Abstract
Background
Corona virus disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus −2 (SARS-CoV-2) has created ruckus throughout the world. Growing epidemiological studies have depicted atherosclerosis as a comorbid factor of COVID-19. Though both these diseases are triggered via inflammatory rage that leads to injury of healthy tissues, the molecular linkage between them and their co-influence in causing fatality is not yet understood.
Methods
We have retrieved the data of differentially expressed genes (DEGs) for both atherosclerosis and COVID-19 from publicly available microarray and RNA-Seq datasets. We then reconstructed the protein-protein interaction networks (PPIN) for these diseases from protein-protein interaction data of corresponding DEGs. Using RegNetwork and TRRUST, we mapped the transcription factors (TFs) in atherosclerosis and their targets (TGs) in COVID-19 PPIN.
Results
From the atherosclerotic PPIN, we have identified 6 hubs (TLR2, TLR4, EGFR, SPI1, MYD88 and IRF8) as differentially expressed TFs that might control the expression of their 17 targets in COVID-19 PPIN. The important target proteins include IL1B, CCL5, ITGAM, IFIT3, CXCL1, CXCL2, CXCL3 and CXCL8. Consequent functional enrichment analysis of these TGs have depicted inflammatory responses to be overrepresented among the gene sets.
Conclusion
Finally, analyzing the DEGs in cardiomyocytes infected with SARS-CoV-2, we have concluded that MYD88 is a crucial linker of atherosclerosis and COVID-19, the co-existence of which lead to fatal outcomes. Anti-inflammatory therapy targeting MYD88 could be a potent strategy for combating this comorbidity.
Keywords: Atherosclerosis, COVID-19, SARS-CoV-2, Transcription factors, Differentially expressed genes, Protein-protein interaction network
Graphical abstract
1. Introduction
The global pandemic due to SARS-CoV-2 scare has brought life to a standstill. It has been named corona virus disease 2019 (COVID-19) and it affects the respiratory mechanism after being inhaled into the lungs [1]. Although a year has passed after the outbreak, as of 10th January 2021, 88, 383, 771 confirmed cases and 1,919,126 deaths with 12,454 new deaths worldwide in the last 24 h had been reported by World Health Organization (WHO). The main symptoms of this disease is dry cough with severe pneumonia and respiratory failure which leads to cardiac arrest [1]. The viral RNA for SARS-CoV-2 is internalized within the cells to produce a greater number of viral progeny and this triggers the primary immune response of the body [2]. As the primary immune response is triggered, large amount of cytokines and chemokines are released from the site of infection to cause inflammation in the alveolar tissue and this results in ARDS (Acute respiratory distress syndrome) [3]. ARDS is a key pathological symptom for viral infection of the lungs that is caused due to rapid apoptosis of alveolar cells infected with the virus thereby leading to severe hypoxia [4]. This inflammation mediated injury to the alveolar cells caused by cytokine release has been popularly termed as “cytokine storm” [3].
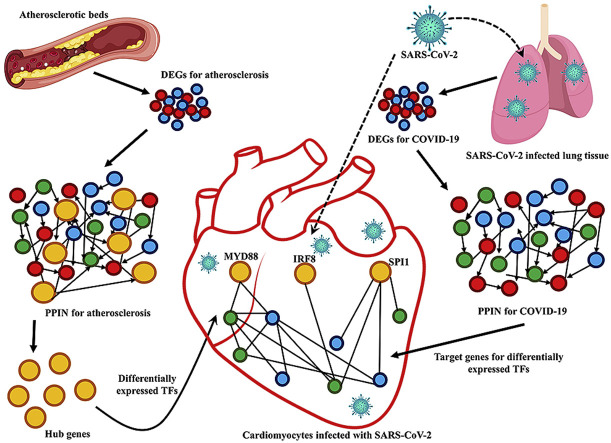
Concurrently, several reports from the epicenter of COVID-19 i.e., Wuhan, China had stated that patients with cardiovascular disease (CVD) have greater risk of COVID-19 mediated implications [[5], [6], [7], [8]]. Several studies from different countries have documented that mortality rate of COVID-19 patients having pre-existing cardiovascular disease varies between 11% and 19% [[9], [10], [11], [12]]. It was found that 50% of the patients with coronary artery calcifications (CAC+) were severely affected by COVID-19, whereas only 17% of the patients with no coronary calcifications were severely affected [13]. Moreover, a possible correlation between COVID-19 and atherosclerosis has been shown to amplify the immune response of the body and instigating acute coronary syndromes [14]. It has also been proposed that atherosclerosis is the most common form of cardiovascular disease (CVD) which is characterized by the buildup of lipid laden macrophages (foam cells) within walls of large arteries like coronary artery [15]. Accumulation of apoB-LPs, that are formed from LDL, attracts monocytes which orchestrate the early inflammatory response against the congregated apoB-LPs in the arterial intima [16]. The activated endothelial (EC) cells of the arterial wall secrete chemokines such as CCL5 and CXCL1 which facilitates entry of monocytes in the intimal space [17,18]. Monocytes, after their conversion to foam cells, secrete various chemokines like IL-6, IL-12 and TNF-α to mediate inflammatory response in this disease [16]. In addition to that, ACE2, a negative regulator of RAS (renin–angiotensin system) has been proved to play an important role in progression of atherosclerosis since it converts angiotensin-II (Ang-II) into the vasodilator Ang-(1–7) [19]. Ang-(1–7) prevents plaque rupture and inflammation in atherosclerosis [19]. Thus, deletion of ACE2 causes vascular inflammation that is mediated by Ang-II [20]. In case of COVID-19, Varga et al. had shown that SARS-CoV-2 viral particles enter into endothelial cells via binding with ACE2 [21]. Binding of the virus with ACE2 impairs its ability to convert Ang-II to Ang-(1–7) which results in increase in Ang-II and a decrease in Ang-(1–7). The increased Ang-II promotes inflammation and oxidative stress that leads to development of atherosclerosis [22]. Several reports have confirmed that SARS-CoV-2 may contribute to endothelial dysfunction, either directly via binding with ACE2 or indirectly via cytokine storm [19,23,24]. Thus, endothelial dysfunction and stress to the cardiomyocytes may be worsened due to COVID-19 in atherosclerotic patients. Kadosh et al., in 2020 hypothesized that both atherosclerosis and SARS-CoV-2 may be amplifying NLRP3 inflammasome response and thereby resulting in hyper inflammation [25]. Endothelial dysfunction has also been found to amplify atherosclerosis in patients infected with other viruses like HCV and HIV [26,27]. The aforementioned fact that SARS-CoV-2 infection has a greater manifestation in patients co-morbid for atherosclerosis made us curious to unravel the molecular events bridging these diseases. For this, we have reconstructed networks for atherosclerosis and COVID-19 by using Protein- Protein Interaction (PPI) data of Differentially Expressed Genes (DEGs) for both the diseases. Then, using network and systems biology approach, we have identified MYD88, SPI1 and IRF8 as differentially expressed TFs during atherosclerosis. These DETFs could regulate expression of an array of cytokines and chemokines which are observed to be differentially expressed in cardiomyocytes infected with SARS-CoV-2. Thus, our study could help in designing anti-inflammatory therapeutics to combat COVID-19 comorbidity.
2. Materials and methods
2.1. Data processing
For microarray datasets of atherosclerosis – GSE28829 [28] and GSE100927 [29], the gene specific raw expression values were log2 transformed and then eBayes and top Table function of limma package of R v3.6.3 [30] were used to determine DEGs. For the three RNA-Seq datasets of COVID-19- GSE147507 [31], GSE150819 [32], GSE150392 [33], the read counts were generated using R subread package in R v3.6.3 [30] and then the raw read counts were analyzed using DESeq2 package in R [34,35]. DESeq2 calculates DEGs using negative binomial distribution.
2.2. Identification of DEGs for atherosclerosis and COVID-19
In order to prepare a list of DEGs for atherosclerosis we have considered DEGs from the study by Sulkava et al. [36] along with the DEGs from two microarray datasets–GSE28829 [28], GSE100927 [29]. For GSE28829 [28], the DEGs were screened by comparing early atherosclerotic plaque samples to advanced atherosclerotic plaque samples from carotid artery. In GSE100927 [29], all the DEGs were screened from three atherosclerotic beds namely-carotid artery, infra-popliteal artery and femoral artery whereas aorta, carotid artery, femoral artery were considered for Sulkava et al. [36]. DEGs were identified by considering |log2FC|>1.0 and adjusted P < 0.05 as the cutoff. The fold change (FC) values for the genes from the study by Sulkava et al. [36]were log2 transformed in order to maintain uniformity of expression quantification across the three datasets. Next, we have created a list of 171 DEGs that are common in all three datasets.
To screen DEGs in SARS-CoV-2 infected lung cells, we have considered common DEGs from two RNA-Seq datasets- GSE147507 [31] and GSE150819 [32]. For GSE147507 [31], we considered primary human lung epithelium (NHBE) cells with 3 replicates each for treated and untreated cells with SARS-CoV-2. For GSE150819 [32], human bronchial organoids (hBO) derived from cryopreserved primary human bronchial epithelial cells (hBEpC) was used with 3 replicates each for treated and untreated cells with SARS-CoV-2. DEGs were identified by considering |log2FC|>1.0 and adjusted P < 0.05 as the cutoff. A total of 100 DEGs were found to be common in these two datasets.
The expression of DETF-TG interactions were crosschecked in RNA-Seq dataset - GSE150392 [33] where, human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) was used as a model to examine the mechanisms of cardiomyocyte-specific infection by SARS-CoV-2.
2.3. Reconstruction of networks for atherosclerosis and COVID-19
In order to construct networks for both atherosclerosis and COVID-19, we have used STRINGv11.0(confidence score >0.4) [37]. These networks contain both physical and functional interactions. Next, in order to increase network size, we have added additional 1st shell of interactors (only direct interactors) to the number of DEGs in both the networks. Following this way, we have added 171 and 100 interactors of the DEGs to the 171 and 100 DEGs for atherosclerotic and COVID-19 network respectively. Finally, we have received an atherosclerotic network with 324 nodes and a COVID-19 network with 190 nodes. The network for atherosclerosis was analyzed using Network Analyzer of Cytoscape 3.8.0 [38,39].To compute the topological parameters of this undirected network, degree was considered to identify the hub genes. Only major and super hubs having degree between 51 and 100 and > 100 respectively [40] were considered for selecting hub genes in the network.
2.4. Functional and pathway enrichment analysis of hub genes and TGs
The functional analysis of hub genes and TGs of DETFs has been done in DAVIDv6.8(Database for Annotation, Visualization and Integrated Discovery) [41] by considering FDR < 0.05. Results from four Pathway enrichment analysis databases-KEGG, BioPlanet, Wiki pathways and Reactome was generated in Enrichr for both hub genes and TGs [42] by considering adj. P value < 0.05.
2.5. Identifying crucial TF-TG interactions that are mediating immunological response in cardiomyocytes infected with SARS-CoV-2
The TFs among the hub genes of atherosclerotic network and their TGs in COVID-19 network were identified using TRRUST v2 [43] and RegNetwork [44] transcription factor regulation databases.
2.6. Statistical analysis
The DEGs were identified by determining fold change cut off of |log2FC|>1.0 and adj. P value < 0.05 for all the microarray or RNA-Seq datasets.
For GO functional enrichment analysis and Pathway enrichment analysis, a one-tailed variant of Fisher's exact test i.e. hypergeometric test was used to identify significant over-represented GO terms and pathways [42,45]. FDR<0.05 and adj. P value < 0.05 were considered as cut off for functional and pathway enrichment analysis respectively.
2.7. Graphical designing
Graphical plots were made using EnhancedVolcano, ggplot2 package of R v3.6.3 [35,46], Seaborn package of Python 3.8.3 [47] and Cytoscape 3.8.0 [38]. The Venn diagrams were created in http://bioinformatics.psb.ugent.be/webtools/Venn/.
3. Results
3.1. Identification of hub genes in atherosclerosis protein-protein interaction network
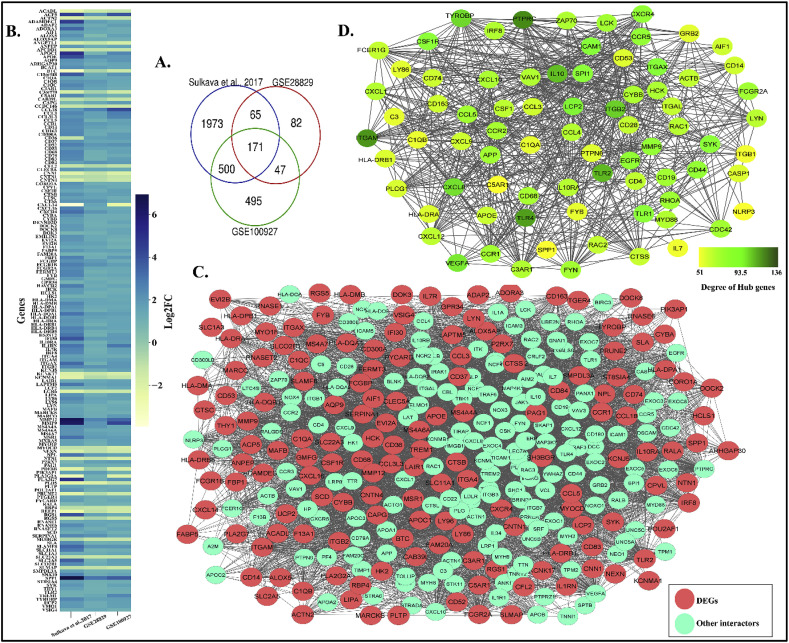
Typically, when a biological system goes into a state of imbalance like in disease condition, the key hub genes in protein-protein interaction network (PPIN) are mostly either up or down-regulated as they attempt to revamp the system from the state of imbalance [48]. To identify the key hub genes associated with atherosclerosis, we initially reconstructed protein-protein interaction network (PPIN) with the differentially expressed genes (DEGs) during the disease state. The DEGs has been screened from different resources [36], GSE28829 [28], GSE100927 [29] by taking |log2FC|>1.0 and adj. P value < 0.05 as cut off. A total of 171 DEGs are found to be common in three datasets (Fig. 1 A and B) (Refer Supplementary File S1). The atherosclerosis PPIN (AtsPPIN) consisting of 324 nodes and 5572 edges has been reconstructed by incorporating 171 1st shell of interactors to DEGs (Fig. 1C) using STRING v11 (confidence score >0.4) [37].
Fig. 1.
DEGs in atherosclerosis, reconstruction of atherosclerosis PPIN and identification of hub genes. (A.) Venn diagram representing DEGs derived from the three atherosclerotic studies. (B.) Heatmap comparing the expression of DEGs for the three atherosclerotic studies. (C.) PPIN for atherosclerosis. The red nodes represent DEGs in the PPIN and other interactors of the network are cyan colored. (D.) Sub network of hub genes from main PPIN for atherosclerosis with nodes having variable color depth to represent the degree centrality of hub genes.
Hub genes are the nodes in a network that are most inter-connected i.e., has the maximum number of edges. To identify the hub genes in AtsPPIN, the degree of the network was calculated using network analyzer in Cytoscape 3.8.0 [38,39]. We considered a total of 78 hub genes (major and super hubs) which have degree > 50 (Fig. 1D) [40]. We have found that PTPRC has the highest degree (v = 136) (Fig. 1D) in the network. PTPRC was earlier known as CD45 and it is one of the requisite factors for T-cell activation in atherosclerotic plaques [49,50]. Simultaneously, it has also been found as a potent marker for distinguishing severe and non-severe pneumonia in COVID-19 patients [51]. Thus, it would be worthwhile to explore the roles of these hub genes in establishing the co-occurrence of atherosclerosis and COVID-19.
3.2. Functional and pathway enrichment analysis of hub genes in AtsPPIN
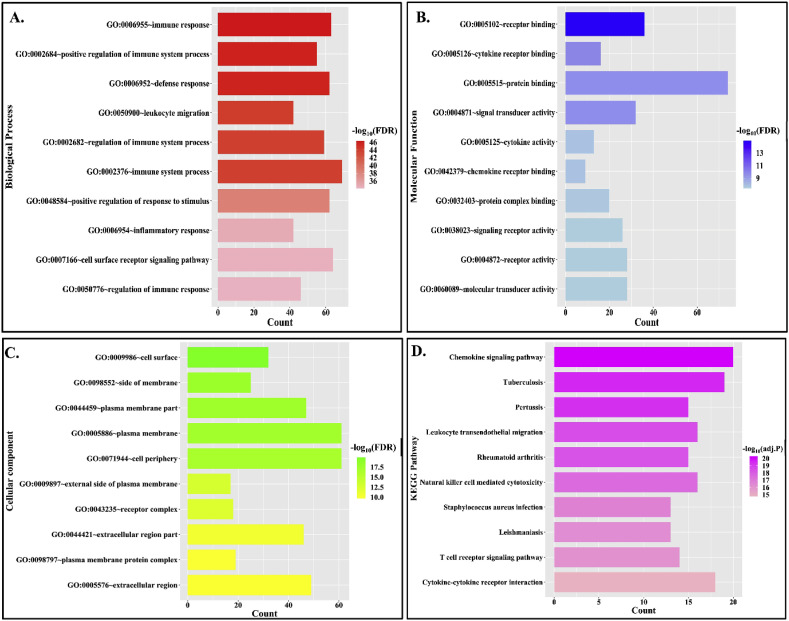
Functional enrichment analysis of hub genes in DAVID v6.8 [41] has shown that GO: 0006955 (immune response) is the most significantly enriched GO biological process in which 63 out of 78 hub genes are found to be enriched with this function (Fig. 2 A) (Refer Supplementary File S2). Consequently, GO:0005102 (receptor binding) and GO:0009986 (Cell surface) are observed to be the most enriched molecular function and cellular component respectively (Fig. 2B and C) (Refer Supplementary File S2). Thus from functional enrichment analysis, it could be stated that cell surface receptors mediating immune response are dominant among the hub genes. The KEGG pathway enrichment has shown “Chemokine signaling pathway” (Fig. 2D) (Refer Supplementary File S2) as the most significant pathway which was also verified by using three more pathway enrichment analyses, such as BioPlanet, WikiPathway and Reactome. All these analyses have depicted immunological response as the most enriched pathway (Refer Supplementary File S2 and Supplementary Fig. F1). Earlier it was reported that immunological inflammation mediated by cytokine release is the driving factor for coronary dysfunction during respiratory tract infection [52]. Moreover, it is considered as the fundamental cause for progression of both atherosclerosis and COVID-19 [3,53]. These results indicate that predisposition of atherosclerosis may amplify the inflammatory response during SARS-CoV-2 infection.
Fig. 2.
Functional and pathway enrichment analysis for hub genes in atherosclerosis PPIN. (A.) GO-biological process enrichment of hub genes. The bar plot represents no. of genes enriched for each function and the color gradient represents significance value (FDR <0.05). (B.) GO-molecular function enrichment of hub genes. The bar plot represents no. of genes enriched for each function and the color gradient represents its significance value (FDR <0.05). (C.) GO- cellular component enrichment of hub genes. The bar plot represents no. of genes enriched for each function and the color gradient represents its significance value (FDR <0.05). (D.) KEGG pathway enrichment of hub genes. The bar plot represents no. of genes enriched for each function and the color gradient represents its significance value (adj. P value < 0.05).
3.3. Identification of differentially expressed transcription factors (DETFs) among the hub genes of AtsPPIN
It was evident earlier that hub proteins in human PPIN could be transcription factors [54]. These hub genes would not only interact but also regulate the expression of its downstream genes. Thus, screening of TFs from hub genes could help to identify effective biomarkers for disease progression [55]. We therefore mapped TFs from 78 hub genes of AtsPPIN with a curated list of transcription factors constructed by integrating TF data of two human regulatory databases-TRRUST v2 and RegNetwork [43,44] (Table 1 ). We identified six hub genes, namely – TLR2, TLR4, EGFR, SPI1, MYD88 and IRF8 as TFs. All these TFs are significantly differentially expressed in at least one of the atherosclerosis datasets taken in our study (Table 1). Among the six TFs, TLR4 is observed to encompass highest connectivity (v = 124) in AtsPPIN. The inflammatory role of TLR4 in atherosclerosis is well established [56]. Interestingly, TLR4 has also been proven to play crucial roles in COVID-19 as it could bind with a multi-epitope vaccine produced from virion's outer surface proteins (E, M and S) [57]. Notable target genes (TGs) of these atherosclerotic DETFs included interleukins like IL18 and IL10 that are key players of the cytokine storm in COVID-19 [58,59]. Thus, investigating the plausible roles of the downstream genes of atherosclerosis DETFs (if any) in triggering severity during SARS-COV-2 infection could give us the possible clues about the molecular link between these two diseases.
Table 1.
Differential expression (log2FC values) of atherosclerotic hub DETFs in atherosclerotic datasets and their TGs in COVID-19 datasets.
| TFs among atherosclerosis hub genes | Degree of the TF in Atherosclerosis network | Expression of TFs in atherosclerosis datasets |
TGs in COVID19 network | Direction in TRRUST/RegNetwork | Degree of the TG in COVID19 network | Expression of TGs in COVID-19 Datasets |
|||
|---|---|---|---|---|---|---|---|---|---|
| Sulkava et al., 2017 | GSE28829 | GSE100927 (Carotid) | GSE147507 (NHBE cells) | GSE150819 | |||||
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | CXCL8 | Activation | 81 | not significant | 2.2 |
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | IL1B | Activation | 73 | 1.1 | 3.4 |
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | CXCL1 | Activation | 55 | 1.4 | 1.6 |
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | CXCL2 | Activation | 42 | 1.4 | 2.2 |
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | CXCL3 | Activation | 26 | 2.3 | 2.6 |
| MYD88 | 72 | 1.1 | 0.4 | 1.0 | TNF | Activation | 109 | 1.9 | not significant |
| IRF8 | 72 | 2.6 | 1.1 | 1.6 | IL1B | Activation | 73 | 1.1 | 3.4 |
| IRF8 | 72 | 2.6 | 1.1 | 1.6 | IL10 | Unknown | 77 | not significant | not significant |
| IRF8 | 72 | 2.6 | 1.1 | 1.6 | TLR3 | Repression | 80 | −0.3 | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | IL1B | Activation | 73 | 1.1 | 3.4 |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | CCL5 | Unknown | 69 | not significant | 3.4 |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | ITGAM | Activation | 59 | not significant | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | IFIT3 | Activation | 34 | 0.7 | 2.2 |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | CD40 | Activation | 55 | 0.3 | 0.9 |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | CSF2RA | Unknown | 16 | not significant | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | CSF3R | Activation | 15 | not significant | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | IL18 | Unknown | 52 | not significant | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | ITGAX | Repression | 46 | not significant | not significant |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | ITGB2 | Activation | 29 | 1.3 | 1.1 |
| SPI1 | 91 | 2.2 | 0.4 | 1.6 | TNF | Unknown | 109 | 1.9 | not significant |
| TLR4 | 128 | 3.7 | nil | 0.6 | TNF | Activation | 109 | 1.9 | not significant |
| TLR2 | 119 | 2.3 | 1.1 | 1.1 | TNF | Activation | 109 | 1.9 | not significant |
| EGFR | 86 | −1.3 | −0.43 | −0.9 | |||||
3.4. Mapping of atherosclerosis DETFs targets in COVID-19 PPIN
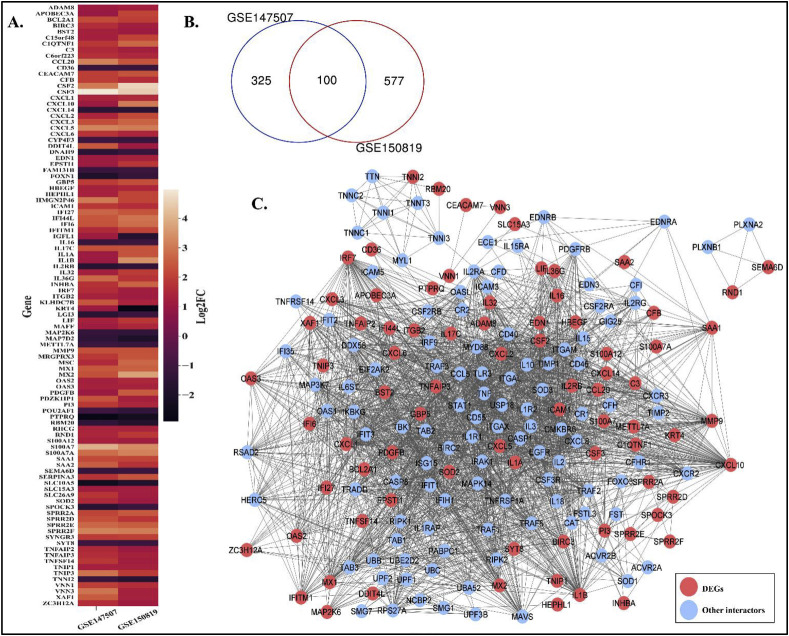
Similar to atherosclerosis, initially we screened out DEGs for COVID-19 from two transcriptome datasets – GSE147507 [31] and GSE150819 [32]. A total of 100 DEGs (Fig. 3 A) (Refer Supplementary File S1) are found to be intersecting across these two COVID-19 studies (Fig. 3B). With these DEGs we have reconstructed COVID-19 PPIN consisting of 190 nodes and 2468 edges (Fig. 3C) by following the protocol that is used for AtsPPIN.
Fig. 3.
DEGs in COVID-19 and the reconstruction of COVID-19 PPIN. (A.) Venn diagram representing DEGs derived from the two COVID-19 studies. (B.) Heatmap comparing the expression of DEGs for the two COVID-19 studies. (C.) PPIN for COVID-19. The red nodes represent DEGs in the PPIN and other interactors of the network are blue colored.
Since deregulated TFs in a disease condition control the expression of their downstream genes, it would be important to find out their targets to unravel their roles in triggering abnormalities in biological system. To explore the efficacy of DETFs in stimulating viral infection in atherosclerotic patients, we mapped the target genes (TGs) of DETFs in COVID-19 network from the TF-TG interaction data received from TRRUST v2 and RegNetwork [43,44]. Five DETFs namely – TLR2, TLR4, SPI1, MYD88 and IRF8 were found to have a total of 17 targets in COVID-19 PPIN (Table 2 ). The number of TGs in COVID-19 PPIN for MYD88, SP11, IRF8 are six, eleven and three respectively (Table 2). Both TLR2 and TLR4 have a common TG i.e. TNF in the COVID-19 network. Moreover, TNF is represented as a common target of four DETFs - TLR2, TLR4, SPI1 and MYD88 (Table 2). We have also calculated the degree of the DETF's targets present in COVID-19 network using network analyzer in Cytoscape 3.8.0 [38,39]. to find out their impact in COVID-19. In this network, TNF is found to hold the highest connectivity (v = 109) indicating its strong influence in the COVID-19 network. Being a pro-inflammatory cytokine, TNF has been shown to mediate inflammatory response in atherosclerotic plaques [60]. Likewise, overexpression of cytokines during SARS-CoV-2 infection is widely reported as severe COVID-19 patient hallmark i.e. so-called “cytokine storm syndrome” [3]. So anti-TNF therapy is also being considered as a strategy for COVID-19 treatment [60,61]. Taken together these evidences indicate that co-occurrence of both atherosclerosis and COVID-19 may cause mayhem in these comorbid patients. Thus, functional categorization of DETFs' targeted genes is a prerequisite to unveil their roles in the progression of SARS-CoV-2 infection in atherosclerotic patients.
Table 2.
Differential expression (log2FC values) of the DETFs and their TGs in cardiomyocytes infected with SARS-CoV-2 (GSE150392).
| TFs among atherosclerosis hub genes | Expression of the TF | TGs in COVID19 network | Expression of the TG |
|---|---|---|---|
| MYD88 | 2.0 | CXCL8 | 3.8 |
| MYD88 | 2.0 | IL1B | 6.6 |
| MYD88 | 2.0 | CXCL1 | 6.1 |
| MYD88 | 2.0 | CXCL2 | 5.6 |
| MYD88 | 2.0 | CXCL3 | 3.9 |
| MYD88 | 2.0 | TNF | not significant |
| IRF8 | 3.7 | IL1B | 6.6 |
| IRF8 | 3.7 | IL10 | not significant |
| IRF8 | 3.7 | TLR3 | not significant |
| SPI1 | not significant | IL1B | 6.6 |
| SPI1 | not significant | CCL5 | 4.4 |
| SPI1 | not significant | ITGAM | 2.8 |
| SPI1 | not significant | IFIT3 | 5.3 |
| SPI1 | not significant | CD40 | not significant |
| SPI1 | not significant | CSF2RA | not significant |
| SPI1 | not significant | CSF3R | not significant |
| SPI1 | not significant | IL18 | not significant |
| SPI1 | not significant | ITGAX | not significant |
| SPI1 | not significant | ITGB2 | not significant |
| SPI1 | not significant | TNF | not significant |
| TLR4 | 3.0 | TNF | not significant |
| TLR2 | 1.9 | TNF | not significant |
| EGFR | 1.3 | ∗∗∗ | ∗∗∗ |
3.5. Functional and pathway analysis of DETFs’ targeted genes
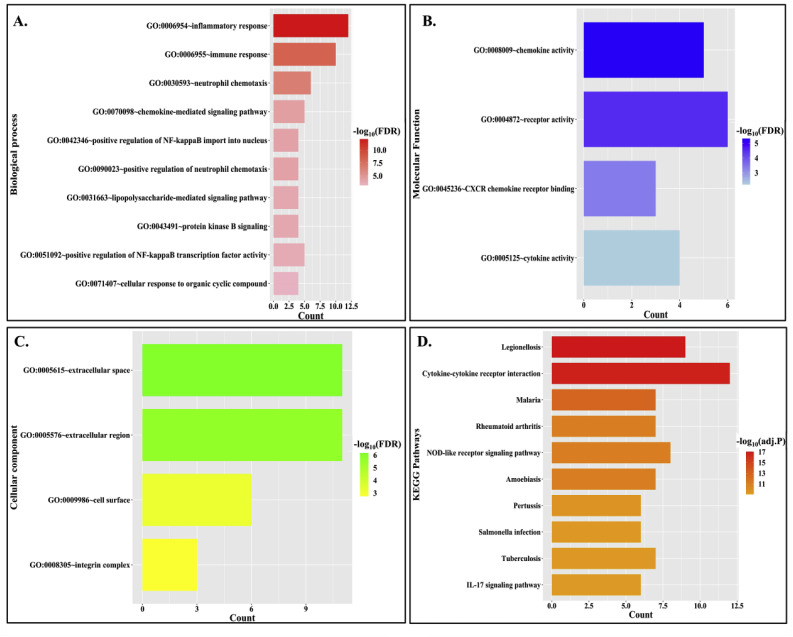
Functional enrichment analysis of atherosclerotic DETFs’ targets present in COVID-19 PPIN using DAVID v6.8 [41] has shown that GO:0006954 (Inflammatory response) is the most significantly enriched GO biological process with 12 TGs (out of 17 TGs) enriched for this GO function (Fig. 4 A) (Refer Supplementary File S2). Subsequently, GO:0008009 (Chemokine activity) and GO:0005615 (Extracellular space) are detected as the most enriched molecular function and cellular component, respectively (Fig. 4B and C) (Refer Supplementary File S2). The KEGG pathway enrichment has shown “Legionellosis” (Fig. 4D) (Refer Supplementary File S2) as the most significant pathway. Legionellosisis a type inflammatory pneumonia caused by the bacteria Legionella pneumophila [62]. The other pathway enrichment analyses (BioPlanet, WikiPathway and Reactome) have also indicated that the immunological response mediated by cytokines as the most enriched pathway (Refer Supplementary File S2 and Supplementary Fig. F2). Thus, it could be inferred from the analysis that deregulated TFs in atherosclerosis might cause either over expression or lower expression of their downstream genes that are mostly involved in inflammatory response. This inflammatory immune turmoil could be the potent reason for increased casualty in these co-morbid patients.
Fig. 4.
Functional and pathway enrichment analysis of atherosclerotic DETFs targeted genes in COVID-19 network. (A.) GO-biological process enrichment of TGs in COVID-19 network for atherosclerotic DETFs. The bar plot represents no. of genes enriched for each function and the color gradient represents significance value (FDR <0.05). (B.) GO-molecular function enrichment of TGs in COVID-19 network for atherosclerotic hub DETFs. The bar plot represents no. of genes enriched for each function and the color gradient represents its significance value (FDR <0.05). (C.) GO- cellular component enrichment of TGs in COVID-19 network for atherosclerotic hub DETFs. The bar plot represents the no. of genes enriched for each function and the color gradient represents its significance value (FDR <0.05). (D.) KEGG pathway enrichment of TGs in COVID-19 network for atherosclerotic hub DETFs. The bar plot represents the no. of genes enriched for each function and the color gradient represents its significance value (adj. P value < 0.05).
3.6. Expression analysis of DETFs and their TGs in cardiomyocytes infected with SARS-CoV-2
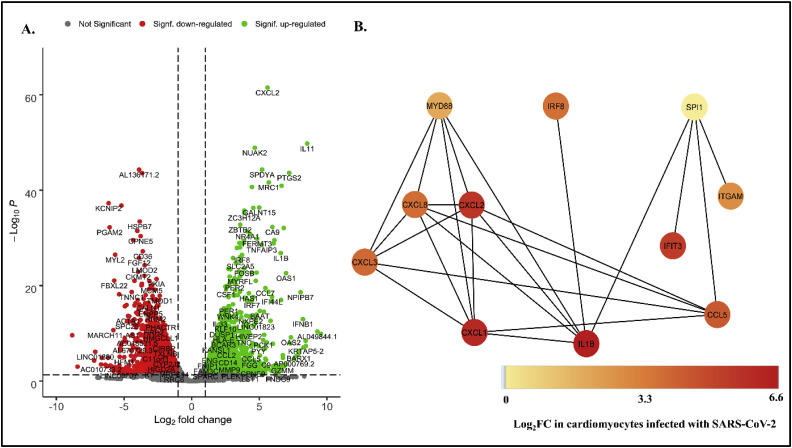
The regulatory circuit in a biological system is tightly regulated and highly orchestrated. Dysregulation in the higher order of a network might cause deregulation at the lower order. Thus the TFs in AtsPPIN and their corresponding TGs in COVID-19 PPIN, if found to be differentially expressed in comorbid patients, it could be stated that DETFs in atherosclerotic patients might be responsible for causing deregulation of their downstream genes, which in turn may be aggravating viral infection via amplified inflammatory response. For this we have checked the expression analogy between TFs and TGs from the RNAseq dataset GSE150392 [32] in which cardiomyocytes were infected with SARS-CoV-2.The DEGs (|log2FC|>1.0; adj.P value < 0.05) were screened (Fig. 5 A) and mapped to candidate TF-TGs. We have noticed that except SPI1, all other four atherosclerotic DETFs (TLR2, TLR4, MYD88 and IRF8) are significantly over expressed in SARS-CoV-2 infected cardiomyocytes (Fig. 5B; Table 2). Moreover, we noticed that the five targets out of six TGs of MYD88 (83.3%) found in COVID-19 PPIN are upregulated in comorbid case (Table 2). Two important TGs that might be upregulated by MYD88 include CXCL8 and IL1B which themselves are potential hub genes in COVID-19 network (Table 1). In RegNetwork, it is found that MYD88 acts as an activator for the expression of the cytokines including CXCL1, CXCL2, CXCL3, CXCL8 (Table 1) and all of these cytokines along with MYD88 are found to be significantly overexpressed in comorbid dataset (Table 2). Moreover, we have observed that IL1B, the common target of three DETFs - IRF8, SPI1 and MYD88, is upregulated in this dataset. Upregulation of IL1B could be influenced by the upregulation of IRF8 and MYD88 because both of them are found to be the activators of IL1B gene expression (Fig. 5B). IL1B which encodes IL-1β is a pro-inflammatory cytokine and one of the important factors for triggering innate immune response mediated inflammation in the lungs of COVID-19 patients and atherosclerotic beds [63]. Though TLR2 and TLR4 are found to be upregulated in comorbid sample but their only target gene TNF is not significantly differentially expressed in cardiomyocytes infected with SARS-CoV-2.
Fig. 5.
Expression and network analysis of DETF- TG interactions. (A.) Volcano plot representing the expression of genes in cardiomyocyte infected with SARS-CoV-2. Genes are considered as significantly expressed only if adj. P value < 0.05 and |log2FC|>1.0. The green dots represent significantly upregulated genes, red dots represent significantly downregulated genes and grey dots represent non-significant DEGs. (B.) TF-TG interaction in cardiomyocytes infected with SARS-CoV-2.
It is known that innate immune response along with increase in macrophage activity is the primary retort for early inflammatory diseases like CVD and SARS-CoV-2 [3,64]. So it could be inferred from our analysis that in case of patients co-morbid for atherosclerosis and COVID-19, MYD88 and IRF8 play crucial roles in innate immune response that may trigger plague rupture, thrombosis with greater amplification leading to the rapid deterioration of the patient.
4. Discussion
The key initiating step in atherosclerosis is the release of cytokines like IFN-γ and TNF-α in response to sub-endothelial retention of ApoB-LPs by increasing permeability between endothelial cells [65,66]. Also, these overlying endothelial cells secrete cytokines or chemokines such as monocyte chemotactic protein-1(MCP-1) and interleukin-8(IL-8) in a manner that leads to recruitment of monocytes [67,68]. Within the sub-endothelial space, the monocytes differentiate into macrophages due to signaling of the cytokine M-CSF (Macrophage colony-stimulating factor) [64]. These macrophages become lipid laden to form “foam cells” and further release pro-inflammatory cytokines like IL-6 and IL-12 to bring in more immune cells to the site of inflammation [53]. Moreover, reduction in blood flow to the heart as a result of atherosclerosis results in myocardial infarction and myocardial infarction itself results in worsening of atherosclerosis by reinforcing macrophage infiltration at the site of atherosclerotic plaque [69]. Also, Myocardial infarction causes apoptosis of cardiomyocytes and the dying cells trigger innate immune response by releasing cytokines in cardiomyocytes [70]. Thus, cytokines are acting as the major stimuli of atherosclerosis.
Interestingly, earlier reports of COVID-19 had shown that maximum deaths occur due to Acute respiratory distress syndrome (ARDS) [1] and in the case of SARS-CoV infection, cytokine storm caused by enormous amounts of pro-inflammatory cytokines and chemokines like IL-1β, IL-16, TGFβ, CXCL8, CCL2, CCL3 results in ARDS [4]. Thus, patients with atherosclerosis if infected with SARS- CoV-2 may be are more prone to COVID-19 mediated death owing to such cytokine outrage. This proposition has also been echoed in the previous reports [5,6,8]. Moreover, endothelial dysfunction is a common factor in COVID-19 and atherosclerosis because constriction of the lumen of large arteries in either lungs or heart can obstruct blood flow [23].
In our study, we have noticed that PTPRC holds the highest degree (v = 136) in Ats PPI network (Fig. 1D). PTPRC was formerly known as CD45 which is an important antigen for activation of T-cells in atherosclerotic lesions [50]. Moreover, a recent study has revealed that CD45 could act as a potent marker for distinguishing severe and non-severe cases of COVID-19 [51]. Thus, CD45 which is found to be upregulated in Atherosclerosis datasets (GSE28829 and GSE100927) is equally important for both atherosclerosis and COVID-19. Also, pathway enrichment analyses of hub genes from atherosclerosis network have shown that “Chemokine signaling pathway” is the most enriched biological process (Fig. 1D, Supplementary File S2). Early inflammatory response to accumulated lipids in sub-endothelial space activates overlying vascular endothelial cells to elicit recruitment of macrophages by releasing cytokines and chemokines in atherosclerotic arteries [71]. Moreover, the cytokine outrage is a common factor for both atherosclerosis and COVID-19 [3,64]. Thus, pathway analysis also has shown the interconnection between two diseases.
We have screened six atherosclerotic DETFs - TLR2, TLR4, EGFR, SPI1, MYD88, IRF8 which are observed to be the regulators of several target genes in COVID-19 network. Among these DETFs, MYD88, SPI1 and IRF8 may control the expression of IL1B, CCL5, ITGAM, IFIT3, CXCL1, CXCL2, CXCL3 and CXCL8 since these genes are differentially expressed in cardiomyocytes infected with SARS-CoV-2. Thus, it could be stated that atherosclerotic DETFs might be responsible for amplifying the immune response in comorbid patients. It was previously reported that TLRs mediate inflammatory immune response in coronary arterial disease via activation of NF-κB pathway [56]. Recent molecular docking and dynamic studies has reported the capability of TLR4 to bind with a multi-epitope vaccine produced from virion's outer surface proteins (E, M and S) [57] and Patra et al. also proposed that TLRs can be effective in combating COVID-19 [72]. In fact, Aboudounya and Heads in their review had proposed a model that TLR4 might be increasing ACE2 expression to facilitate SARS-CoV-2 entry. Metabolites released from apoptotic cells after first myocardial infraction also triggers TLR4 to initiate innate immune response [70]. Interestingly, in our analysis we have found that TLR4 is upregulated in SARS-CoV-2 infected cardiomyocyte (Table 2). Thus, further experimental validation will help to confirm the role of TLR4 in eliciting inflammatory response in these co-morbid patients. We found that EGFR encoding epidermal growth factor receptor is marginally downregulated in most of the atherosclerotic datasets and only one COVID-19 dataset (GSE150819) of our study (Table 1), but it is significantly upregulated in SARS-CoV-2 infected cardiomyocytes (Table 2). Interestingly, none of its target genes are found to be differentially expressed in infected cardiomyocytes. Rather we have noted a substantial number of targets of MYD88, SPI1 and IRF8 in COVID-19 network. Thus, dysregulation of these TFs may result in the upregulation of their corresponding TGs (IL1B, CCL5, ITGAM, IFIT3, CXCL1, CXCL2, CXCL3 and CXCL8) in the SARS-CoV-2 infected cardiomyocytes (Fig. 5B). Although SPI1 is not differentially expressed in cardiomyocytes, some TGs of SPI1 or its protein product known as PU.1 are found to be upregulated in cardiomyocytes and is critical for regulating inflammatory response in both atherosclerosis and COVID-19 progression. These upregulated TGs include ITGAM, CCL5, IFIT3 and IL1B, which encode CD11b, C–C motif chemokine 5, Interferon-induced protein with tetratricopeptide repeats 3 and interleukin–1β respectively (Table 2). CCL5 is an important pro-inflammatory chemokine that has been shown to be triggered in both primary respiratory syncytial virus (RSV) infection and atherosclerosis [73,74]. It has been reported that CD11b encoded by ITGAM is expressed by macrophages to mediate their adhesion to the surface of endothelial cells in both SARS-CoV-2 infection and coronary thrombosis [75]. SoCD11b could be proposed as a connecting link between COVID-19 and thrombosis. The upregulated TG IFIT3 may be an important mediator of antiviral response against influenza as it was found that airway and lung cells with low IFIT3 are more prone to influenza infection [76]. Moreover, IFIT3 was also reported to be a biomarker for SARS-CoV-2 infection in in-silico studies [77]. Interestingly, it was found that pro-inflammatory response in atherosclerotic lesions is mediated by M1 macrophages through IFIT3 upregulation [78]. So, SPI1 might be magnifying the inflammatory response by differentially regulating its TGs and thus causing havoc in patients co-morbid for both atherosclerosis and COVID-19.
IRF8 is responsible for regulating important inflammatory mediators including type 1 IFN concentrations in CVD and thus it has been projected as biomarker for vascular inflammations [79]. Moreover, IRF8 from dendritic cells has roles in elevating the adaptive immune response in response to pulmonary viral infections [80]. We have found that IRF8 might be responsible for upregulated expression of IL1B along with SPI1 and MYD88 (Fig. 5B) which was also echoed in an earlier study [63] indicating that IL-1β, the pro-inflammatory cytokine, triggers innate immune response in both COVID-19 and in atherosclerosis.
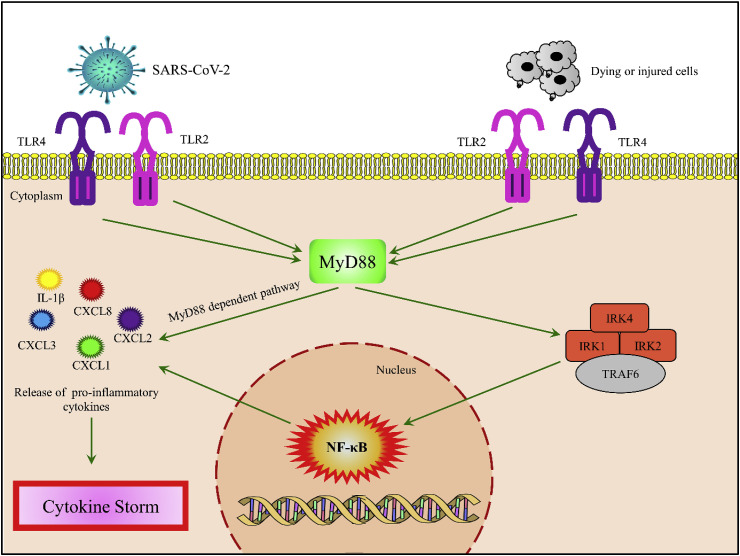
MYD88 has already been projected as a potential target for inflammatory lung diseases and atherosclerosis [81,82]. It was reported that during Rhinoviral (RV) infection interleukin–1β, encoded by IL1B, is triggered by MYD88 to elicit production of CXCL8, which is a chemo-attractant for neutrophil in RV-induced airway inflammation [83]. In case of atherosclerosis, the engulfment of oxidized low-density lipoprotein (ox-LDL) results in transformation of macrophages to foam cells and the main pathway which needs to be activated to trigger inflammatory response in foam cells is NF-κB [84,85]. It was also reported that dying cells after first myocardial infraction also activates NF- κB signaling pathway to release cytokines in cardiomyocytes [70]. Both TLR4 and TLR2 trigger the adaptor molecule MYD88 to elicit inflammatory response via NF-κB signaling pathway in response to either oxidized LDL in atherosclerosis or dying cells in cardiomyocytes [56,70]. NF- κB in turn activates transcription of various pro-inflammatory genes including cytokines like IL-1β [86,87]. We have found MYD88 might regulate expression of CXCL1, CXCL2, CXCL3 and CXCL8 which are basically cytokine or chemokine encoding genes. It is also noteworthy to mention that MYD88 dependent pathways activate macrophages, cytokines and chemokines not only in atherosclerotic lesions and injured cardiomyocytes, but also in mice infected with SARS-CoV [[88], [89], [90]]. Reports published earlier had stated that neutrophil trafficking is mediated by CXCL1 and CXCL2 in collateral lung damage [91]. Thus deregulation of MYD88 may be one of the crucial factors for rapid worsening of a comorbid patient. As both TLR4 and TLR2 was found to bind to SARS-CoV-2 and SARS-CoV respectively [57,92] and both the TLRs are found to be upregulated in infected cardiomyocytes (Table 2), we propose that MyD88 might be the central player which is being activated by both TLR2 and TLR4 in response to SARS-CoV-2 and dying cells in cardiomyocytes (Fig. 6 ). Activation of MyD88 might be triggering cytokine storm either by MyD88 dependent pathway or by activation of NF-κB pathway (Fig. 6).
Fig. 6.
Overview of the major immunological signaling events linking atherosclerosis and COVID-19 in cardiomyocytes. In response to both dying cells and SARS-CoV-2, TLR4 and TLR2 might be initiating inflammatory response by activating MyD88. MyD88 might be either activating NF-κB pathway or by itself initiating release of pro-inflammatory cytokines – CXCL1, CXCL2, CXCL3, CXCL8 and IL-1β which leads to cytokine storm in cardiac tissue.
Only CD11b encoded by ITGAM was found to activate inflammatory response in atherosclerosis and COVID-19 co-morbidity [75]. Some of the TF/TGs, like CXCL8, TNF, TLR4, are found to be the mediators of SARS-CoV-2 associated co-morbidities, but not atherosclerosis alone [93].
5. Conclusions
Of the cases reported worldwide, it is evident that individuals with pre-existing comorbidities like hypertension, obesity, chronic lung disease, diabetes, and cardiovascular disease are at a much greater risk of dying from COVID-19. It becomes a major challenge for global healthcare systems to follow appropriate medical interventions for the survival of comorbid patients. Thus, exploring the genetic intersection between COVID-19 and the co-existing diseases is an imperative step towards the identification of therapeutic interventions. In this study, we have analyzed differential gene expression and regulation in three types of cases- COVID-19 patient, atherosclerosis patient and COVID-19 infection in cardiomyocytes to portray plausible molecular events which occur in co-morbid patients. Though there were myriad of inflammatory markers which have been individually found to be responsible for causing havoc either in SARS-CoV-2 infection or atherosclerosis alone, however, their role in causing fatality in patients co-morbid for both diseases was missing. We hereby propose that the augmentation of inflammatory responses in comorbid cases succeed via deregulation of an array of transcription factors and their corresponding target genes. We have also suggested MYD88 as a crucial linker of major cross-talking tract concerned for two disorders. Thus, anti-inflammatory therapies targeting MYD88 could serve as a good measure to combat the comorbidity. The limitation of this study includes the used datasets which are devoid of significant patient heterogeneity, for example, male-female ratio and age groups. Overall the investigation offers a range of immunological markers for further experimental validation to confirm their role in the progression of magnified inflammatory response that leads to death in comorbid patients.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contribution
SP designed the study. DD executed all the experiments and wrote the manuscript. SP helped in drafting and editing the manuscript.
Declaration of competing interest
The authors declare that there isn't any conflict of interest with any organization or financial entity.
Acknowledgement
We are grateful to Raiganj University for providing the infrastructure to carry out this work. DD is thankful for SVMCM (Swami Vivekananda Merit Cum Means) fellowship by Government of West Bengal, India. We are also grateful to Prof. Pinak Chakrabarti, JC Bose National Fellow, Department of Biochemistry, Bose Institute for critical reading and valuable suggestions in improving the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104459.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020;35:266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. Cytokine and Growth Factor Reviews the cytokine storm in COVID-19 : an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., Huang H., Yang B., Huang C. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020;5:802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Javanmardi F., Keshavarzi A., Akbari A., Emami A., Pirbonyeh N. Prevalence of underlying diseases in died cases of COVID-19: a systematic review and meta-analysis. PLoS One. 2020;15:1–13. doi: 10.1371/journal.pone.0241265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan S., Pant M., Upadhyay S.K. The case fatality rate in COVID-19 patients with cardiovascular disease: global Health challenge and paradigm in the current pandemic. Curr. Pharmacol. Reports. 2020:1–10. doi: 10.1007/s40495-020-00239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan Q., Yang K., Wang W., Jiang L., Song J. Correction to: clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:1294–1297. doi: 10.1007/s00134-020-06028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W. And the northwell COVID-19 research consortium, presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. J. Am. Med. Assoc. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillinger J.G., Benmessaoud F.A., Pezel T., Voicu S., Sideris G., Chergui N., Hamzi L., Chauvin A., Leroy P., Gautier J.F., Sène D., Henry P. Null null, coronary artery calcification and complications in patients with COVID-19. JACC Cardiovasc. Imaging. 2020;13:2468–2470. doi: 10.1016/j.jcmg.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grzegorowska O., Lorkowski J. Possible correlations between atherosclerosis, acute coronary syndromes and COVID-19. J. Clin. Med. 2020;9 doi: 10.3390/jcm9113746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanepps J.S., Vorp D.A. Mechano-pathobiology of atherogenesis: a review. J. Surg. Res. 2007;142:202–217. doi: 10.1016/j.jss.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Moore K.J., Sheedy F.J., Fisher E.A. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo H., Lai T., Li C., Wu W. TNF-α induces CXCL1 chemokine expression and release in human vascular endothelial cells in vitro via two distinct signaling pathways. Acta Pharmacol. Sin. 2014;35:339–350. doi: 10.1038/aps.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surmi B.K., Hasty A.H. The role of chemokines in recruitment of immune cells to the artery wall and adipose tissue. Vasc. Pharmacol. 2010;52:27–36. doi: 10.1016/j.vph.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Zhang H.-G. Vigilance on new-onset atherosclerosis following SARS-CoV-2 infection. Front. Med. 2021;7:1135. doi: 10.3389/fmed.2020.629413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahara M., Ikutomi M., Morita T., Minami Y., Nakajima T., Hirata Y., Nagai R., Sata M. Deletion of angiotensin-converting enzyme 2 promotes the development of atherosclerosis and arterial neointima formation. Cardiovasc. Res. 2014;101:236–246. doi: 10.1093/cvr/cvt245. [DOI] [PubMed] [Google Scholar]
- 21.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet (London, England) 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt-Ott K.M., Kagiyama S., Phillips M.I. The multiple actions of angiotensin II in atherosclerosis. Regul. Pept. 2000;93:65–77. doi: 10.1016/s0167-0115(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 23.Nägele M.P., Haubner B., Tanner F.C., Ruschitzka F., Flammer A.J. Endothelial dysfunction in COVID-19: current findings and therapeutic implications. Atherosclerosis. 2020;314:58–62. doi: 10.1016/j.atherosclerosis.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans P.C., Rainger G.E., Mason J.C., Guzik T.J., Osto E., Stamataki Z., Neil D., Hoefer I.E., Fragiadaki M., Waltenberger J., Weber C. Endothelial dysfunction in COVID-19 : a position paper of the ESC working group for atherosclerosis and vascular biology , and the ESC Council of basic cardiovascular science. Cardiovasc. Res. 2020;116:2177–2184. doi: 10.1093/cvr/cvaa230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadosh B.S., Garshick M.S., Gaztanaga J., Moore K.J., Newman J.D., Pillinger M., Ramasamy R., Reynolds H.R., Shah B., Hochman J., Fishman G.I., Katz S.D. COVID-19 and the heart and vasculature. Arterioscler. Thromb. Vasc. Biol. 2020;40:2045–2053. doi: 10.1161/ATVBAHA.120.314513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anand A.R., Rachel G., Parthasarathy D. HIV proteins and endothelial dysfunction: implications in cardiovascular disease. Front. Cardiovasc. Med. 2018;5:185. doi: 10.3389/fcvm.2018.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adinolfi L.E., Restivo L., Zampino R., Guerrera B., Lonardo A., Ruggiero L., Riello F., Loria P., Florio A. Chronic HCV infection is a risk of atherosclerosis. Role of HCV and HCV-related steatosis. Atherosclerosis. 2012;221:496–502. doi: 10.1016/j.atherosclerosis.2012.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Döring Y., Manthey H.D., Drechsler M., Lievens D., Megens R.T.A., Soehnlein O., Busch M., Manca M., Koenen R.R., Pelisek J., Daemen M.J., Lutgens E., Zenke M., Binder C.J., Weber C., Zernecke A. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–1683. doi: 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 29.Steenman M., Espitia O., Maurel B., Guyomarch B., Heymann M.-F., Pistorius M.-A., Ory B., Heymann D., Houlgatte R., Gouëffic Y., Quillard T. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci. Rep. 2018;8:3940. doi: 10.1038/s41598-018-22292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:1–12. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatsuya S., Itoh Y., Sakai Y., Saito A., Okuzaki D., Motooka D., Minami S., Kobayashi T., Yamamoto T., Okamoto T., Takayama K. 2020. Generation of Human Bronchial Organoids for SARS-CoV-2 Research. BioRxiv. [DOI] [Google Scholar]
- 33.Sharma A., Garcia G.J., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep. Med. 2020;1:100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team. Language R.:A. 2020. And Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- 36.Sulkava M., Raitoharju E., Levula M., Seppälä I., Lyytikaïnen L.P., Mennander A., Järvinen O., Zeitlin R., Salenius J.P., Illig T., Klopp N., Mononen N., Laaksonen R., Kähönen M., Oksala N., Lehtimäki T. Differentially expressed genes and canonical pathway expression in human atherosclerotic plaques-Tampere Vascular Study. Sci. Rep. 2017;7:1–10. doi: 10.1038/srep41483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303.metabolite. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assenov Y., Ramírez F., Schelhorn S.-E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 40.Vandereyken K., Van Leene J., De Coninck B., Cammue B.P.A. Hub protein controversy: taking a closer look at plant stress response hubs. Front. Plant Sci. 2018;9:1–24. doi: 10.3389/fpls.2018.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A., David W.S. A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han H., Cho J.-W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E., Lee S., Kang B., Jeong D., Kim Y., Jeon H.-N., Jung H., Nam S., Chung M., Kim J.-H., Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z.-P., Wu C., Miao H., Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database. 2015 doi: 10.1093/database/bav095. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14 : more genomes , a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blighe K., Rana S., Lewis M. 2020. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling.https://github.com/kevinblighe/EnhancedVolcano [Google Scholar]
- 47.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 48.Subudhi A.K., Boopathi P.A., Pandey I., Kaur R., Middha S., Acharya J., Kochar S.K., Kochar D.K., Das A. Disease specific modules and hub genes for intervention strategies: a co-expression network based approach for Plasmodium falciparum clinical isolates. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015;35:96–108. doi: 10.1016/j.meegid.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Grivel J.-C., Ivanova O., Pinegina N., Blank P.S., Shpektor A., Margolis L.B., Vasilieva E. Activation of T lymphocytes in atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 2011;31:2929–2937. doi: 10.1161/ATVBAHA.111.237081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander D., Shiroo M., Robinson A., Biffen M., Shivnan E. The role of CD45 in T-cell activation resolving the paradoxes ? Immunol. Today. 1992;13:477–481. doi: 10.1016/0167-5699(92)90021-X. [DOI] [PubMed] [Google Scholar]
- 51.Jin M., Shi N., Wang M., Shi C., Lu S., Chang Q., Sha S., Lin Y., Chen Y., Zhou H., Liang K., Huang X., Shi Y., Huang G. CD45: a critical regulator in immune cells to predict severe and non-severe COVID-19 patients. Aging (Albany. NY) 2020;12:19867–19879. doi: 10.18632/aging.103941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bazaz R., Marriott H.M., Francis S.E., Dockrell D.H. Mechanistic links between acute respiratory tract infections and acute coronary syndromes. J. Infect. 2013;66:1–17. doi: 10.1016/j.jinf.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Ramji D.P., Davies T.S. Cytokines in atherosclerosis: key players in all stages of disease and promising therapeutic targets. Cytokine Growth Factor Rev. 2015;26:673–685. doi: 10.1016/j.cytogfr.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiran M., Nagarajaram H.A. Interaction and localization diversities of global and local hubs in human protein-protein interaction networks. Mol. Biosyst. 2016;12:2875–2882. doi: 10.1039/c6mb00104a. [DOI] [PubMed] [Google Scholar]
- 55.Rahman M.R., Islam T., Zaman T., Shahjaman M., Karim M.R., Huq F., Quinn J.M.W., Holsinger R.M.D., Gov E., Moni M.A. Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: insights from a systems biomedicine perspective. Genomics. 2020;112:1290–1299. doi: 10.1016/j.ygeno.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 56.Roshan M.H.K., Tambo A., Pace N.P. The role of TLR2, TLR4, and TLR9 in the pathogenesis of atherosclerosis. Int. J. Inflamm. 2016;2016:1532832. doi: 10.1155/2016/1532832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet Ul Alam A.S.M., Siddique M.A., Saha O., Rahaman M.M., Sultana M., Crandall K.A., Hossain M.A. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2, the etiologic agent of COVID-19 pandemic: an in silico approach. PeerJ. 2020;8 doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu L., Zhang H., Dauphars D.J., He Y.-W. A potential role of interleukin 10 in COVID-19 pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satış H., Özger H.S., Aysert Yıldız P., Hızel K., Gulbahar Ö., Erbaş G., Aygencel G., Guzel Tunccan O., Öztürk M.A., Dizbay M., Tufan A. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine. 2021;137:155302. doi: 10.1016/j.cyto.2020.155302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sbarsi I., Falcone C., Boiocchi C., Campo I., Zorzetto M., De Silvestri A., Cuccia M. Inflammation and atherosclerosis: the role of TNF and TNF receptors polymorphisms in coronary artery disease. Int. J. Immunopathol. Pharmacol. 2007;20:145–154. doi: 10.1177/039463200702000117. [DOI] [PubMed] [Google Scholar]
- 61.Robinson P.C., Richards D., Tanner H.L., Feldmann M. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2:e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castillo N.E., Rajasekaran A., Ali S.K. Legionnaires' disease: a review. Infect. Dis. Clin. Pract. 2016;24 https://journals.lww.com/infectdis/Fulltext/2016/09000/Legionnaires__Disease__A_Review.2.aspx [Google Scholar]
- 63.Conti P., Ronconi G., Caraffa A., Gallenga C., Ross R., Frydas I., Kritas S. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 64.McLaren J.E., Michael D.R., Ashlin T.G., Ramji D.P. Cytokines, macrophage lipid metabolism and foam cells: implications for cardiovascular disease therapy. Prog. Lipid Res. 2011;50:331–347. doi: 10.1016/j.plipres.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Pober J.S., Sessa W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 66.Williams K.J., Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler. Thromb. Vasc. Biol. 1995;15:551–562. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelken N.A., Coughlin S.R., Gordon D., Wilcox J.N. Monocyte chemoattractant protein-1 in human atheromatous plaques. J. Clin. Invest. 1991;88:1121–1127. doi: 10.1172/JCI115411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boisvert W.A., Santiago R., Curtiss L.K., Terkeltaub R.A. A leukocyte homologue of the IL-8 receptor CXCR-2 mediates the accumulation of macrophages in atherosclerotic lesions of LDL receptor-deficient mice. J. Clin. Invest. 1998;101:353–363. doi: 10.1172/JCI1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dutta P., Courties G., Wei Y., Leuschner F., Gorbatov R., Robbins C.S., Iwamoto Y., Thompson B., Carlson A.L., Heidt T., Majmudar M.D., Lasitschka F., Etzrodt M., Waterman P., Waring M.T., Chicoine A.T., van der Laan A.M., Niessen H.W.M., Piek J.J., Rubin B.B., Butany J., Stone J.R., Katus H.A., Murphy S.A., Morrow D.A., Sabatine M.S., Vinegoni C., Moskowitz M.A., Pittet M.J., Libby P., Lin C.P., Swirski F.K., Weissleder R., Nahrendorf M. Myocardial infarction accelerates atherosclerosis. Nature. 2012;487:325–329. doi: 10.1038/nature11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fioranelli M., Bottaccioli A.G., Bottaccioli F., Bianchi M., Rovesti M., Roccia M.G. Stress and inflammation in coronary artery disease: a review psychoneuro endocrine immunology-based. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glass C.K., Witztum J.L. Atherosclerosis : the road ahead review. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 72.Patra R., Chandra Das N., Mukherjee S. Targeting human TLRs to combat COVID-19: a solution? J. Med. Virol. 2021;93:615–617. doi: 10.1002/jmv.26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Culley F.J., Pennycook A.M.J., Tregoning J.S., Dodd J.S., Walzl G., Wells T.N., Hussell T., Openshaw P.J.M. Role of CCL5 ( RANTES ) in viral lung disease. J. Virol. 2006;80:8151–8157. doi: 10.1128/JVI.00496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bakogiannis C., Sachse M., Stamatelopoulos K., Stellos K. 2017. Cytokine Platelet-Derived Chemokines in in Fl Ammation and Atherosclerosis. [DOI] [PubMed] [Google Scholar]
- 75.Mazzone A., Castelnovo L., Tamburello A., Gatti A., Brando B., Faggioli P., Mumoli N. Monocytes could be a bridge from in fl ammation to thrombosis on COVID-19 injury : a case report. Thromb. Updat. 2020;1:100007. doi: 10.1016/j.tru.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wellington D., Laurenson-schafer H., Abdel-haq A., Dong T. IFITM3 : how genetics influence influenza infection demographically. Biomed. J. 2019;42:19–26. doi: 10.1016/j.bj.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hachim M.Y., Al Heialy S., Hachim I.Y., Halwani R., Senok A.C., Maghazachi A.A., Hamid Q. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front. Immunol. 2020;11:1372. doi: 10.3389/fimmu.2020.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang C., Lewis C., Borg N.A., Canals M., Diep H., Drummond G., Goode R.J., Schittenhelm R.B., Vinh A., Zhu M., Kemp-harper B., Kleifeld O., Stone M.J. Proteomic identification of interferon-induced proteins with tetratricopeptide repeats ( IFITs ) as markers of M1 macrophage polarization infection and immunity program , Monash biomedicine discovery Institute & Department of cardiovascular disease Progra. J. Proteome Res. 2018;17:1485–1499. doi: 10.1021/acs.jproteome.7b00828. [DOI] [PubMed] [Google Scholar]
- 79.Chmielewski S., Piaszyk-borychowska A., Wesoly J., Bluyssen H.A.R. STAT1 and IRF8 in vascular inflammation and cardiovascular disease : diagnostic and therapeutic potential. Int. Rev. Immunol. 2016;35:434–454. doi: 10.3109/08830185.2015.1087519. ISSN. [DOI] [PubMed] [Google Scholar]
- 80.Bosteels C., Neyt K., Vanheerswynghels M., van Helden M.J., Sichien D., Debeuf N., De Prijck S., Bosteels V., Vandamme N., Martens L., Saeys Y., Louagie E., Lesage M., Williams D.L., Tang S.C., Mayer J.U., Ronchese F., Scott C.L., Hammad H., Guilliams M., Lambrecht B.N. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity. 2020;52:1039–1056. doi: 10.1016/j.immuni.2020.04.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Padova F., Quesniaux V.F.J., Ryffel B. Expert Opinion on Therapeutic Targets MyD88 as a therapeutic target for inflammatory lung diseases. Expert Opin. Ther. Targets. 2018;25:401–408. doi: 10.1080/14728222.2018.1464139. [DOI] [PubMed] [Google Scholar]
- 82.Michelsen K.S., Wong M.H., Shah P.K., Zhang W., Yano J., Doherty T.M., Akira S., Rajavashisth T.B., Arditi M. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10679–10684. doi: 10.1073/pnas.0403249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stokes C.A., Ismail S., Dick E.P., Bennett J.A., Johnston S.L., Edwards M.R., Sabroe I., Parker L.C. Role of interleukin-1 and MyD88-dependent signaling in Rhinovirus infection †. J. Virol. 2011;85:7912–7921. doi: 10.1128/JVI.02649-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kita T., Kume N., Minami M., Hayashida K., Murayama T., Sano H., Moriwaki H., Kataoka H., Nishi E., Horiuchi H., Arai H., Yokode M. Role of oxidized LDL in atherosclerosis. Ann. N. Y. Acad. Sci. 2001;947:196–199. doi: 10.1111/j.1749-6632.2001.tb03941.x. [DOI] [PubMed] [Google Scholar]
- 85.Gareus R., Kotsaki E., Xanthoulea S., van der Made I., Gijbels M.J.J., Kardakaris R., Polykratis A., Kollias G., de Winther M.P.J., Pasparakis M. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metabol. 2008;8:372–383. doi: 10.1016/j.cmet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 86.Smoak K.A., Aloor J.J., Madenspacher J., Merrick B.A., Collins J.B., Zhu X., Cavigiolio G., Oda M.N., Parks J.S., Fessler M.B. Myeloid differentiation primary response protein 88 couples reverse cholesterol transport to inflammation. Cell Metabol. 2010;11:493–502. doi: 10.1016/j.cmet.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sugiyama K.I., Muroi M., Kinoshita M., Hamada O., Minai Y., Sugita-Konishi Y., Kamata Y., Tanamoto K.I. NF-κB activation via MyD88-dependent Toll-like receptor signaling is inhibited by trichothecene mycotoxin deoxynivalenol. J. Toxicol. Sci. 2016;41:273–279. doi: 10.2131/jts.41.273. [DOI] [PubMed] [Google Scholar]
- 88.Li B., Xia Y., Hu B. Infection and atherosclerosis: TLR-dependent pathways. Cell. Mol. Life Sci. 2020;77:2751–2769. doi: 10.1007/s00018-020-03453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from Lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Su Q., Li L., Sun Y., Yang H., Ye Z., Zhao J. Effects of the TLR4/Myd88/NF-κB signaling pathway on NLRP3 inflammasome in coronary Microembolization-induced myocardial injury. Cell. Physiol. Biochem. 2018;47:1497–1508. doi: 10.1159/000490866. [DOI] [PubMed] [Google Scholar]
- 91.Sawant K.V., Xu R., Cox R., Hawkins H., Sbrana E., Kolli D., Garofalo R.P., Rajarathnam K. Chemokine CXCL1-mediated neutrophil trafficking in the lung: role of CXCR2 activation. J. Innate Immun. 2015;7:647–658. doi: 10.1159/000430914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dosch S.F., Mahajan S.D., Collins A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dolan M.E., Hill D.P., Mukherjee G., McAndrews M.S., Chesler E.J., Blake J.A. Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Sci. Rep. 2020;10:20848. doi: 10.1038/s41598-020-77632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.