Abstract
The neoplastic Hodgkin/Reed-Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL) depend on chronic activation of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signalling pathways to maintain survival and proliferation. Accumulating reports highlight the importance of the inactivation or reduced expression of negative JAK/STAT regulators such as the protein-tyrosine phosphatase 1B (PTP1B/PTPN1) in this process. Various PTPN1 mRNA variants as well as truncated PTP1B proteins were identified in cHL cell lines and primary cHL tumour samples. These PTPN1 mRNA variants lack either one or several exon sequences and therefore render these PTP1B variants catalytically inactive. Here, we show that one of these mutants, PTP1B∆2–4, is not only a catalytically inactive variant, but also augmented the IL-4-induced JAK/STAT activity similar to the recently reported PTP1B∆6 splice variant. Moreover, while PTP1B∆6 diminished the activity and protein levels of PTP1BWT, PTP1BWT remained unaffected by PTP1B∆2–4, arguing for different molecular mechanisms of JAK/STAT modulation by PTP1B∆6 and PTP1B∆2–4. Collectively, these data indicate that PTPN1 variants missing one or more exon sequences originated either from alternative splicing or from gene mutation, create PTP1B gain-of-function variants with oncogenic potential by augmenting JAK/STAT signalling in cHL.
We describe the impact of the deletion of exon 2–4 of PTP1B, rendering PTP1B∆2–4 into a positive JAK/STAT regulator similar to PTP1B∆6, highlighting the relevance of exon-deleting alterations to convert a tumour-suppressor into a protein with pro-oncogenic properties.
Introduction
The hallmark of classical Hodgkin lymphoma (cHL) are malignant mononucleated Hodgkin cells and the characteristic bi- or multinucleated Reed-Sternberg cells (HRS cells), in combination with a reactive infiltrate of different cell types (1,2). The HRS cells are characterized by the constitutive activation of the Janus kinase/Signal Transducers and Activators of Transcription (JAK/STAT) signalling cascade (3–5). JAK/STAT signalling is induced upon binding of a cytokine/growth factor to its cognate receptor. Subsequently, members of the Janus kinase (JAK) family, JAK1, JAK2, JAK3 and TYK2 are recruited to the cytoplasmic part of the receptor, followed by the phosphorylation of the JAKs on specific tyrosine residues. In turn, JAK-mediated phosphorylation of the receptor creates binding sites for the Src homology 2 (SH2) domains of the STATs. Hereupon, recruited STATs are phosphorylated at specific tyrosine residues by the JAKs causing dimerization of STATs. STAT dimers translocate into the nucleus, bind to specific promoter regions and induce the expression of specific target genes involved in cell–cycle control (e.g. cyclin D1, c-myc, p21) and cell survival (e.g. BCLXL, MCL1, BCL2) consequently highlighting the important role of JAK/STAT signalling in oncogenesis (6,7).
PTP1B, encoded by the protein tyrosine phosphatase, non-receptor type 1 (PTPN1) gene on chromosome 20 (20q13.13), is a crucial negative regulator of JAK/STAT signalling. PTP1B consists of a catalytic domain (30–278), a proline-rich domain (279–401) and a hydrophobic C-terminal domain (402–435) targeting the enzyme to the membrane of the endoplasmatic reticulum (ER). The phosphatase has inhibitory effects on different cytokine receptors and receptor protein tyrosine kinases (RPTKs) as well as the JAK and STAT proteins by direct dephosphorylation of these proteins (8). The role of PTP1B in oncogenesis is diverse and depends on the tumour entity, yet, acting as tumour-suppressor in B cell lymphoma (9–11). Given the essential role of PTP1B in JAK/STAT signalling, the identification and characterization of the mechanisms controlling the activity of this negative JAK/STAT regulator are crucial for the understanding of its role in the pathogenesis of cHL. For instance, various mutations in PTPN1 were identified in different cHL cell lines and samples from primary mediastinal B cell lymphoma (PMBL) and cHL patients, including nonsense, missense or frameshift mutations, but also the loss of one or several exons was described (12,13). Some of these mutants display a diminished phosphatase activity and are less capable of reducing STAT activity in IL-4 stimulated cells. However, the functional impact of these catalytically inactive PTP1B mutants has not been analysed in detail.
We recently characterized PTP1B∆6, a PTP1B variant lacking the exon 6, in detail (13). Additionally, we found the PTP1B variants PTP1B∆2–4, lacking exons 2–4 and PTP1∆B2-8, lacking exons 2–8, which were identified in individual cHL cases and in the cell lines Sup-HD1 and U-HO1, respectively. By investigating the functional relevance, only PTP1B∆2–4 augmented IL-4-induced JAK/STAT activity, comparable to PTP1B∆6 (13). However, PTP1B∆6, but not PTP1B∆2–4, caused a decrease in PTP1BWT activity and degradation of the wild-type protein. Taken together, we show that the deletion of exon 2–4 renders PTP1B∆2–4 into a positive JAK/STAT regulator.
Materials and methods
Patients, tissue samples and immunohistochemistry
One hundred and nine tumour samples were collected from patients with cHL, who underwent surgical resection at the University Hospital Ulm. Histological type was classified according to the criteria of the World Health Organization (WHO) (14). Informed consent was obtained from all patients before surgery, and the Ethics Committee of the University of Ulm (242/12) authorized this study. Patient characteristics are presented in Supplemental Table 1, available at Carcinogenesis Online. Immunohistochemistry was performed using DAKO Real Detection System Alkaline Phosphatase/RED rabbit/mouse (K5005, Agilent Technologies, Santa Clara, USA). In brief, paraffin wax embedded tissue sections were deparaffinized in xylene for 15 min and dehydrated with graded ethanol washes (100–70%). Antigen retrieval was performed by pre-treatment with citrate buffer pH 6.0 using a pressure cooker. Thereafter, slides were cooled to room temperature (RT), washed with PBS for 1 min and incubated at RT with anti-PTP1B primary monoclonal antibody [Ab-1 (FG6-1G), 1:100 dilution, Merck4Biosciences (Calbiochem®)] for 30 min. The sections were rinsed with PBS for 1 min, incubated with LINK biotinylated secondary antibody for 30 min at RT, followed by a wash with PBS for 1 min and incubation with Streptavidin Alkaline Phosphatase antibody for 30 min at RT. After another wash, the slides were incubated with RED chromogen for 16 min at RT and counterstained with DAKO REAL haematoxylin for 5 min RT. Staining intensity of PTP1B expression in HRS cells was scored in 6 different categories (Figure 2A).
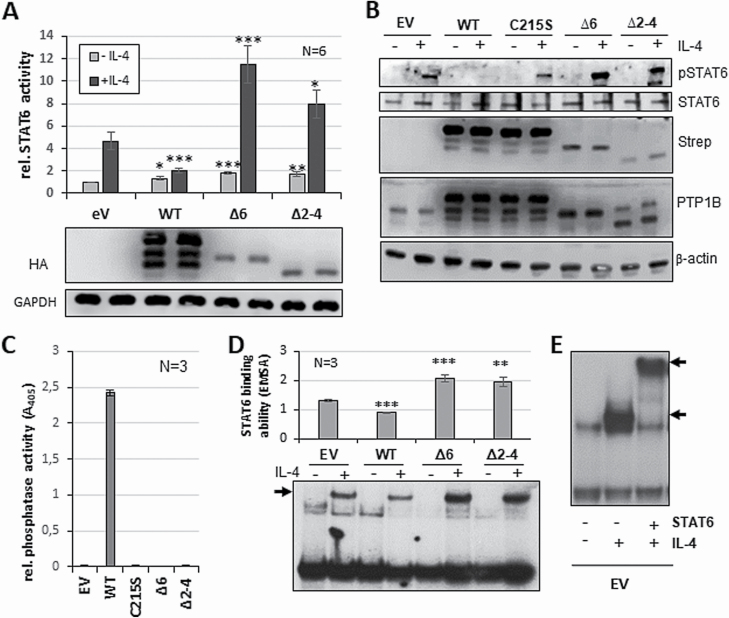
Figure 2.
PTP1B∆2–4 has a positive impact on STAT6 activity. (A) Luciferase assay for STAT6 activity with and without (control) stimulation with 5 ng/ml IL-4 for 18 h (upper part). HEK293-STAT6 cells were transiently transfected with either pcDNA3.1 (EV), or with HA-PTP1BWT, HA-PTP1B∆6, HA-PTP1B∆2–4 or HA-PTP1B∆2–8 vectors. Immunoblot analysis of WCE of one exemplary luciferase measurements (lower part) using the indicated antibodies. (B) Immunoblot analysis of WCE of HEK293-STAT6 cells either transfected with pcDNA3.1 (EV), or with HA-PTP1BWT, HA-PTP1BC215S, HA-PTP1B∆6 or HA-PTP1B∆2–4, each either with or without (control) stimulation with 5 ng/ml IL-4 for 30 min. Antibodies against pSTAT6, STAT6, Strep-tag, PTP1B and β-actin are used. C) Phosphatase assay with Strep-tagged PTP1BWT, PTP1BC215S, PTP1B∆6 or PTP1B∆2–4 ectopically expressed in HEK293 cells. pEXPR-IBA105 (EV) serves as control. Mean of three independent experiments is depicted. (D) Electrophoretic mobility shift assay to determine STAT6 DNA binding activity with whole cell extracts from HEK293-STAT6 cells either transfected with pcDNA3.1 (EV), or with HA-PTP1BWT, HA-PTP1B∆6 or HA-PTP1B∆2–4, each either with or without (control) stimulation with 5 ng/ml IL-4 for 30 min (bottom part). Quantification of STAT6 DNA binding levels of three independent experiments (upper part). (E) Electrophoretic mobility supershift using EV transfected HEK-STAT6 cells with (shift, lower arrow) and without IL-4 stimulation and additional STAT6 antibody (STAT6 (S20)-X, Santa Cruz) depicting the STAT6 supershift (upper arrow). Mean values and standard error of mean (SEM) are depicted. Comparisons were performed between empty vector measurement and individual treatments/transfections. Significances are calculated using student’s t-test (P < 0.05 was regarded as significant).
Cell culture, transfection and treatment
Most of the cell lines (HEK293, L-428, L-1236, KM-H2, SUP-HD1) were purchased from the DSMZ more than 20 years ago. The cHL cell line U-HO1 (15) and the LCL cell lines (N,T,H) and HEK293-STAT6, were generated in our lab. All cell lines were authenticated by STR profiling (in 2020/U-HO1 in 2017/HEK293 in 2016). Cell lines were grown in IMDM/RPMI (4:1) supplemented with 10% fetal bovine serum, glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humified atmosphere with 5% CO2. Reagents were purchased from Lonza (Basel, Switzerland) and Biochrom (Berlin, Germany). HEK293 cells were transfected via the calcium-phosphate method as described previously (16). HEK293-STAT6 cells were generated by transfecting a STAT6 expression vector and selected with serial dilution on medium laced with G418 antibiotics. L-428 cells were transfected by Amaxa® Cell Line Nucleofector® Kit as indicated by the manufacturer. Stably transfected L-428 cells were selected with serial dilution on medium laced with G418 antibiotics. Stable L-428 cells were treated with indicated amounts of etoposide, gemcitabine, doxorubicin (Internal pharmacy, University hospital Ulm) and the STAT6 inhibitor AS1516499 (Hycultec GmbH, Beutelsbach).
Plasmid construction
PTPN1WT, PTPN1C215S, PTPN1∆6, PTPN1∆2–4 and PTPN1∆2–8 cDNAs were integrated into the pcDNA3.1 vector (HA-tag) and into the pEXPR-IBA105 vector (Strep-tag). T4 DNA ligase, buffers and restriction enzymes were purchased from New England Biolabs (Ipswich, USA).
Preparation of whole-cell extracts
Whole-cell extracts (WCE) were prepared using TNT buffer (20 mM Tris pH 8.0, 200 mM NaCl, 1% Triton-X 100, 1 mM DTT, phosphatase and protease inhibitors) or Dignam C buffer (EMSA: 20 mM HEPES pH7.9, 25% Glycerol, 0.42 M NaCl, 1.5 mM MgCL2, 0.2 mM EDTA, 1 mM DTT and protease inhibitors). Extracts were cleared by centrifugation at 20 000 × g for 10 min. Protein levels were determined using Bradford Reagent (Bio-Rad, Hercules, USA) at an absorbance of 595 nm.
Luciferase reporter assay
Cells were transfected with PTPN1-plasmids encoding PTPN1WT, PTPN1C215S, PTPN1∆6 and PTPN1∆2–4 variants, STAT6-luciferase reporter plasmid and actin-renilla plasmid, as described above, and stimulated with IL-4 after 24 h incubation. Total cell proteins were extracted after another overnight incubation period. Measurements were performed on a FB 12 Luminometer (Berthold Detection Systems, Pforzheim, Germany) using the Dual-Luciferase® Reporter Assay System (Promega, Madison, USA).
Immunoblotting
Proteins were separated by 10% SDS-Page and transferred to nitrocellulose membranes (0.2 µm; GE Healthcare, Chicago, USA). Immunoblots were performed with the following antibodies: rabbit anti-pSTAT6(Y641) antibody (Cell Signaling, Danvers, USA), rabbit polyclonal anti-STAT6 (C-20) antibody (Santa Cruz, Dallas, USA), rabbit anti-PTPN1 (Sigma, St. Louis, USA), rat anti-HA antibody (Roche, Basel, Suisse), StrepMAB-HRP (IBA LifeScience, Göttingen, Germany) and mouse anti-β-actin antibody (Sigma, St. Louis, USA). Signal development was performed using horseradish peroxide-conjugated secondary antibodies: anti-rabbit HRP (Sigma, St. Louis, USA), anti-mouse HRP (Sigma, St. Louis, USA) and anti-rat HRP (Cell Signaling, Danvers, USA). Protein bands were visualized by WesternSure® PREMIUM Chemiluminescent substrate (Li-cor, Lincoln, USA) using the C-DiGit ® Blot Scanner (Li-cor, Lincoln, USA) and Image Studio Digits Ver.5.2 software.
RNA isolation and semi-quantitative RT-PCR analysis
Isolation of the neoplastic HRS cells was already described in Zahn et al. 2017 (Method S1). Extraction of RNA was employed by RNeasy Micro Kit (Qiagen, Hilden, Germany) and reverse transcribed by SuperScript II cDNA synthesis Kit (Invitrogen, Carlsbad, USA) following the instruction of the manufacturer. Semi-quantitative real-time PCR (sqPCR) was conducted with FastStart Universal SYBR Green Master (Rox) from Roche on a Rotor-Gene Q PCR machine. PCR thermocycling parameters were 95°C for 5 min, 40 cycles of 95°C for 30 s, 53°C for 45 s and 72°C for 15 s. Each sample was run in triplicate and GAPDH and ACTB served as housekeeping genes. Fold induction was determined in a ∆∆Ct-based fold change calculation (17). (Primer Sequences (5′–3′): ptpn1 forward: TGGGTGAAGGAAGAGACCCA and reverse: CCCACGACCCGACTTCTAAC; ptpn1Δ6 forward: CGCACAATACTGGCCACAAAA and reverse: GGGTCTTTCCTCTTGTCCATTGT; ptpn1Δ2–4 forward: GGGCGGCC ATTTACCA GTTA and reverse: TCCAATTCTAGCTGTCGCACT; ptpn1Δ2–8 forward: GCGGCCATTTACCAGCATGA and reverse: AACTCAGTGCAT GGTCCTCG; GAPDH forward: GCCAAAAGGGTCATCATCTC and reverse: TGTGGTCATGAGTCCTTCCA; ACTB forward: TGTGGCATCCACGAAACTAC and reverse: GGAGCAATGATCTTGATCTTCA).
Strep-tag immunoprecipitation and phosphatase activity assay
TNT extracts (0.5–1 mg protein) of HEK293 cells ectopically expressing Strep-tagged PTP1B variants (for phosphatase assay without phosphatase inhibitors) were incubated with 20 µl StrepTactin-Sepharose (IBA LifeScience, Göttingen, Germany) or magnetic Streptavidin Beads (New England Biolabs, Ipswich, USA) on a rotor at 4°C overnight. Isolated Strep-tagged PTP1B variants were subsequently washed with TNT buffer and 1×TBS. Precipitates were used either for immunoblotting or were split up in two for phosphatase assay. For this, pelleted sepharose was suspended in phosphatase buffer with PNPP and one sample was incubated with an additional phosphatase inhibitor. The enzymatic activity of PTP1B was measured by hydrolysation of p-nitrophenyl phosphate (=PNPP) to p-nitrophenol at an absorbance of 405 nm in all samples. Samples with phosphatase inhibitors were used for background extraction. [Phosphatase buffer: 10 mM TRIS, pH7.5; 50 mM NaCl; 1 mM MnCl2; 2 mM DTT; 10 mM PNPP (NEB, Ipswich, USA).]
Electrophoretic mobility shift assay
Double-stranded DNA probes were generated using a radioactively labelled 32p-ddCTP and polymerase I. Oligonucleotides for STAT6: 5′-AGGTCGACTTCCCAAGAACAGAG-3′ (sense) and 5′-AGGCTCTGTTCTTGGGAAGTCGA-3′ (antisense). Six micrograms of WCE were combined with radioactively labelled DNA probe and protein: DNA complexes were separated by gel electrophoresis and visualized by Fuji Medical X-RAY film (Tokio, Japan). Quantification was performed with ImageJ software (https://imagej.net/Welcome).
MTS proliferation assay
To measure the survival and proliferation of cells, the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) from Promega was used according to the manufacturer’s protocol.
Statistical analyses
Student’s t-test was used to compare differences of means between two groups. For all analyses, significance was inferred at P < 0.05 (* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001). Graphpad Quick Calcs online statistical software (GraphPad Software, San Diego, CA, USA) or Social Science Statistics (https://www.socscistatistics.com/about/) was used.
Results
PTP1B expression is variable in primary cHL cases
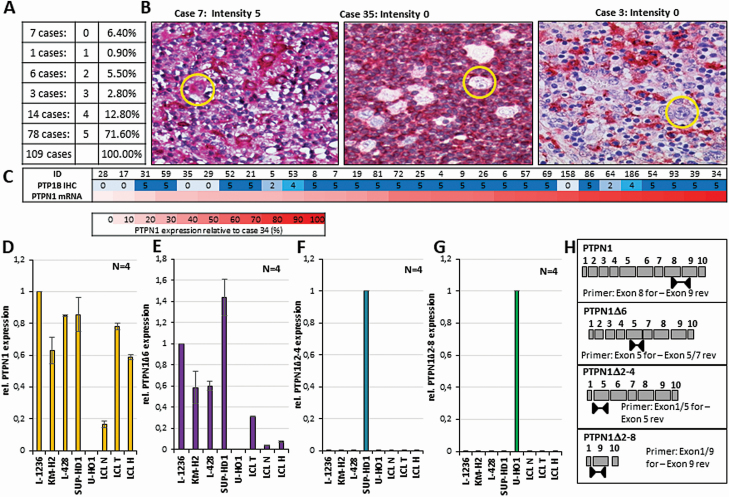
Expression of PTP1B in cHL cell lines is highly variable, with U-HO1 being devoid of PTP1B expression and with Sup-HD1 and HDLM-2 displaying low PTP1B expression levels, for example (12,13). To explore whether the PTP1B expression in primary HRS cells shows a similar variation, we conducted immunohistochemical stainings of PTP1B in a cohort of clinical cHL cases consisting of 109 cases using a scoring system based on the PTP1B signal intensity in the HRS cells (characteristics of the cHL cohort is given in Supplemental Table 1, available at Carcinogenesis Online). Three examples for the staining intensity 0 and 5 of the malignant HRS cells used for evaluation of the PTP1B expression levels are depicted in Figure 1B. About 71% of the HRS cells of the cases are positive for PTP1B expression, 15.6% show an intermediate or heterogenous protein level, 6.4% show very weak PTP1B levels, and 6.4% were negative for PTP1B (Figure 1A). No correlation of PTP1B staining intensity with the relapse status, age at diagnosis, staging or subtype was detected (Supplemental Figure 1A–D, available at Carcinogenesis Online). A heterogeneous expression was also seen comparing the PTPN1 mRNA levels with the PTP1B protein levels in the HRS cells of a set of cHL cases (Figure 1C). Here, some of the cases show high PTP1B protein levels with low PTPN1 mRNA levels (cases 31, 59 and 52), while others show high PTPN1 mRNA, but low PTP1B protein levels (cases 158, 64 and 186), suggesting that mRNA and protein expression of PTP1B is only weakly correlated in HRS cells (r = +0.37, Supplemental Figure 1E, available at Carcinogenesis Online). Collectively, these results support the idea that PTP1B expression in HRS cells is not strictly dependent on PTPN1 transcription, but appears to be also regulated at post-transcriptional levels.
Figure 1.
Expression of PTP1B and PTPN1 mRNA in primary cHL cases and analysis of PTPN1 mRNA variants in cHL cell lines. (A) Percentage of the different PTP1B expression levels in HRS cells determined by immunohistochemical staining of 109 classical Hodgkin lymphoma cases. The intensity was classified as (0) negative, (1) HRS cells weakly positive and negative, (2) all HRS cells weakly positive, (3) heterogeneous signal, (4) all HRS cells either strong or weakly positive and (5) strong PTP1B signal in HRS cells. (B) Immunohistochemical staining of PTP1B in three different cHL cases, staining intensity 5 and 0 exemplarily depicted. Single HRS cells are marked by a yellow circle. (C) Relative PTPN1 mRNA expression levels (lower part), normalized against case 34 (in %) and the corresponding PTP1B staining intensity (upper part). (D–G) Relative value of the mRNA expression of the total PTPN1 (D), the PTPN1∆6 (E), the PTPN1∆2–4 (F) and the PTPN1∆2–8 (G) variants in the different cHL cell lines and three different LCL cell lines measured by semi-quantitative real-time PCR (sqPCR) analyses. CT values normalized against the mRNA expression levels of the two housekeeper genes β-actin and GAPDH. (Expression of PTPN1WT (D) or of PTPN1∆6 (E) in L-1236 was arbitrary set to 1, expression of PTPN1∆2–4 in SUP-HD1 (F) was arbitrary set to 1, expression of PTPN1∆2–8 in U-HO1 (G) was arbitrary set to 1.) (H) Schematic presentation of the position of exon spanning primers used for the indicated sqPCR depicted in (D–G).
Identification of PTP1B variants
Recently, we and others identified PTPN1 mutations and PTPN1 mRNA variants in cHL cell lines and primary cHL cases (12,13,18,19). Moreover, we demonstrated that one of the PTPN1 variants, lacking exon 6 (PTPN1∆6), is a PTPN1 splice variant capable of augmenting JAK/STAT activity. PTPN1∆6 expression is detectable in a wide variety of B cells and B cell lymphomas at low levels, but found to be increased in HRS cells of a defined set of cHL cases and in all cHL cell lines (Figure 1E) with the exception of U-HO1 (13). Additionally, we identified two mRNA variants lacking either exons 2–4 (PTPN1∆2–4) from HRS cells of case 35 or exons 2–8 (PTPN1∆2–8) from HRS cells of case 3, both negative for PTP1B protein expression (13). These variants were also seen in the cell lines U-HO1 and SUP-HD1, and are already described to be caused by mutations in PTPN1 (12,13). To determine whether PTPN1∆2–4 and PTPN1∆2–8 are individual variants only detectable in the cHL cell lines Sup-HD1 and U-HO1, semiquantitative real-time PCR (sqPCR) analysis with mutation-specific primer sets for total PTPN1 mRNA, PTPN1∆6, PTPN1∆2–4 and PTPN1∆2–8 mRNA (Figure 1H) was performed. While a signal for total PTPN1 mRNA was detected in all analysed cell lines, with the exception of the U-HO1 (Figure 1D, (13)), a PTPN1∆2–4 mRNA was only detectable in Sup-HD1 (Figure 1F) and PTPN1∆2–8 only in U-HO1 (Figure 1G). Moreover, expression of PTPN1∆2–4 mRNA was not detected in HRS cells of a set of cHL cases (Supplementary Figure 2C, available at Carcinogenesis Online). In contrast, PTPN1∆6 mRNA was detectable in all cHL and LCL cell lines analysed at variable levels with the exception of U-HO1. These results imply that the mutation-based PTPN1∆2–4 and PTPN1∆2–8 mRNA variants are personal variants for the defined cHL cell line.
PTP1B∆2–4 is a positive regulator of STAT6 signalling
Gunawardana et al. characterized a set of PTP1B mutant proteins, which displayed a gradual loss of phosphatase activity and a diminished negative impact on JAK/STAT activity (12). However, as only SNVs and truncating mutants were analysed, we aimed to functionally characterize PTP1B exon-deleted mutants. Here, we focussed on the exon-deleted PTP1B mutant PTP1B∆2–4 since PTP1B∆2–8 was neither detectable in immunoblot analyses nor did we observe a change in JAK/STAT activation upon co-expression of the variant (data not shown). To explore whether PTP1B∆2–4 exerts a positive effect on JAK/STAT activity similar to PTP1B∆6 (13), PTP1BWT, PTP1B∆6 or PTP1B∆2–4 were ectopically expressed in HEK293-STAT6 cells and were subsequently stimulated with IL-4 (Figure 2A). Ectopic expression of PTP1BWT, as expected, attenuated the IL-4-induced STAT6 activity, whereas expression of either PTP1B∆6 or PTP1B∆2–4 augmented STAT6 activity significantly. To determine whether the loss of exons in PTP1B∆2–4 affects its phosphatase activity similar to PTP1B∆6, an in vitro phosphatase analysis was conducted. Both, PTP1B∆2–4 and PTP1B∆6, displayed a complete loss of phosphatase activity comparable to the catalytically inactive PTP1BC215S mutant (Figure 2C).
To further compare the characteristics of PTP1B∆2–4 and PTP1B∆6, we analysed the IL-4-induced STAT6 phosphorylation in HEK293-STAT6 cells. As expected, IL-4-induced STAT6 phosphorylation seen in empty vector-transfected HEK293-STAT6 cells (Figure 2B) was distinctively decreased upon ectopic PTP1BWT expression. In contrast, PTP1B∆6 or PTP1B∆2–4 clearly increased the IL-4-induced STAT6 phosphorylation. Moreover, ectopically expressed PTP1BWT diminished STAT6 DNA binding whereas either PTP1B∆6 or PTP1B∆2–4 augmented STAT6 DNA binding (Figure 2D/E). Together, these results imply that the loss of exon sequences converted PTP1B∆2–4 from a negative into a positive JAK/STAT regulator similar to PTP1B∆6.
PTP1B∆6 and PTP1B∆2–4 augment JAK/STAT activity in HEK293-STAT6 cells by different modes of action
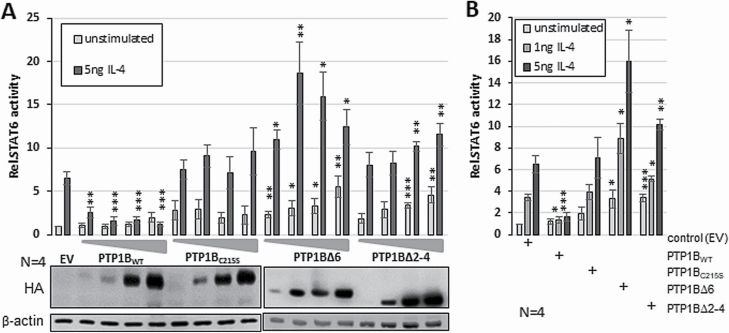
One specific hallmark of PTP1B∆6 is its ability to augment STAT6 activity at very low amounts or in case of sub-optimal stimulation conditions (13). To investigate whether PTP1B∆2–4 behaves in a similar fashion, we performed STAT6 luciferase assays in HEK293-STAT6 cells using increasing amounts of PTP1B variants with and without stimulation with 5 ng of IL-4 (Figure 3A) and compared the effect of small amounts of the PTP1B variants upon stimulation with 1 or 5 ng/ml IL-4 (Figure 3B). While ectopic PTP1BWT consistently diminished IL-4-induced STAT6 activity, ectopic expression of PTP1BC215S only moderately enhanced IL-4-induced STAT6 activity (Figure 3A). Ectopic expression of increasing amounts of PTP1B∆6 augmented basal STAT6 activity significantly. Moreover, the strongest effect was observed upon transfection of as little as 5 ng PTP1B∆6 vector in HEK293-STAT6 cells stimulated with 5 ng IL-4. While PTP1B∆6-based augmentation of IL-4-induced STAT6 activity peaked with low amounts and dropped thereafter, PTP1B∆2–4 augmented STAT6 activity in a concentration-dependent manner. Similarly, PTP1B∆6 co-expression boosted the JAK-STAT activity even at the sub-optimal concentration of 1ng IL-4, while this effect was less pronounced by PTP1B∆2–4, underscoring the idea of PTP1B∆6 being a positive JAK/STAT regulator even under suboptimal conditions. Taken together, these results suggest that PTP1B∆6 and PTP1B∆2–4 augment JAK/STAT activity in HEK293-STAT6 cells by different modes of action.
Figure 3.
Different kinetics of the impact of PTP1B variants on STAT6 activity. (A) Luciferase reporter assay stimulated with 5ng/ml IL-4 for 18 h. HEK293-STAT6 cells transiently transfected with either 1, 5, 10 or 50 ng of HA-PTP1BWT, PTP1BC215S, HA-PTP1B∆6 or HA-PTP1B∆2–4 - pcDNA3.1 (EV) served as control (upper part). Mean of four independent experiments is depicted. Immunoblot analysis of WCE of luciferase measurements (lower part) using the indicated antibodies. (B) Luciferase reporter assay stimulated with either 1 or 5 ng/ml IL-4 for 18 h. HEK293-STAT6 cells transiently transfected with 10 ng of pcDNA3.1 (EV), HA-PTP1BWT, PTP1BC215S, HA-PTP1B∆6 or HA-PTP1B∆2–4. Mean of four independent experiments is depicted. (A+B) Mean values and standard error of mean (SEM) are depicted. Comparisons were performed between empty vector measurement and individual treatments/transfections. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant).
PTPB∆2–4 supports proliferation and survival of L-428 cells
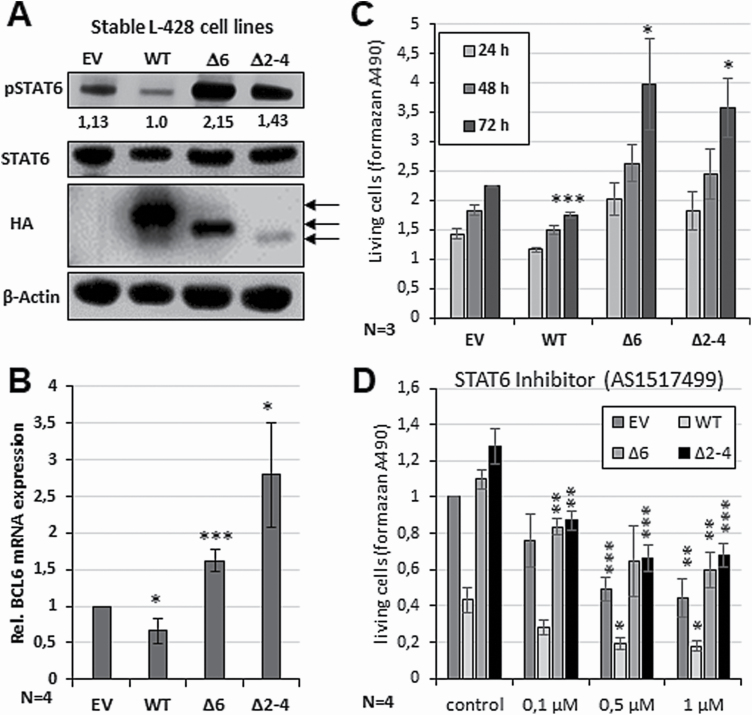
To explore the impact of the different PTP1B variants on the JAK/STAT pathway in a cHL background, L-428 cells were stably transfected with either PTP1BWT, PTP1B∆6, PTP1B∆2–4 or the empty vector and gathered after growth on G-418 selection medium in serial dilutions. Increased STAT6 phosphorylation was seen in L-428 cells stably expressing either PTP1B∆6 or PTP1B∆2–4, whereas the PTP1BWT expression caused diminished STAT6 phosphorylation, as expected (Figure 4A). The positive impact of PTP1B∆6 or PTP1B∆2–4 on STAT6 activity is also supported by an increased expression of the STAT6 target gene BCL6 (Figure 4B). While PTP1BWT diminished BCL6 mRNA levels, PTP1B∆6 or PTP1B∆2–4 augmented BCL6 expression. As a constitutive JAK/STAT activity is crucial for proliferation in cHL cell lines (20–25), the impact of the different PTP1B variants on the cell growth of the stably transfected L-428 cell clones was analysed by MTS assays (Figure 4C). Here, the ectopic expression of PTP1B∆2–4 increased L-428 proliferation similar to PTP1B∆6. To delineate whether STAT6 is involved in the altered proliferation in the L-428-PTP1B cell lines, the different cell lines were treated with the STAT6 inhibitor AS1517499. Here, all cell lines displayed a decreased proliferation upon increasing AS1517499 concentrations (Figure 4D).
Figure 4.
Impact of PTP1B∆ variants on the proliferation and survival of the cHL cell line L-428. (A) Immunoblot analysis of WCE of L-428 cells stably transfected with either pcDNA3.1 (EV) or HA-PTP1BWT, HA-PTP1B∆6 or HA-PTP1B∆2–4. Antibodies against pSTAT6, HA and β-actin are used. Numbers indicate relative pSTAT6 levels normalized against β-actin value (Supplemental Figure 3B). (B) Altered BCL6 mRNA expression levels of the L-428 cells stably transfected with the PTP1BWT, PTP1B∆6 or PTP1B∆2–4 determined by sqPCR analysis. Measurements were performed in triplicates and the mean of four independent experiments and standard error of mean (SEM) are depicted. Comparisons were performed between EV measurement and individual stable cell line. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant). (C) Altered proliferation of the L-428 cells stably transfected with the PTP1BWT, PTP1B∆6, PTP1B∆2–4 variant or EV control over time (24, 48 and 72 h). Mean of three independent MTS assay measurements and SEM are depicted. Comparisons were performed between control measurement and individual stable cell line. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant). (D) Altered proliferation of the L-428 cells stably transfected with the PTP1BWT, PTP1B∆6, PTP1B∆2–4 variant or EV control and incubated with the indicated amounts of the STAT6 inhibitor AS1517499 for 72 h. Mean of four independent MTS assay measurements and SEM are depicted. Comparisons were performed between control measurement and individual stable cell line. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant).
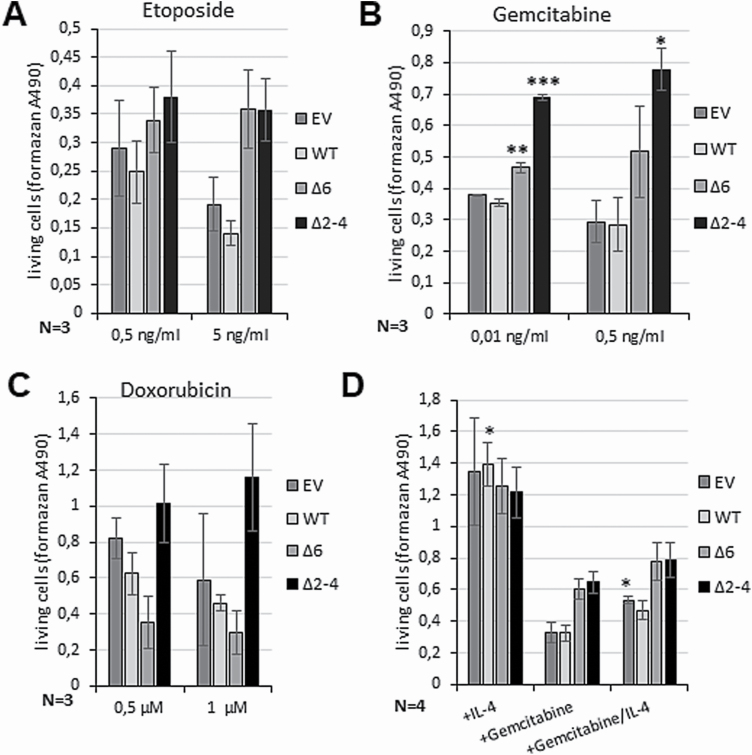
As the activity of the JAK/STAT pathway has been shown to be involved in resistance mechanisms against chemotherapeutical agents (13,26–28), the efficacy of the in cHL treatment commonly used drugs etoposide (Figure 5A), gemcitabine (Figure 5B) and doxorubicin (Figure 5C) on the different stably transfected L-428 cells was analysed. The agents show a lethal effect on L-428 control cells leading to an approximately 60–70% (gemcitabine and etoposide) or 50–60% (doxorubicin) reduced viability already at low concentrations (Supplemental Figure 3, available at Carcinogenesis Online). Overexpression of PTP1BWT only slightly augmented the cytotoxic effect on L-428, whereas ectopically expressed PTP1BΔ2–4 displayed reduced cytotoxicity depicted by an increased number of living cells after treatment with 0.5 and 5 ng/ml etoposide (Figure 5A) or 0.01 or 0.5 ng/ml gemcitabine (Figure 5B) in comparison to the control. Similarly, PTP1BΔ6 also partially protected the L-428 cells after etoposide and gemcitabine treatment (Figure 5A and B), but with lower efficiency than PTP1B∆2–4. In case of doxorubicin (0.5 and 1 µM) addition (Figure 5C) a moderate protective effect was only visible in the case of PTP1BΔ2–4, but not in case of PTP1BΔ6. To determine whether an IL-4-induced STAT6 activation might revert the cytotoxic effect of gemcitabine, stable transfected L-428-PTP1B cell lines were either treated with IL-4 or gemcitabine alone, or in combination (Figure 5D). IL-4 stimulation alone increased proliferation of all L-428-PTP1B cell lines compared to the untreated controls. Gemcitabine treatment alone, on the other hand, diminished cell proliferation, but appeared to be less effective in L-428-PTP1B∆6 and -PTP1B∆2–4 cells, as expected. Co-treatment with IL-4 partially rescued the cells against the cytotoxic effect of gemcitabine suggesting that JAK/STAT inhibition is part of the adverse effects of gemcitabine in cHL, which can be reverted by an additional IL-4 signal. A similar effect of gemcitabine was described by Chen et al. 2018 in a pancreatic cancer growth mouse model. The authors determined an inhibited tumour growth and induced apoptosis after treatment with a combination of gemcitabine and erlotinib in vivo by downregulated phosphorylation levels of JAKs and STATs (29). Furthermore, upregulation of STAT1 activity was described to confer etoposide resistance in lung cancer (30) and resistance to apoptosis induced by doxorubicin in colon cancer cells (31). Taken together, the pro-proliferative effect and the protective function of PTP1B∆2–4 highlight the pro-oncogenic function of this PTP1B mutant.
Figure 5.
PTP1B∆2–4 has protective effects against cytotoxic agents in the cHL cell line L-428. (A–C) Altered proliferation of the L-428 cells stably transfected with pcDNA3.1 (EV), or with HA-PTP1BWT, HA-PTP1B∆6 or HA-PTP1B∆2–4 after treatment with the indicated amount of etoposide (A), gemcitabine (B) and doxorubicin (C) measured by MTS proliferation assay. The untreated cells served as control. Mean of three independent MTS assay measurements is depicted. (D) Proliferation of L-428 cells stably transfected with pcDNA3.1 (EV), or with HA-PTP1BWT, HA-PTP1B∆6 or HA-PTP1B∆2–4 either stimulated with 20 ng/ml IL-4 for 48 h, laced with 1 ng/ml gemcitabine or laced with 1 ng/ml gemcitabine in combination with IL-4 stimulation (20 ng/ml, 48 h) measured by MTS proliferation assay. The untreated cells served as control. Mean of four independent MTS assay measurements is depicted. (A–D) Mean values and standard error of mean (SEM) are depicted. Comparisons were performed between untreated control and individual treatment for each stable cell line. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant).
PTP1B∆6, but not PTP1B∆2–4, attenuates PTP1BWT activity and protein stability
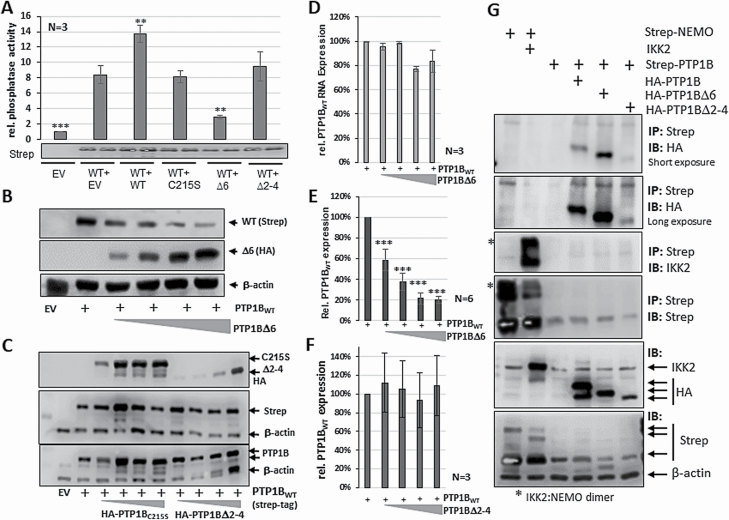
The molecular basis for the positive effect of PTP1B∆6 is its capability to impair the phosphatase activity of PTP1BWT by formation of PTP1B∆6:PTP1BWT heterodimers (13). Therefore, the impact of the co-expression of the PTP1B∆2–4 variant on PTP1BWT phosphatase activity was compared to the effect of co-expressed PTP1B∆6 and PTP1BC215S by in vitro phosphatase assays (Figure 6A). Elevated phosphatase activity was observed upon Strep-PTP1BWT expression, which was increased even further by adding HA-PTP1BWT. Co-expression of the HA-PTP1B∆2–4 variant or the co-expression of the enzymatic inactive HA-PTP1BC215S mutant had no impact on the Strep-PTP1BWT activity. In contrast, the co-expression of HA-PTP1B∆6 had a distinct inhibitory effect on Strep-PTP1BWT activity. Notably, the co-expression of PTP1BWT and PTP1B∆6 was not only leading to a decrease in the PTP1BWT phosphatase activity, but also to lower PTP1BWT protein levels determined by immunoblot analysis. To get a further insight into this effect, ectopically expressed PTP1BWT mRNA and protein levels were analysed upon co-expression of increasing amounts of PTP1B∆6 (Figure 6B/E). Diminishing PTP1BWT protein levels were observed upon co-expression of increasing amounts of PTP1B∆6, even when PTP1B∆6 was only slightly expressed. In contrast, the sqPCR results indicate that PTP1BWT mRNA expression was affected only with high amounts of PTP1B∆6 (Figure 6D). Together, these results imply that PTP1B∆6 impairs the activity of PTP1BWT at least in part by induced proteolysis of PTP1BWT. In contrast, the co-expression of the PTP1B∆2–4 variant or the enzymatic inactive PTP1BC215S caused no change in PTP1BWT expression (Figure 6C and F). The missing effect of PTP1B∆2–4 on the stability of PTP1BWT is also mirrored by a minor PTP1B∆2–4:PTP1BWT heterodimer formation in contrast to PTP1B∆6 (Figure 6G), which was analysed in comparison with the known high-affinity IKK2:NEMO interaction. While PTP1B∆6 shows an increased PTP1BWT binding, as expected, PTP1B∆2–4:PTP1BWT dimer formation was similar to PTP1BWT homodimer formation. Collectively, these results further support the notion that the positive impact of PTP1B∆2–4 on JAK/STAT signalling is unique and differs from the PTP1B∆6-mediated mechanism.
Figure 6.
PTP1BWT activity and stability remains unaltered upon co-expression of PTP1B∆2–4. (A) Phosphatase assay with Strep-tagged PTP1BWT alone or together with HA-PTP1BWT, HA-PTP1BC215S, HA-PTP1B∆6 or HA-PTP1B∆2–4 ectopically expressed in HEK293 cells, as indicated. pEXPR-IBA105 (EV) serves as control. Mean values of three independent experiments and standard error of mean (SEM) are depicted. Comparisons were performed between PTP1BWT measurement and individual transfections. Significances are calculated using Student’s t-test (P < 0.05 was regarded as significant). (B) Immunoblot analysis of whole cell extracts from HEK293 cells either transfected with pcDNA3.1 (EV), or with Strep-PTP1BWT and increasing amounts of HA-PTP1B∆6 (1:50, 1:5, 1:2, 1:1). Antibodies against PTP1B and β-actin used to determine amounts of PTP1BWT (endogen and exogen), PTP1B∆6 and the housekeeper β-actin. (C) Immunoblot analysis of whole cell extracts from HEK293 cells either transfected with pcDNA3.1 (EV), with Strep-PTP1BWT or with Strep-PTP1BWT and increasing amounts of HA-PTP1BC215S or HA-PTP1B∆2–4 (1:50, 1:5, 1:2, 1:1). Antibodies against Strep-tag used to determine amounts of exogen PTP1BWT, against HA-tag to determine amounts of PTP1BC215S or HA-PTP1B∆2–4, antibodies against PTP1B used to determine amounts of whole PTP1B and antibodies against the housekeeper β-actin. (D) Relative mRNA expression of the PTPN1WT RNA levels expressed in HEK293 cells with or without increasing amounts of PTP1B∆6 (1:50, 1:5, 1:2, 1:1) measured by sqPCR. Values normalized against the mRNA expression levels of the two housekeeper genes β-actin and GAPDH and calculated in comparison to the ectopic PTP1BWT expression alone. Mean and SEM of three independent experiments depicted. (E) Quantification of the amount of PTP1BWT protein as depicted in (B). Mean value and SEM of six independent experiments is shown and significance was determined in comparison to the PTP1BWT alone measurements. (F) Quantification of the amount of PTP1BWT protein as depicted in (C). Mean value and SEM of three independent experiments is shown and significances are calculated using Student’s t-test determined in comparison to the PTP1BWT alone measurement (P < 0.05 was regarded as significant). (G) Strep-tag immunoprecipitation (IP, upper 4 blots) using WCE of HEK293 cells transfected with Strep-tagged PTP1BWT alone or together with HA-PTP1BWT, HA-PTP1B∆6 or HA-PTP1B∆2–4. HEK293 cells transfected with Strep-tagged NEMO alone and in combination with untagged IKK2 served as IP positive control. Immunoblots (IB, lower 2 blots) using the same WCE served as intake control. Protein bands visualized using indicated antibodies.
Discussion
Inactivating mutations in the genes encoding negative regulators of the JAK/STAT pathway have been seen in several types of B cell lymphoma highlighting the importance of JAK/STAT signalling for the pathogenesis of B cell lymphoma. Besides SOCS1 mutations, which have been identified in cHL, diffused large B cell lymphoma (DLBCL), and (PMBL), also mutations in PTPN2, encoding TC-PTP, and PTPN1 were observed (9,12,15,32–35). Gunawardana et al., for instance, reported a set of PTPN1 mutations in cHL and PMBL cases and cell lines and found a correlation of the mutation status of PTPN1 and its expression in HRS cells of clinical cHL cases (12). However, the authors also demonstrated that PTP1B levels in HRS were very variable suggesting that this is not a black-and-white picture, and therefore additional mechanisms controlling PTP1B protein levels and activity take hold. Consistently, the overall decreased activity of PTP1B previously demonstrated in the cHL cell lines (13) might be also caused by such additional mechanisms. An AKT-mediated PTP1B phosphorylation and a reversible oxidation of Cys215 are only two examples for mechanisms potentially involved in PTP1B control might be in place also in cHL (36–38). We observed reduced levels of PTP1B in 29% of the HRS cells in the cHL cohort (Figure 1A+B). However, PTPN1 mRNA and PTP1B protein levels were only weakly correlated in our cohort (Supplementary Figure 1E, available at Carcinogenesis Online), which could be explained by such mechanisms affecting either PTPN1 mRNA translation or PTP1B protein stability. While the reduction in PTP1B expression could be caused by various mechanisms, we speculated that in some cases a lack of PTP1B level is due to an inactivating mutation in PTPN1 similar to the situation in the cHL cell lines U-HO1 and SUP-HD1 (12). However, in contrast to the recently identified PTPN1∆6 splice variant, which is expressed in nearly all cHL cell lines (Figure 1E, (13)), PTPN1∆2–4 and PTPN1∆2–8 expression is limited to SUP-HD1 and U-HO1, respectively (Figure 1D-G). Moreover, sequencing of the PTPN1 mRNA indeed showed that both mutants, PTPN1∆2–4 and PTPN1∆2–8, are also expressed in HRS cells of single cHL cases (Supplementary Figure 2C, available at Carcinogenesis Online and (13)).
Altered PTP1B proteins lacking one or more exons like PTP1B∆2–4 might be generated by a deletion of the respective DNA section or by mutations in splice sites as been identified by Gunawardana et al. (12,19) and Tiacci et al. (9). However, a functional characterization of these altered PTP1B proteins has not been conducted. Especially in the light of steadily emerging efforts to establish personalized therapy regimens on basis of the individual mutational profile, it is essential to determine the functional consequences of such an exon deleted PTP1B mutant. In a first approach to unravel the functional relevance of exon deletion mutants, we analysed the impact of the exon deletion in PTP1B∆2–4 on the IL-4-induced JAK/STAT activation in comparison to the already known positive effect of the PTP1B∆6 splice variant. The PTP1B∆2–8 mutant, in contrast, was excluded from analysis as it was not detectable by immunoblot analyses probably either due to the small size or due to an instability of the resulting PTP1B peptide of 28 amino acids, which lack functional domains (Supplementary Figure 4A, available at Carcinogenesis Online). Indeed, the PTP1B∆2–4 mutant is able to augment IL-4-induced STAT6 activity in HEK293-STAT6 cells as measured by reporter gene analyses (Figure 2A), phospho-STAT6 status (Figure 2B), and DNA binding capability (Figure 2D/E). Collectively, these experiments point to the fact that PTP1B∆2–4 could be involved to maintain the constitutive activation of the JAK/STAT signalling pathway in cHL (3,39).
To address the role of PTP1B∆2–4 in cHL, we used L-428 cells stably expressing PTP1B∆2–4 and compared the consequences for STAT activation, target gene expression and changes in proliferation with PTP1BWT, PTP1BC215S, PTP1B∆6 expressing L-428 cells. Again, PTP1B∆2–4 augmented STAT6 activity leading to an increased expression of the STAT target gene BCL6 to a level comparable to PTP1B∆6, while PTP1BWT diminished both, STAT6 phosphorylation and BCL6 target gene expression (Figure 4A and B). The oncogenic potential of PTP1B∆2–4 is underscored by the increased proliferation of L-428 cells stably expressing ectopic PTP1B∆2–4 (Figure 4C) as well as the increased resistance against the chemotherapeutic agents such as etoposide, gemcitabine and doxorubicin (Figure 5A–D). Interestingly, a protective effect of an augmented JAK/STAT signalling against the cytotoxic effect of all three chemotherapeutic agents has already been shown (29–31). This oncogenic and protective effect suggests that exon deleted PTP1B mutants, similar to PTP1B∆2–4, could be involved in the negative treatment response observed in relapsed or refractory HL.
How PTP1B∆2–4 exerts it stimulatory and protective effects are not completely understood. However, we hypothesize that PTP1B∆2–4 exerts its positive effects on cHL proliferation and survival by several mechanisms. One potential mechanism might be a competition of PTP1B∆2–4 with PTP1BWT for binding to its interaction partners or to its substrates like JAK2 or TYK2, which would explain the dose-dependency of the PTP1B∆2–4 effect on the JAK/STAT signalling (Figure 3). The dose-dependency of the PTP1B∆2-4-mediated effects on JAK/STAT activity is in contrast to the low expression levels at which PTP1B∆6 has its functional optimum (Figure 3). This difference is based on the lack of an increased PTP1BWT binding and subsequently unaltered PTP1BWT phosphatase activity, as demonstrated in vitro phosphatase experiments (Figure 6B). Moreover, PTP1BWT protein levels remain unaffected upon PTP1B∆2–4 co-expression (Figure 6C/D). Therefore, PTP1B∆2–4 appears to act at least partially by competitive means while PTP1B∆6 seems to catalyse a drop in PTP1BWT activity and protein stability even when expressed at low levels. As PTP1B modulates various signalling pathways including its effect on AKT, RAS or SRC (8,40–42), higher PTP1B∆2–4 protein levels might additionally interfere with pathways other than JAK/STAT signalling. Moreover, PTP1B∆2–4 might also form novel specific protein interactions due to its altered structure (Supplementary Figure 4A, available at Carcinogenesis Online). Of note, in the stably transfected L-428 cells protein levels of ectopic PTP1B∆2–4 is recurrently lower than ectopic PTP1B∆6 or PTP1BWT (Figure 4A). Although this might be due to increased instability of PTP1B∆2–4, we hypothesize that this is rather caused by a toxic effect of high amounts of PTP1B∆2–4 interfering with protein complexes essential for cell survival. Similarly, a toxic effect of higher PTP1B∆2–4 levels could also explain the lack of a PTP1B∆2–4 protein signal in SUP-HD1 cells (12,13). The suggested mode of action of the PTP1B∆6 and PTP1B∆2–4 variant is summarized in a schematic model (Supplementary Figure 4B, available at Carcinogenesis Online).
Taken together, the data presented here illustrate the complexity of the mechanisms controlling the JAK/STAT pathway in cancer, especially in B cell lymphoma. The complexity is further underscored by the fact that co-mutations in SOCS1 and PTPN1, STAT6 and JAK2 have been detected in DLBCL, PMBL and cHL (9,12), but these potentially combinatorial effects have not been explored yet. Moreover, whether the oncogenic potential of PTP1B∆6 and PTP1B∆2–4 is mediated solely through the impact on JAK/STAT signalling or also by additional mechanisms is currently unknown and needs to be determined in future analyses. However, the partial rescue of gemcitabine treated L-428 PTP1B cells by IL-4 stimulation and the reduced proliferation upon treatment with the STAT6 inhibitor clearly argue for an essential role of the STAT6 signalling for proliferation and survival of L-428 cells, although controversial results regarding the role of STAT6 in cHL cell lines have been published (9). Thus, unravelling the functional impact of the different altered negative JAK/STAT regulators will not only improve the insight into this pro-oncogenic pathway, but might also lead the path towards novel therapeutic targets. In sum, our results reveal a common oncogenic potential of a PTP1B exon deleted mutant and a PTP1B splice variant emphasizing the necessity for a detailed biochemical analysis of protein variants found in cancer to correctly estimate their functional role in oncogenesis.
Supplementary Material
Acknowledgements
The authors like to thank Dr Kevin Mellert, Dr Ulrike Kostetzka, Dr Ingo Melzner, Karola Dorsch and Dr Stephanie Weißinger for their support, help with the cHL cohort and the use of the PALM microbeam.
Glossary
Abbreviations
- cHL
classical Hodgkin lymphoma;
- DLBCL
diffused large B cell lymphoma;
- ER
endoplasmatic reticulum
- HRS
Hodgkin/Reed-Sternberg;
- JAK
Janus kinase;
- PMBL
primary mediastinal B cell lymphoma;
- RPTKs
receptor protein tyrosine kinases;
- RT
room temperature;
- SEM
standard error of mean;
- STAT
signal transducer and activator of transcription;
- WCE
whole-cell extracts
Funding
This work was supported by funds from the Deutsche Forschungsgemeinschaft (MO 384/6-1 to Peter Möller and MA2367/6-1 to Ralf Marienfeld).
Conflict of Interest Statement: The authors declare no conflict of interest. The funders had no role in the analysis of the data, in the writing of the manuscript or in the decision to publish the results.
References
- 1. Bräuninger, A., et al. (2006) Molecular biology of Hodgkin’s and Reed/Sternberg cells in Hodgkin’s lymphoma. Int. J. Cancer, 118, 1853–1861. [DOI] [PubMed] [Google Scholar]
- 2. Ansell, S.M. (2016) Hodgkin lymphoma: 2016 update on diagnosis, risk-stratification, and management. Am. J. Hematol., 91, 434–442. [DOI] [PubMed] [Google Scholar]
- 3. Küppers, R., et al. (2012) Hodgkin lymphoma. J. Clin. Invest., 122, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vockerodt, M., et al. (2015) The Epstein-Barr virus and the pathogenesis of lymphoma. J. Pathol., 235, 312–322. [DOI] [PubMed] [Google Scholar]
- 5. Scott, D.W., et al. (2014) The tumour microenvironment in B cell lymphomas. Nat. Rev. Cancer, 14, 517–534. [DOI] [PubMed] [Google Scholar]
- 6. Murray, P.J. (2007) The JAK-STAT signaling pathway: input and output integration. J. Immunol., 178, 2623–2629. [DOI] [PubMed] [Google Scholar]
- 7. Thomas, S.J., et al. (2015) The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer, 113, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yip, S.C., et al. (2010) PTP1B: a double agent in metabolism and oncogenesis. Trends Biochem. Sci., 35, 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tiacci, E., et al. (2018) Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood, 131, 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanquart, C., et al. (2009) Implication of protein tyrosine phosphatase 1B in MCF-7 cell proliferation and resistance to 4-OH tamoxifen. Biochem. Biophys. Res. Commun., 387, 748–753. [DOI] [PubMed] [Google Scholar]
- 11. Hoekstra, E., et al. (2016). Increased PTP1B expression and phosphatase activity in colorectal cancer results in a more invasive phenotype and worse patient outcome. Oncotarget, 7(16), 21922–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gunawardana, J., et al. (2014) Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet., 46, 329–335. [DOI] [PubMed] [Google Scholar]
- 13. Zahn, M., et al. (2017) A novel PTPN1 splice variant upregulates JAK/STAT activity in classical Hodgkin lymphoma cells. Blood, 129, 1480–1490. [DOI] [PubMed] [Google Scholar]
- 14. Union for International Cancer Control. (2014) HODGKIN LYMPHOMA (ADULT) [Internet]. Medicines for treatment of the following cancers – review – EML and EMLc. 20th Expert Committee on the Selection and Use of Essential Medicines. 2014. p. 1–9. [Google Scholar]
- 15. Mader, A., et al. (2007) U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma. Cytogenet. Genome Res., 119, 204–210. [DOI] [PubMed] [Google Scholar]
- 16. Palkowitsch, L., et al. (2011) The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-kappaB activation. J. Biol. Chem., 286, 7522–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao, X., et al. (2013) An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath., 3, 71–85. [PMC free article] [PubMed] [Google Scholar]
- 18. Knecht, H., et al. (2010) 3D nuclear organization of telomeres in the Hodgkin cell lines U-HO1 and U-HO1-PTPN1: PTPN1 expression prevents the formation of very short telomeres including “t-stumps”. BMC Cell Biol., 11, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gunawardana, J., et al. (2013) Protein tyrosine phosphatase type-1 (PTPN1) is frequently mutated in primary mediastinal B cell lymphoma and Hodgkin lymphoma No Title. Blood, 122(21), 242. [Google Scholar]
- 20. Skinnider, B.F., et al. (2002) Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood, 99, 618–626. [DOI] [PubMed] [Google Scholar]
- 21. Skinnider, B.F., et al. (2002) The role of cytokines in classical Hodgkin lymphoma. Blood, 99, 4283–4297. [DOI] [PubMed] [Google Scholar]
- 22. Skinnider, B.F., et al. (2001) Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood, 97, 250–255. [DOI] [PubMed] [Google Scholar]
- 23. Baus, D., et al. (2006) Specific function of STAT3, SOCS1, and SOCS3 in the regulation of proliferation and survival of classical Hodgkin lymphoma cells. Int. J. Cancer, 118, 1404–1413. [DOI] [PubMed] [Google Scholar]
- 24. Kube, D., et al. (2001) STAT3 is constitutively activated in Hodgkin cell lines. Blood, 98, 762–770. [DOI] [PubMed] [Google Scholar]
- 25. Weniger, M.A., et al. (2006) Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene, 25, 2679–2684. [DOI] [PubMed] [Google Scholar]
- 26. Delgado-Martin, C., et al. (2017) JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias. Leukemia, 31, 2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heuck, F., et al. (2004) Combination of the human anti-CD30 antibody 5F11 with cytostatic drugs enhances its antitumor activity against Hodgkin and anaplastic large cell lymphoma cell lines. J. Immunother., 27, 347–353. [DOI] [PubMed] [Google Scholar]
- 28. Tan, F.H., et al. (2014) The role of STAT3 signaling in mediating tumor resistance to cancer therapy. Curr. Drug Targets, 15, 1341–1353. [DOI] [PubMed] [Google Scholar]
- 29. Chen, L., et al. (2018) Combination of gemcitabine and erlotinib inhibits recurrent pancreatic cancer growth in mice via the JAK-STAT pathway. Oncol Rep., 39, 1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaewpiboon, C., et al. (2015) Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol. Med. Rep., 11, 2315–2321. [DOI] [PubMed] [Google Scholar]
- 31. Meissl, K., et al. (2017) The good and the bad faces of STAT1 in solid tumours. Cytokine, 89, 12–20. [DOI] [PubMed] [Google Scholar]
- 32. Pike, K.A., et al. (2016) TC-PTP and PTP1B: Regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine, 82, 52–57. [DOI] [PubMed] [Google Scholar]
- 33. Mottok, A., et al. (2007) Somatic hypermutation of SOCS1 in lymphocyte-predominant Hodgkin lymphoma is accompanied by high JAK2 expression and activation of STAT6. Blood, 110, 3387–3390. [DOI] [PubMed] [Google Scholar]
- 34. Chikuma, S., et al. (2017) Suppressors of cytokine signaling: potential immune checkpoint molecules for cancer immunotherapy. Cancer Sci., 108, 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kleppe, M., et al. (2011) Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin’s lymphoma and T-cell non-Hodgkin’s lymphoma. Haematologica, 96, 1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barrett, W.C., et al. (1999) Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry, 38, 6699–6705. [DOI] [PubMed] [Google Scholar]
- 37. Lee, S.R., et al. (1997) Reversible inactivation of protein tyrosine phosphatase IB by hydrogen peroxide generated in A431 cells stimulated with epidermal growth factor. FASEB J., 11(9), 15366–72. [DOI] [PubMed] [Google Scholar]
- 38. Ravichandran, L.V., et al. (2001) Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor. Mol. Endocrinol., 15, 1768–1780. [DOI] [PubMed] [Google Scholar]
- 39. Vainchenker, W., et al. (2013) JAK/STAT signaling in hematological malignancies. Oncogene, 32347, 2601–1310. [DOI] [PubMed] [Google Scholar]
- 40. Lessard, L., et al. (2010) The two faces of PTP1B in cancer. Biochim. Biophys. Acta - Proteins Proteomics, 1804, 613–619. [DOI] [PubMed] [Google Scholar]
- 41. Dubé, N., et al. (2004) The role of protein tyrosine phosphatase 1B in Ras signaling. Proc. Natl. Acad. Sci. U. S. A., 101, 1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tiganis, T. (2013) PTP1B and TCPTP–nonredundant phosphatases in insulin signaling and glucose homeostasis. FEBS J., 280, 445–458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.