Little is known about the mechanism by which LCTs are secreted. Since LCTs are major virulence factors in clostridial pathogens, we wanted to define the mechanism by which an LCT in C. perfringens, TpeL, is secreted by a protein (TpeE) lacking homology to previously described secretion-associated holins.
KEYWORDS: Clostridium, molecular genetics, perfringens, secretion systems, toxin
ABSTRACT
Large clostridial toxins (LCTs) are secreted virulence factors found in several species, including Clostridioides difficile, Clostridium perfringens, Paeniclostridium sordellii, and Clostridium novyi. LCTs are large toxins that lack a secretion signal sequence, and studies by others have shown that the LCTs of C. difficile, TcdA and TcdB, require a holin-like protein, TcdE, for secretion. The TcdE gene is located on the pathogenicity locus (PaLoc) of C. difficile, and holin-encoding genes are also present in the LCT-encoded PaLocs from P. sordellii and C. perfringens. However, the holin (TpeE) associated with the C. perfringens LCT TpeL has no homology and a different membrane topology than TcdE. In addition, TpeE has a membrane topology identical to that of the TatA protein, which is the core of the twin-arginine translocation (Tat) secretion system. To determine if TpeE was necessary and sufficient to secrete TpeL, the genes from a type C strain of C. perfringens were expressed in a type A strain of C. perfringens, HN13, and secretion was measured using Western blot methods. We found that TpeE was required for TpeL secretion and that secretion was not due to cell lysis. Mutant forms of TpeE lacking an amphipathic helix and a charged C-terminal domain failed to secrete TpeL, and mutations that deleted conserved LCT domains in TpeL indicated that only the full-length protein could be secreted. In summary, we have identified a novel family of holin-like proteins that can function, in some cases, as a system of protein secretion for proteins that need to fold in the cytoplasm.
IMPORTANCE Little is known about the mechanism by which LCTs are secreted. Since LCTs are major virulence factors in clostridial pathogens, we wanted to define the mechanism by which an LCT in C. perfringens, TpeL, is secreted by a protein (TpeE) lacking homology to previously described secretion-associated holins. We discovered that TpeE is a member of a widely dispersed class of holin proteins, and TpeE is necessary for the secretion of TpeL. TpeE bears a high degree of similarity in membrane topology to TatA proteins, which form the pore through which Tat secretion substrates pass through the cytoplasmic membrane. Thus, the TpeE-TpeL secretion system may be a model for understanding not only holin-dependent secretion but also how TatA proteins function in the secretion process.
INTRODUCTION
The Clostridia, which include many pathogenic species of humans and animals, secrete a wide variety of toxins as important virulence factors in causing disease (1). One family of secreted factors is the large clostridial toxins (LCTs), which can inactivate host cell Rho family GTPases by glycosylation with either glucose or N-acetylglucosamine (2–5). Each LCT has a specific subset of GTPases that it uses as the substrates, but inactivation of the GTPases usually leads to disruption of the actin cytoskeleton, causing cells to round up and undergo apoptosis (2). Currently known LCTs include TcdA and TcdB of Clostridioides difficile (previously Clostridium difficile), alpha-toxin (TcnA) of Clostridium novyi, lethal toxin (TcsL) and hemorrhagic toxin (TcsH) of Paeniclostridium (formerly Clostridium) sordellii, and the TpeL toxin of Clostridium perfringens (Fig. 1A). The well-studied TcdA and TcdB toxins of C. difficile have been shown to be directly linked to virulence in C. difficile-associated infections using animal models (6). P. sordellii has been associated with postabortion toxic shock syndrome in humans (7, 8), and TcsL has been shown to be important for virulence in an animal model (9). C. perfringens TpeL has been linked to increased virulence in chicken necrotic enteritis (NE) infections (10–12) but has not been shown to be essential for virulence in NE infections or other animal diseases.
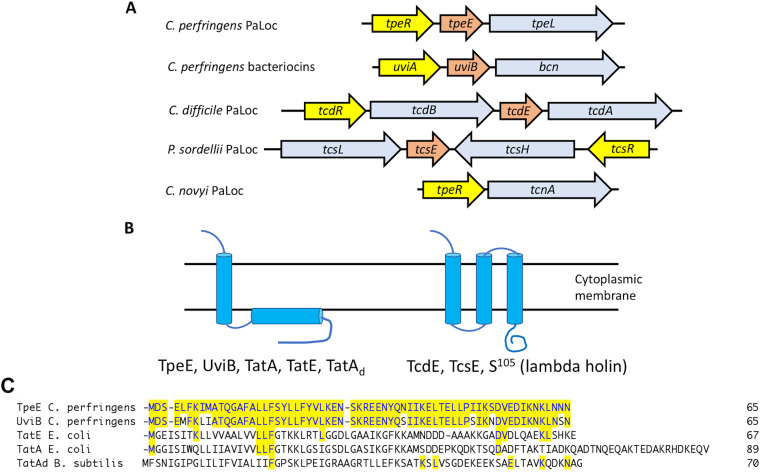
FIG 1.
Gene synteny, membrane topology, and sequence homology of LCT-associated holin-like proteins in clostridia. (A) Gene organization in LCT PaLocs and a bacteriocin-containing operon. Light blue, LCTs and bacteriocin; yellow, sigma factors; orange, holin-like proteins. Gene lengths are not to scale. (B) Proposed membrane topology of holins and holin-like proteins from panel A and twin-arginine transport (Tat) proteins. TatA and TatE are from E. coli, and TatAd is from B. subtilis. (C) Sequence alignments of holin-like proteins. The proteins were aligned using the ClustalW algorithm in the MegAlign software suite, part of the Lasergene 16 DNA analysis package from DNAStar. Residues that match the TpeE sequence are highlighted in yellow shading. Note that there are 7 residue differences between TpeE and UviB. GenBank sequence accession numbers are as follows: WP_003453321.1 for C. perfringens TpeE, ABG87870.1 for C. perfringens UviB, WP_105463372.1 for E. coli TatE, and QNN28895.1 for B. subtilis TatAd. The PDB ID for E. coli TatA is 2MN7_A.
The genes encoding LCTs are located in pathogenicity loci, which vary from species to species (Fig. 1A). Three genes comprise the core members of each pathogenicity locus (PaLoc), the toxin-encoding, sigma factor-encoding, and holin-like protein-encoding genes. The exception is the C. novyi PaLoc, in which no holin-encoding genes have been found adjacent to the tcnA gene (Fig. 1A). In a previous report (13), it was noted that the uviA-uviB-bcn locus, which is involved in bacteriocin production in C. perfringens, has a gene synteny identical to PaLocs encoding LCTs (Fig. 1A). For each of these loci, the sigma factors have been shown to be important for the transcriptional regulation of the LCT-encoding genes, and the same is true for the uviA gene in the bcn locus (14, 15). The holins have been proposed to mediate secretion, and one of them, TpeE, is the subject of this work.
C. difficile TcdE is the only holin for which there is definitive evidence that it is essential for secretion (16) (Fig. 1A). TcdE is proposed to have a membrane topology similar to that of the λ phage holin S105, including three transmembrane helices and a cytoplasmic C-terminal domain (Fig. 1B), and TcdE was able to function as a holin in λ phage (16). Using the TMHMM Web-based server (http://www.cbs.dtu.dk/services/TMHMM-2.0/), a similar membrane topology is predicted for the P. sordellii TcsE holin-like protein (Fig. 1B). Mutation of the tcdE gene led to a very significant decrease in extracellular TcdA in the medium, and complementation of the mutant restored secretion to levels above that of the wild-type strain (16). Assays to detect cytoplasmic enzymes in the supernatant and membrane permeability using propidium iodide indicated that little cell lysis was occurring during the period in which TcdA was being secreted (16). However, just prior to this, a report by Olling et al. (17) indicated that, in fact, there was considerable cell lysis occurring during stationary phase, resulting in toxin release. This seeming contradiction was resolved by a later report showing that there was an induction of an autolytic transglycosylase, Cwp19, when C. difficile cells were grown in brain heart infusion (BHI) broth, as was done in the report by Olling et al. (17), but not in trypticase-yeast extract (TY) medium, which was used to show that TcdE was required for secretion (16). Together, these results indicated that C. difficile possesses both a holin-dependent and a holin-independent (i.e., lysis) mechanism for TcdA/TcdB secretion.
In contrast to TcdE and TcsE, the holin-like protein TpeE from C. perfringens is predicted to have a single-transmembrane helix, an amphipathic helix (APH) located on the inner side of the cytoplasmic membrane (CM), and a hydrophilic C-terminal domain (Fig. 1B; see also Results). A BLAST analysis (18) of the TpeE protein from the type C strain JGS1495 (GenBank accession number WP_003453321.1) shows 1,255 matches in bacteria, of which 1,252 are in the Firmicutes, with most being clustered in the clostridia (859). A conserved domain analysis (19) puts TpeE in the BhlA holin family, which places it in superfamily VI (DUF 2762) according to the classification of Reddy and Saier (20, 21). TpeE homologs are often annotated as bacteriocins; this may be due to the early annotation of UviB from plasmid pIP404 from C. perfringens (22), which is a homolog, as a bacteriocin. UviB proteins from many species, including the two homologs found in the small plasmids carried by C. perfringens strain SM101 (23), are predicted to have the same membrane topology as TpeE as well as a high level of sequence homology to TpeE (Fig. 1C). Despite their putative substrates, TpeL and Bcn have little sequence or predicted structural homology. These findings bring up an interesting biological question: How do completely different holins (TcdE and TpeL) secrete similar substrates (TcdA/TcdB and, presumably, TpeL), while nearly identical holins, TpeE and UviB, secrete structurally different proteins, TpeL and Bcn?
We noted that despite little sequence homology, the predicted membrane topology for TpeE is the same as those for the TatA and TatE proteins from Escherichia coli and TatAd from Bacillus subtilis (24, 25) (Fig. 1B and C). TatA comprises the core of the twin-arginine translocation (Tat) system, which is widely found in Gram-positive bacteria, Gram-negative bacteria, and plants (26) but is absent in C. perfringens. The Tat complex is involved in the translocation of folded proteins across the CM, which is similar in function to that proposed for the holins described above. The Tat system in E. coli is composed of three proteins, TatA, TatB (referred to as TatA-like protein), and the integral membrane protein TatC (27). Substrates with a Tat recognition sequence at the N terminus, which contains the twin-arginine residues that gave the system its name, are bound by TatBC and transferred to a TatA complex, an oligomer of TatA, which translocates the substrate across the CM (26, 27). TatE is a TatA-like protein in E. coli that can functionally replace TatA in E. coli (28).
Given the differences in membrane topology between TcdE and TpeE and TpeE’s similarity in topology to TatA, we decided to determine if TpeE is necessary for the secretion of TpeL in C. perfringens. We focused on the TpeL PaLoc found in the type C strain JGS1495 (29). However, this strain is recalcitrant to genetic manipulation (our unpublished data). To have a more workable genetic system, the tpeE and tpeL genes from strain JGS1495 were transferred to strain HN13, a derivative of the genetically manipulatable strain 13 (30). By removing the two genes from their native host, we were also able to address the question of whether TpeE was necessary and sufficient for the secretion of TpeL. We also took advantage of the topological similarity between TpeE and the TatA protein to guide us in making specific mutations in regions of TpeE that have been shown to be important for TatA function. In this sense, the well-studied TatA system provides a framework for analyzing the functions and characteristics of the TpeE holin translocation system.
RESULTS
Cloning the tpeE and tpeL genes from the type C strain JGS1495 and heterologous expression in C. perfringens strain HN13.
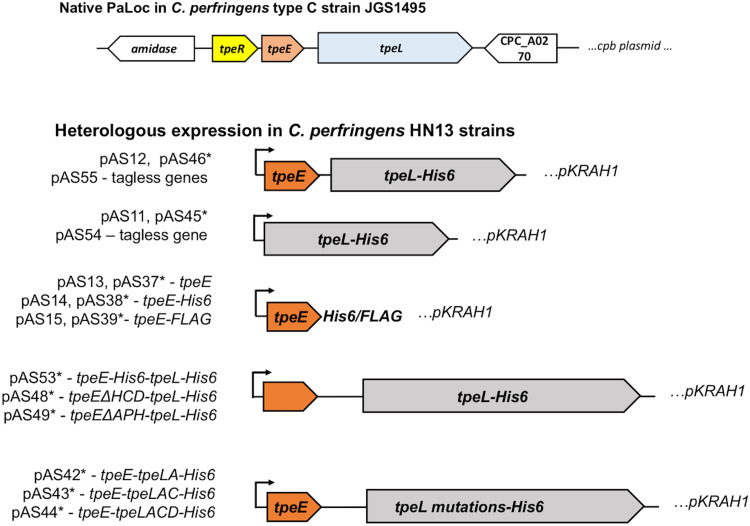
Genes encoding small holin-like proteins are often found adjacent to LCT-encoding genes in some pathogenic clostridia (Fig. 1). Govind and Dupuy demonstrated that the holin-like protein TcdE is required for the secretion of the toxins TcdA and TcdB in C. difficile (16). For C. perfringens, we set out to determine if the holin-like protein TpeE was necessary and sufficient to mediate the secretion of its cognate toxin, TpeL. To accomplish this, we amplified tpeE and tpeL from type C strain JGS1495 and expressed the genes using the lactose-inducible expression plasmid pKRAH1 (31) in C. perfringens strain HN13 (30) (Fig. 2). This removed these genes from being under the control of TpeR, an alternative sigma factor shown to be involved in regulating these genes (15). This approach allowed us to study the secretion of TpeL free from factors that may be specific to the type C strain and the large plasmid on which the tpeE and tpeL genes are carried (29, 32). Additionally, strain HN13 is genetically manipulatable, and there are multiple genetic molecular tools developed for use in this strain. Peptide antigen tags (FLAG and His6) were fused to both proteins for identification by immunoblotting (Fig. 2).
FIG 2.
Schematic diagram showing plasmid constructs used for heterologous expression of tpeE and tpeL in C. perfringens strain HN13. pKRAH1 has a lactose-inducible promoter for regulated expression (31), which is indicated by arrows. ΔHCD, deletion of the high charge density domain at the C terminus; ΔAPH, deletion of the amphipathic helix. TpeLA, TpeLAC, and TpeLACD are constructs with deletions of nested C-terminal domains of full-length TpeL. All constructs have a SacII site at the 5′ end and a BamHI site at the 3′ end of the inserts. Some, indicated with an asterisk, have an additional SalI site inserted between the tpeE and tpeL genes to allow efficient cloning. Each plasmid and its description are also listed in Table 1.
TpeL-His6 secretion is TpeE dependent.
We cultured strains expressing TpeL-His6 in TY broth supplemented with lactose to induce the expression of the tpeE and/or tpeL gene. Large quantities of TpeL-His6 were detected in the supernatants only if TpeE was coexpressed (Fig. 3A). Whole-cell lysates were prepared from the same cultures as the supernatants, subjected to immunoblotting, and found to have qualitatively equivalent levels of TpeL-His6 between strains expressing the toxin alone and strains coexpressing TpeE (Fig. 3A).
FIG 3.
Efficient TpeL secretion is TpeE dependent in C. perfringens. (A) Western blotting using an antibody direct against the His6 tag on TpeL to indicate the location (supernatant or intracellular) of TpeL. pKRAH, empty vector control; pAS11, toxin only; pAS12, toxin and holin; pAS15, holin alone. For the lysis control and loading controls, BCP was identified by fluorescently labeled streptavidin (see Materials and Methods). (B) A C-terminal polyhistidine tag does not significantly affect TpeL expression or secretion in C. perfringens. (Top) Coomassie-stained protein gels demonstrating similar amounts of TpeL in concentrated culture supernatants from strains overexpressing TpeE (no tag) and TpeL with and without a C-terminal His6 tag. Note that there is a protein in the supernatant that is somewhat larger than TpeL; this can be seen clearly in the lane with pKRAH1 and no TpeE (leftmost lane). (Bottom) Anti-His6 immunoblot combined with Ponceau S staining of the PVDF membrane showing similar levels of TpeL in the supernatants (white arrows) whether the His6 tag was present or not. Strains expressing TpeL with and without the His6 tag in the absence of TpeE are shown as secretion negative controls. Whole-cell lysate samples were included to control for differences in TpeL and TpeL-His6 expression. Strains expressing the empty vector and TpeE alone were included to control for effects related to the expression vector and TpeE. Note the increased levels of TpeL in the supernatant when the TpeE protein was also present. The results are representative of data from three independent experiments. (C) Sensitivity of lysis detection by the conjugated streptavidin probe. The image shows a Western blot using an antibody against Tpe-His6 and a fluorescent streptavidin conjugate to detect the cytoplasmic BCP of C. perfringens. When the lysate from the toxin-only strain was added to the filtered supernatant and concentrated, lysis was reliably detected at levels as low as 2% of total cell lysis (see Materials and Methods). +C represents a positive control, using pAS11 to express the toxin, and −C represents a negative control with the empty vector, pKRAH1. The image is representative of results from two separate biological experiments. MW, molecular weight marker.
For whole-cell lysates, we used fluorescence-conjugated streptavidin to bind a cytoplasmic protein and visually assess loading among the stains, as previously described (33). This gave a signal at roughly 20 kDa and demonstrates no significant differential sample loading between the strains expressing the toxin (Fig. 3A). A search for biotinylated proteins in C. perfringens suggests that this is most likely the biotin carboxyl carrier protein (BCP) (34), which is predicted to have a molecular weight of 18 kDa (35).
To determine if the C-terminal His6 tag on TpeL affected the efficiency of secretion, we compared intracellular and extracellular TpeL levels in strains expressing tagged and untagged versions of TpeL and with and without TpeE. TpeL levels in the supernatants were not statistically different between strains expressing TpeL and those expressing TpeL-His6 alone or between strains expressing TpeL and those expressing TpeL-His6 with TpeE (Fig. 3B). Conversely, we also tested whether the presence of the FLAG tag on TpeE affected TpeL secretion and found that the FLAG tag did not inhibit secretion (see Fig. S1 in the supplemental material).
Secretion of TpeL was not due to cell lysis by TpeE.
Small amounts of extracellular TpeL-His6 were occasionally detected in the strain lacking coexpressed TpeE (Fig. 3A). Reasoning that the presence of the cytoplasmic BCP in culture supernatants would indicate cell lysis, the conjugated streptavidin probe was also applied to supernatant samples as an indicator of lysis, but no signal was detected (Fig. 3A). To measure the lowest level of detection of the BCP in the supernatants, the culture supernatants were spiked with whole-cell lysates at increasing levels, analogous to measuring the percentage of cell lysis (see Materials and Methods). This method was able to reliably detect cytoplasmic BCP in concentrated supernatant samples containing 2% lysate and higher (Fig. 3C), which represents the upper boundary for lysis in these experiments. As an additional test for cell lysis, total cell counts and CFU per milliliter were not found to be statistically different between the strains expressing the toxin with and without TpeE (Fig. S2A). Finally, we performed a third assay for cell lysis by comparing the activities of another cytoplasmic protein, lactate dehydrogenase (LDH), in the culture supernatants and did not detect LDH activity in the supernatants of the strains with or without the holin (Fig. S2B).
The very low level or even the absence of lysis suggests that the toxin may be secreted at low levels by a TpeE-independent method. C. perfringens can release membrane vesicles (36), and the possibility of TpeL secretion by the release of membrane vesicles was assessed by immunoblotting and probing for TpeL in supernatant sample fractions that were spun in an ultracentrifuge for 1 h at 100,000 × g to pellet membrane vesicles (36). TpeL-His6 was not detected in the pelleted supernatant fraction (data not shown), suggesting that either TpeL-His6 was not secreted by membrane blebbing or the centrifugation was not sufficient to pellet the vesicles.
When expressed in C. perfringens, TpeE is not toxic, while TpeL expression decreases the growth rate.
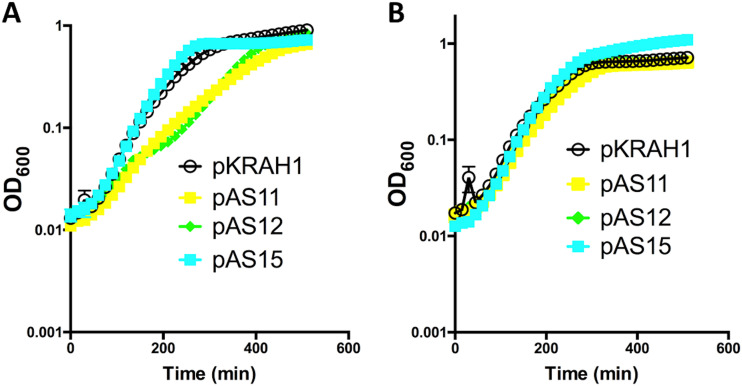
We next set out to understand the physiological effects of expressing TpeE and TpeL in C. perfringens. During growth in liquid TY medium, expressing TpeE alone had no effect on cell growth (Fig. 4A). However, expressing TpeL resulted in a lower growth rate, as did coexpressing TpeL with TpeE (Fig. 4A). In the absence of the inducer (lactose), there were no detectable differences in growth rates (Fig. 4B). As noted above, the lower growth rates did not result in significant levels of lysis, suggesting that they were due to some metabolic limitation that lowered the maximum growth rate achievable by that strain.
FIG 4.
Expression of tpeE in C. perfringens strain HN13 did not negatively affect the growth rate, while overexpression of tpeL decreased the growth rate. Shown are results for pKRAH1 (vector control), pAS15 (tpeE), pAS11 (tpeL), and pAS12 (tpeE-tpeL). (A) Results after induction with lactose; (B) controls with no lactose added. The means and standard deviations (SD) from five replicate samples are shown. The curves are representative of results from three independent biological replicates.
TpeE localizes to the membrane in C. perfringens.
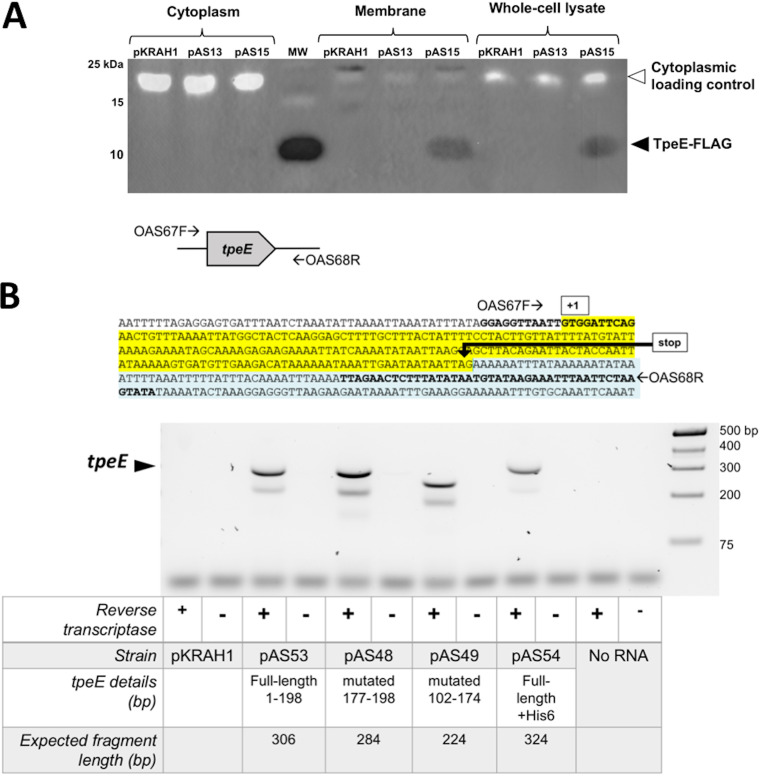
Given the holin-like attributes predicted in TpeE (Fig. 1), we set out to determine whether it collected in the membrane of C. perfringens. Since inducing the cultures at the time of inoculation resulted in variable growth rates (Fig. 4A), the strains were instead induced in late log phase for these experiments, and we allowed 3 to 4 h for expression and secretion to occur before harvesting samples. By dual-color immunoblotting, we detected TpeE-FLAG in the membrane and whole-cell fractions but not the cytoplasmic fractions (Fig. 5A). Furthermore, these results indicate that TpeE-FLAG does not form a covalently linked multimer since the protein is predicted to be 8.6 kDa in mass (37) and was detected near the 10-kDa molecular weight marker (Fig. 5A).
FIG 5.
(A) TpeE localizes to the membrane in C. perfringens. Western blotting was performed using anti-FLAG antibodies to identify the cellular location of TpeE-FLAG. Plasmids pKRAH1 (vector control), pAS13 (tpeE), and pAS15 (tpeE-FLAG) were used. The cytoplasmic loading control consisted of a fluorescent streptavidin conjugate to detect the cytoplasmic BCP of C. perfringens. (B) Detection of tpeE transcripts in C. perfringens using RT-PCR. (Top) Location of primers used for RT-PCR. (Bottom) Agarose gel showing the relative sizes of RT-PCR products and the conditions used for each sample.
Despite success in detecting TpeE by Western blotting, we were not able to detect TpeE expression in C. perfringens by direct protein staining even with considerable effort using multiple types of SDS-PAGE. Instead, we validated the transcription of the version of the gene with no tag by reverse transcription-PCR (RT-PCR) (Fig. 5B, lanes 3 and 4).
Expression of TpeE in E. coli is toxic.
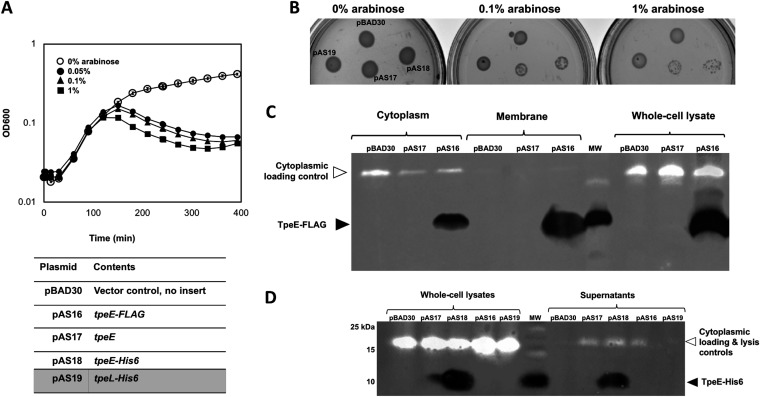
Given that TpeE demonstrated no lytic or holin-like activity in C. perfringens, we expressed TpeE in the unrelated Gram-negative bacterium E. coli to determine if holin-like activity could be detected in this species, as was found to be the case with TcdE from C. difficile (16). A dramatic, dose-dependent decrease in the optical density at 600 nm (OD600) of the cultures began about 150 min after the addition of the inducer arabinose (Fig. 6A). To see if this effect could be repeated under different environmental conditions, E. coli cultures were spotted onto solid medium with different concentrations of arabinose. Again, dramatic dose-dependent inhibition of growth was seen in strains expressing TpeE (Fig. 6B).
FIG 6.
TpeE is toxic to and localizes to the membrane and cytoplasm of E. coli. (A) Growth curves with E. coli showing the toxic effects of adding increasing concentrations of the inducer arabinose at the time of inoculation. The means and SD from five replicate samples are shown. The curve is representative of data from three independent biological replicates. (B) Arabinose induction inhibits the growth of colonies containing the tpeE gene (pAS17 and pAS18) on agar plates in E. coli. (C) Western blot showing that TpeE-FLAG was detected in whole-cell lysates, the cytoplasm, and membranes when expressed in E. coli. The cytoplasmic loading controls represent the use of a fluorescent streptavidin conjugate to detect the cytoplasmic biotin carboxyl carrier protein AccB (22.5 kDa) in E. coli. (D) Western blot showing the strains of E. coli in which TpeE appeared in the supernatant after lysis along with the AccB cytoplasmic protein (detected using a fluorescent streptavidin conjugate).
We determined the cellular location of TpeE in E. coli by dual-color immunoblotting and probing for C-terminal antigen tags. Analogous to the experiments in C. perfringens, expression was induced in the late logarithmic growth phase, known to be ∼2 h postinoculation by previous growth curve analyses with E. coli. In contrast to C. perfringens cultures, large amounts of TpeE were detected not only in membrane fractions but also in the cytoplasmic fraction (Fig. 6C). The conjugated streptavidin probe was again employed to control for loading and lysis and likely bound the cytoplasmic, biotinylated protein AccB in E. coli (38). Significant levels of lysis were detected in E. coli supernatants but only in strains expressing TpeE (Fig. 6D).
Mutagenesis of the C-terminal highly charged domain and the amphipathic helix in TpeE abolishes TpeE-dependent TpeL secretion by C. perfringens.
Full-length TpeE is predicted to be 65 amino acids (aa) long, a predominantly alpha-helical protein with one transmembrane domain (39). As depicted in Fig. 1 and Fig. 7A, the N terminus of TpeE is likely extracellular (40), followed by a membrane-spanning alpha-helix, while the cytoplasmic portion contains an amphipathic region followed by a string of asparagine and lysine residues (KNKLNNN) at the C terminus (Fig. 7A). To confirm the presence and extent of the amphipathic helix (APH), we used the helical wheel projection software Heliquest (https://heliquest.ipmc.cnrs.fr/), which predicted that the helix extended from residues 34 to 58 with a pronounced amphipathicity (Fig. 7A). We hypothesized that the negatively charged residues of the amphipathic helix may form salt bridges with complementary charges at the C terminus of the holin (Fig. 7A), similar to an experimentally tested model for the TatA protein in the Tat secretion system (41). To test this hypothesis, we deleted the region encoding the amphipathic helix (Δ34–58) and, separately, the C-terminal hydrophilic region of TpeE (Δ59–65) and coexpressed them with TpeL. In dual-color immunoblots, TpeL-His6 was not detected in the culture supernatants of strains coexpressing the mutated TpeE forms, although it was seen in the positive-control strain expressing full-length TpeE (Fig. 7B). Whole-cell lysate samples demonstrated that strains carrying the mutated TpeE were able to produce the full-length toxin and full-length TpeE alone as negative controls, even though some degradation of the TpeL protein occurs in the intracellular environment (Fig. 7B). This indicates that both the C-terminal hydrophilic domain and the amphipathic helix are required for TpeL secretion. Additionally, the expression of TpeE during TpeL secretion was validated in a strain expressing both proteins with a His6 tag fused at their C termini (pAS53 in Fig. 7B). A strain expressing FLAG-tagged TpeE without the toxin was included to determine if we could detect both the His6-tagged and FLAG-tagged versions on the same membrane. After immunoblotting with anti-His6, the membrane was imaged, rewetted, probed with an anti-FLAG antibody, and reimaged. Both tagged versions were then visible on the membrane (Fig. 7B).
FIG 7.
Conserved domains in TpeE are required for secretion of TpeL by C. perfringens. (A) Diagram showing the predicted cellular location of each domain in TpeE. The amphipathic nature of the central alpha-helix is shown using a helical wheel projection of hydrophobic (yellow) and hydrophilic (all other colors) residues. TMD, transmembrane domain. (B) Western blots showing the presence of TpeL-His6 and TpeE-FLAG in whole-cell lysates and supernatants. Note the lack of a TpeL-His6 signal in the supernatants of the strains expressing the TpeE protein with the amphipathic helix or C-terminal domain deleted, even though no cell lysis could be detected with the streptavidin-BCP control.
We did not fuse antigen tags to the mutated forms of TpeE since both His6 and FLAG tags are highly charged and could interfere with the mutagenesis effects that we desired to study. Instead, we validated the transcription of full-length TpeE mRNA and its mutated forms by RT-PCR (Fig. 5B, lanes 5 to 8). As controls, PCR of DNA collected from each strain was performed using the same primers as those used for RT-PCR, resulting in a banding pattern (Fig. S2) identical to those seen with RT-PCR (Fig. 5B).
Although we were able to detect His6- and FLAG-tagged TpeE by immunoblotting, we were not able to detect TpeE or its mutant forms in C. perfringens by SDS-PAGE and protein staining. We believe that this was because the proteins are expressed at moderate levels, and the mutated TpeE forms have very low molecular weights. Therefore, we used peptide mass spectrometry methods to detect the presence of the highly charged domain (HCD) and APH deletion proteins (see primers and methods in the supplemental material). The TpeE-specific peptides EENYQNIIK and ELTELLPIIK (Fig. 1C) were detected in SDS-PAGE gels when extracts from plasmid-expressed FLAG-tagged (pAS58) and HCD-deleted (pAH48) TpeE were analyzed, but no TpeE peptides were found with the APH-deleted TpeE (pAS49) and the empty vector control (pKRAH1). This suggests that the HCD-deleted form of TpeE was present but that the APH-deleted TpeE was absent or present at only very low levels in the cell.
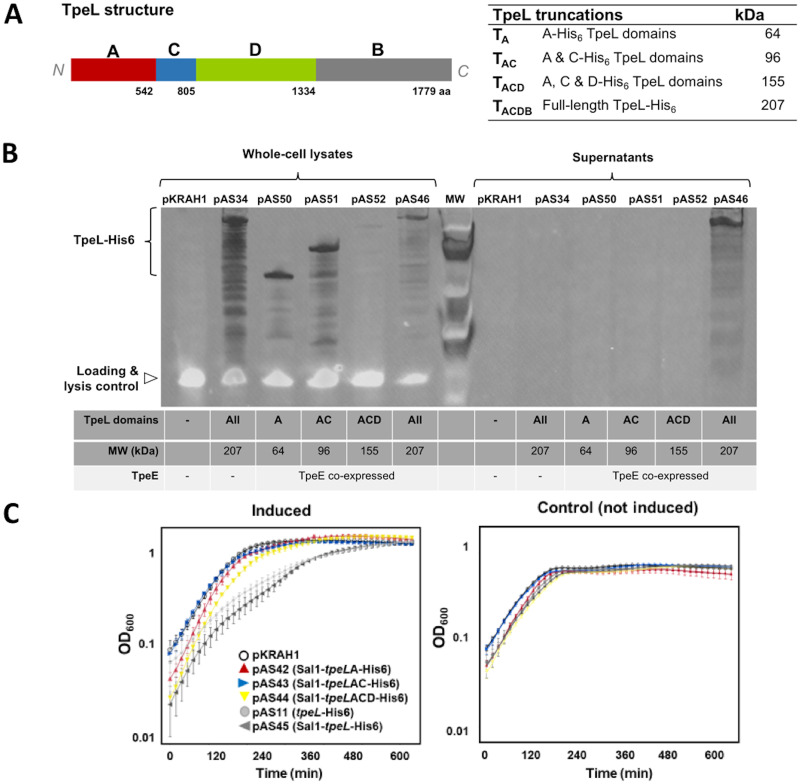
C-terminal truncations of TpeL are not secreted by TpeE.
TpeL is a large clostridial toxin (LCT) that shares a common ACDB structure with other members of this class, where A is the glycosyltransferase domain, C is the autocatalytic protease domain, D is the delivery domain, and B is the receptor domain, but lacks the C-terminal CROPs (combined repetitive oligopeptides) domain (Fig. 8A). In order to determine which domains are needed for secretion, we truncated the toxin by deleting regions encoding domains from the most C-terminal region (B domain) and each additional domain in succession. Each truncated form was coexpressed with full-length TpeE in C. perfringens. Dual-color immunoblotting demonstrated that none of the TpeL truncations were secreted when coexpressed with TpeE compared to a full-length TpeL control (Fig. 8B). Whole-cell lysates verified that each strain expressed TpeL in its truncated form, although TpeL lacking just the B domain showed lower levels of intracellular protein than the other forms (Fig. 8B). This may be due to the hydrophobicity of the D domain, which could make the protein unstable when expressed without the B domain. These results suggest that either the binding domain or all domains of TpeL are needed for TpeE-dependent secretion.
FIG 8.
Truncations of the TpeL toxin lead to loss of secretion. (A) Diagram showing the location of specific domains in the TpeL protein. A, glycosyltransferase domain; C, autocatalytic protease domain; D, delivery domain; B, receptor domain. A table listing the TpeL truncations used and their respective molecular weights is included. (B) Western blot showing the intracellular levels and secretion of full-length and truncated TpeL. An anti-His6 antibody was used to detect each form of the protein. Note the absence of the streptavidin-BCP lysis control signal in the supernatant fractions, indicating that very little lysis had occurred. (C) Growth curves showing that strains expressing truncated TpeL proteins (pAS42, pAS43, and pAS44) do not have defects in their growth in comparison to strains expressing full-length TpeL (pAS11 and pAS45). The means and standard errors of the means (SEM) from five replicate samples are shown. The curves are representative of results from three independent biological replicates.
Growth curves generated with the strains expressing the TpeL truncations suggest that while the expression of the complete protein containing the B domain retards growth early in the exponential phase, strains expressing the truncated versions grew similarly to the one expressing an empty vector control (Fig. 8C). The strain expressing the TpeL truncation lacking the B domain cannot be directly compared to the other forms since its intracellular concentration was shown to be lower in immunoblot experiments. However, taken together, the results of the immunoblot and growth curve experiments (Fig. 8B and C) suggest that the B domain of TpeL is necessary for TpeE-dependent TpeL secretion.
DISCUSSION
Our experimental approach was to determine if the holin TpeE was necessary and sufficient for the secretion of the LCT TpeL in C. perfringens. To simplify the system, we moved the tpeE and tpeL genes out of the host type C strain and into the genetically manipulatable strain HN13. We detected large amounts of the toxin TpeL-His6 in culture supernatants only in C. perfringens strains coexpressing TpeE, even though both strains had similar amounts of intracellular TpeL (Fig. 3A). We demonstrated that the antigen tags present on the C termini of TpeE and TpeL, which were used to locate the proteins to specific locations in the cell by Western blotting, did not play a role in the efficiency of secretion of TpeL (Fig. 3B). Significant levels of cell lysis by the strains expressing the holin, toxin, or holin-toxin combination could not be detected using three independent methods: (i) comparative amounts of CFU per milliliter and direct microscopic counts of bacteria (see Fig. S2A in the supplemental material), (ii) the presence of the cytoplasmic enzyme LDH in culture supernatants (Fig. S2B), and (iii) the detection of a biotin-conjugated cytoplasmic protein (the biotin carboxyl carrier protein [BCP]) in culture supernatants (Fig. 3A). We used a cytoplasmic extract titration method to estimate the lowest level of detection of the BCP while simultaneously detecting the presence of TpeL in culture supernatants and determined that we could detect <2% lysis of cells in our assay (Fig. 3C). Added together, these results indicate that TpeE is indeed necessary and sufficient for the secretion of TpeL by C. perfringens and does not use cell lysis as a mechanism for toxin release. These findings align with previous reports that the LCTs TcdA and TcdB required the holin TcdE for secretion in C. difficile (16, 42). However, there may be other features, in particular regulatory functions, that were lost when we transferred the holin-toxin system from its native host to a heterologous strain.
Low levels of TpeL in the absence of TpeE coexpression (Fig. 3A) were occasionally detected, similar to results seen with TcdE-independent secretion of TcdA in C. difficile (16). While it seems that cell lysis and shedding of membrane vesicles are unlikely, secretion could be due to the presence of an endogenous holin, CPE0383, found in strain 13, the parent of strain HN13 (43). For CPE0383, transcriptome sequencing (RNA-seq) data from a recent study from our laboratory (44) suggest that the gene encoding this holin is transcribed only at very low levels, and the holin is predicted to have 4 transmembrane domains using the TMHMM Web-based software (http://www.cbs.dtu.dk/services/TMHMM-2.0/), so if it provides a low level of TpeL secretion, it is likely using a different mechanism than TpeE. Therefore, the mechanism underlying the small amounts of holin-independent secretion of TpeL and TcdA is still not understood.
The expression of TpeE alone showed no negative effect on growth in C. perfringens (Fig. 4A), which is in contrast to that observed with TcdE in C. difficile, in which expression in the absence of TcdA was toxic to the cells (16, 42), which may indicate different mechanisms of action between TpeE and TcdE. The expression of TpeE was lethal in E. coli (Fig. 6A), which correlates with results from a homolog of TpeE, the holin BhlA from Bacillus pumilus, which also showed significant toxicity when expressed in E. coli (45). This indicates that C. perfringens has a protective mechanism against the toxic effects of TpeE, presumably due to the formation of pores in the membrane, which is lacking in E. coli. Some of the differences in lethality may be due to lower TpeE levels in C. perfringens than in E. coli (Fig. 5 and 6), as evidenced by the lower signal in Western blots, possibly due to differences in promoter strength between pBAD30 (E. coli) and pKRAH1 (C. perfringens). However, the lethal effect in E. coli was delayed; although expression was induced at inoculation, the loss in optical density was not seen until ∼2.5 h later, in the late logarithmic phase of growth (Fig. 6A). This may indicate that the levels of TpeE in the cytoplasmic membrane need to build to a critical level before pore formation is initiated, much like a phage holin such as the lambda phage holin S105 (46). It was found that tcdE, like S105, may be transcribed and translated into multiple isoforms, which can affect the secretion of TcdA (42). We did not see evidence of similar TpeE isoforms in C. perfringens using RT-PCR (Fig. 5B) and SDS-PAGE and Western blotting.
The TMHMM Web-based software (http://www.cbs.dtu.dk/services/TMHMM-2.0/) predicted that TpeE had a single transmembrane helix (data not shown). Since we suspected that TpeE may have a membrane topology similar to that of the TatA family of proteins, we used helical wheel projections to identify an amphipathic helix (APH) in the middle of the sequence (Fig. 7A), similar to that found in the structure of TatAd from B. subtilis (24, 25). There is also a 7-residue C-terminal domain containing mostly charged and hydrophilic residues (HCD), which we postulated may interact with the charged residues on the amphipathic helix. To begin to understand how TpeE contributes to TpeL secretion, we removed both the C-terminal residues and the predicted amphipathic helix of the protein (Fig. 7A). Each mutation abolished the rapid TpeE-dependent secretion seen with the full-length protein (Fig. 7B). While we could detect the HCD-deleted protein using mass spectrometry methods (see primers and methods in the supplemental material), we could not detect the APH-deleted TpeE, so we cannot confidently state that the APH domain is essential for TpeL secretion. Despite the requirement for both of the deleted domains, the addition of an antigen tag to the C terminus of TpeE did not have an effect on TpeL secretion (Fig. 3B), which suggests that the amphipathic helix and C-terminal domains are needed for specific functions in the TpeE-mediated secretion of TpeL.
Nested deletions of the four domains (ACDB) starting from the C terminus of TpeL also resulted in a complete loss of secretion, although there was too little of the stable intracellular ACD form to be able to conclude if secretion was actually defective. The instability of the ACD form could be due to the exposure of a hydrophobic domain within the delivery domain of the protein that would lead to rapid turnover by intracellular proteases. But clearly, the A and AB domains were not sufficient for secretion. This is the first time that the role of individual domains in LCT secretion has been examined.
The actual mechanism for the holin-dependent transport of TpeL and the other LCTs is not known. Even though they lack sequence homology, the TatA family of proteins has a membrane topology very similar to that of the holin family represented by TpeE and UviB. The precise mechanism by which TatA forms a pore to transport the TatA substrates is not known, although three early models were proposed: an iris mechanism (47), a gated pore mechanism (48), and a membrane-weakening mechanism (49). More recently, a charge zipper mechanism was proposed, where a pore is formed by TatA in which the amphipathic helix becomes inserted into the membrane antiparallel to the transmembrane helix, and this conformation is stabilized by salt bridges between the charges on the C-terminal domain and the amphipathic helix (41, 50). Support for this model was provided by the determination of the solution nuclear magnetic resonance (NMR) structure of TatAd from B. subtilis (24), which also provided support for the TpeE membrane topology shown in Fig. 1B. Solid-state NMR of a truncated form of TatAd (residues 2 to 45) that contained the transmembrane helix and the amphipathic helix suggested that the amphipathic helix was pulled into the membrane by the short (14-residue) transmembrane helix and was partially stabilized by intramolecular salt bridges (25). Using the charged zipper mechanism for the TatA secretion system as the basis for a model, we have developed a hypothesis for how TpeE might function to secrete TpeL (Fig. 9). In this model, TpeE exists as nonoligomerized monomers in the membrane since pore formation in the absence of the substrate would lead to the collapse of the proton motive force (PMF) and death of the cell. In the next step, TpeL is inserted into the membrane using a surface-exposed hydrophobic patch (Fig. 9) that is analogous to a conserved hydrophobic domain that is part of the pore-forming domain found in TcdA that helps the protein traverse the endosomal membrane and gain access to the cytoplasm of mammalian cells (51). The insertion of TpeL into the membrane acts as the initiator of the amphipathic helix of TpeE to fold into the membrane, followed by oligomerization around the TpeL protein, forming a pore through which the toxin is transported (Fig. 9). Evidence in support of this model includes the requirement for both the APH domain and the HCD of TpeE for secretion as well as the absence of secretion when just the A or AC domains of TpeL were present since they do not include the residues encoding the major portion of the hydrophobic patch (residues 1081 to 1101 in TpeL), which is conserved among all of the LCTs (51).
FIG 9.
Model showing a potential mechanism of TpeE-dependent secretion of TpeL. Hydrophobic residues are shown in orange in the TpeL model. The sequence shown is based on the charged zipper mechanism proposed for the Tat secretion system (41), where TatA is a membrane topology model for TpeE. The main features of the model are described in Discussion. The asterisks represent potential salt bridges between charged residues in the amphipathic helix (APH) and high charge density (HCD) domains. The model for the TpeL structure was made using the TcdA structure as a template with the Swiss Model software package. The hydrophobic surface residues were identified using the PyMOL molecular graphics system. TMH, transmembrane helix.
Proteins destined for secretion that require a holin-like protein for transport across the cytoplasmic membrane fall into three classes, those requiring a cofactor that needs to be added in the cytoplasm, proteins that require the cytoplasmic environment to achieve proper folding, or oligomeric proteins that cannot utilize the Sec secretion pathway (26, 27). This is the case for both the holin-dependent substrates such as the LCTs discussed here as well as the Tat system. Except for TcnA in C. novyi, all of the LCTs have an associated holin (Fig. 1A). This suggests that they too require folding in the cytoplasm and are secreted via holin-mediated mechanisms. There are two possible reasons why the LCTs need to fold in the cytoplasm: (i) their large size and complex folds (51) require additional factors such as chaperones to fold properly, or (ii) the addition of a zinc atom in the C (autoprotease) domain requires cytoplasmic conditions. Although the members of the TcdA/TcdB/TcsL/TcsH family of LCTs have a holin associated with them with a very different membrane topology than that of the TpeE, UviB, and TatA proteins, it seems likely that they too require the holin for secretion because they need to fold in the cytoplasm. So why do they use a holin with a different membrane topology from that of TpeL? One possible answer is that TpeL is unique among LCTs in lacking a large CROPs (combined repetitive oligopeptides) domain at the C terminus. The CROPs domain of TcdB (residues 1834 to 2367) is connected to the rest of TcdB by a hinge region, which allows the CROPs domain to be flexible and interact with the other domains of the toxin. The presence of such a flexible feature may make the other LCTs unable to utilize a TpeE-like holin for secretion.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1, and primers are listed in Table S1 in the supplemental material. E. coli strain DH10B was grown in Luria-Bertani (LB) broth (1% tryptone and sodium chloride plus 0.5% yeast extract) at 37°C for all experiments. C. perfringens strains HN13 and JGS1495 were cultured anaerobically with either brain heart infusion (BHI) broth (Thermo Fisher) or TY medium (3% tryptone, 2% yeast extract, 0.1% sodium thioglycolate) at 37°C in an anaerobic chamber (Coy Laboratory Products, Inc.) under anaerobic conditions (approximately 85% nitrogen, 5% hydrogen, and 10% carbon dioxide). One hundred micrograms per microliter of kanamycin was added to media for E. coli strains containing pCRBluntIITOPO, and 100 μg/ml ampicillin was added for strains carrying pBAD30. Twenty micrograms per microliter of chloramphenicol was added to the culture media for C. perfringens strains carrying pKRAH1.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Clostridium perfringens type A strain HN13 | ΔgalKT | 30 |
| Clostridium perfringens type C strain JGS1495 | Contains tpeE-tpeL genes | J. Glenn Songer |
| Escherichia coli strain DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG λ− | 59 |
| Escherichia coli strain LOBSTR | Derivative of strain BL21; designed for low background binding to Ni affinity columns | 58 |
| Plasmids | ||
| pKRAH1 | Cmr; lactose-inducible expression system plasmid for use in C. perfringens | 31 |
| pCRBluntIITOPO | PCR cloning vector | Invitrogen |
| pBAD30 | Ampr; arabinose-inducible expression system plasmid for use in E. coli | 56 |
| pAS6 | Kmr; SacII tpeL-His6 BamHI in pCRBluntIITOPO | This study |
| pAS7 | Kmr; SacII tpeE and tpeL-His6 BamHI in pCRBluntIITOPO | This study |
| pAS8 | Kmr; SacII tpeE BamHI | This study |
| pAS9 | Kmr; SacII tpeE-His6 BamHI | This study |
| pAS10 | Kmr; SacII tpeE-FLAG BamHI in pCRBluntIITOPO | This study |
| pAS11 | Cmr; tpeL-His6 in pKRAH1 | This study |
| pAS12 | Cmr; tpeE-tpeL-His6 in pKRAH1 | This study |
| pAS13 | Cmr; tpeE in pKRAH1 | This study |
| pAS14 | Cmr; tpeE-His6 in pKRAH1 | This study |
| pAS15 | Cmr; tpeE-FLAG in pKRAH1 | This study |
| pAS16 | Ampr; tpeE-FLAG in pBAD30 | This study |
| pAS17 | Ampr; tpeE in pBAD30 | This study |
| pAS18 | Ampr; tpeE-His6 in pBAD30 | This study |
| pAS19 | Amp; tpeL-His6 in pBAD30 | This study |
| pAS25 | Kmr; SacII-tpeE-SalI in pCRBluntIITOPO | This study |
| pAS26 | Kmr; SacII-tpeE-His6-SalI in pCRBluntIITOPO | This study |
| pAS27 | Kmr; SacII-tpeE-FLAG-SalI in pCRBluntIITOPO | This study |
| pAS28 | Kmr; SacII-tpeE1–58-SalI in pCRBluntIITOPO | This study |
| pAS29 | Kmr; SacII-tpeEΔhelix-SalI in pCRBluntIITOPO | This study |
| pAS30 | Kmr; SalI-tpeLA-His6-BamHI in pCRBluntIITOPO | This study |
| pAS31 | Kmr; SalI-tpeLAC-His6-BamHI in pCRBluntIITOPO | This study |
| pAS32 | Kmr; SalI-tpeLACD-His6-BamHI in pCRBluntIITOPO | This study |
| pAS33 | Kmr; SalI-tpeLACDB-His6-BamHI in pCRBluntIITOPO | This study |
| pAS34 | Kmr; tpeE-SalI-tpeLACDB-His6-BamHI in pCRBluntIITOPO | This study |
| pAS35 | Kmr; SacII-tpeL-BamHI in pCRBluntIITOPO | This study |
| pAS36 | Kmr; SacII-tpeE-tpeL-BamHI in pCRBluntIITOPO | This study |
| pAS37 | Cmr; tpeE (SalI site added) in pKRAH1 | This study |
| pAS38 | Cmr; tpeE-His6 (SalI site added) in pKRAH1 | This study |
| pAS39 | Cmr; tpeE-FLAG (SalI site added) in pKRAH1 | This study |
| pAS40 | Cmr; tpeE1–58 (SalI site added) in pKRAH1 | This study |
| pAS41 | Cmr; tpeE1–34,59–65 (SalI site added) in pKRAH1 | This study |
| pAS42 | Cmr; (SalI site added) tpeL1–542-His6 in pKRAH1 | This study |
| pAS43 | Cmr; (SalI site added) tpeL1–805-His6 in pKRAH1 | This study |
| pAS44 | Cmr; (SalI site added) tpeL1–1334-His6 in pKRAH1 | This study |
| pAS45 | Cmr; (SalI site added) tpeL-His6 in pKRAH1 | This study |
| pAS46 | Cmr; tpeE-(SalI site added)-tpeL-His6 in pKRAH1 | This study |
| pAS48 | Cmr; tpeE1–58-(SalI site added)-tpeL-His6 in pKRAH1 | This study |
| pAS49 | Cmr; tpeE1–34,59–65-(SalI site added)-tpeL-His6 in pKRAH1 | This study |
| pAS50 | Cmr; tpeE-(SalI site added)-tpeL1–542-His6 in pKRAH1 | This study |
| pAS51 | Cmr; tpeE-(SalI site added)-tpeL1–805-His6 in pKRAH1 | This study |
| pAS52 | Cmr; tpeE-(SalI site added)-tpeL1–1334-His6 in pKRAH1 | This study |
| pAS53 | Cmr; tpeE-His6-(SalI site added)-tpeL-His6 in pKRAH1 | This study |
| pAS54 | Cmr; tpeL in pKRAH1 | This study |
| pAS55 | Cmr; tpeE-(SalI site added)-tpeL-His6 in pKRAH1 | This study |
| pAS58 | Cmr; tpeE-FLAG-tpeL-His6 in pKRAH1 | This study |
Kmr represents kanamycin resistance, Ampr represents ampicillin resistance, and Cmr represents chloramphenicol resistance.
For protein expression studies, C. perfringens strains were cultured under anaerobic conditions in an anaerobic chamber at 37°C in TY medium. Cultures grown overnight were used to inoculate experimental cultures at a 1:50 dilution. Lactose was added to a final concentration of 20 or 30 mM to induce expression from the lactose-inducible promoter in pKRAH1 (31) at the time of inoculation for the growth curve analyses and in the late logarithmic growth phase (3.75 h postinoculation) for protein expression experiments. Culture samples for SDS-PAGE analysis were harvested 3 h (Fig. 3A) or 4 h (Fig. 3B, Fig. 5A, Fig. 7B, and Fig. 8A) following induction.
For E. coli, cultures grown overnight from frozen stocks were used to inoculate, at a 1:50 dilution, experimental cultures. Arabinose was added to induce expression from pBAD30. Inducing sugar was added either at inoculation or in the late logarithmic phase of growth (2 h, as determined by growth curve analysis). E. coli culture samples for SDS-PAGE analysis were harvested 2 h following induction in order to capture a time where expression and effects could be observed before the culture cleared from lysis (Fig. 6A).
For measuring growth, 200 μl of the cell culture was aliquoted into clear, flat-bottomed, 96-well microtiter plates (Costar; Corning). For C. perfringens experiments, 30 mM lactose was added at inoculation, and anaerobic conditions were maintained by sealing the plates with clear adhesive film. For E. coli experiments, various amounts of arabinose were added at inoculation. For both organisms, the optical density was monitored over time at 600 nm at 37°C in a Tecan microplate reader, with sampling and agitation every 15 to 30 min. For the spot plate assays, 5 μl of a culture grown overnight was placed onto agar plates with increasing amounts of arabinose and incubated overnight.
Constructs, cloning, and mutagenesis.
Genomic DNA was isolated from C. perfringens strain JGS1495 by phenol-chloroform extraction and used as the template in full-length, antigen-tagged, and mutated isoforms for tpeE and tpeL amplifications. Primer OAS28F was used to flank tpeE with a 5′ SacII site, OAS36R with a 3′ BamHI site, and OAS57R with a 3′ SalI restriction site, as indicated. C-terminal His6 sequences were fused to tpeE with primers OAS51R (3′ BamHI) and OAS62R (3′ SalI), and FLAG sequences were fused with primers OAS55R (3′ BamHI) and OAS63R (3′ Sal). tpeE-tpeL with 5′ SacII and 3′ BamHI were amplified with primers OAS28F and OAS31R, and overlapping primers OAS29 and OAS30 were used to fuse polyhistidine tag sequences to the C terminus of tpeL, with additional Gly-Ser-Gly sequences between the gene and histidine-encoding codons. All constructs were confirmed by sequencing using primers OAS37 to OAS49.
To mutate the amphipathic helix of tpeE, Heliquest (52) was used to predict the boundaries of the helix, which included nucleotides 103 to 177. From this, a reverse primer, OAS64R, was made to truncate tpeE at nucleotide 102, and the remaining 21 bases were included in the primer itself to add the terminal residues. To delete the 7 C-terminal codons from tpeE, a reverse primer truncating it to nucleotide 174 was used to amplify the remaining portion of the gene (OAS64R).
The ACDB domain boundaries of TpeL were determined by comparison to the structure of TcdA (51) and the sequence of TpeL, with the A domain being amino acids 1 to 542 (4, 5, 53), the C domain of TpeL being amino acids 543 to 805 (4), and the B domain being amino acids 1335 to 1779 (54), leaving the D domain as amino acids 806 to 1334. Therefore, tpeL was amplified to nucleotide 1626 for the A domain construct with primers OAS59F and OAS61R, to nucleotide 2415 for the AC construct with OAS65R, and to nucleotide 4002 for the ACD construct with OAS66R.
SeqBuilder software was used for primer design (SeqBuilder Pro version 10.0; DNAStar, Madison, WI). PCR products were blunt-end ligated into pCRBluntIITOPO and electroporated into E. coli DH10B (55). Constructs were verified by Sanger sequencing (Genomics Sequencing Center, Virginia Tech, Blacksburg, VA, and Eton Biosciences, Eton, NC) and alignment to JGS1495 sequences (SeqMan NGen version 10.0; DNAStar, Madison, WI). Constructs were then digested from pCRBluntIITOPO and ligated into pKRAH1 (31) or pBAD30 (56). Due to lethality and other off-target effects, plasmids containing full-length tpeE and tpeL in pKRAH1 were not transformed into E. coli, and instead, ligation mixes were transformed directly into C. perfringens strain HN13 by electroporation (57). Plasmids comprising mutated tpeE and full-length tpeL-His6 were assembled by digesting previously constructed plasmids of each tpeE isoform in pKRAH1 with SalI and BamHI and ligating this product with tpeL-His6 in pCRBluntIITOPO digested with SalI and BamHI. Similarly, plasmids comprising full-length tpeE and mutated tpeL isoforms were assembled by digesting pAS37 (full-length tpeE in pKRAH1) with SalI and BamHI and ligating it with each mutated tpeL isoform digested with the same restriction enzymes in pCRBluntIITOPO.
E. coli DH10B and LOBSTR (58) strains with tpeE, tpeE-His6, tpeE-FLAG, and tpeL-His6 constructs were generated by isolating the constructs from previously constructed plasmids and ligating the constructs into pBAD30 (56).
SDS-PAGE, staining, and immunoblotting.
Culture samples (1 to 1.5 ml) were separated into supernatant and pellet fractions by centrifugation (14,000 × g for 2 min) and stored at −80°C until used. Cell pellets were suspended in Dulbecco’s phosphate-buffered saline (DPBS) and lysed by agitation with 0.1-mm zircon beads for 120 s, with 60-s intervals on ice to prevent overheating. In TpeE localization studies, whole-cell lysate samples were prepared, and the lysate was further fractionated into membrane and cytoplasmic samples by ultracentrifugation at 40,000 rpm for 1 h at 4°C. Following separation, cytoplasmic fractions were concentrated with trichloroacetic acid (TCA) precipitation and washed twice in acetone. Supernatant fractions were similarly concentrated after filtration through 0.45-μm membranes. Positive lysis controls were generated by adding the lysate from the empty vector strain back to the clarified supernatant and then concentrating the proteins as described above. All fractions were suspended in SDS-PAGE buffer with 100 mM dithiothreitol and heated to 95°C for at least 10 min. Tris base was added as needed to return the pH to neutral in TCA-concentrated samples.
Proteins were separated by SDS-PAGE and either stained or immunoblotted. Proteins were stained with Coomassie brilliant blue in 40% methanol and 10% acetic acid. For immunoblots, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane in chilled buffer containing 10 mM NaHCO3 and 3 mM Na2CO3 with 20% methanol. Membranes were transferred to the Snap i.d. 2.0 system (Millipore Sigma) and blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS) (pH 7.4) with 1% Tween 20. Antibodies were diluted into TBS-Tween 20 with 1% BSA and applied to membranes for 10 min. Primary antibodies used were His-H8 (1:250 dilution) to detect polyhistidine tags and OctA-probe H-5 (1:250 dilution) for FLAG detection (both from Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody used was StarBright700 anti-mouse (Bio-Rad) (1:10,000 dilution), and conjugated streptavidin (IRDye 800 CW; Li-Cor, Lincoln, NE) at a dilution of 1:5,000 was added to the secondary antibody mixture for the detection of biotinylated, cytoplasmic protein controls (BCP [18 kDa] in C. perfringens and AccB [22.5 kDa] in E. coli). Fluorescent blot images were captured with a ChemiDoc MP imaging system (Bio-Rad).
RNA isolation and RT-PCR.
RNA was extracted from the same cells as the ones used in the immunoblot experiments. Amplification by PCR was done with and without reverse transcriptase. Primers OAS67 and OAS68 were designed to amplify cDNA from mRNA of TpeE and the TpeE isoforms generated in this study from a probable ribosomal binding site 11 nucleotides upstream of the +1 site to 97 nucleotides after the translation stop sequence. Four-hour-postinduction culture samples from the TpeE mutant experiments (Fig. 7) were harvested, pelleted by centrifugation, and stored at −80°C. RNA was isolated from these samples with the Direct-Zol RNA MiniPrep Plus kit (Zymo Research), and in addition to the on-column kit digestion step, contaminating DNA was further digested with a second DNase I treatment (New England BioLabs). The RNA concentration was assessed by nanospectrometry, quality was measured by TapeStation analysis (Genomics Sequencing Center, Virginia Tech, Blacksburg, VA), and only RNA with integrity values above 7.7 were used. RT-PCR was carried out with the OneTaq one-step RT-PCR kit (New England BioLabs), with equivalent amounts of RNA added to each reaction mixture and using the recommended reagent concentrations and thermocycler settings. Equal volumes of each reaction mixture were loaded onto a 2% agarose gel prestained with GelRed nucleic acid stain (Biotium, Fremont, CA), and images were captured using a Bio-Rad ChemiDoc MP system.
Statistics.
Means and standard error were determined and graphed with JMP software (version 11.0, 1989 to 2019; SAS Institute, Inc., Cary, NC) or Prism 6 (GraphPad Software).
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Keith Ray, Richard Helm, and the Virginia Tech Proteomics Incubator for assistance with mass spectrometry. We thank Jaime Jensen, Vanderbilt University, for assistance with modeling the structure of TpeL and Emily Byrd and Nirmala Sharma for their help generating E. coli strains.
Research reported in this publication was supported by the Institute of Allergy and Infectious Disease of the National Institutes of Health under award number R21AI109391 to S.B.M.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Navarro MA, McClane BA, Uzal FA. 2018. Mechanisms of action and cell death associated with Clostridium perfringens toxins. Toxins (Basel) 10:212. doi: 10.3390/toxins10050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schirmer J, Aktories K. 2004. Large clostridial cytotoxins: cellular biology of Rho/Ras-glucosylating toxins. Biochim Biophys Acta 1673:66–74. doi: 10.1016/j.bbagen.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Pauillac S, D’Allayer J, Lenormand P, Rousselle JC, Bouvet P, Popoff MR. 2013. Characterization of the enzymatic activity of Clostridium perfringens TpeL. Toxicon 75:136–143. doi: 10.1016/j.toxicon.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Guttenberg G, Hornei S, Jank T, Schwan C, Lu W, Einsle O, Papatheodorou P, Aktories K. 2012. Molecular characteristics of Clostridium perfringens TpeL toxin and consequences of mono-O-GlcNAcylation of Ras in living cells. J Biol Chem 287:24929–24940. doi: 10.1074/jbc.M112.347773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagahama M, Ohkubo A, Oda M, Kobayashi K, Amimoto K, Miyamoto K, Sakurai J. 2011. Clostridium perfringens TpeL glycosylates the Rac and Ras subfamily proteins. Infect Immun 79:905–910. doi: 10.1128/IAI.01019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551-15. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen AL, Bhatnagar J, Reagan S, Zane SB, D’Angeli MA, Fischer M, Killgore G, Kwan-Gett TS, Blossom DB, Shieh WJ, Guarner J, Jernigan J, Duchin JS, Zaki SR, McDonald LC. 2007. Toxic shock associated with Clostridium sordellii and Clostridium perfringens after medical and spontaneous abortion. Obstet Gynecol 110:1027–1033. doi: 10.1097/01.AOG.0000287291.19230.ba. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2005. Clostridium sordellii toxic shock syndrome after medical abortion with mifepristone and intravaginal misoprostol—United States and Canada, 2001–2005. MMWR Morb Mortal Wkly Rep 54:724. [PubMed] [Google Scholar]
- 9.Carter GP, Awad MM, Hao Y, Thelen T, Bergin IL, Howarth PM, Seemann T, Rood JI, Aronoff DM, Lyras D. 2011. TcsL is an essential virulence factor in Clostridium sordellii ATCC 9714. Infect Immun 79:1025–1032. doi: 10.1128/IAI.00968-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park M, Rafii F. 2019. The prevalence of plasmid-coded cpe enterotoxin, beta2 toxin, tpeL toxin, and tetracycline resistance in Clostridium perfringens strains isolated from different sources. Anaerobe 56:124–129. doi: 10.1016/j.anaerobe.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Gu C, Lillehoj HS, Sun Z, Lee Y, Zhao H, Xianyu Z, Yan X, Wang Y, Lin S, Liu L, Li C. 2019. Characterization of virulent netB+/tpeL+ Clostridium perfringens strains from necrotic enteritis-affected broiler chicken farms. Avian Dis 63:461–467. doi: 10.1637/11973-092018-Reg.1. [DOI] [PubMed] [Google Scholar]
- 12.Coursodon CF, Glock RD, Moore KL, Cooper KK, Songer JG. 2012. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 18:117–121. doi: 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy B, Mani N, Katayama S, Sonenshein AL. 2005. Transcription activation of a UV-inducible Clostridium perfringens bacteriocin gene by a novel sigma factor. Mol Microbiol 55:1196–1206. doi: 10.1111/j.1365-2958.2004.04456.x. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy B, Matamouros S. 2006. Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res Microbiol 157:201–205. doi: 10.1016/j.resmic.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Carter GP, Larcombe S, Li L, Jayawardena D, Awad MM, Songer JG, Lyras D. 2014. Expression of the large clostridial toxins is controlled by conserved regulatory mechanisms. Int J Med Microbiol 304:1147–1159. doi: 10.1016/j.ijmm.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, Pich A, Just I, Gerhard R. 2012. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy BL, Saier MH, Jr. 2013. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim Biophys Acta 1828:2654–2671. doi: 10.1016/j.bbamem.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier MH, Jr., Reddy BL. 2015. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol 197:7–17. doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garnier T, Cole ST. 1988. Complete nucleotide sequence and genetic organization of the bacteriocinogenic plasmid, pIP404, from Clostridium perfringens. Plasmid 19:134–150. doi: 10.1016/0147-619X(88)90052-2. [DOI] [PubMed] [Google Scholar]
- 23.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res 16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Zhao E, Li H, Xia B, Jin C. 2010. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J Am Chem Soc 132:15942–15944. doi: 10.1021/ja1053785. [DOI] [PubMed] [Google Scholar]
- 25.Walther TH, Grage SL, Roth N, Ulrich AS. 2010. Membrane alignment of the pore-forming component TatA(d) of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J Am Chem Soc 132:15945–15956. doi: 10.1021/ja106963s. [DOI] [PubMed] [Google Scholar]
- 26.Goosens VJ, van Dijl JM. 2017. Twin-arginine protein translocation. Curr Top Microbiol Immunol 404:69–94. doi: 10.1007/82_2016_7. [DOI] [PubMed] [Google Scholar]
- 27.Frain KM, van Dijl JM, Robinson C. 19 June 2019. The twin-arginine pathway for protein secretion. EcoSal Plus 2019 10.1128/ecosalplus.ESP-0040-2018. [DOI] [PubMed]
- 28.Eimer E, Frobel J, Blummel AS, Muller M. 2015. TatE as a regular constituent of bacterial twin-arginine protein translocases. J Biol Chem 290:29281–29289. doi: 10.1074/jbc.M115.696005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan KA, Elbourne LDH, Tetu SG, Melville SB, Rood JI, Paulsen IT. 2015. Genomic analyses of Clostridium perfringens isolates from five toxinotypes. Res Microbiol 166:255–263. doi: 10.1016/j.resmic.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Nariya H, Miyata S, Suzuki M, Tamai E, Okabe A. 2011. Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl Environ Microbiol 77:1375–1382. doi: 10.1128/AEM.01572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartman AH, Liu H, Melville SB. 2011. Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl Environ Microbiol 77:471–478. doi: 10.1128/AEM.01536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewerke LT, Kies PJ, Muh U, Ellermeier CD. 2018. Bacterial sensing: a putative amphipathic helix in RsiV is the switch for activating sigmaV in response to lysozyme. PLoS Genet 14:e1007527. doi: 10.1371/journal.pgen.1007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, Funk K, Ketter A, Kim S, Kimchi A, Kitts PA, Kuznetsov A, Lathrop S, Lu Z, McGarvey K, Madden TL, Murphy TD, O’Leary N, Phan L, Schneider VA, Thibaud-Nissen F, Trawick BW, Pruitt KD, Ostell J. 2020. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 48:D9–D16. doi: 10.1093/nar/gkz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Kong Q, Roland KL, Curtiss R, III. 2014. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 304:431–443. doi: 10.1016/j.ijmm.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Paredes-Sabja D, Sarker MR, McClane BA. 2016. Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol Spectr 4:TBS-0022-2015. doi: 10.1128/microbiolspec.TBS-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li SJ, Cronan JE, Jr. 1992. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267:16841–16847. doi: 10.1016/S0021-9258(18)41860-1. [DOI] [PubMed] [Google Scholar]
- 39.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Walther TH, Gottselig C, Grage SL, Wolf M, Vargiu AV, Klein MJ, Vollmer S, Prock S, Hartmann M, Afonin S, Stockwald E, Heinzmann H, Nolandt OV, Wenzel W, Ruggerone P, Ulrich AS. 2013. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell 152:316–326. doi: 10.1016/j.cell.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Govind R, Fitzwater L, Nichols R. 2015. Observations on the role of TcdE isoforms in Clostridium difficile toxin secretion. J Bacteriol 197:2600–2609. doi: 10.1128/JB.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A 99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soncini SR, Hartman AH, Gallagher TM, Camper GJ, Jensen RV, Melville SB. 2020. Changes in the expression of genes encoding type IV pili-associated proteins are seen when Clostridium perfringens is grown in liquid or on surfaces. BMC Genomics 21:45. doi: 10.1186/s12864-020-6453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aunpad R, Panbangred W. 2012. Evidence for two putative holin-like peptides encoding genes of Bacillus pumilus strain WAPB4. Curr Microbiol 64:343–348. doi: 10.1007/s00284-011-0074-3. [DOI] [PubMed] [Google Scholar]
- 46.Savva CG, Dewey JS, Deaton J, White RL, Struck DK, Holzenburg A, Young R. 2008. The holin of bacteriophage lambda forms rings with large diameter. Mol Microbiol 69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berks BC, Sargent F, Palmer T. 2000. The Tat protein export pathway. Mol Microbiol 35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 48.Musser SM, Theg SM. 2000. Characterization of the early steps of OE17 precursor transport by the thylakoid DeltapH/Tat machinery. Eur J Biochem 267:2588–2598. doi: 10.1046/j.1432-1327.2000.01269.x. [DOI] [PubMed] [Google Scholar]
- 49.Bruser T, Sanders C. 2003. An alternative model of the twin arginine translocation system. Microbiol Res 158:7–17. doi: 10.1078/0944-5013-00176. [DOI] [PubMed] [Google Scholar]
- 50.Walther TH, Ulrich AS. 2014. Transmembrane helix assembly and the role of salt bridges. Curr Opin Struct Biol 27:63–68. doi: 10.1016/j.sbi.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Chumbler NM, Rutherford SA, Zhang Z, Farrow MA, Lisher JP, Farquhar E, Giedroc DP, Spiller BW, Melnyk RA, Lacy DB. 2016. Crystal structure of Clostridium difficile toxin A. Nat Microbiol 1:15002. doi: 10.1038/nmicrobiol.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gautier R, Douguet D, Antonny B, Drin G. 2008. HELIQUEST: a Web server to screen sequences with specific alpha-helical properties. Bioinformatics 24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 53.Amimoto K, Noro T, Oishi E, Shimizu M. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology (Reading) 153:1198–1206. doi: 10.1099/mic.0.2006/002287-0. [DOI] [PubMed] [Google Scholar]
- 54.Schorch B, Song S, van Diemen FR, Bock HH, May P, Herz J, Brummelkamp TR, Papatheodorou P, Aktories K. 2014. LRP1 is a receptor for Clostridium perfringens TpeL toxin indicating a two-receptor model of clostridial glycosylating toxins. Proc Natl Acad Sci U S A 111:6431–6436. doi: 10.1073/pnas.1323790111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durfee T, Nelson R, Baldwin S, Plunkett G, III., Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgo B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melville SB, Labbe R, Sonenshein AL. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect Immun 62:5550–5558. doi: 10.1128/IAI.62.12.5550-5558.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen KR, Leksa NC, Schwartz TU. 2013. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81:1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.