholander F, and Sjoberg B (2012) Discovery of antimicrobial ribonucleotide reductase inhibitors by screening in microwell format, Proc. Natl. Acad. Sci. USA 109, 9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine R, and Taylor M (1982) Mechanism of adenine toxicity in Escherichia coli, J. Bacteriol 149, 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoniani D, Rossi E, Rinaldo S, Bocci P, Lolicato M, Paiardini A, Raffaelli N, Cutruzzola F, and Landini P (2013) The immunosuppressive drug azathioprine inhibits biosynthesis of the bacterial signal molecule cyclic-di-GMP by interfering with intracellular nucleotide pool availability, Appl. Microbiol. Biotechnol 97, 7325–7336. [DOI] [PubMed] [Google Scholar]
- 41.Attila C, Ueda A, and Wood T (2009) 5-Fluorouracil reduces biofilm formation in Escherichia coli K-12 through global regulator AriR as an antivirulence compound, Appl. Microbiol. Biotechnol 82, 525–533. [DOI] [PubMed] [Google Scholar]
- 42.Garavaglia M, Rossi E, and Landini P (2012) The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli, PLoS One 7, doi: 10.1371/journal.pone.0031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solopova A, Formosa-Dague C, Courtin P, Furlan S, Veiga P, Pechoux C, Armalyte J, Sadauskas M, Kok J, Hols P, Dufrene Y, Kuipers O, Chapot-Chartier M, and Kulakauskas S (2016) Regulation of cell wall plasticity by nucleotide metabolism in Lactococcus lactis, J. Biol. Chem 291, 11323–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaal T, Bartlett M, Ross W, Turnbough C, and Gourse R (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria, Science 278, 2092–2097. [DOI] [PubMed] [Google Scholar]
- 45.Kanjee U, Houry W, and Gottesman S (2013) Mechanisms of acid resistance in Escherichia coli, Annu. Rev. Microbiol 67, 65–81. [DOI] [PubMed] [Google Scholar]
- 46.Fisher J, Meroueh S, and Mobashery S (2005) Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity, Chem. Rev 105, 395–424. [DOI] [PubMed] [Google Scholar]
- 47.Rosenbusch J, and Weber K (1971) Subunit structure of aspartate transcarbamylase from Escherichia coli, J. Biol. Chem 246, 1644–1657. [PubMed] [Google Scholar]
- 48.Thoden J, Raushel F, Benning M, Rayment I, and Holden H (1999) The structure of carbamoyl phosphate synthetase determined to 2.1 Angstrom resolution, Acta Cryst. D-Biol. Cryst 55, 8–24. [DOI] [PubMed] [Google Scholar]
- 49.Samanovic M, Ding C, Thiele D, and Darwin K (2012) Copper in Microbial Pathogenesis: Meddling with the Metal, Cell Host Microbe 11, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arguello J, Raimunda D, and Padilla-Benavides T (2013) Mechanisms of copper homeostasis in bacteria, Front. Cell. Infect. Microbiol 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent M, Hartemann P, and Engels-Deutsch M (2016) Antimicrobial applications of copper, Int. J. Hyg. Environ. Health 219, 585–591. [DOI] [PubMed] [Google Scholar]
- 52.Rademacher C, and Masepohl B (2012) Copper-responsive gene regulation in bacteria, Microbiology 158, 2451–2464. [DOI] [PubMed] [Google Scholar]
- 53.Routh MD, Zalucki Y, Su CC, Long F, Zhang Q, Shafer WM, and Yu EW (2011) Efflux pumps of the resistance-nodulation-division family: A perspective of their structure, function and regulation in gram-negative bacteria, Adv. Enzymol. Relat. Areas Mol. Biol 77, 109–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long F, Su CC, Lei HT, Bolla JR, Do SV, and Yu EW (2012) Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex, In Philos. Trans. R. Soc. Lond. B Biol. Sci, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franke S, Grass G, Rensing C, and Nies DH (2003) Molecular Analysis of the Copper-Transporting Efflux System CusCFBA of Escherichia coli, In J. Bacteriol, pp 3804–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mermod M, Magnani D, Solioz M, and Stoyanov JV (2012) The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli, BioMetals 25, 33–43. [DOI] [PubMed] [Google Scholar]
- 57.Rahman M, Patching SG, Ismat F, Henderson PJ, Herbert RB, Baldwin SA, and McPherson MJ (2008) Probing metal ion substrate-binding to the E. coli ZitB exporter in native membranes by solid state NMR, Mol. Membr. Biol 25, 683–690. [DOI] [PubMed] [Google Scholar]
- 58.Locatelli FM, Goo K-S, and Ulanova D (2016) Effects of trace metal ions on secondary metabolism and the morphological development of streptomycetes, Metallomics 8, 469–480. [DOI] [PubMed] [Google Scholar]
- 59.Chandrangsu P, Rensing C, and Helmann JD (2017) Metal Homeostasis and Resistance in Bacteria, Nat. Rev. Microbiol 15, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sourjik V (2004) Receptor clustering and signal processing in E. coli chemotaxis, Trends Microbiol 12, 569–576. [DOI] [PubMed] [Google Scholar]
- 61.Chilcott G, and Hughes K (2000) Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli, Microbiol. Mol. Biol. Rev 64, 694-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minamino T, and Imada K (2015) The bacterial flagellar motor and its structural diversity, Trends Microbiol 23, 267–274. [DOI] [PubMed] [Google Scholar]
- 63.Adler J, Hazelbauer G, and Dahl M (1973) Chemotaxis toward sugars in Escherichia coli, J. Bacteriol 115, 824–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X, and Parales R (2008) Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer Tap, J. Bacteriol 190, 972–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konovalova A, Kahne DE, and Silhavy TJ (2017) Outer Membrane Biogenesis, Annu. Rev. Microbiol 71, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Player MR, and Torrence PF (1999) Phosphorothioate oligodeoxyribonucleotides inhibit ribonuclease L thereby disabling a mechanism of interferon action, Bioorg. Med. Chem. Lett 9, 891–894. [DOI] [PubMed] [Google Scholar]
- 67.James DA, and Woolley GA (1998) A fluorescence-based assay for ribonuclease A activity, Anal. Biochem 264, 26–33. [DOI] [PubMed] [Google Scholar]
- 68.Parniak MA, Min KL, Budihas SR, Le Grice SF, and Beutler JA (2003) A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity, Anal. Biochem 322, 33–39. [DOI] [PubMed] [Google Scholar]
- 69.Abrell JW (1971) Ribonuclease I Released from Escherichia coli by Osmotic Shock, Arch. Biochem. Biophys 142, 693–700. [DOI] [PubMed] [Google Scholar]
- 70.Kilstrup M, Hammer K, Ruhdal Jensen P, and Martinussen J (2005) Nucleotide metabolism and its control in lactic acid bacteria, FEMS Microbiol. Rev 29, 555–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


