Abstract
Coronavirus disease 2019 (COVID-19) has spread rapidly throughout the globe. The spectrum of disease is broad but among hospitalized patients with COVID-19, respiratory failure from acute respiratory distress syndrome is the leading cause of mortality. There is an urgent need for an effective treatment. The current focus has been developing novel therapeutics, including antivirals, protease inhibitors, vaccines and targeting the overactive cytokine response with anti-cytokine therapy. The overproduction of early response proinflammatory cytokines results in what has been described as a “cytokine storm” is leading eventually to death when the cells fail to terminate the inflammatory response. Accumulating evidence shows that inflammatory cytokines induce selectin ligands that play a crucial role in the pathogenesis of inflammatory diseases by mediating leukocyte migration from the blood into the tissue. Thus, the selectins and selectin ligands represent a promising therapeutic target for the treatment of COVID-19. In this paper, potential pan-selectin inhibitors were identified employing a virtual screening using a docking procedure. For this purpose, the Asinex and ZINC databases of ligands, including approved drugs, biogenic compounds and glycomimetics, altogether 923,602 compounds, were screened against the P-, L- and E-selectin. At first, the experimentally confirmed inhibitors were docked into all three selectins’ carbohydrate recognition domains to assess the suitability of the screening procedure. Finally, based on the evaluation of ligands binding, we propose 10 purchasable pan-selectin inhibitors to develop COVID-19 therapeutics.
Keywords: COVID-19, docking, pan-selectin inhibitors, selectins, virtual screening
Introduction
Selectin-mediated cell migration and recognition play a crucial role in various physiological as well as pathological processes (Tvaroška et al. 2020). They mediate the immune response to infections or inflammation (Kansas 1996) and are involved in the trafficking of stem cells (Mazo et al. 1998; Ruster et al. 2006; Sackstein et al. 2008; Karp and Teol 2009; Merzaban et al. 2011; Sahin and Buitenhuis 2012), but also the formation of cancer metastases (Mannori et al. 1995; Biancone et al. 1996; Kim and Varki 1997; Yang et al. 1999). Moreover, there are indications that selectins could play a crucial role in one of the significant complications of COVID-19 disease, acute respiratory distress syndrome (ARDS). There is a large body of experimental data demonstrating the role of excessive inflammation in the pathophysiology of COVID-19 and showing the connection between inflammation and the severity of the disease. Three separate stages of COVID-19 development can be identified (Reyes et al. 2020). In the first, the virus penetrates into lung host cells. In the second, a viral spread initiates lung tissue injury and leads to the host immune system’s response. In the third phase, the inflammatory cascade is activated. This activation often leads to cytokine release syndrome, also referred to as the “cytokine storm” (Merad and Martin 2020). Under such conditions, leukocytes activated through receptor-ligand signaling release early response proinflammatory cytokines, amplifying the immune reaction through the further recruitment of leukocytes. Such a process can lead to life-threatening hyper-inflammatory conditions, manifesting as pneumonia, sepsis, respiratory failure and ARDS (Ruan et al. 2020; Zhou et al. 2020) and might ultimately result in multi-organ failure and death (Meduri et al. 1995).
The recent development of novel therapeutics for COVID-19 has focused on antivirals, protease inhibitors, vaccines or anti-cytokine therapy. Novel approaches are badly needed because current attempts to repurpose existing anti-virals as treatments for COVID-19 have miserably failed. Nonetheless, accumulating evidence shows that inflammatory cytokines also induce selectin ligands’ expression (Ebel et al. 2015), which play a vital role in the pathogenesis of inflammatory diseases. Numerous studies have demonstrated the essential role of selectins and their ligands in inflammatory lung diseases, including ARDS, chronic obstructive pulmonary disease and asthma (Romano and Slee 2001; Tvaroška et al. 2020), and the potential of using inhibitors of selectins in their treatment (Woodside and Vanderslice 2008; Aydt et al. 2010). These observations suggest that anti-inflammatory agents based on the inhibition of selectin-ligand interactions can inhibit the selectin-based inflammatory response accompanying lung diseases and represent potential therapeutics that might be useful in treating COVID-19 and promoting improved patient outcomes.
The potential of targeting selectins to alleviate the complications of COVID-19 is further supported by the promising results obtained on dexamethasone and Colchicine. Recently, researchers from Oxford University (Horby et al. 2020) showed that “Low-cost dexamethasone reduces death by up to one third in hospitalized patients with severe respiratory complications of COVID-19.” Dexamethasone is a synthetic corticosteroid inhibiting E-selectin expression (Brostjan et al. 1997) induced by proinflammatory stimuli. Similarly, several studies demonstrated that Colchicine decreases the severity of the disease (Merad and Martin 2020). A clinical trial of 6000 people with COVID-19 infection, funded by the Government of Quebec, began in March 2020 to test the potential efficacy of using Colchicine over 30 days to reduce disease symptoms (Montreal Heart Institute 2020). Colchicine is also used as an anti-inflammatory agent, which inhibits E-selectin and L-selectin expression on the surfaces of neutrophil and endothelial cells (Cronstein et al. 1995). These results support that an anti-inflammatory strategy based on selectin antagonists may be beneficial for the treatment of COVID-19.
The family of selectins involves three isoforms: E-selectin, L-selectin and P-selectin, which play slightly different roles in the human body (Drickamer 1993). E-selectin is inducibly expressed on the surface of activated cells of the vascular endothelium; its expression peaks 3–6 hours after stimulation. P-selectin occurs on the surface of activated platelets and activated endothelial cells. It is stored in α-granules of platelets and Weibel-Palade bodies of endothelial cells; thus, it appears on the surface within minutes after stimulation. L-selectin is constitutively expressed on the surface of certain types of leukocytes.
Selectins are C-type mammalian lectins that bind carbohydrate ligands, tetrasaccharide sialyl Lewis x (sLex) and its modification being the minimal recognition determinants. Recently, 3D structures of several selectin-ligand complexes were solved. E-selectin was complexed with sLex (Graves et al. 1994; Somers et al. 2000), P-selectin with sLex and PSGL-1 (P-selectin glycoprotein ligand) model (Preston et al. 2016). For L-selectin, only the structure of the lectin and EGF domains complexed with a glycomimetic is available (Mehta-D'souza et al. 2017). Various strategies were used to inhibit selectin functions and were the subject of many reviews (Kaila and Thomas 2002; Kaila and Thomas 2003; Barthel et al. 2007; Ludwig et al. 2007; Ernst and Magnani 2009; Aydt et al. 2010; Bedard and Kaila 2010; Wang and Vidal 2013; Videira et al. 2017; Valverde et al. 2019; Tvaroška et al. 2020). Though significant efforts have been devoted to developing selectin inhibitors and numerous compounds were prepared, only a few compounds entered clinical trials.
This paper aims to identify the inhibitors of the interactions of selectins with their ligands as potential therapeutics against COVID-19. As discussed above, suitable selectin inhibitors that might serve as efficient therapeutic agents are still being searched for. This is caused by a partial functional redundancy of the selectins (Aydt et al. 2010) and a low affinity of the inhibitors to at least one of the three selectins. All three selectins have a high sequential and structural similarity of the lectin domain (Lasky 1995), with only subtle distinctions responsible for the different binding affinities of the selectins toward their ligands. They bind to sialyl Lewis X (sLex) with millimolar, yet one order of different magnitude affinities (Poppe et al. 1997) (E-selectin: 0.72 mM; L-selectin: 3.9 mM; P-selectin: 7.8 mM). This tetrasaccharide was identified as a common epitope in various natural selectin ligands; thus, it became one of the key templates in the recent development of novel selectin inhibitors.
In this study, suitable inhibitors are searched for with the use of virtual screening. The docking algorithms have been successfully used for receptors with sufficiently deep cavities to restrict ligands’ orientation and geometry. Since selectins have a flat and mostly hydrophilic interaction surface, searching for suitable inhibitors can be challenging for the docking algorithms (Kranich et al. 2007). Therefore, three different docking algorithms were used and evaluated by comparing experimental data with the predicted values. The XP algorithm that showed the best results had been selected for the final screening against selectins. Based on the evaluation of binding properties and drug-like characteristics, we have identified 10 purchasable compounds that are potential pan-selectin inhibitors and suitable candidates for further experimental investigation of their use as COVID-19 therapeutics.
Results and discussion
Potential inhibitors of selectins were searched for by screening the Asinex (Asinex 2020) and the ZINC (Irwin and Shoichet 2005; Irwin et al. 2012; Sterling and Irwin 2015) databases of compounds with the use of Glide (Friesner et al. 2004; Halgren et al. 2004; Friesner et al. 2006). Asinex database (518,081 compounds) contains drug-like compounds with good ADMET properties. The selected subset of compounds from the ZINC database is compounds with in vitro activity (407,602 compounds) or biogenic compounds (518,081 compounds), including all FDA approved drugs. Thus, most of the screened compounds exhibited drug-like properties. To assess the suitability and performance of the three docking algorithms (high-throughput virtual screening (HTVS), SP and XP) for the use for the selectins, a set of compounds with experimentally confirmed inhibition activity was docked. In the subsequent phase, the best selected docking algorithm has been employed for screening selected databases.
Docking of experimentally confirmed ligands
The experimentally confirmed inhibitors published in the articles by Calosso et al. (2014)) and Gouge-Ibert et al. (2010)) were docked into the carbohydrate recognition domains of the E-, P- and L-selectin, respectively, using two different algorithms, namely SP and XP. The docked compounds, together with the experimental IC50 values, are listed in Supplementary data (Tables SI and SII).
IC 50 values are commonly used as a measure of the activity of inhibitors. In this paper, the logarithm of the values is used, as it is proportional to the binding energy predicted by Glide. Nonetheless, while IC50 values allow for a straightforward comparison of the inhibitors within a single study, they cannot be used to compare inhibitors among the different studies due to the various experimental conditions. To deal with this issue, the logarithm of the ratio of IC50 of a compound to IC50 of sLex was used (Calosso et al. 2014)
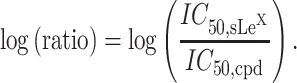 |
(1) |
Unfortunately, this measure is not available for all the experimental data used due to the limited data sets and low affinity of sLex to P-selectin in the study by Gouge-Ibert et al. (2010)).
For E-selectin and P-selectin, the results with the XP algorithm are shown in Figure 1 (the results are provided in Supplementary data, Table SI). The compounds with available log(ratio) are compared with the experimental activity in Figure 1A. However, for some of the compounds with P-selectin, the values are not available. Hence, the comparison for P-selectin is shown separately in Figure 1B. Since the structures of selectins were optimized in complex with sLex, the predicted Glide Energy of sLex is significantly lower than the other compounds due to perfect shape complementarity. The presented correlations imply that with an appropriate set-up, the XP docking algorithm provides the results in qualitative agreement with the experiments. The comparison of the results obtained at the XP level with the experimental data will be discussed in more detail later in the text. The results obtained with the SP algorithm are provided in Figure 2 (the results are provided in Supplementary data in Table SII). The SP algorithm shows a slightly worse correlation with experimental data than the XP algorithm. Nonetheless, the computational costs for screening at the SP level of precision are ~10-fold lower than the XP algorithm. Therefore, the SP algorithm seems to be more suitable for crude screening of the large databases of ligands. A much faster HTVS algorithm was also examined (Figure 5A). However, the algorithm failed to reproduce the experimental data. Consequently, it was not used in further docking studies in screening databases.
Fig. 1.
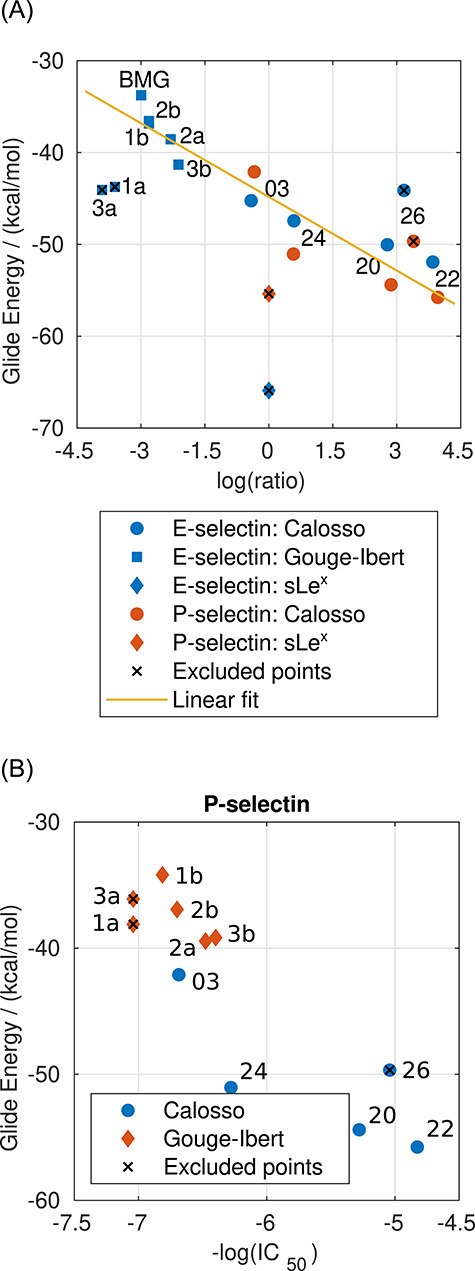
Comparison of the results from docking with the XP algorithm with the experimental data for E- and P-selectin. Results for compounds with available log(ratio) are shown in (A), and results for all compounds and P-selectin are shown in (B). This figure is available in black and white in print and in color at Glycobiology online.
Fig. 2.
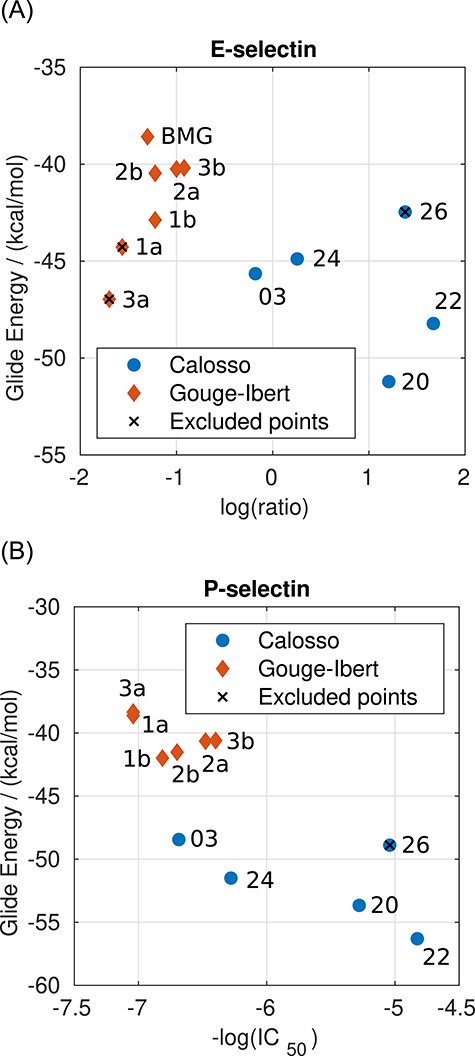
Comparison of the results from docking with the SP algorithm with the experimental data for (A) E-selectin and (B) P-selectin. This figure is available in black and white in print and in color at Glycobiology online.
Fig. 5.
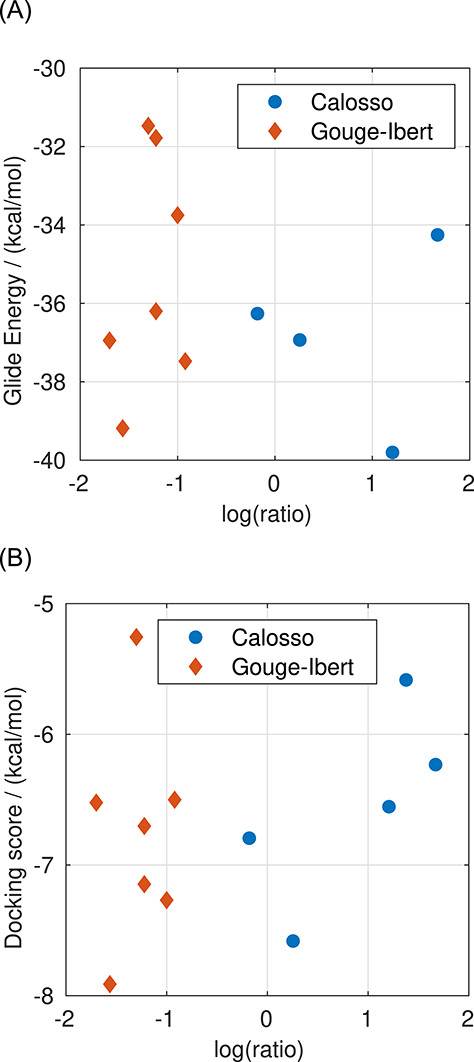
(A) Comparison of the results from docking with the HTVS algorithm with the experimental data for E-selectin. The compound 26 from the paper by Calosso et al. (2014)) was dropped by the docking algorithm. (B) Comparison of the docking score calculated at the XP level of precision with the experimental data for E-selectin. This figure is available in black and white in print and in color at Glycobiology online.
For L-selectin, there are not enough experimental data available. However, based on the selectins’ structural similarity, with a 60–70% identity of the lectin-like domain and the same binding mode of sLex (Figure 6), a very similar correlation to those presented for E- and P-selectin can be assumed.
Fig. 6.
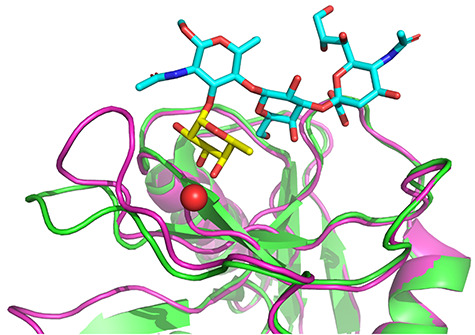
Overview of E- and P-selectin interactions with sLeX in the binding site (Somers et al. 2000; Woelke et al. 2013). The tetrasaccharide sLeX binds in a Ca2+-dependent manner via interactions with fucose hydroxyls. The fucose residue is shown in yellow and the Ca2+ cation as a red sphere. This figure is available in black and white in print and in color at Glycobiology online.
The Glide algorithm chooses the best pose based on the model energy score—the parameter “Glide Emodel,” which is a combination of empirical GlideScore, molecular mechanics interaction energy (Coulombic and van der Waals interactions) and the ligand internal strain energy (Friesner et al. 2004). However, Glide also calculates the Coulomb–van der Waals interaction energy score, referred to as “Glide Energy.” When applied to the selectins, we find that Glide Energy is the best of these parameters at reproducing the experimental activity. The comparison of the docking score to E-selectin’s experimental data is shown in Figure 5B for illustration. The good correlation obtained with the molecular mechanic “Glide Energy” scoring function corresponds with the findings of Woelke et al. that the interactions between selectins and their ligands are mostly electrostatic (Woelke et al. 2013).
To shed some light onto the possible concerns and issues of the selected docking algorithms, the comparison presented in Figure 1A needs to be discussed in more detail. The linear fit (R2 = 0.904) shown in Figure 1A has the following form:
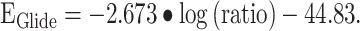 |
(2) |
As stated above, comparing the results obtained with the XP algorithm to experimental data implies that with an appropriate set-up, the XP docking algorithm provides results in qualitative agreement with the experiments. Still, there are some ligands for which the predicted energy does not correlate with the experimental data. The calculated Glide energies range from −66 to −33 kcal/mol. At first sight, the predicted Glide Energy values of sLex are noticeably higher than expected and come out as outliers in the presented correlation. We assume that this discrepancy reflects the perfect shape complementarity of the receptors with sLex, resulting from the optimization of the geometry of complexes with sLex. Noteworthy, the observed IC50 value of sLex toward the P-selectin was higher than 5 mM, and it was not determined precisely. Therefore, the log(ratio) of the compounds from this study cannot be calculated for P-selectin. The other outliers are common to both selectins and likely originate from the rigid receptor docking and the Glide Energy being sensitive to the ligands’ net charge (Friesner et al. 2004).
The issues can lead to both false-positive and false-negative predictions. To reduce the chance of identifying false positive hits as potential inhibitors in the subsequent screening of the databases of compounds, a tool for identifying of pan-assay interference compounds (PAINS) was employed (Baell and Holloway 2010; Baell and Nissink 2018). Since the tool does not ensure that all false-positive hits are omitted, the inhibitors’ structural properties and the complexes’ predicted structures were assessed. The inhibitors were required to interact with the Ca2+ cation of the lectin domain. Figure 6 shows the binding mode of sLex to the E-selectin. Our investigation revealed that any potential ligand must exhibit the same interactions (including the coordination mode) with Ca2+ as the native ligands. On the other hand, to reduce the chance of omitting potentially useful compounds, a more extensive set of ligands was employed. Yet, some of the potentially valuable compounds could have been omitted due to rigid receptor docking.
To demonstrate the rigid receptor issue, docking of compounds 3 and 26 from the paper by Calosso et al. (2014)) will be discussed in more detail. A comparison of the structures of the two compounds is shown in Figure 4, revealing that compound 26 features two benzoyl functional groups instead of hydroxyl groups, which increase the size of the molecule. Therefore, compound 3 can be docked correctly in E-selectin’s binding site, while compound 26 cannot be due to the receptor’s rigidity and the resulting steric clashes.
Fig. 4.
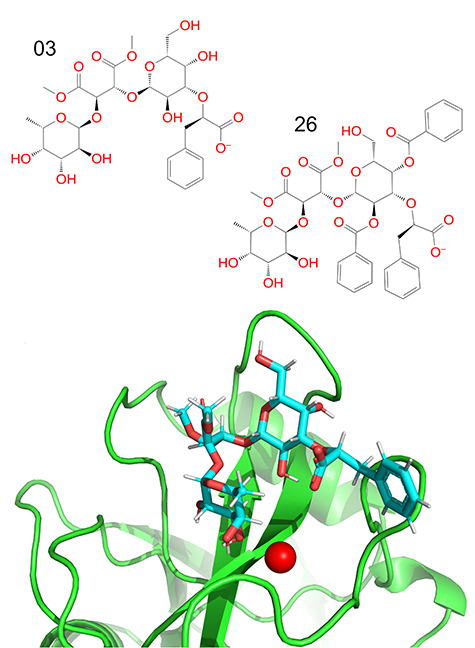
Compound 3 from the paper by Calosso et al. (2014)) docked in the binding site of E-selectin using the XP algorithm (left side), comparison of the structure of compounds 3 and 26 from the same paper (right side). The ligands are expected to bind in a Ca2+-dependent manner. This figure is available in black and white in print and in color at Glycobiology online.
Equation (2) was later used to predict the experimental activity relative to the activity of sLex, allowing to put the proposed inhibitors into the context of known/tested selectin antagonists. The set of inhibitors used to verify the method covers the values of log(ratio) ranging from −3 to 4.5. Thus, the applicability of some of the proposed inhibitors is limited.
Screening of the ligand databases
The previously verified methods were used for docking of compounds from the Assinex and ZINC databases. At first, for efficient screening of compounds, the compounds from selected databases were docked using the SP algorithm. The distributions of predicted binding energies with the SP algorithm are shown in Figure 3. The maximum of the distribution is located around −30 kcal/mol for all three selectins. Based on the data for SP docking algorithm presented in Figure 2, the majority of the compounds have a low or negligible affinity toward selectins. Hence, only a small portion of the compounds need to be docked at the XP level of precision.
Fig. 3.
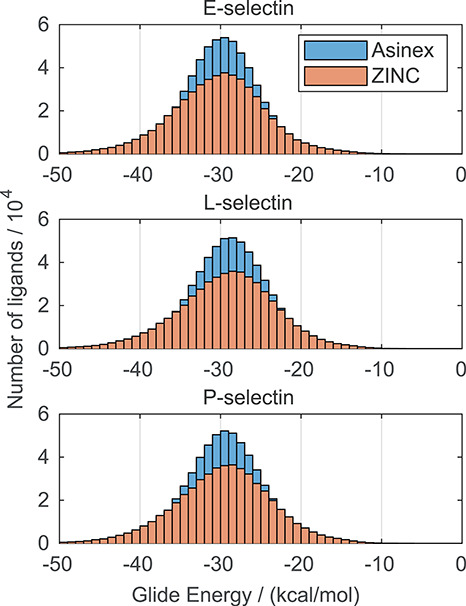
Distribution of Glide Energy of compounds from the Asinex and ZINC database for the different receptors predicted by the SP algorithm. This figure is available in black and white in print and in color at Glycobiology online.
For each selectin, the top 300 compounds from the ZINC database and 500 compounds from the Asinex databases were subsequently redocked at the XP level of precision. Since the SP algorithm may provide false-positive and false-negative results (more details are provided in Supplementary data, Figure S2), the results obtained with the SP algorithm were reviewed, and the number of redocked compounds from Asinex database was increased to enrich the set in glycomimetics and macrocyclic glycomimetics, which are included in Asinex database. In addition, the top 300 glycomimetics and macrocyclic glycomimetics from Asinex database were also docked at the XP level of precision to further explore the chemical space of glycomimetics. The distribution of Glide Energy for the glycomimetics at the SP level of accuracy is provided in Supplementary data, Figure S1.
The potential inhibitors of all three selectins were searched for. Numerous studies on selectin inhibitors indicate that an effective selectin antagonist has to inhibit at least two selectins (Tvaroška et al. 2020). Thus, for each compound, the average Glide energy for the three receptors that define a pan-selectin inhibitor was calculated
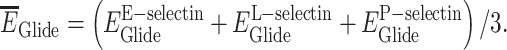 |
(3) |
The crystal structures and NMR studies revealed the bound conformation of sLex in selectin complexes (Egger et al. 2013, Poppe et al. 1997, Preston et al. 2016, Somers et al. 2000). These studies showed that crucial interactions for binding are the fucose residue interactions with the Ca2+ cation and the galactose residue interactions with amino acids in the binding site. Figure 6 shows the binding mode of sLex to the E- and P-selectin. Based on this information, several E-selectin inhibitors were prepared with potential for anti-inflammatory therapeutics (Egger et al. 2013). These investigations suggested that any potential inhibitor must exhibit the same interactions (including the coordination mode) with Ca2+ as the minimal carbohydrate determinant. Therefore, to eliminate the poses corresponding to binding to different locations or non-specific binding, the inhibitors were required to interact with the Ca2+ cation of the lectin domain. Thus, compounds that did not meet these conditions were excluded.
To filter out potential false-positive hits, compounds having a structural motive characteristic for PAINS were identified. For this purpose, PAINS-Remover was employed (Baell and Holloway 2010; Baell and Nissink 2018). Out of the 800 compounds used in the screening at the XP level of precision, 27 compounds were marked as PAINS. The predicted interactions of these compounds were revised, and the compounds with potentially nonspecific interactions were excluded. In the presented results, compound 7 was marked by the tool as a potentially false-positive hit due to catechol functional group characteristics for PAINS. Nonetheless, the catechol hydroxyl groups were predicted to interact with Ca2+ cation in the required orientation. Hence, the compound was not excluded.
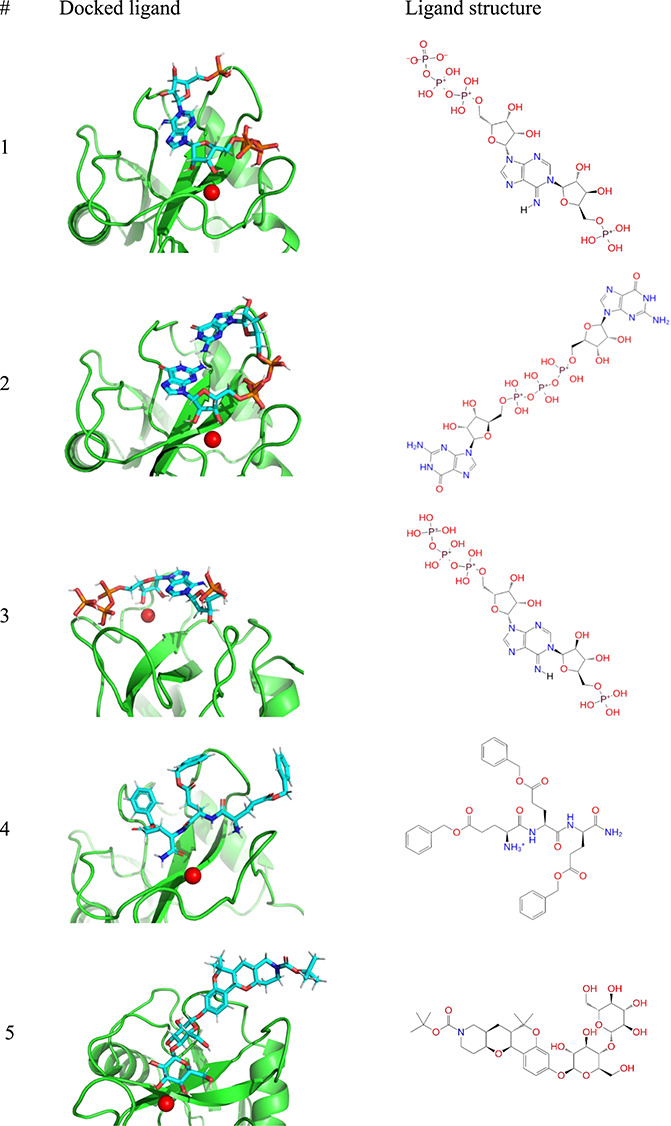
The compounds were sorted by the average Glide energy, and the list of the top 10 compounds is presented in Table I. The structures of the compounds and their binding poses for E-selectin are shown in Table II. Since the binding poses for P- and L-selectin are very similar to those for E-selectin (see Figure 6), they are not shown. Compounds 1, 2 and 3 are nucleotide-like molecules originating from the ZINC databases. Like the native selectin ligands, they interact with the Ca2+ cation through two ribose hydroxyl groups. Compound 4 is a tripeptide-like compound that interacts with the Ca2+ cation through the C-terminal amide group. Interactions of macrocyclic compound 6 with the Ca2+ involve a hydroxyl group and a carbonyl group (part of an amide group). Compound 7 exhibits the interaction of the two catechol hydroxyl groups with Ca2+. Recently, the catechols were shown to bind to calcium(II)-binding lectins, the binding mode was confirmed by the crystal structure (PDB ID 6YO3) (Kuhaudomlarp et al. 2021). Compounds 5, 8 and 9 are glycomimetics, which exhibit interactions of two hydroxyl groups with the Ca2+ cation. Their conformation matches the binding mode observed with the native ligands. Compound 10 also shows interactions of two hydroxyl groups with the Ca2+ cation and suitable orientation of the functional groups.
Table I.
Glide energy of the top 10 hits for all three selectins. Glide energy was calculated at the XP level of precision
| Glide energy/(kcal/mol) | ||||||
|---|---|---|---|---|---|---|
| Ligand code | # | E-selectin | L-selectin | P-selectin | Average | |
| ZINC000261496860 | 1 | −64.7 | −55.2 | −61.3 | −60.4 | |
| ZINC000085426282 | 2 | −71.1 | −53.6 | −53.0 | −59.2 | |
| ZINC000261496857 | 3 | −71.2 | −55.0 | −44.9 | −57.1 | |
| BAS 00380341 | 4 | −54.0 | −57.7 | −53.2 | −55.0 | |
| LAS 33898721 | 5 | −51.8 | −53.6 | −51.3 | −52.3 | |
| LAS 52112123 | 6 | −51.3 | −50.4 | −48.9 | −50.2 | |
| ASN 03798802 | 7 | −47.3 | −51.7 | −50.1 | −49.7 | |
| BDG 34018174 | 8 | −48.7 | −41.2 | −47.5 | −45.8 | |
| BDG 33898632 | 9 | −41.6 | −47.7 | −47.2 | −45.5 | |
| BAS 00060295 | 10 | −41.5 | −40.9 | −50.2 | −44.2 | |
Table II.
The top 10 compounds docked into the binding site of E-selectin. The structures of the compounds are shown on the right
|
The binding Glide energy of selected compounds ranges from −71 to −41 kcal/mol, and the average Glide energy, representing the assumed potency of a pan-selectin inhibitor, ranges from −59 to −44 kcal/mol. Compared with the Glide Energy of minimal carbohydrate determinant, sLex − 44.8 kcal/mol, obtained based on the linear fit (2), most of the presented compounds show better predicted binding affinity. The linear fit has also been employed for the estimation of their log(ratio), which describes their binding properties relative to sLex. The values of the estimated log(ratio) are provided in Table III. To put the docking results into the context of currently tested pan-selectin antagonists, GMI-1070 was also docked at the XP level of precision. Obtained values of Glide Energy are −59.9 kcal/mol with E-selectin, −70.4 kcal/mol with L-selectin and −68.9 kcal/mol with P-selectin, giving an average of −66.4 kcal/mol. This value is better compared with the top hits from the screening. Nonetheless, the impact of the high net charge of the compound on Glide Energy’s values needs to be questioned.
Table III.
Estimated log(ratio) values of proposed potential pan-selectin inhibitors for E- and P-selectin
| Ligand code | log(ratio) toward E-selectin | log(ratio) toward P-selectin |
|---|---|---|
| ZINC000261496860 | >4.5 | >4.5 |
| ZINC000085426282 | >4.5 | 3.1 |
| ZINC000261496857 | >4.5 | 0.0 |
| BAS 00380341 | 3.4 | 3.1 |
| LAS 33898721 | 2.6 | 2.4 |
| LAS 52112123 | 2.4 | 1.5 |
| ASN 03798802 | 0.9 | 2.0 |
| BDG 34018174 | 1.4 | 1.0 |
| BDG 33898632 | --1.2 | 0.9 |
| BAS 00060295 | --1.2 | 2.0 |
For comparison with other glycomimetics from Asinex database, the distribution of XP pan-selectin Glide Energy is shown in Figure 7. It suggests that most of the glycomimetics from the database have a similar or lower affinity to selectins than sLex.
Fig. 7.
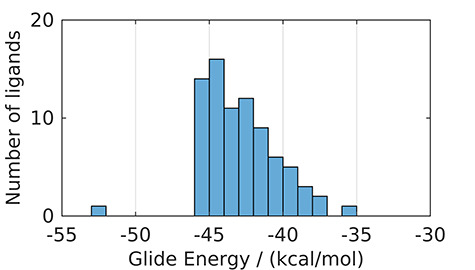
Distribution of pan-selectin Glide Energy for selected glycomimetics from the Asinex database predicted at the XP level of precision. This figure is available in black and white in print and in color at Glycobiology online.
For the predicted inhibitors, ADMET criteria were evaluated using the QikProp tool (Schrödinger Release 2017–2: QikProp, Schrödinger, LLC, New York, NY, 2017). Table IV shows predictions for some of the critical parameters for the assessment of the suitability of the compound for being used as a drug. However, since the predicted antagonists’ target is located outside cells, not all criteria necessarily need to be fulfilled. Because all selected compounds are required to bind to the binding site in an N-terminal calcium-dependent lectin domain responsible for sLex recognition, they will act as competitive inhibitors. Moreover, the use of compounds from Asinex and ZINC databases has the advantage of assuring the potential for medical use as they contain compounds with good ADMET properties, compounds having in vitro activity or biogenic compounds, including all FDA-approved drugs.
Table IV.
Parameters for assessing the suitability of the compound for being used as a drug predicted by QikProp
| Compound | Mw | P oct/W | Oral absorption/% | # Of violations of rule of 5 | # Of violations of rule of 3 |
|---|---|---|---|---|---|
| BAS 00380341 | 674.75 | 2.6 | 12.1 | 3 | 2 |
| LAS 52112123 | 646.74 | 3.8 | 57.6 | 2 | 2 |
| BDG 34018174 | 473.53 | 0.7 | 36.4 | 0 | 1 |
| ASN 03798802 | 713.86 | 2.7 | 37.1 | 2 | 2 |
| BDG 33898632 | 542.58 | --1.3 | 5.8 | 3 | 1 |
| BAS 00060295 | 592.55 | --0.8 | 1.1 | 3 | 2 |
| LAS 33898721 | 713.78 | 0.0 | 7.2 | 3 | 2 |
Elevated levels of selectins are associated with various pathophysiological processes, but selectins are also crucial in healing processes. Therefore, selectins’ blocking must be very carefully designed to avoid a potentially detrimental effect. Thus, selectin inhibitors require a very balanced mode of action. Consequently, the development of selectin inhibitors is a difficult task. For example, though the sLex is the minimal structural determinant recognized by all three selectins, the designed sLex analog celexin (Kerr et al. 2000) or sLex mimetics (Binder and Ernst 2011; Binder et al. 2012; Egger et al. 2013) were not developed into drug candidates. The selected compounds from Asinex and ZINC databases exhibit a good predicted pan-selectin activity and drug-like properties. Another advantage of these 10 compounds is that they can be commercially obtained. Therefore, the results suggest that some of the chosen compounds seem to be possible candidates for in vivo testing to speed up the development of therapeutics against COVID-19.
Our in silico screening results are supported by recently published data where virtual screening of the National Cancer Institute Diversity IV database led to a novel class of non-carbohydrate glycomimetics, catechols, that binds calcium(II)-binding lectins (Kuhaudomlarp et al. 2021). The binding modes of best catechols were confirmed by various experimental techniques and solving the crystal structure that established their binding mode.
Conclusions
The pandemic of coronavirus disease 2019 (COVID-19) represents a significant threat to human health. With a high mortality rate and the lack of effective COVID-19 therapy, novel approaches for developing efficient therapeutics are urgently needed. The recent clinical data highlight that COVID-19 is associated with severe acute respiratory symptoms that causes lung injury resulting from the overreactive immune system. Breaking this vicious cycle by blocking the mediators of the “cytokine storm” presents a possible strategy. Accumulating evidence shows that inflammatory cytokines induce selectin ligands that play a crucial role in the pathogenesis of inflammatory diseases by mediating leukocyte migration from the blood into the tissue. Thus, the selectins and selectin ligands represent a promising therapeutic target for the treatment of COVID-19.
The virtual screening of potential inhibitors recently became a popular method for screening the different proteins related to the spreading and virulence of COVID-19 due to its low cost and time requirements. In this paper, the Asinex and ZINC databases of compounds, altogether 923,602 compounds, were screened against the P-, L- and E-selectin to search for pan-selectin inhibitors. Since these databases contain approved drugs, biogenic compounds and glycomimetics, it may be assumed that the selected compounds have good drug-like properties.
At first, the experimentally confirmed inhibitors were docked into all three selectins’ carbohydrate recognition domains to assess the suitability screening procedure. The suitability of the docking procedure was verified against the experimental data published by Calosso et al. (2014)) and Gouge-Ibert et al. (2010)). It was found for the SP, and especially the XP docking algorithm, that Glide Energy correlates well with the experimental activity.
This study led to identifying 10 purchasable compounds that are potential pan-selectin inhibitors and suitable candidates for further experimental investigation of their use as COVID-19 therapeutics. Moreover, due to selectins’ involvement in the adhesion cascade of leukocytes and the promotion of metastases, these molecules might also be potential candidates for the development of anti-inflammatory and anti-cancer drugs.
Materials and methods
Receptor
The structures of the receptors (selectins) were taken from the PDB database: 1G1S for the P-selectin complex with the PSGL-1 fragment (obtained with resolution 1.90 Å), 3CFW for L-selectin (resolution 2.20 Å) and 1G1T for the E-selectin complex with sLex (resolution 1.50 Å) (Somers et al. 2000; Mehta-D'souza et al. 2017). The selectins were reported to adopt high-affinity and low-affinity conformations (Somers et al. 2000; Preston et al. 2016). Therefore, the corresponding residues in the L- and E-selectin structures were pulled toward their conformation in P-selectin (i.e. the high-affinity conformation) (Woelke et al. 2013), and the structures were optimized with sLex ligand with the use of AMBER 16 (details are provided in Supplementary data). For E-selectin, X-ray structure of high-affinity conformation is also available (PDB ID 4CSY, obtained with resolution 2.41 Å) (Preston et al. 2016); the comparison of the E-selectin structure used for docking with the structures published by Somers et al. and Preston et al. is shown in Figure 8. Prepared high-affinity conformation shares high structural similarity with the published high-affinity conformation. Nonetheless, the prepared structure was used for docking to ensure compatibility with our preliminary MD simulations and conformational analyses. Subsequently, the structures for docking were processed with Protein Preparation Wizard (Sastry et al. 2013) in Maestro. The grids were generated with the use of the Receptor Grid Generation tool, the dimensions of the grid box were set proportionally to sLex and the center of the box was placed at the center of the sLex tetrasaccharide in the optimized structures.
Fig. 8.
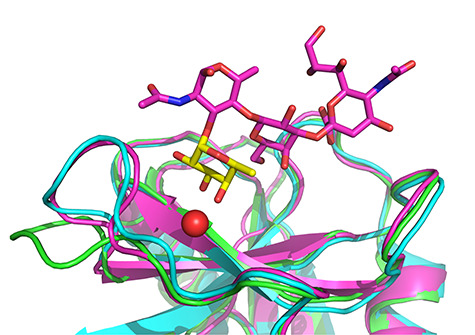
Comparison of structure of E-selectin (low-affinity conformation) soaked with sLeX published by Somers et al. (green), structure of E-selectin in high-affinity conformation co-crystalized with sLeX published by Preston et al. (cyan) and prepared high-affinity conformation used for docking (magenta) (Somers et al. 2000; Woelke et al. 2013; Preston et al. 2016). The fucose residue is shown in yellow and the Ca2+ cation as a red sphere. This figure is available in black and white in print and in color at Glycobiology online.
The side chains of the protein at the binding site are expected to have high conformational flexibility in solution, as they are mostly hydrophilic. Hence, the receptor scaling factors of van der Waals radii were set to 0.9, and the partial charge cutoff to 0.10 (determined by the preceding optimization of the method). These parameters partially compensate for the rigid receptor docking issues by downscaling the van der Waals radii of the nonpolar parts of the receptor. However, using a rigid receptor might still result in false-negative (a good binder requires slightly different conformation of the receptor) and false-positive results (the penalization of good binders resulting from their suboptimal conformation vs. a worse binder in its optimal conformation). However, a different study (Chaput and Mouawad 2017) showed that Glide could provide the correct top-ranked ligands for more than 60% of targets.
Ligands
The structures of experimentally confirmed ligands were obtained from the BindingDB (Gilson et al. 2016) database. Calosso et al. (2014)) developed acyclic inhibitors of selectins as analogs to the polysaccharide ligands, and Gouge-Ibert et al. (2010)) developed fluorinated inhibitors of selectins that mimicked sLex. The structures for virtual screening were obtained from the Asinex and ZINC databases of ligands. To reduce the chance of omitting potentially useful compounds, a more extensive set of ligands was employed.
Each compound was subjected to the ligand preparation protocol by LigPrep in Maestro: optimization with the use of the OPLS3e (Harder et al. 2016) force field and the generation of possible ionization states at a pH of 7.0 ± 2.0 using Epik (Shelley et al. 2007; Greenwood et al. 2010).
Asinex Elite Library® and Asinex Synergy Library contain compounds “screened against a panel of early ADMET tests to make sure screening hits are devoid of potential ADMET problems and are amenable for rapid hit-to-lead optimization” (Asinex 2020). Asinex Gold & Platinum Collections contain drug-like compounds for cost-effective screening. Asinex Signature Libraries include “cutting-edge chemistry combined with in silico and in vitro screening validation” (Asinex 2020). A total of 405,508 compounds from Asinex libraries (including Asinex Elite Library® and Asinex Synergy Library, Asinex Gold & Platinum Collections, Asinex Macrocycle Pre-Plated Set, PPI Pre-Plated Set, Glycomimetics Pre-Plated Set, Glycomimetics, Macrocyclic Glycomimetics and Macrocycles) were screened.
The screened ZINC libraries included compounds labeled as having in vitro activity (407,602 compounds) or tagged as biogenic (518,081 compounds). These also include all drugs approved by the FDA or other authorities and various natural products. The redundant compounds among the libraries were docked only once.
Docking
For an efficient screening of compounds, two docking algorithms were employed: (1) a standard precision (SP) algorithm; (2) the top 300 compounds from the ZINC libraries and top 500 compounds from the Asinex libraries for each receptor were docked using an extra precision (XP) algorithm, which was shown to provide a good correlation between the predicted and experimentally confirmed activity. The HTVS algorithm struggled to reproduce the experimental data with selectin ligands/inhibitors. Therefore, the HTVS algorithm was only tested and not used for screening the databases of ligands.
When applied to the selectins, we find that Glide Energy is the best of these parameters at reproducing the experimental activity. The following scaling parameters of van der Waals radii of ligands were used: scaling factor 0.90 and partial charge cutoff 0.15 (determined by the preceding optimization). Any competitive inhibitor of interactions of a selectin must bind into the same site as the native ligand. Thus, poses that did not comply the requirement were excluded. Moreover, PAINS-Remover (Baell and Holloway 2010; Baell and Nissink 2018) was employed to ensure that maximum of potential false positive hits are excluded.
The structures of complexes were visualized using PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Funding
Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601); Large Infrastructures for Research, Experimental Development and Innovations project “e-Infractructure CZ—LM2018140”; European Regional Development Fund—Project “MSCAfellow@MUNI” (No. CZ.02.2.69/0.0/0.0/17_050/0008496); Scientific Grant Agency of the Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences (grant VEGA-02/0024/16 to I.T.).
Supplementary Material
Contributor Information
Pavel Šmak, National Centre for Biomolecular Research, Faculty of Science, Masaryk University, 625 00 Brno, Czech Republic.
Selvaraj Chandrabose, Central European Institute of Technology (CEITEC), Masaryk University, 625 00 Brno, Czech Republic.
Igor Tvaroška, National Centre for Biomolecular Research, Faculty of Science, Masaryk University, 625 00 Brno, Czech Republic; Institute of Chemistry, Slovak Academy of Sciences, 845 38 Bratislava, Slovak Republic.
Jaroslav Koča, National Centre for Biomolecular Research, Faculty of Science, Masaryk University, 625 00 Brno, Czech Republic; Central European Institute of Technology (CEITEC), Masaryk University, 625 00 Brno, Czech Republic.
References
- Asinex . 2020. Screening Libraries (All Libraries): Asinex Corporation. [Google Scholar]
- Aydt EM, Bock D, Wolff G. 2010. Selectin antagonists and their potential impact for the treatment of inflammatory lung diseases. In: Hansel TT, editor. New drugs and targets for asthma and copd. Basel: Karger. p. 175–184. [Google Scholar]
- Baell JB, Holloway GA. 2010. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 53:2719–2740. [DOI] [PubMed] [Google Scholar]
- Baell JB, Nissink JWM. 2018. Seven year itch: pan-assay interference compounds (PAINS) in 2017-utility and limitations. ACS Chem Biol. 13:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. 2007. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. 11:1473–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard PW, Kaila N. 2010. Selectin inhibitors: A patent review. Expert Opin Ther Pat. 20:781–793. [DOI] [PubMed] [Google Scholar]
- Biancone L, Araki M, Araki K, Vassalli P, Stamenkovic I. 1996. Redirection of tumor metastasis by expression of E-selectin in vivo. J Exp Med. 183:581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder FPC, Ernst B. 2011. E- and P-selectin: Differences, similarities and implications for the design of P-selectin antagonists. Chimia. 65:210–213. [DOI] [PubMed] [Google Scholar]
- Binder FPC, Lemme K, Preston RC, Ernst B. 2012. Sialyl Lewisx: A "pre-organized water oligomer"? Angewandte Chemie-International Edition. 51:7327–7331. [DOI] [PubMed] [Google Scholar]
- Brostjan C, Anrather J, Csizmadia V, Natarajan G, Winkler H. 1997. Glucocorticoids inhibit E-selectin expression by targeting NF-kappa B and not ATF/c-Jun. J Immunol. 158:3836–3844. [PubMed] [Google Scholar]
- Calosso M, Tambutet G, Charpentier D, St-Pierre G, Vaillancourt M, Bencheqroun M, Gratton JP, Prevost M, Guindon Y. 2014. Acyclic tethers mimicking subunits of polysaccharide ligands: Selectin antagonists. ACS Med Chem Lett. 5:1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput L, Mouawad L. 2017. Efficient conformational sampling and weak scoring in docking programs? Strategy of the wisdom of crowds. J Chem. 9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronstein BN, Molad Y, Reibman J, Balakhane E, Levin RI, Weissmann G. 1995. Colchicine alters the quantitative and qualitative display of selectins on endothelial cells and neutrophils. J Clin Investig. 96:994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K. 1993. Evolution of Ca2+-dependent animal lectins. Prog Nucleic Acid Res Mol Biol. 45(45):207–232. [PubMed] [Google Scholar]
- Ebel ME, Awe O, Kaplan MH, Kansas GS. 2015. Diverse inflammatory cytokines induce selectin ligand expression on murine CD4 T cells via p38 alpha MAPK. J Immunol. 194:5781–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger J, Weckerle C, Cutting B, Schwardt O, Rabbani S, Lemme K, Ernst B. 2013. Nanomolar E-selectin antagonists with prolonged half-lives by a fragment-based approach. J Am Chem Soc. 135:9820–9828. [DOI] [PubMed] [Google Scholar]
- Ernst B, Magnani JL. 2009. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discov. 8:661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK et al. 2004. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 47:1739–1749. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. 2006. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 49:6177–6196. [DOI] [PubMed] [Google Scholar]
- Gilson MK, Liu TQ, Baitaluk M, Nicola G, Hwang L, Chong J. 2016. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouge-Ibert V, Pierry C, Poulain F, Serre AL, Largeau C, Escriou V, Scherman D, Jubault P, Quirion JC, Leclerc E. 2010. Synthesis of fluorinated C-mannopeptides as sialyl Lewis(x) mimics for E- and P-selectin inhibition. Bioorg Med Chem Lett. 20:1957–1960. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Crowther RL, Chandran C, Rumberger JM, Li S, Huang KS, Presky DH, Familletti PC, Wolitzky BA, Burns DK. 1994. Insight into E-selectin/ligand interaction from the crystal structure and mutagenesis of the lec/EGF domains. Nature. 367:532–538. [DOI] [PubMed] [Google Scholar]
- Greenwood JR, Calkins D, Sullivan AP, Shelley JC. 2010. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 24:591–604. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. 2004. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 47:1750–1759. [DOI] [PubMed] [Google Scholar]
- Harder E, Damm W, Maple J, Wu CJ, Reboul M, Xiang JY, Wang LL, Lupyan D, Dahlgren MK, Knight JL et al. 2016. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 12:281–296. [DOI] [PubMed] [Google Scholar]
- Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E et al. 2020. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 384:693–704. doi: 10.1101/2020.06.22.20137273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. 2005. ZINC - a free database of commercially available compounds for virtual screening. J Chem Inf Model. 45:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. 2012. ZINC: A free tool to discover chemistry for biology. J Chem Inf Model. 52:1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila N, Thomas BE. 2002. Design and synthesis of sialyl Lewis(x) mimics as E- and P-selectin inhibitors. Med Res Rev. 22:566–601. [DOI] [PubMed] [Google Scholar]
- Kaila N, Thomas BE. 2003. Selectin inhibitors. Expert Opin Ther Pat. 13:305–317. [DOI] [PubMed] [Google Scholar]
- Kansas GS. 1996. Selectins and their ligands: Current concepts and controversies. Blood. 88:3259–3287. [PubMed] [Google Scholar]
- Karp JM, Teol GSL. 2009. Mesenchymal stem cell homing: The devil is in the details. Cell Stem Cell. 4:206–216. [DOI] [PubMed] [Google Scholar]
- Kerr KM, Auger WR, Marsh JJ, Comito RM, Fedullo RL, Smits GJ, Kapelanski DP, Fedullo PF, Channick RN, Jamieson SW et al. 2000. The use of cylexin (CY-1503) in prevention of reperfusion lung injury in patients undergoing pulmonary thromboendarterectomy. Am J Respir Crit Care Med. 162:14–20. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Varki A. 1997. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj J. 14:569–576. [DOI] [PubMed] [Google Scholar]
- Kranich R, Busemann AS, Bock D, Schroeter-Maas S, Beyer D, Heinemann B, Meyer M, Schierhorn K, Zahlten R, Wolff G et al. 2007. Rational design of novel, potent small molecule pan-selectin antagonists. J Med Chem. 50:1101–1115. [DOI] [PubMed] [Google Scholar]
- Kuhaudomlarp S, Siebs E, Shanina E, Topin J, Joachim I, Silva Figueiredo Celestino Gomes P, Varrot A, Rognan D, Rademacher C, Imberty A et al. 2021. Non-carbohydrate glycomimetics as inhibitors of calcium(II)-binding lectins. Angew Chem Int Ed. doi: 10.1002/anie.202013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky LA. 1995. Selectin-carbohydrate interactions and the initiation of the inflammatory response. Annu Rev Biochem. 64:113–139. [DOI] [PubMed] [Google Scholar]
- Ludwig RJ, Schon MP, Boehncke WH. 2007. P-selectin: A common therapeutic target for cardiovascular disorders, inflammation and tumour metastasis. Expert Opin Ther Targets. 11:1103–1117. [DOI] [PubMed] [Google Scholar]
- Mannori G, Crottet P, Cecconi O, Hanasaki K, Aruffo A, Nelson RM, Varki A, Bevilacqua MP. 1995. Differential colon-cancer cell-adhesion to E-selectin, P-selectin, and L-selectin - role of mucin type glycoproteins. Cancer Res. 55:4425–4431. [PubMed] [Google Scholar]
- Mazo IB, Gutierrez-Ramos JC, Frenette PS, Hynes RO, Wagner DD, von Andrian UH. 1998. Hematopoietic progenitor cell rolling in bone marrow microvessels: Parallel contributions by endothelial selectins and vascular cell adhesion molecule 1. J Exp Med, 188:465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meduri GU, Kohler G, Hendley S, Tolley E, Stentz F, Postlethwaite A. 1995. Inflammatory cytokines in the bal of patients with ARDS - persistent elevation over time predicts poor outcome. Chest. 108:1303–1314. [DOI] [PubMed] [Google Scholar]
- Mehta-D'souza P, Klopocki AG, Oganesyan V, Terzyan S, Mather T, Li ZH, Panicker SR, Zhu C, McEver RP. 2017. Glycan bound to the selectin low affinity state engages Glu-88 to stabilize the high affinity state under force. J Biol Chem. 292:2510–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Martin JC. 2020. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat Rev Immunol. 20:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzaban JS, Burdick MM, Gadhoum SZ, Dagia NM, Chu JT, Fuhlbrigge RC, Sackstein R. 2011. Analysis of glycoprotein E-selectin ligands on human and mouse marrow cells enriched for hematopoietic stem/progenitor cells. Blood. 118:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montreal Heart Institute . 2020. New clinical study: Potential treatment for coronavirus will be tested in Canada as of today: BioSpace. [Google Scholar]
- Poppe L, Brown GS, Philo JS, Nikrad PV, Shah BH. 1997. Conformation of sLe(x) tetrasaccharide, free in solution and bound to E-, P-, and L-selectin. J Am Chem Soc. 119:1727–1736. [Google Scholar]
- Preston RC, Jakob RP, Binder FPC, Sager CP, Ernst B, Maier T. 2016. E-selectin ligand complexes adopt an extended high-affinity conformation. J Mol Cell Biol. 8:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes AZ, Hu KA, Teperman J, Wampler Muskardin TL, Tardif JC, Shah B, Pillinger MH. 2020. Anti-inflammatory therapy for COVID-19 infection: The case for colchicine. Ann Rheum Dis. 1–8. doi: 10.1136/annrheumdis-2020-219174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano SJ, Slee DH. 2001. Targeting selectins for the treatment of respiratory diseases. Curr Opin Investig Drugs. 2:907–913. [PubMed] [Google Scholar]
- Ruan QR, Yang K, Wang WX, Jiang LY, Song JX. 2020. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China (vol 17, pg 851, 2020). Intensive Care Med. 46:1294–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruster B, Gottig S, Ludwig RJ, Bistrian R, Muller S, Seifried E, Gille J, Henschler R. 2006. Mesenchymal stem cells display coordinated rolling and adhesion behavior on endothelial cells. Blood. 108:3938–3944. [DOI] [PubMed] [Google Scholar]
- Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, Wohlgemuth R. 2008. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med. 14:181–187. [DOI] [PubMed] [Google Scholar]
- Sahin AO, Buitenhuis M. 2012. Molecular mechanisms underlying adhesion and migration of hematopoietic stem cells. Cell Adh Migr. 6:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. 2013. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 27:221–234. [DOI] [PubMed] [Google Scholar]
- Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. 2007. Epik: A software program for pK (a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 21:681–691. [DOI] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT. 2000. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 103:467–479. [DOI] [PubMed] [Google Scholar]
- Sterling T, Irwin JJ. 2015. ZINC 15-ligand discovery for everyone. J Chem Inf Model. 55:2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvaroška I, Chandrabose S, Koča J. 2020. Selectins—The two Dr. Jekyll and Mr. Hyde faces of adhesion molecules—A review. Molecules. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P, Arda A, Reichardt NC, Jimenez-Barbero J, Gimeno A. 2019. Glycans in drug discovery. Fortschr Med. 10:1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira PA, Marcelo F, Grewal RK. 2017. Glycosyltransferase inhibitors: A promising strategy to pave a path from laboratory to therapy. In: Carbohydrate chemistry: Chemical and biological approaches. London (UK): The Royal Society of Chemistry.
- Wang S, Vidal S. 2013. Recent design of glycosyltransferase inhibitors. In: Rauter AP, Lindhorst TK, editors. Carbohydrate Chemistry: Chemical and Biological Approaches, Vol. 39. Cambridge: Royal Society of Chemistry. p. 78–101. [Google Scholar]
- Woelke AL, Kuehne C, Meyer T, Galstyan G, Dernedde J, Knapp EW. 2013. Understanding selectin counter-receptor binding from electrostatic energy computations and experimental binding studies. J Phys Chem B. 117:16443–16454. [DOI] [PubMed] [Google Scholar]
- Woodside DG, Vanderslice P. 2008. Cell adhesion antagonists - therapeutic potential in asthma and chronic obstructive pulmonary disease. BioDrugs. 22:85–100. [DOI] [PubMed] [Google Scholar]
- Yang J, Hirata T, Croce K, Merrill-Skoloff G, Tchernychev B, Williams E, Flaumenhaft R, Furie BC, Furie B. 1999. Targeted gene disruption demonstrates that P-selectin glycoprotein ligand 1 (PSGL-1) is required for P-selectin-mediated but not E-selectin-mediated neutrophil rolling and migration. J Exp Med. 190:1769–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du RH, Fan GH, Liu Y, Liu ZB, Xiang J, Wang YM, Song B, Gu XY et al. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 395:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


