Abstract
Alterations in breast cancer gene 1 (BRCA1), a tumor suppressor gene, increase the risk of breast and ovarian cancers. BRCA1 forms a heterodimer with BRCA1‐associated RING domain protein 1 (BARD1) and functions in multiple cellular processes, including DNA repair and centrosome regulation. BRCA1 acts as a tumor suppressor by promoting homologous recombination (HR) repair, and alterations in BRCA1 cause HR deficiency, not only in breast and ovarian tissues but also in other tissues. The molecular mechanisms underlying BRCA1 alteration‐induced carcinogenesis remain unclear. Centrosomes are the major microtubule‐organizing centers and function in bipolar spindle formation. The regulation of centrosome number is critical for chromosome segregation in mitosis, which maintains genomic stability. BRCA1/BARD1 function in centrosome regulation together with Obg‐like ATPase (OLA1) and receptor for activating protein C kinase 1 (RACK1). Cancer‐derived variants of BRCA1, BARD1, OLA1, and RACK1 do not interact, and aberrant expression of these proteins results in abnormal centrosome duplication in mammary‐derived cells, and rarely in other cell types. RACK1 is involved in centriole duplication in the S phase by promoting polo‐like kinase 1 activation by Aurora A, which is critical for centrosome duplication. Centriole number is higher in cells derived from mammary tissues compared with in those derived from other tissues, suggesting that tissue‐specific centrosome characterization may shed light on the tissue specificity of BRCA1‐associated carcinogenesis. Here, we explored the role of the BRCA1‐containing complex in centrosome regulation and the effect of its deficiency on tissue‐specific carcinogenesis.
Keywords: BRCA1, breast cancer, centrosome, centrosome amplification, HBOC
Alterations of BRCA1, a tumor suppressor gene, increase the risk of breast and ovarian cancers. Recently, we found that BRCA1 functions in the regulation of centrosome number together with Obg‐like ATPase (OLA1) and receptor for activating protein kinase C 1 (RACK1). In this review, we explored the role of the BRCA1‐containing complex in centrosome regulation and the effect of its deficiency on tissue‐specific carcinogenesis.
1. INTRODUCTION
Breast cancer gene 1 (BRCA1) on chromosome 17q is the gene responsible for hereditary breast and ovarian cancer syndrome. 1 Women carrying pathogenic germline variants of BRCA1 are at high risk of breast (57%) and ovarian cancers (40%). 2 Biallelic alterations of BRCA1 are observed in Fanconi anemia, a syndrome characterized by progressive bone marrow failure and increased risk of cancer. 3 Mice with homozygous null alleles of Brca1 show embryonically lethal between E6 and E13.5 because of growth retardation, apoptosis, and aneuploidy. 4 Mammary‐specific knockout of Brca1 in combination with p53 deficiency leads to basal‐like breast cancer with a propensity for the triple‐negative subtype, which is the most frequent subtype in BRCA1‐associated hereditary breast cancer in humans. 4
The BRCA1 gene product is a 1863 amino acid (aa) protein with a RING domain in the amino (N)‐terminus and 2 tandem BRCT domains in the carboxy (C)‐terminus (Figure 1A). 5 BRCA1‐associated RING domain 1 (BARD1), which was identified as a binding protein to the N‐terminal fragment (aa 1‐304) of BRCA1, also has a RING domain in its N‐terminus and BRCT domains in the C‐terminus (Figure 1B). 6 BRCA1 and BARD1 form a stable complex through their RING domains 7 and interact with various proteins involved in DNA damage repair, centrosome regulation, chromatin remodeling, and transcriptional regulation. 7 , 8
FIGURE 1.

Structure of BRCA1 and BARD1. BRCA1 and BARD1 have a RING domain and nuclear export signal (NES) in the N‐terminus and 2 BRCT domains in the C‐terminus. BRCA1 includes a nuclear localization signal (NLS) and a coiled‐coil domain. BARD1 contains ankyrin (ANK) repeats
DNA double‐stranded breaks (DSBs) are repaired by homologous recombination (HR) or non‐homologous end joining. HR is an error‐free system that repairs DSBs using undamaged DNA homologous to the damaged DNA as a template. BRCA1 is an essential factor for HR. 8 Some BRCA1 variants that are deficient in HR have been identified in hereditary breast cancer and by high‐throughput analyses 9 , 10 , 11 , 12 , 13 In addition, alterations in several HR‐related factors other than BRCA1 have been reported in hereditary and sporadic breast cancers. 14 , 15 Therefore, HR activities are important for tumor suppression in breast tissues. However, BRCA1 alterations induce HR deficiency not only in mammary epithelial cells but also in other tissue‐derived cells. 8 , 9 , 11 , 13 These data suggest that BRCA1 alterations cause additional defects in cellular functions, which may contribute to its mammary tissue‐specific carcinogenesis.
Recently, we and other groups reported that deficiencies of BRCA1 and its interacting proteins induce centrosome aberrations only in mammary epithelium‐derived cells. 16 , 17 , 18 , 19 In this review, we focus on the function of BRCA1 in centrosome regulation and discuss how the function of BRCA1 contributes to tumor suppression.
2. THE CENTROSOME
2.1. Structure and function of the centrosome
The centrosome is a non‐membranous organelle that contains a pair of centrioles surrounded by proteinaceous pericentriolar material (PCM) (Figure 2A). 20 , 21 The centriole is a barrel‐shaped structure composed of 9 microtubule triplets (Figure 2A). A daughter centriole is attached to the side wall of the mother centriole. The PCM is composed of several layers of proteins. 21 The outer layer of the PCM contains abundant γ‐tubulin ring complexes, which are required for microtubule nucleation. Therefore, the centrosome acts as a major microtubule‐organizing center that is involved in the regulation of the cytoskeleton, cell polarity, intracellular trafficking, and cell division. 22 During mitosis, microtubules sprouting from centrosomes are attached to the centromere of chromosomes and form mitotic spindles. 20
FIGURE 2.
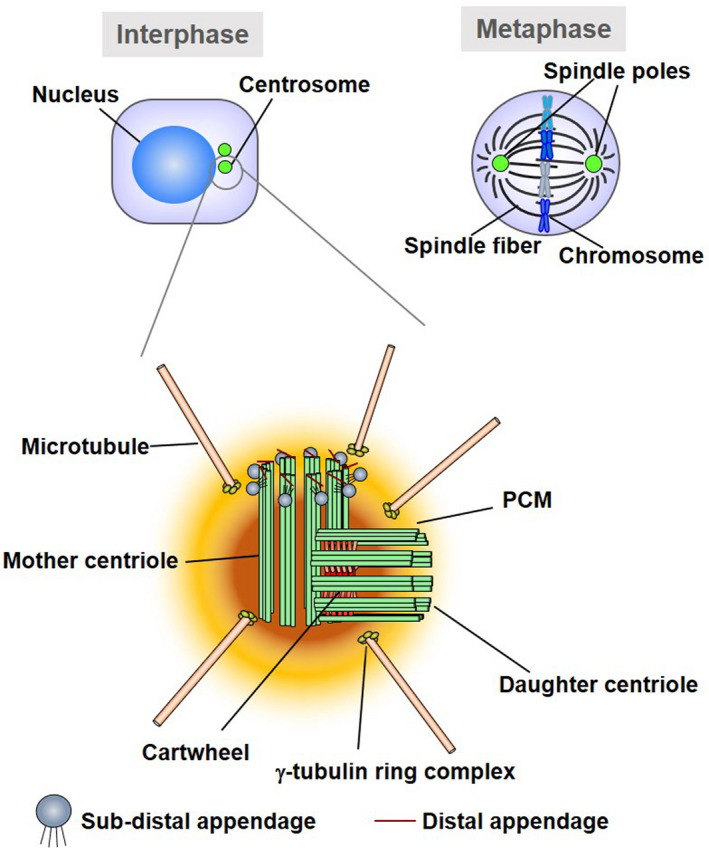
Schematic diagram of the centrosomes. The centrosome consists of mother centriole and daughter centriole, embedded in the pericentriolar matrix (PCM). The PCM contains γ‐tubulin ring complexes required for microtubule nucleation
2.2. Regulation of centrosome number
The centrosome is duplicated only once during each cell cycle 23 (Figure 3). After mitosis, a daughter cell inherits 1 centrosome from the mother cell. In G1 phase, the centrosome has 2 mother centrioles interconnected by a linker. At the end of G1 phase, polo‐like kinase 4 (PLK4) is activated and phosphorylates several centrosomal proteins, including STIL, triggering the formation of a procentriole containing cartwheel on the side wall of the mother centriole. 24 During S phase, a daughter centriole is generated from the procentriole. In late G2 phase, the linker is degraded by the activation of NIMA‐related kinase 2 (NEK2), which allows the movement of each centrosome to opposite poles of the cell. 25 Before mitosis, the daughter centriole is tightly connected to the mother centriole (called engagement). Engagement is disrupted by the cleavage of several proteins, including pericentrin and cohesion, by separase during mitosis (called disengagement), and a new linker is formed between the disengaged centrioles. 26 , 27 The engagement of centrioles inhibits further procentriole formation. 28 Therefore, disengagement is the licensing step for centriole duplication. 27 If the disengagement occurs before mitosis, which is known as premature disengagement, centrioles can undergo additional duplication (called reduplication), resulting in extra‐centrioles. 29 , 30 Disengagement is promoted by polo‐like kinase 1 (PLK1) through the phosphorylation of pericentrin, sSgo1, or other centrosomal proteins. 31 , 32 The activity of PLK1 is regulated by phosphorylation catalyzed by several kinases, among which Aurora A phosphorylates PLK1 at Thr‐210 with scaffold proteins, thereby regulating PLK1 activity spatio‐temporally. 33 , 34 , 35 , 36 , 37 , 38
FIGURE 3.
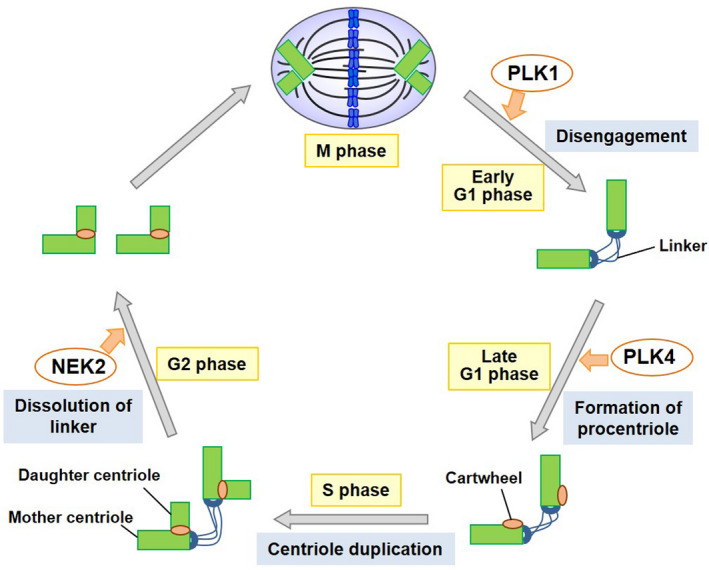
Schematic diagram of the centrosome duplication cycle. The mother and daughter centrioles are disengaged in late mitosis‐early G1 phase. After centriole disengagement, linkers are established between the 2 centrioles. In late G1 phase, a procentriole containing cartwheel is formed at each centriole. The building of the new centriole starts in the early S phase and 1 new daughter centriole forms perpendicularly to each mother centriole during the S phase. The new daughter centriole gradually elongates during the S and G2 phases. In the G2 phase, the 2 centrosomes separate through the dissolution of the linker and move to opposite sides of the cell to form the spindle poles
2.3. Centrosome aberrations in cancer
Structural and numerical aberrations of centrosomes were reported in various malignancies. 39 Regulators of centrosome duplication, including Aurora A and PLK1, are frequently dysregulated or mutated in cancers including breast cancer. 40 In breast tissues, centrosome aberrations are present in pre‐malignant lesions. 41 , 42 , 43 The frequency of centrosome aberrations increases in correlation with progressive tumor stage 42 and is associated with the degree of chromosomal instability. 42 Estrogen stimulation is associated with the induction of centrosome aberrations in breast tissues, suggesting that estrogen promotes tumor development via centrosome aberrations in estrogen‐dependent tissues. 44 In mouse models, overexpression of Plk4 induces centrosome amplification resulting in carcinogenesis. 45 These data suggest that centrosome aberrations contribute to carcinogenesis at an early stage by inducing chromosomal instability.
Excess centrosomes promote remodeling of the cytoskeleton by increasing Rac1 activity 46 and affecting cell‐cell adhesion, 47 which increase the invasiveness of cancer cells. In addition, centrosome aberrations or upregulation of centrosome regulators, including NEK2 and PLK4 in cancer cells, are related to the resistance to chemotherapeutics. 48 , 49 These processes may be involved in the poor prognosis of cancer patients with centrosome aberrations. 50
3. ROLES OF BRCA1 IN CENTROSOME REGULATION
3.1. BRCA1 dysfunction induces centrosome aberration
In invasive breast cancers, centrosome aberrations are more common in the triple‐negative subtype than in other histological subtypes. 50 Centrosome aberrations are frequently observed in BRCA1‐associated hereditary breast cancers and sporadic breast cancers lacking BRCA1 expression, 51 , 52 and are found even in non‐cancerous mammary epithelia in carriers of pathogenic BRCA1 variants. 53 These findings suggest that BRCA1 dysfunction induces centrosome aberrations during carcinogenesis in breast tissues.
BRCA1 localizes to centrosomes during interphase and to spindle poles during mitosis. 54 , 55 , 56 , 57 Inhibition or knockdown of BRCA1 induces centrosome amplification in cell lines derived from mammary epithelia. 16 Centrosome amplification induced by BRCA1 knockdown is rescued by wild‐type BRCA1, but not by several BRCA1 variants reported in hereditary breast cancer cases. 58 Missense variants of BRCA1, including I42V and L52F, are proficient in HR, but deficient in centrosome regulation. 58 Therefore, BRCA1 contributes to centrosome regulation independently of its DNA repair activity.
3.2. Regulation of BRCA1 localization to centrosomes
BRCA1 localizes to the mother centriole and is recruited to the new mother centriole in late G1 phase before procentriole formation. 55 The centrosomal localization of BRCA1 is decreased by disruption of the nuclear export signal (NES) of BRCA1 or inhibition of nucleus‐to‐cytosol transport by leptomycin B, 56 suggesting that centrosomal BRCA1 is derived from the nucleus.
Both the N‐terminus and C‐terminus of BRCA1 are required for its centrosomal localization, whereas the RING domain is dispensable. 56 There are 2 pools of centrosomal BRCA1, the slow turnover pool and the fast turnover pool, which is enhanced by association with BARD1. 56 However, the differences in the function and regulatory mechanisms between the 2 pools are not clear. 56
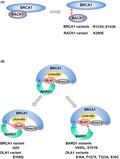
Receptor for activated protein C kinase 1 (RACK1) consists of 7 WD40 motifs exhibiting β‐propeller structures. 59 RACK1 directly binds to the N‐terminal region of BRCA1 (aa 70‐304), although the RING domain of BRCA1 is dispensable for the interaction. 19 Knockdown of RACK1 changes the centrosomal localization of BRCA1 from a discrete dot‐like pattern colocalizing with centrioles to a dispersed pattern with spike‐like protrusions. 19 Conversely, RACK1 overexpression intensifies centrosomal BRCA1 signals. 19 The BRCA1 variants R133H and E143K and the RACK1 variant K280E, which decrease the affinity of BRCA1 for RACK1 (Figure 4A), decrease its centrosomal localization. 19 These findings suggest that the N‐terminus of BRCA1 contributes to the regulation of BRCA1 localization by mediating the interaction with RACK1. Given that RACK1 functions as a scaffold protein, 59 other unknown proteins might also associate with BRCA1 via RACK1 to regulate BRCA1 localization.
FIGURE 4.
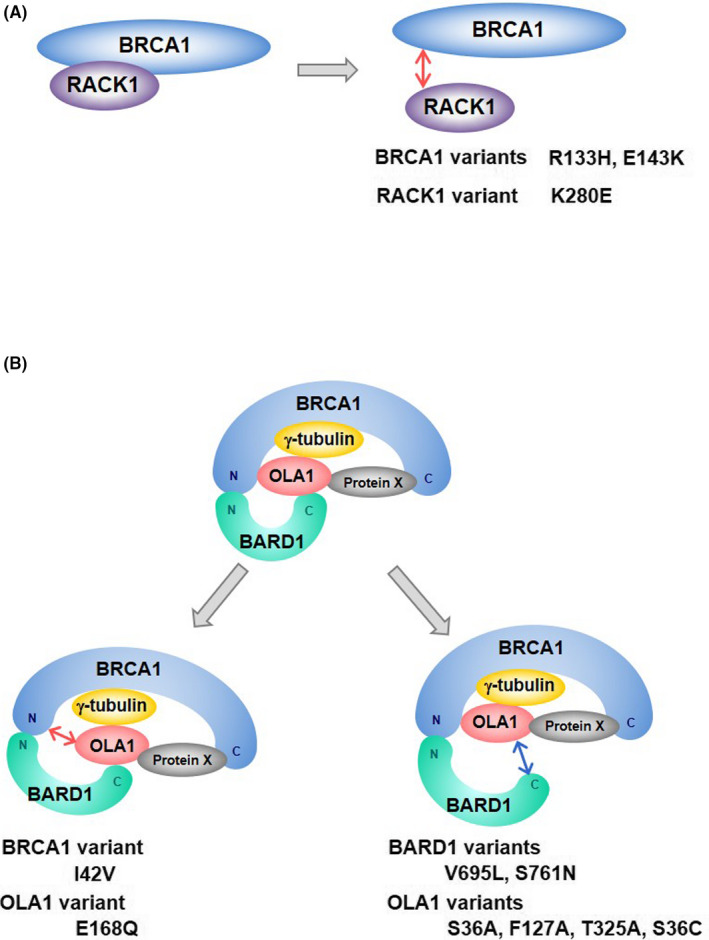
BRCA1 complex. A, The BRCA1‐RACK1 interaction and the effects of the BRCA1 and RACK1 variants. B, BRCA1 complex and abnormal complexes containing variants of BRCA1, BARD1, or OLA1. “N” indicates the N‐terminus. “C” indicates the C‐terminus
The role of the C‐terminus of BRCA1 in its centrosomal localization is poorly understood. The 2 BRCT domains in the C‐terminus interact with various phospho‐proteins. 5 Fanconi anemia group J protein (FANCJ, also called BACH1 or BRIP1) binds to the BRCT domains of BRCA1 and promotes the centrosomal localization of BRCA1 after DNA damage. 60 However, the role of FANCJ in the physiological BRCA1 localization to the centrosome remains undetermined. Furthermore, although the second BRCT domain of BRCA1 binds to p53, which functions in centrosome regulation, 61 , 62 whether p53 contributes to the regulation of BRCA1 localization to centrosomes is not clear.
3.3. The centrosomal BRCA1 complex contributes to the regulation of centrosome number
The BRCA1/BARD1 heterodimer interacts with various proteins, forming distinct complexes with corresponding molecular functions. Obg‐like ATPase (OLA1) was isolated by immunoprecipitation using the C‐terminus of BARD1 as bait. 17 In addition to the C‐terminus of BARD1, OLA1 directly binds to the N‐terminus of BRCA1 and γ‐tubulin to form BRCA1 complex (Figure 4B). 17
Upregulation of OLA1 is observed in various cancers, 63 and is associated with poor prognosis in breast and hepatocellular carcinomas. 64 , 65 Although OLA1 has ATPase or GTPase activity and contributes to stress responses in cells, 64 , 66 the precise role of OLA1 in carcinogenesis remains to be elucidated.
OLA1 localizes to centrosomes during interphase and to mitotic spindles and spindle poles during mitosis. 17 Deficiency and excess of OLA1 increase centrosome number in mammary epithelium‐derived cells but not in non‐breast cancer cells. 17 , 18 Conversely, OLA1 knockdown suppresses cell proliferation even in non‐breast cancer cell lines (unpublished data), suggesting that other activities of OLA1 affect cell proliferation irrespective of tissue type.
The proper formation of the complex composed of BRCA1, BARD1, and OLA1 is critical for the regulation of centrosome number (Figure 4B). 67 BRCA1 I42V decreases the interaction with OLA1. 17 Conversely, OLA1 E168Q, a substitution found in a breast cancer cell line, abrogates the interaction with BRCA1. 17 These variants do not rescue centrosome amplification in BRCA1 or OLA1 knockdown cells. 17 Substitutions of amino acids located at the interaction surface between BARD1 and OLA1, such as S761N of BARD1 and T325A of OLA1, abolish the interaction between the 2 proteins. 18 In addition to those variants, the BARD1 V695L variant and OLA1 S36A and F127A variants are deficient in binding of BARD1‐OLA1 and centrosome regulation. 18 We recently found that the OLA1 S36A variant decrease the centrosomal localization of OLA1 (unpublished data). Ser‐36 of OLA1 faces the ATP binding pocket of OLA1. 66 Binding of ATP to OLA1 may contribute to the loading of OLA1 into the centrosomal BRCA1 complex. The OLA1 S36C variant, which was found in cervical cancer, is also deficient in binding to BARD1 and in the regulation of centrosome number. 18 Collectively, these findings indicate that formation of the centrosomal BRCA1 complex composed of BRCA1, BARD1, and OLA1 is necessary for centrosome regulation (Figure 4).
3.4. Molecular mechanism of centrosome regulation by the BRCA1 complex
Centrosome amplification induced by BRCA1 deficiency is associated with centriole overduplication and premature disengagement. 68 Centriole overduplication is also induced by abnormal expression of OLA1 17 , 18 and can occur through several pathways, including concomitant overduplication, reduplication, elongation, and fragmentation of centrioles. The pathways contributing to centriole overduplication induced by the deficiency of BRCA1, BARD1, or OLA1 remain to be elucidated.
Overexpression of RACK1 induces centriole reduplication following premature centriole disengagement. 19 , 38 RACK1 acts as a scaffold protein to facilitate the interaction of Aurora A with PLK1, resulting in the Thr‐210 phosphorylation and activation of PLK1 in S phase (Figure 5, upper panel). 38 The Aurora A/PLK1 interaction can also be mediated by Bora, CEP192, and Furry in G2 and M phases. 33 , 34 , 35 , 36 , 37 , 38 RACK1 overexpression results in the accumulation of phosphorylated PLK1 at the centrosome before G2 and M phases, when PLK1 activity physiologically is maximal. Overactivation of PLK1 in S phase induces premature disengagement resulting in centriole overduplication (Figure 5, lower panel). Functional BRCA1 is required for centriole overduplication in RACK1‐overexpressing cells, 19 suggesting that the BRCA1 complex contributes to the regulation of PLK1 activity.
FIGURE 5.
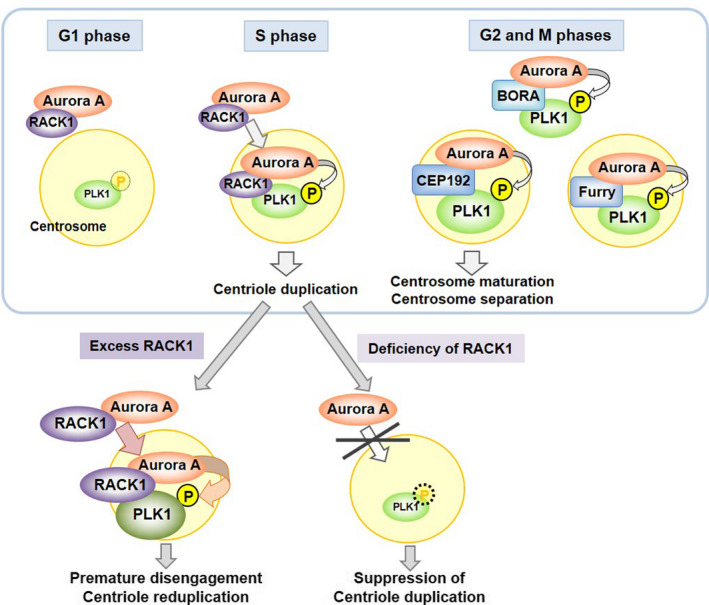
Regulation of PLK1 activity by RACK1. Under physiological conditions, RACK1 recruits Aurora A to centrosomes, thereby facilitating Aurora A binding to and phosphorylating PLK1 in the S phase. In G2 and M phases, activation of PLK1 is supported by Bora, CEP192, and Furry. Excess RACK1 induces overactivation of PLK1 in the S phase, resulting in premature centriole disengagement and centriole reduplication. Deficiency of RACK1 suppresses centriole duplication due to inadequate activation of PLK1
BRCA1 binds to and ubiquitinates γ‐tubulin to regulate centrosome activity. 16 , 54 BRCA1 monoubiquitinates γ‐tubulin at Lys‐48 and Lys‐344. 16 Forced expression of γ‐tubulin with the K48R, K344R, and K48R/K344R mutations, which abolish ubiquitination, induces centrosome amplification. 16 In addition, BRCA1 suppresses the formation of microtubule asters from centrosomes by inhibiting microtubule anchoring. 55 BRCA1 knockdown and forced expression of non‐ubiquitinatable γ‐tubulin mutants enhance the formation of large microtubule asters from centrosomes. 57 These data suggest that BRCA1 deficiency causes centrosome aberrations by altering the mechanical forces that centriole enable disengagement. 69
3.5. BRCA1 regulates the centrosome in a tissue‐specific manner
BRCA1 localizes to centrosomes in various cells derived from breast and non‐breast tissues. 19 , 54 , 55 , 56 , 57 Starita et al reported that inhibition of BRCA1 activity induces centrosome amplification in breast cancer cell lines (Hs578T and T47D) and immortalized mammary epithelial cell lines (MCF10A and MTSV1‐7), but not in cells derived from non‐breast tissues (DLD1, HeLa, PC3, and U2OS). 16 The effect of BRCA1 knockdown on inducing centrosome amplification was confirmed in Hs578T and MCF10A cells. 58 , 68
Abnormalities of other components of the centrosomal BRCA1 complex also induce centrosome aberrations specifically in mammary epithelium‐derived cells. 17 , 18 , 19 Overexpression of OLA1 induces centrosome amplification in Hs578T, MCF7, T47D, and MCF10A cells, but not in HeLa, PC3, Saos‐2, and U2OS cells. 18 Overexpression of BARD1 induces centrosome amplification in Hs578T and T47D cells but not in HeLa cells. 18 Overexpression of RACK1 induces centrosome amplification in Hs578T, MCF7, T47D, and MCF10A but not in 6 cell lines derived from non‐breast tissues. 19 Although Zou et al reported that BRCA1 knockdown causes centrosome amplification in U2OS cells, dysfunction of the centrosomal BRCA1 complex tends to cause centrosome aberrations in a tissue‐specific manner (Table 1).
TABLE 1.
Summary of the effects of BRCA1 complex deficiency in various cell lines
| References | Proteins | Treatments | Cell lines from breast tissue | Cell lines from non‐breast tissue |
|---|---|---|---|---|
| Starita et al, 2004 16 | BRCA1 | Inhibition by BIF | Hs578T, T47D, MCF10A, MTSV1‐7, ZR75‐1 | DLD1, HeLa, PC3, U2OS |
| Knockdown | Hs578T | U2OS | ||
| Ko et al, 2006 68 | BRCA1 | Knockdown | Hs578T, MCF10A | |
| Kais et al, 2012 58) | BRCA1 | Knockdown | Hs578T | |
| Zou et al, 2014 60 | BRCA1 | Knockdown | U2OS | |
| Matsuzawa et al, 2014 17 | OLA1 | Knockdown | Hs578T, T47D, HCC1937 | HeLa |
| Yoshino et al, 2018 18 | OLA1 | Overexpression | Hs578T, MCF‐7, T47D, MCF10A | HeLa, PC3, Saos‐2, U2OS |
| BARD1 | Overexpression | Hs578T, T47D | HeLa | |
| Yoshino et al, 2019 19 | RACK1 | Knockdown | MCF7 | HeLa |
| RACK1 | Overexpression | Hs578T, MCF7, T47D, MCF10A |
HeLa, KMST‐6, PC3, OVCAR‐3, Saos‐2, U2OS |
In the cell line columns, cell lines presented in red characters show centrosome amplification; the cell line presented in blue characters show suppression of centriole duplication; and cell lines presented in black characters do not show centrosome abnormalities. BIF; a fragment of RNA helicase A that binds to the C‐terminus of BRCA1.
However, the mechanism underlying the tissue specificity of centrosome regulation by the BRCA1 complex is not understood. The expression and localization of BRCA1, BARD1, OLA1, and RACK1 to centrosomes is not tissue specific. 17 , 18 , 19 , 54 , 55 , 56 , 57 To the best of our knowledge, a centrosomal protein that is specifically expressed in breast tissues has not been reported. However, mammary epithelium‐derived cells contain a larger fraction of cells with duplicated centrioles than non‐breast tissue‐derived cells. 19 As reduplication of centrioles occurs after the first duplication, mammary epithelium‐derived cells may be more amenable to centriole reduplication. The differences in the mechanisms of centrosome regulation among tissues should be investigated in future studies.
3.6. BRCA1 contributes to DNA damage‐induced centrosome amplification
DNA damage‐induced centrosome amplification (DDICA) is induced by various insults including ionizing radiation, mitomycin C, and hydroxyurea. 60 , 61 , 70 , 71 , 72 Although the precise mechanism underlying DDICA is unclear, the involvement of BRCA1 in DDICA has been reported. 60 Mitomycin C treatment increases BRCA1 localization to centrosomes, thereby increasing centrosomal PLK1. FANCJ is involved in the recruitment of BRCA1 to the centrosome in mitomycin C‐treated cells. Knockdown of either BRCA1 or FANCJ suppresses the increase of centrosomal PLK1 and the induction of DDICA.
The biological significance of DDICA is not well understood. When cells with extra centrosomes enter into mitosis, some cells undergo abnormal cell division, such as multipolar cell division, and finally die by apoptosis or mitotic catastrophe. 73 , 74 Some cells complete mitosis by forming pseudo‐bipolar spindles through centrosome clustering to evade cell death during mitosis. 73 , 75 However, these cells usually undergo cell death or cell cycle arrest after cell division. 73 Therefore, DDICA may be a mechanism to eliminate cells after prolonged, intolerable DNA damage.
By contrast, DDICA may be beneficial for the survival of cancer cells. Extra centrosomes increase chromosome mis‐segregation during mitosis, which could confer advantages for cell survival and proliferation. 73 Loss of p53 spares cells with extra centrosomes from cell death or cell cycle arrest during and after mitosis 76 , 77 , 78 , therefore p53‐deficient cancer cells may particularly benefit from extra centrosomes for cell proliferation.
In addition, various DNA repair factors localize to centrosomes. 79 DDICA may contribute to DNA damage repair by amplifying centrosomes, which act as signaling hubs for these DNA repair factors. DDICA has been detected in various cell lines 71 , 72 , 80 and is not tissue specific (unpublished data). However, the regulatory mechanism of DDICA and tissue specificity of BRCA1 deficiency in DDICA should be investigated in the future.
4. CONCLUSION
The centrosomal BRCA1 complex contributes to the maintenance of chromosomal stability by regulating centrosomes. Dysfunction of the BRCA1 complex causes centrosome aberrations in mammary epithelium‐derived cells. Concomitant alterations in the regulation of centrosomes and other functions including HR caused by BRCA1 deficiency may especially increase the risk of cancer in the breast tissue of pathogenic BRCA1 variant carriers. The precise mechanism by which the BRCA1 complex regulates centrosomes, and the specific effect of BRCA1 complex dysfunction on inducing tumors in mammary epithelial cells, are issues that remain to be investigated. Improving our understanding of these mechanisms will facilitate the diagnosis of hereditary breast cancer and enable the development of novel therapies targeting centrosome aberrations.
CONFLICT OF INTEREST
The authors declare no conflicts of interest for this article.
ACKNOWLEDGMENTS
This study was supported by grants‐in‐aid from JSPS KAKENHI Grant Numbers JP18K15233 (to Y. Yoshino), JP19H03493 (to N. Chiba and Y. Yoshino), a Research Grant of the Princess Takamatsu Cancer Research Fund, and a Research Program of the Smart‐Aging Research Center, Tohoku University (to N. Chiba).
Yoshino Y, Fang Z, Qi H, Kobayashi A, Chiba N. Dysregulation of the centrosome induced by BRCA1 deficiency contributes to tissue‐specific carcinogenesis. Cancer Sci. 2021;112:1679–1687. 10.1111/cas.14859
REFERENCES
- 1. Miki Y, Swensen J, Shattuck‐Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science (80). 1994;266:66‐71. [DOI] [PubMed] [Google Scholar]
- 2. Chen S, Parmigiani G. Meta‐Analysis of BRCA1 and BRCA2 Penetrance. J Clin Oncol. 2007;25:1329‐1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sawyer SL, Tian L, Kähkönen M, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dine J, Deng C‐X. Mouse models of BRCA1 and their application to breast cancer research. Cancer Metastasis Rev. 2013;32:25‐37. [DOI] [PubMed] [Google Scholar]
- 5. Takaoka M, Miki Y. BRCA1 gene: function and deficiency. Int J Clin Oncol. 2018;23:36‐44. [DOI] [PubMed] [Google Scholar]
- 6. Wu LC, Wang ZW, Tsan JT, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430‐440. [DOI] [PubMed] [Google Scholar]
- 7. Tarsounas M, Sung P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol. 2020;21:284‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao W, Wiese C, Kwon Y, et al. The brca tumor suppressor network in chromosome damage repair by homologous recombination. Annu Rev Biochem. 2019;88:221‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ransburgh DJR, Chiba N, Ishioka C, et al. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010;70:988‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Starita LM, Islam MM, Banerjee T, et al. A multiplex homology‐directed DNA repair assay reveals the impact of more than 1,000 BRCA1 missense substitution variants on protein function. Am J Hum Genet. 2018;103:498‐508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Towler WI, Zhang J, Ransburgh DJR, et al. Analysis of BRCA1 variants in double‐strand break repair by homologous recombination and single‐strand annealing. Hum Mutat. 2013;34:439‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Findlay GM, Daza RM, Martin B, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouwman P, van der Gulden H, van der Heijden I, et al. A high‐throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142‐1155. [DOI] [PubMed] [Google Scholar]
- 14. Nielsen FC, van Overeem HT, Sørensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16:599‐612. [DOI] [PubMed] [Google Scholar]
- 15. Nik‐Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole‐genome sequences. Nature. 2016;534:47‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starita LM, Machida Y, Sankaran S, et al. BRCA1‐dependent ubiquitination of gamma‐tubulin regulates centrosome number. Mol Cell Biol. 2004;24:8457‐8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsuzawa A, Kanno S‐I, Nakayama M, et al. The BRCA1/BARD1‐interacting protein OLA1 functions in centrosome regulation. Mol Cell. 2014;53:101‐114. [DOI] [PubMed] [Google Scholar]
- 18. Yoshino Y, Qi H, Fujita H, et al. BRCA1‐interacting protein OLA1 requires interaction with BARD1 to regulate centrosome number. Mol cancer Res. 2018;16:1499‐1511. [DOI] [PubMed] [Google Scholar]
- 19. Yoshino Y, Qi H, Kanazawa R, et al. RACK1 regulates centriole duplication by controlling localization of BRCA1 to the centrosome in mammary tissue‐derived cells. Oncogene. 2019;38:3077‐3092. [DOI] [PubMed] [Google Scholar]
- 20. Uzbekov RE, Avidor‐Reiss T. Principal postulates of centrosomal biology. Version 2020. Cells. 2020;9(10):2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc B Biol Sci. 2014;369:20130459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chavali PL, Putz M, Gergely F. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos Trans R Soc B Biol Sci. 2014;369:20130468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujita H, Yoshino Y, Chiba N. Regulation of the centrosome cycle. Mol Cell Oncol. 2016;3:e1075643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ong JY, Bradley MC, Torres JZ. Phospho‐regulation of mitotic spindle assembly. Cytoskeleton. 2020;77(12):558‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hardy T, Lee M, Hames RS, et al. Multisite phosphorylation of C‐Nap1 releases it from Cep135 to trigger centrosome disjunction. J Cell Sci. 2014;127:2493‐2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuo K, Ohsumi K, Iwabuchi M, et al. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol. 2012;22:915‐921. [DOI] [PubMed] [Google Scholar]
- 27. Tsou M‐FB, Wang W‐JJ, George KA, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fry AM. Solving the centriole disengagement puzzle. Nat Cell Biol. 2014;17:3‐5. [DOI] [PubMed] [Google Scholar]
- 29. Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by plk1. Curr Biol. 2010;20:1277‐1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karki M, Keyhaninejad N, Shuster CB. Precocious centriole disengagement and centrosome fragmentation induced by mitotic delay. Nat Commun. 2017;8:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J, Lee K, Rhee K. PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat Commun. 2015;6:10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Yang Y, Duan Q, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008;14:331‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seki A, Coppinger J, Jang C. Bora and Aurora A Cooperatively Activate Plk1 and Control the Entry into Mitosis. Science (80). 2008;320:1655‐1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macůrek L, Lindqvist A, Lim D, et al. Polo‐like kinase‐1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119‐123. [DOI] [PubMed] [Google Scholar]
- 35. Ikeda M, Chiba S, Ohashi K, et al. Furry protein promotes aurora A‐mediated Polo‐like kinase 1 activation. J Biol Chem. 2012;287:27670‐27681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joukov V, Walter JC, De Nicolo A. The Cep192‐Organized Aurora A‐Plk1 Cascade Is Essential for Centrosome Cycle and Bipolar Spindle Assembly. Mol Cell. 2014;55:578‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruinsma W, Macůrek L, Freire R, et al. Bora and Aurora‐A continue to activate Plk1 in mitosis. J Cell Sci. 2014;127:801‐811. [DOI] [PubMed] [Google Scholar]
- 38. Yoshino Y, Kobayashi A, Qi H, et al. RACK1 regulates centriole duplication through promoting the activation of polo‐like kinase 1 by Aurora A. J Cell Sci. 2020;133(17):jcs238931. [DOI] [PubMed] [Google Scholar]
- 39. Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7:1122‐1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Almeida BP, Vieira AF, Paredes J, et al. Pan‐cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLOS Comput Biol. 2019;15(3):e1006832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pihan GA, Wallace J, Zhou Y, et al. Centrosome abnormalities and chromosome instability occur together in pre‐invasive carcinomas. Cancer Res. 2003;63:1398‐1404. [PubMed] [Google Scholar]
- 42. Lingle WL, Barrett SL, Negron VC, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978‐1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lopes CAM, Mesquita M, Cunha AI, et al. Centrosome amplification arises before neoplasia and increases upon p53 loss in tumorigenesis. J Cell Biol. 2018;217:2353‐2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li JJ, Weroha SJ, Lingle WL, et al. Estrogen mediates Aurora‐A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc Natl Acad Sci U S A. 2004;101:18123‐18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levine MS, Bakker B, Boeckx B, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell. 2017;40(3):313‐322.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Godinho SA, Picone R, Burute M, et al. Oncogene‐like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ganier O, Schnerch D, Oertle P, et al. Structural centrosome aberrations promote non‐cell‐autonomous invasiveness. EMBO J. 2018;e98576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marina M, Saavedra HI. Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Front Biosci. 2014;19:352‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D’Assoro AB, Barrett SL, Folk C, et al. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat. 2002;75:25‐34. [DOI] [PubMed] [Google Scholar]
- 50. Denu RA, Zasadil LM, Kanugh C, et al. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer. 2016;16:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watanabe G, Chiba N, Nomizu T, et al. Increased centrosome number in BRCA‐related breast cancer specimens determined by immunofluorescence analysis. Cancer Sci. 2018;109:2027‐2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimomura A, Miyoshi Y, Taguchi T, et al. Association of loss of BRCA1 expression with centrosome aberration in human breast cancer. J Cancer Res Clin Oncol. 2009;135:421‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martins FC, De S, Almendro V, et al. Evolutionary pathways in BRCA1‐associated breast tumors. Cancer Discov. 2012;2:503‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hsu L‐C, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci. 1998;95:12983‐12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tarapore P, Hanashiro K, Fukasawa K. Analysis of centrosome localization of BRCA1 and its activity in suppressing centrosomal aster formation. Cell Cycle. 2012;11:2931‐2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brodie KM, Henderson BR. Characterization of BRCA1 Protein Targeting, Dynamics, and Function at the Centrosome: A role for the nuclear export signal, CRM1, and Aurora A kinase. J Biol Chem. 2012;287:7701‐7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sankaran S, Starita LM, Groen AC, et al. Centrosomal microtubule nucleation activity is inhibited by BRCA1‐dependent ubiquitination. Mol Cell Biol. 2005;25:8656‐8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kais Z, Chiba N, Ishioka C, et al. Functional differences among BRCA1 missense mutations in the control of centrosome duplication. Oncogene. 2012;31:799‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal. 2011;9(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zou J, Zhang D, Qin G, et al. BRCA1 and FancJ cooperatively promote interstrand crosslinker induced centrosome amplification through the activation of polo‐like kinase 1. Cell Cycle. 2014;13:3685‐3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mullee LI, Morrison CG. Centrosomes in the DNA damage response—the hub outside the centre. Chromosom Res. 2016;24:35‐51. [DOI] [PubMed] [Google Scholar]
- 62. Chai Y, Cui J, Shao N, et al. The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene. 1999;18:263‐268. [DOI] [PubMed] [Google Scholar]
- 63. Sun H, Luo X, Montalbano J, et al. DOC45, a Novel DNA damage‐regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol Cancer Res. 2010;8:57‐66. [DOI] [PubMed] [Google Scholar]
- 64. Chen H, Song R, Wang G, et al. OLA1 regulates protein synthesis and integrated stress response by inhibiting eIF2 ternary complex formation. Sci Rep. 2015;5:13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang S, Zhang C, Sun C, et al. Obg‐like ATPase 1 (OLA1) overexpression predicts poor prognosis and promotes tumor progression by regulating P21/CDK2 in hepatocellular carcinoma. Aging. 2020;12:3025‐3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koller‐Eichhorn R, Marquardt T, Gail R, et al. Human OLA1 defines an ATPase subfamily in the Obg family of GTP‐binding proteins. J Biol Chem. 2007;282:19928‐19937. [DOI] [PubMed] [Google Scholar]
- 67. Otsuka K, Yoshino Y, Qi H, et al. The Function of BARD1 in Centrosome Regulation in Cooperation with BRCA1/OLA1/RACK1. Genes. 2020;11:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ko MJ, Murata K, Hwang D‐S, et al. Inhibition of BRCA1 in breast cell lines causes the centrosome duplication cycle to be disconnected from the cell cycle. Oncogene. 2006;25:298‐303. [DOI] [PubMed] [Google Scholar]
- 69. Cabral G, Sanegre Sans S, Cowan CR, et al. Multiple mechanisms contribute to centriole separation in C. elegans. Curr Biol. 2013;23:1380‐1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shimada M, Komatsu K. Emerging connection between centrosome and DNA repair machinery. J Radiat Res. 2009;50:295‐301. [DOI] [PubMed] [Google Scholar]
- 71. Wang C‐Y, Huang EY‐H, Huang S, et al. DNA‐PK/Chk2 induces centrosome amplification during prolonged replication stress. Oncogene. 2015;34:1263‐1269. [DOI] [PubMed] [Google Scholar]
- 72. Sugihara E, Kanai M, Saito S, et al. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer Res. 2006;66:4020‐4029. [DOI] [PubMed] [Google Scholar]
- 73. Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoshino Y, Ishioka C. Inhibition of glycogen synthase kinase‐3 beta induces apoptosis and mitotic catastrophe by disrupting centrosome regulation in cancer cells. Sci Rep. 2015;5:13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Baudoin NC, Nicholson JM, Soto K, et al. Asymmetric clustering of centrosomes defines the early evolution of tetraploid cells. Elife. 2020;9.e54565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lambrus BG, Uetake Y, Clutario KM, et al. P53 protects against genome instability following centriole duplication failure. J Cell Biol. 2015;210:63‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Serçin Ö, Larsimont J‐C, Karambelas AE, et al. Transient PLK4 overexpression accelerates tumorigenesis in p53‐deficient epidermis. Nat Cell Biol. 2015;18:100‐110. [DOI] [PubMed] [Google Scholar]
- 78. Cuomo ME, Knebel A, Morrice N, et al. p53‐Driven apoptosis limits centrosome amplification and genomic instability downstream of NPM1 phosphorylation. Nat Cell Biol. 2008;10:723‐730. [DOI] [PubMed] [Google Scholar]
- 79. Lerit DA, Poulton JS. Centrosomes are multifunctional regulators of genome stability. Chromosom Res. 2016;24:5‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Löffler H, Fechter A, Liu FY, et al. DNA damage‐induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2013;32:2963‐2972. [DOI] [PubMed] [Google Scholar]


