Abstract
Although spikelet-related traits such as size of anther, spikelet, style, and stigma are associated with sexual reproduction in grasses, no QTLs have been reported in sorghum. Additionally, there are only a few reports on sorghum QTLs related to grain size, such as grain length, width, and thickness. In this study, we performed QTL analyses of nine spikelet-related traits (length of sessile spikelet, pedicellate spikelet, pedicel, anther, style, and stigma; width of sessile spikelet and stigma; and stigma pigmentation) and six grain-related traits (length, width, thickness, length/width ratio, length/thickness ratio, and width/thickness ratio) using sorghum recombinant inbred lines. We identified 36 and 7 QTLs for spikelet-related traits and grain-related traits, respectively, and found that most sorghum spikelet organ length- and width-related traits were partially controlled by the dwarf genes Dw1 and Dw3. Conversely, we found that these Dw genes were not strongly involved in the regulation of grain size. The QTLs identified in this study aid in understanding the genetic basis of spikelet- and grain-related traits in sorghum.
Subject terms: Developmental biology, Genetics, Plant sciences
Introduction
Sorghum [Sorghum bicolor (L.) Moench] is the world’s fifth most important C4 cereal crop (faostat.fao.org), and is a promising crop with higher stress tolerance than other major cereals1,2. Because sorghum is rich in morphological diversity and has a relatively small genome (~ 800 Mb3,4) when compared with other C4 grasses5, it is a suitable genetic model for C4 grasses.
Sorghum has a wide range of uses, including the production of grains for food (grain sorghum), forage (forage sorghum), and biomass for bioenergy (biomass/sweet sorghum). Therefore, there is a need to breed sorghum varieties that correspond to their particular use. For example, when considering plant height phenotypes, each favorable trait is used in the breeding process; semi-dwarf phenotypes are important for grain production because they reduce the risk of lodging and make mechanical harvesting more efficient, while non-dwarf phenotypes are important for increased biomass yields. Four major plant height-controlling loci (Dw1-Dw4) were identified in 19546 and these have mainly been used for sorghum breeding for plant height. The plant height-responsible genes have been identified for three of these four loci (Dw1: Sobic.009G2308007,8, Dw2: Sobic.006G0677009, Dw3: Sobic.007G16380010), which have advanced our knowledge of the molecular mechanisms of plant height regulation in sorghum.
Since sorghum grains are important as food, numerous studies have been conducted on traits related to grain weight, and more than 100 quantitative trait loci (QTLs) for grain weight have been identified11–27. However, there are only a few reports on sorghum QTLs related to grain size, such as grain length, width, and thickness28–30. Sorghum grains, like those of other grasses, are enfolded into floral bracts (glumes, lemmas, and paleas) that form a terminal unit called a spikelet. Sorghum has two types of spikelets: one is a sessile spikelet (SS), and the other is a pedicellate spikelet (PS; Fig. 1a,b). Of these, only SSs are fertile. Recently, it was reported that the PS contributes to seed weight by translocating its photosynthetic products to the SS31, suggesting that the PS is not a “useless” organ in sorghum.
Figure 1.
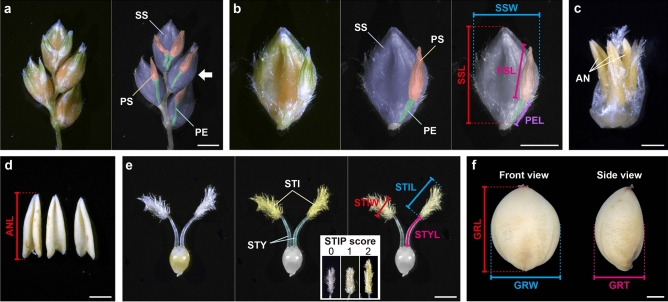
Examples of spikelet-related organs and a mature grain of BTx623. (a) The adaxial view of a secondary branch from the BTx623 panicles. The arrow indicates the second-top spikelet pair. (b) Enlarged view of the second-top spikelet pair. (c) An isolated mature floret from the sessile spikelet. (d) Anthers isolated from the floret. (e) An isolated pistil from the floret. The inset image shows an example of stigma pigmentation values; we scored white as 0 (BTx623), an intermediate pale yellow color as 1 (RIL065), and yellow as 2 (NOG). (f) Front and side views of a mature grain. SS sessile spikelet; PS pedicellate spikelet; PE pedicel; SSL sessile spikelet length; SSW sessile spikelet width; PSL pedicellate spikelet length; PEL pedicel length; AN anther; ANL anther length; STY style; STI stigma; STYL style length; STIL stigma length; STIW stigma width; STIP stigma pigmentation; GRL grain length; GRW grain width; GRT grain thickness. Scale bar = 2 mm in (a, b) and 1 mm in (c–f).
Morphologies of spikelet-related organs, such as the glume, lemma, palea, lodicule, anther, and pistil (ovule with style and stigma), are important for sexual reproduction in grasses. For example, it is known that the efficiency of hybrid seed production can be improved by increasing the percentage of exserted stigma, which is closely related to the balance between spikelet size and stigma length in rice32. In contrast, cleistogamy, which means autonomous self-pollination caused by the failure of spikelets to open, is a useful genetic tool for the prevention of possible gene transfer in transgenic crops33, which maintains the genetic purity of inbreds across generations34.
Because of the importance of these morphologies, QTLs and genes that control spikelet-related traits, such as anther length, spikelet length, width, style length, and stigma length, are particularly well studied in rice32,35–39. For example, genome-wide association studies for spikelet length and width and style length found that the well-known genes, GS3 (GRAIN SIZE 3, for spikelet length and style length) and GW5 (GRAIN WIDTH 5, for spikelet width) were involved in these traits in rice36. GS3 (Os03g0407400) encodes a putative transmembrane protein and has been identified as an evolutionarily important gene that controls grain size and has major effects on rice grain length and weight40,41. GW5/qSW5 (QTL for seed width on chromosome 5, Os05g0187500/LOC_Os05g09520) encodes a novel nuclear protein that was identified as a rice domestication-related gene involved in grain width and weight42,43. In addition, SG6 (SHORT GRAIN 6, Os06g0666100/LOC_Os06g45540) controls grain size by regulating spikelet hull (lemma) cell division37. SG6 encodes an uncharacterized plant A/T-rich sequence and zinc-binding protein, and its overexpression resulted in increased plant height and significantly larger and heavier grains. Liu et al. found that LOC_Os03g14850 was the gene responsible for qSTL3 (QTL for stigma length on chromosome 3), which controls stigma length32, while LOC_Os03g14850 encodes a MADS-box family gene with an M-alpha type-box that affects both stigma length and grain size. Dang et al. reported a novel style length regulator gene, OsSYL2 (Os02g0733900/LOC_Os02g50110), which encodes an 80-amino-acid protein with no putative conserved domains38. Zhang et al. reported that the grain size (both spikelet length and width) was positively regulated by the Rho-like GTPase OsRac1 (Os01g0229400/LOC_Os01g12900), which is thought to promote cell division39.
While much is known about the genes that regulate spikelets in rice, no genes or QTLs have been reported for spikelet-related traits in sorghum, except for a few reports of Multiseeded genes (MSD1-3) that control PS fertility44–46. To our knowledge, the genetic regulation of spikelet-related organs in sorghum is far behind that of rice.
Recently, we established and reported a sorghum recombinant inbred line (RIL) derived from a parental cross between BTx623 and the Japanese landrace NOG47. Each parent seemed to represent diversified sorghum accessions that were classified into three groups; NOG belonged to the subgroup that represented Asian sorghums and was only distantly related to the American/African accessions, including BTx623. We constructed a high-density linkage map based on 3,710 single nucleotide polymorphisms (SNPs) obtained by restriction-site-associated DNA sequencing (RAD-seq) of 213 RIL individuals. This population was suitable for various QTL analyses because, in addition to detecting Dw3 and Dw1 as plant height QTLs, other traits showed distinct differences47,48. It will be meaningful to improve our knowledge about the sorghum QTLs that control spikelet-related traits and grain size for future sorghum research. In this study, we performed QTL analyses of nine spikelet-related traits and six grain-related traits in sorghum using the RILs to enhance our knowledge of the genetic control of spikelet- and grain-related traits in sorghum.
Results
Morphologies of sorghum spikelets, spikelet-related organs, and grain
The panicle of sorghum forms a primary branch at each node, with subsequent secondary or tertiary branches developing from the primary branch22. Each inflorescence branch has three terminal spikelets: one sessile spikelet (SS) directly attached to the inflorescence branch and two pedicellate spikelets (PSs) attached to the inflorescence branch via a pedicel (PE). Several spikelet pairs, one SS, and one PS each developed below the terminal spikelets (Fig. 1a,b). Only the SS forms a complete floret consisting of a lemma, palea, two lodicules, three anthers, and a pistil (Fig. 1c–e), and developed into a seed (fertile); whereas, the PS is sterile because it lacks a complete floret structure49. Preliminary experiments showed that there were some differences in spikelet-related organs between BTx623 and NOG, and we performed QTL analysis using the RILs derived from a cross between BTx623 and NOG47 for genetic dissection of these traits. In addition, to enrich our knowledge of grain size regulation in sorghum, we also performed QTL analysis for grain-related traits. We measured nine spikelet-related traits: sessile spikelet length (SSL), sessile spikelet width (SSW), pedicellate spikelet length (PSL), pedicel length (PEL) (Fig. 1b), anther length (ANL) (Fig. 1d), style length (STYL), stigma length (STIL), stigma width (STIW), and stigma pigmentation (STIP) (Fig. 1e), using spikelets from each RIL collected just before flowering. We also measured six grain-related traits: grain length (GRL), grain width (GRW), grain thickness (GRT) (Fig. 1f), grain length/width ratio (GLWR), grain length/thickness ratio (GLTR), and grain width/thickness ratio (GWTR) of mature seeds in each RIL.
Phenotypic evaluation of spikelet-related and grain-related traits in the RIL population
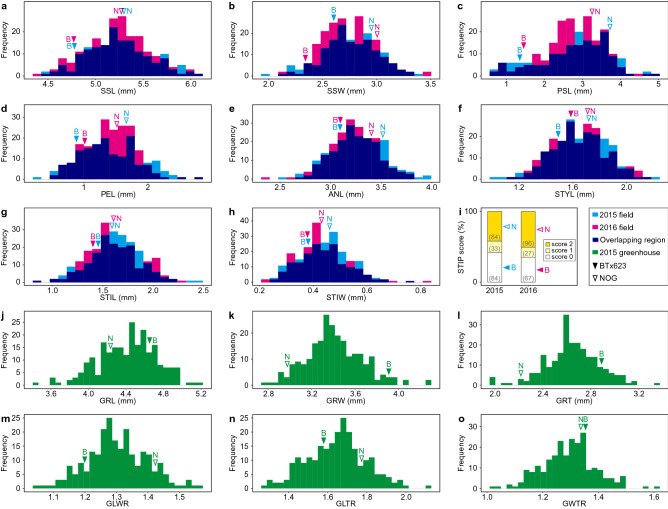
To validate the reproducibility of our results, the parents (BTx623 and NOG) and RILs in successive generations (F7 and F8) were cultivated in Tokyo (field) for two consecutive years (2015 and 2016) for the analysis of spikelet-related traits. First, we evaluated the distribution of phenotypic data between experimental cultivations from different years. Focusing on the parents of the RILs, we found that NOG showed higher trait values than BTx623 for all spikelet-related traits (Fig. 2a–i). Although there was variation between years, each phenotype showed a similar distribution, and the phenotypic distribution among the RILs compared to their parents revealed transgressive segregations for most of the traits. For grain-related traits, BTx623 showed higher trait values than NOG with regard to GRL, GRW, and GRT (Fig. 2j–l), indicating that NOG produces smaller grains. No significant difference was detected in GWTR between the parents; however, GLWR and GLTR were larger in NOG, indicating that NOG produces more slender and flatter grains (Fig. 2m–o). Phenotypic distribution among the RILs also showed transgressive segregations for grain-related traits when compared to their parents.
Figure 2.
Frequency distribution of nine spikelet-related traits and six grain-related traits in the RIL population. (a) Sessile spikelet length (SSL), (b) sessile spikelet width (SSW), (c) pedicellate spikelet length (PSL), (d) pedicel length (PEL), (e) anther length (ANL), (f) style length (STYL), (g) stigma length (STIL), (h) stigma width (STIW), (i) stigma pigmentation (STIP), (j) grain length (GRL), (k) grain width (GRW), (l) grain thickness (GRT), (m) grain length/width ratio (GLWR), (n) grain length/thickness ratio (GLTR), and (o) grain width/thickness ratio (GWTR). Filled arrowheads (B) and open arrowheads (N) indicate phenotypic values for BTx623 and NOG, respectively.
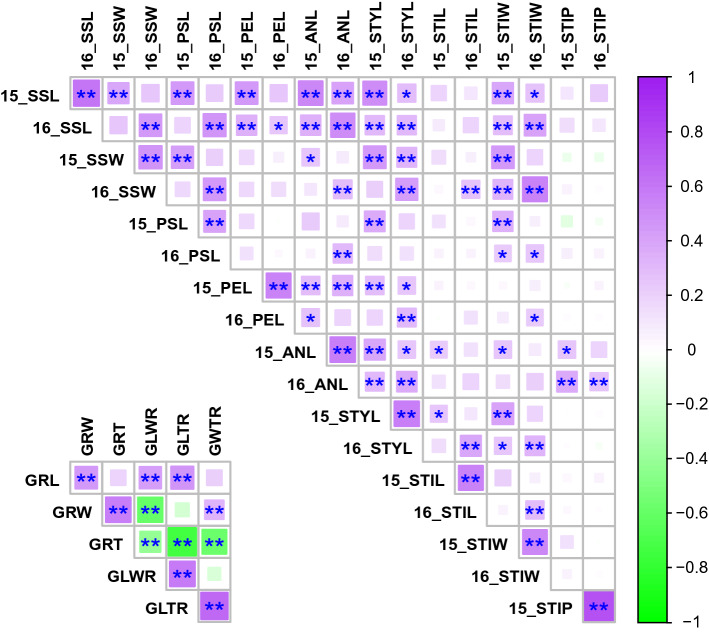
Next, we evaluated the correlation between each trait (Fig. 3). For spikelet-related traits, each dataset was named according to the year and trait name; for example, the SSL phenotypic data collected in 2015 was 15_SSL. Although no negative correlation was observed, significant positive correlations between years were observed for all spikelet-related traits (SSL: r > 0.62, P < 0.0001; SSW: r > 0.49, P < 0.0001; PSL: r > 0.42, P < 0.0001; PEL: r > 0.56, P < 0.0001; ANL: r > 0.59, P < 0.0001; STYL: r > 0.59, P < 0.0001; STIL: r > 0.57, P < 0.0001; STIW: r > 0.55, P < 0.0001; and STIP: r > 0.78, P < 0.0001). We found that STIP had little correlation with other spikelet-related traits. However, STIP was only slightly correlated with ANL. For spikelet-related traits, the correlation among length- and width-related traits tended to exhibit relatively strong correlations except for STIL, suggesting that there are some common genetic mechanisms for the regulation of these spikelet-related traits. For grain-related traits, there were positive correlations between GRL and GRW (r > 0.46, P < 0.0001) and GRW and GRT (r > 0.59, P < 0.0001).
Figure 3.
Phenotypic correlations for the nine spikelet-related and six grain-related traits measured in this study. Pearson’s correlation coefficients for each trait measured in the RIL population. Purple and green colors represent positive and negative correlations (r) between traits, respectively. SSL sessile spikelet length; SSW sessile spikelet width; PSL pedicellate spikelet length; PEL pedicel length; ANL anther length; STYL style length; STIL stigma length; STIW stigma width; STIP stigma pigmentation; GRL grain length; GRW grain width; GRT grain thickness; GLWR grain length/width ratio; GLTR grain length/thickness ratio; and GWTR grain width/thickness ratio. For the spikelet-related traits, “15_” or “16_” indicate the results from 2015 or 2016. *P < 0.001, **P < 0.0001.
QTL analysis of the spikelet-related and grain-related traits in the RIL population
For the genetic dissection of spikelet-related and grain-related traits, we performed QTL analysis using phenotypic data with 3710 SNP markers identified from RAD-seq data47. Composite interval mapping was carried out using the data for each trait, and as a result, we identified 36 and 7 QTLs for spikelet-related traits and grain-related traits, respectively (Table 1).
Table 1.
Summary of the spikelet- and grain-related QTLs detected in this study. Marker intervals were estimated based on confidence intervals (2.0-LOD).
| Trait | Year | Chr | QTL ID | Position (cM) | Nearest marker | Marker interval | LOD | PVE (%) | AE |
|---|---|---|---|---|---|---|---|---|---|
| SSL | 2015 | 6 | 15_qSSL6 | 79.9 | Chr06.49894350 | Chr06.48086748-Chr06.50331377 | 4.59 | 10.17 | 0.11 |
| 9 | 15_qSSL9 | 103.8 | Chr09.57273964 | Chr09.56467290-Chr09.57917278 | 6.28 | 14.27 | 0.13 | ||
| 2016 | 6 | 16_qSSL6 | 78.4 | Chr06.48677734 | Chr06.48086748-Chr06.50250487 | 3.92 | 7.09 | 0.09 | |
| 9 | 16_qSSL9 | 103.5 | Chr09.57166357 | Chr09.56467290-Chr09.57917278 | 8.87 | 17.03 | 0.13 | ||
| SSW | 2015 | 1 | 15_qSSW1 | 139.9 | Chr01.73492838 | Chr01.72086062-Chr01.74542240 | 6.31 | 9.81 | -0.08 |
| 6 | 15_qSSW6 | 68.3 | Chr06.46616258 | Chr06.45449430-Chr06.47716228 | 7.51 | 11.89 | 0.09 | ||
| 7 | 15_qSSW7 | 114.1 | Chr07.64706648 | Chr07.63878301-Chr07.65440280 | 4.63 | 7.03 | -0.07 | ||
| 9 | 15_qSSW9 | 99.6 | Chr09.56047912 | Chr09.54927200-Chr09.56880571 | 15.09 | 26.76 | 0.13 | ||
| 2016 | 6 | 16_qSSW6 | 74.5 | Chr06.47921285 | Chr06.47546161-Chr06.48677734 | 6.59 | 12.13 | 0.08 | |
| 9 | 16_qSSW9 | 102.0 | Chr09.56571248 | Chr09.55806970-Chr09.57681027 | 6.83 | 12.62 | 0.09 | ||
| PSL | 2015 | 6 | 15_qPSL6 | 70.3 | Chr06.46819000 | Chr06.45449430-Chr06.47716228 | 6.50 | 16.87 | 0.39 |
| 2016 | 6 | 16_qPSL6 | 79.4 | Chr06.49336636 | Chr06.48086748-Chr06.50331377 | 5.34 | 11.73 | 0.27 | |
| PEL | 2015 | 7 | 15_PEL7 | 86.8 | Chr07.59727807 | Chr07.59407685-Chr07.60042098 | 11.38 | 24.21 | 0.23 |
| 9 | 15_PEL9 | 104.2 | Chr09.57599415 | Chr09.56571248-Chr09.58014412 | 5.12 | 9.93 | 0.15 | ||
| 2016 | 7 | 16_PEL7 | 86.1 | Chr07.59407685 | Chr07.58788758-Chr07.59727807 | 9.34 | 19.53 | 0.17 | |
| ANL | 2015 | 1 | 15_qANL1 | 165.5 | Chr01.78907937 | Chr01.77228064-Chr01.79222517 | 5.10 | 9.48 | 0.08 |
| 9 | 15_qANL6 | 103.5 | Chr09.57166357 | Chr09.56467290-Chr09.57917278 | 7.73 | 14.81 | 0.10 | ||
| 2016 | 1 | 16_qANL1 | 122.6 | Chr01.65134709 | Chr01.64608936-Chr01.65571022 | 4.34 | 8.35 | 0.07 | |
| 9 | 16_qANL9 | 103.5 | Chr09.57166357 | Chr09.56571248-Chr09.57917278 | 5.92 | 11.60 | 0.08 | ||
| STYL | 2015 | 6 | 15_qSTYL6 | 76.0 | Chr06.48086748 | Chr06.47546161-Chr06.48677734 | 7.35 | 13.68 | 0.08 |
| 7 | 15_qSTYL7 | 86.1 | Chr07.59727807 | Chr07.59407685-Chr07.60042098 | 6.69 | 12.36 | 0.07 | ||
| 2016 | 2 | 16_qSTYL2 | 116.5 | Chr02.67908557 | Chr02.67181119-Chr02.68205478 | 5.35 | 8.71 | -0.06 | |
| 6 | 16_qSTYL6 | 76.0 | Chr06.48086748 | Chr06.47921285-Chr06.48677734 | 9.42 | 16.12 | 0.08 | ||
| 7 | 16_qSTYL7 | 88.1 | Chr07.60042098 | Chr07.59407685-Chr07.60649532 | 7.79 | 13.07 | 0.07 | ||
| STIL | 2015 | 1 | 15_qSTIL1 | 168.8 | Chr01.80075044 | Chr01.78907937-Chr01.80451024 | 5.76 | 9.64 | 0.08 |
| 3 | 15_qSTIL3 | 144.2 | Chr03.73116039 | Chr03.72187849-Chr03.73272999 | 12.71 | 23.14 | 0.12 | ||
| 2016 | 1 | 16_qSTIL1 | 169.0 | Chr01.80451024 | Chr01.78907937-Chr01.80451024 | 4.12 | 6.84 | 0.07 | |
| 3 | 16_qSTIL3 | 144.2 | Chr03.73116039 | Chr03.72187849-Chr03.73584465 | 13.77 | 25.69 | 0.14 | ||
| STIW | 2015 | 3 | 15_qSTIW3 | 1.2 | Chr03.1014391 | Chr03.196942-Chr03.1842334 | 9.59 | 14.51 | -0.04 |
| 6 | 15_qSTIW6 | 76.0 | Chr06.48086748 | Chr06.47740711-Chr06.49157376 | 6.35 | 9.24 | 0.03 | ||
| 9 | 15_qSTIW9 | 102.0 | Chr09.56571248 | Chr09.55806970-Chr09.57681027 | 8.07 | 11.98 | 0.03 | ||
| 2016 | 3 | 16_qSTIW3 | 1.2 | Chr03.1014391 | Chr03.196942-Chr03.1842334 | 9.07 | 14.14 | -0.04 | |
| 6 | 16_qSTIW6 | 76.3 | Chr06.48086748 | Chr06.47546161-Chr06.49157376 | 5.77 | 8.66 | 0.03 | ||
| 9 | 16_qSTIW9 | 103.5 | Chr09.57166357 | Chr09.56467290-Chr09.57681027 | 7.63 | 11.69 | 0.03 | ||
| STIP | 2015 | 1 | 15_qSTIP1 | 126.6 | Chr01.67767009 | Chr01.67509497-Chr01.68618079 | 50.62 | 68.83 | 0.77 |
| 2016 | 1 | 16_qSTIP1 | 126.6 | Chr01.67767009 | Chr01.67509497-Chr01.68618079 | 39.41 | 61.72 | 0.72 | |
| GRL | 2015 | 4 | qGRL4 | 98.6 | Chr04.59140841 | Chr04.57959040-Chr04.60473635 | 7.92 | 16.82 | -0.12 |
| GRT | 2015 | 8 | qGRT8 | 24.2 | Chr08.3019891 | Chr08.3019891-Chr08.4069903 | 5.43 | 11.86 | -0.08 |
| GLWR | 2015 | 8 | qGLWR8 | 50.2 | Chr08.6382451 | Chr08.5764378-Chr08.7916525 | 3.77 | 8.39 | 0.03 |
| GLTR | 2015 | 4 | qGLTR4 | 96.1 | Chr04.57959040 | Chr04.57115462-Chr04.58794505 | 15.92 | 28.84 | -0.08 |
| 8 | qGLTR8.1 | 23.6 | Chr08.3019891 | Chr08.2790929-Chr08.3185552 | 3.49 | 5.71 | 0.03 | ||
| 8 | qGLTR8.2 | 83.3 | Chr08.57258936 | Chr08.56896649-Chr08.57745670 | 3.99 | 6.52 | 0.03 | ||
| GWTR | 2015 | 4 | qGWTR4 | 96.1 | Chr04.57959040 | Chr04.57115462-Chr04.58794505 | 6.44 | 13.92 | -0.04 |
PVE phenotypic variance explained. For the additive effects (AE), positive values indicate that alleles from NOG increased the trait score. SSL sessile spikelet length, SSW sessile spikelet width, PSL pedicellate spikelet length, PEL pedicel length, ANL anther length, STYL style length, STIL stigma length, STIW stigma width, STIP stigma pigmentation, GRL grain length, GRT grain thickness, GLWR grain length/width ratio, GLTR grain length/thickness ratio, GWTR grain width/thickness ratio.
Spikelet-related traits
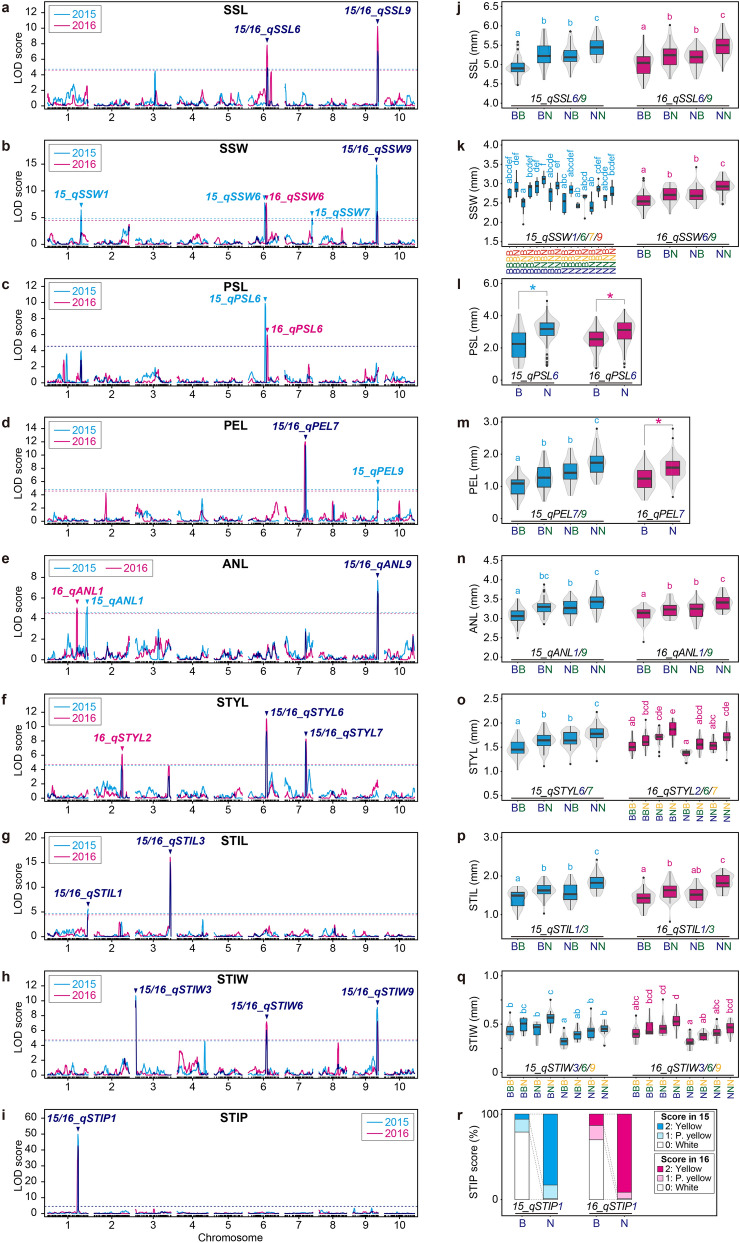
For sessile spikelet length (SSL), two QTLs were detected on chromosomes 6 and 9 in both 2015 and 2016 trials (15_qSSL6, 15_qSSL9, 16_qSSL6, and 16_qSSL9) with the logarithm of the odds (LOD) scores ranging from 3.92 to 8.87, and the percentage of phenotypic variation explained by each QTL (PVE) values from 7.09 to 17.03% (Fig. 4a, Table 1). Comparisons of confidence intervals (2.0-LOD) of each QTL between 2015 and 2016 suggested that qSSL6 and qSSL9 were highly reproducible and reliable QTLs. qSSL9 had a higher LOD score (6.28 in 2015, 8.87 in 2016) and was relatively robust in each cultivation compared to qSSL6. The effects of increasing SSL were attributed to the NOG alleles for qSSL6 and qSSL9 (Fig. S1a, Fig. 4j).
Figure 4.
Results of QTL analysis for the nine spikelet-related traits (a–i) and their allelic effects (j–r). (a,j) Sessile spikelet length, (b,k) sessile spikelet width, (c,l) pedicellate spikelet length, (d,m) pedicel length, (e,n) anther length, (f,o) style length, (g,p) stigma length, (h,q) stigma width, and (i,r) stigma pigmentation. (a–i) LOD profiles obtained from composite interval mapping (CIM). Cyan and magenta horizontal dotted lines represent a threshold of the 1000 × permutation test (P < 0.05) in 2015 and 2016, respectively. (j–r) Contributions of SNP genotypes for significant QTLs. Box and violin plots (j–q) and a 100% stacked column chart (r) show the effects of the nearest marker genotypes for each QTL or allelic combinations of QTLs. “15_” or “16_” indicate the results from 2015 or 2016. Different letters denote significant differences according to the Tukey–Kramer test (P < 0.05). Asterisks indicate significant differences between genotypes (Welch’s t-test, P < 0.01).
For sessile spikelet width (SSW), four QTLs were detected on chromosomes 1, 6, 7, and 9 in 2015 (15_qSSW1, 15_qSSW6, 15_qSSW7, and 15_qSSW9), and two QTLs were detected on chromosomes 6 and 9 in 2016 (16_qSSW6 and 16_qSSW9), with LOD scores ranging from 4.63 to 15.09, and PVE values from 7.03% to 26.76% (Fig. 4b, Table 1). Comparisons of confidence intervals suggested that qSSW9 had high reproducibility, while the other QTLs were more susceptible and unstable due to environmental factors. qSSW9 had a higher LOD score (15.09 in 2015 and 6.83 in 2016) and was relatively robust in each cultivation compared with the other QTLs. The effects of increasing SSW were attributed to the NOG alleles for 15/16_qSSW6 and 15/16_qSSW9, but to the BTx623 allele for 15_qSSW1 and 15_qSSW7 (Fig. S1b, Fig. 4k).
For pedicellate spikelet length (PSL), a single QTL was detected on chromosome 6 in 2015 (15_qPSL6; LOD score 6.50, PVE 16.87%) and 2016 (16_qPSL6; LOD score 5.34, PVE 11.73%) (Fig. 4c, Table 1). Although two QTLs were detected in close proximity, their confidence intervals did not overlap, suggesting that PSL is unstable in response to environmental factors. The effects of increasing PSL were attributed to the NOG alleles for both 15_qPSL6 and 16_qPSL6 (Fig. 4l).
For pedicel length (PEL), two QTLs were detected on chromosomes 7 and 9 in 2015 (15_qPEL7 and 15_qPEL9), and a single QTL was detected on chromosome 7 in 2016 (16_qPEL7) with LOD scores ranging from 5.12 to 11.38 and PVE values from 9.93 to 24.21% (Fig. 4d, Table 1). Although the peak markers were slightly different, 15_qPEL7 and 16_qPEL7 had overlapping confidence intervals, suggesting that qPEL7 is a reproducible and reliable QTL. The effects of increasing PEL were attributed to the NOG alleles at all QTLs (Fig. S1c, Fig. 4m).
For anther length (ANL), two QTLs were detected on chromosomes 1 and 9 in both 2015 and 2016 (15_qANL1, 15_qANL9, 16_qANL1, and 16_qANL9), with LOD scores ranging from 4.34 to 7.73 and PVE values from 8.35% to 14.81% (Fig. 4e, Table 1). Comparisons of confidence intervals suggested that qANL9 showed high reproducibility, while qANL1 was unstable to environmental factors. The effects of increasing ANL were attributed to the NOG alleles for all QTLs (Fig. S1d, Fig. 4n).
For style length (STYL), two QTLs were detected on chromosomes 6 and 7 in 2015 (15_qSTYL6 and 15_qSTYL7), and three QTLs were detected on chromosomes 2, 6, and 7 in 2016 (16_qSTYL2, 16_qSTYL6, and 16_qSTYL7) with LOD scores ranging from 5.35 to 9.42, and PVE values from 8.71% to 16.12% (Fig. 4f, Table 1). Both 15/16_qSTYL6 and 15/16_qSTYL7 had overlapping confidence intervals, suggesting that qSTYL6 and qSTYL7 are robust and reliable QTLs, whereas qSTYL2 is unstable to environmental factors. The effects of increasing STYL were attributed to the NOG alleles in 15/16_qSTYL6 and 15/16_qSTYL7, but to the BTx623 allele in 16_qSTYL2 (Fig. S1e, Fig. 4o).
For stigma length (STIL), two QTLs were detected on chromosomes 1 and 3 in both 2015 and 2016 (15_qSTIL1, 15_qSTIL3, 16_qSTIL1, and 16_qSTIL3) with LOD scores ranging from 4.12 to 13.77 and PVE values from 6.84% to 25.69% (Fig. 4g, Table 1). Both 15/16_qSTIL1 and 15/16_qSTIL3 had overlapping confidence intervals, suggesting that qSTIL1 and qSTIL3 are highly reproducible and reliable QTLs. The effects of increasing STIL were attributed to the NOG alleles for all QTLs (Fig. S1f, Fig. 4p).
For stigma width (STIW), three QTLs were detected on chromosomes 3, 6, and 9 in both 2015 and 2016 (15_qSTIW3, 15_qSTIW6, 15_qSTIW9, 16_qSTIW3, 16_qSTIW6, and 16_qSTIW9) with LOD scores ranging from 5.77 to 9.59, and PVE values from 8.66% to 14.51% (Fig. 4h, Table 1). Comparisons of the confidence intervals suggested that qSTIW3, qSTIW6, and qSTIW9 are highly reproducible and reliable QTLs. The effects of increasing STIW were attributed to the NOG alleles for qSTIW6 and qSTIW9 and to the BTx623 allele for qSTIW3 (Fig. S1g, Fig. 4q).
For stigma pigmentation (STIP), an extremely significant QTL was detected on chromosome 1 in both 2015 (15_qSTIP1; LOD score 50.62, PVE 68.83%) and 2016 (16_qSTIP1; LOD score 39.41, PVE 61.72%) (Fig. 4i, Table 1). The complete match of the confidence intervals for 15_qSTIP1 and 16_qSTIP1 indicated that qSTIP1 is a highly reproducible and reliable QTL. The effect of increasing STIP (to be yellow) was attributed to the NOG alleles for qSTIP1 (Fig. 4r).
Grain-related traits
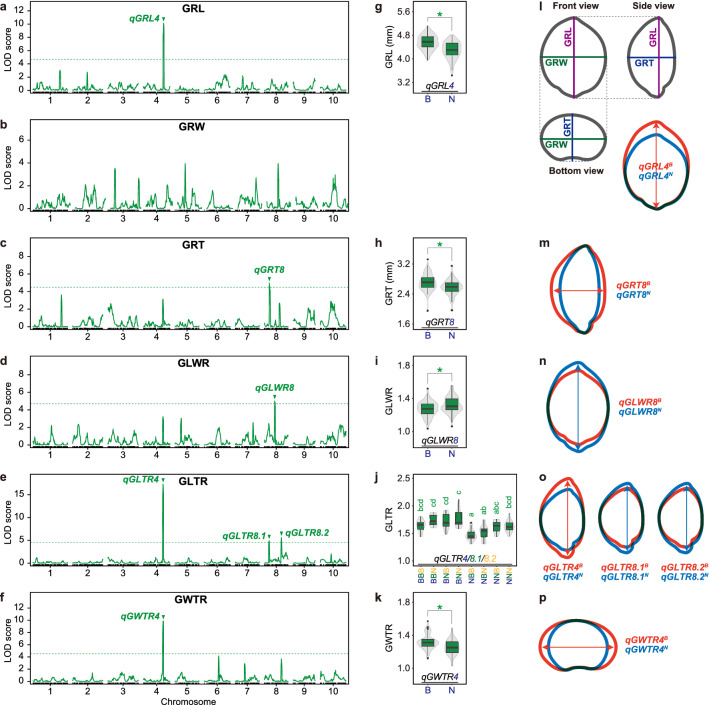
A single QTL on chromosome 4 (qGRL4; LOD score 7.92, PVE 16.82%) was detected for grain length (GRL) (Fig. 5a, Table 1). The effect of increasing GRL was attributed to the BTx623 allele (Fig. 5g,l). Although we could not identify any QTLs for grain width (Fig. 5b), a single QTL was detected for grain thickness (GRT) on chromosome 4 (qGRT8; LOD score 5.43, PVE 11.86%) (Fig. 5c, Table 1). The effect of increasing GRT and GRL effects was attributed to the BTx623 alleles (Fig. 5h,m).
Figure 5.
Results of QTL analysis for the six grain-related traits (a–f), their allelic effects (g–k), and schematics of the allelic effects (l-p). (a,g,l) Grain length, (b) grain width, (c,h,m) grain thickness, (d,i,n) grain length/width ratio, (e,j,o) grain length/thickness ratio, and (f,k,p) grain width/thickness ratio. (a–f) LOD profiles obtained from composite interval mapping (CIM). Horizontal dotted lines represent a threshold of the 1000 × permutation test (P < 0.05). (g–k) Contributions of SNP genotypes for significant QTLs. Box and violin plots show the effects of the nearest marker genotypes for each QTL or allelic combinations of QTLs. Different letters denote significant differences according to the Tukey–Kramer test (P < 0.05). Asterisks indicate significant differences between genotypes (Welch’s t-test, P < 0.01).
For the grain length/width ratio (GLWR), a single QTL was detected on chromosome 8 (qGLWR8; LOD score 3.77, PVE 8.39%) (Fig. 5d, Table 1). The effect of increasing GLWR was attributed to the NOG allele (Fig. 5i,n). For the grain length/thickness ratio (GLTR), three QTLs were detected on chromosomes 4 and 8 (qGLTR4, qGLTR8.1, and qGLTR8.2) with LOD scores ranging from 3.49 to 15.92 and PVE values from 5.71% to 28.84% (Fig. 5e, Table 1). The effects of increasing GLTR were attributed to the BTx623 alleles for qGLTR4 and to the NOG allele for qGLTR8.1 and qGLTR8.2 (Fig. S1h, Fig. 5j,o).
For the grain width/thickness ratio (GWTR), a single QTL was detected on chromosome 4 (qGWTR4; LOD score 6.44, PVE 13.92%) (Fig. 5f, Table 1), and the effect of increasing GWTR was attributed to the BTx623 allele (Fig. 5k,p).
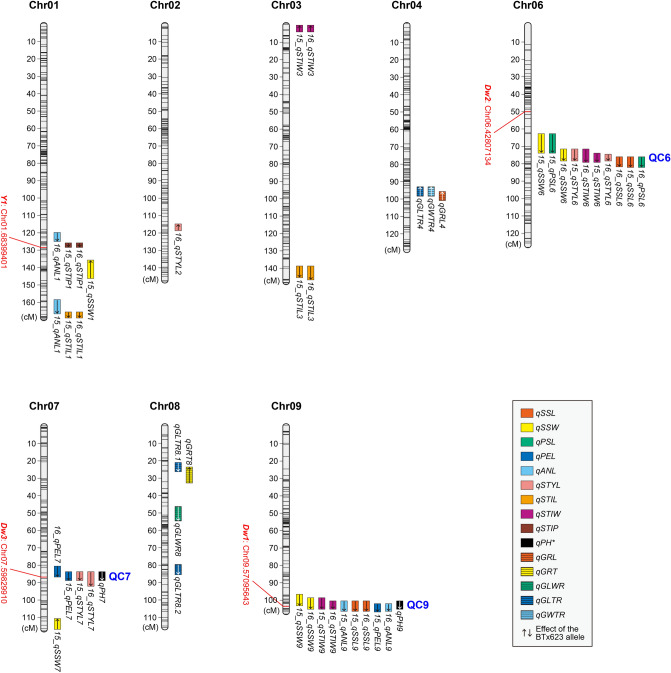
The positions of the confidence intervals on the chromosomes for all the QTLs detected in this study are summarized in Fig. 6. The detected QTLs were distributed across eight chromosomes, which did not include chromosomes 5 and 10. QTL clusters with four or more QTLs detected in similar positions were found on chromosomes 6, 7, and 9 (QC6, QC7, and QC9 in Fig. 6). Among these QTL clusters, QC6 affected SSL, SSW, PSL, STYL, and STIW; QC7 affected PEL and STYL; and QC9 affected SSL, SSW, PEL, ANL, and STIW.
Figure 6.
Graphical representations of the QTLs detected in this study. Colored boxes indicate the confidence intervals (2.0-LOD) of significant QTLs, with each color representing a different trait. Arrows in each box indicate the direction of the phenotypic effect of the BTx623 allele (up, increasing; down, decreasing). *Positions of qPH7/9 were taken from the results obtained by our previous study using the same population47. For the spikelet-related traits, “15_” or “16_” indicate results from 2015 or 2016. QC6, QC7, and QC9 indicate QTL clusters found on chromosomes 6, 7, and 9, respectively. Transcriptional start points for known related genes in BTx623 are shown in red on the left side of the chromosome.
Comparison between our QTLs and those of previous studies and a search for the responsible genes
In our QTL analysis of nine spikelet-related traits, three QTL clusters were identified on chromosomes 6 (QC6, Chr06:47546161–50331377), 7 (QC7, Chr07:58788758–60649532), and 9 (QC9, Chr09:54927200–57917278). Whether the formation of these QTL clusters was due to the pleiotropic effects of a single gene or simply the physical linkage of multiple genes is unclear; however, it will be interesting to identify the genes responsible for these QTL clusters.
For QC7 and QC9, each cluster corresponded to plant height-related QTLs obtained in our previous report using the same populations and genotype data47 (Fig. 6; qPH7 and qPH9). Comparing these QTLs with those reported previously using the QTL Atlas50, both QC7 and QC9 were supported by numerous previous reports of plant height- and panicle length-related QTLs in sorghum27,51–57 (Table S1). In sorghum, four major loci controlling plant height, Dw1, Dw2, Dw3, and Dw4, have been extensively characterized and reported that their recessive alleles reduce internode length6. The Dw1 locus was mapped to chromosome 9, Dw2 to 6, Dw3 to 7, and Dw4 to 458. Dw1 (Sobic.009G230800, Chr09:57093313–57095643), which is known to regulate the length of internodes by controlling cell proliferation, encodes a positive regulator gene of brassinosteroid signaling7,8,59. Dw2 (Sobic.006G067700, Chr06:42803037–42807134) encodes a protein kinase9, and Dw3 (Sobic.007G163800, Chr07:59821905–59829910) encodes an ABCB1 auxin efflux transporter10. In addition to internode length, pleiotropic effects of Dw3, including seed weight, panicle size, tiller number, and leaf angle, have also been reported60–62. We previously showed that Dw1 and Dw3 are likely the genes responsible for qPH9 and qPH7 based on sequencing analysis, which revealed that NOG had functional alleles of both Dw1 and Dw3 genes, while BTx623 had loss-of-function alleles of both genes47 (Fig. S2a and b). Based on the functional aspects of Dw1 and Dw3 (e.g., controlling organ size) and the direction of the allelic effects, in which the size of spikelet-related organs and plant height were increased in the NOG allele, QC7 and QC9 likely corresponded to Dw3 and Dw1, respectively.
For the gene responsible for QC6, we initially hypothesized that Dw2 is also involved in this cluster because Dw2 is located on chromosome 69. However, Dw2 was found to be located much further upstream than the QC6 region (Dw2, Chr06:42803037–42807134; QC6, Chr06:47546161–50331377; Fig. 6). In addition, the lack of QTLs detected around the Dw2 region in our previous QTL analysis for plant height using the same populations47 indicated that there were no functional differences in Dw2 between the parents of our RILs (BTx623 and NOG). Therefore, we concluded that Dw2 was not responsible for QC6. Although no organ size-related genes have been identified around the QC6 region to date, we searched for reported QTLs around QC6 using the QTL Atlas (Table S1). As a result, QTLs for plant height53,63, fresh biomass64, and panicle length65,66 were found within the QC6 region, suggesting that minor organ size-related gene(s) other than the major Dw genes are located in the QC6 region.
For qSTIP1, we found that QTLs for grain color and proanthocyanidins were reported around this locus67,68 (Table S1). Furthermore, we also found that the Y1 gene (Sobic.001G398100, Chr01:68399401–68400602), an MYB transcription factor that controls seed pericarp color by regulating flavonoid biosynthesis69,70, is encoded within the confidence interval of qSTP1. Y1 is the ortholog of maze P1 (Pericarp color 1), a transcriptional regulator involved in flavonoid-related red pigmentation in the maize pericarp71. Given Y1 as a strong candidate for qSTIP1, we confirmed polymorphisms of the Y1 locus between BTx623 and NOG by PCR and sequencing in this study. Consistent with the previously reported pattern of functional and loss-of-function alleles70, BTx623 has a loss-of-function allele with a 3.2 kb deletion, while NOG has a functional allele with no deletion (Fig. S2c). These results suggest that Y1 is responsible for both seed and stigma colors.
For spikelet-related traits, all other QTLs detected in this study were explained in previous reports. For the confidence interval of 16_qANL1, the QTLs for panicle length22 and dry matter growth72; 15_qSSW1, the QTLs for fresh biomass73, grain weight74, and panicle width75; 15_qANL1, the QTLs for leaf length and panicle length26; 15_qSTIL1, the QTLs for panicle width76 and fresh biomass51; 16_qSTYL2, the QTLs for plant height53 and panicle length17; 15_qSTIW3, the QTLs for shoot length77 and panicle length78; 15_qSTIL3, the QTL for stem circumference79; 15_qPSL6, the QTLs for plant height63,80 and panicle length25; and 15_qSSW7, the QTLs for panicle length and grain yield25 have been reported (Table S1). Hence, QTLs for organ size regulators have been reported within the confidence interval of the length- and width- related QTLs detected in this study, confirming the reliability of our results. However, the details of these QTLs remain unclear, and further research is required to identify the genes and corresponding mutations responsible for these traits.
We tested whether sorghum orthologous genes of the spikelet-related organ regulator genes identified in rice could be responsible for the QTLs detected in our analysis. Putative orthologous genes in sorghum were defined using BLASTP results and synteny information. The putative orthologue of GS3: Sobic.001G341700 (Chr01:62910779–62916258), GW5: Sobic.009G070000 (Chr09:8184500–8186318), SG6: Sobic.010G220800 (Chr10:56310472–56315161), qSTL3 (LOC_Os03g14850): Sobic.007G086200 (Chr07:11383927–11384670), OsSYL2: Sobic.004G247700 (Chr04:59522591–59523507), and OsRac1: Sobic.009G084100 (Chr09:13296136–13300901) were identified from our analysis. However, we found that none of the putative orthologs of the spikelet-related organ regulator genes in rice were included in the confidence intervals of the QTLs for sorghum spikelet-related traits (Table 1 and Table S1).
We expected that Multiseeded genes (MSD1-3)44–46 may affect PSL, however, no QTLs were detected around MSD1 (Sobic.007G135700, Chr07: 56156125–56157410), a TCP transcription factor44; and MSD3 (Sobic.001G407600, Chr01:69162600..69165731), a plastidial ω-3 fatty acid desaturase46. On the other hand, we found that MSD2 (Sobic.006G095600, Chr06: 46567412–46571064), a lipoxygenase45, was encoded within the confidence interval of 15_qPSL6 (Table 1 and Table S1). However, there were no polymorphisms in the ORF of MSD2 between the parents of our RILs (data not shown).
For the grain-related traits, all the QTLs detected in this study were also supported by previous studies that reported grain size and weight QTLs17–21 (Table S1). We found that orthologous genes for rice PGL2 (Sobic.004G237000, Chr04: 58488864–58490438) and maize Gln-4 (Sobic.004G247000, Chr04: 59472640–59476805) were encoded within the confidence interval of qGRL4. PGL2 encodes an atypical bHLH protein that controls grain length and weight in rice81, while Gln-4 encodes a cytosolic glutamine synthetase related to seed weight in maize82. We expected that PGL2 or Gln4 orthologs in sorghum would be responsible for qGRL4; however, there were no polymorphisms in ORFs in either gene between the parents of our RILs (data not shown). The OsSYL2 ortholog (Sobic.004G247700) is also encoded within the confidence interval of qGRL4; however, there were no polymorphisms in the ORF of the gene between the parents of our RILs (data not shown).
Discussion
Phenotypic distributions among the RILs showed transgressive segregation for almost all of the traits, suggesting a polygenic inheritance of spikelet- and grain-related traits in sorghum. As expected, multiple QTLs were detected for many of these traits; however, considering the phenotypic distribution pattern and sum of PVE values, it is likely that there are still quite a few minor QTLs that could not be detected in this study.
We initially expected that orthologs of genes known to regulate spikelet-related organs in rice would also be involved in the regulation of the sorghum spikelet; however, no such orthologs were detected as QTLs in this population. These results suggest that rice and sorghum may utilize widely different systems to control spikelet-related traits. However, further research using a large number of sorghum varieties is required to validate this hypothesis, since the present results were obtained from analyses using only a single biparental population. For PSL and the grain-related traits, we found that MSD2 was encoded within the confidence interval of 15_qPSL6; and orthologous genes for rice PGL2 and maize Gln-4 were encoded within the confidence interval of qGRL4. Although there were no polymorphisms in ORFs in either gene between the parents of our RILs, further investigation is needed to determine whether these orthologs are involved in sorghum pedicellate spikelet length and grain size, as polymorphisms in the promoter regions of these genes may also alter these traits.
Because we could not measure the spikelet- and grain-related traits under the same environment in this study, we cannot conclusively discuss the correlation between these traits; however, we examined it only as a guide. In our population, we found that grain- and spikelet-related traits showed weak correlations, except for SSW and STIW (Fig. S3). Based on these results and the fact that the QTLs detected for spikelet-related traits and grain-related traits were completely different (Figs. 4, 5, 6), we hypothesized that spikelet size and grain size are not mutually associated in our sorghum population, unlike what has been reported in rice42, barley83,84, and wheat85,86. These differences might be due to both parents of our population belonging to the “grain sorghum type” in which the grains protrude from the glumes; thus, grain size may not be limited by spikelet (glume) size.
It is noteworthy that Dw3 and Dw1, which have significant effects on the spikelet-related organ size, were not detected for grain-related traits in our population, even though dw3 was previously reported to reduce grain yield by reducing grain size in sorghum87. Similarly, RNAi-mediated repression of the Dw1 ortholog reduces grain size in rice59, but it has not yet been reported whether Dw1 affects grain size in sorghum. To validate these points, we re-examined whether Dw3 and Dw1 affect the grain size parameters (GRL, GRW, and GRT) (Fig. S4). We found that both Dw3 and Dw1 had little or no effect on the grain size parameters. For the Dw1 allele, grain length and thickness were larger in the functional allele (Dw1); however, the grain width was larger in the dw3 mutant allele than in the functional Dw3 allele (Fig. S4a). Contrary to previous reports87, dw3 had no effect on the grain length and thickness but rather increased the grain width, suggesting that dw3 does not reduce grain size in this population. We showed that grain length and thickness slightly decreased in dw1, which is consistent with the results reported in rice59. This result suggests that a reduction in grain size in dw1 may reduce grain yield. Considering that dw3 and dw1 have been widely used in grain sorghum breeding, we hypothesized that the degree of reduction in grain size (disadvantage) due to dw3 and dw1 was much smaller than the degree of reduction in plant height (advantage), which is why dw3 and dw1 are convenient for breeding programs. We also checked the influence of dw3 and dw1 on plant height and found that they reduced plant height by 45.8% and 32.8%, respectively, but the reduction in grain size was at most 3% (Fig. S4b). These results support our hypothesis in this study.
By enriching the knowledge about the genetic regulation of spikelet- and grain-related traits using a large number of sorghum varieties in the future, we may be able to improve these traits without worrying about trade-offs; for example, breeders may be able to improve grain-related traits of elite varieties of grain sorghum without changing their plant height by introducing grain-related QTLs, such as qGRL4 and qGRT8, which are not related to plant height.
The QTLs identified in this study will be informative for understanding the genetic basis of spikelet-related organ morphologies and future breeding of spikelet- and grain-related traits in sorghum.
Methods
Plant materials
Recently, we established and reported a sorghum recombinant inbred line (RIL), derived from a parental cross between BTx623 and the Japanese landrace NOG47. This population was suitable for various QTL analyses because, in addition to detecting Dw3 and Dw1 as plant height QTLs, other traits showed distinct differences47,48. Therefore, we believe that this population might also be suitable for the analysis of spikelet- and grain-related traits in this study. Seeds of BTx623 were kindly provided by John Mullet and Bill Rooney of Texas A&M University. NOG seeds, originally from Iwate Prefecture, Japan, were purchased from Noguchi Seeds (Hannou, Saitama, Japan). In total, 213 RILs were generated by recurrent selfing of progeny derived from a cross with BTx623 and NOG at the Institute of Plant Science and Resources (IPSR) at Okayama University, as previously reported47. All seed stocks were stored at 15 °C until further use. All applicable international, national, and institutional guidelines for the use of plants in the present study were followed.
Genotype data
Genotype data of the RILs were obtained using the restriction site-associated DNA sequencing (RAD-seq) method88. Briefly, each genome of the 213 RILs from the F6 generation was subjected to RAD-seq to detect genome-wide SNPs, and 3710 high-quality SNPs were selected to construct the linkage map. The map covered a total length of 1299.7 cM and had an average marker density of 0.4 cM. Detailed methods and information for genotyping have been described in our previous reports47,89.
Cultivation
Field trials were performed in a green field at the University of Tokyo (latitude: 35°42′58.5′′ N, longitude: 139°45′44.5′′ E), Yayoi, Bunkyo-Ku, Tokyo, Japan, from May to September in 2015 and 2016. Plants were germinated in 200-cell trays filled with synthetic culture soil (BONSOL 1, Sumitomo Chemical Co., Ltd., Japan) for two weeks, and then transplanted into the field (n = 2 per line). The space between ridges was 60 cm, with a 15 cm distance between individuals in the same ridge. The field was fertilized using N:P:K = 10:10:10 (kg ha–1) fertilizer before the trials in each year. Cultivation and phenotyping for spikelet-related traits were conducted in 2015 and 2016, using the F7 and F8 generations, respectively. For grain-related traits, cultivation was performed in an open-air greenhouse at the experimental farm of the IPSR (latitude: 34°35′30.5′′ N, longitude: 133°46′08.2′′ E) in Kurashiki, Okayama, Japan, from June to September 2015 with the F7 generation. Plants were germinated in small trays filled with vermiculite for approximately two weeks and then transplanted to 30-cm diameter pots filled with soil from the IPSR field (n = 2 per line). Mature grains (F8 seeds) were harvested from one healthy plant per line (F7 generation) to phenotype grain-related traits.
Phenotyping
Sessile spikelet length (SSL), sessile spikelet width (SSW), pedicellate spikelet length (PSL), pedicel length (PEL), anther length (ANL), style length (STYL), stigma length (STIL), stigma width (STIW), and stigma pigmentation (STIP) were measured. To standardize the developmental stages of spikelets among the RILs, we sampled spikelets at the just-before-flowering stage in each RIL. In the inflorescence that began flowering at the top, we defined spikelets at the just-before-flowering stage (spikelets that would flower within a day) as those attached to the primary branch just below the flowered primary branch. In each RIL, we sampled three second-top spikelet pairs (Fig. 1a arrow, one SS, and one PS each) on the secondary branches developing in the middle portion of the inflorescence from one healthier plant per line at the just-before-flowering stage. Sampled spikelets were dissected and photographed under a stereomicroscope (M125, Leica, Germany) with a CCD camera (MC170 HD, Leica). Length—and width-related traits were measured using Photoshop CS5 (Adobe, USA). The definitions of each length- and width-related trait were as follows: SSL, length of SS (outer glume) from base to tip, SSW: length of the part of the SS where the width of the SS was the largest, PSL: length of PS from base to tip, PEL: length of PE from base to the beginning of PS, ANL: longitudinal length of AN, STYL: length of STY from the upper end of the ovary to the beginning of STI; STIL, longitudinal length of STI; and STIW, length of the STI, where the width of the STI was the largest. We subjectively scored STIP values on a three-step scale, with white as 0, intermediate color (pale yellow) as 1, and yellow as 2. For all spikelet-related traits, the average value of three independent spikelets was used as the phenotypic value of an individual plant.
Grain length (GRL), grain width (GRW), and grain thickness (GRT) were measured. We used mature grains collected from the whole inflorescence of a single individual for analysis. Briefly, we captured an image of 10 mature grains for each RIL using a flatbed scanner (GT-X820, EPSON, Japan), and quantified these three traits as grain size parameters using the MATLAB Image Processing Toolbox (MATLAB_R2015a). Detailed methods and information for the acquisition and analysis of grain images have been described in our previous reports90. The grain length/width ratio (GLWR), grain length/thickness ratio (GLTR), and grain width/thickness ratio (GWTR) were calculated from the quantified GRL, GRW, and GRT. Correlation analysis was performed using the R91/corrplot package92 and modified using Illustrator CS5 (Adobe).
QTL mapping
Genotype probabilities were calculated using the calc.genoprobility function with a step size of 1 cM and an assumed genotyping error probability of 0.05, using the Kosambi map function93 as implemented in the R/qtl package94. QTL analysis was performed using the composite interval mapping (CIM) function of the R/qtl package with the Haley–Knott regression method95. Linkage analysis was performed using the R/qtl package in R version 3.5.2. The LOD significance threshold for detecting QTLs was calculated by performing 1000 iterations using the R/qtl permutation test. Confidence intervals (CIs) for the QTLs were estimated based on the 2.0-LOD support interval, and the nearest flanking markers located outside the boundary of each CI were defined as both ends of the marker intervals. The additive effect and the percentage of phenotypic variance explained (PVE) by each QTL were obtained using the fitqtl function of the R/qtl package. Box and violin plots were created using the R/ggplot2 package96. A graphical representation of QTLs with linkage groups and markers was produced using the MapChart software package97 and modified using Illustrator CS5.
Comparison between our QTLs and those of previous studies and a search for the responsible genes
Comparisons between detected QTLs in this study and previously reported sorghum QTLs were performed using the QTL Atlas50 (https://aussorgm.org.au/sorghum-qtl-atlas/). To search for sorghum orthologs of rice or maize responsible genes, we used the BLASTP program in the Phytozome database98 (https://phytozome.jgi.doe.gov/pz/portal.html) and synteny information obtained from SynMap299 (http://genomevolution.org/coge/SynMap.pl). We defined the sorghum genes supported by both BLASTP results (< E-10) and syntenic information as putative orthologs of these genes. PCR and subsequent sequencing were performed according to previous reports8,70. Primers used for the validation of polymorphisms in the candidate genes are listed in Supplementary Table S2.
Supplementary Information
Acknowledgements
This research was partly supported by the Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (to N.T. and W.S.) and KAKENHI grants [17H01457 from the Japanese Society for the Promotion of Science (JSPS) to N.T.]. We would like to thank Editage (https://www.editage.com) for English language editing.
Author contributions
H.T., W.S., and N.T. designed the experiments. H.T., M.S., L.S., KK.H., and H.I. conducted the experiments and analyzed the results. H.T. and W.S wrote the final versions of the paper. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hideki Takanashi, Email: atakana@g.ecc.u-tokyo.ac.jp.
Nobuhiro Tsutsumi, Email: atsutsu@g.ecc.u-tokyo.ac.jp.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88917-x.
References
- 1.Tuinstra, M. R., Grote, E. M., Goldsbrough, P. B. & Ejeta, G. Genetic analysis of post-flowering drought tolerance and components of grain development in Sorghum bicolor (L.) Moench. Mol. Breed.3, 439–448, 10.1023/A:1009673126345 (1997).
- 2.Ogbaga CC, Stepien P, Johnson GN. Sorghum (Sorghum bicolor) varieties adopt strongly contrasting strategies in response to drought. Physiol. Plant. 2014;152:389–401. doi: 10.1111/ppl.12196. [DOI] [PubMed] [Google Scholar]
- 3.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 4.McCormick RF, et al. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018;93:338–354. doi: 10.1111/tpj.13781. [DOI] [PubMed] [Google Scholar]
- 5.Price HJ, et al. Genome evolution in the genus Sorghum (Poaceae) Ann. Bot. 2005;95:219–227. doi: 10.1093/aob/mci015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinby JR, Karper RE. Inheritance of height in Sorghum. Agron. J. 1954;46:211–216. doi: 10.2134/agronj1954.00021962004600050007x. [DOI] [Google Scholar]
- 7.Hilley J, Truong S, Olson S, Morishige D, Mullet J. Identification of Dw1, a regulator of Sorghum stem internode length. PLoS ONE. 2016;11:e0151271. doi: 10.1371/journal.pone.0151271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi M, et al. Sorghum Dw1, an agronomically important gene for lodging resistance, encodes a novel protein involved in cell proliferation. Sci. Rep.-Uk. 2016;6:28366. doi: 10.1038/srep28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilley, J. L. et al. Sorghum Dw2 encodes a protein kinase regulator of stem internode length. Sci. Rep.-UK7, 10.1038/s41598-017-04609-5 (2017). [DOI] [PMC free article] [PubMed]
- 10.Multani DS, et al. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- 11.Spagnolli FC, Mace E, Jordan D, Borras L, Gambin BL. Quantitative trait loci of plant attributes related to Sorghum grain number determination. Crop Sci. 2016;56:3046–3054. doi: 10.2135/cropsci2016.03.0185. [DOI] [Google Scholar]
- 12.Rajkumar et al. Molecular mapping of genomic regions harbouring QTLs for root and yield traits in sorghum (Sorghum bicolor L. Moench). Physiol. Mol. Biol. Plants19, 409–419, 10.1007/s12298-013-0188-0 (2013). [DOI] [PMC free article] [PubMed]
- 13.Mocoeur A, et al. Stability and genetic control of morphological, biomass and biofuel traits under temperate maritime and continental conditions in sweet sorghum (Sorghum bicolour) Theor. Appl. Genet. 2015;128:1685–1701. doi: 10.1007/s00122-015-2538-5. [DOI] [PubMed] [Google Scholar]
- 14.Han LJ, et al. Fine mapping of qGW1, a major QTL for grain weight in sorghum. Theor. Appl. Genet. 2015;128:1813–1825. doi: 10.1007/s00122-015-2549-2. [DOI] [PubMed] [Google Scholar]
- 15.Gelli, M. et al. Mapping QTLs and association of differentially expressed gene transcripts for multiple agronomic traits under different nitrogen levels in sorghum. BMC Plant Biol.16, 10.1186/s12870-015-0696-x (2016). [DOI] [PMC free article] [PubMed]
- 16.Bai, C. M. et al. QTL mapping of agronomically important traits in sorghum (Sorghum bicolor L.). Euphytica213, 10.1007/S10681-017-2075-1 (2017).
- 17.Sakhi, S., Shehzad, T., Rehman, S. & Okuno, K. Mapping the QTLs underlying drought stress at developmental stage of sorghum (Sorghum bicolor (L.) Moench) by association analysis. Euphytica193, 433–450, 10.1007/s10681-013-0963-6 (2013).
- 18.Tao, Y. F. et al. Novel grain weight loci revealed in a cross between cultivated and wild Sorghum. Plant Genome-US11, 10.3835/plantgenome2017.10.0089 (2018). [DOI] [PMC free article] [PubMed]
- 19.Paterson AH, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic-loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 20.Boyles RE, et al. Quantitative trait loci mapping of agronomic and yield traits in two grain sorghum biparental families. Crop Sci. 2017;57:2443–2456. doi: 10.2135/cropsci2016.12.0988. [DOI] [Google Scholar]
- 21.Tao YF, et al. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 2020;18:1093–1105. doi: 10.1111/pbi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown PJ, et al. Inheritance of inflorescence architecture in sorghum. Theor. Appl. Genet. 2006;113:931–942. doi: 10.1007/s00122-006-0352-9. [DOI] [PubMed] [Google Scholar]
- 23.Feltus FA, et al. Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor. Appl. Genet. 2006;112:1295–1305. doi: 10.1007/s00122-006-0232-3. [DOI] [PubMed] [Google Scholar]
- 24.Murray, S. C. et al. Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci.48, 2165–2179, 10.2135/cropsci2008.01.0016 (2008).
- 25.Reddy, R. N. et al. Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet.126, 1921–1939, 10.1007/s00122-013-2107-8 (2013). [DOI] [PubMed]
- 26.Shehzad, T. & Okuno, K. QTL mapping for yield and yield-contributing traits in sorghum (Sorghum bicolor (L.) Moench) with genome-based SSR markers. Euphytica203, 17–31, 10.1007/s10681-014-1243-9 (2015).
- 27.Srinivas G, et al. Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor. Appl. Genet. 2009;118:1439–1454. doi: 10.1007/s00122-009-0993-6. [DOI] [PubMed] [Google Scholar]
- 28.Tao YF, et al. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 2019 doi: 10.1111/pbi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindo D, et al. Quantitative trait loci for sorghum grain morphology and quality traits: Toward breeding for a traditional food preparation of West-Africa. J. Cereal Sci. 2019;85:256–272. doi: 10.1016/j.jcs.2018.11.012. [DOI] [Google Scholar]
- 30.Zou GH, et al. Sorghum qTGW1a encodes a G-protein subunit and acts as a negative regulator of grain size. J. Exp. Bot. 2020;71:5389–5401. doi: 10.1093/jxb/eraa277. [DOI] [PubMed] [Google Scholar]
- 31.AuBuchon-Elder T, et al. Sterile spikelets contribute to yield in sorghum and related grasses ([OPEN]) Plant Cell. 2020;32:3500–3518. doi: 10.1105/tpc.20.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Q. M. et al. Fine mapping and candidate gene analysis of qSTL3, a stigma length-conditioning locus in rice (Oryza sativa L.). Plos One10, 10.1371/journal.pone.0127938 (2015). [DOI] [PMC free article] [PubMed]
- 33.Daniell H. Molecular strategies for gene containment in transgenic crops. Nat. Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena KB, Singh L, Ariyanayagam RP. Role of partial cleistogamy in maintaining genetic purity of pigeonpea. Euphytica. 1993;66:225–229. doi: 10.1007/Bf00025307. [DOI] [Google Scholar]
- 35.Ogami, T., Yasui, H., Yoshimura, A. & Yamagata, Y. Identification of anther length QTL and construction of chromosome segment substitution lines of Oryza longistaminata. Plants-Basel8, 10.3390/plants8100388 (2019). [DOI] [PMC free article] [PubMed]
- 36.Zhou H, et al. Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol. Plant. 2017;10:634–644. doi: 10.1016/j.molp.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Zhou SR, Xue HW. The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division. J. Integr. Plant Biol. 2020;62:847–864. doi: 10.1111/jipb.12851. [DOI] [PubMed] [Google Scholar]
- 38.Dang, X. et al. OsSYL2AA, an allele identified by gene-based association, increases style length in rice (Oryza sativa L.). Plant J.104, 1491–1503, 10.1111/tpj.15013 (2020). [DOI] [PMC free article] [PubMed]
- 39.Zhang Y, Xiong Y, Liu RY, Xue HW, Yang ZB. The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division. Proc. Natl. Acad. Sci. USA. 2019;116:16121–16126. doi: 10.1073/pnas.1902321116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan CH, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 41.Mao HL, et al. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 43.Weng JF, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 44.Jiao, Y. P. et al. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun.9, 10.1038/s41467-018-03238-4 (2018). [DOI] [PMC free article] [PubMed]
- 45.Gladman, N. et al. Fertility of pedicellate spikelets in sorghum is controlled by a jasmonic acid regulatory module. Int. J. Mol. Sci.20, 10.3390/ijms20194951 (2019). [DOI] [PMC free article] [PubMed]
- 46.Dampanaboina L, et al. Sorghum MSD3 encodes an ω-3 fatty acid desaturase that increases grain number by reducing jasmonic acid levels. Int. J. Mol. Sci. 2019;20:5359. doi: 10.3390/ijms20215359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kajiya-Kanegae H, et al. RAD-seq-based high-density linkage map construction and QTL mapping of biomass-related traits in Sorghum using the Japanese landrace Takakibi NOG. Plant Cell Physiol. 2020;61:1262–1272. doi: 10.1093/pcp/pcaa056. [DOI] [PubMed] [Google Scholar]
- 48.Ohnishi N, Wacera WF, Sakamoto W. Photosynthetic responses to high temperature and strong light suggest potential post-flowering drought tolerance of Sorghum Japanese landrace Takakibi. Plant Cell Physiol. 2019;60:2086–2099. doi: 10.1093/pcp/pcz107. [DOI] [PubMed] [Google Scholar]
- 49.Casady AJ, Miller FR. Inheritance of hermaphrodite pedicelled spikelets of sorghum. Crop Sci. 1970;10:612–613. doi: 10.2135/cropsci1970.0011183X001000050053x. [DOI] [Google Scholar]
- 50.Mace E, et al. The Sorghum QTL atlas: A powerful tool for trait dissection, comparative genomics and crop improvement. Theor. Appl. Genet. 2019;132:751–766. doi: 10.1007/s00122-018-3212-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang HL, et al. Identification and validation of QTLs controlling multiple traits in sorghum. Crop Pasture Sci. 2016;67:193–203. doi: 10.1071/CP15239. [DOI] [Google Scholar]
- 52.Murray, S. C. et al. Genetic improvement of sorghum as a biofuel feedstock: II. QTL for stem and leaf structural carbohydrates. Crop Sci.48, 2180–2193, 10.2135/cropsci2008.01.0068 (2008).
- 53.Bouchet S, et al. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics. 2017;206:573–585. doi: 10.1534/genetics.116.198499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felderhoff, T. J. et al. QTLs for energy-related traits in a sweet x grain sorghum [Sorghum bicolor (L.) Moench] mapping population. Crop Sci.52, 2040–2049, 10.2135/cropsci2011.11.0618 (2012).
- 55.Wang XM, et al. Two distinct classes of QTL determine rust resistance in sorghum. BMC Plant Biol. 2014;14:366. doi: 10.1186/s12870-014-0366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown PJ, Rooney WL, Franks C, Kresovich S. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics. 2008;180:629–637. doi: 10.1534/genetics.108.092239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai T, Yonemaru J, Kaidai H, Kasuga S. Quantitative trait locus analysis for days-to-heading and morphological traits in an RIL population derived from an extremely late flowering F-1 hybrid of sorghum. Euphytica. 2012;187:411–420. doi: 10.1007/s10681-012-0727-8. [DOI] [Google Scholar]
- 58.Li X, Li XR, Fridman E, Tesso TT, Yu JM. Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc. Natl. Acad. Sci. USA. 2015;112:11823–11828. doi: 10.1073/pnas.1509229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirano K, et al. Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep.-UK. 2017;7:126. doi: 10.1038/s41598-017-00096-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira MG, Lee M. Identification of genomic regions affecting plant height in sorghum and maize. Theor. Appl. Genet. 1995;90:380–388. doi: 10.1007/Bf00221980. [DOI] [PubMed] [Google Scholar]
- 61.Cassady AJ. Effect of a single height (Dw) gene of sorghum on grain yield, grain yield components, and test weight1. Crop Sci. 1965;5:385–388. doi: 10.2135/cropsci1965.0011183X000500050002x. [DOI] [Google Scholar]
- 62.Truong SK, McCormick RF, Rooney WL, Mullet JE. Harnessing genetic variation in leaf angle to increase productivity of Sorghum bicolor. Genetics. 2015;201:1229–U1809. doi: 10.1534/genetics.115.178608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madhusudhana, R. & Patil, J. V. A major QTL for plant height is linked with bloom locus in sorghum [Sorghum bicolor (L.) Moench]. Euphytica191, 259–268, 10.1007/s10681-012-0812-z (2013).
- 64.Wang HL, et al. Identification of QTLs for salt tolerance at germination and seedling stage of Sorghum bicolor L. Moench. Euphytica. 2014;196:117–127. doi: 10.1007/s10681-013-1019-7. [DOI] [Google Scholar]
- 65.Zou GH, et al. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J. Exp. Bot. 2012;63:5451–5462. doi: 10.1093/jxb/ers205. [DOI] [PubMed] [Google Scholar]
- 66.Hmon, K. P. W., Shehzad, T. & Okuno, K. QTLs underlying inflorescence architecture in sorghum (Sorghum bicolor (L.) Moench) as detected by association analysis. Genet. Resour. Crop Evol.61, 1545–1564, 10.1007/s10722-014-0129-y (2014).
- 67.Mace, E. S. & Jordan, D. R. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet.121, 1339–1356, 10.1007/s00122-010-1392-8 (2010). [DOI] [PubMed]
- 68.Rhodes, D. H. et al. Genome-wide association study of grain polyphenol concentrations in global sorghum [Sorghum bicolor (L.) Moench] germplasm. J. Agric. Food Chem.62, 10916–10927, 10.1021/jf503651t (2014). [DOI] [PubMed]
- 69.Boddu J, Svabek C, Ibraheem F, Jones AD, Chopra S. Characterization of a deletion allele of a sorghum Myb gene, yellow seed1 showing loss of 3-deoxyflavonoids. Plant Sci. 2005;169:542–552. doi: 10.1016/j.plantsci.2005.05.007. [DOI] [Google Scholar]
- 70.Ibraheem F, Gaffoor I, Chopra S. Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor. Genetics. 2010;184:915–926. doi: 10.1534/genetics.109.111831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grotewold E, Athma P, Peterson T. Alternatively spliced products of the maize P-gene encode proteins with homology to the DNA-binding domain of Myb-like transcription factors. Proc. Natl. Acad. Sci. USA. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fiedler K, et al. Genetic dissection of temperature-dependent sorghum growth during juvenile development. Theor. Appl. Genet. 2014;127:1935–1948. doi: 10.1007/s00122-014-2350-7. [DOI] [PubMed] [Google Scholar]
- 73.Guan YA, et al. QTL mapping of bio-energy related traits in Sorghum. Euphytica. 2011;182:431–440. doi: 10.1007/s10681-011-0528-5. [DOI] [Google Scholar]
- 74.Boyles, R. E. et al. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome-US9, 10.3835/plantgenome2015.09.0091 (2016). [DOI] [PubMed]
- 75.Hart, G. E., Schertz, K. F., Peng, Y. & Syed, N. H. Genetic mapping of Sorghum bicolor (L.) Moench QTLs that control variation in tillering and other morphological characters. Theor. Appl. Genet.103, 1232–1242, 10.1007/s001220100582 (2001).
- 76.Zhang D, et al. Genetic analysis of inflorescence and plant height components in sorghum (Panicoidae) and comparative genetics with rice (Oryzoidae) BMC Plant Biol. 2015;15:107. doi: 10.1186/s12870-015-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chopra, R., Burow, G., Burke, J. J., Gladman, N. & Xin, Z. G. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol.17, 10.1186/s12870-016-0966-2 (2017). [DOI] [PMC free article] [PubMed]
- 78.Morris GP, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao, J., Perez, M. B. M., Hu, J. Y. & Fernandez, M. G. S. Genome-wide association study for nine plant architecture traits in Sorghum. Plant Genome-US9, 10.3835/plantgenome2015.06.0044 (2016). [DOI] [PubMed]
- 80.Higgins, R. H., Thurber, C. S., Assaranurak, I. & Brown, P. J. Multiparental mapping of plant height and flowering time QTL in partially isogenic sorghum families. G3-Genes Genom. Genet.4, 1593–1602, 10.1534/g3.114.013318 (2014). [DOI] [PMC free article] [PubMed]
- 81.Heang D, Sassa H. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 2012;62:133–141. doi: 10.1270/jsbbs.62.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin A, et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006;18:3252–3274. doi: 10.1105/tpc.106.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Habgood RM, Uddin MR. Some effects of artificial variation in light interception, number of grains and husk constriction on the development of grain weight in normal and high-lysine barley. J. Agric. Sci. 1983;101:301–309. doi: 10.1017/S002185960003759x. [DOI] [Google Scholar]
- 84.Scott WR, Appleyard M, Fellowes G, Kirby EJM. Effect of genotype and position in the ear on carpel and grain-growth and mature grain weight of spring barley. J. Agric. Sci. 1983;100:383–390. doi: 10.1017/S0021859600033530. [DOI] [Google Scholar]
- 85.Brinton J, Uauy C. A reductionist approach to dissecting grain weight and yield in wheat. J. Integr. Plant Biol. 2019;61:337–358. doi: 10.1111/jipb.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okamoto Y, Takumi S. Pleiotropic effects of the elongated glume gene P1 on grain and spikelet shape-related traits in tetraploid wheat. Euphytica. 2013;194:207–218. doi: 10.1007/s10681-013-0916-0. [DOI] [Google Scholar]
- 87.George-Jaeggli B, Jordan DR, van Oosterom EJ, Hammer GL. Decrease in sorghum grain yield due to the dw3 dwarfing gene is caused by reduction in shoot biomass. Field Crops Res. 2011;124:231–239. doi: 10.1016/j.fcr.2011.07.005. [DOI] [Google Scholar]
- 88.Baird, N. A. et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. Plos One3, 10.1371/journal.pone.0003376 (2008). [DOI] [PMC free article] [PubMed]
- 89.Kobayashi M, et al. Heap: A highly sensitive and accurate SNP detection tool for low-coverage high-throughput sequencing data. DNA Res. 2017;24:397–405. doi: 10.1093/dnares/dsx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakamoto, L. et al. Comparison of shape quantification methods for genomic prediction, and genome-wide association study of sorghum seed morphology. Plos One14, 10.1371/journal.pone.0224695 (2019). [DOI] [PMC free article] [PubMed]
- 91.R-Development-Core-Team. R: A Language and Environment for Statistical Computing. (R-Development-Core-Team, 2018).
- 92.Taiyun, W.V.S. R Package "corrplot": Visualization of a Correlation Matrix (Version 0.84). https://github.com/taiyun/corrplot (2017).
- 93.Kosambi DD. The estimation of map distances from recombination values. Ann. Eugenic. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 94.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 95.Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/Hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 96.Villanueva, R. A. M. & Chen, Z. J. ggplot2: Elegant graphics for data analysis, 2nd edition. Meas.-Interdiscip. Res.17, 160–167, 10.1080/15366367.2019.1565254 (2019).
- 97.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 98.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics. 2017;33:2197–2198. doi: 10.1093/bioinformatics/btx144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.