Summary
Glycans are one of the fundamental classes of macromolecules and are involved in a broad range of biological phenomena. A large variety of glycan structures can be synthesized depending on tissue or cell types and environmental changes. Here, we developed a comprehensive glycosylation mapping tool, termed GlycoMaple, to visualize and estimate glycan structures based on gene expression. We informatically selected 950 genes involved in glycosylation and its regulation. Expression profiles of these genes were mapped onto global glycan metabolic pathways to predict glycan structures, which were confirmed using glycomic analyses. Based on the predictions of N-glycan processing, we constructed 40 knockout HEK293 cell lines and analyzed the effects of gene knockout on glycan structures. Finally, the glycan structures of 64 cell lines, 37 tissues, and primary colon tumor tissues were estimated and compared using publicly available databases. Our systematic approach can accelerate glycan analyses and engineering in mammalian cells.
eTOC Blurb
Diverse glycan structures are synthesized depending on cell types. Huang et al. have developed a comprehensive glycosylation mapping tool based on gene expression, named GlycoMaple, for visualizing glycosylation pathways and estimating glycan structures synthesized in cells. The tool could contribute to supporting glycan analysis, biomarker development, and glycoengineering.
Graphical Abstract
Introduction
Glycans are one of the major macromolecules that make up cells where they exist in free forms or as glycoconjugates bound to proteins and lipids. They are recognized by intrinsic and extrinsic glycan-binding proteins and play key roles in both intracellular and intercellular events that influence multiple biological functions and processes (Ohtsubo and Marth, 2006; Varki, 2017). Glycans on proteins regulate protein folding, selective transport, stability, and activity (Helenius and Aebi, 2004; Kamiya et al., 2008). In addition to physiological functions, glycan structures can affect pathological conditions. Several glycans on the cell surface act as receptors for toxins and virus infections (Raman et al., 2016; Thompson et al., 2019). Inherited pathological mutations in genes involved in glycan synthesis and degradation cause severe developmental disorders (Ng and Freeze, 2018). Glycosylation also defines the oncogenic stage and metastatic potential during tumor progression (Munkley and Elliott, 2016; Pinho and Reis, 2015). Therefore, determining the structures of complex glycans can lead to both the identification of glycan biomarkers and the development of vaccines and therapeutics in biomedical and biotechnological contexts (Hutter and Lepenies, 2015; Mereiter et al., 2019).
Glycan structures are synthesized by a series of reactions mediated by glycosyltransferases or glycoside hydrolases (Bourne and Henrissat, 2001). These enzymes need to be correctly translated, folded, and localized to become active. In addition, these enzyme activities are regulated by substrate or product concentrations and environmental conditions. Although many factors are involved in the glycosylation pathway, glycan structural diversity could be predicted from gene expression data if a model could be generated (Narimatsu et al., 2019). So far, a great deal of efforts have been made to construct simulation models to predict glycan structures on proteins using kinetic, transcriptomic, and mass spectrometric information (Jimenez del Val et al., 2011; Krambeck et al., 2009; Krambeck and Betenbaugh, 2005; Kremkow and Lee, 2018; Liu et al., 2013; Umaña and Bailey, 1997). Probabilistic modeling frameworks were untilized to develop a low-parameter approach to predict the effects of gene engineering (Liang et al., 2020; Spahn et al., 2016). Moreover, artificial neural networks with minimal parameter estimation are emerging to describe N-glycosylation (Kotidis and Kontoravdi, 2020). Despite transcription levels do not always reflect enzyme levels directly, prediction of glycan structures included gene expression data show strong consistency compared with mass spectra alone (Bennun et al., 2013). However, all the previous works only focused on a single glycosylation pathway such as the N-glycosylation. It is desired to show a more bird’s-eye view of glycan metabolic pathways. In the present study, we constructed a global glycosylation mapping tool, termed GlycoMaple. Our systematic approach will contribute to supporting glycan analysis, the exploration of disease biomarkers, and glycan engineering to produce biopharmaceutical proteins.
Results
Construction of a comprehensive glycosylation mapping tool, GlycoMaple
The ability to predictively estimate the profile of glycan structures produced by a cell or tissue would be highly useful for understanding cellular properties and regulation, as well as for engineering cells. We developed a comprehensive glycosylation mapping tool, termed GlycoMaple, based on gene expression profiles. This tool enabled us to visualize glycosylation pathways and estimate glycan structures in cells of interest. We first generated a comprehensive list of glycan-related genes, which included genes encoding glycosyltransferases (GTs), glycoside hydrolases (GHs), glycan-binding proteins (lectins), and genes required for substrate synthesis and transport, regulators of activity or localization of GTs, and Golgi homeostasis. We first added the 200, 78, and 15 human genes encoding GTs, GHs, and carbohydrate-binding modules, respectively, that are registered in the CAZy database (Lombard et al., 2014). Subsequently, genes encoding proteins other than GTs or GHs were also added, such as genes encoding sulfotransferases, regulatory factors, and sugar nucleotide synthesis and transport, and genes involved in Golgi homeostasis for glycoconjugate synthesis and degradation. The aim of GlycoMaple is not only to visualize glycosylation pathways, but also to overview the expression of glycan-related genes by category. Therefore, 189 genes encoding lectins or glycan-binding proteins were accumulated from genetic resources for animal lectins (http://www.imperial.ac.uk/research/animallectins/default.html) or were added manually (Kilpatrick, 2002; Taylor et al., 2015). For protein modification by a glycosylphosphatidylinositol (GPI) or glycosaminoglycans (GAGs), genes encoding carrier proteins were added to the list (Esko and Selleck, 2002; Liu et al., 2018; Mizumoto et al., 2014). We compared our list with published papers that also collected glycan-related genes (Nairn et al., 2012; Nairn et al., 2008; Schjoldager et al., 2020), and manually updated our list with the most accurate information. In total, 950 genes were collected and categorized into 30 groups (Tables S1 and S2). We next drew 19 maps for the biosynthesis and catabolic pathways of glycans in human. To visualize the glycan structures, symbol nomenclature for glycans (SNFG) was used (Neelamegham et al., 2019). The genes required for each reaction were then input into the pathway maps (Table S3). The substrate specificities of each GT, GH, or sulfotransferase were curated based on handbooks and original papers (Taniguchi et al., 2011).
Glycomic analysis of HEK293 cells
To verify the usefulness of GlycoMaple, we used the human embryonic kidney 293 (HEK293) cell line as a model. The HEK293 cell line is one of the most commonly cultured cell lines in basic biomedical researches (Stepanenko and Dmitrenko, 2015) as well as in biotechnological contexts (Thomas and Smart, 2005). We first analyzed N- and O-glycan structures and GSLs in HEK293 cells using mass spectrometry (Figure 1 and S1A).
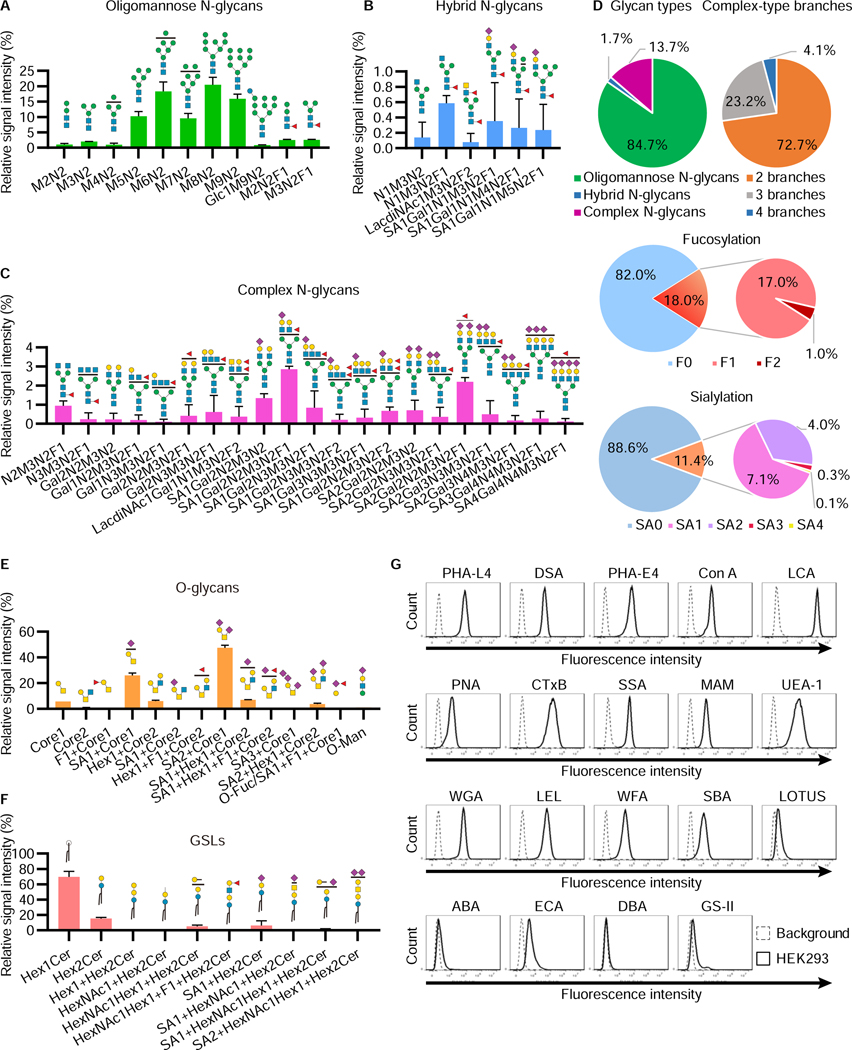
Figure 1. Glycomic analysis of HEK293 cells.
A–D, Relative abundance of N-glycans in HEK293 cells. Permethylated N-glycan structures in HEK293 cells were analyzed using NSI-MS. The relative abundances of oligomannose (A), hybrid (B), and complex (C) N-glycan structures are shown. The symbol of each sugar follows the SNFG (Neelamegham et al., 2019). Fucoses represented on the N-glycan structures are attached to either the core or branches on N-glycans. Those fucosylated structures were mixtures of both structures (See Figure S1B). The distribution of N-glycan-type structures and the branch number of complex structures are shown, as well as the number of fucoses and sialic acids on N-glycans (D). The data represent the means ± errors from two independent experiments.
E, F, Relative abundance of O-glycans (E) and GSLs (F) in HEK293 cells. The symbol of each sugar follows the SNFG. The blank circles and squares represent hexoses (Hex) and N-acetyl-hexosamines (HexNAc), respectively. The sugars such as fucose and sialic acid depicted at the top of each structure have multiple binding sites, which are represented as compositions (See Figure S1C, S1D, and Table S4). The data represent the means ± errors from two independent experiments.
G, Lectin staining of glycan structures on the cell surface. HEK293 cells were incubated with 19 lectins conjugated with fluorescein or biotin and analyzed by flow cytometry. The data show representative results from at least three independent experiments.
In the N-glycan analysis, 38 composition structures were detected (Figures 1A–C and Table S4). The percentages of oligomannose, hybrid, and complex N-glycan structures were 84.7%, 1.7%, and 13.7%, respectively (Figure 1D). In the complex N-glycans, tetra-antennary structures were observed (4.1%), although biantennary structures were the most abundant (72.7%). N-glycan structures containing LacdiNAc structures were also detected (Figures 1B, C and S1B). Fucosylated and sialylated N-glycans were detected as 18.0% and 11.4% of total glycan structures, respectively.
In the O-glycan analysis, 14 compositions including 23 structures were observed (Figure 1E). Most were mucin-type O-N-acetylgalactosamine (GalNAc) structures (21 structures), although O-Man and O-Fuc structures were also detected (Figure S1C and Table S4). The major structures were disialyl-T and monosialyl-T antigens. Only the structures based on core 1 and core 2 were detected in mass spectrometry at MS1 (i.e., without fragmentation that increases sensitivity). Core 3 and core 4 structures were only assigned from MS2 signature ions, suggesting that these core structures are present at low abundance or contaminated from serum (Table S4). O-Glycan structures with a fucose modified to either Gal or GlcNAc were also detected (Figure S1D).
In the GSL analysis, ceramide structures with 10 different sugar compositions were detected (Figure 1F). Hexosylceramides that were either glucosylceramides or galactosylceramides were the most abundant structures. In addition, lactosylceramides, globo-series (Gb3 and Gb4), and ganglio-series glycosphingolipids were detected. Lacto- or neolacto-series glycosphingolipids were also observed (Table S4).
Mass spectrometric analysis of glycans has several limitations, including in the analyses of linkage, branches, isomers, and glycans with high molecular sizes. Thus, to further characterize glycan signatures on the cell surface, lectin-based analyses were also carried out. We used 19 fluorescent-conjugated lectins for cell staining (Figure 1G). The staining with lectins that detect N-glycans suggested the existence of oligomannose structures (ConA staining), GlcNAc-β1,4-(GlcNAc-β1,2-)Man branches (DSA staining), GlcNAc-β1,6-(GlcNAc-β1,2-)Man branches (PHA-L4 staining), bisected structures (PHA-E4 staining), LacdiNAc structures (WFA staining), and core-fucose (LCA staining) structures. Staining with SSA, MAM, and UEA-1 represented the structures modified by α−2,6-sialic acid, α−2,3-sialic acid, and α−1,2-Fuc, respectively. CtxB staining suggested that ganglioside GM1a or fucosylated Lewis structures exist in HEK293 cells (Wands et al., 2018). Together, HEK293 cells can produce various types of glycan structures, which is useful for the validation of glycosylation pathways using GlycoMaple analysis.
Evaluation of GlycoMaple using O-glycomics data in HEK293 cells
We next analyzed gene expression profiles in HEK293 cells using RNA-seq to predict glycosylation pathways in GlycoMaple. Each gene expression value was calculated as transcripts per million (TPM) (Wagner et al., 2012). We detected the expression of 950 glycan-related genes (Table S5) to integrate the information with the pathway maps. To evaluate GlycoMaple, we focused on mucin-type O-glycan structures. Among the 53 mucin-type O-glycan structures in the GlycoMaple pathway map, 21 glycan structures detected in our glycomics data and previous reports (O-glycan structures on MUC-1 (Razawi et al., 2013) expressed in HEK293 cells) were used as the actual detected structures (Figure 2A).
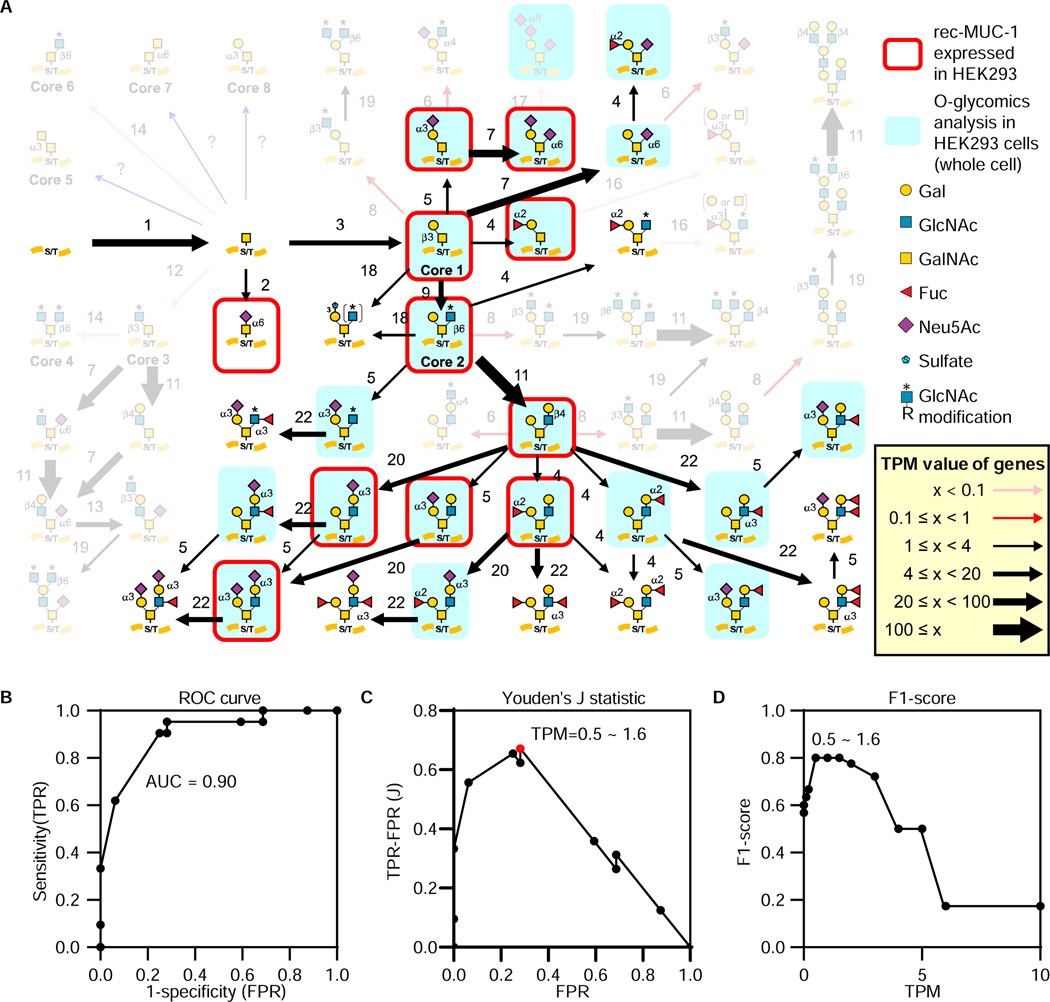
Figure 2. Validation of GlycoMaple using mucin-type O-glycan structures in HEK293 cells.
A, Mucin-type O-glycans were compared between GlycoMaple estimation and MS analysis. Gene expression in HEK293 cells was analyzed using RNA-seq. Based on the gene expression profile (TPM values), the mucin-type O-glycan biosynthetic pathways were visualized. Each arrow represents the expression of genes responsible for the reaction. Thin pink arrows (TPM < 0.1) or red arrows (0.1 ≤ TPM < 1) indicate that the responsible genes for the pathways are not expressed or rarely expressed in the cells, respectively. The black arrows (1 ≤ TPM) indicate that the genes in the pathways are considered to be expressed in the cells. The thickness of these arrows shows the expression levels of the genes: thin black arrows, 1 ≤ TPM < 4; normal black arrows, 4 ≤ TPM < 20; thick black arrows, 20 ≤ TPM < 100; very thick black arrows, 100 ≤ TPM. If several genes overlapped in a reaction, the maximum TPM value among the values of overlapped genes was used. When several gene products make a complex for a reaction, the minimum TPM value of the subunit genes was used. Blue arrows indicate the reactions for which the responsible genes are not clear. The O-glycans identified by MS glycomic analyses in HEK293 cells (Table S4) or on MUC-1 from HEK293 (Razawi et al., 2013) are shaded blue and/or outlined in red, respectively. Those not predicted by GlycoMaple are rendered opaque. Each numbered reaction and the responsible genes are listed in Table S3 and S5, respectively.
B, Relationship between False Positive Rate (FPR) and True Positive Rate (TPR) obtained in Figure S2A was plotted as a ROC curve. The AUC value was calculated.
C, D, Youden’s J statistic (C) and F1-score (D) at various TPM thresholds were plotted. The highest score of the Youden’s J statistic (highlighted as a red dot) was corresponding to the TPM thresholds from 0.5–1.6. F1-score also showed that TPM values at 0.5–1.6 were the best.
We then mapped the gene expression profile of HEK293 cells onto the mucin-type O-glycan biosynthesis pathway. The predicted glycan structures were estimated at various TPM threshold values (0 to 10). We collected the number of true-positive, false-positive, true-negative, and false-negative predictions, in order to calculate the accuracy, precision, sensitivity (recall), and specificity under the different thresholds (Figure S2A). Based on the calculated values, we plotted the receiver operating characteristic (ROC) curve, and measured the area under the curve (AUC), which gave a performance measurement for our classification model at various threshold settings (Figure 2B). If AUC is greater than 0.7, the model generally shows acceptable discrimination (Mandrekar, 2010). When we used the predicted O-glycan structures by GlycoMaple and the detected O-glycan structures, the AUC was 0.90, indicating that the GlycoMaple analysis shows high performance and provides excellent discrimination.
We next validated GlycoMaple with other cell types using published data. For detected O-glycan structures, data in a published paper were utilized (Fujitani et al., 2013), which reported glycomic analyses in various human cell lines. Among these, RNA-seq data in 6 cell lines (A549, Caco2, HeLa, HEK293, HepG2, and HL60) were available in the Human Protein Atlas. Using these data, the ROC curve and AUC were measured (Figure S2B). The AUC values in A549, Caco2, HeLa, HEK293, HepG2, and HL60 cells were 0.78, 0.74, 0.82, 0.80, 0.89, and 0.85, respectively. The lower AUC values compared with our HEK293 data would be due to the different culture condition and limitations in O-glycan analysis (Zaia, 2010). Nevertheless, these positive results indicate that, using the expression profile of glycan-related genes, GlycoMaple can be employed for the estimation of glycan biosynthesis.
Threshold setting of the TPM value to estimate glycosylation pathways
We next evaluated the TPM threshold in the reaction pathways of GlycoMaple using mucin-type O-glycan structures in HEK293 cells. We measured the Youden’s J statistic which determines the cut-off values for prediction (Figure 2C). The highest score of this statistic corresponded to TPM values from 0.5–1.6. The F1-score, which is the harmonic means of sensitivity and precision, also showed TPM values from 0.5–1.6 having the best performance (Figure 2D). We next analyzed the 6 human cell lines used in the above to determine the optimal TPM threshold (Figure S2C). Since the public glycomics data have restrictions as described above, false-positive rate was increased at low TPM threshold in some cell lines. Nevertheless, the average F1-score of the 6 cell lines were highest at TPM = 1. Therefore, we set the threshold of the TPM value as 1. When the TPM value was < 1, we defined the gene as not expressed or rarely expressed in the cells. When the TPM value was ≥ 1, the gene was considered to be expressed (Figure 2A).
Based on the TPM threshold (TPM = 1), we estimated the mucin-type O-glycan structures in HEK293 (Figure 2A). Core 1 and core 2 should be expressed as a result of the expression of C1GALT1 and C1GALT1C1 (step 3) and GCNT1, 3, and 4 (step 9), respectively, but HEK293 cells could not form core 3 and core 4 because of the lack of B3GNT6 expression (step 12) (Iwai et al., 2002). Furthermore, because there was a very limited expression of A4GNT (step 6), B3GNT3 (step 8), GCNT3 (step 14), and ABO (step 16), the glycan structures formed by these reactions would rarely occur in HEK293 cells. These estimations matched the glycomics data (Figure 1E and Table S4). On the other hand, a tri-sialylated core 1 structure (Sia-Sia-Gal-(Sia-)GalNAc), which was detected by mass spectrometry (Figure 1E), was not estimated in GlycoMaple. The pathways in GlycoMaple are drawn based on the reported knowledge. So far, only ST8SIA6 has been reported to mediate the synthesis of the tri-sialylated core 1 from the Sia-Gal-(Sia-)GalNAc structure (Teintenier-Lelièvre et al., 2005). Because of the low expression of ST8SIA6 (TPM = 0.09) in HEK293 cells, it was estimated that this structure would not be synthesized by GlycoMaple. Although this could be viewed as a false negative judgement, this result also suggests that enzymes other than ST8SIA6 might contribute to the synthesis of this structure.
Visualization of glycosylation pathways and estimation of glycan structures in HEK293 cells
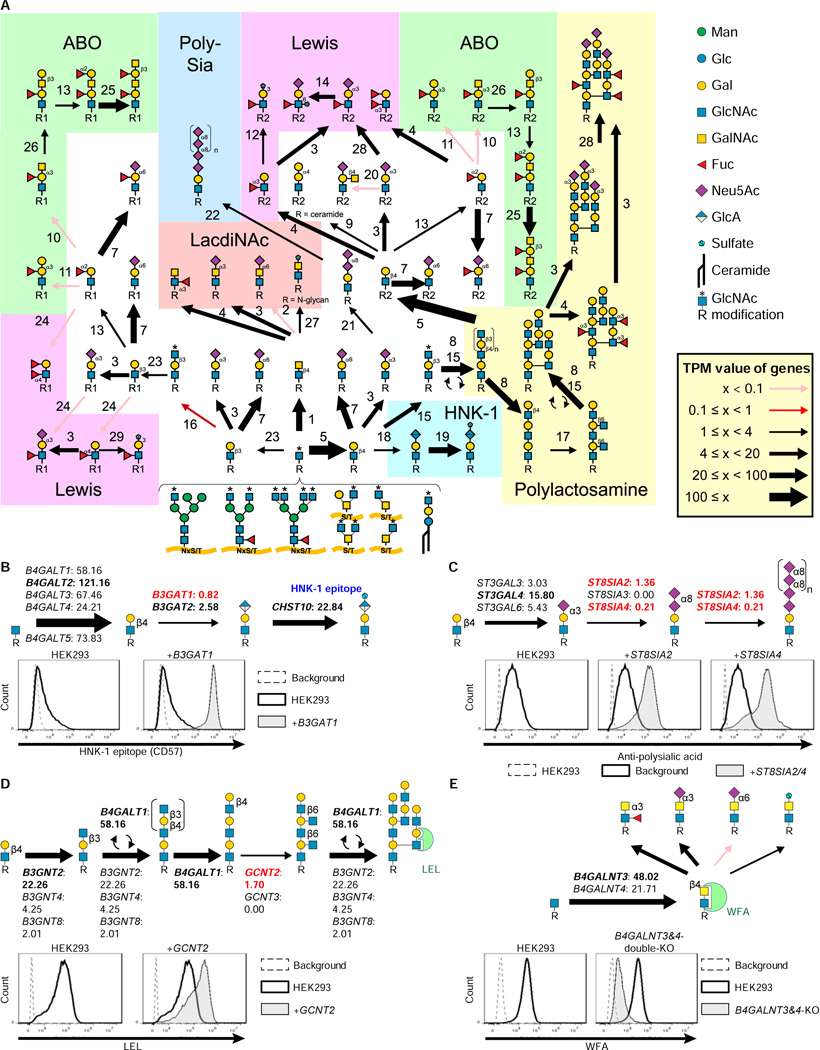
We input gene expression information to all the 19 pathways for glycan synthesis and degradation (Figure S3). GlycoMaple could visualize the target glycan synthetic pathway and estimate the structures based on the expression levels of genes. After the core structures of N- and O-glycans and GSLs are synthesized, for example, GlcNAc residues on the core structures are modified to form more complex capping structures, such as polysialic acid, LacdiNAc, ABO-type antigens, Lewis-type antigens, and polylactosamine (Morise et al., 2017; Stowell and Stowell, 2019; Ujita et al., 1999). Mapping the gene expression profiles of HEK293 cells estimated that glycans such as ABO- and Lewis a- and b-type antigens would not be formed in HEK293 cells (Figure 3A). Among the capping structures, we picked up glycan structures including the human natural killer-1 (HNK-1) epitope, polysialic acid, and branched polylactosamine structures, which were expected to be low because of the limited expression of one or more genes responsible for these reactions. The HNK-1 epitope is a unique trisaccharide that possesses sulfated glucuronic acid (GlcA) at the non-reducing terminus (SO3−−3-GlcA-β1,3-Gal-β1,4-GlcNAc-) (Morise et al., 2017). The biosynthesis of the HNK-1 epitope requires the expression of genes encoding β1,4-galactosyltransferases (the first step), β1,3-glucuronyltransferases (the second step), and carbohydrate sulfotransferases (the third step). Although the genes responsible for the first and third steps were well-expressed, the expressions of B3GAT1 (TPM: 0.82 ± 0.07) and B3GAT2 (TPM: 2.58 ± 0.24), which are responsible for the second step, were low (Figure 3B). In particular, B3GAT1 encodes the major GlcA transferase for HNK-1 synthesis (Morise et al., 2017). Consistent with our estimation, the HNK-1 epitope was only weakly expressed in HEK293 cells. When B3GAT1 was overexpressed, HEK293 cells began to produce glycans with HNK-1 (Figure 3B). Polysialic acid is found predominantly on neuronal cell adhesion molecules (NCAMs) of neurons and immune cells (Colley et al., 2014). To generate polysialic acid-containing glycans, ST8SIA2 or ST8SIA4 is required (Kanehisa et al., 2017). In HEK293 cells, NCAM1 was expressed (TPM: 12.90 ± 0.22; Table S5), but the expression of both ST8SIA2 and ST8SIA4 was limited (Figure 3C). Therefore, the cells cannot produce polysialic acid structures. By overexpressing ST8SIA2 or 4 in HEK293 cells, polysialic acid structures were able to be generated (Figure 3C). In the polylactosamine synthesis, we found that the expression of GCNT2, which encodes an enzyme required for I-branched polylactosamine structure formation, was low. When GCNT2 was overexpressed, LEL staining was increased (Figure 3D), suggesting that its expression is the limiting step for polylactosamine formation in HEK293 cells. In contrast, LacdiNAc structures were observed in HEK293 cells (Figure 1C and S1B). B4GALNT3 and 4, which are responsible for LacdiNAc formation (Figure 3E), were expressed in HEK293 cells. By knocking out these two genes, we were able to eliminate LacdiNAc structures. These results indicate that the GlycoMaple contributes to finding the limiting or critical steps in glycosylation pathways, and can predict changes in glycans by genetic manipulations.
Figure 3. Visualization and estimation of capping structures in HEK293 cells.
A, Visualization of the biosynthetic pathway for capping structures in HEK293 cells. The setting of arrows was described in Figure 2A. The TPM values of each gene are presented in Table S5.
B, The biosynthesis pathway of the HNK-1 epitope. The expression of genes involved in HNK-1 epitope synthesis is shown. The TPM values (means from triplicated RNA-seq data) were used. HEK293 cells and HEK293 cells stably expressing B3GAT1 were stained with anti-CD57 (HNK-1) antibody followed by Alexa488-conjugated anti-mouse IgM. Cells were analyzed using flow cytometry. Background, without primary antibody.
C, The biosynthesis pathway of polysialic acid. The expression of the genes involved in polysialic acid synthesis is shown. HEK293 cells and HEK293 cells stably expressing ST8SIA2 or ST8SIA4 were stained with anti-polysialic acid antibodies followed by PE-conjugated anti-mouse antibodies.
D, The biosynthesis pathway of polylactosamine. The expression of genes required for polylactosamine structure synthesis is shown. HEK293 cells and HEK293 cells stably expressing GCNT2 were stained with FITC-conjugated LEL lectin and analyzed using flow cytometry.
E, The biosynthesis pathway of LacdiNAc. The gene expression of B4GALNT3 and 4 in HEK293 cells is shown. HEK293 B4GALNT3 and 4 double-KO cells were stained with WFA lectin and analyzed using flow cytometry.
Construction of a cell library deficient in genes related to the N-glycosylation pathway
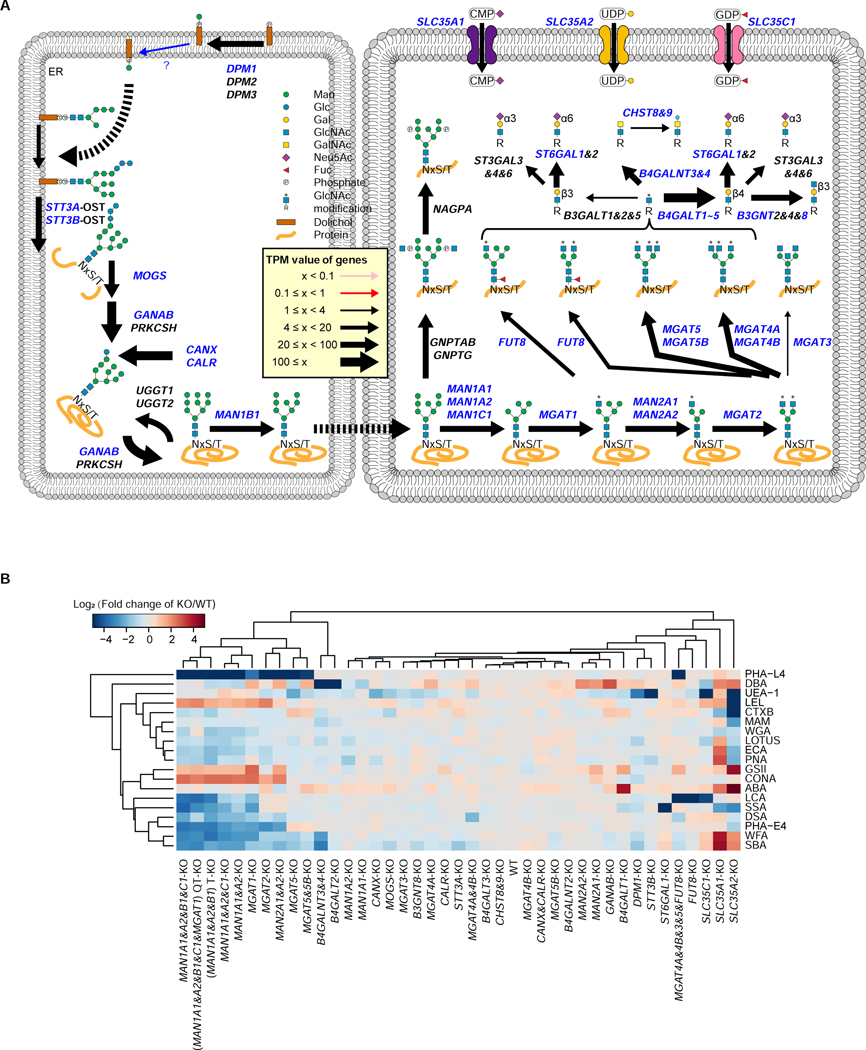
We next selected genes involved in N-linked glycan processing and capping, based on the gene expression profiles measured in HEK293 cells, and generated a HEK293 cell library in which genes related to N-linked glycosylation were knocked out (Figure 4A). Forty different kinds of KO cells were established (Table S6), and we characterized the glycan changes in these cells using 19 different lectins (Figure 4B). The KO cells deficient in various mannosidase-I genes clustered together in the heatmap, since their N-glycan structures were unable to be processed beyond oligomannoses (Jin et al., 2018; Ren et al., 2019). Consistent with this expectation, staining with a Man-binding lectin, ConA, was increased, whereas staining with lectins that bind to complex N-glycans, such as PHA-L4, PHA-E4, and DSA, was decreased (Figure 4B). MGAT1-KO, MGAT2-KO, and MAN2A1- and A2-double-KO cells showed similar staining patterns to those of the multiple mannosidase-I-KO cells. In MGAT5-KO cells, PHA-E4 staining was increased, suggesting that N-glycans containing bisecting GlcNAc were upregulated, which was confirmed by mass spectrometric analysis (Figure S4A).
Figure 4. Construction of the KO cell library for the N-glycosylation pathway.
A, Processing of the N-glycosylation pathway. Genes required for the N-glycosylation pathway are illustrated. Arrow thicknesses represent expression values as described in Figure 2A. The gene products highlighted in blue were knocked out in HEK293 cells.
B, Lectin staining profiles of the KO cell library. Forty different KO cells were constructed in HEK293 cells and analyzed by staining with 19 lectins. The staining of HEK293 cells by each lectin was set as 1. The staining intensities of each lectin in the cells were compared with those of the HEK293 cells, and were calculated as relative intensities. The log2 values of the data are visualized as a clustered heatmap. The data used the mean value from two independent experiments.
New insights into glycan regulation revealed by combining GlycoMaple and glycomic analysis
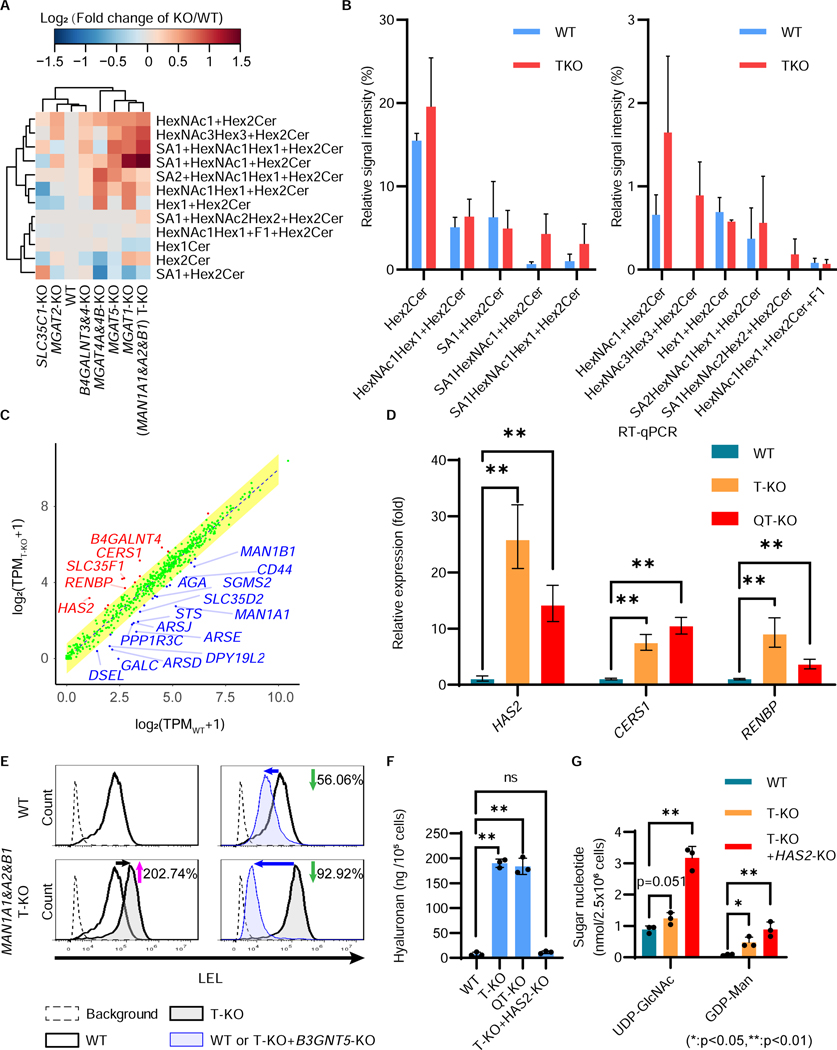
During the lectin analysis of our KO cell library, we found that LEL staining was increased in cells deficient in multiple mannosidase-I genes as well as MGAT1 or MGAT2 (Figure 4B). We expected that LEL staining would be decreased if N-glycan structures were changed to oligomannoses. However, we observed the opposite result. To understand why LEL staining increased, seven representative KO cells (MAN1A1-, A2-, and B1-triple-KO (T-KO); MGAT1-KO; MGAT2-KO; MGAT4-KO; MGAT5-KO; SLC35C1-KO; and B4GALNT3- and 4-double-KO) were subjected to comparative glycomic analysis by mass spectrometry. As we expected, the N-glycan structures were changed in these KO cells compared with the HEK293 parental wild-type (WT) cells (Figures S4B and S4C). In addition, the KO of genes related to N-glycan processing also affected the structures of O-glycans and GSLs, suggesting that disruption of a glycan-related gene in one pathway causes global changes in glycan structures (Figures S4B, 5A and S4D).
Figure 5. Increase of LacNAc-containing GSLs and hyaluronan in MAN1A1&A2&B1-T-KO cells.
A, Comparative GSL analysis of seven KO cells. Relative abundance of GSLs in HEK293 MAN1A1-, A2-, and B1-triple-KO (T-KO), MGAT1-KO, MGAT2-KO, MGAT4A&4B-KO, MGAT5-KO, B4GALNT3- and 4-KO, and SLC35C1-KO cells. The data are visualized as clustered heatmaps. The relative amounts of glycan structures in each cell type were calculated, and were compared with those of HEK293 cells. The log2 values of the data are visualized as heatmaps. The data used the mean value from two independent experiments.
B, Relative abundance of GSL species in HEK293 WT and T-KO cells. The data represent the means ± errors from two independent experiments.
C, Comparison of the expression of glycan-related genes in WT versus T-KO cells. TPM values (averages of triplicated data) were calculated and plotted as log2(TPM + 1) values. The yellow area represents the predicted interval expression in WT or T-KO cells. Representative examples of genes with higher expressions in WT or T-KO cells are indicated by blue or red text, respectively. It is noted that expression levels of genes MAN1A1 (0.16 fold), MAN1A2 (0.50 fold), and MAN1B1 (0.43 fold) were significantly down-regulated in T-KO cells, probably as results of nonsense-mediated mRNA decay.
D, Quantitative RT-PCR analysis of CERS1, HAS2, and RENBP mRNA levels mRNA levels in HEK293 WT, T-KO, and QT-KO cells. HPRT (hypoxanthine phosphoribosyltransferase) was used to normalize the data. The bars represent RQ (relative quantification) values ± RQmax and RQmin (error bars), from triplicate samples.
E, Enhanced LEL staining in MAN1A1&A2&B1-T-KO (T-KO) cells was due to an increase of GSLs. Flow cytometric analysis of cells stained by fluorescent-conjugated LEL. HEK293 wild-type (WT), T-KO, B3GNT5-KO, and T-KO+B3GNT5-KO cells were stained by fluorescent-conjugated LEL, and analyzed by flow cytometry. Background, without lectin staining.
F, The levels of hyaluronan in the culture media of WT, T-KO, and QT-KO cells were quantified. The bars represent the means ± SD from triplicate samples. P-values (two-tailed, student’s t-test) are shown.
G, Levels of UDP-GlcNAc and GDP-Man were analyzed using high-performance anion-exchange chromatography. The bars represent the means ± SD from triplicate samples. P-values (two-tailed, student’s t-test) are shown.
In glycomic analysis using mass spectrometry, several GSLs were elevated in MAN1A1&A2&B1-triple-KO (T-KO) cells; GM2 and (neo)lacto-series GSLs, such as Lc3 and polylactosamine-containing GSLs, were particularly elevated (Figures 5B, S5A and S5B). We compared the gene expression profiles of T-KO and MAN1A1&A2&B1&C1&MGAT1-quintuple-KO (QT-KO) cells with WT cells. Among the glycan-related genes, the TPM value of CERS1, which encodes ceramide synthase 1, was 3.48 and 3.09 times upregulated in the T-KO and QT-KO cells, respectively (Figures 5C, S5C and S5D). This increase was confirmed by quantitative PCR (Figure 5D) and correlates closely with upregulated ceramide species (Figure S5E). In addition, the KDSR, CERS4, B4GALT6, and B4GALT4 genes, whose encoded enzymes are required for the GSL biosynthesis, were significantly upregulated in both T-KO and QT-KO cells (Figure S5D). To analyze whether the increased LEL staining was caused by polylactosamine extension on GSLs, we disrupted B3GNT5, which encodes Lc3 synthase, required for synthesizing lacto- and neolacto-series GSLs (Kuan et al., 2010). When B3GNT5 was knocked out in the WT cells, LEL staining decreased by 56%, which was presumed to be the contribution of GSLs to polylactosamine structures under normal conditions (Figure 5E). In the T-KO cells, LEL staining was increased by 203%, compared with WT cells. In contrast, LEL staining was decreased by 93%, when B3GNT5 was knocked out in the T-KO cells (Figure 5E). Because there were almost no polylactosamine structures on N-glycans in the T-KO cells, these decreases indicate that polylactosamine-containing GSLs are increased in T-KO cells. A similar phenotype was observed in cells treated with a mannosidase-I inhibitor, kifunensine, suggesting that N-glycan changes to oligomannose structures generally affect GSLs (Figure S5F). It is possible that polylactosamine-containing GSLs may compensate for the lack of polylactosamines on N-glycan structures in T-KO cells.
The gene expression profiles revealed that, in addition to CERS1, the expression of HAS2 and RENBP were highly upregulated in T-KO and QT-KO cells (Figures 5C, 5D, and S5C). HAS2 encodes hyaluronan synthase 2. Hyaluronan is a GAG consisting of GlcA and GlcNAc (Figure S5G) (Weigel and DeAngelis, 2007). Compared with WT cells, T-KO and QT-KO cells produced 24.8-fold and 24.0-fold more hyaluronan, respectively (Figure 5F). The increase was eliminated by knocking out HAS2 in T-KO cells. RENBP encodes GlcNAc-2-epimerase, which converts GlcNAc to N-acetylmannosamine for neuraminic acid synthesis (Figure S5H). Both upregulated genes, HAS2, RENBP, and CERS1, contribute to the consumption of GlcNAc as UDP-GlcNAc. When nucleotide sugar levels were analyzed, UDP-GlcNAc and GDP-Man levels were slightly increased in the T-KO cells (Figures 5G and S5I). Knocking out HAS2 in T-KO cells further elevated the UDP-GlcNAc and GDP-Man levels. Because UDP-GlcNAc was not used for N-glycan processing in the T-KO cells, these cells might adapt to reduce GlcNAc or UDP-GlcNAc levels by overexpressing HAS2, RENBP, and CERS1. These unexpected impacts on orthogonal biosynthetic pathways demonstrate the capability of GlycoMaple, when combined with glycomic analysis, to provide new insights into the cross-cutting mechanisms that regulate glycan synthesis.
Automatic mapping and visualization of glycosylation pathways
To expand the utility of GlycoMaple, we developed a web-based automatic mapping tool. When RNA-seq data are uploaded, the web-based tool automatically maps the gene expression information onto 19 glycosylation pathways. In addition, the expressions of 950 glycan-related genes can be displayed as bar plots categorized by function. We used RNA-seq data from the Human Protein Atlas, which contains RNA-seq data from 64 human cell lines and 37 tissue types (Colwill and Graslund, 2011). We then visualized glycan pathways based on gene expression profiles (See GlycoMaple website). As an example, we focused on mucin-type O-glycans. In the liver, some genes involved in mucin-type O-glycan biosynthesis were not expressed, resulting in the formation of only simple structures (Figure 6A). In contrast, many genes involved in mucin-type O-glycan biosynthesis were expressed in the small intestine, and a variety of O-glycan structures could be synthesized (Figure 6B). In the mucin-type O-glycan biosynthesis pathway, 48 genes were categorized. We detected these genes in 37 tissue types and compared gene expression profiles. The tissues and genes were clustered using heatmap analysis (Figure 6C). Digestive organs, including the stomach, duodenum, small intestine, colon, and rectum were clustered in the same tree. In these tissues, some genes were highly expressed compared with other tissue types, such as B3GNT6, GCNT3, ST6GALNC1, B3GNT3, and FUT2. This finding is consistent with the fact that digestive organs express high levels of mucin characterized by branching, sialylation, and fucosylation (Jin et al., 2017; Robbe et al., 2004).
Figure 6. Comparison of glycosylation pathways among various tissues.
A and B, Mucin-type O-glycosylation pathways in the liver (A) and small intestine (B) were visualized using GlycoMaple, based on gene expression profiles in the Human Protein Atlas (HPA). Each numbered reaction is listed in Table S3.
C, Expression profiles of genes involved in mucin-type O-glycosylation in tissues. The TPM values of 48 genes required for mucin-type O-glycosylation in 37 human tissues deposited in the HPA were normalized (z-score) and are visualized as a clustered heatmap.
D and E, Comparison of pathways using GlycoMaple. Gene expression profiles (median TPM values) in primary tumor (N = 288) and normal (N = 304) colon tissues were used to show the pathways. Fold changes that were > 2 and < 0.5 are shown as pink and green arrows, respectively. Each numbered reaction is listed in Table S3.
F, Different expressions of glycan-related genes that were upregulated or downregulated in colon tumor tissue. N = 288 for primary tumor and N = 304 for normal tissues.
Prediction of changed glycan structures in colon cancer cells using GlycoMaple
GlycoMaple enabled us to compare glycan structures between normal and diseased tissues. Using RNA-seq data of 288 colon primary tumor samples and 304 normal colon tissue samples from two public databases, TCGA and GTEx (Table S7), we analyzed glycosylation pathways using GlycoMaple, which predicted glycan changes during carcinogenesis (Figure S6). In the N-glycan processing pathways, upregulation of core fucose (Figure 6D, step 17), β−1,4-GlcNAc on α−1,3-Man branch (step 19), and β−1,6-GlcNAc on α−1,6-Man branch (step 20) were predicted in the cancer tissues. In addition, bisecting GlcNAc N-glycans were predicted to be decreased in the cancer tissues because of competition between MGAT5 and MGAT3 in the pathway (Bubka et al., 2014; Nagae et al., 2018). The expression of MGAT5, which transfers β−1,6-GlcNAc, was increased in the cancer tissues, whereas MGAT3 expression, which adds bisecting GlcNAc, was unchanged (Figure 6F). The predicted glycan structures matched the reported structures that are upregulated in colorectal cancer (Balog et al., 2012).
For the structures that cap glycans, a clear competition was observed between the Sda and Lewis antigens (Figure 6E). It is well known that Sda antigens are decreased and Lewis antigens are increased in colorectal cancer (Malagolini et al., 2007). The gene expression levels of FUT3 and FUT6, which encode enzymes responsible for producing Lewis x, were upregulated (Figure 6F), whereas B4GALNT2, which encodes β1,4-GalNAc transferase responsible for the construction of Sda, remained at low levels, resulting in increased cancer-associated Lewis x or sialyl-Lewis x antigens. In addition, the GlycoMaple analysis predicted that LacdiNAc (Figure 6E, step 1), I-branched polylactosamine (steps 15 and 17), α−2,6-sialic acid (step 7), type-I lactosamine structures (step 16), and α−1,2-fucose structures were increased, whereas HNK-1 (step 19) and α−2,3-sialic acid (step 3) structures were decreased. The prediction was consistent with the results of actual glycan analysis (Fernández-Rodríguez et al., 2000; Holst et al., 2015; Muinelo-Romay et al., 2010), indicating that the GlycoMaple analysis has the potential to explore and provide leads for identifying glycan-based biomarkers for disease progression, including tumorigenesis and metastasis.
Discussion
Currently, several webtools are available to analyze cellular metabolic pathways, such as KEGG (Kanehisa et al., 2017) and Recon (Noronha et al., 2017). These are very useful tools for displaying comprehensive metabolic pathways in various species. However, because these tools cover all metabolic pathways, they cannot describe the glycosylation pathways in all of their rich and cross-interacting detail. In some pathways, the precise substrate specificities of glycosyltransferases are not reflected or the most recent data has not yet been deposited in appropriate repositories. Moreover, because character strings or complicated chemical structures are used to represent glycan structures, it can be difficult to develop an intuitive understanding of the visualized pathways. We focused on creating glycan metabolic pathways that incorporate the latest knowledge and enzyme substrate properties. To facilitate intuitive browsing by glycobiology researchers and by the broader biomedical community, we have adopted the SNFG representations of glycans (Neelamegham et al., 2019). It covers almost 1000 genes involved in glycosylation and its regulation; no other pathway database contains the same level of detailed information about glycosylation. We believe that GlycoMaple combining glycosylation pathways with gene expression data would contribute to Glycobiology from a wide range of research fields.
GlycoMaple potentially informs conventional glycan analyses. Compared with glycan analyses using mass spectrometry or high-performance liquid chromatography, GlycoMaple analysis using RNA-seq can be performed with a small amount of starting material. In addition, conventional analyses sometimes have difficulties distinguishing glycan isomer structures. Because GlycoMaple allows the estimation of glycan structures based on gene expression, it also supports the annotation of glycan isomers detected in a glycomic analysis based on known enzyme specificities. Conversly, when GlycoMaple analysis predicts that a unique glycan structure exists in the cell of interest, its existence can be confirmed by targeted mass spectrometry.
In the present study, we constructed a glycogene KO cell library consisting of 40 different KO cells. We unepectively found the compensatory mechanisms between glycan biosynthetic pathways in HEK293 cells. In multiple α1,2-mannosidase-I-KO cells, which synthesize only oligomannose N-glycans, the expressions of CERS1, RENBP, and HAS2 were significantly increased. All of these genes are thought to contribute to the reduction of cytoplasmic GlcNAc or UDP-GlcNAc levels. The amount of glycolipids containing polylactosamine was increased, as were hyaluronan levels. Whereas our finding in HEK293 cells might be dependent upon cell lines, it may connect the two independent glycan changes in other cell-types. Interestingly, it is reported that both oligomannose N-glycans and hyaluronan are increased in some cancer cells (McCarthy et al., 2018; Park et al., 2020). Changes in N-glycan structures affect the synthesis of other glycans, which could be regulated by both gene expression and accumulated substrate levels. Since it is possible to see glycan metabolic pathways comprehensively, GlycoMaple could find unexpected and novel cross-talk between mechanisms.
GlycoMaple can contribute to the discovery of novel glycan biomarkers and therapeutic targets by identifying differences in glycan structures between normal and diseased tissues. Because the expression of glycosyltransferases is generally low, they are often overlooked when all gene expression levels are compared between samples. However, by narrowing the genes to be analyzed down to 950 glycan-related genes, sensitivity for detecting altered expression levels is increased. We used public datasets to estimate the glycans that were elevated or decreased in colon cancer tissues compared with normal tissues. The glycan structures that were estimated to be upregulated or downregulated were consistent with actual glycomic data.
Limitations
Although GlycoMaple analysis can be used to estimate the presence or absence of glycan structures, it has limitations. This is an “estimation” of the glycosylation pathways using RNA-seq. After the initial clues of glycan structural information were obtained in GlycoMaple, it is necessary to confirm and determine the structures using conventional glycan analyses. Besides, the estimation of glycans from GlycoMaple is not quantitative. Although we could estimate whether the target glycan structures could be synthesized or not (exists or not), the amounts of the glycan structures are not reflected in the pathways. Whereas the validation of GlycoMaple showed that gene expression is an important factor to correlate with the synthesized glycan structures, gene expression level and glycosylation activity of the gene product are not linearly related. In the mucin O-glycan biosynthesis pathway in HEK293 cells, 9 false positive glycan structures were predicted in GlycoMaple. Seven of the 9 glycan structures have been confirmed by MS2. Two predicted glycan structures including a sulfated O-glycan were not detected in our MS analysis. Sulfated glycans were technically difficult to detect under the conditions of the measured glycomics. It is thought that false positive predictions result primarily from the limitation of MS-based glycomics data. On the other hand, GlycoMaple analysis does not consider the localization of enzymes within the Golgi compartment, limitation of donor and accepter substrates, and the Golgi environment, which might lead to false positive predictions. To predict the quantities of glycans with high accuracy, the development of a simulation of glycosylation pathways, which considers many parameters including substrate concentration, enzyme kinetics, and localization, will be required. Second, the analysis can only provide information about glycans in the whole cell, but not glycan structures added onto specific proteins. If it becomes possible to broadly understand the mechanism by which specific glycans are found on specific sites of specific proteins, it would be of extreme usefulness for the production of pharmaceutical glycoproteins. However, significantly larger glycoprotein-specific glycosylation datasets will be required to build this knowledge. Nonetheless, the ability of GlycoMaple to simulate whole cell glycan expression provides a platform upon which future tools, that simulate individual protein glycosylation, can be assembled.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Morihisa Fujita (fujita@jiangnan.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study. The code generated during this study are available at the GlycoMaple website (https://glycosmos.org/glycomaples/index).
Experimental model and subject details
Cell culture
HEK293 cells (ATCC, CRL-1573), and their derivative HEK293FF6 cells (Hirata et al., 2015), were cultured in Dulbecco’s modified Eagle medium (DMEM; Biological Industries, Kibbutz Beit Haemek, Israel) containing 10% fetal bovine serum (FBS; Biological Industries) in a humidified 37°C incubator with 5% CO2. Appropriate antibiotic concentrations were used when necessary: hygromycin (400 mg/mL), puromycin (1 μg/mL), and streptomycin (100 U/mL)/penicillin (100 μg/mL).
Method Details
Establishment of gene KO cell lines
Genes for the KO cell library were deleted using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system. For each gene, a pair of single-guide RNA targets were designed using the E-CRISP website (Heigwer et al., 2014) (http://www.e-crisp.org/E-CRISP/) and were ligated into the KO vector pX330-EGFP (Hirata et al., 2015) digested with BpiI (Thermo Fisher Scientific, Waltham, MA, USA). All oligonucleotides used for gene KOs in this study are listed in Table S8. A pair of KO pX330-EGFP plasmids were transfected into HEK293 cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The GFP-positive cells were sorted using an S3e Cell Sorter (BioRad, Hercules, CA, USA) at 2.5 days after transfection. The sorted cells were cultured for > 10 days and further subjected to limiting dilution to obtain clonal KO cells. The genes of the clonal KO cells were confirmed by PCR and sequencing. A clone with no WT allele was selected. The DNA sequence of each KO cell line are listed in Table S8.
Establishment of cells stably expressing genes
To establish cell lines stably expressing genes, cDNAs amplified by PCR were ligated into pME-hyg or pME-puro vector vectors (Ohishi et al., 2003). B3GNT1 was ligated into the EcoRI and NotI sites of pME-hyg; GCNT2, ST8SIA2, and ST8SIA4 were ligated into the XhoI and NotI sites of pME-hyg; and ST8SIA1 was ligated into a XhoI site of pME-puro. All restriction enzymes and the In-Fusion HD Cloning Kit were purchased from Takara (Shiga, Japan). Approximately 7 days after transfecting the plasmids, culture media were changed to DMEM with 10% FBS containing antibiotics (hygromycin or puromycin). Cells were selected for 2 weeks, and surviving cells were used for the analysis.
Preparation of GSLs and enrichment of glycoproteins from cells
Method for glycan preparation and condition for mass spectrometry were as previously described (Aoki et al., 2007; Aoki et al., 2008; Boccuto et al., 2014). Cells (~1×107 cells) were resuspended in 50% methanol, disrupted with tissue homogenizer on ice. The solvent composition was adjusted to chloroform/methanol/water (30:60:8, v/v/v) and agitated for at least 2 hours at room temperature. The homogenates were then centrifuged to collect precipitated proteins, and the supernatant was kept as lipid extract. The precipitated proteins were re-extracted three times with the same solvent mix, and the original and all three re-extractions were combined and dried under nitrogen gas. Contaminating glycerophospholipids were removed from the dried lipid extract by saponification. Briefly, 0.5 M potassium hydroxide in methanol/water (95:5, v/v) was incubated with the dried extract for at least 6 h at 37°C. The saponification reaction was terminated with an equal volume of 5% acetic acid to make the final solution 50% aqueous in methanol. GSLs were desalted and separated from released fatty acids on tC18 Sep-Pak cartridges (Waters) pre-equilibrated with methanol and 1% acetic acid. GSLs were analyzed by separation on HPTLC silica gel 60 plates (EMD Millipore) using a mobile phase of chloroform/methanol/water (60:35:8 or 60:40:10, v/v/v) and visualized by spraying with 0.25% orcinol (Sigma) in 3 M aqueous H2SO4/ethanol (1:1, v/v). Standards of neutral GSLs and gangliosides were obtained from Matreya LLC. GSL abundance was normalized to cell numbers.
Protein-linked glycan analysis
N-glycans released from glycoproteins by PNGaseF were permethylated and quantified as described previously (Aoki et al., 2007). Briefly, glycoproteins were digested with trypsin and chymotrypsin (Sigma), and glycopeptides were enriched from the digests by Sep-Pak C18 cartridge chromatography (Waters). N-glycans were released by PNGaseF (NEB) and recovered by passing through Sep-Pak C18. Glycopeptides carrying O-glycans were recovered from the C18 Sep-Pak and used for O-glycan analysis (Kumagai et al., 2013; Morelle et al., 2009). For reductive β-elimination, O-linked glycopeptides was resuspended in a solution of 100 mM sodium hydroxide and 1 M sodium borohydride, and incubated for 18 hrs at 45°C in a glass tube sealed with a Teflon-lined screw top. The tube was transferred to ice and acetic acid added to 10% to neutralize the reaction. The sample was then loaded onto a column of AG 50W-X8 cation exchange resin (Bio-Rad) to desalt. Released oligosaccharide were eluted from the resin with three bed-volumes 5% acetic acid and lyophilized to dryness. To remove borate from the sample, a solution of 10% acetic acid in methanol was added and the sample then dried under a stream of nitrogen gas at 37°; this was repeated for a total of five times. The sample was then resuspended in 5% acetic acid and loaded onto a C18 cartridge column (Waters) that was previously washed with acetonitrile and pre-quilibrated with 5% acetic acid. Run-through from the column was collected after loading; the column was then washed a total of five times with 5% acetic acid. The run-through and washes were combined and evaporated to dryness.
Mass spectrometry of GSLs and glycoprotein glycans
The preparation of GSLs and glycoprotein glycans for quantitative mass spectrometry was performed as described previously (Boccuto et al., 2014; Ferreira et al., 2018; Kumagai et al., 2013). Briefly, intact GSLs and released glycoprotein glycans were permethylated with 12C-methyliodide prior to MS analysis according to the method of Anumula and Taylor (Anumula and Taylor, 1992). Known amounts of a maltotetraose oligosaccharide (Dp4) were permethylated with 13C-methyliodide for use as reference standards for GSL and glycoprotein glycan quantification (Rohrer et al., 2016). N-glycans, O-glycans and GSLs were permethylated and analyzed from two biological replicates for HEK293 cell lines. Glycan analysis was carried out by nanospray ionization (NSI)-MS in positive ion mode and quantified relative to a known amount of external standard (Dp4, permethylated with heavy methyliodide) supplemented into the sample prior to injection (Aoki et al., 2019). Relative abundance of each glycan componets were also obtained. Glycan structures carrying unique oligosaccharide sequence, e.g., polylactosamine chain, bisecting GlcNAc, etc, were further subjected to manual MSn analysis for complete structural determination (Nairn et al., 2012; Nairn et al., 2008). The MS-based glycomics data generated in these analyses and the associated annotations are presented in accordance with the MIRAGE standards and the Athens Guidelines, which include explicit descriptions of instrument settings, fragmentation strategies, and quantification parameters (MIRAGE).
Analysis of nucleotide sugars by HPAEC-PAD HPLC
Nucleotide sugars were extracted and analyzed using HPLC system equipped with high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) as decribed previously (Kamiya et al., 2008; Tomiya et al., 2001). Nucleotide sugars were separated on a CarboPac PA-1 column equipped with a PA-1 guard column, BorateTrap column and detected by absorbance at 260 nm. Nucleotide sugar standrads were obtained from Sigma-Aldrich.
Construction of the GlycoMaple web tool
The GlycoMaple web tool consists of data files, pathway images in Scalable Vector Graphics (SVG) format and a python script to process input and output data. Data files consist of gene information that map to the pathway images; each arrow in the pathway image is assigned to an enzyme(s) so that expression values for each gene name are mapped to arrows in the SVG pathway image. The python script thus reads in expression values from either the user’s upload file or from predefined files based on the RNA-seq data available from the Human Protein Atlas (HPA) web site (Pontén et al., 2008), then adjusts the width of the arrows in the SVG pathway images to reflect the expression value. Pathway images were generated by first drawing the pathways in Microsoft Powerpoint and exporting them into pdf files. These pdf files were then converted to SVG vector images using the Inkscape software (Bah, 2007). Then, the SVG files were edited using Inkscape so that each arrow that corresponds to an enzyme could be assigned an ID value. These IDs were mapped to enzyme names in one of the data files. Finally, a Flask server was set up in a Docker container so that the web server, data files, python script and SVG images could be packaged together and easily distributed. The GlycoMaple is available at https://glycosmos.org/glycomaples/index. A tutorial video and an instruction for GlycoMaple are deposited as Movie S1 and Methods S1, respectively.
RNA sequencing of HEK293 cells
Total RNA was extracted using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA). Samples with RNA integrity number (RIN) ≥ 7 were considered to be of high quality, and were processed further and subjected to the subsequent analysis. Total RNA-seq libraries were generated using 4 μg of total RNA, which was analyzed using the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA). These libraries were then sequenced using the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten), and 125 bp/150 bp paired-end reads were generated. The raw reads containing adaptors and the low-quality reads from the raw data were removed using Trimmomatic, to obtain the clean reads. The transcriptome sequencing was conducted by OE Biotech Co., Ltd. (Shanghai, China), and the clean reads were provided. The clean reads were mapped to the hg38 reference genome using hisat2 (version 2.1.0) (Kim et al., 2015). The output BAM files were converted to SAM files using SAMtools 1.9 (Li et al., 2009). The final TPM values were obtained using Stringtie 1.3.5 (Pertea et al., 2016; Pertea et al., 2015).
Validation of GlycoMaple using the mucin-type O-glycan biosynthetic pathway
To validate the GlycoMaple tool, statistical analysis was performed using the mucin-type O-glycan biosynthetic pathway, in which the glycan structures estimated by GlycoMaple were compared against those detected from mass spectrometry. Among the 53 mucin-type O-glycan structures in the GlycoMaple pathway map, 21 glycan structures detected in the glycomics data and previous reports (Razawi et al., 2013) were used as the actual detected structures in HEK293 cells. Then, the gene expression (TPM) values obtained using RNA-seq analysis in HEK293 cells were input into the mucin-type O-glycan pathway. By changing the threshold of TPM values (0 to 10), the predicted glycan structures were estimated. If there is a reaction called A -> B -> C -> D and the TPM value of a gene involved in the reaction B -> C is lower than the threshold value, it is assumed that the products C and D will not be synthesized regardless of whether the TPM value of the genes involved in the C -> D reaction is higher than the threshold. True-positive, false-positive, true-negative, and false-negative predictions were collected, and the accuracy, precision, sensitivity (recall), and specificity under the different thresholds were calculated. Based on the calculated values, the receiver operating characteristic (ROC) curve was calculated, and the area under the curve (AUC) was measured. To set the threshold of the TPM value, the Youden’s J statistic and the F1-score were used. The same approach was applied to another cell types (A549, Caco2, HeLa, HEK293, HepG2, and HL60 cells). MS data for O-glycan structures (Fujitani et al., 2013) and RNA-seq data available in the human protein atlas (HPA) were used for the analysis.
Gene expression analysis of colon tissue using public databases
The Xena Browser from the University of California, Santa Cruz, was used to compare the gene expression levels between colon primary tumor and normal tissues. The datasets of TPM values from colon primary tumor (N = 288) and normal (N = 304)) tissues were obtained from the TCGA and databases, respectively.
Hyaluronan ELISA Assay
To measure hyaluronan levels, the Hyaluronan Quantikine ELISA Kit from (R&D Systems, DHYAL0) was used according to the manufacturer’s instructions. Briefly, Cells (5 × 105) were cultured with 1 mL DMEM medium containing 10% FBS in a 24-well plate 3 d before analysis. 190 μL dilution buffer was used to dilute 10 μL medium to 200 μL samples, 50 of which was used for the assay.
Lectins and antibodies
ABA (Agaricus bisporus)-fluorescein isothiocyanate (FITC), ConA (Canavalia ensiformis)-biotin, DBA (Dolichos biflorus)-FITC, DSA (Datura stramonium)-FITC/biotin, ECA (Erythrina crista-galli)-FITC, LCA (Lens culinaris)-biotin, Lotus (Lotus tetragonolobus)-FITC, MAM (Maackia amurensis)-FITC, PHA-E4 (Phaseolus vulgaris)-FITC, PHA-L4 (Phaseolus vulgaris)-FITC, PNA (Arachis hypogaea)-FITC, SBA (Glycine max)-FITC, SSA (Sambucus sieboldiana)-biotin, UEA-I (Ulex europaeus)-FITC, and WGA (Triticum vulgaris)-FITC were purchased from J-Chemical (Tokyo, Japan). CtxB (cholera toxin B)-Alexa Fluor 488 and GSII (Griffonia simplicifolia)-FITC were purchased from Thermo Fisher Scientific. DyLight 488-labeled LEL (Lycopersicon esculentum) and fluorescein-labeled WFA (Wisteria floribunda) were purchased from Vector Laboratories (Burlingame, CA, USA). The anti-CD57 monoclonal antibody, FITC, and streptavidin PE conjugate were purchased from Thermo Fisher Scientific.
Flow cytometry
Cells (5 × 105) were cultured in a 12-well plate 1 day before analysis and harvested with 300 μL FACS buffer (phosphate-buffered saline containing 1% bovine serum albumin and 0.1% NaN3). After washing with phosphate-buffered saline, cells were stained with 10 μg/mL lectins or 10 μg/mL antibodies on ice for 15 or 30 min, respectively. For biotinylated lectins and primary antibodies, cells were stained with fluorescent-conjugated streptavidin and secondary antibodies, respectively. After staining, cells were washed twice with 100 μL FACS buffer and analyzed using Accuri C6 (BD, Franklin Lakes, NJ, USA). The resulting data were analyzed using FlowJo software (BD). For kifunensine treatment, cells (5 × 105) were cultured in a 6-well plate with DMEM medium containing 10% FBS and 1 mg/ml Kifunensine or DMSO. After 3 days of treatment, cells were stained with fluorescent-conjugated LEL, and subjected to flow cytometric analysis.
Quantitative real-time (RT)-PCR analysis
Cells (1 × 106 cells) were prepared 1 day before RNA extraction. RNA was extracted according to the manufacturer’s protocol (Promega, Madison, WI, USA) and resuspended in 100 μL RNase-free water. The cDNA was synthesized from 500 ng RNA using PrimeScript RT Master Mix for quantitative RT-PCR (Takara). RT-PCR was performed according to the manufacturer’s instructions, using standard parameters, in a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). Triplicate reactions were run for each cDNA and primer pair.
Supplementary Material
(Related to Figure 1) List of glycan-related genes
(Related to Figure 1) Category of glycan metabolic pathways
(Related to Figure 2) List of mapped genes
(Related to Figure 2) Glycomic analysis
(Related to Figure 3) RNA-seq data in HEK293 cells
(Related to Figure 4) List of KO cell library
(Related to Figure 6) TPM data of colon cancer tissues
(Related to STAR Methods) Information of knockout cells
(Related to STAR Methods) Tutorials for GlycoMaple
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD57 Monoclonal Antibody (TB01 (TBO1)), FITC | eBioscience | Cat# 11–0577-42, RRID:AB_1311193 |
| F(ab’)2-Goat anti-Mouse IgG (H+L) Secondary Antibody, PE | eBioscience | Cat# 12–4010-82, RRID:AB_11063706 |
| IgM Monoclonal Antibody (eB121–15F9), FITC | eBioscience | Cat# 11–5890-81, RRID:AB_465291 |
| Streptavidin PE Conjugate | eBioscience | Cat# 12–4317-87 |
| Biological Samples | ||
| Cholera Toxin Subunit B (CtxB) (Recombinant), Alexa Fluor 488 Conjugate | Invitrogen | Cat# C34775 |
| Lectin GS-II From Griffonia simplicifolia, Alexa Fluor 488 Conjugate | Invitrogen | Cat# L21415 |
| ABA (Agaricus bisporus)-FITC (fluorescein isothiocyanate) | J-Chemical | Cat# 10601C |
| ConA (Canavalia ensiformis)-Biotin | J-Chemical | Cat# 03102C |
| DBA (Dolichos biflorus)-FITC | J-Chemical | Cat# 42204C |
| DSA (Datura stramonium)-FITC/Biotin | J-Chemical | Cat# 31211C |
| ECA (Erythrina cristagalli)-FITC | J-Chemical | Cat# 12204C |
| LCA (Lens culinaris)-Biotin | J-Chemical | Cat# 41213C |
| Lotus (Lotus tetragonolobus)-FITC | J-Chemical | Cat# 52500C |
| MAM (Maackia amurensis)-FITC | J-Chemical | Cat# 92600C |
| PHA-E4 (Phaseolus vulgaris)-FITC | J-Chemical | Cat# 81503C |
| PHA-L4 (Phaseolus vulgaris)-FITC | J-Chemical | Cat# 61210C |
| PNA (Arachis hypogaea)-FITC | J-Chemical | Cat# 61213C |
| SBA (Glycine max)-FITC | J-Chemical | Cat# 71600C |
| SSA (Sambucus sieboldiana)-Biotin | J-Chemical | Cat# 51213C |
| UEA-I (Ulex europaeus)-FITC | J-Chemical | Cat# 21503C |
| WGA (Triticum vulgaris)-FITC | J-Chemical | Cat# 02101C |
| Lycopersicon Esculentum (Tomato) Lectin (LEL, TL), DyLight 488 | Vector Laboratories | Cat# DL-1174, RRID:AB_2336404 |
| Wisteria Floribunda Lectin (WFA, WFL), Fluorescein | Vector Laboratories | Cat# FL-1351, RRID:AB_2336875 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Certified Fetal Bovine Serum (FBS) | Biological Industries | Cat# 04–001-1ACS |
| DMEM, high glucose | Biological Industries | Cat# 01–052-1A |
| Kifunensine | Cayman Chemical | Cat#10009437 |
| Opti-MEM Reduced Serum Medium | Gibco | Cat# 31985062 |
| Lipofectamine™ 2000 Transfection Reagent | Invitrogen | Cat# 11668–019 |
| Hygromycin B Gold | Invitrogen | Cat# ant-hg-5 |
| Puromycin | Invitrogen | Cat# ant-pr-5 |
| Blasticidin | Invitrogen | Cat# ant-bl-5 |
| PEI MAX - Transfection Grade Linear Polyethylenimine Hydrochloride (MW 40,000) | Polysciences | Cat# 24765 |
| D-PBS | Sangon Biotech | Cat# E607009–0500 |
| Trypsin-EDTA Solution, With Phenol Red | Sangon Biotech | Cat# E607002–0100 |
| Critical Commercial Assays | ||
| SV Total RNA Isolation System | Promega | Cat# Z3100 |
| Hyaluronan Quantikine ELISA Kit | R&D Systems | Cat# DHYAL0 |
| SanPrep Column DNA Gel Extraction Kit | Sangon Biotech | Cat# B518131 |
| SanPrep Column PCR Product Purification Kit | Sangon Biotech | Cat# B518141 |
| SanPrep Column Plasmid Mini-Preps Kit | Sangon Biotech | Cat# B518191 |
| PrimeSTAR® GXL DNA Polymerase | Takara | Cat# R050A |
| DNA Ligation Kit <Mighty Mix> | Takara | Cat# 6023 |
| PrimeScript™ RT Master Mix (Perfect Real Time) | Takara | Cat# RR036A |
| TB Green® Premix Ex Taq™ II (Tli RNaseH Plus), Bulk | Takara | Cat# RR820L |
| EcoRI | Takara | Cat# 1040S |
| NotI | Takara | Cat# 1166S |
| XhoI | Takara | Cat# 1094A |
| In-Fusion HD Cloning Kit | Takara | Cat# 639649 |
| FastDigest BpiI (IIs class) | Thermo Fisher Scientific | Cat# FD1014 |
| KOD FX Neo | TOYOBO | Cat# KFX-201 |
| Deposited Data | ||
| RNA seq of HEK293 cells | This study | SRA: SRR12213061; SRR12213062; SRR12213063 |
| RNA seq of T-KO cells | This study | SRA: SRR12213058; SRR12213059; SRR12213060 |
| RNA seq of QT-KO cells | This study | SRA: SRR12213055; SRR12213056; SRR12213057 |
| Glycomics MS raw data | This study | https://glycopost.gly cosmos.org (ID: GPST000125) |
| RNA seq of 288 colon primary tumor samples and 304 normal colon tissue samples | Chang et al., 2019/ UCSC Xena platform | https://xenabrowser. net/ |
| Experimental Models: Cell Lines | ||
| HEK293 cells (parental HEK293) | Jin et al., 2018 | N/A |
| HEK293 FF6 cells | Hirata et al., 2015 | N/A |
| DPM1-KO cells | This paper | N/A |
| STT3A-KO cells | Toshihiko Kitajima et., 2018 | STT3A-knockout cell lines |
| STT3B-KO cells | Toshihiko Kitajima et., 2019 | STT3B-knockout cell lines |
| MOGS-KO cells | Liu et al., 2018 | MOGS-KO cells |
| GANAB-KO cells | Liu et al., 2018 | GANAB-KO cells |
| CANX-KO cells | Liu et al., 2018 | CANX-KO cells |
| CALR-KO cells | Liu et al., 2018 | CALR-KO cells |
| CANX&CALR-KO cells | Liu et al., 2018 | CANX/CALR double-KO |
| MAN1A1-KO cells | Jin et al., 2018 | A1-KO24 |
| MAN1A2-KO cells | Jin et al., 2018 | A2-KO37 |
| MAN1A1&A2-KO cells | Jin et al., 2018 | D-KO35 |
| T-KO (MAN1A1&A2&B1) cells | Jin et al., 2018 | T-KO |
| MAN1A1&A2&C1-KO cells | Jin et al., 2018 | N/A |
| MAN1A1&A2&B1&C1-KO cells | Ren et al., 2019 | QD-KO |
| QT-KO (MAN1A1&A2&B1&C1&MGAT1) cells | Ren et al., 2019 | QT-KO |
| MAN2A1-KO cells | This study | N/A |
| MAN2A2-KO cells | This study | N/A |
| MAN2A1&A2-KO cells | This study | N/A |
| MGAT1-KO cells | This study | N/A |
| MGAT2-KO cells | This study | N/A |
| MGAT3-KO cells | This study | N/A |
| MGAT4A-KO cells | This study | N/A |
| MGAT4B-KO cells | This study | N/A |
| MGAT4A&4B-KO cells | This study | N/A |
| MGAT5-KO cells | This study | N/A |
| MGAT5B-KO cells | This study | N/A |
| MGAT5&5B-KO cells | This study | N/A |
| FUT8-KO cells | This study | N/A |
| MGAT4A&4B&3&5&FUT8-KO cells | This study | N/A |
| B4GALT1-KO cells | This study | N/A |
| B4GALT2-KO cells | This study | N/A |
| B4GALT3-KO cells | This study | N/A |
| B3GNT8-KO cells | This study | N/A |
| ST6GAL1-KO cells | This study | N/A |
| B4GALNT3&4-KO cells | This study | N/A |
| CHST8&9-KO cells | This study | N/A |
| B4GALNT2-KO cells | This study | N/A |
| SLC35A1-KO cells | This study | N/A |
| SLC35A2-KO cells | This study | N/A |
| SLC35C1-KO cells | This study | N/A |
| B3GNT5-KO | This study | N/A |
| Oligonucleotides | ||
| qPCR Primers for the HPRT gene: 5′-TGGCGTCGTGATTAGTGATG-3′ 5′-TCCAGCAGGTCAGCAAAGAA-3′ |
Liu et al., 2018 | N/A |
| qPCR Primers for the CERS1 gene: 5′-TCCATCTACGCTACGCTATACA-3′ 5′-GCACAAGGATGCCCACATTG-3′ |
PrimerBank | 11641421a2 |
| qPCR Primers for the HAS2 gene: 5′-CTCTTTTGGACTGTATGGTGCC-3′ 5′-AGGGTAGGTTAGCCTTTTCACA-3′ |
PrimerBank | 169791020c1 |
| qPCR Primers for the RENBP gene: 5′-CTGCTCCGTCATTGCATTCG-3′ 5′-AGGGCAACAATAGGAACTTGTC-3′ |
PrimerBank | 213417819c3 |
| sgRNA sequence of KO cells | This study | Table SX |
| DNA and Plasmids | ||
| pME-hyg | Ohishi et al., 2003 | pMEhyg |
| pME-hyg-B3GAT1 | This study | N/A |
| pME-hyg-GCNT2 | Thermo Fisher Scientific | Cat# A11613 |
| pME-hyg-ST8SIA2 | Thermo Fisher Scientific | Cat# A11613 |
| pME-hyg-ST8SIA4 | Thermo Fisher Scientific | Cat# A11613 |
| pME-hyg-B3GAT1 | This study | N/A |
| pME-hyg-ST8SIA2 | This study | N/A |
| pME-hyg-ST8SIA4 | This study | N/A |
| pME-hyg-GCNT2 | This study | N/A |
| pME-puro | Ohishi et al., 2003 | pMEpuro |
| pME-puro-ST8SIA1 | Tashima et al,. 2006 | N/A |
| pME-puro-ST8SIA1 | This study | N/A |
| pX330-mEGFP | Hirata et al., 2015 | N/A |
| Software and Algorithms | ||
| FlowJo software | https://www.flowjo.com/ | Version 7.6 |
| R | https://www.r-project.org/ | Version 3.6.2 |
| HISAT2 | http://ccb.jhu.edu/software/hisat2/manual.shtml | Version 2.1.0 |
| SAMtools | http://samtools.sourceforge.net/ | Version 1.9 |
| Stringtie | http://ccb.jhu.edu/software/stringtie/ | Version 1.3.5 |
| BD Accuri C6 software | http://www.bdbiosciences.com/accuri/ | Version 1.0.264.21 |
| Bio-Rad S3e cell Sorter & ProSort Software | https://www.bio-rad.com/ | Version 1.4 |
| Inkscape | https://inkscape.org/ | Version 0.92.5 |
| Other | ||
| MicroAmp™ Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL | Applied Biosystems | 4346906 |
| StepOnePlus Real-Time PCR system | Applied Biosystems | 4376692 |
| Flow Cytometry | BD biosciences | C6 |
| Cell sorter | Bio-Rad | S3e |
| T100 Thermal Cycler | Bio-Rad | T100 |
| iMark Microplate Absorbance Reader | Bio-Rad | N/A |
| PNGaseF | Promega | Cat#V4831 |
| Sep-Pak Vac C18 1cc, 100mg | Waters | WAT023590 |
| Sep-Pak Plus Light tC18 | Waters | WAT036805 |
| Supelclean ENVI-Carb spe | Spelco | 57109-U |
| LTQ XL, Orbitrap Discovery | Thermo Fisher Scientific | |
| Iodomethane | Sigma-Aldrich | 289566–100G |
| Sodium hydroxide solution, 50% w/w | Fisher | SS254–1 |
| Anyhydrous DMSO | Sigma-Aldrich | |
| Oligosaccharides Kit | Supelco | 47265 |
| Dowex® 50WX8 hydrogen form 100–200 mesh | Sigma-Aldrich | 217506–500G |
| Sodium borohydride | Sigma-Aldrich | 213462–25G |
| Xcalibur software | Thermo Fisher Scientific | Ver 3.0.63 |
Highlights.
A comprehensive glycosylation mapping tool, termed GlycoMaple, was developed.
GlycoMaple could visualize and estimate glycan structures based on gene expression.
A cell library knocked out genes related to N-linked glycosylation was constructed.
Glycan changes between normal and diseased tissues were estimated using GlycoMaple.
Acknowledgement
We thank Ji-Xiong Leng for establishment of MAN2A1 and MAN2A2 knockout cell lines, and Drs Hideki Nakanishi and Ning Wang (Jiangnan University) for discussion. This work was supported by grants from the National Natural Science Foundation of China 32071278 and 31770853 (MF), the Program of Introducing Talents of Discipline to Universities 111–2-06, National first-class discipline program of Light Industry Technology and Engineering LITE2018–015, Top-notch Academic Programs Project of Jiangsu Higher Education Institutions, the International Joint Research Laboratory for Investigation of Glycoprotein Biosynthesis at Jiangnan University, and the GaLSIC collaborative research fund, Soka University, Japan. This work has also been supported by the Integration Promotion Program of the Japan Science and Technology Agency and the National Bioscience Database Center (NBDC) 17934031 (KFAK), National Institutes of Health (NIH) Common Fund Grant R21AI129873 (KA), P41GM103490 (KA and MT), and U01GM125267 (KA and MT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of Interests
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anumula KR, and Taylor PB (1992). A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem 203, 101–108. [DOI] [PubMed] [Google Scholar]
- Aoki K, Heaps AD, Strauss KA, and Tiemeyer M. (2019). Mass spectrometric quantification of plasma glycosphingolipids in human GM3 ganglioside deficiency. Clinical Mass Spectrometry 14, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim J-M, Cantu R, Wells L, and Tiemeyer M. (2007). Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem 282, 9127–9142. [DOI] [PubMed] [Google Scholar]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, and Tiemeyer M. (2008). The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem 283, 30385–30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah T. (2007). Inkscape: guide to a vector drawing program (prentice hall press; ). [Google Scholar]
- Balog CI, Stavenhagen K, Fung WL, Koeleman CA, McDonnell LA, Verhoeven A, Mesker WE, Tollenaar RA, Deelder AM, and Wuhrer M. (2012). N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol Cell Proteomics 11, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennun SV, Yarema KJ, Betenbaugh MJ, and Krambeck FJ (2013). Integration of the transcriptome and glycome for identification of glycan cell signatures. PLoS Comput Biol 9, e1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuto L, Aoki K, Flanagan-Steet H, Chen CF, Fan X, Bartel F, Petukh M, Pittman A, Saul R, Chaubey A, et al. (2014). A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Hum Mol Genet 23, 418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, and Henrissat B. (2001). Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol 11, 593–600. [DOI] [PubMed] [Google Scholar]
- Bubka M, Link-Lenczowski P, Janik M, Pocheć E, and Lityńska A. (2014). Overexpression of N-acetylglucosaminyltransferases III and V in human melanoma cells. Implications for MCAM N-glycosylation. Biochimie 103, 37–49. [DOI] [PubMed] [Google Scholar]
- Colley KJ, Kitajima K, and Sato C. (2014). Polysialic acid: biosynthesis, novel functions and applications. Crit Rev Biochem Mol Biol 49, 498–532. [DOI] [PubMed] [Google Scholar]
- Colwill K, and Graslund S. (2011). A roadmap to generate renewable protein binders to the human proteome. Nat Methods 8, 551–558. [DOI] [PubMed] [Google Scholar]
- Esko JD, and Selleck SB (2002). Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71, 435–471. [DOI] [PubMed] [Google Scholar]
- Fernández-Rodríguez J, Feijoo-Carnero C, Merino-Trigo A, Páez de la Cadena M, Rodríguez-Berrocal FJ, de Carlos A, Butrón M, and Martínez-Zorzano VS (2000). Immunohistochemical analysis of sialic acid and fucose composition in human colorectal adenocarcinoma. Tumour Biol 21, 153–164. [DOI] [PubMed] [Google Scholar]
- Ferreira CR, Xia ZJ, Clement A, Parry DA, Davids M, Taylan F, Sharma P, Turgeon CT, Blanco-Sanchez B, Ng BG, et al. (2018). A Recurrent De Novo Heterozygous COG4 Substitution Leads to Saul-Wilson Syndrome, Disrupted Vesicular Trafficking, and Altered Proteoglycan Glycosylation. Am J Hum Genet 103, 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani N, Furukawa J, Araki K, Fujioka T, Takegawa Y, Piao J, Nishioka T, Tamura T, Nikaido T, Ito M, et al. (2013). Total cellular glycomics allows characterizing cells and streamlining the discovery process for cellular biomarkers. Proc Natl Acad Sci U S A 110, 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, and Boutros M. (2014). E-CRISP: fast CRISPR target site identification. Nat Methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Helenius A, and Aebi M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73, 1019–1049. [DOI] [PubMed] [Google Scholar]
- Hirata T, Fujita M, Nakamura S, Gotoh K, Motooka D, Murakami Y, Maeda Y, and Kinoshita T. (2015). Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol Biol Cell 26, 3071–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst S, Wuhrer M, and Rombouts Y. (2015). Glycosylation characteristics of colorectal cancer. Adv Cancer Res 126, 203–256. [DOI] [PubMed] [Google Scholar]
- Hutter J, and Lepenies B. (2015). Carbohydrate-Based Vaccines: An Overview. Methods Mol Biol 1331, 1–10. [DOI] [PubMed] [Google Scholar]
- Iwai T, Inaba N, Naundorf A, Zhang Y, Gotoh M, Iwasaki H, Kudo T, Togayachi A, Ishizuka Y, Nakanishi H, et al. (2002). Molecular cloning and characterization of a novel UDP-GlcNAc:GalNAc-peptide beta1,3-N-acetylglucosaminyltransferase (beta 3Gn-T6), an enzyme synthesizing the core 3 structure of O-glycans. J Biol Chem 277, 12802–12809. [DOI] [PubMed] [Google Scholar]
- Jimenez del Val I, Nagy JM, and Kontoravdi C. (2011). A dynamic mathematical model for monoclonal antibody N-linked glycosylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnol Prog 27, 1730–1743. [DOI] [PubMed] [Google Scholar]
- Jin C, Kenny DT, Skoog EC, Padra M, Adamczyk B, Vitizeva V, Thorell A, Venkatakrishnan V, Lindén SK, and Karlsson NG (2017). Structural Diversity of Human Gastric Mucin Glycans. Mol Cell Proteomics 16, 743–758. [DOI] [PubMed] [Google Scholar]
- Jin ZC, Kitajima T, Dong W, Huang YF, Ren WW, Guan F, Chiba Y, Gao XD, and Fujita M. (2018). Genetic disruption of multiple alpha1,2-mannosidases generates mammalian cells producing recombinant proteins with high-mannose-type N-glycans. J Biol Chem 293, 5572–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Kamiya D, Yamamoto K, Nyfeler B, Hauri HP, and Kato K. (2008). Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. J Biol Chem 283, 1857–1861. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, and Morishima K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–d361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick DC (2002). Animal lectins: a historical introduction and overview. Biochimica et Biophysica Acta (BBA)-General Subjects 1572, 187–197. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotidis P, and Kontoravdi C. (2020). Harnessing the potential of artificial neural networks for predicting protein glycosylation. Metabolic engineering communications 10, e00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krambeck FJ, Bennun SV, Narang S, Choi S, Yarema KJ, and Betenbaugh MJ (2009). A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology 19, 1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krambeck FJ, and Betenbaugh MJ (2005). A mathematical model of N-linked glycosylation. Biotechnol Bioeng 92, 711–728. [DOI] [PubMed] [Google Scholar]
- Kremkow BG, and Lee KH (2018). Glyco-Mapper: A Chinese hamster ovary (CHO) genome-specific glycosylation prediction tool. Metab Eng 47, 134–142. [DOI] [PubMed] [Google Scholar]
- Kuan CT, Chang J, Mansson JE, Li J, Pegram C, Fredman P, McLendon RE, and Bigner DD (2010). Multiple phenotypic changes in mice after knockout of the B3gnt5 gene, encoding Lc3 synthase--a key enzyme in lacto-neolacto ganglioside synthesis. BMC Dev Biol 10, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Katoh T, Nix DB, Tiemeyer M, and Aoki K. (2013). In-gel β-elimination and aqueous-organic partition for improved O- and sulfoglycomics. Anal Chem 85, 8692–8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Chiang AWT, Hansen AH, Arnsdorf J, Schoffelen S, Sorrentino JT, Kellman BP, Bao B, Voldborg BG, and Lewis NE (2020). A Markov model of glycosylation elucidates isozyme specificity and glycosyltransferase interactions for glycoengineering. Current research in biotechnology 2, 22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Puri A, and Neelamegham S. (2013). Glycosylation Network Analysis Toolbox: a MATLAB-based environment for systems glycobiology. Bioinformatics 29, 404–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Guo XY, Hirata T, Rong Y, Motooka D, Kitajima T, Murakami Y, Gao XD, Nakamura S, Kinoshita T, et al. (2018). N-Glycan-dependent protein folding and endoplasmic reticulum retention regulate GPI-anchor processing. J Cell Biol 217, 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, and Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42, D490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagolini N, Santini D, Chiricolo M, and Dall’Olio F. (2007). Biosynthesis and expression of the Sda and sialyl Lewis x antigens in normal and cancer colon. Glycobiology 17, 688–697. [DOI] [PubMed] [Google Scholar]
- Mandrekar JN (2010). Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 5, 1315–1316. [DOI] [PubMed] [Google Scholar]
- McCarthy JB, El-Ashry D, and Turley EA (2018). Hyaluronan, Cancer-Associated Fibroblasts and the Tumor Microenvironment in Malignant Progression. Frontiers in cell and developmental biology 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereiter S, Balmana M, Campos D, Gomes J, and Reis CA (2019). Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 36, 6–16. [DOI] [PubMed] [Google Scholar]
- Mizumoto S, Yamada S, and Sugahara K. (2014). Human genetic disorders and knockout mice deficient in glycosaminoglycan. Biomed Res Int 2014, 495764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle W, Faid V, Chirat F, and Michalski JC (2009). Analysis of N- and O-linked glycans from glycoproteins using MALDI-TOF mass spectrometry. Methods Mol Biol 534, 5–21. [DOI] [PubMed] [Google Scholar]
- Morise J, Takematsu H, and Oka S. (2017). The role of human natural killer-1 (HNK-1) carbohydrate in neuronal plasticity and disease. Biochimica et Biophysica Acta (BBA)-General Subjects 1861, 2455–2461. [DOI] [PubMed] [Google Scholar]
- Muinelo-Romay L, Gil-Martín E, and Fernández-Briera A. (2010). α(1,2)fucosylation in human colorectal carcinoma. Oncol Lett 1, 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J, and Elliott DJ (2016). Hallmarks of glycosylation in cancer. Oncotarget 7, 35478–35489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Kizuka Y, Mihara E, Kitago Y, Hanashima S, Ito Y, Takagi J, Taniguchi N, and Yamaguchi Y. (2018). Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V. Nat Commun 9, 3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn AV, Aoki K, dela Rosa M, Porterfield M, Lim JM, Kulik M, Pierce JM, Wells L, Dalton S, Tiemeyer M, et al. (2012). Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. J Biol Chem 287, 37835–37856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn AV, York WS, Harris K, Hall EM, Pierce JM, and Moremen KW (2008). Regulation of glycan structures in animal tissues transcript profiling of glycan-related genes. J Biol Chem 283, 17298–17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen YH, Schjoldager KT, Steentoft C, et al. (2019). An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol Cell 75, 394–407.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lütteke T, O’Boyle N, Packer NH, Stanley P, Toukach P, et al. (2019). Updates to the Symbol Nomenclature for Glycans guidelines. Glycobiology 29, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, and Freeze HH (2018). Perspectives on Glycosylation and Its Congenital Disorders. Trends Genet 34, 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha A, Danielsdottir AD, Gawron P, Johannsson F, Jonsdottir S, Jarlsson S, Gunnarsson JP, Brynjolfsson S, Schneider R, Thiele I, et al. (2017). ReconMap: an interactive visualization of human metabolism. Bioinformatics 33, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Nagamune K, Maeda Y, and Kinoshita T. (2003). Two subunits of glycosylphosphatidylinositol transamidase, GPI8 and PIG-T, form a functionally important intermolecular disulfide bridge. J Biol Chem 278, 13959–13967. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, and Marth JD (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867. [DOI] [PubMed] [Google Scholar]
- Park DD, Phoomak C, Xu G, Olney LP, Tran KA, Park SS, Haigh NE, Luxardi G, Lert-Itthiporn W, Shimoda M, et al. (2020). Metastasis of cholangiocarcinoma is promoted by extended high-mannose glycans. Proc Natl Acad Sci U S A 117, 7633–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho SS, and Reis CA (2015). Glycosylation in cancer: mechanisms and clinical implications. Nature Reviews Cancer 15, 540–555. [DOI] [PubMed] [Google Scholar]
- Pontén F, Jirström K, and Uhlen M. (2008). The Human Protein Atlas—a tool for pathology. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 216, 387–393. [DOI] [PubMed] [Google Scholar]
- Raman R, Tharakaraman K, Sasisekharan V, and Sasisekharan R. (2016). Glycan–protein interactions in viral pathogenesis. Curr Opin Struct Biol 40, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razawi H, Kinlough CL, Staubach S, Poland PA, Rbaibi Y, Weisz OA, Hughey RP, and Hanisch FG (2013). Evidence for core 2 to core 1 O-glycan remodeling during the recycling of MUC1. Glycobiology 23, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren WW, Jin ZC, Dong W, Kitajima T, Gao XD, and Fujita M. (2019). Glycoengineering of HEK293 cells to produce high-mannose-type N-glycan structures. J Biochem 166, 245–258. [DOI] [PubMed] [Google Scholar]
- Robbe C, Capon C, Coddeville B, and Michalski JC (2004). Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J 384, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JS, Basumallick L, and Hurum DC (2016). Profiling N-linked oligosaccharides from IgG by high-performance anion-exchange chromatography with pulsed amperometric detection. Glycobiology 26, 582–591. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Narimatsu Y, Joshi HJ, and Clausen H. (2020). Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol 21, 729–749. [DOI] [PubMed] [Google Scholar]
- Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, and Lewis NE (2016). A Markov chain model for N-linked protein glycosylation--towards a low-parameter tool for model-driven glycoengineering. Metab Eng 33, 52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanenko AA, and Dmitrenko VV (2015). HEK293 in cell biology and cancer research: phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution. Gene 569, 182–190. [DOI] [PubMed] [Google Scholar]
- Stowell CP, and Stowell SR (2019). Biologic roles of the ABH and Lewis histo-blood group antigens Part I: infection and immunity. Vox Sang 114, 426–442. [DOI] [PubMed] [Google Scholar]
- Taniguchi N, Honke K, and Fukuda M. (2011). Handbook of glycosyltransferases and related genes (Springer Science & Business Media; ). [Google Scholar]
- Taylor ME, Drickamer K, Schnaar RL, Etzler ME, and Varki A. (2015). Discovery and Classification of Glycan-Binding Proteins. In Essentials of Glycobiology, rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, et al. , eds. (Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; [PubMed] [Google Scholar]
- Copyright 2015-2017. by The Consortium of Glycobiology Editors, La Jolla, California. All rights reserved.), pp. 361–372.
- Teintenier-Lelièvre M, Julien S, Juliant S, Guerardel Y, Duonor-Cérutti M, Delannoy P, and Harduin-Lepers A. (2005). Molecular cloning and expression of a human hST8Sia VI (alpha2,8-sialyltransferase) responsible for the synthesis of the diSia motif on O-glycosylproteins. Biochem J 392, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, and Smart TG (2005). HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods 51, 187–200. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, de Vries RP, and Paulson JC (2019). Virus recognition of glycan receptors. Curr Opin Virol 34, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, and Lee YC (2001). Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem 293, 129–137. [DOI] [PubMed] [Google Scholar]
- Ujita M, McAuliffe J, Hindsgaul O, Sasaki K, Fukuda MN, and Fukuda M. (1999). Poly-N-acetyllactosamine Synthesis in BranchedN-Glycans Is Controlled by Complemental Branch Specificity of i-Extension Enzyme and β1, 4-Galactosyltransferase I. J Biol Chem 274, 16717–16726. [DOI] [PubMed] [Google Scholar]
- Umaña P, and Bailey JE (1997). A mathematical model of N-linked glycoform biosynthesis. Biotechnol Bioeng 55, 890–908. [DOI] [PubMed] [Google Scholar]
- Varki A. (2017). Biological roles of glycans. Glycobiology 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, and Lynch VJ (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131, 281–285. [DOI] [PubMed] [Google Scholar]
- Wands AM, Cervin J, Huang H, Zhang Y, Youn G, Brautigam CA, Matson Dzebo M, Björklund P, Wallenius V, Bright DK, et al. (2018). Fucosylated Molecules Competitively Interfere with Cholera Toxin Binding to Host Cells. ACS Infect Dis 4, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, and DeAngelis PL (2007). Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem 282, 36777–36781. [DOI] [PubMed] [Google Scholar]
- Zaia J. (2010). Mass spectrometry and glycomics. OMICS 14, 401–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Related to Figure 1) List of glycan-related genes
(Related to Figure 1) Category of glycan metabolic pathways
(Related to Figure 2) List of mapped genes
(Related to Figure 2) Glycomic analysis
(Related to Figure 3) RNA-seq data in HEK293 cells
(Related to Figure 4) List of KO cell library
(Related to Figure 6) TPM data of colon cancer tissues
(Related to STAR Methods) Information of knockout cells
(Related to STAR Methods) Tutorials for GlycoMaple
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. The code generated during this study are available at the GlycoMaple website (https://glycosmos.org/glycomaples/index).