Abstract
Studying the genome of centenarians may give insights into the molecular mechanisms underlying extreme human longevity and the escape of age-related diseases. Here, we set out to construct polygenic risk scores (PRSs) for longevity and to investigate the functions of longevity-associated variants. Using a cohort of centenarians with maintained cognitive health (N = 343), a population-matched cohort of older adults from 5 cohorts (N = 2905), and summary statistics data from genome-wide association studies on parental longevity, we constructed a PRS including 330 variants that significantly discriminated between centenarians and older adults. This PRS was also associated with longer survival in an independent sample of younger individuals (p = .02), leading up to a 4-year difference in survival based on common genetic factors only. We show that this PRS was, in part, able to compensate for the deleterious effect of the APOE-ε4 allele. Using an integrative framework, we annotated the 330 variants included in this PRS by the genes they associate with. We find that they are enriched with genes associated with cellular differentiation, developmental processes, and cellular response to stress. Together, our results indicate that an extended human life span is, in part, the result of a constellation of variants each exerting small advantageous effects on aging-related biological mechanisms that maintain overall health and decrease the risk of age-related diseases.
Keywords: Centenarians, Cognitive health, Genetics, Healthy aging, Longevity
The human aging process is influenced by genetic and environmental factors, which makes it one of the most complex traits to study (1,2). Previous studies estimated that the heritability of life span up to approximately 70 years of age ranges from 10% to 25% (3,4). However, to reach higher ages, we become increasingly dependent on the favorable genetic elements of our genomes. In fact, the heritability of becoming a centenarian has been estimated to be approximately 60% (5). Interestingly, centenarian genomes are depleted of single-nucleotide polymorphisms (SNPs) associated with age-related diseases, while they are enriched with protective SNPs (6,7). Therefore, studying the genetic variants enriched in centenarians may give insights into the underlying etiology of extreme human longevity (6,7).
The research of SNPs that influence the human life span has focused mainly on the replication of candidate genes discovered in model organisms (8,9). Recently, genome-wide association studies (GWAS) have been performed to identify genetic loci associated with longevity. GWAS of longevity, in which the frequency of genetic variants is compared between long-lived persons and the average population, do not require prior knowledge and have the potential to discover new genetic determinants (10). These studies have identified a constellation of SNPs associated with a longer life span across a wide range of populations (11–16). However, the association of the identified genetic loci has typically a low replication rate across independent studies, with only the APOE-ε4 allele (variant rs429358) and genetic variants in CDKN2A/B gene consistently associated with reduced life span (11,14,15,17). The difficulty in replicating longevity-associated SNPs may be attributable to different measures of survival and longevity, different statistical methods, and population dynamics (8,15,18). For example, some studies used a dichotomous longevity phenotype based on the survival to age older than 90 or 100 years, others used the top 10% or 1% of survivors in a population (12,14), while other studies modeled age at death as a continuous variable and yet others used more sophisticated statistical models (13,15). On top of methodological and phenotypical divergencies between studies, population dynamics including gene–environmental interactions and population biases may potentially have a large effect on longevity (18) and might explain the poor replication rate in independent cohorts. Lastly, the genetic variants identified thus far carry small effects, such that large sample sizes are required for an association with longevity to reach statistical significance in a GWAS setting (8).
Although poorly replicated, 29 genomic regions have been associated with a longer life span in the most recent GWAS (11,12,14–16,18). The genes that harbor these variants have been implicated in age-related diseases including cardiovascular diseases (eg, APOE, ANRIL), type 1 diabetes (eg, FOXO3, LPA), cancer (CDKN2B, BEND4), and neurological diseases (APOE, GPR78, GRIK2) (13,15). Together, this suggests that an extended human life span is associated with a lower genetic risk of age-related diseases (8,15,19). Indeed, centenarians across populations have been shown to compress their disability period to the very end of their lives, escaping or delaying age-related diseases until extreme ages (5,20–22).
We hypothesize that variants associated with longevity are maximally enriched in cognitively healthy centenarians because, in addition to reaching at least 100 years (~1% of the population), these centenarians are cognitively healthy and represent an even smaller percentage of the general population (~0.1%) (20). We previously found that the selection for cognitive health next to being 100 years or older is associated with prolonged longevity in this cohort compared to centenarians from the general population (20,23). Therefore, the centenarians in this cohort represent the ideal group to construct and test polygenic risk scores (PRSs) for longevity. A PRS is a weighted score of independent variants representative of the risk to develop a phenotypic trait and can be used to study the combined influence of genetic factors on a certain trait. Although a PRS of parental longevity was previously associated with survival, validation in a cohort of extremely old individuals is missing. Besides, to prioritize SNPs to include in the PRS using a cohort of cognitively healthy agers may improve association statistics of the PRS.
In this study, we started from 29 genomic regions previously associated with longevity: We annotated SNPs to likely affected genes and sought to detect significant associations using gene-based tests as opposed to single-variant associations. Importantly, we constructed PRSs combining the effect of multiple variants and tested the association of the risk scores (a) with becoming a cognitively healthy centenarian and (b) with survival in a subset of controls with follow-up data. We further explored the relationship between the PRS and the deleterious effect of APOE-ε4 allele, and using an innovative framework, we functionally annotate the variants included in the best PRS model.
Materials and Methods
Study Population
As cases, we used a sample of 358 participants from the 100-plus Study cohort (20). This study includes Dutch-speaking individuals who can provide official evidence for being aged 100 years or older and self-report to be cognitively healthy. As controls, we used (a) a sample of 1779 Dutch older adults from the Longitudinal Aging Study of Amsterdam (24), (b) a sample of 1206 older adults with subjective cognitive decline who visited the memory clinic of the Alzheimer Center Amsterdam and SCIENCe project, who were labeled cognitively normal after the extensive examination (25), (c) a sample of 40 healthy controls from the Netherlands Brain Bank (26), (d) a sample of 201 individuals from the twin study (27), and (e) a sample of 86 older adults from the 100-plus Study (partners of centenarian’s children). Individuals with subjective cognitive decline were followed up over time in the SCIENCe project, and only individuals who did not convert to mild cognitive impairment or dementia during follow-up were included in this study. We checked whether the inclusion of controls from cohorts with different inclusion criteria was problematic in terms of cohort-specific associations both at the single-variant level (Supplementary Table S10) and at the PRS level (Supplementary Figure S7). The Medical Ethics Committee of the Amsterdam UMC approved all studies. All participants and/or their legal representatives provided written informed consent for participation in clinical and genetic studies.
Genotyping and Imputation Procedures
Genetic variants in our cohort were determined by standard genotyping or imputation methods and we applied established quality control methods. All individuals were genotyped using Illumina Global Screening Array (GSAsharedCUSTOM_20018389_A2). We used high-quality genotyping in all individuals (individual call rate >98%, variant call rate >98%), individuals with sex mismatches were excluded and departure from Hardy–Weinberg equilibrium was considered significant at p < 1 × 10−6. Genotypes were prepared for imputation using available scripts (HRC-1000G-check-bim.pl) to compare variant ID, strand, and allele frequencies to the Haplotype Reference Panel (HRC v1.1, April 2016) (28). All autosomal variants were submitted to the Sanger imputation server (https://imputation.sanger.ac.uk). The server uses MACH to phase data and imputation to the reference panel was performed with positional Burrows Wheeler transform (PBWT). A total of 3312 population participants and 358 centenarians passed quality control. Prior to analysis, we excluded individuals of non-European ancestry based on 1000Genomes clustering and individuals with a family relationship based on identity-by-descent >0.2 (29). This led to the exclusion of 8 centenarians and 197 controls (non-European) and 7 centenarians and 210 controls (family relations), leaving 2905 older adults and 343 cognitively healthy centenarians for the analyses.
Mapping Genetic Variants to Affected Genes
We selected 29 genetic variants for which there was evidence of a significant association with longevity from previous GWAS and candidate-gene studies (Supplementary Table S2), and we linked these variants to their likely affected genes (variant–gene mapping). To do so, we combined annotation from Combined Annotation Dependent Depletion (CADD, v1.3) (30,31), expression quantitative trait loci (eQTL) in blood from Genotype-Tissue Expression consortium (v8) (32), and positional mapping up to 500 kb from the reported variants (RefSeq build 98) (33). CADD annotation was used to inspect each variant’s consequences: In the case of coding variants, we confidently associated the variant with the corresponding gene. For noncoding variants, we first considered possible eQTLs and in case these were not available, we included all genes at increasing distance d from the variant (starting with d ≤ 50 kb, up to d ≤ 500 kb, increasing by 50 kb until at least one match is found).
Gene-Based Association
At the gene level, we combined multiple variants in a gene-based test using MAGMA (v1.06) (34). As genes, we used those that were associated with our variant–gene mapping, and as variants, we used those with minor allele frequency more than 1% in our population. In MAGMA, genetic variants located within 2 kb around each gene were considered for the gene-based test, and as gene model we adopted the snp-wise top model (--gene- model snp-wise=top), which is most sensible when only a small proportion of SNPs in a gene shows association (34). Associations were adjusted for population substructure (principal components [PCs] 1–5) and association p-values were corrected for multiple tests (false discovery rate [FDR], correction for the number of genes tested). The number of PCs used as covariates was arbitrarily chosen: given the homogeneous population that we used in this study, we believe this should account for any major population effects. However, we repeated the main associations of the PRS including 5 additional PCs as covariates (Supplementary Table S9). Before analyses, we explored inflation in MAGMA association statistics: We ran MAGMA with the stated settings for 5000 randomly selected genes and compared the observed p-value distribution with an expected uniform distribution. The deviation between the median values of the observed and expected distributions is indicative of test inflation: We found that inflation was 1.1.
Polygenic Risk Scores
We calculated a PRS for each sample in our cohort. As weights for the PRS, we used variant effect sizes (log of odds ratios [ORs]) available in the summary statistics of the GWAS on parental longevity (15). We did not use weights from a case–control GWAS as the most recent included our cohort, thus the resulting variant effect sizes would be biased. Due to the study setting, parental longevity effect sizes are in general smaller than case–control GWAS of longevity (14,15). This would affect the ORs of our associations, but not the significance, as it would just shift the distribution of the PRS while keeping the same distance between the groups (older adults and centenarians, in our case). It is then the power of the parental longevity study, due to the large sample size, that determines replicability and predictability of the PRS (35). Therefore, we believe that using effect sizes from a parental longevity study has not affected our findings. The PRSs were Z-standardized and regressed against case–control status (with centenarians as cases and older adults as controls), correcting for population substructure (PCs 1–5). P values were corrected using FDR. The resulting OR can be interpreted as OR difference per one standard deviation increase in the PRS. We calculated a set of different PRSs: First, using the set of 29 previously identified variants, then we recursively included in the PRS independent variants that associated subsignificantly with longevity. The inclusion of variants was based on the reported significance in the GWAS summary statistics: PRS-8: p < 5 × 10−8, PRS-7: p < 5 × 10−7, PRS-6: p < 5 × 10−6, PRS-5: p < 5 × 10−5, PRS-4: p < 5 × 10−4, PRS-3: p < .005, PRS-2: p < .05, and PRS-1: p < .5. The selection of independent variants to include in each PRS was performed with linkage disequilibrium (LD)-based clumping (R2 < 0.001 within 750 kb window) using the genotypes of the European samples from the 1000Genome project (Phase 3, N = 503) (29). Due to their large effect size, we stratified all PRSs by APOE variants, that is, we calculated PRSs with and without APOE variants.
Survival Analysis
We investigated whether the PRS was predictive for survival in a subset of the older adults for which follow-up data were available. A total of 1620 participants (mean age 62.7 ± 6.4, 53% female) were eligible for the survival analysis. The age at study inclusion was regarded as T1, while the age at last visit, death, or loss to follow-up was regarded as T2, with the survival time calculated as T2−T1. The mean follow-up time was 10.4 ± 6.9 years, and at the time of analyses, 380 individuals had deceased (23%). Survival analysis was performed implementing left truncation as we anticipated selection bias at old ages, and using the function Surv(T1, T2, death, type=”counting”) as implemented in R-package survival. We performed a survival analysis using the (Z-standardized) PRS without APOE variants with the highest evidence of association in our cohort (Supplementary Figure S6). Resulting hazard ratios (HRs) have to be interpreted with respect to a 1 unit increase in the PRS. First, we used a multivariate Cox regression to investigate the association of the PRS after correcting for APOE-ε4 status (dichotomized), gender, and population stratification (PCs 1–5). For visualization purposes, we split the population into high PRS and low PRS categories based on the median PRS value of the individuals with age younger than 65 years. We then calculated survival differences between the individuals with low PRS and those with high PRS (stratifying for APOE-ε4 status) in a univariate analysis and displayed survival probabilities over age with Kaplan–Meier curves. We calculated differences in years at 50% survival probability between the PRSs. We tested the interaction effect of (a) PRS and gender and (b) PRS and APOE-ε4 status on survival by adding an interaction term in the Cox regression model. To evaluate gender-specific effects of the PRS on survival, we repeated the multivariate Cox regression analyses separately in males and females.
Functional Annotation of Variants Comprising the Best PRS
We inspected the functional consequences of the variants included in the best PRS model. First, we investigated these variants in the GWAS catalog seeking for previous associations with any trait (36). Similarly, we looked at whether the genes associated with these variants were previously reported to associate with any trait in the GWAS catalog. To do so, we linked variants to genes as done for the previously identified variants. However, we realized that allowing multiple genes to associate with a variant could result in an enrichment bias, as neighboring genes are often functionally related. To control for this, we implemented sampling techniques (1000 iterations): At each iteration, we (a) sampled one gene from the pool of genes associated with each variant (thus allowing only a 1:1 relationship between variants and genes), and (b) looked whether the resulting genes were previously reported in the GWAS catalog. Averaging by the number of iterations, we obtained an unbiased estimation of the overlap of the PRS-associated genes with each trait in the GWAS catalog.
Finally, we investigated the molecular pathways enriched in the PRS-associated genes. Again, we used sampling techniques: At each iteration, we (a) sampled one gene from the pool of genes associated with each variant and (b) performed gene-set overlap analysis with the resulting list of genes. Gene-set enrichment analysis was performed with GOSt function as implemented in R-package gprofiler2, with Biological Processes (GO:BP) as background, excluding electronic annotations and correcting p values using FDR (37). Finally, we averaged p values for each enriched term over the iterations (N = 1000). To reduce the complexity of the resulting enriched biological processes, we exploited the tool REVIGO (38). This tool summarizes enrichment results by removing redundant terms based on a semantic similarity measure and displays remaining terms in an embedded space via eigenvalue decomposition of the pairwise distance matrix. We chose Lin as a semantic distance measure and allowed small similarities among terms to be clustered (39). Last, we compared results from our sampling-based approach with a traditional gene-set enrichment approach, by applying both methods to the full set of genes associated with all variants.
Gene Expression of Longevity-Associated Genes
We investigated the expression of the longevity-associated genes using the publicly available data set GSE11882, which comprises RNA expression from the hippocampus region in the brain. We selected samples reported to be cognitively healthy and aged 30–65 years (young, N = 13) and samples aged 80 years or older (old, N = 16). We performed differential analysis (old vs young) on (a) the set of genes associated with the previously reported variants and (b) the set of PRS-associated genes. Sample selection and differential analysis were performed using the GEO2R platform (40). We corrected p values for multiple tests (FDR) and displayed results with the Volcano plot.
Implementation
Quality control of genotype data, population stratification analysis, relatedness analysis, and association analysis were performed with PLINK (v2.00a2LM and v1.90b4.6), whereas PRS analysis, functional enrichment analysis, and plots were performed with a mixture of homemade R (v3.5.2), bash and Python (v2.7.14) scripts. All scripts are available at https://github.com/TesiNicco/CentenAssoc. Variant–gene annotation and gene-set enrichment analysis are implemented in a package available at https://github.com/TesiNicco/AnnotateMe and can be run at http://snpxplorer.eu.ngrok.io.
Results
Study Population
We studied the genetics underlying extreme human longevity in a case–control setting using as cases individuals who reached at least 100 years of age and who self-reported as cognitively healthy. As controls, we used a sample of population-matched, older-adults drawn from 5 different studies (see the Methods section). After establishing quality control of the genotyping data, 343 cognitively healthy centenarians (mean age at inclusion 101.4 ± 1.8, 71.7% females) and 2905 controls (mean age 68.3 ± 11.5, 48.2% females) were included in the analyses (Supplementary Table S1).
Linking Genetic Variants With Genes
We linked genetic variants previously associated with longevity (Supplementary Table S2) to their likely affected genes. However, for noncoding variants, the closest gene is not necessarily the affected gene. Of the 29 investigated variants, only a few are coding (N = 5), while most are intronic (N = 16) or intergenic (N = 8), for which variant consequences are unclear. To investigate the variant effect on gene function, we combined variant consequences as predicted by the CADD (30), eQTL in blood from the Genotype-Tissue Expression consortium (32), and positional information to associate each variant to the gene(s) it likely affects. This allows each genetic variant to associate with one or more genes, depending on annotation certainties. With this procedure, the 29 genetic variants mapped to 65 unique genes: 16 SNPs mapped to 1 gene, while 6 mapped to 2 genes, 4 to 3 genes, 1 to 6 genes, 1 to 8 genes, and 1 to 12 genes (Supplementary Figure S1 and Supplementary Table S3). This annotation tool is freely accessible to the community at http://snpxplorer.eu.ngrok.io.
Combined Association of Multiple Variants at the Gene Level
While single-variant associations represent the standard procedure for GWAS, we hypothesized that testing the aggregated association of multiple variants across a gene might improve association statistics. In total, we tested the joint association of variants at the gene level for 53/65 genes using the MAGMA statistical framework (see the Methods section) (34). After correction for multiple tests (FDR), the association of APOE and CDKN2B genes remained significant at FDR less than 10% (p = 3.14 × 10−12 and p = .002, respectively; Supplementary Figure S2 and Supplementary Table S4).
Polygenic Risk Scores
A PRS is a weighted score of independent variants that quantifies the genetic risk to develop a certain trait. As weights for the PRS, we used effect sizes as found in the summary statistics of the largest GWAS on parental longevity (15). First, we constructed a PRS using the previously identified longevity variants and tested the association of the PRS with becoming a cognitively healthy centenarian. We found a significant association of the PRS (OR = 1.42, 95% confidence interval [CI] = 1.26–1.60, p = 6.59 × 10−9), mainly driven by APOE variants (when excluding the APOE variants: OR = 1.07, 95% CI = 0.96–1.20, and p = .22; Figure 1 and Supplementary Table S5). Single-variant association of these variants is available in Supplementary Table S6.
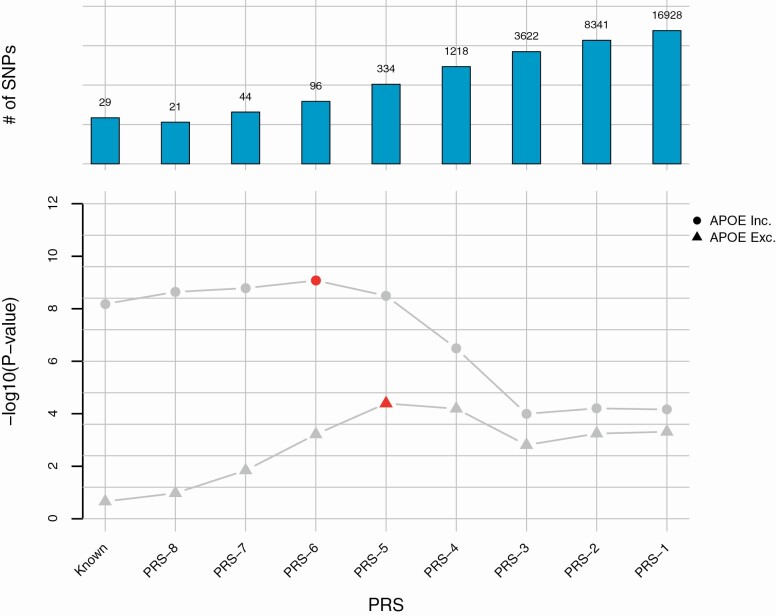
Figure 1.
Association of the PRSs with becoming a cognitively healthy centenarian and the effect of inclusion of subsignificant variants. The top panel shows the number of variants (in log10 scale) included in each PRS (including APOE variants). The bottom panel shows the p value of the difference in PRS between the cognitively healthy centenarians and controls. Circles denote PRSs including the APOE variants and triangles denote PRSs without the APOE variants. Darker points refer to the most significant models including and excluding APOE, respectively. Known refers to the PRS including the 29 previously identified variants associated with longevity. PRS-x refers to the PRS including additional subsignificant variants depending on the p value of association of the variants in the by-proxy GWAS on longevity: PRS-8 for variants with p < 5 × 10−8; PRS-7 for variants with p < 5 × 10−7; PRS-6 for variants with p < 5 × 10−6, etc. GWAS = genome-wide association studies; PRS = polygenic risk score.
Next, we investigated whether the addition of subsignificant, independent longevity variants increased the association of the PRS with becoming a cognitively healthy centenarian (see the Methods section). The number of additionally included variants to the PRS was based on the association p-value as found in the summary statistics provided by Timmers et al. (15): PRS-8 (p < 5 × 10−8, 19 variants in total), PRS-7 (p < 5 × 10−7, 42 variants in total), PRS-6 (p < 5 × 10−6, 94 variants), PRS-5 (p < 5 × 10−5, 332 variants), PRS-4 (p < .0005, 1216 variants), PRS-3 (p < .005, 3620 variants), PRS-2 (p < .05, 8339 variants), and PRS-1 (p < .5, 16 926 variants; Figure 1, Supplementary Tables S5 and S7). For all these PRSs, we tested the difference between cognitively healthy centenarians and population controls. We observed a consistent direction of the effect for all PRSs, with centenarians having on average a higher score than population controls. Including APOE variants, we found that the most predictive PRS was the PRS-6, which comprised 96 independent variants (OR = 1.44, 95% CI = 1.28–1.61, p = 8.39 × 10−10). Excluding APOE variants, the most predictive PRS was the PRS-5, comprising 330 independent variants (OR = 1.27, 95% CI = 1.13–1.42, p = 4.05 × 10-5; Figure 1, Supplementary Figure S3 and Supplementary Tables S5 and S7). Single-variant association for all variants is available in Supplementary Table S8. A more stringent correction for population effects, including 5 additional PCs as covariates, did not change our findings (Supplementary Table S9). Of note, while controls were a combination of different cohorts, we did not observe cohort-specific associations (Supplementary Table S10 and Supplementary Figure S7).
Survival Analysis
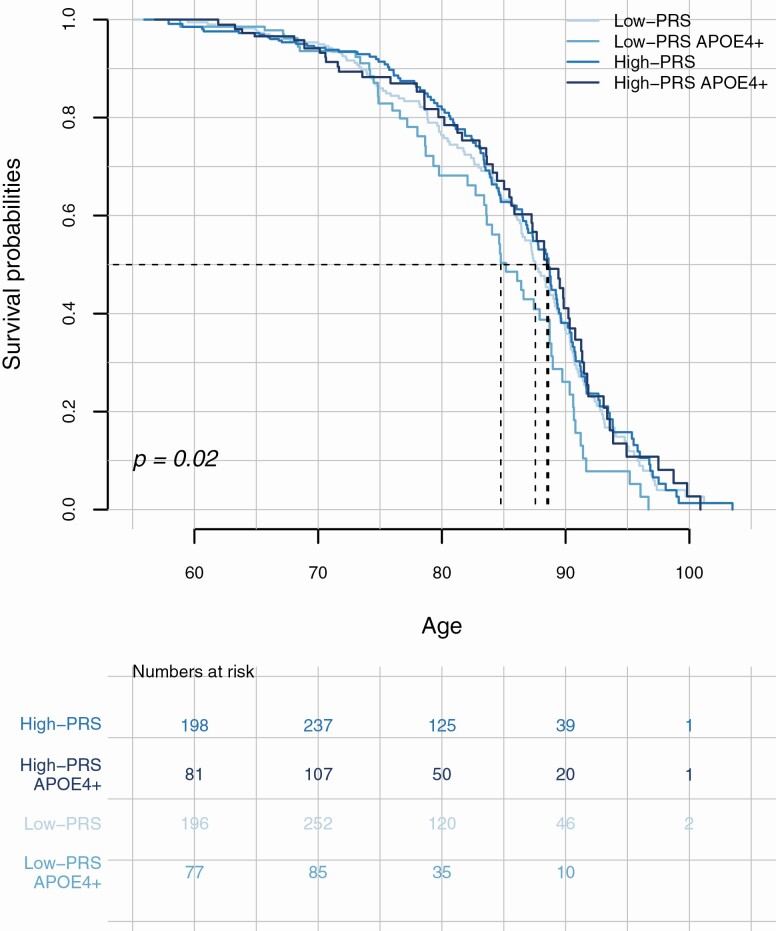
We investigated whether the PRS could predict survival in a subset of the population controls for which follow-up data were available. To investigate the association of the PRS with survival considering APOE variants, we performed a survival analysis using the PRS without APOE variants with the highest evidence of association in our cohort, that is, PRS-5. We performed a multivariate Cox regression to estimate the association of the PRS-5 with survival while adjusting for age at inclusion, gender, population substructure, and APOE-ε4 carriership. The PRS-5 was significantly associated with survival in the expected direction (HR = 0.89, 95% CI = 0.80–0.98, p = .02), that is, having a higher PRS corresponded to reduced mortality. At 50% survival probability (P50), this resulted in a 3.86-year difference in survival between individuals with low PRS who were APOE-ε4 carriers, and those with high-PRS who were not APOE-ε4 carriers (Figure 2). We observed that APOE-ε4 carriers with a low PRS had the shortest survival (P50 CI = 0.39–0.65 at age 84.7), followed by non-APOE-ε4 carriers with low PRS (P50 CI = 0.43–0.58 at age 87.5), then, APOE-ε4 carriers with high PRS (P50 CI = 0.38–0.63 at age 88.5) while individuals who were non-APOE-ε4 carriers with high PRS survived longest (P50 CI = 0.43–0.59 at age 88.6; Figure 2 and Supplementary Table S11). However, we did not observe a significant interaction effect of PRS and APOE-ε4 status (p = .27). In line with the known difference in longevity between males and females, gender was significantly associated with survival (HR = 1.82 for males compared to females, 95% CI = 1.48–2.26, p = 2.72 × 10−8). A separate analysis in males and females suggested that the PRS was more strongly associated with survival in males than in females (HRM = 0.88, 95% CIM = 0.75–1.03 and pM = 0.11; HRF = 0.93, 95% CIF = 0.80–1.05 and pF =0.24; Supplementary Figure S4). However, we did not find a significant interaction effect between PRS and gender (p = .60).
Figure 2.
Association of the PRS-5 model without APOE on survival, stratified by APOE-ε4 status. The top panel shows the survival curves for individuals with high PRS and low PRS, stratified by APOE-ε4 status (dichotomized in “carriers” of APOE-ε4 allele and noncarriers) in the population controls for which follow-up data were available (N = 1620). The p value refers to the association of the PRS-5 in the multivariate Cox regression model while adjusting for gender, APOE-ε4 carriership (dichotomized), and population substructure (PCs 1–5). The lower panel shows the number of samples in each category. PRS = polygenic risk score.
Functional Annotation of PRS
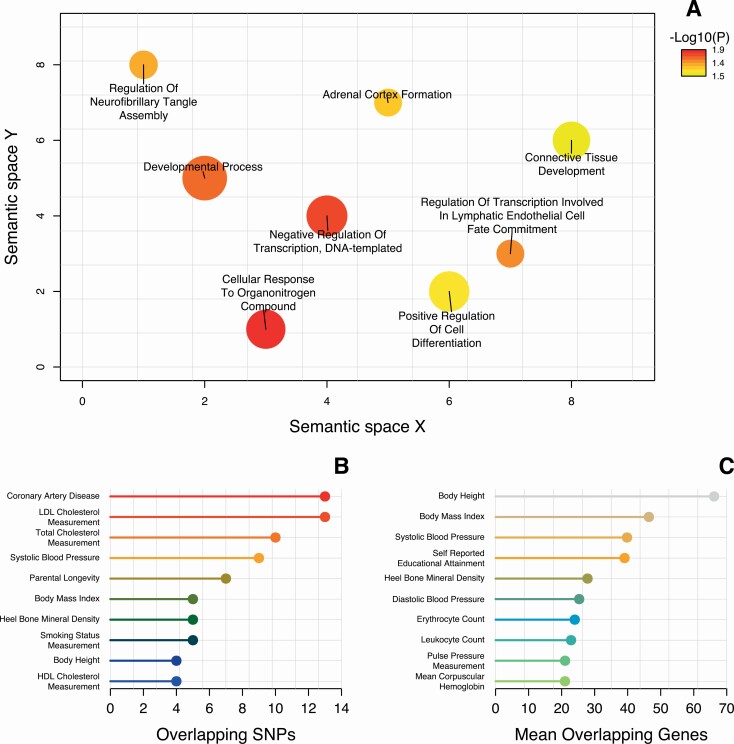
We studied the functional implications of the 330 variants included in PRS-5. First, we linked these variants to 471 unique genes (see the Methods section, Supplementary Figure S5 and Supplementary Table S12). Then, we looked in the GWAS catalog which variants and associated genes, included in our PRS-5, were previously found to associate with any trait. At the variant level, of the 330 unique variants, 46 were reported to associate with the total 115 previously analyzed traits, including diseases such as coronary artery disease (CAD, NSNPs = 13), blood pressure (NSNPs = 9), and cardiovascular diseases (NSNPs = 13), but also smoking (NSNPs = 5) and parental longevity (NSNPs = 7; Figure 3B). At the gene level, 300 of the 471 genes in our list were previously associated with lipid metabolism, CAD, neurological traits, and immunological signatures (Figure 3C).
Figure 3.
Functional enrichment analysis of variants included in PRS-5 and likely associated genes. (A) Significantly enriched biological processes after merging similar terms based on semantic similarity. Color: encodes significance. Dot size represents the total number of genes associated with that term. (B) The 10 GWAS catalog terms that have the most overlapping variants in PRS-5. (C) The top GWAS catalog terms that have the most overlapping genes that associate with the variants in PRS-5. GWAS = genome-wide association studies; PRS = polygenic risk score.
Next, we performed a gene-set enrichment analysis to explore the biological processes enriched in the 471 PRS-5-associated genes (see the Methods section, also available at http://snpxplorer.eu.ngrok.io). We found 48 biological processes significantly enriched after correction for multiple tests (FDR <5%, Supplementary Table S13), which we reduce to 8 by clustering similar terms together based on semantic similarity measures. These terms pointed toward regulatory and differentiation processes, cellular response to stress, and nervous system development (Figure 3A and Supplementary Table S14). To evaluate the performance of our novel sampling-based method with respect to a traditional gene-set enrichment analysis, we applied the latter to the same 471 genes and compared the results of both methods. The traditional gene-set enrichment analysis yielded 122 significantly enriched pathways, of which 45 pathways overlap with the 48 significant pathways identified using the sampling-based approach (Supplementary Table S15). This suggests that our sampling-based approach may be considered conservative compared to traditional gene-set enrichment analyses.
Gene Expression of Longevity-Associated Genes
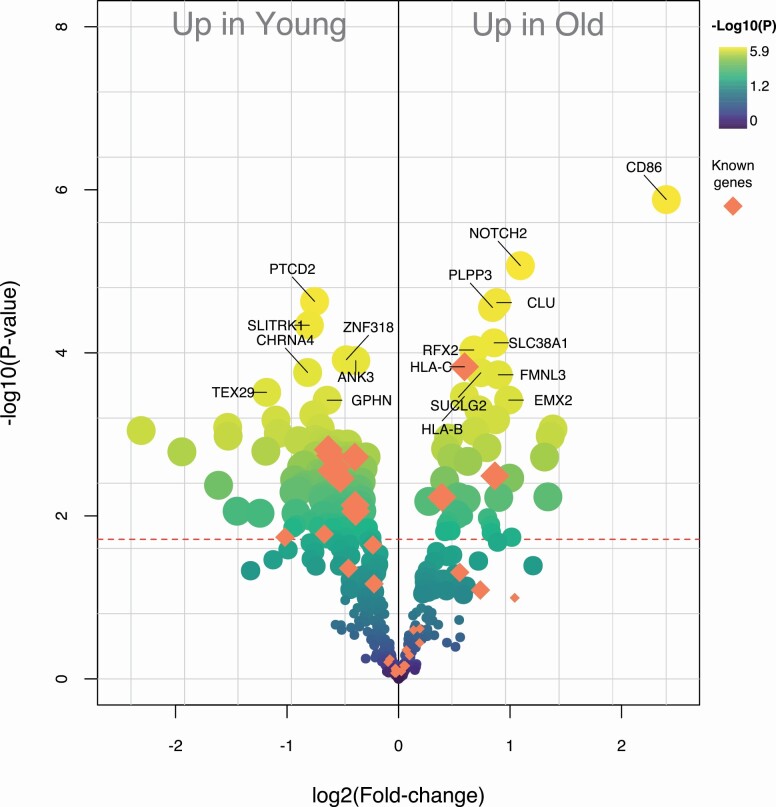
Finally, we studied the expression of the genes linked with the previously identified longevity variants as well as with the PRS-5-associated variants, using a publicly available data set comprising RNA expression from the hippocampus region in the brain. We compared the RNA expression in individuals aged 30–65 years (young, N = 13) as opposed to those aged older than 80 years (old, N = 16). We found that 174 of 432 available genes were differentially expressed after correction for multiple tests (FDR <5%, Figure 4 and Supplementary Table S16): 41 genes were over-expressed in old individuals, while 133 were over-expressed in young individuals.
Figure 4.
Volcano plot of 432 genes associated with the PRS-5 variants and with the previously identified variants, representing the difference in RNA expression in the hippocampus region of the brain between young individuals (N = 16) and old individuals (N = 13; publicly available data set GSE11882). All individuals were reported to be cognitively healthy at the time of death. Diamonds refer to 33 genes associated with the previously identified variants. Annotations are shown for the 15 most significant genes as well as for the 3 most significant genes that overlap between the 2 gene sets. PRS = polygenic risk score.
Discussion
In this study, we investigated the SNPs underlying extreme human longevity using a sample of cognitively healthy centenarians from the 100-plus Study cohort and a sample of population-matched older adults. We constructed a PRS comprising 330 variants that were capable of distinguishing between cognitively healthy centenarians and population controls. This PRS was significantly associated with survival in an independent sample of individuals and may compensate, in part, for the increased mortality risk associated with the APOE-ε4 allele. Using a novel framework, we functionally annotated the variants included in the PRS, which indicated that these were previously associated with cardiometabolic, immunological, oncological, and neurodegenerative conditions. Functional annotation of the genes most likely affected by these variants revealed significant enrichment for regulatory and differentiation processes, cellular response to stress, and nervous system development.
We constructed a PRS that was associated with becoming a cognitively healthy centenarian and also with prolonged survival across an age continuum, even after excluding the 2 APOE alleles which associated strongest with longevity. Including APOE alleles, the PRS comprising 29 previously associated variants significantly associated with becoming a cognitively healthy centenarian, and association statistics only slightly improved upon the addition of variants that subsignificantly associated with longevity. After excluding APOE variants, the association of this PRS was not significant, likely due to the different populations and study designs in which the longevity association of the 29 variants was identified, their low number, and the small effect sizes. However, the inclusion of subsignificant variants boosted the predictive performance of the PRS, which indicated that these subsignificant variants provide additional distinguishing power, but in aggregate, this is relatively little compared to the strong APOE effect. The predictive power of the PRS including 330 variants was highest, and having a high PRS score was associated with longer survival in an independent sample of older adults. We did not identify single variants driving the increase in distinguishing power effect, such that we assume that all variants contributed similarly. Adding even more variants with lower significance to the PRS decreased association statistics, which eventually stabilized, likely due to random fluctuation of the data.
We explored the relationship between PRS and APOE-ε4 carriership: Fully according to expectations, APOE-ε4 carriers with a low PRS had the lowest survival, while as expected, non-APOE-ε4 carriers with a high PRS survived longest, on average 3.86 years longer. Between these extremes, non-APOE-ε4 carriers with low PRS had lower survival compared to APOE-ε4 carriers with a high PRS. This suggests that the variants in the PRS may compensate for the strong disease/mortality risk-increasing effect exerted by the APOE-ε4 allele; however, replication in a large and independent data set is needed to confirm this finding. A number of studies described this effect in dementia, and although the results did not strongly replicate across different studies, several variants (eg, rs5882 in the CETP gene and rs4934 in the SERPINA3 gene) were reported to exhibit buffering effects with respect to APOE-ε4 (41,42).
The majority of the variants included in the best PRS were previously associated with age-related conditions and parental longevity. Given that the variants included were selected from a study on parental longevity, this was not surprising. Functionally, genetic variants were associated with metabolite and lipid measurements (serum metabolites, total cholesterol, high- and low-density lipoproteins), cardiovascular-related traits (blood pressure, CADs, obesity, smoking), neurological conditions (multiple sclerosis, schizophrenia, bipolar disorder), and immunological signatures (IgG glycosylation levels, Crohn’s disease, celiac disease). These traits have been associated with longevity either directly, as part of known hallmarks of aging, or indirectly, through their effect on age-related diseases (1,8). Likewise, when we investigated the genes associated with the variants in the PRS, we observed an enrichment for mechanisms associated with the aging individual: chronic low-grade inflammation, cellular stress, and a reduced speed of cell replacement, development, and differentiation (1).
Recently, increased parental life span was associated with a lower PRS of low-density lipoprotein cholesterol levels, systolic blood pressure, and body mass index (15). We previously showed that cognitively healthy centenarians have a significantly lower PRS of Alzheimer’s disease (AD) compared to population controls (43). The overlap between the variants that contribute to the AD PRS and our best longevity PRS is limited: Apart from APOE variants, the longevity-associated variant rs9665907 is in LD with the known AD variant rs11218343 (in/near SORL1, R2 = 0.39) (44), and the variant rs6558008 is in low LD with the known AD variant rs9331896 (in/near CLU, R2 = 0.05) (45). This suggests that, in addition to the effect of APOE alleles, the SORL1- and CLU-associated signals may partly overlap in the genetic association of AD and longevity (in opposite directions). Two other studies investigated the relationship between longevity and risk alleles for several age-related diseases: One was able to discriminate between long-lived individuals and controls (46), while the other did not find significant differences between centenarians and controls (47). We speculate that the main reason for this discrepancy is that our PRS was constructed based on the association statistics from a well-powered GWAS, which was not available when the previous studies were performed. Additionally, the stricter selection criteria of the centenarians from the 100-plus Study may have contributed to the discriminative power of the PRS.
Across populations, extreme longevity is known to be more prevalent among females than males, which likely reflects gender differences of environmental exposure, disease predisposition, and genetics (48). In our study, we found that the effect size of the PRS on male survival was larger compared to female survival, suggesting that males depend more on having advantageous genetic variants to reach extreme ages than females. In the cohort investigated, an important environmental gender difference is smoking behavior: In accordance with the smoking behaviors in their birth cohort, 76% of the centenarian males had smoked regularly during their lifetime, compared to only 15% of the females (49). Biological differences may also play a role: Estrogens protect females from cardiovascular diseases during their fertile period (48,50), produce more vigorous cellular and humoral immune reactions, and are more resistant to infections caused by viruses and other pathogens (48). From a genetic perspective, impairments in DNA-repair mechanisms become more prevalent with increasing ages, but there are indications that this effect starts a decade earlier in males compared to females (50). Also, several studies reported that women have longer telomeres compared to men (50). Together, these studies suggest that females may be more inherently predisposed to live longer than males and that differential exposure to hazardous environments may lead to selective survival of resilient males. Although conclusive evidence that explains the gender differences in longevity is still lacking, these aspects may in part explain our finding that males are more dependent on an advantageous genetic background to reach extreme ages than females. Note that we did not find a significant interaction effect between PRS and gender, therefore these findings will have to be replicated in a larger cohort.
Strengths and Limitations
Linking variants with genes likely affected is difficult: As such, exploiting diverse sources of variant annotations, such as predicted variant consequences, eQTLs, and genomic position, is essential to pinpoint the genes likely associated with a variant. We designed a novel framework that allows multiple genes to associate with each variant, in which we consider the annotation certainties when performing gene-set enrichment analyses. A limitation of our analysis is that our cohort of centenarians is relatively small compared to the sample sizes of previous GWAS. Due to the rarity of this phenotype in the general population, the collection of large cohorts is prohibitive (20). As a consequence of the limited size, we could not perform exhaustive sex-stratified analyses and thus we cannot exclude that we failed to identify sex-specific associations. Centenarians were compared with a sample of controls combined from different cohorts, yet from the same Dutch population, which may be considered as a strength of our study. While the inclusion of different cohorts with different inclusion criteria had maximized the available sample size in our study, this could potentially result in confounding effects. However, we assessed that no significant cohort-specific association or population effect affected our results, both at the single variant and PRS level. We note that our cohort of centenarians was collected in a specific area during a specific time such that location- and period effects may influence genetic associations. This may in part challenge the replication of the current findings in long-lived individuals from other populations or collected at different times.
Conclusions
We showed that a longevity PRS comprising 330 variants is significantly associated with cognitively healthy aging and with prolonged survival. We found suggestive evidence that the PRS compensates for the deleterious effect of high-impact APOE-ε4 allele and with a novel approach, we functionally annotated the variants in this PRS, showing that many of these variants were previously associated with age-related diseases and with aging-related cellular mechanisms.
Funding
The Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc Fonds. The clinical database structure was developed with funding from Stichting Dioraphte. The SCIENCe project is supported by a research grant from Gieskes Strijbis Fonds. Genotyping of the Dutch case–control samples was performed in the context of EADB (European Alzheimer DNA Biobank), funded by the JPco-fuND FP-829-029 (ZonMW project number, 733051061). The 100-plus Study was supported by Stichting Alzheimer Nederland (WE09.2014-03), Stichting Diorapthe, horstingstuit foundation, Memorabel (ZonMW project number 733050814), and Stichting VUmc Fonds. Genotyping of the 100-plus Study was performed in the context of EADB (European Alzheimer DNA biobank) funded by the JPco-fuND FP-829-029 (ZonMW project number, 33051061). LASA is largely supported by a grant from the Netherlands Ministry of Health, Welfare and Sports, Directorate of Long-Term Care.
Supplementary Material
Acknowledgments
The following studies and consortia have contributed to this manuscript. Amsterdam Dementia Cohort (ADC): Research at the Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. 100-plus Study: we are grateful for the collaborative efforts of all participating centenarians and their family members and/or relatives. Wiesje van der Flier holds the Pasman chair. Longitudinal Aging Study of Amsterdam (LASA): the authors are grateful to all LASA participants, the fieldwork team, and all researchers for their ongoing commitment to the study.
Conflict of Interest
All the authors in the study declared no conflict of interest. The funders had no role in the design of the study at any stage.
References
- 1. Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature. 2018;561:45–56. doi: 10.1038/s41586-018-0457-8 [DOI] [PubMed] [Google Scholar]
- 2. Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet. 2013;132:1323–1338. doi: 10.1007/s00439-013-1342-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruby JG, Wright KM, Rand KA, et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics. 2018;210:1109–1124. doi: 10.1534/genetics.118.301613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaplanis J, Gordon A, Shor T, et al. Quantitative analysis of population-scale family trees with millions of relatives. Science. 2018;360(6385):171–175. doi: 10.1126/science.aam9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sebastiani P, Perls TT. The genetics of extreme longevity: lessons from the new England centenarian study. Front Genet. 2012;3:277. doi: 10.3389/fgene.2012.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garagnani P, Giuliani C, Pirazzini C, et al. Centenarians as super-controls to assess the biological relevance of genetic risk factors for common age-related diseases: a proof of principle on type 2 diabetes. Aging (Albany NY). 2013;5:373–385. doi: 10.18632/aging.100562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tesi N, van der Lee SJ, Hulsman M, et al. Centenarian controls increase variant effect sizes by an average twofold in an extreme case–extreme control analysis of Alzheimer’s disease. Eur J Hum Genet. 2019;27:244–253. doi: 10.1038/s41431-018-0273-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melzer D, Pilling LC, Ferrucci L. The genetics of human ageing. Nat Rev Genet. 2020;21(2):88–101. doi: 10.1038/s41576-019-0183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh PP, Demmitt BA, Nath RD, Brunet A. The genetics of aging: a vertebrate perspective. Cell. 2019;177:200–220. doi: 10.1016/j.cell.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- 11. Broer L, Buchman AS, Deelen J, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70:110–118. doi: 10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sebastiani P, Gurinovich A, Bae H, et al. Four genome-wide association studies identify new extreme longevity variants. J Gerontol A Biol Sci Med Sci. 2017;72:1453–1464. doi: 10.1093/gerona/glx027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fortney K, Dobriban E, Garagnani P, et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet. 2015;11:e1005728. doi: 10.1371/journal.pgen.1005728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deelen J, Evans DS, Arking DE, et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun. 2019;10:3669. doi: 10.1038/s41467-019-11558-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Timmers PR, Mounier N, Lall K, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife. 2019;8:e39856. doi: 10.7554/eLife.39856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeng Y, Nie C, Min J, et al. Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6:21243. doi: 10.1038/srep21243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garatachea N, Emanuele E, Calero M, et al. ApoE gene and exceptional longevity: insights from three independent cohorts. Exp Gerontol. 2014;53:16–23. doi: 10.1016/j.exger.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 18. Giuliani C, Garagnani P, Franceschi C. Genetics of human longevity within an eco-evolutionary nature-nurture framework. Circ Res. 2018;123:745–772. doi: 10.1161/CIRCRESAHA.118.312562 [DOI] [PubMed] [Google Scholar]
- 19. Giuliani C, Pirazzini C, Delledonne M, et al. Centenarians as extreme phenotypes: an ecological perspective to get insight into the relationship between the genetics of longevity and age-associated diseases. Mech Ageing Dev. 2017;165:195–201. doi: 10.1016/j.mad.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 20. Holstege H, Beker N, Dijkstra T, et al. The 100-plus Study of cognitively healthy centenarians: rationale, design and cohort description. Eur J Epidemiol. 2018;33:1229–1249. doi: 10.1007/s10654-018-0451-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perls T. Dementia-free centenarians. Exp Gerontol. 2004;39:1587–1593. doi: 10.1016/j.exger.2004.08.015 [DOI] [PubMed] [Google Scholar]
- 22. Beker N, Sikkes SAM, Hulsman M, et al. Longitudinal maintenance of cognitive health in centenarians in the 100-plus study. JAMA Netw Open. 2020;3:e200094. doi: 10.1001/jamanetworkopen.2020.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beker N, Sikkes SAM, Hulsman M, Schmand B, Scheltens P, Holstege H. Neuropsychological test performance of cognitively healthy centenarians: normative data from the Dutch 100-plus study. J Am Geriatr Soc. 2019;67:759–767. doi: 10.1111/jgs.15729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoogendijk EO, Deeg DJ, Poppelaars J, et al. The Longitudinal Aging Study Amsterdam: cohort update 2016 and major findings. Eur J Epidemiol. 2016;31:927–945. doi: 10.1007/s10654-016-0192-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Flier WM, Scheltens P. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis. 2018;62:1091–1111. doi: 10.3233/JAD-170850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rademaker MC, de Lange GM, Palmen SJMC. The Netherlands brain bank for psychiatry. Handbook of Clinical Neurology. 2018;150:3–16. doi: 10.1016/B978-0-444-63639-3.00001-3 [DOI] [PubMed] [Google Scholar]
- 27. Willemsen G, de Geus EJ, Bartels M, et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231 [DOI] [PubMed] [Google Scholar]
- 28. McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McInnes BT, Pedersen T. Evaluating measures of semantic similarity and relatedness to disambiguate terms in biomedical text. J Biomed Inform. 2013;46:1116–1124. doi: 10.1016/j.jbi.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sundermann EE, Wang C, Katz M, et al. Cholesteryl ester transfer protein genotype modifies the effect of apolipoprotein ε4 on memory decline in older adults. Neurobiol Aging. 2016;41:200.e7–200.e12. doi: 10.1016/j.neurobiolaging.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huq AJ, Fransquet P, Laws SM, et al. Genetic resilience to Alzheimer’s disease in APOE ε4 homozygotes: a systematic review. Alzheimers Dement. 2019;15:1612–1623. doi: 10.1016/j.jalz.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 43. Tesi N, van der Lee SJ, Hulsman M, et al. Immune response and endocytosis pathways are associated with the resilience against Alzheimer’s disease. Transl Psychiatry. 2020;10:332. doi: 10.1038/s41398-020-01018-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sebastiani P, Solovieff N, Dewan AT, et al. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beekman M, Nederstigt C, Suchiman HE, et al. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc Natl Acad Sci U S A. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ostan R, Monti D, Gueresi P, Bussolotto M, Franceschi C, Baggio G. Gender, aging and longevity in humans: an update of an intriguing/neglected scenario paving the way to a gender-specific medicine. Clin Sci (Lond). 2016;130:1711–1725. doi: 10.1042/CS20160004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Poppel F, Reher D, Sanz-Gimeno A, Sanchez-Dominguez M, Beekink E. Mortality decline and reproductive change during the Dutch demographic transition: revisiting a traditional debate with new data. Demogr Res. 2012;27:299–338. doi: 10.4054/DemRes.2012.27.11 [DOI] [Google Scholar]
- 50. Fischer KE, Riddle NC. Sex differences in aging: genomic instability. J Gerontol A Biol Sci Med Sci. 2018;73:166–174. doi: 10.1093/gerona/glx105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.