The fitness of an individual bacterial cell is highly dependent upon the temporal tuning of gene expression levels when subjected to different environmental cues. Kinetic regulation of transcription initiation is a key step in modulating the levels of transcribed genes to promote bacterial survival.
KEYWORDS: RNA polymerase, gene regulation, kinetics, promoter motifs, transcription initiation
ABSTRACT
The fitness of an individual bacterial cell is highly dependent upon the temporal tuning of gene expression levels when subjected to different environmental cues. Kinetic regulation of transcription initiation is a key step in modulating the levels of transcribed genes to promote bacterial survival. The initiation phase encompasses the binding of RNA polymerase (RNAP) to promoter DNA and a series of coupled protein-DNA conformational changes prior to entry into processive elongation. The time required to complete the initiation phase can vary by orders of magnitude and is ultimately dictated by the DNA sequence of the promoter. In this review, we aim to provide the required background to understand how promoter sequence motifs may affect initiation kinetics during promoter recognition and binding, subsequent conformational changes which lead to DNA opening around the transcription start site, and promoter escape. By calculating the steady-state flux of RNA production as a function of these effects, we illustrate that the presence/absence of a consensus promoter motif cannot be used in isolation to make conclusions regarding promoter strength. Instead, the entire series of linked, sequence-dependent structural transitions must be considered holistically. Finally, we describe how individual transcription factors take advantage of the broad distribution of sequence-dependent basal kinetics to either increase or decrease RNA flux.
TRANSCRIPTION INITIATION OVERVIEW
The bacterial RNA polymerase (RNAP) core enzyme, composed of β, β′, and ω subunits, along with an α dimer subunit, represents the catalytic machinery responsible for DNA-templated RNA synthesis (1, 2). For RNAP to initiate promoter specific transcription, it must first assemble with a σ factor to form RNAP holoenzyme (3, 4). Bacterial σ factors are classified into two families based on homology, called σ70 and σ54 (5). This review focuses on mechanisms specific to Escherichia coli σ70 (6); those of σ54 are distinct and require ATP-dependent remodeling by bacterial enhancer-binding proteins (7). Within the σ70 family, a further group classification is made based on the presence or absence of four structural domains (σ1.1, σ2, σ3, and σ4) (8, 9). Group 1 includes the essential housekeeping σ factor (σ70) and contains all four structural domains; group 2 includes σ38 (σS) which lacks σ1.1 and plays important roles in stress responses and survival but can also transcribe housekeeping genes (10–13); group 3 (usually containing σ2, σ3, σ4) and group 4 (containing only σ2 and σ4) generally transcribe smaller sets of genes in response to specific stresses. In the case of group 4, these stress signals are often generated outside the cell, leading to their designation of extracytoplasmic function (ECF) σ factors (14–16). Expression of σ factors can be regulated based on environmental conditions either at the gene or protein level (13, 17), resulting in temporal concentration changes and permitting competition in binding of RNAP core to form σ-specific holoenzymes (18–21). This review will focus on mechanisms specific to group 1 σ-containing holoenzymes.
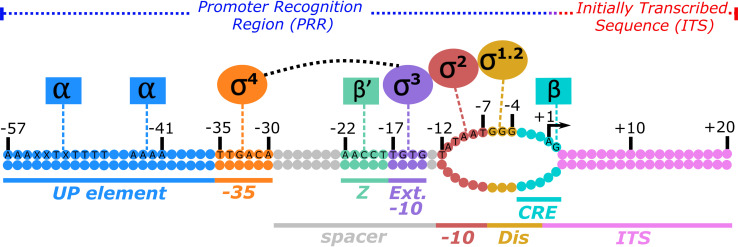
Initiation begins when the RNAP holoenzyme (R) binds to a fully duplexed promoter DNA sequence (P) to form a closed complex (RPc). Sequence positions within a promoter, defined as a DNA segment essential for holoenzyme-specific initiation (22), are numbered relative to the transcription start site (TSS) occurring at +1 position. Sequences upstream of the TSS are negative and those downstream are positive. Promoter recognition is primarily driven by protein-DNA contacts between σ2, σ3, and σ4 with the −10, the extended −10, and the −35 regions of the promoter, respectively (23–25). Contacts between the C-terminal domain of the α-subunits (αCTDs) and upstream (UP) elements in the DNA also contribute (26, 27) (Fig. 1). Contacts within the −10 region nucleate DNA unwinding around the TSS, resulting in an open complex (RPo) (25, 28–30) after passing through a variety of structural intermediates. These intermediates involve conformational changes in the DNA (i.e., wrapping of upstream DNA and loading downstream DNA into the RNAP cleft) (31–35) and mobile RNAP structural elements that facilitate DNA opening and stabilize the RPo (30, 36–41). Subsequent to DNA opening and binding of the initiating nucleotide, DNA template-directed nucleoside triphosphate (NTP) condensation reactions begin the stepwise polymerization of the nascent RNA transcript (42, 43). Within these initially transcribing complexes (RPitc), holoenzyme contacts upstream of the TSS remain fixed, while downstream DNA is translocated toward the active site leading to DNA “scrunching” and an increase in the size of the DNA bubble (44, 45).
FIG 1.
Promoter sequence elements and the RNA polymerase holoenzyme subunits that recognize them. Shown is a promoter under the control of the E. coli σ70 holoenzyme. The promoter recognition region (PRR) and the initially transcribed sequence (ITS) are delineated by the transcription start site (TSS) at nucleotide position +1 (arrow). While it varies depending upon NTP availability and promoter context, a bias for purines (A or G) is typically observed for the TSS (157, 228–231). An open DNA bubble is formed around the TSS, corresponding to positions −11 to +2. Specific sequences of the nontemplate strand are listed and correspond to either the optimal and/or consensus sequence for a given element. PRRs include (i) the full upstream (UP) element (A−57AAXXTXTTTTnnAAAA−41) (101), where X is an A or T nucleotide and n is no preference, contacting the carboxyl-terminal domain of the α-subunits (αCTDs, blue), (ii) the consensus −35 hexamer (T−35TGACA−30) contacting σ4 (orange), (iii) the spacer region between −35 and −10 sites with an optimal length of 17 bp (gray) containing both (iv) the zipper (Z) element (A−22ACCT−18) (170) contacting β′ (light green) and (v) an optimal extended (Ext) −10 element (T−17GTG−14) contacting σ3 (purple), (vi) the consensus −10 hexamer (T−12ATAAT−7) contacting σ2, where the −12 bp remains double stranded (red), (vii) an optimal discriminator (Dis) element (G−6GG–4) contacting σ1.2 (yellow), and (viii) the core recognition element (CRE) (54) contacting β (teal). The CRE also makes up part of the ITS (pink), extending to the +2G position.
For RPitc to enter into processive transcription elongation, the holoenzyme must break contacts made with the DNA promoter in a process termed promoter escape (reviewed in references 46 and 47). RPitc complexes typically require synthesis of an RNA-DNA hybrid of at least nine nucleotides to escape and form a stable elongation complex (48). Variations in this length have been correlated with RPo stability (49). Not all RPitc complexes go on to escape the promoter but instead become stuck in abortive cycles, where short RNA transcripts are repeatedly generated and released (50–52). One major determinant that dictates the probability of escape versus abortive cycling is mobile region 3.2 of σ which makes contacts with the template DNA in RPo but must be displaced for an RNA transcript longer than ∼5 nucleotides to emerge (53–56). This steric clash between σ3.2 and the RNA can induce RNAP pausing/backtracking (57–59) and, along with conformational changes in the template strand during RPitc scrunching (60, 61), can control the release of abortive transcripts or lead to σ3.2 repositioning and escape (62–65). While the classic model dictates that σ dissociates during promoter escape (66), updates have been proposed to account for experimental observations of σ being retained throughout elongation (reviewed in reference 67). Such complexes may even reinitiate transcription following termination if the RNAP remains bound to the DNA and diffuses to the original or a new promoter (68, 69).
For more on mechanisms of bacterial transcription initiation, we direct the reader to published reviews (70–75). Taken together, the transcription cycle depicts the holoenzyme as a molecular isomerization machine under the regulatory control of the σ factor. The many conformations and intermediate states passed through on the way to the production of an RNA transcript provide ample opportunities for kinetic regulation.
QUANTITATIVELY MODELING TRANSCRIPTION INITIATION IN BACTERIA
From a biological perspective, the purpose of transcription is to generate a concentration of RNA suitable for downstream processes, including RNA-dependent regulation and protein translation. Cellular RNA concentration is dictated by the rate of production, the rate of degradation, and the cell volume, all of which are dynamic variables that can be affected by cell growth rate and division (reviewed in references 76, to ,78). When considering the rate of RNA production, transcription initiation represents the kinetic bottleneck, where only one RNAP can initiate on a promoter at a time. This is in contrast to transcription elongation, where multiple polymerases act simultaneously on the DNA template.
In the simplest model of initiation, the process is described using Michaelis-Menten enzyme kinetics (79, 80), where RNAP is the enzyme, promoter DNA is the substrate, and full-length RNA transcript is the product (81). This analysis assumes a nonequilibrium steady state where a new RNAP-promoter complex is formed for each one lost by dissociation or escape, leading to a constant concentration of the RNAP-promoter complex and a constant velocity (V) or rate of RNA production. When DNA is present in large excess relative to RNAP and the free RNAP concentration is well below the Km (defined as the concentration of RNAP that yields the half-maximal rate), V becomes independent of DNA concentration and proportional to the free RNAP concentration (82–84). Use of this model can differentiate between regulatory mechanisms of constitutively active and environmentally responsive promoters. Specifically, variations in transcript production in constitutive promoters arise from growth rate-dependent changes in the free RNAP concentration without changes in intrinsic promoter parameters Km and Vmax (i.e., the maximum velocity obtained at saturation), whereas the activity of environmentally responsive promoters depends on factors that effectively change Km and/or Vmax to increase or decrease transcript production under conditions of constant RNAP concentration (85, 86).
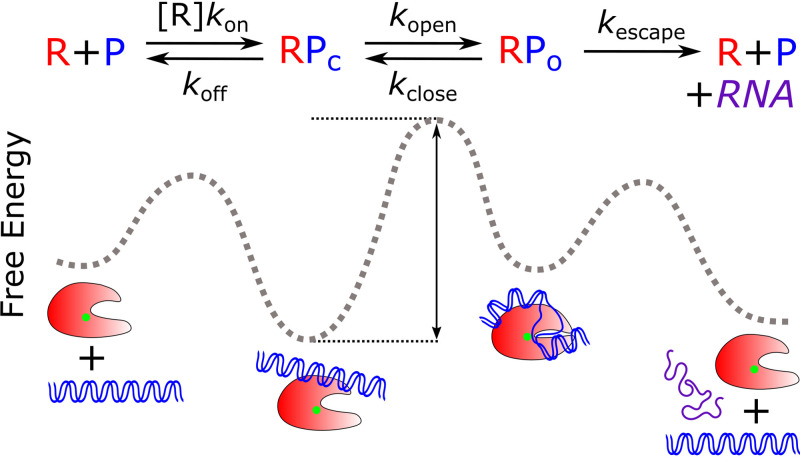
While Km and Vmax can be empirically determined, they depend on the underlying kinetic rate constants that describe transitions between intermediates on the path to escape. As a result, mechanistic understanding of transcription initiation and its regulation is enhanced by the use of free energy models (87), where kinetic conversions are modeled using transition-state theory and scale with the free energy barrier between two states. We recently developed a computational resource that allows one to calculate the RNA flux (i.e., the steady-state rate or velocity of RNA transcript synthesis) as a function of varied initiation rate constants (88). This overall initiation rate is calculated in the context of a minimal three-state pathway used extensively in formative analysis of transcription initiation kinetics (89). The kinetic scheme is represented by one concentration-dependent equilibrium binding step that describes initial promoter recognition, one reversible transition that represents all interconversions required for formation of the RPo, and one irreversible step representing promoter escape. In the free energy diagram, the stability of each intermediate is represented by an energy well, whereas the barrier height between an intermediate and its transition state (i.e., activation energy) determines the interconversion rates between two intermediates (Fig. 2). As a result, changes in interconversion rates brought on by a regulatory factor are represented by changes in the magnitude of the energy barrier and/or the stabilities of different intermediates (88). To be clear, in the model described in Fig. 2, these rates often do not represent single microscopic rate constants that would describe one molecular transition (i.e., DNA opening, closing, escape, etc.). Rather, they represent composite functions of all the intermediate kinetic steps involved within those given transitions (90, 91). As the number of significantly populated intermediates changes depending on the promoter, this model provides a useful representation of the general characteristics of all promoters rather than the most accurate depiction of an individual promoter’s kinetics. Below, after describing the properties of promoter sequence motifs, we use this computational model to illustrate how promoter sequence can affect the kinetics of initiation and how these effects are dependent on the other rate constants within the initiation pathway.
FIG 2.
Kinetic scheme of initiation used for calculation of free energy landscapes. In this kinetic model, RNAP (R; red) and promoter DNA (P; blue) form a closed promoter complex (RPc) with a concentration-dependent association rate (kon) and dissociate with rate koff. The equilibrium between the open promoter complex (RPo) and RPc is depicted by the composite forward and reverse isomerization rates, kopen and kclose, respectively. RPo formation involves the wrapping of upstream DNA and loading into the RNAP cleft, coupled with RNAP conformational changes. The DNA is opened around the TSS, positioned near the active site (green dot). Promoter escape is modeled as an irreversible transition by rate kescape, leading to RNAP dissociation from the promoter template and the generation of one full-length RNA transcript. These individual rate constants are used to calculate an overall initiation rate, which we use as a readout of the steady-state rate of RNA production. The stability of and transitions between these initiation intermediates can be depicted on a free energy reaction coordinate diagram. Here, the height of the barrier between an intermediate’s energy well and its transition state (black arrow) determines the interconversion rates, and the depth of an intermediate’s well determines its stability.
PROMOTER SEQUENCE AND EFFECTS ON INITIATION KINETICS
Initiation rates are highly sequence dependent and vary 10,000-fold in vivo, partially explained by similar variations in RPo formation rates and lifetimes measured in vitro (49, 81, 92). Diverse promoter sequences lead to different rate-limiting transitions during initiation, resulting in individual promoters being kinetically controlled at distinct steps. The kinetics at each step in the pathway are determined both by sequence-specific contacts between the promoter and the holoenzyme as well as nonspecific interactions made with the DNA phosphate backbone. In addition, purine-pyrimidine base preferences and nearest-neighbor effects dictate the energetic stabilities of both the duplex DNA and DNA/RNA hybrids (93–97) which can, in turn, exert effects independent of sequence motif conservation. Generally, DNA duplex stability affects RPo formation (98), and DNA/RNA duplex stability and base stacking interactions between the incoming NTP and the 3′ end of the RNA affect escape kinetics (43, 99).
Promoter sequences can be functionally separated by location: sequences upstream of the TSS in the promoter recognition region (PRR) and those downstream of the TSS in the initially transcribed sequence (ITS) (Fig. 1). Broadly speaking, promoter sequences upstream of and including portions of the −10 region affect initial binding, subsequent isomerizations, and rates of RPo formation. In contrast, sequences in the −10 region and further downstream have larger effects on RPo stability (72). Additionally, both the PRR and ITS can affect promoter escape rates. In the sections that follow, we summarize decades of work on how a promoter sequence affects initiation kinetics. All DNA sequences listed below refer to the nontemplate strand (5′ to 3′ direction) unless otherwise specified.
Promoter recognition region.
(i) UP elements and αCTDs. The furthest sequence-specific contact upstream of the TSS is the recognition of the UP element by the αCTDs (reviewed in reference 27). UP elements are found at positions −60 to −40 relative to the TSS, where an AT-rich sequence is favorable for the interaction (26, 100). An optimal UP element sequence is able to increase transcript levels >300-fold in vivo (101) (Fig. 1), and further analysis identified two individual subsite sequences proximal and distal relative to the −35 region (102). Structurally, the αCTDs can adopt different conformations during UP element recognition (30, 103, 104), where transcriptional activation requires recognition by both αCTDs at the distal site but only a single αCTD is needed at the proximal site (102). Initial in vitro characterization on the rRNA promoter rrnBP1 indicated that the presence of the UP element increases both the association rate constant (kon), approaching the theoretical diffusion limit, and the composite isomerization rate constant (kopen) (105). These effects have been understood as an energetic coupling between upstream DNA wrapping and the conformational changes required for loading DNA into the RNAP active-site cleft (reviewed in references 71 and 72) and not due to direct effects on the DNA opening step (34). In contrast, recent single-molecule work on rrnBP1 only observed the UP-dependent effect on kon (106). Either way, by changing initiation kinetics, UP elements are an important determinant in the activation of rRNA transcription (107).
Not all αCTD-DNA contacts are sequence specific. The presence of upstream DNA lacking a UP element sequence can also lead to increased promoter activity (108, 109). Work on lacUV5, which lacks a UP element, and λPR, which contains a distal UP element, confirmed that, in both cases, the αCTD-DNA interactions increase kon and kopen without affecting the dissociation rate of the complex (109, 110).
(ii) −35 hexamer and σ4. The consensus sequence for the −35 hexamer is T−35TGACA−30, where the TTG motif is the most highly conserved (111–114) and is important for sequence-specific binding (115, 116). By assessing transcriptional output in vitro and in vivo, experimental studies established hierarchies of base preferences, where changes to the −30 base are the least detrimental to promoter activity (117, 118), and confirmed that a consensus −35 site yields the highest output on constitutive promoters (114). The −35 hexamer is recognized by a helix-turn-helix DNA binding motif within σ4 (24), and these interactions, along with those of the UP element, represent the first sequence-specific interactions formed upon holoenzyme binding (30, 33, 119). Bending of the DNA occurs just upstream of the −35 site upon promoter recognition (104, 110) and is likely the result of conformational coupling with upstream DNA wrapping (see previous section) and/or effects of protein-protein interactions of σ4 with an αCTD bound to a UP element proximal site (120, 121). This αCTD-σ4 interaction primarily facilitates association kinetics (kon) (120). The −35 site is not essential for RPo formation, and strand separation can still occur in the complete absence of this motif (122).
(iii) Spacer region between −35 and −10. No consensus sequence of the spacer region has been identified (111), although nonrandom distributions of bases have been noted (123). While shorter lengths can be accommodated, a spacer length of 17 base pairs (bp) is structurally ideal for making both −35-σ4 and −10-σ2 interactions (23, 124). Accordingly, 17 bp is also the most common spacer length (111, 113, 114) and leads to the highest transcriptional output from a variety of promoters (125–128). Changes in both RNAP affinity (116, 127) and the isomerization rate (kopen) (127, 129) have been observed by altering the spacer length. Addition of an AT-rich sequence just upstream of −10 (termed the −15 sequence) enhanced RNAP binding and subsequent RPo formation (116, 130), consistent with studies that showed replacing the spacer region with G and C stretches leads to an inhibition of promoter activity (128) and those with T (131) and A (132) stretches increased promoter activity. This AT preference, like what is observed for UP elements, likely facilitates helix deformations (133) that have been proposed to link the binding energetics of −35 and −10 site recognition during promoter opening (134).
(iv) Extended −10 region and σ3. It was first noted that the presence of −15T and −14G was important for a promoter designed to be constitutively active in the absence of an activator protein (135). Subsequent sequence analysis on a collection of 300 E. coli σ70 promoters identified that the extended −10 consensus sequence (T−15G−14) is present in ∼20% of promoters (136) and is enriched in promoters with longer spacer lengths and less consensus in the −35 site (113, 137). On some promoters, the extended −10 can compensate for nonconsensus −35 or −10 motifs (reviewed in reference 138). The extended −10 region contacts σ3 (23, 139, 140), where perpendicular α-helices insert into the major groove, causing the promoter to bend toward the σ factor (124, 141). Further addition of another TG motif directly upstream at −17/−16 contacts both core and σ subunits (140) and can lead to increased promoter activity (136, 137). The extended −10 motif has been reported to increase the association (kon) and isomerization (kopen) rates as well as increase RPo lifetime by slowing its dissociation (kclose) (142–144).
Some suggest that the definition of the extended −10 motif should be reevaluated as T−15GnT−12, termed the −15 motif (138) (note that this is different than the AT-rich −15 sequence discussed above). This change would account for the fact that the −12T (the first base in the −10 motif) remains double stranded (ds), whereas the rest of the −10 hexamer is opened to form the single-stranded DNA (ssDNA) bubble during promoter recognition (discussed below). This naming would split up the −10 hexamer to account for its different functional roles in binding and isomerization/RPo lifetime (138, 145).
(v) −10 hexamer and σ2. The consensus sequence for the −10 hexamer is T−12ATAAT−7, with −11A and −7T being the most highly conserved and the most sensitive to nucleotide substitution (111, 113, 114, 118, 146). Like the consensus −35, a consensus −10 region yields the highest transcriptional output on constitutive promoters (114). In RPo, the −12 bp defines the upstream edge of the DNA bubble and is thought to be recognized as dsDNA, where conserved tryptophans in σ2 bracket the −12T (25, 30, 54, 124, 140). Promoter contacts with this tryptophan “chair” are proposed to stabilize RPo by preventing reannealing of the single strands, replacing the stacking interactions lost when −11A flips out of the DNA helix (25, 30, 140). However, an intriguing recent structural study indicates that the −12 bp transiently melts in the early steps of forming the stacking interaction (30), perhaps providing a rational for the conservation of the more easily melted A-T bp, and experimental studies that have suggested σ2 interacts with a single-stranded −12 site (reviewed in reference 138). Bases in both the nontemplate and template strands of the ssDNA region of the −10 are flipped out (reviewed in reference 147), where the −11A and −7T bind within pockets of σ while the template strand −9T interacts with β subunit of RNAP (25, 30, 54, 124, 140). This −9T–β-protrusion interaction is thought to stabilize a pre-RPo intermediate and allow for inhibition by the transcription factor TraR (30). Consistent with this hypothesis, the combination of DksA and ppGpp, which mimics the effects of TraR (148), represses transcription on promoters that show enrichment for the template strand −9T (149).
The −11A nucleates DNA unwinding (25, 29, 150, 151), although the exact mechanism, including the order of events and whether DNA is unwound before or after it is bent into the cleft toward the active site, has been subject of debate (reviewed in reference 74). In addition to the −11A, the −7T appears to play a critical role in the kinetics of RPo formation, and as a result, it has been suggested that nucleation may be more delocalized within the −10 hexamer (98). RNAP binding affinity to forked-junction templates is dependent on the entire −10 sequence, not just the −11A and the −7T (150, 152), and these conserved positions are not absolutely required for RPo formation, as an AT-rich region lacking those specific bases also yields fast promoter melting kinetics (98). This later result is likely due to DNA duplex instability (93), as promoter melting activity directly correlates to the −11 bp stability (29), and a C-rich −10 region exhibits very little promoter melting (98).
Thus, the −10 region serves multiple functions in initiation, including promoter recognition of the dsDNA −12 bp, nucleating DNA unwinding, and specific ssDNA contacts maintained throughout RPo formation. These effects combined can increase the association (kon) and isomerization (kopen) rates and decrease kclose to facilitate an increase in RPo lifetime, with the largest kinetic effects typically observed on the forward isomerization rate (29, 116, 144, 153–156).
(vi) Discriminator and σ1.2. The discriminator sequence lies between the −10 and the TSS, with six-base discriminators being most common (157). The discriminator sequence can affect TSS selection (158), where purine-rich discriminators favor TSSs closer to the −10 than promoters containing pyrimidine-rich discriminators (159). Originally defined as a GC-rich region commonly found in rRNA promoters (160), it was later shown that a short sequence immediately downstream of the −10 (5′-GGG-3′) binds optimally to σ (161) and leads to an increase in RPo stability, decreasing the reverse isomerization rate (kclose) (144, 162). Region 1.2 of σ interacts with this sequence (144, 162, 163), creating a binding pocket for the G positioned one base downstream of the −10 site (54). On rrnBP1, mutating the native C two bases downstream of the −10 to G leads to a large stabilization of RPo (144). The presence of this C is common at rRNA promoters (160), contributing to their relatively unstable RPo (144). This instability permits rRNA promoters to be regulatory targets of the initiating nucleotide, DksA/ppGpp, and TraR (144, 148, 149, 164–166).
Mechanistically, the discriminator is thought to drive a series of in-cleft and downstream conformational changes in the holoenzyme that stabilize RPo (see the discussion in reference 39). Additionally, analogous to NTP-dependent scrunching that occurs during initial transcription (44, 45), NTP-independent scrunching in RPo has been observed, leading to changes in the bubble size that correlate with variability in TSS position (158, 159, 167). The G two bases downstream of the −10-σ1.2 interaction prevents RPo scrunching, explaining the lack of this G at rRNA promoters, which require RPo scrunching to accommodate their unusually long eight-base discriminator sequences (158). On promoters containing a C at this position, such as rrnBP1, the complementary G on the template strand forms a binding pocket the with β′ lid, σ3.2, and the neighboring base (104), likely representing a key interaction in the RPo scrunched intermediate (168). In addition, it has been proposed that RPo scrunching facilitates promoter escape by reducing the abortive pathway (158, 169). Consistent with this model, discriminator sequences affect the kinetics, length, and probability of abortive RNA production, where a more stable RPo leads to longer abortive RNAs prior to escape (49). While the energetic costs of RPo scrunching have been determined (167), it is unknown whether starting from a scrunched state favors subsequent nucleotide addition steps to bias the system toward the productive synthesis pathway (169). However, recent structural work has suggested that RPo scrunching potentially reduces abortive synthesis by permitting one extra nucleotide to be incorporated into the nascent RNA before sterically clashing with σ3.2 (104).
(vii) Promoter elements that interact with RNAP core—Z and CRE. Not all promoter elements interact with σ. For instance, the β′ zipper region makes contacts with a “zipper or Z-element” corresponding to bases −22 to −18 within the spacer region (170) (Fig. 1). The Z-element facilitates RPo formation and can serve as a substitute for −35 recognition by σ4, although it is not clear whether this interaction with the core is sequence specific (170). Changes in the spacer conformation were suggested to be partially dependent on the presence of a −18T (171), and subsequent structural studies indicated that highly conserved residues in the β′ zipper make contacts at the −18 and −17 positions (30, 140).
Another RNAP core-promoter interaction is that of the β-subunit with the ssDNA of −4 to +2, termed the core recognition element (CRE) (54). Contacts are made with all but the −1 nucleotide, where the +2G is specifically bound in a pocket on the face of β, leading to an increased lifetime of the complex (54, 172). In the case of both CRE and Z, additional effects subsequent to RPo have been observed. Both modulate pausing properties (170, 172, 173), and CRE can also modulate TSS selection (174) and promoter escape (99, 175). Lastly, as the polymerase changes register during elongation and termination, CRE-like sequences encountered at these positions can affect the rates of these processes as well (172).
Initially transcribed sequence.
(i) “Downstream DNA.” In the context of RPo, the ITS is commonly referred to as downstream DNA. Nonspecific DNA contacts are made with RNAP mobile elements in β and β′ that lead to stabilization of RPo due to changes in both the forward (kopen) (37) and reverse (kclose) isomerization rates (37, 39, 176–178). While increasing the length of downstream duplex leads to higher RNAP affinity (179), the effects of downstream DNA in RPo formation past +6 appear to be dependent on the sequence of the discriminator, being amplified in the context of a GC-rich sequence (180). Combined, these results suggest that the RNAP-downstream DNA contacts cannot be energetically decoupled from upstream PRRs. This is likely due to the coupled conformational changes required for DNA loading into the cleft (37, 71) instead of downstream DNA sequence-dependent effects on RPo stability (99).
(ii) “Escape region.” In the context of escape, the ITS is the sequence that is transcribed during RPitc. Originally defined as bases +1 to +20 (181), it is now known that this region is variable in length, where escape points occur between +3 (predicted for rrnBP1 [49]) to +19 on phage variant promoters (182, 183). Sequences within the ITS can affect both the propensity for abortive transcription and the escape rate (99, 181, 182, 184). These effects can in part be explained by the presence or absence of pause-inducing sequences. Pausing was originally observed during transcription elongation (reviewed in reference 185) and later confirmed during initiation, occurring both on- and off-pathway to productive transcription and, in some cases, inducing backtracking (57, 58, 186). The Y−1G+1 sequence (Y is T or C) was shown to be enriched for slow escape kinetics, especially when found as repeats (99). Pausing at this sequence occurs frequently when positioned at ITS positions +6 to +7, where the steric clash of the emerging RNA with σ3.2 presents an additional energy barrier to escape (57–59). Subsequent in vivo studies defined an ITS pause sequence to contain a T two bases upstream of the Y−1G+1 motif (187), and this extended sequence has proved even more detrimental to escape kinetics (99). Observations on the effects of general base composition on escape propensity have also been made. A nontemplate purine-rich ITS favors productive transcription and fast escape kinetics, whereas a T-rich ITS promotes abortive transcription and slow escape (99, 184, 188, 189). Importantly, promoter escape can represent the rate-limiting step for some promoters (190), explained in part by ITS effects on abortive probabilities, escape kinetics, and pausing.
THE WHOLE IS GREATER THAN THE SUM OF ITS PARTS: USING SEQUENCE TO PREDICT TRANSCRIPTIONAL ACTIVITY
While we have presented an overview of how individual sequence motifs affect certain rate constants, using this information to predict transcriptional activity is not trivial. The first attempts to correlate activity to sequence only analyzed the steps up to RPo formation, using the product of the initial binding equilibrium constant and the forward isomerization rate constant (KBk2). Here, a linear correlation was observed between the log of KBk2 and a promoter’s similarity to consensus, focusing on the sequences around and including both the −35 and −10 regions and the length of the spacer region (191). However, it was not determined how either similarity to consensus or KBk2 correlates to overall RNA production, which would require accounting for contributions of promoter escape. Subsequent studies that compared a promoter’s association rate (kon) measured in vitro to in vivo promoter strength (192, 193) found no correlation, suggesting that simply evaluating only part of the initiation pathway is not sufficient to predict transcriptional activity.
Advances in high-throughput methods have provided large data sets for modeling the effect of promoter and regulatory sequences on transcriptional activity (194–196). A first approximation to modeling protein-DNA interactions is based on an “additivity” approach, where the free energy contributions of each base pair within the binding site are treated independent of one another and added together (92, 195, 197–199). However, this approach is limited in that it does not account for multivalent binding interactions and thermodynamic linkage (200). Models that include multivalent binding between the −35 and −10 hexamers and that account for promoter context (i.e., background sequences outside the −35 and −10 regions, spacer length, presence/absence of UP elements, etc.) have been able to account for >90% of the sequence-dependent variance in transcriptional activity (196, 200). These results clearly demonstrate that, for an accurate prediction of transcriptional activity, the effect of a single sequence motif cannot be isolated from the rest of the promoter sequence.
THE EFFECT OF A SEQUENCE MOTIF DEPENDS ON PROMOTER CONTEXT
Studies evaluating the effect of a sequence motif in the context of different promoters show large variations in resultant changes to individual rate constants and overall transcriptional activity (49, 99, 137, 184, 201, 202). This calls into question if a generalized functional outcome can actually be prescribed to an individual sequence motif. Figure 3 illustrates this idea by presenting free energy diagrams for two hypothetical promoters: promoter number 1, where RPc is more stable than RPo, and promoter number 2, where RPo is more stable than RPc (Fig. 3A). These two promoters have individual rate constants that vary orders of magnitude, yet the overall initiation rates are similar, each generating ∼5 RNAs min−1 (Fig. 3A). This example emphasizes the potential downfalls in using the stability of one individual intermediate to predict transcriptional activity. For instance, without consideration of the entire pathway, one would predict that promoter 1 would be more active based on the initial binding equilibrium constant and that promoter 2 would be more active based on RPo stability, even when the rates of escape for both promoters are the same. Using the starting free energy diagrams from Fig. 3A as a representation of the kinetics in the absence of a sequence motif (−), we test how the addition of a sequence motif (+) might affect the overall RNA production rate. Here, we treat the added motif like a transcription factor and codify its effect in terms of fold changes to a specific rate constant(s). By applying the same fold changes in rate constant(s) to both promoters, we can see that the added motif can lead to different outcomes on RNA production simply due to differences in the starting (i.e., basal) kinetics for each promoter. For instance, the addition of a UP element or a consensus −35 region, modeled by increasing kon, results in a larger change in RNA flux on promoter 2 as a result of it not already being near the diffusion limit like promoter 1 (Fig. 3B). In contrast, the addition an optimal discriminator, modeled by decreasing kclose, has a larger effect on promoter 1, which started with a relatively unstable RPo (Fig. 3C). The addition of a pause sequence to the ITS, modeled as slowing kescape, is also promoter specific (Fig. 3D), in line with experimental evidence that has indicated the Y−1G+1 sequence exerts promoter-specific effects (99). While often understood in qualitative terms, quantitively illustrating these three basic examples directly shows that sequence motifs have the largest effect on promoters that are rate limited at the kinetic steps they control.
FIG 3.
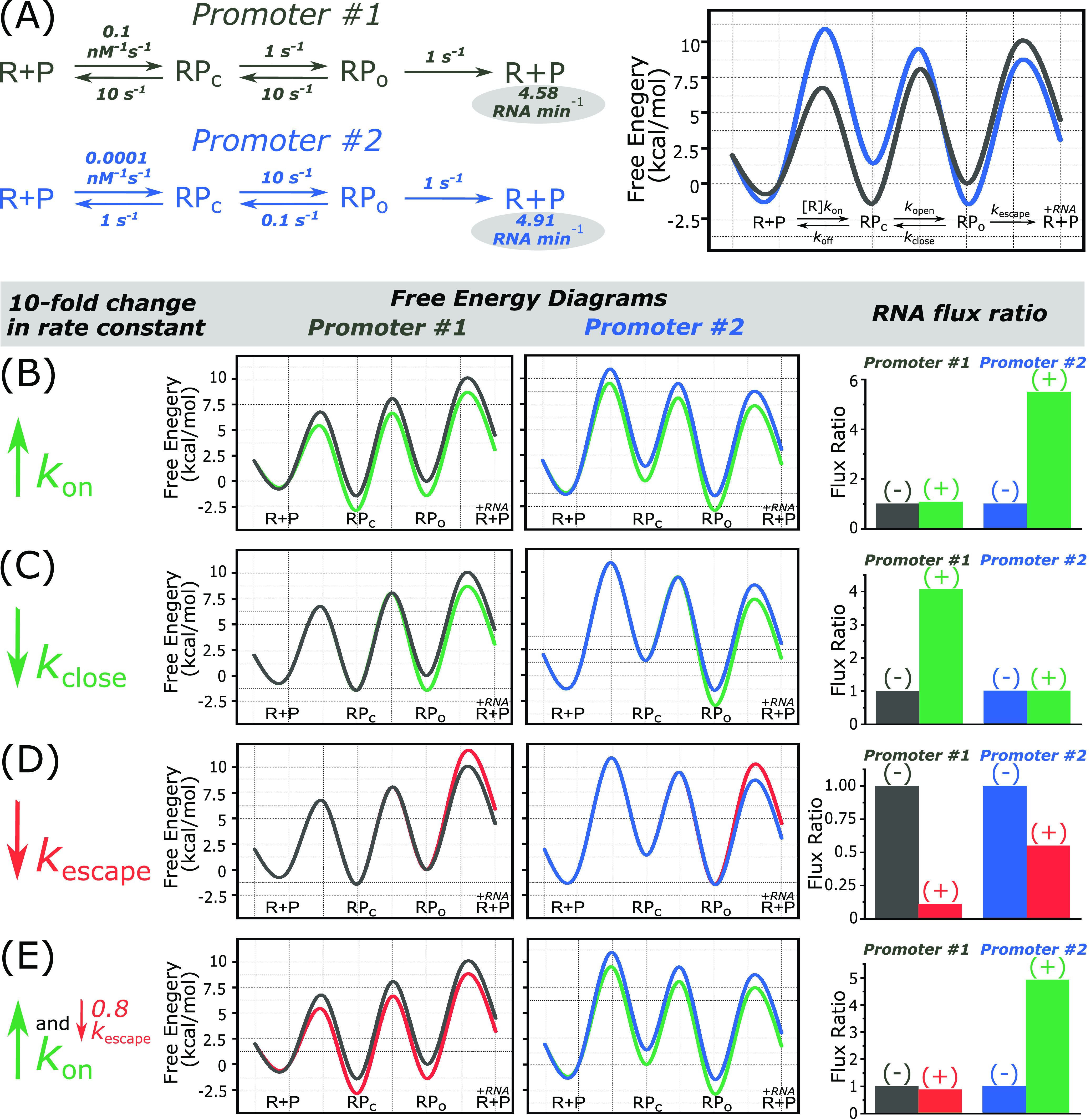
The effect of a sequence motif on RNA flux is promoter context dependent. (A) Two different promoters are used to illustrate how changes in rate constants due to the addition of a sequence motif affect transcription. Using the rate constants listed at an RNAP concentration of 1 μM, free energy diagrams and the resultant steady-state rate of RNA synthesis in units of RNA per minute were calculated with an online resource we developed (88), available at https://egalburt.github.io/transcript-flux-calculator/fluxcalc.html. Using these sets of rate constants, promoter 1 forms a more stable RPc but less stable RPo than promoter 2, but both promoters yield similar RNA production rates. To simulate the addition of different consensus motifs, RNA flux was calculated for both promoters by applying a 10-fold increase in rate on kon (B), a 10-fold decrease on kclose (C), a 10-fold decrease on kescape (D), and a 10-fold increase on kon in addition to a 0.8-fold (20%) decrease on kescape (E). In each panel, the gray and blue diagrams represent those calculated in panel A, and the free energy diagrams obtained by increasing or decreasing the rate constant(s) are plotted in green when leading to an increase in RNA flux or red when leading to a decrease in RNA flux. The resultant changes in flux upon changing a rate constant(s) (+) are plotted as a ratio of the flux values obtained in panel A (−), such that no change yields a ratio of 1, an increase in transcript rate yields a ratio greater than 1, and a decrease in transcript rate yields a ratio less than 1.
The above-mentioned examples are simplified, where addition of a motif only affects one rate constant. For instance, while generally thought to affect early steps in initiation such as binding and isomerization, PRRs have also been shown to have effects on promoter escape and abortive transcript production (99, 182, 183, 201, 203–205). This effect is not direct per se, as new sequence contacts are typically not formed during escape, although exceptions have been noted during the generation of long abortive transcripts (183). Rather, it is an energetic effect linked to the relative stability of RPo (204, 205), the intermediate preceding nucleotide incorporation. As a result, a sequence motif that may be functionally characterized as being important in binding, such as a UP element, can indirectly encode effects on subsequent rate constants, where the overall effect on flux depends on a promoter’s coupled transitions (201). To illustrate this, we use the same case presented in Fig. 3B, where increasing kon 10-fold models the effects of adding a motif important for the initial binding step, decreasing the free energies of both RPc and RPo. As a more stable RPo frequently leads to a lower rate of promoter escape (204, 205), we now assume that addition of this sequence motif also affects escape kinetics. A 10-fold increase in kon coupled with a 20% decrease in kescape still leads to activation on promoter 2 but actually leads to repression on promoter 1, a promoter that already contained optimal binding kinetics (Fig. 3E). While we only observe an ∼10% decrease in RNA flux, this overall effect can be magnified by slowing escape kinetics even further. Consistent with this idea, nucleotide changes in the ITS have the largest effect for promoters containing consensus PRRs, which are rate limited at escape (184, 206). Thus, the repressive effect on escape outweighs the activating effect on association in the context of promoter 1, leading to an overall reduction in transcription. This result illustrates that promoter context cannot only dictate the magnitude but also dictate the direction (i.e., up or down) of a change in flux resulting from the addition of a sequence motif.
THE REGULATORY EFFECT OF A TRANSCRIPTION FACTOR DEPENDS ON PROMOTER CONTEXT
Analogous to how the effect of a sequence motif is dependent on the entire promoter sequence (Fig. 3), the extent of regulation encoded by transcription factors is determined by the kinetic variations made possible by an individual promoter sequence. Many transcription factors regulate initiation by associating directly with a specific DNA sequence to either cooperatively recruit or competitively occlude RNAP binding to the promoter (reviewed in references 207 and 208). However, not all transcription factors directly recognize a specific DNA sequence (here termed DNA site independent) and are instead recruited to initiation complexes through protein-protein interactions with σ, RNAP core, or both (reviewed in references 70 and 209, to ,211). In Mycobacterium tuberculosis, two essential DNA site-independent transcription factors called CarD and RbpA are recruited to promoter regions via interaction with RNAP holoenzyme to regulate transcription (reviewed in references 75, 211, and 212). RNA-sequencing experiments suggest that in vivo, both CarD and RbpA can activate transcription on some promoters but repress transcription on others (213, 214). In vitro kinetic experiments indicate both factors increase the forward isomerization rate and, in the case of CarD, decrease the reverse isomerization rate, leading to an increase in RPo stability (215–218). While these kinetic effects by themselves would be a mechanism for transcriptional activation, we also observed CarD and RbpA to slow promoter escape kinetics (219). As in Fig. 3E, where differential changes in multiple rate constants can lead to activation or repression depending on the basal kinetics of the promoter, we have proposed a model where these factors can activate transcription at promoters rate limited at RPo formation but can repress transcription at promoters rate limited at escape (88, 219).
The prototypical examples of DNA site-independent transcription factors are E. coli DksA, in combination with ppGpp, and its homolog TraR that bind the RNAP secondary channel (reviewed in reference 220). Recent structural studies indicate that these factors induce conformational changes in RNAP that may facilitate bubble nucleation and/or σ ejection from the RNAP channel—a mechanism for activation (30, 221)—but also may stabilize DNA contacts within the channel of an intermediate prior to RPo, promote a clash with the position of the DNA template strand near the active site, and/or promote DNA melting outside the RNAP cleft—a mechanism for repression (30, 104, 221, 222). Kinetically, these structural changes have been linked to increasing the forward isomerization rate but also reducing the lifetime of RPo by increasing the reverse rate. Combined, these kinetic changes activate amino acid promoters that form RPo slowly but have long RPo lifetimes while inhibiting rRNA promoters that form RPo quickly but have short RPo lifetimes (148, 166, 221, 223, 224). Here, the same RNAP contacts that lead to the same fold changes in rate constants can lead to different regulatory outcomes depending on the rate-limiting step of a given promoter (88).
WHAT IS THE CONSENSUS ON CONSENSUS SEQUENCES?
A promoter sequence containing each motif in its consensus form, as depicted in Fig. 1, deviates significantly from real promoters found in the genome. A study tracking promoter evolution from randomized sequences indicated that recently evolved promoters primarily contain only −35 and −10 consensus-like motifs (225). This suggests that loss of consensus sequence and the addition of sequence motifs is driven by adjustments in initiation kinetics that result in increased fitness. DNA site-independent transcription factors may further exploit these kinetic variations to confer differential regulation. Alternatively, as newly evolved promoters lack transcription factor binding sites (225), promoter sequence evolution could be driven by a pressure to adjust basal promoter kinetics to take advantage of the effects of existing DNA site-independent factors.
Not all promoters require both the −35 and −10 motifs (122), where a “mix-and-match” approach has been taken by nature with regard to which PRRs are used for RNAP recruitment (reviewed in reference 138). In fact, the bacterial RNAP can bind and initiate transcription from PRRs that are far from consensus (225), perhaps explaining pervasive transcription (226) and antisense transcription following secondary initiation (68). Furthermore, experimental studies have indicated bringing a promoter closer to consensus, either through sequence mutation or entire addition, can actually result in lower transcriptional output (196, 201, 227), likely explained by the “over”-stabilization of initiation intermediates and the slowing of escape (88, 200) (Fig. 3E). Combined, these results suggest that fully consensus promoters are likely not favorable due to “self-inhibition” brought on by inefficient promoter escape and that promoters evolve to have specific kinetics that allow them to be subject to regulation.
CONCLUDING REMARKS
Here, we have summarized how PRRs often have redundant functional roles with regard to the specific initiation rate constant(s) they affect, permitting variation of motif combinations for active promoters (138). These motifs often affect more than one rate constant, and the outcome of those effects (both in magnitude and in direction) is dependent on the entire sequence context of the promoter. As a result, quantitative predictions about how sequence affects kinetics require consideration of the entire kinetic landscape. As a final thought, we caution against use of broad statements such as “the presence of consensus motif X facilities transcription” or “transcription factor X represses transcription.” Without specifying the promoter context, these statements will not always hold true.
ACKNOWLEDGMENTS
We thank Ruth Saecker for helpful comments and stimulating discussions and the reviewers for their constructive feedback regarding this review. Our apologies go to those in the field whose work was not included due to space limitations.
Work related to this review in the Galburt lab is supported by NIGMS (5R01GM134362). Drake Jensen is in part supported by the Gary K. Ackers Fellowship and Elliot L. Elson Education and Training Fellowship.
Biographies

Drake Jensen is currently a Ph.D. student at Washington University in St. Louis. In 2013, he received a B.S. in biology and a B.A. in chemistry from Southern Illinois University Edwardsville. He stayed on in the laboratory of Chin-Chuan Wei until 2015, receiving an M.S. in chemistry. There, he used calorimetric approaches to study thermodynamic properties of Ca2+-binding proteins and for developing teaching protocols in undergraduate lab courses. He joined Eric Galburt’s lab at Washington University in 2016, where he studies transcription initiation kinetics. His focus is on defining transcriptional regulatory mechanisms for the Mycobacterium tuberculosis transcription factors CarD and RbpA. Relevant to this review, these factors lack DNA binding specificity and, as a result, the authors proposed that their activity is indirectly encoded by different promoter sequences due to changes in initiation kinetics.

Eric Galburt is an associate professor in the Biochemistry and Molecular Biophysics Department at Washington University in St. Louis and has been there since 2009. He received his B.S. in physics at Brown University and earned his Ph.D. in bio chemistry at the University of Washington in the Fred Hutchinson Cancer Research Center in the laboratory of Barry Stoddard studying the structures of T4 PNK and homing endonucleases. His interest in transcription was born during his postdoctoral work with Carlos Bustamante at UC Berkeley, where he used single-molecule approaches to follow eukaryotic RNAP II elongation, and continued during his visiting scientist position at the Max Planck Institute in Dresden, Germany, with Stephan Grill. He is broadly interested in the kinetic mechanisms of transcription initiation in both bacteria and eukaryotes, as these processes represent the integration and transfer of biological information and underly the complexity of living organisms.
REFERENCES
- 1.Borukhov S, Nudler E. 2008. RNA polymerase: the vehicle of transcription. Trends Microbiol 16:126–134. 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland C, Murakami KS. 2018. An introduction to the structure and function of the catalytic core enzyme of Escherichia coli RNA polymerase. EcoSal Plus 8:ESP-004-2018. 10.1128/ecosalplus.ESP-0004-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess RR, Travers AA, Dunn JJ, Bautz EKF. 1969. Factor stimulating transcription by RNA polymerase. Nature 221:43–46. 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 4.Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39. 10.1016/S0959-440X(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 5.Wösten MMSM. 1998. Eubacterial sigma-factors. FEMS Microbiol Rev 22:127–150. [DOI] [PubMed] [Google Scholar]
- 6.Feklistov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 7.Danson AE, Jovanovic M, Buck M, Zhang X. 2019. Mechanisms of σ54-dependent transcription initiation and regulation. J Mol Biol 431:3960–3974. 10.1016/j.jmb.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber TM, Gross CA. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57:441–466. 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 9.Paget MS. 2015. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5:1245–1265. 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge-Aronis R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr Opin Microbiol 5:591–595. 10.1016/s1369-5274(02)00372-7. [DOI] [PubMed] [Google Scholar]
- 11.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada T, Tanaka K, Ishihama A. 2017. The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One 12:e0179181. 10.1371/journal.pone.0179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci U S A 91:7573–7577. 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol Microbiol 74:557–581. 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 16.Todor H, Osadnik H, Campbell EA, Myers KS, Donohue TJ, Gross CA. 2020. Rewiring the specificity of extra-cytoplasmic function sigma factors. Proc Natl Acad Sci U S A 117:33496–33506. 10.1073/pnas.2020204117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S. 2019. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294:11685–11700. 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda H, Fujita N, Ishihama A. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 28:3497–3503. 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihama A. 2000. Functional modulation of Escherichia Coli RNA polymerase. Annu Rev Microbiol 54:499–518. 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 20.Nyström T. 2004. MicroReview: Growth versus maintenance: a trade‐off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54:855–862. 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 21.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. 2006. Insights into transcriptional regulation and σ competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci U S A 103:5332–5337. 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mejía-Almonte C, Busby SJW, Wade JT, van Helden J, Arkin AP, Stormo GD, Eilbeck K, Palsson BO, Galagan JE, Collado-Vides J. 2020. Redefining fundamental concepts of transcription initiation in bacteria. Nat Rev Genet 21:699–616. 10.1038/s41576-020-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285–1290. 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 24.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol Cell 9:527–539. 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 25.Feklistov A, Darst SA. 2011. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147:1257–1269. 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407–1413. 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 27.Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol 37:687–695. 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 28.Roberts JW, Roberts CW. 1996. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell 86:495–501. 10.1016/S0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 29.Heyduk E, Kuznedelov K, Severinov K, Heyduk T. 2006. A consensus adenine at position –11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J Biol Chem 281:12362–12369. 10.1074/jbc.M601364200. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Chiu C, Gopalkrishnan S, Chen AY, Olinares PDB, Saecker RM, Winkelman JT, Maloney MF, Chait BT, Ross W, Gourse RL, Campbell EA, Darst SA. 2020. Stepwise promoter melting by bacterial RNA polymerase. Mol Cell 78:275.e6–288.e6. 10.1016/j.molcel.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivetti C, Guthold M, Bustamante C. 1999. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J 18:4464–4475. 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Capp MW, Record MT, Jr. 2002. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: large scale conformational changes in forming the kinetically significant intermediates. J Mol Biol 319:649–671. 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 33.Davis CA, Bingman CA, Landick R, Record MT, Jr, Saecker RM. 2007. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 104:7833–7838. 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreenivasan R, Shkel IA, Chhabra M, Drennan A, Heitkamp S, Wang H-C, Sridevi MA, Plaskon D, McNerney C, Callies K, Cimperman CK, Record MT, Jr. 2020. Fluorescence-detected conformational changes in duplex DNA in open complex formation by Escherichia coli RNA polymerase: upstream wrapping and downstream bending precede clamp opening and insertion of the downstream duplex. Biochemistry 59:1565–1581. 10.1021/acs.biochem.0c00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosa RP, Florez-Ariza AJ, Díaz-Celis C, Onoa B, Cassago A, de Oliveira PSL, Portugal RV, Guerra DG, Bustamante CJ. 14 May 2020. Interactions of upstream and downstream promoter regions with RNA polymerase are energetically coupled and a target of regulation in transcription initiation. BioRxiv 10.1101/2020.05.13.070375. [DOI]
- 36.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT, Jr. 2006. Solute probes of conformational changes in open complex (RPo) formation by Escherichia coli RNA polymerase at the λPR promoter: evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. Biochemistry 45:2161–2177. 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drennan A, Kraemer M, Capp M, Gries T, Ruff E, Sheppard C, Wigneshweraraj S, Artsimovitch I, Record MT, Jr. 2012. Key roles of the downstream mobile jaw of Escherichia coli RNA polymerase in transcription initiation. Biochemistry 51:9447–9459. 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH. 2012. Opening and closing of the bacterial RNA polymerase clamp. Science 337:591–595. 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruff EF, Drennan AC, Capp MW, Poulos MA, Artsimovitch I, Record MT, Jr. 2015. E. coli RNA polymerase determinants of open complex lifetime and structure. J Mol Biol 427:2435–2450. 10.1016/j.jmb.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feklistov A, Bae B, Hauver J, Lass-Napiorkowska A, Kalesse M, Glaus F, Altmann K-H, Heyduk T, Landick R, Darst SA. 2017. RNA polymerase motions during promoter melting. Science 356:863–866. 10.1126/science.aam7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyaci H, Chen J, Jansen R, Darst SA, Campbell EA. 2019. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature 565:382–385. 10.1038/s41586-018-0840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson KL, Evensen CE, Molzahn CM, Felth LC, Dyke S, Liao G, Shkel IA, Record MT, Jr. 2019. RNA polymerase: step-by-step kinetics and mechanism of transcription initiation. Biochemistry 58:2339–2352. 10.1021/acs.biochem.9b00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plaskon D, Henderson K, Felth L, Molzahn C, Evensen C, Dyke S, Shkel I, Record MT, Jr. 11 September 2020. Kinetic-mechanistic evidence for which E. coli RNA polymerase-λPR open promoter complex initiates and for stepwise disruption of contacts in bubble collapse. BioRxiv 10.1101/2020.09.11.293670. [DOI]
- 44.Revyakin A, Liu C, Ebright RH, Strick TR. 2006. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314:1139–1143. 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. 2006. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314:1144–1147. 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu LM. 2002. Promoter clearance and escape in prokaryotes. Biochim Biophys Acta 1577:191–207. 10.1016/S0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 47.Hsu LM. 2008. Promoter escape by Escherichia coli RNA polymerase. EcoSal Plus 3:e4.5.2.2. 10.1128/ecosalplus.4.5.2.2. [DOI] [PubMed] [Google Scholar]
- 48.Sidorenkov I, Komissarova N, Kashlev M. 1998. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell 2:55–64. 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 49.Henderson KL, Felth LC, Molzahn CM, Shkel I, Wang S, Chhabra M, Ruff EF, Bieter L, Kraft JE, Record MT, Jr. 2017. Mechanism of transcription initiation and promoter escape by E. coli RNA polymerase. Proc Natl Acad Sci U S A 114:E3032–E3040. 10.1073/pnas.1618675114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpousis AJ, Gralla JD. 1980. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry 19:3245–3253. 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 51.Susa M, Sen R, Shimamoto N. 2002. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J Biol Chem 277:15407–15412. 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- 52.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. 2003. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 2. formation and characterization of two distinct classes of initial transcribing complexes. Biochemistry 42:3787–3797. 10.1021/bi0269613. [DOI] [PubMed] [Google Scholar]
- 53.Murakami KS, Masuda S, Darst SA. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280–1284. 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. 2012. Structural basis of transcription initiation. Science 338:1076–1080. 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernández-Tornero C, Kulbachinskiy A, Murakami KS. 2014. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem 289:24549–24559. 10.1074/jbc.M114.584037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Molodtsov V, Lin W, Ebright RH, Zhang Y. 2020. RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription. Proc Natl Acad Sci U S A 117:5801–5809. 10.1073/pnas.1920747117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duchi D, Bauer DLV, Fernandez L, Evans G, Robb N, Hwang LC, Gryte K, Tomescu A, Zawadzki P, Morichaud Z, Brodolin K, Kapanidis AN. 2016. RNA polymerase pausing during initial transcription. Mol Cell 63:939–950. 10.1016/j.molcel.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lerner E, Chung S, Allen BL, Wang S, Lee J, Lu SW, Grimaud LW, Ingargiola A, Michalet X, Alhadid Y, Borukhov S, Strick TR, Taatjes DJ, Weiss S. 2016. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 113:E6562–E6571. 10.1073/pnas.1605038113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dulin D, Bauer DLV, Malinen AM, Bakermans JJW, Kaller M, Morichaud Z, Petushkov I, Depken M, Brodolin K, Kulbachinskiy A, Kapanidis AN. 2018. Pausing controls branching between productive and non-productive pathways during initial transcription in bacteria. Nat Commun 9:1478. 10.1038/s41467-018-03902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winkelman JT, Winkelman BT, Boyce J, Maloney MF, Chen AY, Ross W, Gourse RL. 2015. Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol Cell 59:768–780. 10.1016/j.molcel.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo Y, De S, Feng Y, Steitz TA. 2020. Structural insights into transcription initiation from de novo RNA synthesis to transitioning into elongation. iScience 23:101445. 10.1016/j.isci.2020.101445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cashel M, Hsu LM, Hernandez VJ. 2003. Changes in conserved region 3 of Escherichia coli σ70 reduce abortive transcription and enhance promoter escape. J Biol Chem 278:5539–5547. 10.1074/jbc.M211430200. [DOI] [PubMed] [Google Scholar]
- 63.Kulbachinskiy A, Mustaev A. 2006. Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J Biol Chem 281:18273–18276. 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 64.Samanta S, Martin CT. 2013. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J Biol Chem 288:31993–32003. 10.1074/jbc.M113.497669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pupov D, Kuzin I, Bass I, Kulbachinskiy A. 2014. Distinct functions of the RNA polymerase σ subunit region 3.2 in RNA priming and promoter escape. Nucleic Acids Res 42:4494–4504. 10.1093/nar/gkt1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Travers AA, Burgess RR. 1969. Cyclic re-use of the RNA polymerase sigma factor. Nature 222:537–540. 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- 67.Mooney RA, Darst SA, Landick R. 2005. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell 20:335–345. 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 68.Harden TT, Herlambang KS, Chamberlain M, Lalanne J-B, Wells CD, Li G-W, Landick R, Hochschild A, Kondev J, Gelles J. 2020. Alternative transcription cycle for bacterial RNA polymerase. Nat Commun 11:448. 10.1038/s41467-019-14208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang W, Ha KS, Uhm H, Park K, Lee JY, Hohng S, Kang C. 2020. Transcription reinitiation by recycling RNA polymerase that diffuses on DNA after releasing terminated RNA. Nat Commun 11:450. 10.1038/s41467-019-14200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6:507–519. 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saecker RM, Record MT, Jr, deHaseth PL. 2011. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412:754–771. 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruff EF, Record MT, Jr, Artsimovitch I. 2015. Initial events in bacterial transcription initiation. Biomolecules 5:1035–1062. 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J, Borukhov S. 2016. Bacterial RNA polymerase-DNA interaction–the driving force of gene expression and the target for drug action. Front Mol Biosci 3:73. 10.3389/fmolb.2016.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazumder A, Kapanidis AN. 2019. Recent advances in understanding σ70-dependent transcription initiation mechanisms. J Mol Biol 431:3947–3959. 10.1016/j.jmb.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boyaci H, Saecker RM, Campbell EA. 2020. Transcription initiation in mycobacteria: a biophysical perspective. Transcription 11:53–65. 10.1080/21541264.2019.1707612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dennis PP, Bremer H. 2008. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. Ecosal Plus 3:e5.2.3. 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 77.Planson A-G, Sauveplane V, Dervyn E, Jules M. 2020. Bacterial growth physiology and RNA metabolism. Biochim Biophys Acta 1863:194502. 10.1016/j.bbagrm.2020.194502. [DOI] [PubMed] [Google Scholar]
- 78.Baptista I, Ribeiro AS. 2020. Stochastic models coupling gene expression and partitioning in cell division in Escherichia coli. Biosystems 193–194:104154. 10.1016/j.biosystems.2020.104154. [DOI] [PubMed] [Google Scholar]
- 79.Johnson KA. 2013. A century of enzyme kinetic analysis, 1913 to 2013. FEBS Lett 587:2753–2766. 10.1016/j.febslet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McClure WR, Cech CL, Johnston DE. 1978. A steady state assay for the RNA polymerase initiation reaction. J Biol Chem 253:8941–8948. 10.1016/S0021-9258(17)34268-0. [DOI] [PubMed] [Google Scholar]
- 81.Record MT, Jr, Reznikoff WS, Craig ML, McQuade KI, Schlax PJ. 1996. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In Neidhardt FC (ed), Escherichia coli and Salmonella. Cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 82.Bremer H, Dennis P, Ehrenberg M. 2003. Free RNA polymerase and modeling global transcription in Escherichia coli. Biochimie 85:597–609. 10.1016/S0300-9084(03)00105-6. [DOI] [PubMed] [Google Scholar]
- 83.Klumpp S, Hwa T. 2008. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc Natl Acad Sci U S A 105:20245–20250. 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patrick M, Dennis PP, Ehrenberg M, Bremer H. 2015. Free RNA polymerase in Escherichia coli. Biochimie 119:80–91. 10.1016/j.biochi.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 85.Liang S-T, Bipatnath M, Xu Y-C, Chen S-L, Dennis P, Ehrenberg M, Bremer H. 1999. Activities of constitutive promoters in Escherichia coli. J Mol Biol 292:19–37. 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, Dennis P, Ehrenberg M, Bremer H. 2002. Kinetic properties of rrn promoters in Escherichia coli. Biochimie 84:981–996. 10.1016/S0300-9084(02)00010-X. [DOI] [PubMed] [Google Scholar]
- 87.Roy S, Garges S, Adhya S. 1998. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem 273:14059–14062. 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 88.Galburt EA. 2018. The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression. Proc Natl Acad Sci U S A 115:E11604–E11613. 10.1073/pnas.1809454115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McClure WR. 1980. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci U S A 77:5634–5638. 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cleland WW. 1975. Partition analysis and the concept of net rate constants as tools in enzyme kinetics. Biochemistry 14:3220–3224. 10.1021/bi00685a029. [DOI] [PubMed] [Google Scholar]
- 91.Tsodikov OV, Record MT, Jr. 1999. General method of analysis of kinetic equations for multistep reversible mechanisms in the single-exponential regime: application to kinetics of open complex formation between Eσ70 RNA polymerase and λPR promoter DNA. Biophys J 76:1320–1329. 10.1016/S0006-3495(99)77294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McClure WR. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54:171–204. 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 93.Margalit H, Shapiro BA, Nussinov R, Owens J, Jernigan RL. 1988. Helix stability in prokaryotic promoter regions. Biochemistry 27:5179–5188. 10.1021/bi00414a035. [DOI] [PubMed] [Google Scholar]
- 94.Vesnaver G, Breslauer KJ. 1991. The contribution of DNA single-stranded order to the thermodynamics of duplex formation. Proc Natl Acad Sci U S A 88:3569–3573. 10.1073/pnas.88.9.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.SantaLucia J, Jr. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A 95:1460–1465. 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holbrook JA, Capp MW, Saecker RM, Record MT, Jr. 1999. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry 38:8409–8422. 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 97.Wu P, Nakano S, Sugimoto N. 2002. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur J Biochem 269:2821–2830. 10.1046/j.1432-1033.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 98.Heyduk E, Heyduk T. 2014. Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry 53:282–292. 10.1021/bi401277w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heyduk E, Heyduk T. 2018. DNA template sequence control of bacterial RNA polymerase escape from the promoter. Nucleic Acids Res 46:4469–4486. 10.1093/nar/gky172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aiyar SE, Gourse RL, Ross W. 1998. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase α subunit. Proc Natl Acad Sci U S A 95:14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Estrem ST, Gaal T, Ross W, Gourse RL. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A 95:9761–9766. 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Estrem ST, Ross W, Gaal T, Chen ZWS, Niu W, Ebright RH, Gourse RL. 1999. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev 13:2134–2147. 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. 2002. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science 297:1562–1566. 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 104.Shin Y, Qayyum MZ, Pupov D, Esyunina D, Kulbachinskiy A, Murakami KS. 2021. Structural basis of ribosomal RNA transcription regulation. Nat Commun 12:528. 10.1038/s41467-020-20776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT, Jr, Gourse RL. 1994. Factor independent activation of rrnB P1: an “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol 235:1421–1435. 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 106.Mumm JP, Friedman LJ, Gelles J. 18 February 2020. Mechanism of upstream promoter element stimulation of transcription at a ribosomal RNA promoter determined by single-molecule imaging. BioRxiv 10.1101/2020.02.17.953182. [DOI]
- 107.Gourse RL, Gaal T, Aiyar SE, Barker MM, Estrem ST, Hirvonen CA, Ross W. 1998. Strength and regulation without transcription factors: lessons from bacterial rRNA promoters. Cold Spring Harbor Symp Quant Biol 63:131–140. 10.1101/sqb.1998.63.131. [DOI] [PubMed] [Google Scholar]
- 108.Tang Y, Murakami K, Ishihama A, deHaseth PL. 1996. Upstream interactions at the lambda pRM promoter are sequence nonspecific and activate the promoter to a lesser extent than an introduced UP element of an rRNA promoter. J Bacteriol 178:6945–6951. 10.1128/jb.178.23.6945-6951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ross W, Gourse RL. 2005. Sequence-independent upstream DNA–αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc Natl Acad Sci U S A 102:291–296. 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davis CA, Capp MW, Record MT, Jr, Saecker RM. 2005. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 102:285–290. 10.1073/pnas.0405779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11:2237–2255. 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lisser S, Margalit H. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res 21:1507–1516. 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. 2007. Anatomy of Escherichia coli σ 70 promoters. Nucleic Acids Res 35:771–788. 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shimada T, Yamazaki Y, Tanaka K, Ishihama A. 2014. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One 9:e90447. 10.1371/journal.pone.0090447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsujikawa L, Tsodikov OV, deHaseth PL. 2002. Interaction of RNA polymerase with forked DNA: evidence for two kinetically significant intermediates on the pathway to the final complex. Proc Natl Acad Sci U S A 99:3493–3498. 10.1073/pnas.062487299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cook VM, deHaseth PL. 2007. Strand opening-deficient Escherichia coli RNA polymerase facilitates investigation of closed complexes with promoter DNA: effects of DNA sequence and temperature. J Biol Chem 282:21319–21326. 10.1074/jbc.M702232200. [DOI] [PubMed] [Google Scholar]
- 117.Kobayashi M, Nagata K, Ishihama A. 1990. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter −35 region on promoter strength. Nucleic Acids Res 18:7367–7372. 10.1093/nar/18.24.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moyle H, Waldburger C, Susskind MM. 1991. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol 173:1944–1950. 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, Heumann H. 2005. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc Natl Acad Sci U S A 102:4706–4711. 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. 2003. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the α C-terminal domain and σ region 4. Genes Dev 17:1293–1307. 10.1101/gad.1079403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen H, Tang H, Ebright RH. 2003. Functional interaction between RNA polymerase α subunit C-terminal domain and σ70 in UP-element- and activator-dependent transcription. Mol Cell 11:1621–1633. 10.1016/s1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 122.Niedziela-Majka A, Heyduk T. 2005. Escherichia coli RNA polymerase contacts outside the –10 promoter element are not essential for promoter melting. J Biol Chem 280:38219–38227. 10.1074/jbc.M507984200. [DOI] [PubMed] [Google Scholar]
- 123.Beutel BA, Record MT, Jr. 1990. E. coli promoter spacer regions contain nonrandom sequences which correlate to spacer length. Nucleic Acids Res 18:3597–3603. 10.1093/nar/18.12.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zuo Y, Steitz TA. 2015. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell 58:534–540. 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stefano JE, Gralla JD. 1982. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci U S A 79:1069–1072. 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aoyama T, Takanami M, Ohtsuka E, Taniyama Y, Marumoto R, Sato H, Ikehara M. 1983. Essential structure of E. coli promoter effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res 11:5855–5864. 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mulligan ME, Brosius J, McClure WR. 1985. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem 260:3529–3538. 10.1016/S0021-9258(19)83654-2. [DOI] [PubMed] [Google Scholar]
- 128.Warne SE, deHaseth PL. 1993. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the -10 and -35 regions are dependent on spacer DNA sequence. Biochemistry 32:6134–6140. 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 129.McKane M, Gussin GN. 2000. Changes in the 17 bp spacer in the PR promoter of bacteriophage λ affect steps in open complex formation that precede DNA strand separation. J Mol Biol 299:337–349. 10.1006/jmbi.2000.3757. [DOI] [PubMed] [Google Scholar]
- 130.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. 2004. A mutant spacer sequence between -35 and -10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc Natl Acad Sci U S A 101:6911–6916. 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lozinski T, Adrych-Rozek K, Markiewicz WT, Wierzchowski KL. 1991. Effect of DNA bending in various regions of a consensus-like Escherichia coli promoter on its strength in vivo and structure of the open complex in vitro. Nucleic Acids Res 19:2947–2953. 10.1093/nar/19.11.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Collis CM, Molloy PL, Both GW, Drew HR. 1989. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res 17:9447–9468. 10.1093/nar/17.22.9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hook-Barnard IG, Hinton DM. 2009. The promoter spacer influences transcription initiation via σ70 region 1.1 of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 106:737–742. 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sztiller-Sikorska M, Heyduk E, Heyduk T. 2011. Promoter spacer DNA plays an active role in integrating the functional consequences of RNA polymerase contacts with −10 and −35 promoter elements. Biophys Chem 159:73–81. 10.1016/j.bpc.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Keilty S, Rosenberg M. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem 262:6389–6395. 10.1016/S0021-9258(18)45582-2. [DOI] [PubMed] [Google Scholar]
- 136.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. 2000. DNA sequence elements located immediately upstream of the –10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res 28:1864–1870. 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mitchell JE, Zheng D, Busby SJW, Minchin SD. 2003. Identification and analysis of ‘extended –10’ promoters in Escherichia coli. Nucleic Acids Res 31:4689–4695. 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hook-Barnard IG, Hinton DM. 2007. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Bio 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 139.Barne KA, Bown JA, Busby SJW, Minchin SD. 1997. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended −10” motif at promoters. EMBO J 16:4034–4040. 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. 2015. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife 4:e08504. 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712–719. 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 142.Camacho A, Salas M. 1999. Effect of mutations in the “extended −10” motif of three Bacillus subtilis σA-RNA polymerase-dependent promoters. J Mol Biol 286:683–693. 10.1006/jmbi.1998.2526. [DOI] [PubMed] [Google Scholar]
- 143.Voskuil MI, Chambliss GH. 2002. The TRTGn motif stabilizes the transcription initiation open complex. J Mol Biol 322:521–532. 10.1016/s0022-2836(02)00802-1. [DOI] [PubMed] [Google Scholar]
- 144.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. 2006. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell 125:1069–1082. 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 145.Djordjevic M. 2011. Redefining Escherichia coli σ70 promoter elements: −15 motif as a complement of the −10 motif. J Bacteriol 193:6305–6314. 10.1128/JB.05947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pribnow D. 1975. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A 72:784–788. 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karpen ME, deHaseth PL. 2015. Base flipping in open complex formation at bacterial promoters. Biomolecules 5:668–678. 10.3390/biom5020668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gopalkrishnan S, Ross W, Chen AY, Gourse RL. 2017. TraR directly regulates transcription initiation by mimicking the combined effects of the global regulators DksA and ppGpp. Proc Natl Acad Sci U S A 114:E5539–E5548. 10.1073/pnas.1704105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, Gourse RL. 2019. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc Natl Acad Sci U S A 116:8310–8319. 10.1073/pnas.1819682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matlock DL, Heyduk T. 2000. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry 39:12274–12283. 10.1021/bi001433h. [DOI] [PubMed] [Google Scholar]
- 151.Lim HM, Lee HJ, Roy S, Adhya S. 2001. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc Natl Acad Sci U S A 98:14849–14852. 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mekler V, Severinov K. 2013. Cooperativity and interaction energy threshold effects in recognition of the −10 promoter element by bacterial RNA polymerase. Nucleic Acids Res 41:7276–7285. 10.1093/nar/gkt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fenton MS, Gralla JD. 2001. Function of the bacterial TATAAT -10 element as single-stranded DNA during RNA polymerase isomerization. Proc Natl Acad Sci U S A 98:9020–9025. 10.1073/pnas.161085798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McKane M, Malone C, Gussin GN. 2001. Mutations at position −10 in the λPR promoter primarily affect conversion of the initial closed complex (RPc) to a stable, closed intermediate (RPi). Biochemistry 40:2023–2031. 10.1021/bi0019085. [DOI] [PubMed] [Google Scholar]
- 155.Schroeder LA, Gries TJ, Saecker RM, Record MT, Jr, Harris ME, deHaseth PL. 2009. Evidence for a tyrosine–adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J Mol Biol 385:339–349. 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Debnath S, Roy NS, Bera I, Ghoshal N, Roy S. 2013. Indirect read-out of the promoter DNA by RNA polymerase in the closed complex. Nucleic Acids Res 41:366–377. 10.1093/nar/gks1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vvedenskaya IO, Zhang Y, Goldman SR, Valenti A, Visone V, Taylor DM, Ebright RH, Nickels BE. 2015. Massively systematic transcript end readout, “MASTER”: transcription start site selection, transcriptional slippage, and transcript yields. Mol Cell 60:953–965. 10.1016/j.molcel.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Winkelman JT, Chandrangsu P, Ross W, Gourse RL. 2016. Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters. Proc Natl Acad Sci U S A 113:E1787–E1795. 10.1073/pnas.1522159113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Winkelman JT, Vvedenskaya IO, Zhang Y, Zhang Y, Bird JG, Taylor DM, Gourse RL, Ebright RH, Nickels BE. 2016. Multiplexed protein-DNA cross-linking: scrunching in transcription start site selection. Science 351:1090–1093. 10.1126/science.aad6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Travers AA. 1980. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol 141:973–976. 10.1128/JB.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, Nikiforov V, Heyduk T, Severinov K, Kulbachinskiy A. 2006. A basal promoter element recognized by free RNA polymerase σ subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell 23:97–107. 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 162.Barinova N, Kuznedelov K, Severinov K, Kulbachinskiy A. 2008. Structural modules of RNA polymerase required for transcription from promoters containing downstream basal promoter element GGGA. J Biol Chem 283:22482–22489. 10.1074/jbc.M802445200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Haugen SP, Ross W, Manrique M, Gourse RL. 2008. Fine structure of the promoter-σ region 1.2 interaction. Proc Natl Acad Sci U S A 105:3292–3297. 10.1073/pnas.0709513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092–2097. 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 165.Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12:125–134. 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 166.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322. 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 167.Yu L, Winkelman JT, Pukhrambam C, Strick TR, Nickels BE, Ebright RH. 2017. The mechanism of variability in transcription start site selection. Elife 6:e32038. 10.7554/eLife.32038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pupov D, Petushkov I, Esyunina D, Murakami KS, Kulbachinskiy A. 2018. Region 3.2 of the σ factor controls the stability of rRNA promoter complexes and potentiates their repression by DksA. Nucleic Acids Res 21:11477–11487. 10.1093/nar/gky919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Winkelman JT, Gourse RL. 2017. Open complex DNA scrunching: a key to transcription start site selection and promoter escape. Bioessays 39:1600193. 10.1002/bies.201600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yuzenkova Y, Tadigotla VR, Severinov K, Zenkin N. 2011. A new basal promoter element recognized by RNA polymerase core enzyme. EMBO J 30:3766–3775. 10.1038/emboj.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Singh SS, Typas A, Hengge R, Grainger DC. 2011. Escherichia coli σ 70 senses sequence and conformation of the promoter spacer region. Nucleic Acids Res 39:5109–5118. 10.1093/nar/gkr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Petushkov I, Pupov D, Bass I, Kulbachinskiy A. 2015. Mutations in the CRE pocket of bacterial RNA polymerase affect multiple steps of transcription. Nucleic Acids Res 43:5798–5809. 10.1093/nar/gkv504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. 2014. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344:1285–1289. 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vvedenskaya IO, Vahedian-Movahed H, Zhang Y, Taylor DM, Ebright RH, Nickels BE. 2016. Interactions between RNA polymerase and the core recognition element are a determinant of transcription start site selection. Proc Natl Acad Sci U S A 113:E2899–E2905. 10.1073/pnas.1603271113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Petushkov IV, Kulbachinskiy AV. 2020. Role of interactions of the CRE region of Escherichia coli RNA polymerase with nontemplate DNA during promoter escape. Biochemistry (Mosc) 85:792–800. 10.1134/S000629792007007X. [DOI] [PubMed] [Google Scholar]
- 176.Bartlett MS, Gaal T, Ross W, Gourse RL. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol 279:331–345. 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 177.Ederth J, Artsimovitch I, Isaksson LA, Landick R. 2002. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem 277:37456–37463. 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 178.Wigneshweraraj Siva R, Burrows PC, Severinov K, Buck M. 2005. Stable DNA opening within open promoter complexes is mediated by the RNA polymerase β′-jaw domain. J Biol Chem 280:36176–36184. 10.1074/jbc.M506416200. [DOI] [PubMed] [Google Scholar]
- 179.Mekler V, Minakhin L, Severinov K. 2011. A critical role of downstream RNA polymerase-promoter interactions in the formation of initiation complex. J Biol Chem 286:22600–22608. 10.1074/jbc.M111.247080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mekler V, Minakhin L, Borukhov S, Mustaev A, Severinov K. 2014. Coupling of downstream RNA polymerase–promoter interactions with formation of catalytically competent transcription initiation complex. J Mol Biol 426:3973–3984. 10.1016/j.jmb.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kammerer W, Deuschle U, Gentz R, Bujard H. 1986. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J 5:2995–3000. 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chander M, Austin KM, Aye-Han N-N, Sircar P, Hsu LM. 2007. An alternate mechanism of abortive release marked by the formation of very long abortive transcripts. Biochemistry 46:12687–12699. 10.1021/bi701236f. [DOI] [PubMed] [Google Scholar]
- 183.Chander M, Lee A, Vallery TK, Thandar M, Jiang Y, Hsu LM. 2015. Mechanisms of very long abortive transcript release during promoter escape. Biochemistry 54:7393–7408. 10.1021/acs.biochem.5b00712. [DOI] [PubMed] [Google Scholar]
- 184.Hsu LM, Cobb IM, Ozmore JR, Khoo M, Nahm G, Xia L, Bao Y, Ahn C. 2006. Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry 45:8841–8854. 10.1021/bi060247u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kang JY, Mishanina TV, Landick R, Darst SA. 2019. Mechanisms of transcriptional pausing in bacteria. J Mol Biol 431:4007–4029. 10.1016/j.jmb.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lerner E, Ingargiola A, Lee JJ, Borukhov S, Michalet X, Weiss S. 2017. Different types of pausing modes during transcription initiation. Transcription 8:242–253. 10.1080/21541264.2017.1308853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Winkelman JT, Pukhrambam C, Vvedenskaya IO, Zhang Y, Taylor DM, Shah P, Ebright RH, Nickels BE. 2020. XACT-Seq comprehensively defines the promoter-position and promoter-sequence determinants for initial-transcription pausing. Mol Cell 79:797–811. 10.1016/j.molcel.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Skancke J, Bar N, Kuiper M, Hsu LM. 2015. Sequence-dependent promoter escape efficiency is strongly influenced by bias for the pretranslocated state during initial transcription. Biochemistry 54:4267–4275. 10.1021/acs.biochem.5b00272. [DOI] [PubMed] [Google Scholar]
- 189.Imashimizu M, Tokunaga Y, Afek A, Takahashi H, Shimamoto N, Lukatsky DB. 2020. Control of transcription initiation by biased thermal fluctuations on repetitive genomic sequences. Biomolecules 10:1299. 10.3390/biom10091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reppas NB, Wade JT, Church GM, Struhl K. 2006. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell 24:747–757. 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 191.Mulligan ME, Hawley DK, Entriken R, McClure WR. 1984. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res 12:789–800. 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Deuschle U, Kammerer W, Gentz R, Bujard H. 1986. Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J 5:2987–2994. 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Brunner M, Bujard H. 1987. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J 6:3139–3144. 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. 2009. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol 27:1173–1175. 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kinney JB, Murugan A, Callan CG, Cox EC. 2010. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci U S A 107:9158–9163. 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Urtecho G, Tripp AD, Insigne KD, Kim H, Kosuri S. 2019. Systematic dissection of sequence elements controlling σ70 promoters using a genomically encoded multiplexed reporter assay in Escherichia coli. Biochemistry 58:1539–1551. 10.1021/acs.biochem.7b01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.von Hippel PH, Berg OG. 1986. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A 83:1608–1612. 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Benos PV, Bulyk ML, Stormo GD. 2002. Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res 30:4442–4451. 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Brewster RC, Jones DL, Phillips R. 2012. Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput Biol 8:e1002811. 10.1371/journal.pcbi.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Einav T, Phillips R. 2019. How the avidity of polymerase binding to the –35/–10 promoter sites affects gene expression. Proc Natl Acad Sci U S A 116:13340–13345. 10.1073/pnas.1905615116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ellinger T, Behnke D, Knaus R, Bujard H, Gralla JD. 1994. Context-dependent Effects of upstream A-tracts stimulation or inhibition of Escherichia coli promoter function. J Mol Biol 239:466–475. 10.1006/jmbi.1994.1389. [DOI] [PubMed] [Google Scholar]
- 202.Ross W, Aiyar SE, Salomon J, Gourse RL. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol 180:5375–5383. 10.1128/JB.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Strainic MG, Sullivan JJ, Velevis A, deHaseth PL. 1998. Promoter recognition by Escherichia coli RNA polymerase: effects of the UP element on open complex formation and promoter clearance. Biochemistry 37:18074–18080. 10.1021/bi9813431. [DOI] [PubMed] [Google Scholar]
- 204.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. 2003. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry 42:3798–3811. 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 205.Ko J, Heyduk T. 2014. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem J 463:135–144. 10.1042/BJ20140179. [DOI] [PubMed] [Google Scholar]
- 206.Hsu LM. 2009. Monitoring abortive initiation. Methods 47:25–36. 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Browning DF, Busby SJW. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 208.Browning DF, Butala M, Busby SJW. 2019. Bacterial transcription factors: regulation by pick ‘n’ mix. J Mol Biol 431:4067–4077. 10.1016/j.jmb.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 209.Zenkin N, Yuzenkova Y. 2015. New insights into the functions of transcription factors that bind the RNA polymerase secondary channel. Biomolecules 5:1195–1209. 10.3390/biom5031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wahl MC, Sen R. 2019. Exploiting phage strategies to modulate bacterial transcription. Transcription 10:222–230. 10.1080/21541264.2019.1684137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vishwakarma RK, Brodolin K. 2020. The σ subunit-remodeling factors: an emerging paradigms of transcription regulation. Front Microbiol 11:1798. 10.3389/fmicb.2020.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Flentie K, Garner AL, Stallings CL. 2016. Mycobacterium tuberculosis transcription machinery: ready to respond to host attacks. J Bacteriol 198:1360–1373. 10.1128/JB.00935-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Prusa J, Jensen D, Santiago-Collazo G, Pope SS, Garner AL, Miller JJ, Manzano AR, Galburt EA, Stallings CL. 2018. Domains within RbpA serve specific functional roles that regulate the expression of distinct mycobacterial gene subsets. J Bacteriol 200:e00690-17. 10.1128/JB.00690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhu DX, Garner AL, Galburt EA, Stallings CL. 2019. CarD contributes to diverse gene expression outcomes throughout the genome of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 116:13573–13581. 10.1073/pnas.1900176116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rammohan J, Ruiz Manzano A, Garner AL, Stallings CL, Galburt EA. 2015. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res 43:3272–3285. 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Davis E, Chen J, Leon K, Darst SA, Campbell EA. 2015. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res 43:433–445. 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rammohan J, Ruiz Manzano A, Garner AL, Prusa J, Stallings CL, Galburt EA. 2016. Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD. Nucleic Acids Res 44:7304–7313. 10.1093/nar/gkw577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hubin EA, Fay A, Xu C, Bean JM, Saecker RM, Glickman MS, Darst SA, Campbell EA. 2017. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife 6:e22520. 10.7554/eLife.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jensen D, Manzano AR, Rammohan J, Stallings CL, Galburt EA. 2019. CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase. Nucleic Acids Res 47:6685–6698. 10.1093/nar/gkz449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A, Ross W. 2018. Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol 72:163–184. 10.1146/annurev-micro-090817-062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chen J, Gopalkrishnan S, Chiu C, Chen AY, Campbell EA, Gourse RL, Ross W, Darst SA. 2019. E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Elife 8:e49375. 10.7554/eLife.49375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades SE, Murakami KS. 2018. Allosteric effector ppGpp Potentiates the inhibition of transcript initiation by DksA. Mol Cell 69:828.e5–839.e5. 10.1016/j.molcel.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Stumper SK, Ravi H, Friedman LJ, Mooney RA, Corrêa IR, Gershenson A, Landick R, Gelles J. 2019. Delayed inhibition mechanism for secondary channel factor regulation of ribosomal RNA transcription. Elife 8:e40576. 10.7554/eLife.40576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yona AH, Alm EJ, Gore J. 2018. Random sequences rapidly evolve into de novo promoters. Nat Commun 9:1530. 10.1038/s41467-018-04026-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wade JT, Grainger DC. 2014. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12:647–653. 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- 227.Graña D, Gardella T, Susskind MM. 1988. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics 120:319–327. 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wilson HR, Archer CD, Liu JK, Turnbough CL. 1992. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol 174:514–524. 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jeong W, Kang C. 1994. Start site selection at lac UV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res 22:4667–4672. 10.1093/nar/22.22.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Walker KA, Osuna R. 2002. Factors affecting start site selection at the Escherichia coli fis promoter. J Bacteriol 184:4783–4791. 10.1128/jb.184.17.4783-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lewis DEA, Adhya S. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol Microbiol 54:692–701. 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]