Abstract
Pirh2 is an E3 ligase belonging to the RING-H2 family and shown to bind, ubiquitinate and downregulate p73 tumor suppressor function without altering p73 protein levels. AIP4, an E3 ligase belonging to the HECT domain family, has been reported to be a negative regulatory protein that promotes p73 ubiquitination and degradation. Herein, we found that Pirh2 is a key regulator of AIP4 that inhibits p73 function. Pirh2 physically interacts with AIP4 and significantly downregulates AIP4 expression. This downregulation is shown to involve the ubiquitination of AIP4 by Pirh2. Importantly, we demonstrated that the ectopic expression of Pirh2 inhibits the AIP4–p73 negative regulatory pathway, which was restored when depleting endogenous Pirh2 utilizing Pirh2-siRNAs. We further observed that Pirh2 decreases AIP4-mediated p73 ubiquitination. At the translational level and specifically regarding p73 cell cycle arrest function, Pirh2 still ensures the arrest of p73-mediated G1 despite AIP4 expression. Our study reveals a novel link between two E3 ligases previously thought to be unrelated in regulating the same effector substrate, p73. These findings open a gateway to explain how E3 ligases differentiate between regulating multiple substrates that may belong to the same family of proteins, as it is the case for the p53 and p73 proteins.
This is the first report that Pirh2 regulates AIP4-mediated p73 ubiquitination and degradation. Our data provide new insight into the mechanism of p73 regulation and may lead to the development of novel anticancer therapies.
Introduction
p73 is a tumor suppressor that belongs to the p53 family known for its apoptosis and cell cycle arrest function (1–3). In early research, less attention was drawn to p73 in comparison to p53, which showed more apoptotic activity and functional cell cycle arrest despite the high structural homology between the two proteins (4–6). Knockout of the p73 gene in mice showed less effect on tumor growth when compared with similar studies in p53 (7,8). p73 was rarely mutated in human cancers (8–14). However, the role of p73 in cancer was highlighted when several studies focusing on mouse models later revealed that mutations in p73 when accompanied by p53 mutants, lead to more aggressive tumors. Also, mice with p73+/− deletions developed spontaneous tumors indicating the role of p73 in tumor suppression (15,16). At the molecular level, it was shown that overexpression of p73 could activate p53 responsive genes and trigger apoptosis (17,18). p73 responds to DNA damage and induces cell cycle arrest through the transactivation of the p21 gene (19,20). In addition, p73 expression is aberrant in several human tumors (20,21). However, knowledge of the regulation of p73 remains unclear. As a member of the p53 family, many E3 ligases previously shown to regulate the p53 protein, such as MDM2, Pirh2, COP1, UBE4B, etc., were considered to be associated with p73 as well (22–26). Wu et al. showed that Pirh2, a RING-H2 E3 ligase initially discovered as a negative regulator to p53, can bind, polyubiquitinate and inhibit p73 transcriptional activity, but it lacks the degradation effect at the protein level (27). The utilization of K63 ubiquitin lysine chains that do not induce proteasomal degradation post-ubiquitination, as it is the case with K48 lysine chains utilized by Pirh2 to regulate p53, provided an explanation (28–31). Despite the polyubiquitination of p73 by Pirh2, the absence of proteasomal degradation raises many questions regarding this regulatory mechanism. In parallel, a study done by Rossi et al. showed that p73 is negatively regulated by a ubiquitin-protein ligase: AIP4. AIP4 belongs to the HECT (Homologous to the E6-AP Carboxyl Terminus) domain proteins and is shown to bind and ubiquitinate p73 (32). Also, AIP4 decreases p73 protein levels and inhibits p73-dependent transcriptional activity.
In this study, we found that Pirh2 physically interacts with AIP4 in cells and in vitro. We observed that Pirh2 downregulates AIP4, whereas AIP4 enhances Pirh2 expression. We confirmed these findings using AIP4-siRNA or Pirh2-siRNA that depleted their endogenous expression. The regulatory mechanism was further defined as Pirh2 decreased AIP4–p73 negative regulation. Our findings demonstrate that AIP4 ubiquitination mediated by Pirh2 results in AIP4 downregulation. Our data reveal that Pirh2 promotes AIP4 self-ubiquitination and reduces AIP4 protein stability. Moreover, our results indicate that the ubiquitination of p73 by AIP4 is significantly decreased when Pirh2 is introduced, and that AIP4 activity is restored when Pirh2 is depleted in cells. We further observe that Pirh2 reduces the inhibition of AIP4-mediated p73 cell cycle arrest (33).
Materials and methods
Cell culture and DNA transfection
Human H1299 and HEK293 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cell lines were cultured and were frozen in liquid nitrogen immediately upon arrival, and were routinely tested by PCR for mycoplasma contamination by using the following primers: Myco_fw1: 5′-ACACCATGGGAGCTGGTAAT-3′, Myco_rev1: 5′-CTTCATCGACTTTCAGACCCAAGGCA-3′. All cell lines were maintained in α-minimal essential medium supplemented with 10% fetal bovine serum. Calcium phosphate transfection methods were utilized, as described earlier (26), or transfection was accomplished using Lipofectamine 2000 (Invitrogen).
Plasmids and antibodies
pcDNA3.1 served as the backbone mammalian expression vector for p73, AIP4 and Pirh2. The pSUPER.neo (VEC-pBS-0004) vector served as the backbone for siRNAs generated for AIP4 and Pirh2. Myc-Pirh2 and mutant constructs were generated by PCR amplification and subcloned into the pcDNA3.1-Myc vector; similarly, GST-Pirh2 was generated by PCR amplification and subcloned into the pGEX-5X-1 vector as described earlier (24). Flag-p73α and Flag-p73β were amplified by PCR and subcloned into the pCMV-Tag1 vector (Stratagene). Also, His-p73α and His-p73β were PCR-amplified and subcloned into pET15b vector (Novagen). Myc-AIP4 and AIP4 mutant (C830A) were provided by Dr T. Pawson. Flag-AIP4 and various AIP4 constructs were provided by Dr Atfi. HECT-AIP4 and four individual WW domains of AIP4 constructs were amplified by PCR and subcloned into the vector pCMV-3xFlag (Sigma). All PCR products have been confirmed by sequencing. Regarding antibodies, three different p73-specific antibodies were used [ER-15, ab17230, Abcam; p73 (GC-15), #558785, BD and p73, sc17823, Santa Cruz Biotechnology]. Three AIP4 antibodies were used (anti-AIP4/ITCH, ab220637 and ab108515, Abcam; and anti-AIP4, sc28367, Sant Cruz Biotechnology). Two Pirh2 antibodies were used (ab57152, Abcam and A300-357A, Bethyl Laboratories). Two p53 antibodies: P1801 and DO-1 were used as described previously (26). An anti-Myc-specific antibody (9E10, Roche), anti-Flag (mouse monoclonal M5, mouse monoclonal M2, rabbit polyclonal M2, Sigma), anti-GST (B-14, Santa Cruz Biotechnology), anti-HA (12CA5, Roche), anti-His (Novagen) and anti-Actin (Sigma) were used according to the manufacturer’s instructions.
siRNA experiments
The Pirh2 target sequences were: CCTTGCTGTGACAAGCTTT and GCTTTAAAGTGAAGGAAGT and GCTTTATACTTGCCGCTTG representing Pirh2-siRNA1, Pirh2-siRNA2 and Pirh2-siRNA3, respectively (33). The AIP4 target sequences were: AAGTGCTTCTCAGAATGATGA and AACCACAACACACGAATTACA, representing AIP4-siRNA1 and AIP4-siRNA2, respectively (32).
Expression and recombinant protein preparation
All GST- or His-tagged recombinant proteins were expressed in the E. coli strain BL21 (DE3, Novagen), treated with isopropyl-β-d-thiogalactoside (1 mM) for 4 h at 30°C while shaking to induce fusion protein expression. Samples were centrifuged at 6000 rpm for 10 min. Proteins were purified using the glutathione Sepharose 4B (Amersham) for GST-fusion proteins (Pirh2, AIP4-WT and mutants, GST-p73α or GST-p73β) or using Ni2+-NTA agarose (Qiagen) for His-fusion proteins (His-p73α or His-p73β). All proteins were tested for purity prior to ubiquitination reactions by separating them on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels and staining with Coomassie blue overnight and then destained for 12 h the following day.
Immunoprecipitation
After 30 h of transfection, cells were lysed using lysis buffer [50 mM Tris–HCl (pH 8.0), 5 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 0.5% NP-40], with the addition of a protease inhibitor tablet (Roche), and immunoprecipitated with the indicated antibodies. The immune complexes were collected with protein (A/G-agarose beads or A beads following the manufacturer’s recommendation) and washed three times with the lysis buffer [50 mM Tris–HCl (pH 8.0), 5 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 0.2% NP-40]. The immunoprecipitates were analyzed by SDS–PAGE, transferred to polyvinylidene difluoride membranes and analyzed by western blotting.
In vitro ubiquitination assay
The in vitro ubiquitination assay was performed as described previously (26,33). Reactions were performed using purified GST-AIP4 or various GST-AIP4 mutant constructs. GST-Pirh2 (0.5–1 µg) alone or in combination with GST-AIP4 (0.5–1 µg) was used as well. Along with the E3 enzyme, E1 (40 ng, Calbiochem), E2 (Ubc-H5b, 100 ng, Calbiochem), Myc-tagged ubiquitin (5 µg, BostonBiochem) and ubiquitination buffer (50 mM Tris–HCl, 2 mM ATP, 5 mM MgCl2, 2 mM dithiothreitol) were added and using distilled water the final volume was adjusted to 40 µl. Reactions were incubated for 90 min in a 30°C water bath. The reactions were then stopped using SDS loading dye and heated at 95°C for 6 min. Proteins were separated on a 10% SDS–PAGE, transferred to polyvinylidene difluoride membranes and analyzed by western blotting. Self-ubiquitination was visualized by immunoblotting with ubiquitin-tagged antibodies. For p73 ubiquitination, purified p73 proteins (300 ng) were added to the mix. Reactions were performed and analyzed as described above. Immunoprecipitation (IP) experiments were performed to avoid false-negative data when studying p73 ubiquitination due to Pirh2 and AIP4 self-ubiquitination. The lysates were incubated with a p73-specific antibody (ER-15) for 1 h on ice. Afterward, samples were rocked for 3 h with protein agarose beads. The beads were then washed with lysis buffer and separated on 10% SDS–PAGE. Analysis for p73 ubiquitination was determined by immunoblotting using a p73-specific antibody. Also, similar experiments were performed using an AIP4-specific antibody for IP.
In cell ubiquitination assay
Cells were transfected with plasmids expressing p73, HA-tagged ubiquitin, in addition to AIP4 and Pirh2 alone, or in combination and at different concentrations depending on the experiment. Cells were collected 30 h post-transfection, lysed in lysis buffer (50 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 0.5% NP-40, 0.1% SDS and the addition of protease inhibitor tablet). Lysates were then sonicated and clarified by spinning at 4°C for 15 min to remove cell debris. Immune complexes recovered with protein A-Sepharose were washed four times with radioimmunoprecipitation assay buffer (RIPA buffer), separated on 10% SDS–PAGE and analyzed by immunoblotting as described previously (26).
Cell cycle analysis
H1299 cells were transfected with p73α, p73β or in combination with indicated expression plasmids. Thirty hours post-transfection, cells were washed twice with phosphate-buffered saline and fixed overnight at −20°C with 70% cold ethanol to a total volume of 3 ml. Pelleted cells were collected. Cells were then dissolved in sodium citrate (3.8 M), 10 mg/ml RNAse and 50 mg/ml propidium iodide to stain the nuclei. Cell cycle arrest (G0/G1; S; G2/M) was examined by flow cytometry using a FACScan Flow cytometer. The relative proportion of cells in each phase was determined using FlowJo software (TreeStar Inc.). In parallel, western blot analysis was performed to assess transfection efficiency.
Results
Pirh2 interacts with AIP4 in cells and in vitro
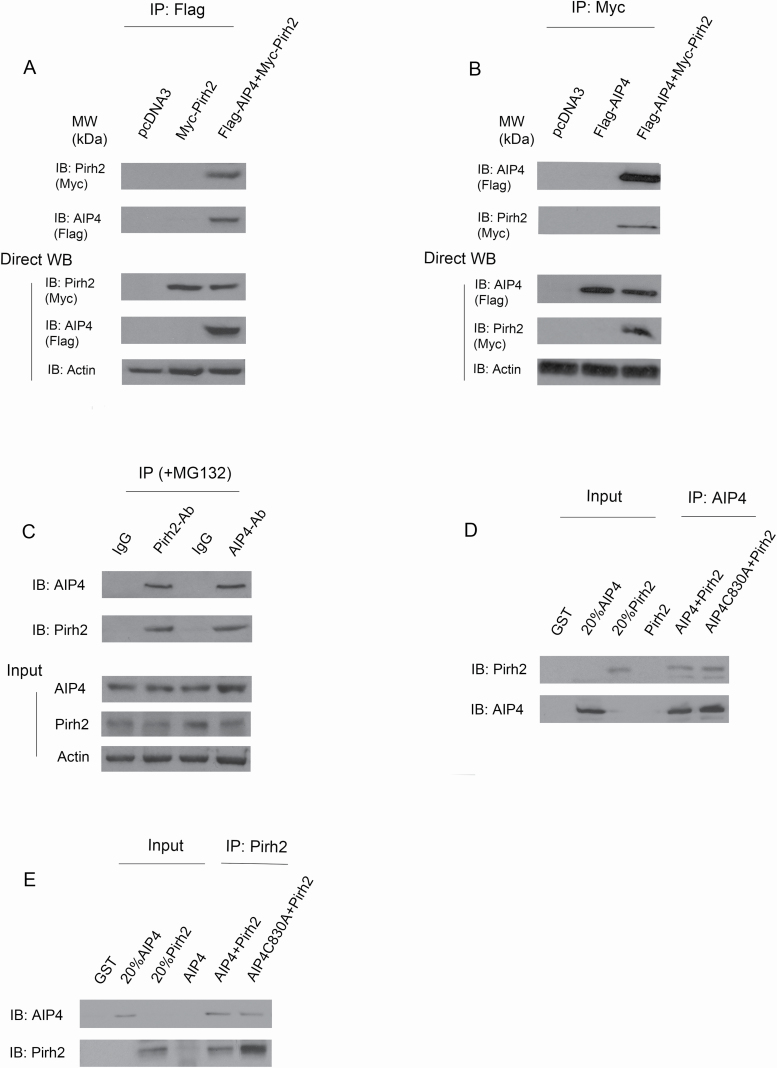
Individual E3s often control the ubiquitination of multiple substrates, and so it is likely that Pirh2 targets other proteins. Hdm2, for example, serves as an E3 ligase for p53 (34), β-arrestin (35), Tip60 (36), IGF-1R (37) and pRb (38). To search for additional Pirh2 substrates, a yeast two-hybrid screen was employed. One candidate clone was found to interact with Pirh2, but not with unrelated proteins. Sequence analysis of the cloned cDNA insert revealed that it was homologous with mouse Itch (atrophin-1 interacting protein 4, AIP4/or ITCH in humans). To determine whether Pirh2 interacts with AIP4 in cells, p53 null-H1299 cells were transfected with expression plasmids pcDNA3-Myc-PIrh2, empty vector or in combination with pcDNA3-Flag-AIP4. The cell extracts were immunoprecipitated with an anti-Flag antibody for AIP4 (M5) and analyzed by immunoblotting. As shown in Figure 1A, Pirh2 was coimmunoprecipitated with AIP4. Similarly, AIP4 was coimmunoprecipitated with Pirh2 when using an anti-Myc antibody for Pirh2 (Figure 1B). To determine whether endogenous Pirh2 and AIP4 interact under more physiological conditions in the absence of overexpression, an IP/western blotting experiment was performed using extracts prepared from human H1299 cells. We detected endogenous AIP4 coimmunoprecipitated with Pirh2 and endogenous Pirh2 coimmunoprecipitated with AIP4 (Figure 1C). To determine whether Pirh2 binds AIP4 directly, GST-Pirh2 and GST-AIP4 proteins were purified from E. coli. A coimmunoprecipitation (co-IP) assay was performed using AIP4 or Pirh2 antibodies as indicated and analyzed by western blotting. Our data revealed that the two E3 ligases interact directly in vitro (Figure 1D and E). Also, an AIP4 construct bearing a point mutation in the HECT domain (C830A) known for lacking HECT catalytic activity (39,40) was added. We observed that the HECT domain’s catalytic activity of AIP4 is not related to its role in binding (Figure 1D and E). Taken together, we demonstrated that Pirh2 interacts with AIP4 in cells and in vitro.
Figure 1.
In cells and in vitro binding of Pirh2 and AIP4. (A) In-cell binding of Pirh2 and AIP4 was assessed. H1299 cells were transfected with plasmids expressing Myc-Pirh2, or in combination with Flag-AIP4, and immunoprecipitated with M5 antibody for Flag-AIP4, and analyzed by western blotting with anti-Myc for Pirh2 and M2 for AIP4 (top panel). Direct western blots for Pirh2, AIP4 and Actin are shown in the lower panel. (B) Similar to (A), except that cell extracts were immunoprecipitated with anti-Myc and analyzed by immunoblotting with anti-Flag as indicated. (C) Cell lysates were prepared from H1299 cells, immunoprecipitated with the indicated antibodies and analyzed by western blotting with antibodies to detect Pirh2 or AIP4 (top panel). Direct western blots for AIP4, Pirh2 and Actin are shown in the lower panel. (D) In vitro interaction of Pirh2 and AIP4 was evaluated. GST-Pirh2 and GST-AIP4 fusion proteins were purified from E. coli. The ability of GST-Pirh2 to bind GST-AIP4 was analyzed by immunoprecipitating with an AIP4-specific antibody and immunoblotting with a Pirh2-specific antibody or with an AIP4-specific antibody, as indicated. (E) Similar to (D), except that protein lysates were immunoprecipitated with a Pirh2-specific antibody and immunoblotting with the indicated antibodies.
Mapping the Pirh2–AIP4-binding domains
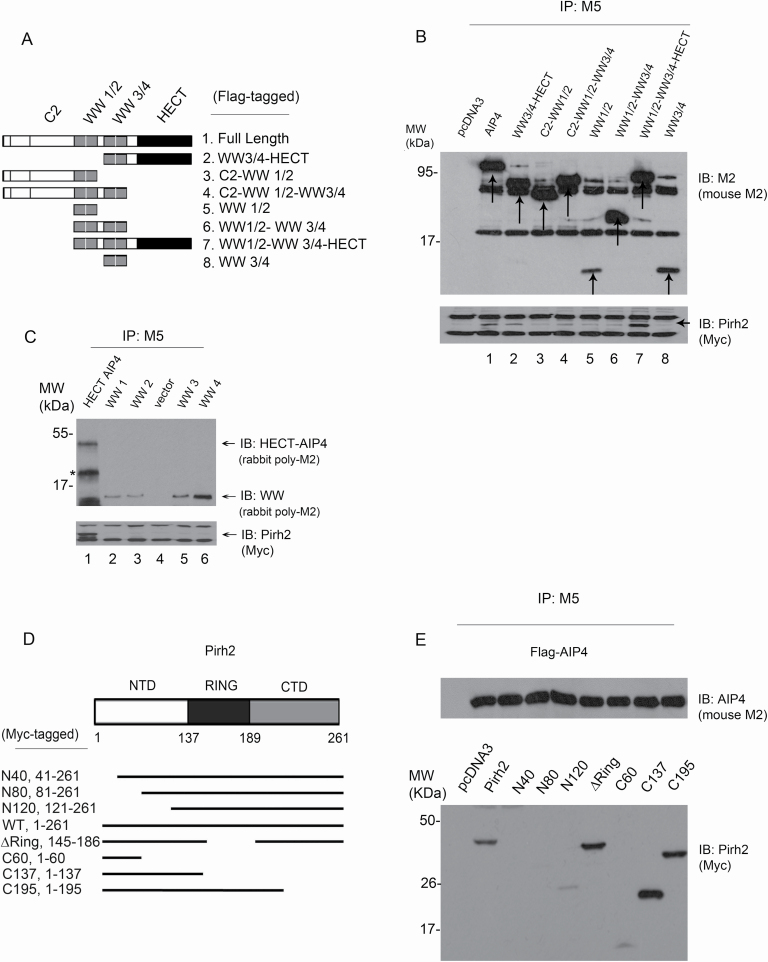
It has been previously shown that Pirh2 binds p53 and p73 proteins (24,27), whereas AIP4 binds p73 exclusively (32) despite the homology (~80%) between p73 and p53 in their DNA-binding sequences (5). We aimed to map the Pirh2 and AIP4 interaction domains. Since these findings may explain how Pirh2 signals binding to p53 or p73. For example, AIP4 binding to Pirh2 may interfere with the same regions of Pirh2 that are utilized in p53 binding. This could also act as a gateway to understand how the binding of AIP4 to other effector proteins could be affected due to interference mediated by Pirh2. Seven AIP4 mutant constructs were designed focusing on the three major domains of AIP4: C2, multiple WW domains (WW1/2, WW3/4) and the HECT domain (Figure 2A). Two terminal mutant constructs, one lacking only the C2 domain and the other lacking only the HECT domain, were generated. In addition, five WW constructs were generated: C2–WW1/2, WW3/4–HECT, WW1/2, WW3/4 and WW1/2–WW3/4 (Figure 2A). Co-IPs were then performed. Wild-type (WT)-AIP4 was used as a positive control for Pirh2 binding. All constructs showed a decrease in binding affinity to Pirh2 except the constructs WW3/4–HECT (#2) and WW1/2–WW3/4–HECT (#7); both contain the HECT domain (Figure 2B). Hence, the deletion of the HECT domain and not the WW domains, which are commonly known for their role in binding (40), inhibited AIP4–Pirh2 binding confirming this domain’s role in the binding mechanism. We next investigated whether the four individual WW domains of AIP4 could interact with Pirh2. As shown in Figure 2C, Pirh2 is bound to HECT-AIP4, but not to the four WW domains of AIP4, confirming that the WW domains of AIP4 are not required to bind Pirh2 in cells. Also, the N-terminal (NTD), RING and C-terminal (CTD) domains of Pirh2 were mapped for AIP4 binding. Seven constructs were designed, as shown in Figure 2D. Co-IP experiments revealed the role of the NTD of Pirh2 in binding AIP4. The deletion of the RING domain, known for its catalytic activity, did not affect AIP4 binding (Figure 2E). We confirmed the binding of both ligases, Pirh2 and AIP4, in cells and in vitro, and that the HECT domain of AIP4 is required for Pirh2 interaction; however, the NTD domain of Pirh2 is required for AIP4 interaction.
Figure 2.
Mapping the interaction sites of Pirh2 and AIP4. (A) Schematic representation of AIP4 different constructs designed for domain mapping. All constructs were expressed in a Flag-tagged vector. This figure was based on information obtained in Lallemand et al. (39). (B) Cell extracts prepared from H1299 cells ectopically expressing Myc-Pirh2 and Flag-AIP4 (full length and truncated constructs) were analyzed by co-IP. Cell lysates were immunoprecipitated using the M5 antibody (for AIP4) to pull down AIP4 constructs. Pirh2 binding was analyzed by immunoblotting using the Myc antibody for Myc-Pirh2 (bottom panel). A mouse monoclonal M2 antibody was used for immunoblotting to detect the expression of AIP4 constructs (top panel). (C) Plasmids expressing 3xFlag-tagged HECT-AIP4, WW1, WW2, WW3 and WW4 of AIP4 were transfected into H1299 cells together with Myc-Pirh2. Lysates were immunoprecipitated with an anti-Flag antibody (M5) to pull down AIP4 constructs, as indicated. Flag-tagged AIP4 constructs were visualized by immunoblotting with a rabbit polyclonal M2 antibody (top panel). The ability of Flag-AIP4 constructs to bind Myc-Pirh2 was analyzed by immunoblotting with an antibody against Myc (Myc-Pirh2, bottom panel). (D) Schematic representation of Pirh2 different constructs designed for mapping. All constructs were expressed in the Myc-tagged vector. (E) Cell extracts prepared from H1299 cells ectopically expressing Flag-AIP4 and Myc-Pirh2 (full length and truncated constructs) were analyzed by co-IP. Cell lysates were immunoprecipitated using the M5 antibody to pull down AIP4. Pirh2 binding was analyzed by immunoblotting using the Myc antibody for Myc-Pirh2 constructs (bottom panel). A mouse monoclonal M2 antibody was also used for immunoblotting to detect AIP4 expression (top panel).
Pirh2 downregulates AIP4 protein expression whereas AIP4 upregulates Pirh2 expression
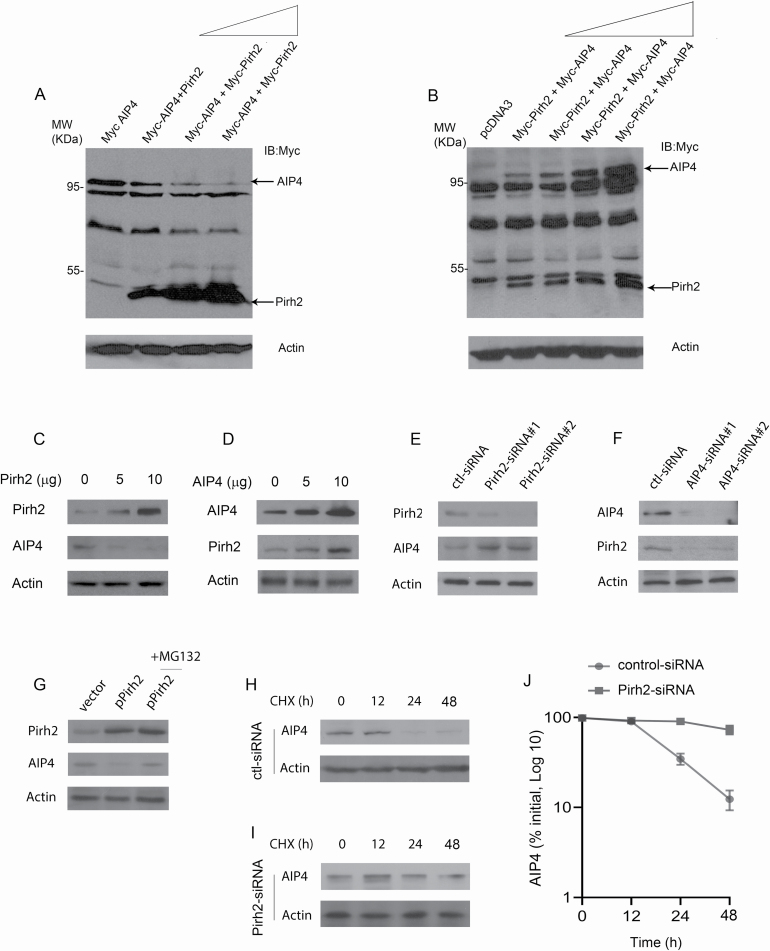
After confirming the binding between the two E3 ligases, we investigated their relationship at the expression level. We studied each ligase’s expression in response to overexpression or knockdown of the other ligase. As a first step, exogenously, AIP4 expression was monitored in response to Pirh2 overexpression in a dose-dependent experiment. Transient overexpression of Pirh2 resulted in a reduction in AIP4 protein levels (Figure 3A). The downregulation of AIP4 by Pirh2 was highly significant, where AIP4 expression was almost completely lost with increased amounts of Pirh2. On the contrary, Pirh2 expression was remarkably elevated when AIP4 was increased in a dose-dependent manner (Figure 3B). This may be explained by the possible existence of a feedback loop between AIP4 and Pirh2. To further confirm our findings, H1299 cells were transfected with increased amounts of the Pirh2 expression construct. As shown in Figure 3C, endogenous AIP4 protein levels were significantly decreased when the cells were transfected with the Pirh2 expression vector. As we expected, endogenous Pirh2 protein levels were increased when AIP4 was overexpressed (Figure 3D). To investigate whether endogenous Pirh2 regulates AIP4 levels in cells, Pirh2 was subjected to ablation by siRNA. Importantly, we observed that endogenous AIP4 protein levels were increased when Pirh2 was depleted by siRNA in cells (Figure 3E). Consistently, the endogenous Pirh2 levels were decreased when AIP4 was depleted by siRNA in cells, suggesting that Pirh2 may be upstream of AIP4 and act as the rate-limiting factor in the AIP4–Pirh2 regulatory mechanism (Figure 3F). Next, we examined whether Pirh2 mediates AIP4 degradation through the ubiquitin–proteasome pathway. Plasmids expressing Pirh2 or empty vector were transfected into H1299 cells in the presence or absence of MG132, a proteasome inhibitor (26). The addition of MG132 significantly increased AIP4 protein levels in the presence of Pirh2, suggesting that Pirh2 promotes AIP4 degradation through the ubiquitin–proteasome pathway (Figure 3G). To further explore whether Pirh2 regulates endogenous AIP4 protein stability, H1299 cells depleted with Pirh2-siRNA or with control-siRNA were treated with cycloheximide to inhibit de novo protein synthesis. The half-life of AIP4 was maintained at approximately 24 h in H1299 cells treated with control-siRNA (Figure 3H). The half-life of endogenous AIP4 was increased to approximately 48 h in H1299 cells treated with Pirh2-siRNA (Figure 3I). The half-life of AIP4 in H1299 cells depleted Pirh2, or control-siRNA, was determined by densitometry of the immunoblots (Figure 3J). Together, these data indicate that Pirh2 regulates the stability of AIP4 in cells.
Figure 3.
Pirh2–AIP4 regulatory pathway. (A) H1299 cells were transfected with a plasmid expressing Myc-AIP4, along with increased amounts of Myc-Pirh2 expression plasmid (5, 10 and 20 µg). Western blot analysis was performed using the Myc antibody to detect both AIP4 and Pirh2 (top panel). An antibody against β-actin was used as a loading control (bottom panel). (B) H1299 cells were transfected with a plasmid expressing Myc-Pirh2, along with increased amounts of Myc-AIP4 expression construct (5, 10, 15 and 20 µg). Western blot analysis was performed using the Myc antibody to detect both AIP4 and Pirh2 (top panel). Actin was used as a loading control (bottom panel). (C) H1299 cells were transfected with increased amounts of Pirh2 expression plasmid (0, 5 and 10 µg). Endogenous Pirh2 and AIP4 protein levels were detected by western blotting. (D) H1299 cells were transfected with increased amounts of AIP4 expression plasmid (0, 5 and 10 µg). Endogenous Pirh2 and AIP4 protein levels were detected by western blotting. (E) H1299 cells were transfected with control-siRNA, Pirh2-siRNA1 or Pirh2-siRNA2 and analyzed by western blotting with the indicated antibodies. (F) H1299 cells were transfected with control-siRNA, AIP4-siRNA1 or AIP4-siRNA2. Endogenous AIP4 and Pirh2 protein levels were detected by western blotting. (G) H1299 cells were transfected with a plasmid expressing Pirh2 in the presence or absence of MG132, a proteasomal inhibitor. The endogenous Pirh2 and AIP4 protein levels were detected by western blotting. (H and I) Pirh2 ablation by siRNA stabilizes AIP4. H1299 cells were transfected with control-siRNA (H) or Pirh2-siRNA (I) in the presence of cycloheximide (20 µg/ml). The endogenous AIP4 protein levels were determined by immunoblotting with an AIP4-specific antibody. An antibody against actin was used as a loading control. (J) As determined by densitometry of the immunoblots in (H) and (I). Error bars indicate SEM (n = 3).
Pirh2 stabilizes p73 by decreasing the levels of AIP4
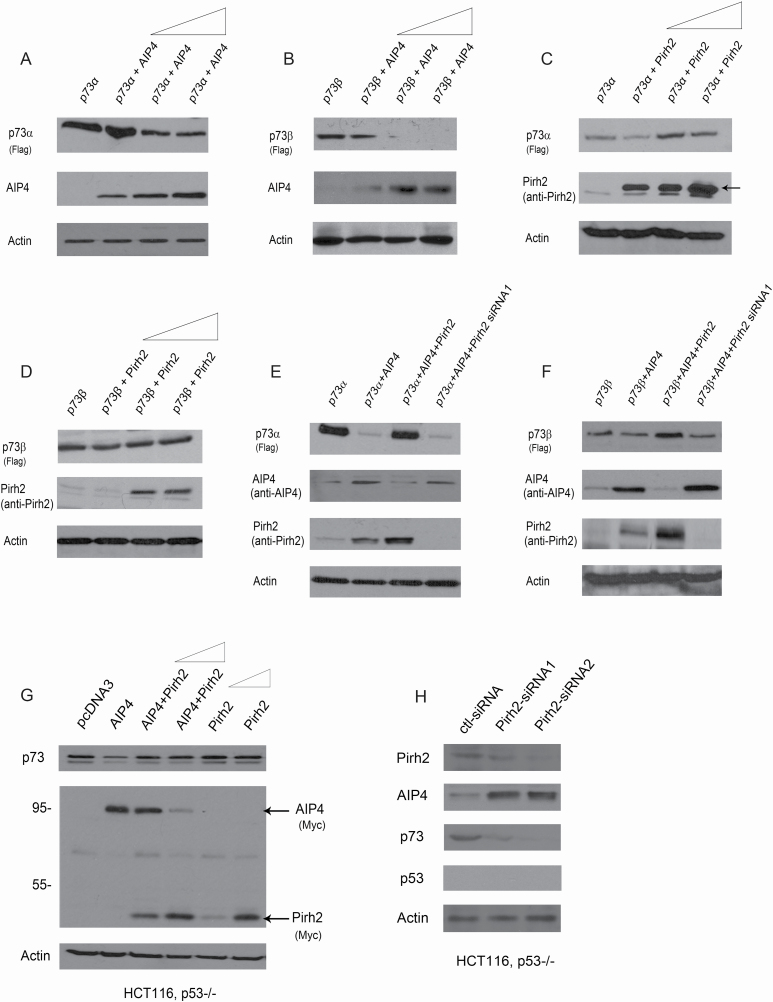
To obtain insight into the effect of the Pirh2–AIP4 regulatory mechanism on p73 proteins, we investigated the consequence of overexpression or silencing of Pirh2 and AIP4 on p73 expression. H1299 cells were transfected with plasmids expressing p73α, or in combination with increased amounts of AIP4. Consistent with previous findings (32), we observed that overexpression of AIP4 decreased p73α protein levels in a dose-dependent manner (Figure 4A). Similar data were obtained for p73β (Figure 4B). Furthermore, the levels of p73α protein were elevated when Pirh2 was overexpressed (Figure 4C); in contrast, p73β protein levels did not change when Pirh2 was overexpressed (Figure 4D). Interestingly, the downregulation of p73α by AIP4 ceased when coexpressed with Pirh2 and restored when Pirh2 was depleted by siRNA in cells (Figure 4E). These data indicated that Pirh2 inhibits AIP4’s negative regulatory effect on p73α. Similar data were obtained for p73β (Figure 4F). Consistently, we found that AIP4 decreased p73 protein levels and Pirh2 inhibits AIP4-mediated p73 degradation in HCT116, p53−/− cells (Figure 4G). We further tested the ability of Pirh2 to regulate the levels of endogenous p73 protein in HCT116, p53−/− cells. The ablation of Pirh2 by siRNA led to significantly increased AIP4 protein levels and was accompanied by decreased levels of p73 protein in HCT116, p53−/− cells (Figure 4H). We demonstrated that Pirh2 regulated p73 via the AIP4–p73 axis.
Figure 4.
Pirh2 and AIP4 both regulate p73 expression. (A) H1299 cells were transfected with plasmids expressing p73α, along with increased amounts of Myc-AIP4 (5, 10 and 20 µg). Western blot analysis was performed using a Flag antibody to detect p73α, and a Myc antibody for AIP4. (B) Similar to (A), except that p73β was used. (C) Similar to (A), except that H1299 cells were transfected with p73α, along with increased amounts of the Pirh2 expression construct (5, 10 and 20 µg). (D) Similar to (C), except that p73β was used. (E) H1299 cells were transfected with plasmids expressing p73α, in combination with AIP4, AIP4 and Pirh2, or transfected p73α and AIP4 in Pirh2-depleted H1299 cells, as indicated. The p73α, AIP4 and Pirh2 protein levels were detected by western blotting with the indicated antibodies. (F) Similar to (E), except that p73β was used. (G) HCT116, p53−/− cells were transfected with plasmids expressing AIP4, or with AIP4 and increased amounts of Pirh2 (5 and 10 µg) or with increased amounts of Pirh2 expression plasmid (5 and 10 µg), and analyzed by western blotting with indicated antibodies. The p73, AIP4 and Pirh2 protein levels were detected by western blotting with the indicated antibodies. (H) HCT116, p53−/− cells were transfected with control-siRNA, or Pirh2-siRNA1 or Pirh2-siRNA2. The levels of endogenous Pirh2, AIP4, p73 and p53 proteins were detected by western blotting. Actin was used as a loading control.
Pirh2 negatively regulates AIP4-mediated p73 ubiquitination in cells and in vitro
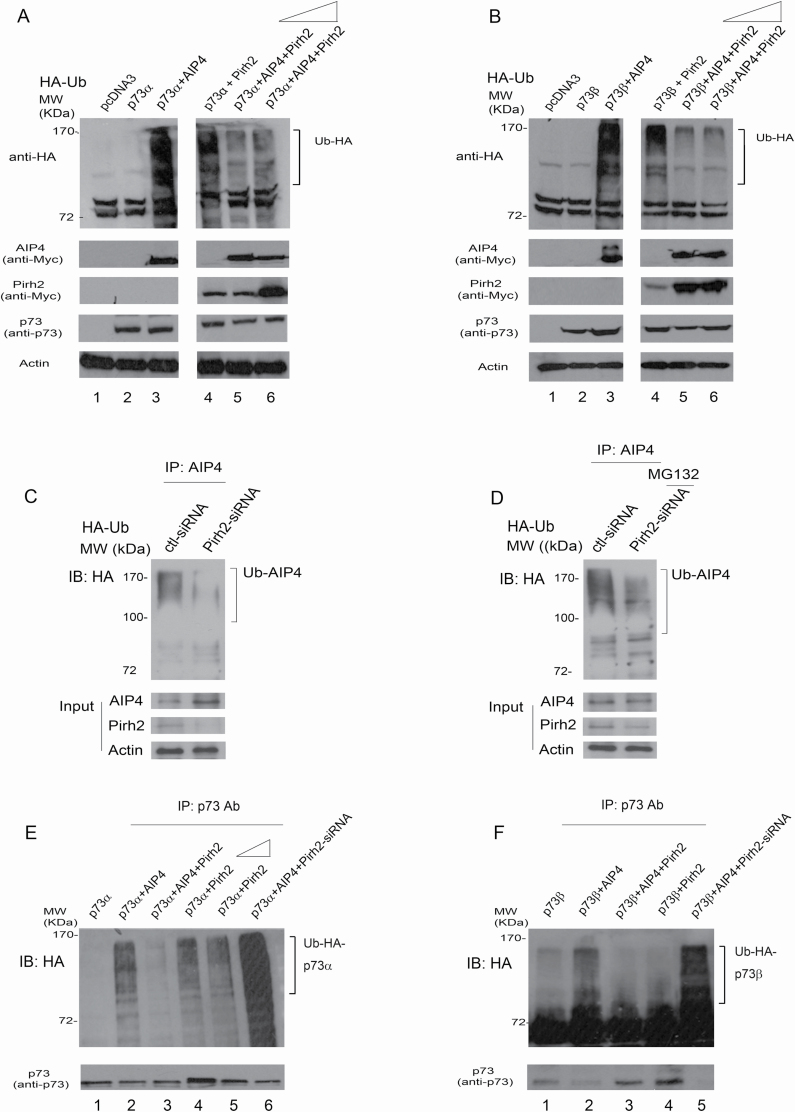
Pirh2 and AIP4 are both shown to ubiquitinate p73 proteins despite having different substrate fate post-ubiquitination. Both are also shown to be regulated by the self-ubiquitination mechanism (24,27,32). However, the consequence of the Pirh2–AIP4 interaction remains unknown for its effect on AIP4 self-ubiquitination and p73 ubiquitination. First, we evaluated AIP4 self-ubiquitination. Then we compared p73 ubiquitination induced by Pirh2 or AIP4, or in combination, knowing that there was an antagonistic relationship between the two ligases. To determine whether Pirh2 regulates AIP4-mediated p73 ubiquitination, p73α and HA-Ub were cotransfected into H1299 cells, or together with AIP4, Pirh2 or both AIP4 and Pirh2, as indicated. We found a major increase in the amount of protein ubiquitination when p73α was coexpressed with AIP4 or with Pirh2 (Figure 5A, lanes 3 and 4). The ubiquitination levels were greatly decreased when p73α was coexpressed with both AIP4 and Pirh2, suggesting that Pirh2 inhibits AIP4-mediated p73α ubiquitination in cells (Figure 5A, lanes 5 and 6). The addition of Pirh2 did not further reduce AIP4-mediated p73 ubiquitination, suggesting that the inhibition of AIP4-mediated p73 ubiquitination by Pirh2 had peaked in this in vitro system. Similar results were obtained for p73β (Figure 5B, lanes 5 and 6). We observed that both ligases ubiquitinated p73 (p73α and p73β); however, Myc-AIP4 showed more vital ubiquitination ability than Myc-Pirh2. To determine whether Pirh2 functions as an E3 ligase for AIP4, H1299 cells were subjected to Pirh2 depletion by siRNA or control-siRNA treatment. The cells were further transfected with an HA-tagged Ub, and immunoprecipitated with an AIP4-specific antibody under denaturing conditions. The ubiquitination of AIP4 was significantly reduced in Pirh2-depleted cells compared with the control-siRNA treatment (Figure 5C, top panel). Western blots confirmed that the depletion of Pirh2 was accompanied by increased levels of AIP4 (Figure 5C, bottom panel). Stronger ubiquitination of AIP4 was observed when cells were treated with Pirh2-siRNA in the presence of MG132, implying that MG132 treatment can increase ubiquitinated AIP4 levels (Figure 5D). We wanted to further investigate whether Pirh2 negatively regulates AIP4-mediated p73 ubiquitination. As shown in Figure 5E, both AIP4 and Pirh2 promote p73α ubiquitination; however, the ubiquitination of p73α was significantly decreased when p73α was coexpressed with AIP4 and Pirh2, confirming that Pirh2 inhibits AIP4-mediated p73α ubiquitination. Furthermore, p73α ubiquitination mediated by AIP4 was greatly increased when the cells were treated with Pirh2-siRNA (Figure 5E, lane 6). As expected, Pirh2 promotes p73α ubiquitination; however, increased amounts of Pirh2 did not increase more p73α ubiquitination (Figure 5E, lanes 4 and 5). Similar data were obtained for p73β ubiquitination (Figure 5F). Our findings revealed that Pirh2 regulates AIP4-mediated p73 ubiquitination in cells.
Figure 5.
Pirh2 inhibited AIP4-mediated p73 ubiquitination in cells. (A) H1299 cells were transfected with plasmids expressing Flag-p73α, in combination with AIP4, Pirh2 or with AIP4 and increased amounts of Pirh2 (5 and 10 µg), in addition to HA-tagged ubiquitin, as indicated. Cell lysates were analyzed by western blotting with anti-HA to detect ubiquitination or with the indicated antibodies. Actin was used as a loading control. (B) Similar to (A), except that p73β was used. (C) H1299 cells were transfected with control-siRNA or Pirh2-siRNA. Forty hours later, cells were further transfected with a plasmid expressing HA-Ub. Lysates were immunoprecipitated with an AIP4-specific antibody and analyzed by immunoblotting with HA-specific, AIP4-specific and Pirh2-specific antibodies. (D) Similar to (C), except that after the second transfection of the HA-Ub expression plasmid, cells were treated with the proteasome inhibitor MG132 (20 µM) for 6 h prior to harvest. (E) H1299 cells were transfected with plasmids expressing p73α, or in combination with AIP4, or with AIP4 and Pirh2, or with increased amounts of Pirh2 (5 and 10 µg) or with AIP4 in Pirh2-depleted H1299 cells, as indicated. Cell lysates were immunoprecipitated with a p73-specific antibody and analyzed by immunoblotting with anti-HA to detect p73 ubiquitination (or protein coimmunoprecipitated with p73, top panel). The levels of p73 were detected by western blotting (bottom panel). (F) Similar to (E), except that p73β was used.
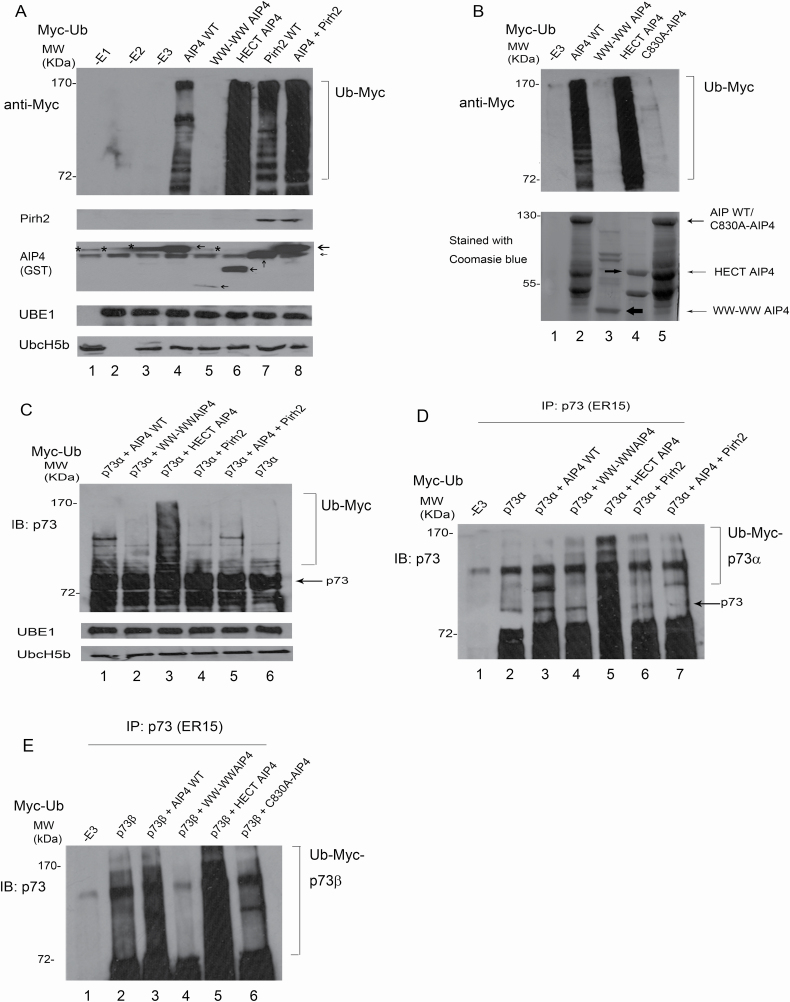
These experiments were performed in cells; we next wanted to determine whether Pirh2 directly inhibits AIP4-mediated p73 ubiquitination in vitro. GST-AIP4, GST-Pirh2, His-p73α and His-p73β were generated and purified from E. coli. Also, an additional three mutant constructs of AIP4 were generated and cloned into the vector pGEX-5X-1: one consisted only of HECT domain known for its catalytic activity, the second constituted only of the WW domains, and the third had a point mutation (C830A) in the HECT domain which has been reported to affect the catalytic activity of the ligase (40). AIP4-mediated in vitro ubiquitination assays were performed (26). Western blots analysis using a Myc-specific antibody targeting Myc-Ub revealed that AIP4-WT promotes ubiquitination in vitro (Figure 6A, lane 4); and surprisingly, the HECT-AIP4 showed more efficient ubiquitination compared with WT-AIP4 (Figure 6A, lane 6). The levels of ubiquitination were significantly increased in the presence of both AIP4 and Pirh2 (Figure 6A, lane 8). Interestingly, the mutant construct C830A completely abolished AIP4 self-ubiquitination, emphasizing the HECT domain’s role in the ubiquitination process (Figure 6B, lane 5, top panel). All GST-AIP4 proteins were run on SDS–PAGE gels and stained with Coomassie blue staining to confirm their purity and correct molecular size (Figure 6B, bottom panel). We sought to determine whether AIP4-WT or AIP4 deleted mutants or Pirh2 regulates p73 ubiquitination in vitro. p73α immunoblot revealed that p73α was heavily ubiquitinated in the presence of HECT-AIP4 compared with AIP4-WT, and Pirh2 reduced AIP4-mediated p73α ubiquitination in vitro (Figure 6C). To eliminate the possible self-ubiquitination of the E3 ligase, coupled in vitro ubiquitination/IP was performed (26). After 2 h reactions, the mixtures were immunoprecipitated with a p73-specific antibody and analyzed by immunoblotting with an anti-p73-specific antibody to detect ubiquitinated p73 (Figure 6D). Consistently, we observed (i) p73α was heavily ubiquitinated in the presence of HECT-AIP4 (lane 5) compared with AIP4-WT (lane 3), implying that WW domains may inhibit HECT-AIP4 functions as an E3 ligase, and (ii) Pirh2-mediated p73α ubiquitination in vitro (Figure 6D, lane 6). Similar data were obtained for p73β ubiquitination (Figure 6E). As expected, the HECT domain of AIP4 showed the highest efficiency in p73 ubiquitination, and the C830A-AIP4 construct lost its catalytic activity. Also, we confirmed that the WW domains are not essential for the ubiquitination of AIP4.
Figure 6.
Pirh2 promotes AIP4 self-ubiquitination and regulates AIP4-mediated p73 ubiquitination in vitro. (A) GST-AIP4 or various GST-AIP4 deletion constructs were evaluated for their E3 ligase ability in the presence of E1, E2 and Myc-Ub, as indicated. The levels of protein ubiquitination were analyzed by western blotting, as shown. The levels of Pirh2, AIP4, UBE1 and Ubc-H5b were detected by western blotting. * shows the background signals of anti-GST antibodies. (B) Similar to (A), except that GST-C830A-AIP4 was used (top panel). GST-AIP4 or GST-AIP4 mutant constructs were purified from E. coli. Prior to ubiquitination assays, constructs were run on SDS–PAGE gel and stained with Coomassie blue (bottom panel). (C) GST HECT-AIP4 was evaluated for its ability to ubiquitinate p73α in vitro. His-p73α, GST-AIP4 or various GST-AIP4 constructs, or GST-Pirh2, E1 and E2 were added to the in vitro ubiquitination reaction and analyzed by western blotting with a p73-specific antibody to detect p73 ubiquitination in vitro. Direct western blots for UBE1 and Ubc-H5b are shown in the lower panel. (D) Similar to (C), except that the reaction after ubiquitination was immunoprecipitated with a p73-specific antibody and analyzed by immunoblotting with a p73-specific antibody as indicated. (E) Similar to (D), except that His-p73β was used.
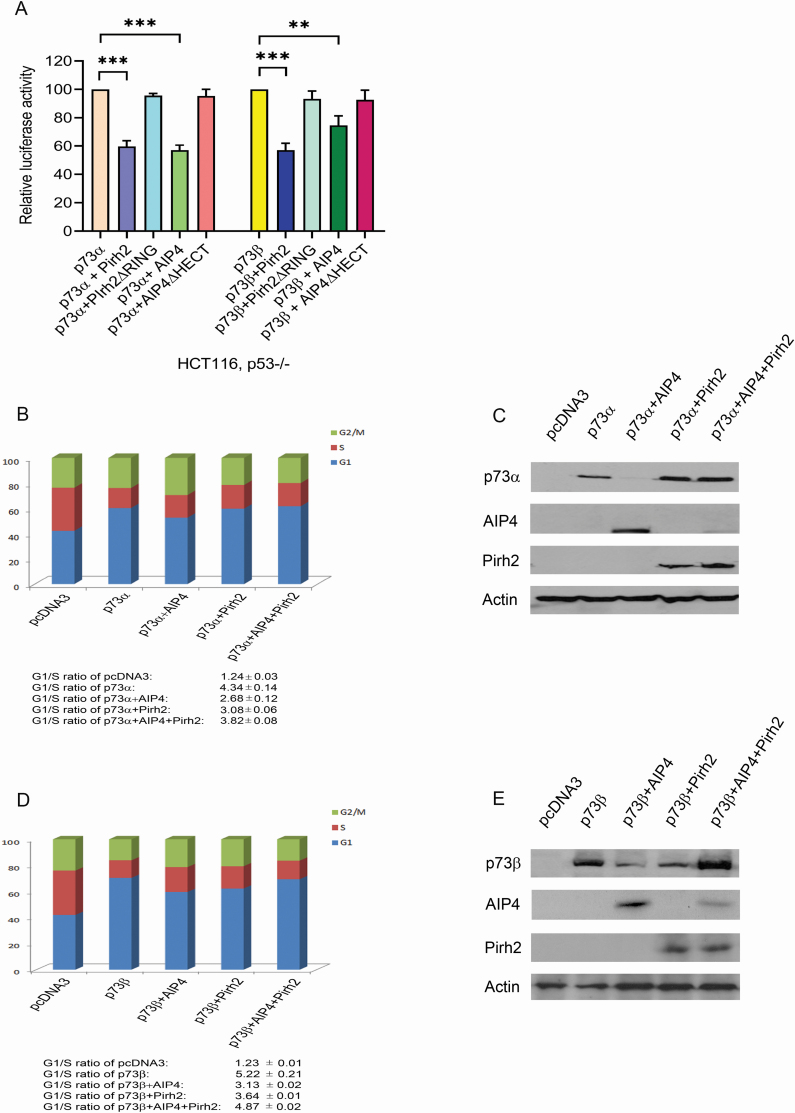
Pirh2 restores p73 cell cycle arrest function
To determine whether Pirh2 or AIP4 affects p73-dependent transcriptional activation in HCT116, p53−/− cells, HCT116, p53−/− cells were transfected with a luciferase reporter construct (p21-Luc) containing the p53-binding site from the p21WAF1 promoter and p73α or p73β alone, or in combination with Pirh2 or with AIP4. As we expected, Pirh2 and AIP4 both repressed p73 transactivation in HCT116, p53−/− cells (Figure 7A). Given that Pirh2 downregulates AIP4 and inhibits AIP4 negative regulation of p73 proteins, we investigated the functional consequences of Pirh2 regulation of AIP4-mediated p73 induced G1 cell cycle arrest. The G1 cell cycle arrest function of both isoforms alpha and beta were analyzed post-overexpression of Myc-Pirh2, or Myc-AIP4 exclusively, or in combination as indicated. As a positive control, overexpression of p73 was used to assess the differences in the cell cycle arrest function after regulation; an empty vector as a negative control (pcDNA3) was used in transfection. As shown in Figure 7B, AIP4 overexpression decreased p73α induced G1 arrest function, which was restored when Pirh2 was introduced. Western blots of isolated portions from the cell lysates were run to confirm transfection efficiency (Figure 7C). Data were consistent between the two isoforms of p73, yet p73β showed a more substantial cell cycle arrest function (Figure 7D). Western blot visualized the transfection efficiency of p73β, or in combination with AIP4, Pirh2 or both AIP4 and Pirh2 (Figure 7E). Our results revealed that p73β had a more substantial cell cycle arrest function where the percentage of cells at G1G0 arrest approached 80%.
Figure 7.
p73 cell cycle arrest function in response to AIP4/Pirh2 regulation. (A) HCT116, p53−/− cells were transfected with plasmids expressing p73α or p73β, or together with Pirh2 or with AIP4 or with their mutants as indicated, and a p21-Luc reporter plasmid. Luc activities were analyzed by post-2-day transfection. Error bars indicate SEM (n = 3). (B) H1299 cells were transfected with plasmids expressing p73α, or in combination with AIP4, or with Pirh2 or AIP4 and Pirh2, as indicated. Empty vector served as the negative control, and p73α alone served as the positive control. The cell cycle profile was determined for p73α by propidium iodide staining and flow cytometry. The results presented are the average of triplicate experiment ± SED. Also, G1/S ratios are presented in the figure. (C) Prior to cell cycle analysis, portions of the cells were analyzed by western blotting with antibodies against the indicated proteins to confirm successful transfection. (D) Similar to (B), except that p73β was used. (E) Similar to (C), except that p73β was used. **P < 0.01, ***P < 0.001.
Discussion
In this study, we proposed a novel relationship between two E3 ligases that share a common regulatory substrate, p73. There are many aspects to this relationship, but to start with, the binding between Pirh2 and AIP4 opens many gateways. Initially, Pirh2 was shown to utilize its amino acid residues 120–137 to bind p53 proteins (24). Similarly, the same residues were essential for p73 binding, highlighting the NTD domain’s role in protein–protein interaction (27). When we mapped Pirh2 domains for AIP4 binding, the NTD domain was also revealed to be important for this function. Thus, Pirh2–AIP4 binding may affect the binding of Pirh2 to its substrates. Regarding AIP4, we report, for the first time, the role of HECT and not the WW domain, as it has always been shown (40), in binding. The HECT domain of AIP4 was investigated earlier and shown not to have an effect on the AIP4–p73 binding (32). Hence, it is very possible that Pirh2–AIP4 forms a complex which may interfere in substrate fate post-ubiquitination. For such reasons, structural analysis regarding Pirh2/AIP4 conformational change post binding and complex formation may reveal whether certain domains are shielded and not exposed for further substrate binding or ubiquitination.
The downregulation of AIP4 by Pirh2 and the upregulation of Pirh2 by AIP4 with consistent findings when monitoring ligase levels proposes a possible feedback loop between these two E3 ligases. Furthermore, ablation of each ligase’s endogenous expression utilizing siRNAs, suggested that Pirh2 is upstream of AIP4. Further investigation is required to confirm the feedback loop. Hakem et al. reported that Pirh2 deficiency mice resulted in higher p53 levels in several tissues in response to DNA damage and despite the presence of other E3 ligases for p53 (41). They showed that Pirh2 is important for the in vivo regulation of p53 stability in response to DNA damage. However, they found that Pirh2 interacted with c-Myc, and Pirh2-mediated c-Myc polyubiquitination and proteolysis (41). They proposed that Pirh2 is a novel tumor suppressor involved in the regulation of both p53 and c-Myc. The two apparently opposite functions of Pirh2, which promote or inhibit tumorigenesis, respectively, are intriguing and suggest a novel cell-growth-regulatory mechanism. Both functions may be necessary at different stages of normal cell growth, and a balance of the two functions is maintained in normal cells. It is worth mentioning that tumor tissues, including breast, head and neck, prostates, etc., show elevated Pirh2 expression (42–48). Proteasomal degradation of AIP4 alone or in complex with Pirh2 could be possible under these circumstances.
Regarding p73, our study showed that at both endogenous and exogenous levels, the degradation of p73 by AIP4 ligases only happens when alone or coexpressed with knockdowns of Pirh2. Pirh2 restored p73 expression by terminating AIP4 negative inhibition. This finding introduces a new insight for therapeutic approaches that may utilize the antagonistic relationship between these two ligases to restore p73 expression. Also, we confirmed that this relationship is moderated through ubiquitination. We detected an apparent decrease in AIP4 induced ubiquitination when Pirh2 was introduced. This ubiquitination did not completely disappear as we showed that Pirh2 ubiquitinates p73 but not as efficiently as does AIP4. However, the lysine chains utilized by both ligases may be different. We previously showed that Pirh2 primarily uses K63-linked chains to ubiquitinate p73 in vitro, but in vivo, Pirh2 employed K11-, K29-, K48- and K63-linked chains to promote p73 ubiquitination (27). p73 protein levels can also be assessed to determine how the utilization of the lysine chains affects substrate fate knowing that Ub moieties, linked via Lys-48, -29 or -11, are targeted by 26S proteasomal degradation while Lys-63-linked chains appear to be used primarily for non-proteasome-dependent regulation of processes such as DNA repair, endocytosis and chromatin remodeling (28–31). Since p73 expression and ubiquitination were both affected, our data showed that the role of p73 tumor suppressors in inducing G1 cell cycle arrest was only restored when AIP4 is accompanied by Pirh2, hence ceasing AIP4 negative regulation of p73.
In summary, our results confirmed that Pirh2 binds and downregulates AIP4, thus inhibiting AIP4-mediated proteasomal degradation of p73 post-ubiquitination and consequently restored p73 partial tumor suppressor activity. The relationship between the two E3 ligases, the domains involved in binding, and p73 ubiquitination, expression and activity all add valuable knowledge to our understanding regarding the p73 regulatory pathway.
Acknowledgements
We gratefully acknowledge Drs Atfi, Marchese and Pawson for providing us with Myc-AIP4, Myc-AIP4 mutant (C830A) and various AIP4 constructs. Conception and design: R.P.L., R.A.Z. and H.H.W.; development of methodology: R.A.Z., H.H.W., Y.A., S.L. and R.P.L.; analysis and interpretation of the data: R.A.Z., H.H.W., Y.A., S.L., C.S., D.D.E. and R.P.L.; writing, review of the manuscript: R.A.Z., H.H.W., Y.A., S.L., C.S., D.D.E. and R.P.L.; revision of the manuscript: D.D.E. and R.P.L. All authors have read the final version.
Abbreviations
- co-IP
coimmunoprecipitation
- IP
immunoprecipitation
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
Funding
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), Women and Children Health Research Institute (WCHRI) and the Canadian Institutes of Health Research (CIHR) to R.P.L. R.A.Z. was supported by an Alberta Cancer Foundation Graduate Studentship and Women and Children’s Health Research Institute Graduate Studentship (WCHRI)/Hair Massacure Award (University of Alberta). D.D.E. holds the Muriel & Ada Hole Kids with Cancer Society Chair in Pediatric Oncology, University of Alberta.
Conflict of Interest Statement: None declared.
References
- 1. Levine, A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 2. Vogelstein, B., et al. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 3. Yang, A., et al. (2000) p63 and p73: p53 mimics, menace and more. Nat. Rev. Mol. Cell Biol., 1, 199–207. [DOI] [PubMed] [Google Scholar]
- 4. Vikhreva, P., et al. (2018) p73 alternative splicing: exploring a biological role for the C-terminal isoforms. J. Mol. Biol., 430, 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dötsch, V., et al. (2010) p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol., 2, a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maas, A.M., et al. (2013) Targeting p73 in cancer. Cancer Lett., 332, 229–236. [DOI] [PubMed] [Google Scholar]
- 7. Yang, A., et al. (2000) p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature, 404, 99–103. [DOI] [PubMed] [Google Scholar]
- 8. Wang, C., et al. (2020) p53-related transcription targets of TAp73 in cancer cells—bona fide or distorted reality? Int. J. Mol. Sci., 21, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uboveja, A., et al. (2020) p73—NAV3 axis plays a critical role in suppression of colon cancer metastasis. Oncogenesis, 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller, P.A.J., et al. (2013) p53 mutations in cancer. Nat. Cell Biol., 15, 2–8. [DOI] [PubMed] [Google Scholar]
- 11. Boominathan, L. (2007) Some facts and thoughts: p73 as a tumor suppressor gene in the network of tumor suppressors. Mol. Cancer, 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Melino, G., et al. (2004) p73 induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem., 279, 8076–8083. [DOI] [PubMed] [Google Scholar]
- 13. Melino, G., et al. (2003) Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci., 28, 663–670. [DOI] [PubMed] [Google Scholar]
- 14. Ethayathulla, A.S., et al. (2012) Structure of p73 DNA-binding domain tetramer modulates p73 transactivation. Proc. Natl. Acad. Sci. USA, 109, 6066–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flores, E.R., et al. (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell, 7, 363–373. [DOI] [PubMed] [Google Scholar]
- 16. Zhu, J., et al. (1998) The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res., 58, 5061–5065. [PubMed] [Google Scholar]
- 17. Jost, C.A., et al. (1997) p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature, 389, 191–194. [DOI] [PubMed] [Google Scholar]
- 18. Putzer, B.M., et al. (2002) Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ., 9, 237–245. [DOI] [PubMed] [Google Scholar]
- 19. Zawacka-Pankau, J., et al. (2010) p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle, 9, 720–728. [DOI] [PubMed] [Google Scholar]
- 20. Ahomadegbe, J.C., et al. (2000) Loss of heterozygosity, allele silencing and decreased expression of p73 gene in breast cancers: prevalence of alterations in inflammatory breast cancers. Oncogene, 19, 5413–5418. [DOI] [PubMed] [Google Scholar]
- 21. Puig, P., et al. (2003) p73 expression in human normal and tumor tissues: loss of p73α expression is associated with tumor progression in bladder cancer. Clin. Cancer Res., 9, 5642–5651. [PubMed] [Google Scholar]
- 22. Momand, J., et al. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 23. Honda, R., et al. (2000) Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene, 19, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 24. Leng, R.P., et al. (2003) Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell, 112, 779–791. [DOI] [PubMed] [Google Scholar]
- 25. Dornan, D., et al. (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature, 429, 86–92. [DOI] [PubMed] [Google Scholar]
- 26. Wu, H., et al. (2011) UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med., 17, 347–355. [DOI] [PubMed] [Google Scholar]
- 27. Wu, H., et al. (2011) Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol. Cancer Res., 9, 1780–1790. [DOI] [PubMed] [Google Scholar]
- 28. Spence, J., et al. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol., 15, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofmann, R.M., et al. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem., 276, 27936–27943. [DOI] [PubMed] [Google Scholar]
- 30. Kim, H.T., et al. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem., 282, 17375–17386. [DOI] [PubMed] [Google Scholar]
- 31. Saeki, Y., et al. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J., 28, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rossi, M., et al. (2005) The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J., 24, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abou Zeinab, R. (2015) The Role of Pirh2 E3 Ligases in Ubiquitination and p73 Regulation. Ph.D. Thesis, University of Alberta, Alberta, Canada. [Google Scholar]
- 34. Fang, S., et al. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem., 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- 35. Shenoy, S.K., et al. (2001) Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science, 294, 1307–1313. [DOI] [PubMed] [Google Scholar]
- 36. Legube, G., et al. (2002) Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J., 21, 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Girnita, L., et al. (2003) Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. USA, 100, 8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uchida, C., et al. (2005) Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J., 24, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lallemand, F., et al. (2005) AIP4 restricts transforming growth factor-β signaling through a ubiquitination-independent mechanism. J. Biol. Chem., 280, 27645–27653. [DOI] [PubMed] [Google Scholar]
- 40. Ingham, R.J., et al. (2005) W.W. domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol., 25, 7092–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hakem, A., et al. (2011) Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genet., 7, e1002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang, C., et al. (2008) The significance of Pirh2 mRNA expression in primary breast cancer. Chin. J. Gen. Surg., 17, 444–448. [Google Scholar]
- 43. Wang, X.M., et al. (2009) p53-induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer, 115, 4554–4563. [DOI] [PubMed] [Google Scholar]
- 44. Shimada, M., et al. (2009) High expression of Pirh2, an E3 ligase for p27, is associated with low expression of p27 and poor prognosis in head and neck cancers. Cancer Sci., 100, 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Logan, I.R., et al. (2006) Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol. Cell. Biol., 26, 6502–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duan, W., et al. (2004) Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J. Natl. Cancer Inst., 96, 1718–1721. [DOI] [PubMed] [Google Scholar]
- 47. Duan, S., et al. (2007) Phosphorylation of Pirh2 by calmodulin-dependent kinase II impairs its ability to ubiquitinate p53. EMBO J., 26, 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hattori, T., et al. (2007) Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res., 67, 10789–10795. [DOI] [PubMed] [Google Scholar]