Cutibacterium acnes is the third most common cause of cerebrospinal fluid (CSF) shunt infection and is likely underdiagnosed due to the difficulty in culturing this pathogen. Shunt infections lead to grave neurologic morbidity for patients especially when there is a delay in diagnosis.
KEYWORDS: shunt infection, biomarker, inflammation, CSF, cytokine, proteome, C. acnes
ABSTRACT
Cutibacterium acnes is the third most common cause of cerebrospinal fluid (CSF) shunt infection and is likely underdiagnosed due to the difficulty in culturing this pathogen. Shunt infections lead to grave neurologic morbidity for patients especially when there is a delay in diagnosis. Currently, the gold standard for identifying CSF shunt infections is microbiologic culture. However, C. acnes infection often results in falsely negative cultures; therefore, new diagnostic methods are needed. To investigate potential CSF biomarkers of C. acnes CSF shunt infection we adapted a rat model of CSF catheter infection to C. acnes. We found elevated levels of interleukin-1β (IL-1β), IL-6, chemokine ligand 2, and IL-10 in the CSF and brain tissues of animals implanted with C. acnes-infected catheters compared to sterile controls at day 1 postinfection. This coincided with modest increases in neutrophils in the CSF and, to a greater extent, in the brain tissues of animals with C. acnes infection, which closely mirrors the clinical findings in patients with C. acnes shunt infection. Mass spectrometry revealed that the CSF proteome is altered during C. acnes shunt infection and changes over the course of disease, typified at day 1 postinfection by an acute-phase and pathogen neutralization response evolving to a response consistent with wound resolution at day 28 compared to a sterile catheter placement. Collectively, these results demonstrate that it is possible to distinguish C. acnes infection from sterile postoperative inflammation and that CSF proteins could be useful in a diagnostic strategy for this pathogen that is difficult to diagnose.
INTRODUCTION
Each year, approximately 18,000 cerebrospinal fluid (CSF) shunts are placed to treat hydrocephalus (1). A common side effect of shunt placement is infection, with a rate ranging from 1.6 to 33%, ultimately resulting in approximately 40,000 infections annually (2–5).
Timely and accurate diagnosis of pediatric CSF shunt infection is critical, since these infections are associated with seizures, reduced IQ scores, decreased school performance, and death (6–10). In addition, treatment is intense, requiring multiple surgeries to remove and replace the shunt, as well as weeks of parenteral antibiotics (5, 10–13). Currently, the diagnosis of these infections relies on culturing a pathogen from the CSF of a patient with a suspected shunt infection (11). However, this is not always reliable because children are frequently pretreated with antibiotics, are infected with fastidious, slow-growing organisms, and often have biofilm infections with few planktonic bacteria in the CSF for culture (14–17). This is especially true in Staphylococcus epidermidis and Cutibacterium acnes (formerly Propionibacterium acnes) shunt infections, since these bacteria are form robust biofilms (14, 18–20). Biofilms are bacterial communities encased in a protective matrix which are recalcitrant to antibiotic therapy (20). C. acnes, the third most common cause of shunt infection, presents an additional challenge as it often takes 1 to 3 weeks to grow in culture, and humans are unable to spontaneously clear this bacterium (16, 21–26). Due to inadequate culture techniques, C. acnes shunt infection has likely been underdiagnosed in the past, since many cases of shunt malfunction are likely related to C. acnes infection (22, 27–29). In the absence of a positive culture, diagnosis of CSF shunt infection relies on CSF indices, which are nonspecific and can be difficult to interpret in the setting of recent shunt placement, which elicits an initial inflammatory response arising from local tissue damage (11, 17, 30–33).
Identifying biomarkers in the CSF of patients with shunt infections would be a significant advance that could improve the ability to diagnose CSF shunt infections, particularly in the case of C. acnes, where cultures are frequently negative. Biomarkers are distinct biochemical, genetic or molecular substances that characterize infection (21, 24). Known biomarkers such as C-reactive protein, erythrocyte sedimentation rate, or procalcitonin are general biomarkers of inflammation/infection and have shown limited utility in diagnosing shunt infection (11, 34, 35). Another approach is to measure cytokines and chemokines, which are substances secreted by immune cells in response to infection. They may provide more specific data regarding the cause of the infection and distinguish infection from other causes of inflammation, such as trauma or recent surgery (36). In addition, CSF proteins elaborated during the infection may serve as novel biomarkers.
To begin to address this gap, our laboratory previously developed a rat model of Staphylococcus epidermidis central nervous system (CNS) catheter infection (37). This model facilitates discrimination between the inflammatory response induced by shunt infection versus that induced by the foreign body alone. This novel model also allowed sampling of the CSF in response to biofilm infection in vivo, as well as clinical signs of illness, reflecting the evolution of infection and the immune response in a way that mimics human disease. Interleukin-1β (IL-1β), (C-C motif) ligand 2 (CCL2), and (C-C motif) ligand 3 (CCL3) were significantly elevated in the CSF of animals with S. epidermidis-infected catheters compared to baseline CSF and postoperative CSF (37).
Although these are promising biomarkers for S. epidermidis shunt infection, further analysis is needed to elucidate potential biomarkers for C. acnes. Both of these Gram-positive organisms are capable of forming biofilm; however, C. acnes is a more indolent bacteria, which may elicit a unique host immune response (14, 18, 20). To date, the host immune response to C. acnes CSF shunt infection has not been investigated.
We adapted our rat model of CNS catheter infection to C. acnes to identify possible CSF biomarkers, since CSF is routinely collected in the evaluation of human shunt infection. We demonstrated preferential biofilm growth with C. acnes consistent with our previous studies (37). CSF and brain tissue samples from animals with C. acnes-infected catheters revealed elevated IL-1β, IL-6, CCL2, and IL-10 at day 1 postinfection compared to controls. The brain tissue of rats with C. acnes demonstrated a greater influx of neutrophils and monocytes/macrophages compared to animals with sterile catheters. Mass spectrometry (MS) revealed distinct changes in the CSF proteome between C. acnes-infected animals compared to controls and an evolution in the proteome over the course of the infection. Specifically, the CSF demonstrated a pathogen neutralization response at day 1 postinfection, which transitioned to more of a wound resolution response at day 28. Collectively, these results suggest that the CSF proteome could be leveraged for the identification of C. acnes shunt infection, which should be investigated in future clinical studies.
RESULTS
C. acnes biofilm infection model.
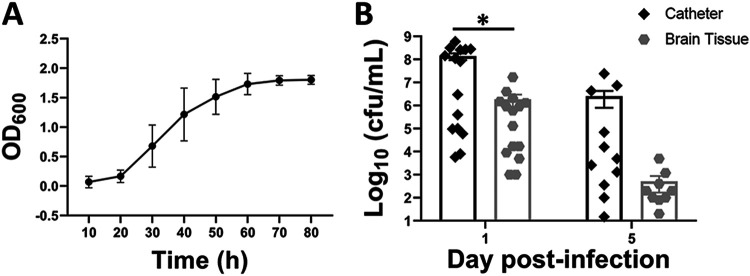
To establish a rat model of C. acnes CNS catheter infection that recapitulates biofilm growth, we first determined growth curves to establish the timing of logarithmic phase growth, which lasted from 20 to 60 h postinoculation (Fig. 1A). Bacteria from the logarithmic phase were used to inoculate silicone catheters and incubated for 24 h to create an adherent biofilm on the catheter, which were implanted into rats (37, 38). This approach was previously successful in our rat model of S. epidermidis CNS catheter infection to avoid confounds from bacterial efflux from the catheter lumen if organisms were injected into the catheter because of the high CSF pressure (37). Catheters demonstrated several-log-higher bacterial burdens than surrounding brain tissues at day 1 postinfection (P = 0.043), reflecting a biofilm model of infection (Fig. 1B) (38, 39). By day 5 postinfection, the bacterial burden on the catheter and in brain tissue began to decline as animals spontaneously cleared the infection (Fig. 1B). By day 28 postinfection, we were unable to recover bacteria from the catheters of 6 of 10 rats and the brain tissues of 8 of the 10 animals, suggesting that most rats had cleared the infection (data not shown). In the few animals where bacteria were recovered from the catheter, the level ranged from 15 to 50 CFU/ml, whereas the levels of bacteria recovered from the brain tissue ranged from 30 to 100 CFU/ml. Animals receiving sterile catheters had no evidence of bacterial growth (data not shown), and we did not quantitate bacteria in the CSF given the limited CSF volumes that were needed for multiplex and MS analysis.
FIG 1.
Kinetics of C. acnes growth in culture (A; n = 5 independent experiments) and on catheters versus surrounding brain tissue (B). Catheters were recovered from brain tissue at the indicated day postinfection and sonicated in PBS, whereupon the surrounding brain tissue was homogenized and bacterial titer was determined (n = 10 to 15). Catheters and brain tissue from rats with sterile catheters were negative and are not presented. (*, P < 0.05; Wilcoxon rank sum two-sample test).
C. acnes infection induces cytokine and chemokine expression in CSF and brain tissue.
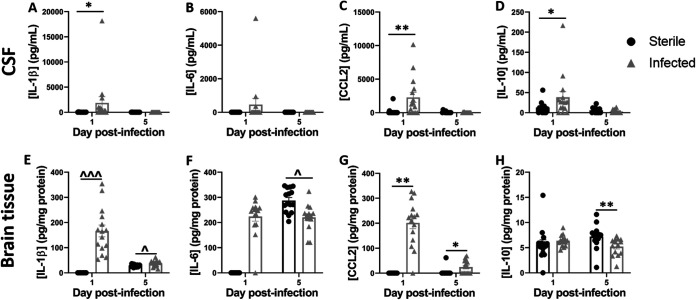
Our prior study with S. epidermidis CNS catheter infection demonstrated elevated expression of IL-10, IL-1β, CCL2, and CCL3, suggesting their potential utility as biomarkers. To assess the possibility that these mediators would also be increased during C. acnes infection and to broaden the array of molecules examined, CSF was analyzed for the presence of 27 chemokines and cytokines by a multianalyte microbead array. Brain tissue homogenates from tissue immediately surrounding the catheter tract were included for comparison. CSF from C. acnes-infected animals demonstrated significantly increased IL-1β (Fig. 2A; P = 0.017), CCL2 (Fig. 2C; P = 0.005), and IL-10 (Fig. 2D; P = 0.048) at day 1 postinfection, with levels reduced at day 5 as bacterial burdens decreased, whereas IL-6 was not significantly induced (Fig. 2B). Similarly, in the brain tissues of animals with C. acnes-infected catheters IL-1β (Fig. 2E; P < 0.001) and CCL2 (Fig. 2G; P = 0.0004) were increased at day 1 postinfection. However, in contrast to the CSF, both IL-1β (P = 0.015) and CCL2 (P = 0.021) remained elevated in brain tissue at day 5 postinfection. IL-6 (Fig. 2F; P = 0.018) and IL-10 (Fig. 2H; P = 0.0004) were higher in the brain tissues of animals that received sterile catheters at day 5 postinfection. No chemokines or cytokines were significantly elevated at day 28 postinfection in either CSF or brain tissue compared to uninfected controls (data not shown).
FIG 2.
C. acnes catheter-associated infection elicits elevated cytokine/chemokine expression compared to sterile injury. Quantitation of IL-1β (A and E), IL-6 (B and F), CCL2 (C and G), and IL-10 (D and H) in the CSF and brain tissues of rats with C. acnes-infected versus sterile catheters (n = 13 to 15). *, P < 0.05; **, P < 0.001 (Wilcoxon rank sum two-sample test). ^, P < 0.05; ^^^, P < 0.0001 (independent two-sample t test).
CSF and brain tissue leukocyte infiltrate during C. acnes infection.
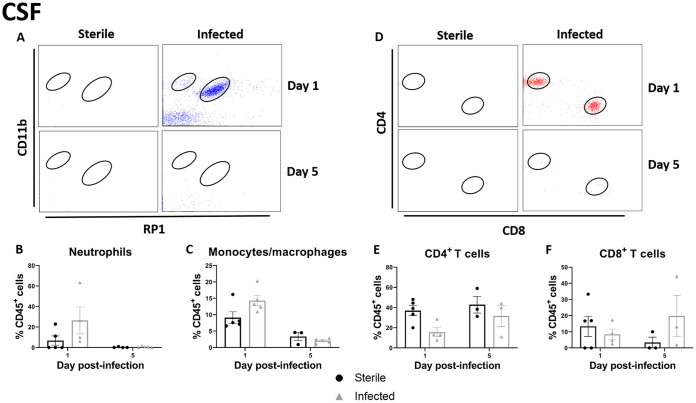
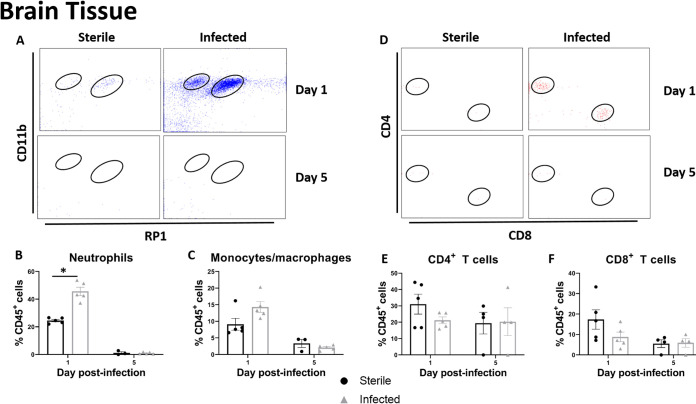
CSF pleocytosis is a clinical indicator of shunt infection; therefore, we examined leukocyte infiltrates in the CSF and brain tissue. There was a trend toward increased neutrophil and macrophage/monocyte influx into the CSF of animals with C. acnes-infected catheters (Fig. 3A to C). Animals with both sterile and C. acnes-infected catheters demonstrated an influx of both CD4+ and CD8+ T cells at day 1 postinfection (Fig. 3D to F). Similar relationships were seen in the brain tissue (Fig. 4), except neutrophil recruitment was significantly increased at day 1 postinfection (P = 0.0335) (Fig. 4B). As the infection progressed and the bacterial burden decreased from day 1 to day 5 postinfection, there appeared to be a shift to a larger CD4+ T cell infiltrate within the CSF (Fig. 3E).
FIG 3.
C. acnes catheter-associated infection elicits heightened leukocyte influx in the CSF compared to sterile postoperative inflammation. (A) Representative dot plots of CD11b and RP1 staining demonstrate CD45+ CD11b+ RP1+ neutrophils and CD45+ CD11b+ RP1– monocytes/macrophages in the CSF of animals with C. acnes-infected versus sterile catheters at days 1 and 5 postinfection. (B and C) Percentages of CD45+ CD11b+ RP1– monocytes/macrophages (B) and CD45+ CD11b+ RP1+ neutrophils (C). (D) Representative dot plots of CD4 and CD8 staining demonstrate CD45+CD3+CD4+ CD4 T cells and CD45+CD3+ CD8+ CD8 T cells in the CSF of animals with C. acnes-infected versus sterile catheters at days 1 and 5 postinfection. (E and F) Percentages of CD45+ CD3+ CD4+ CD4 T cells (E) and CD45+ CD3+ CD8+ CD8 T cells in the CSF (F).
FIG 4.
C. acnes catheter-associated infection elicits heightened leukocyte influx in the brain tissue compared to sterile postoperative inflammation. (A) Representative dot plots of CD11b and RP1 staining demonstrate CD45+ CD11b+ RP1+ neutrophils and CD45+ CD11b+ RP1– monocytes/macrophages in brain tissues of animals with C. acnes-infected versus sterile catheters at days 1 and 5 postinfection. (B and C) Percentages of CD45+ CD11b+ RP1– monocytes/macrophages (B) and CD45+ CD11b+ RP1+ neutrophils (C). (D) Representative dot plots of CD4 and CD8 staining demonstrate CD45+ CD3+ CD4+ CD4 T cells and CD45+ CD3+ CD8+ CD8 T cells in the brain tissue of animals with C. acnes-infected versus sterile catheters at days 1 and 5 postinfection. (E and F) Percentages of CD45+ CD3+ CD4+ CD4 T cells (E) and CD45+ CD3+ CD8+ CD8 T cells in the brain tissue (F). *, P < 0.05 (Wilcoxon rank sum two-sample test).
Differential expression of proteins in the CSF over the course of C. acnes CNS catheter infection.
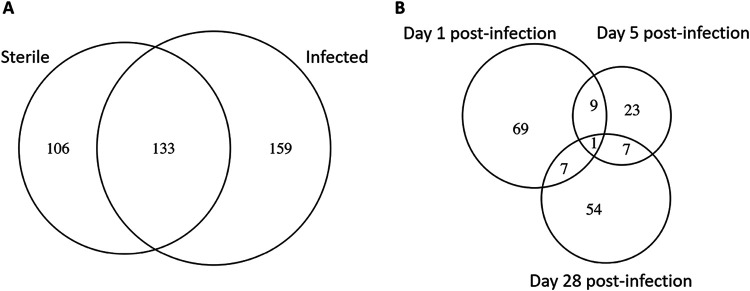
To characterize the CSF proteome throughout active C. acnes infection and convalescence (days 1, 5, and 28 postinfection), CSF was sampled from infected animals versus those that received sterile catheters. Examination of infected versus sterile groups across all time points revealed 106 proteins that were unique to the sterile group, 159 proteins that were unique to the infected group, and 133 proteins that were expressed by both groups (Fig. 5). In comparing sterile and infected groups by time point there were 69 unique proteins that were differentially expressed at day 1 postinfection, 23 at day 5 postinfection and 54 at day 28 postinfection (see Tables S1 to 3 in the supplemental material). Days 1 and 5 postinfection shared 9 proteins that were differentially expressed between the groups (actin cytoplasmic 1, aldo-keto reductase family 1 member B1, dihydropyrimidinase-related protein 2, growth arrest-specific protein 6, insulin-like growth factor-binding protein 3, leucine-rich repeat-containing protein 4B, neurosecretory protein VGF, phosphatidylcholine-sterol acyltransferase, and tubulin α-1B chain). Days 1 and 28 postinfection shared 7 proteins (coagulation factor XII, insulin-like growth factor-binding protein complex acid labile subunit, multifunctional procollagen lysine hydroxylase and glycosyltransferase LH3, protein piccolo, soluble calcium-activated nucleotidase 1, tubulin α-1C chain, and ubiquitin-40S ribosomal protein S27a). A total of 7 proteins were shared between days 5 and 28 postinfection (aspartate aminotransferase, mitochondrial elongation factor 1-α1, integrin β-like protein 1, plectin, receptor-type tyrosine-protein phosphatase F, ribose-phosphate pyrophosphokinase 2, and spectrin alpha chain nonerythrocytic 1). One protein (elongation factor 1-α2) was differentially expressed between sterile and infected groups at all time points.
FIG 5.
Relationship between differentially expressed proteins at days 1, 5, and 28 postinfection in the CSF of C. acnes- infected animals. (A) A Venn diagram depicts the similarities and differences between significantly differentially expressed proteins in the CSF of C. acnes-infected animals compared to sterile catheters at all time points, as determined by MS analysis. Significant proteins for each time point are presented in Tables S1 to S3 in the supplemental material. (B) A Venn diagram dipicts the overlap between significantly differentially expressed proteins in the CSF at days 1, 5, and 28 postinfection.
Finally, pathway analysis of differentially expressed proteins at each of the time points identified biological processes that were active during C. acnes CNS catheter infection. At day 1 postinfection, 12 different biologic processes were enriched with a range of 2 to 14 proteins in each of the pathways (Table 1). At day 5 postinfection, 20 different processes were upregulated with 2 to 29 proteins active in each category (Table 2). At day 28 postinfection, 10 different processes were active with anywhere from 4 to 9 proteins in each category (Table 3). There appeared to be a pathogen neutralization response at day 1 postinfection, as the majority of the enhanced biologic pathways involve catabolic processes as well as reactive oxygen species. Peroxidredoxin-1, peroxidredoxin-2, extracellular superoxide dismutase, catalase, glyceraldehyde-3-phosphate dehydrogenase, and thioredoxin were upregulated during early infection, which would correspond to an early immune response to C. acnes. At day 5 postinfection, elevated complement component C6 and proteins involved in the synaptic vesicle cycle such as dynamin-2 and dynamic-3 were observed. At day 28 when infection is resolved, the CSF proteome demonstrated an upregulation of metabolic and biosynthetic responses. At this time proteins such as very long-chain acyl-CoA synthetase, phosphoglycerate kinase 1, ATP synthase subunits, and pyruvate dehydrogenase subunits were elevated compared to acute infection. Full proteome data are available from the EBI PRIDE database, accession number PXD020527.
TABLE 1.
Day 1 postinfection functional enrichment for sterile versus infected groups
| PANTHER GO-Slim biological process | No. of reference proteins | No. of proteins | Raw P value | FDRa |
|---|---|---|---|---|
| Catabolic process (GO:0009056) | 888 | 14 | 8.51E−06 | 2.51E−03 |
| Cellular catabolic process (GO:0044248) | 764 | 13 | 8.37E−06 | 2.88E−03 |
| Cofactor metabolic process (GO:0051186) | 172 | 8 | 4.78E−07 | 3.29E−04 |
| Reactive oxygen species metabolic process (GO:0072593) | 40 | 6 | 2.28E−08 | 4.71E−05 |
| Drug metabolic process (GO:0017144) | 195 | 6 | 1.29E−04 | 2.41E−02 |
| Response to inorganic substance (GO:0010035) | 58 | 5 | 4.48E−06 | 2.31E−03 |
| Response to reactive oxygen species (GO:0000302) | 12 | 4 | 3.63E−07 | 3.74E−04 |
| Response to oxidative stress (GO:0006979) | 28 | 4 | 6.84E−06 | 2.82E−03 |
| Response to toxic substance (GO:0009636) | 35 | 4 | 1.53E−05 | 3.95E−03 |
| Cellular response to oxidative stress (GO:0034599) | 18 | 3 | 7.02E−05 | 1.61E−02 |
| Superoxide metabolic process (GO:0006801) | 19 | 3 | 8.10E−05 | 1.67E−02 |
| Positive regulation of cytokine secretion (GO:0050715) | 7 | 2 | 5.21E−04 | 8.95E−02 |
FDR, false discovery rate.
TABLE 2.
Day 5 postinfection functional enrichment for sterile versus infected groups
| PANTHER GO-Slim biological process | No. of reference proteins | No. of proteins | Raw P value | FDR |
|---|---|---|---|---|
| Biological process (GO:0008150) | 9320 | 29 | 1.86E−04 | 3.85E−02 |
| Cellular process (GO:0009987) | 7870 | 26 | 3.81E−04 | 5.24E−02 |
| Cellular component organization or biogenesis (GO:0071840) | 2471 | 14 | 8.16E−05 | 2.10E−02 |
| Cellular component organization (GO:0016043) | 2296 | 13 | 1.62E−04 | 3.72E−02 |
| Unclassified (UNCLASSIFIED) | 12357 | 11 | 1.86E−04 | 3.50E−02 |
| Organelle organization (GO:0006996) | 1757 | 11 | 2.55E−04 | 4.38E−02 |
| Establishment of localization in cell (GO:0051649) | 754 | 7 | 4.29E−04 | 5.53E−02 |
| Organelle fission (GO:0048285) | 327 | 6 | 3.07E−05 | 1.27E−02 |
| Microtubule-based process (GO:0007017) | 382 | 6 | 7.19E−05 | 2.12E−02 |
| Cytoskeleton organization (GO:0007010) | 554 | 6 | 5.21E−04 | 6.32E−02 |
| Protein-containing complex localization (GO:0031503) | 176 | 5 | 1.98E−05 | 1.02E−02 |
| Establishment of organelle localization (GO:0051656) | 172 | 5 | 1.77E−05 | 1.22E−02 |
| Organelle localization (GO:0051640) | 218 | 5 | 5.36E−05 | 1.84E−02 |
| Synaptic vesicle transport (GO:0048489) | 14 | 3 | 3.87E−06 | 7.98E−03 |
| Synaptic vesicle endocytosis (GO:0048488) | 23 | 3 | 1.46E−05 | 1.51E−02 |
| Vesicle-mediated transport in synapse (GO:0099003) | 64 | 3 | 2.56E−04 | 3.77E−02 |
| Synaptic vesicle cycle (GO:0099504) | 64 | 3 | 2.56E−04 | 4.06E−02 |
| Mitochondrial fission (GO:0000266) | 18 | 2 | 6.15E−04 | 7.05E−02 |
| Receptor internalization (GO:0031623) | 20 | 2 | 7.46E−04 | 8.11E−02 |
| Regulation of synapse structure or activity (GO:0050803) | 21 | 2 | 8.17E−04 | 8.42E−02 |
TABLE 3.
Day 28 postinfection functional enrichment for sterile versus infected groups
| PANTHER GO-Slim biological process | No. of reference proteins | No. of proteins | Raw P value | FDR |
|---|---|---|---|---|
| Organonitrogen compound biosynthetic process (GO:1901566) | 712 | 9 | 4.41E−04 | 8.27E−02 |
| Organophosphate metabolic process (GO:0019637) | 325 | 6 | 6.36E−04 | 9.38E−02 |
| Nucleoside phosphate metabolic process (GO:0006753) | 178 | 5 | 2.90E−04 | 6.66E−02 |
| Organophosphate biosynthetic process (GO:0090407) | 185 | 5 | 3.45E−04 | 7.12E−02 |
| Nucleotide metabolic process (GO:0009117) | 173 | 5 | 2.56E−04 | 8.79E−02 |
| Nucleobase-containing small molecule metabolic process (GO:0055086) | 214 | 5 | 6.59E−04 | 9.06E−02 |
| Purine-containing compound biosynthetic process (GO:0072522) | 95 | 4 | 2.78E−04 | 7.18E−02 |
| Ribose phosphate biosynthetic process (GO:0046390) | 95 | 4 | 2.78E−04 | 8.21E−02 |
| Nucleoside phosphate biosynthetic process (GO:1901293) | 116 | 4 | 5.78E−04 | 9.17E−02 |
| Nucleotide biosynthetic process (GO:0009165) | 115 | 4 | 5.60E−04 | 9.63E−02 |
DISCUSSION
CSF shunt infections can create serious long-term neurologic consequences, such as seizures and deceased IQ (6–10). With delayed diagnosis it is possible these consequences could be exacerbated. Pointing to the need for improved diagnostic modalities that are both rapid and accurate, especially in the case of C. acnes, which is difficult to grow in culture and can form biofilm (18, 21, 23, 24, 40). While detection of bacteria by nucleic acid application assays is becoming more common in clinical microbiology laboratories, detection of C. acnes genetic material may not always be indicative of infection since it comprises part of the normal skin flora (16, 26, 40–43).
We recently characterized the CSF inflammatory milieu in a rat model of S. epidermidis shunt infection (37). To expand upon this work, we adapted this rat model to C. acnes, which we show here accurately exhibits biofilm growth, as typified by a higher bacterial burden on implanted catheters compared to surrounding brain tissue. C. acnes elicited heightened levels of IL-1β, IL-10, and CCL2 in the CSF, in agreement with our prior S. epidermidis studies. Since these inflammatory mediators are conserved in C. acnes and S. epidermidis catheter infection they represent promising CSF diagnostic markers (37). However, achieving diagnostic specificity requires future studies to determine whether IL-1β, IL-10, and CCL2 are also elevated in Gram-negative CSF shunt infections.
When cultures are negative, clinicians must rely on other CSF indices such as CSF pleocytosis and cell count to determine whether a shunt infection is present. However, the absence of pleocytosis does not rule out shunt infection, and this is especially true in the case of C. acnes infection (11). Although neutrophils were significantly increased in the brain parenchyma surrounding C. acnes-infected catheters, they were minimal in the CSF. This finding supports clinical dogma that infection may not translate to robust neutrophil influx or CSF pleocytosis (11, 30–32, 44). Therefore, clinicians should retain a high index of suspicion for shunt infection despite the absence of pleocytosis, especially if patient symptoms are consistent with infection.
The number of CSF proteins that were differentially expressed between sterile and infected groups at days 1 and 28 postinfection roughly doubled that at day 5, demonstrating that there are long-term changes in the CSF proteome despite resolution of infection at day 28 postinfection. At day 1 postinfection, functional enrichment analysis identified pathways related to the acute-phase response and pathogen neutralization. This analysis included proteins involved in catabolic processes, reactive oxygen species, and responses to reactive oxygen species and oxidative stress. At day 5 postinfection, there was a shift from the acute phase response to proteins involved in synaptic vesicle transport, fusion, and endocytosis. Although speculative, this suggests that synapse destruction and/or loss may occur during infection, which may be responsible for the deleterious neurologic sequelae associated with shunt infection. Another shift in enriched biological processes occurred at day 28 postinfection, when the CSF proteome was dominated by proteins corresponding to metabolic and biosynthetic processes, which likely reflects the postinfection resolution response. Collectively, these unique changes in the CSF proteome suggest that CSF proteins could be used to monitor the course of C. acnes CNS catheter infection and potentially inform antibiotic treatment courses. Unlike in our previous S. epidermidis work, we did not detect significantly different levels of many complement cascade proteins at day 5 postinfection. However, we did observe a significant difference in C6 at day 5 postinfection, despite low bacterial burdens, supporting our earlier hypothesis that complement may contribute to the deleterious neurologic effects associated with shunt infection.
There are several limitations to this study. The limited available immunologic reagents for rats led to the inability to discriminate between monocytes and macrophages in the CSF and brain tissue. However, our results are consistent with our prior study, as well as those from human shunt infection (36, 37, 44). Second, this is an animal model and the results may not be immediately translatable to human studies; however, this model allows us to establish an informed basis for future clinical trials. We only examined a single strain of C. acnes CNS catheter infection in this report, and shunt infections can be caused by a wide variety of organisms, including S. epidermidis, which was investigated in our prior study (37). Additional studies are under way to directly compare the CSF inflammatory profiles associated with a spectrum of Gram-positive and Gram-negative bacteria common in shunt infections. It is likely that the biomarkers for Gram-negative infection could differ from the two Gram-positive organisms we have examined, which was suggested by our previous pilot study (36).
This study describes a rat model of C. acnes catheter infection, which shares immunological attributes with human disease and allows for repeated CSF sampling to explore potential diagnostic biomarkers and inform human studies. We have demonstrated elevated levels of IL-1β, IL-10, and CCL2 in the CSF of animals with C. acnes infection, which aligned with our previous findings in S. epidermidis CNS catheter infection (37). We also found that the CSF proteome evolves over the course of the infection, which could be leveraged for diagnosis and to better define antibiotic treatment courses. Findings from our preclinical studies can be explored in future human studies and validated as diagnostic biomarkers for human shunt infection.
MATERIALS AND METHODS
Animals.
Experiments were performed using equal numbers of 8-week-old male and female Lewis rats (Charles River Laboratories, Wilmington, MA). The Institutional Animal Care and Use Committee at the University of Nebraska Medical Center approved the protocol for animal use (protocol 16-091-09-FC) and is compliant with National Institutes of Health guidelines for the use of rodents. Animals were housed 3 to 4 per cage with species appropriate enrichment in a 12-h light-dark cycle. Food and water were provided ad libitum. Upon shipment, animals had a 3-day acclimation period prior to performance of any procedures. Group sizes were as follows: for quantification of bacterial burden, n = 10 to 15 per group; for chemokine/cytokine analysis, n = 13 to 15 per group; for flow cytometry n = 3 to 5 per group; and for mass spectrometry analysis, n = 9 to 13 per group.
C. acnes and in vitro propagation.
A clinical strain of C. acnes isolated from CSF at the University of Nebraska Medical Center clinical microbiology laboratory was kindly provided by Paul Fey. This isolate has not been laboratory adapted or modified. For growth curves, a single colony of C. acnes was inoculated into 40 ml of reinforced clostridial medium (BD, Franklin Lakes, NJ) and cultured in an anaerobic chamber with shaking at 250 rpm. For animal experiments, a single colony of C. acnes was inoculated into 40 ml of reinforced clostridial medium and incubated into an anaerobic chamber with shaking at 250 rpm for 48 h to achieve log-phase growth.
Catheter preparation and implantation.
Hollow silicone catheters (4-mm length, 1-mm diameter; Cook Medical, Inc., Bloomington, IN) were incubated overnight with 250 μl of log-phase C. acnes to ensure bacterial adherence to the catheter and prevent bacterial efflux as previously described (37, 38, 45). Rats were anesthetized with an intraperitoneal injection of ketamine and xylazine (87 and 13 mg/kg, respectively) and received a subcutaneous injection of bupivacaine (1 mg/kg) and buprenorphine (0.01 mg/kg) to mitigate any potential pain. The head was shaved, and a 1-cm longitudinal incision was made in the scalp. Each animal was then positioned in a stereotactic apparatus, and a burr hole was made in the skull using a 16-gauge needle at the following coordinates to direct catheter placement into the left lateral ventricle of the brain: anterior-posterior = −1.0 mm, medial-lateral = 2.4 mm, and dorsal-ventral = 4.0 mm (46). To secure the catheter in place and prevent bacterial efflux or bleeding, the burr hole was sealed with bone wax, and the scalp incision was closed using Vetbond surgical glue (3M, St. Paul, MN) (37, 38).
CSF collection.
At days 1, 5, and 28 after catheter insertion, rats were anesthetized with an intraperitoneal injection of ketamine and xylazine (87 and 13 mg/kg, respectively), the back of the neck was shaved, and the animals were placed in a stereotactic apparatus as previously described (37). Once positioned, the back of the neck was swabbed with betadine, and a percutaneous cisterna magna puncture was performed using a 25-gauge winged infusion set (Terumo Corporation, Somerset, NJ) with a guard to leave 4 mm of the needle tip exposed. A total of 100 to 120 μl of CSF was withdrawn and stored at –80°C until analysis.
Bacterial enumeration from catheters and brain parenchyma.
Catheters and brain tissue were collected at days 1, 5, and 28 postinfection, as previously described (37, 38, 45). Catheters were sonicated for 5 min in 500 μl of phosphate-buffered saline (PBS) to dislodge adherent bacteria. Surrounding brain tissue from within 2 mm of the catheter tract was homogenized in 500 μl of sterile PBS supplemented with a complete protease inhibitor cocktail tablet (Roche, Basel, Switzerland) and RNase inhibitor (Promega, Madison, WI) using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY). A 100-μl aliquot of brain homogenate and supernatant from sonicated catheters were used to quantitate bacterial burden by 10-fold serial dilution on blood agar plates in an anaerobic chamber for 4 days. Brain tissue homogenates were also used to determine cytokine/chemokine levels by Milliplex analysis, as outlined below (37, 38, 45).
Cytokine/chemokine analysis.
Inflammatory mediators in the CSF and brain tissue were measured with a rat microbead array system according to the manufacturer’s instructions (RECYTMAG-65K, Milliplex; EMD Millipore Corp., Billerica, MA). This assay allowed for simultaneous measurement of 27 different cytokines and chemokines in a single 50-μl CSF or brain supernatant sample, including epidermal growth factor, (C-C) motif cytokine-11 (CCL11), fractalkine (CX3CL1), granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, chemokine (C-X-C) motif ligand 1 (CXCL1), gamma interferon (IFN-γ), IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12 (p70), IL-13, IL-17A, IL-18, chemokine (C-X-C) motif ligand 10 (CXCL10), leptin, chemokine (C-X-C) motif ligand 5 (CXCL5), CCL2, CCL3, chemokine (C-X-C) motif ligand 2 (CXCL2), chemokine (C-C motif) ligand 5 (CCL5), tumor necrosis factor alpha, and vascular endothelial growth factor. The results were analyzed using multiplex assay analysis software (EMD Millipore Corp.) and normalized to sample volume for CSF or total protein as measured by BCA in the case of brain tissue.
Flow cytometry.
To characterize leukocyte populations associated with C. acnes catheter infection, CSF and brain tissue were collected as described above. Brain tissue within 2 mm of the catheter tract was collected and passed through a 70-μm tissue strainer, incubated with digestion medium (HBSS with calcium and magnesium, DNase I, and collagenase), purified with a 22.5% Percoll gradient, and finally passed over myelin removal beads (Miltenyi Biotech, Auburn, CA). Immune cell populations from CSF and brain tissue were identified by fluorescence-activated cell sorting based on the expression of characteristic cell markers: monocytes/macrophages (CD45+ CD11b+ RP-1–), neutrophils (CD45+ CD11b+ RP-1+), CD4+ T cells (CD45+ CD3+ CD4+), and CD8+ T cells (CD45+ CD3+ CD8+) (47, 48).
Label-free mass spectrometry for CSF.
CSF samples were processed by electrophoretic protein separation. Gels were stained with Coomassie brilliant blue G-250 dye (Thermo Fisher) for 2 h and destained overnight in a solution of 10% acetic acid–20% methanol. The gel pieces were then excised, washed with HPLC water and dehydrated with 100% acetonitrile (ACN). Proteins were reduced with 2 mM Tris(2-carboxyethyl)phosphine (TCEP) in 50 mM ammonium bicarbonate (NH4HCO3 [AmBic]) for 1 h at 37°C and dehydrated with ACN. Reduced proteins were then alkylated with 50 mM iodoacetamide–50 mM AmBic, for 20 min in the dark with rotation. Gel pieces were then dehydrated for a second time with ACN to remove all reagents. MS-grade trypsin (15 ng/μl; Promega) was added to the samples, followed by incubation for 30 min on ice. After the removal of excess trypsin, 25 mM AmBic was added to immerse the gel pieces, which were incubated overnight at 37°C. Digested peptides were then extracted from the gel with 50% CAN–0.1% trifluoroacetic acid solution. Samples were dried in a SpeedVac, dissolved in 15 μl of 0.1% formic acid (FA) and subjected to liquid chromatography-tandem mass spectrometry analysis.
In-gel-digested peptide samples were analyzed using an Orbitrap Fusion Lumos coupled with the UltiMate 3000 HPLC system (Thermo Scientific). A 5-μl (500 ng) portion of each sample was loaded onto the trap column (Acclaim PepMap 100, 75 μm × 2 cm; nanoViper; Thermo Scientific) using FA (0.1%) and resolved in a rapid separation liquid chromatography (RSLC) column (Acclaim PepMap RSLC; 75 μm × 15 cm; nanoViper, Thermo Scientific). Samples were eluted using a 90-min linear gradient of ACN (4 to 45%) in 0.1% FA. The parameters for all experiments were as follows: nanospray needle voltage in positive mode, 2,000 V; column flow rate, 0.3 μl/min; loading pump flow, 4 μl/min; and inject mode, μl PickUp. Orbitrap scan mode was used for MS/MS, with a resolution of 120.000 and scan range of 375 to 1,500 m/z. Peptides were placed into dynamic exclusion for 60 s after detection one time. The detector type for MS/MS was set as to Orbitrap, with a resolution of 30,000, isolation mode quadrupole (isolation window, 0.7 Da), activation type HCD (higher-energy collisional dissociation), HCD collision energy of 40%, and first mass of 110 m/z. Stainless steel emitters were purchased from Thermo Fisher (outer diameter, 150 μm; inner diameter, 30 μm; 40-mm length; inserted in a 1/32 microsleeve for installation).
Data analysis.
For bacterial burden, chemokine/cytokine measurements and leukocyte infiltrates, descriptive analysis (mean median, standard deviation, Skewness, Kurtosis, and normality test) was used to determine the distribution of each indicator. P values were reported using a two-sample independent t test when meeting the assumption of normality. When the normality assumption was not met, the P values for the group comparison are from the Wilcoxon rank sum two-sample test. All statistical analyses were conducted in SAS 9.4 (SAS, Cary, NC), and figures were created in Prism 8 (GraphPad Software, San Diego, CA). A P value of <0.05 was considered statistically significant.
The MS data were organized into six tables, each representing protein expression data containing values for sterile and infected groups at days 1, 5, and 28 postinfection. For samples with no abundance information for a protein, that protein was assigned a zero value. We applied “quantile normalization” on the log2 abundance values and removed proteins that had no abundance values for all samples. The abundance profile for each protein in the sterile group was compared to the infected group. The statistical significance of the differentially abundant proteins was determined at days 1, 5, and 28 postinfection using a nonparametric Wilcoxon signed-rank test. Similarly, differentially abundant proteins for each group (sterile and infected) across days were also obtained by using the nonparametric Wilcoxon signed-rank test. The VennDiagram package were used in R (R Foundation for Statistical Computing, Vienna, Austria) for generating a Venn diagram (49, 50). We used the PANTHER functional enrichment tool to identify biological processes enriched in the differentially expressed proteins (51).
Data availability.
The data sets generated during and/or analyzed during the currents study are available in the EBI PRIDE repository, accession number PXD020527. All other data sets generated or analyzed during this study are included here and in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Fey and Barbara Cabrera at the University of Nebraska Medical Center Clinical Microbiology Laboratory for generously providing the C. acnes isolate. We thank Jessica Snowden for her critical review of the manuscript.
This study was supported by the Lageshulte and Wiese Fund at the University of Nebraska Medical Center.
We declare that there are no competing interests.
Experiments were designed by G.L.S., D.L., and T.K. Experiments were performed by G.L.S., M.B., and D.L. Data were analyzed by G.L.S., J.L., I.T., H.A., and T.K. Major contributors to the manuscript were G.L.S. and T.K. All authors have read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.McGirt MJ, Zaas A, Fuchs HE, George TM, Kaye K, Sexton DJ. 2003. Risk factors for pediatric ventriculoperitoneal shunt infection and predictors of infectious pathogens. Clin Infect Dis 36:858–862. doi: 10.1086/368191. [DOI] [PubMed] [Google Scholar]
- 2.Bondurant CP, Jimenez DF. 1995. Epidemiology of cerebrospinal fluid shunting. Pediatr Neurosurg 23:254–259. doi: 10.1159/000120968. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen J, Williams C, Sarang-Sieminski A. 2016. Hydrocephalus and ventriculoperitoneal shunts: modes of failure and opportunities for improvement. Crit Rev Biomed Eng 44:91–97. doi: 10.1615/CritRevBiomedEng.2016017149. [DOI] [PubMed] [Google Scholar]
- 4.Kanev PM, Sheehan JM. 2003. Reflections on shunt infection. Pediatr Neurosurg 39:285–290. doi: 10.1159/000075255. [DOI] [PubMed] [Google Scholar]
- 5.Simon TD, Kronman MP, Whitlock KB, Gove N, Browd SR, Holubkov R, Kestle JR, Kulkarni AV, Langley M, Limbrick DD, Jr, Luerssen TG, Oakes J, Riva-Cambrin J, Rozzelle C, Shannon C, Tamber M, Wellons JC, III, Whitehead WE, Mayer-Hamblett N, Hydrocephalus Clinical Research Network (HCRN). 2016. Variability in management of first cerebrospinal fluid shunt infection: a prospective multi-institutional observational cohort study. J Pediatr 179:185–191.e2. doi: S0022-3476(16)30877-0 [pii]. doi: 10.1016/j.jpeds.2016.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadduck W, Adametz J. 1988. Incidence of seizures in patients with myelomeningocele: a multifactorial analysis. Surg Neurol 30:281–285. doi: 10.1016/0090-3019(88)90300-X. [DOI] [PubMed] [Google Scholar]
- 7.Hunt GM, Holmes AE. 2008. Some factors relating to intelligence in treated children with spina bifida cystica. Dev Med Child Neurol Suppl 17:65–70. doi: 10.1111/j.1469-8749.1975.tb03581.x. [DOI] [PubMed] [Google Scholar]
- 8.Vinchon M, Rekate H, Kulkarni AV. 2012. Pediatric hydrocephalus outcomes: a review. Fluids Barriers CNS 9:18. doi: 10.1186/2045-8118-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinchon M, Dhellemmes P. 2006. Cerebrospinal fluid shunt infection: risk factors and long-term follow-up. Childs Nerv Syst 22:692–697. doi: 10.1007/s00381-005-0037-8. [DOI] [PubMed] [Google Scholar]
- 10.Walters BC, Hoffman HJ, Hendrick EB, Humphreys RP. 1984. Cerebrospinal fluid shunt infection. Influences on initial management and subsequent outcome. J Neurosurg 60:1014–1021. doi: 10.3171/jns.1984.60.5.1014. [DOI] [PubMed] [Google Scholar]
- 11.Tunkel AR, Hasbun R, Bhimraj A, Byers K, Kaplan SL, Michael Scheld W, van de Beek D, Bleck TP, Garton HJ, Zunt JR. 2017. 2017 Infectious Diseases Society of America’s clinical practice guidelines for healthcare-associated ventriculitis and meningitis. Clin Infect Dis 64:e34–e65. doi: 10.1093/cid/ciw861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon TD, Kronman MP, Whitlock KB, Browd SR, Holubkov R, Kestle JRW, Kulkarni AV, Langley M, Limbrick DD, Luerssen TG, Oakes J, Riva-Cambrin J, Rozzelle C, Shannon CN, Tamber M, Wellons JC, Whitehead WE, Mayer-Hamblett N, Hydrocephalus Clinical Research Network. 2019. Patient and treatment characteristics by infecting organism in cerebrospinal fluid shunt infection. J Pediatric Infect Dis Soc 8:235–243. doi: 10.1093/jpids/piy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conen A, Fux CA, Vajkoczy P, Trampuz A. 2017. Management of infections associated with neurosurgical implanted devices. Expert Rev Anti Infect Ther 15:241–255. doi: 10.1080/14787210.2017.1267563. [DOI] [PubMed] [Google Scholar]
- 14.Fux CA, Quigley M, Worel AM, Post C, Zimmerli S, Ehrlich G, Veeh RH. 2006. Biofilm-related infections of cerebrospinal fluid shunts. Clin Microbiol Infect 12:331–337. doi: 10.1111/j.1469-0691.2006.01361.x. [DOI] [PubMed] [Google Scholar]
- 15.Lan CC, Wong TT, Chen SJ, Liang ML, Tang RB. 2003. Early diagnosis of ventriculoperitoneal shunt infections and malfunctions in children with hydrocephalus. J Microbiol Immunol Infect 36:47–50. [PubMed] [Google Scholar]
- 16.Simon TD, Pope CE, Browd SR, Ojemann JG, Riva-Cambrin J, Mayer-Hamblett N, Rosenfeld M, Zerr DM, Hoffman L. 2014. Evaluation of microbial bacterial and fungal diversity in cerebrospinal fluid shunt infection. PLoS One 9:e83229. doi: 10.1371/journal.pone.0083229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Beek D, Drake JM, Tunkel AR. 2010. Nosocomial bacterial meningitis. N Engl J Med 362:146–154. doi: 10.1056/NEJMra0804573. [DOI] [PubMed] [Google Scholar]
- 18.Holmberg A, Lood R, Morgelin M, Soderquist B, Holst E, Collin M, Christensson B, Rasmussen M. 2009. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect 15:787–795. doi: 10.1111/j.1469-0691.2009.02747.x. [DOI] [PubMed] [Google Scholar]
- 19.Mack D, Haeder M, Siemssen N, Laufs R. 1996. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis 174:881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- 20.Otto M. 2009. Staphylococcus epidermidis: the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Achermann Y, Goldstein EJ, Coenye T, Shirtliff ME. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 27:419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubin GG, Portillo ME, Trampuz A, Corvec S. 2014. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Med Mal Infect 44:241–250. doi: 10.1016/j.medmal.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Arnell K, Cesarini K, Lagerqvist-Widh A, Wester T, Sjolin J. 2008. Cerebrospinal fluid shunt infections in children over a 13-year period: anaerobic cultures and comparison of clinical signs of infection with Propionibacterium acnes and with other bacteria. J Neurosurg Pediatr 1:366–372. doi: 10.3171/PED/2008/1/5/366. [DOI] [PubMed] [Google Scholar]
- 24.Nisbet M, Briggs S, Ellis-Pegler R, Thomas M, Holland D. 2007. Propionibacterium acnes: an under-appreciated cause of post-neurosurgical infection. J Antimicrob Chemother 60:1097–1103. doi: 10.1093/jac/dkm351. [DOI] [PubMed] [Google Scholar]
- 25.Viraraghavan R, Jantausch B, Campos J. 2004. Late-onset central nervous system shunt infections with Propionibacterium acnes: diagnosis and management. Clin Pediatr 43:393–397. doi: 10.1177/000992280404300413. [DOI] [PubMed] [Google Scholar]
- 26.Carneiro IM, Pereira AS, Pinto S, Prata F, Faria CC, Marques JG. 2018. Propionibacterium acnes: cause of cerebrospinal fluid shunt infection. Pediatr Infect Dis J 37:e168–e169. doi: 10.1097/INF.0000000000001786. [DOI] [PubMed] [Google Scholar]
- 27.Bayston R, Nuradeen B, Ashraf W, Freeman BJ. 2007. Antibiotics for the eradication of Propionibacterium acnes biofilms in surgical infection. J Antimicrob Chemother 60:1298–1301. doi: 10.1093/jac/dkm408. [DOI] [PubMed] [Google Scholar]
- 28.Portillo ME, Corvec S, Borens O, Trampuz A. 2013. Propionibacterium acnes: an underestimated pathogen in implant-associated infections. Biomed Res Int 2013:804391. doi: 10.1155/2013/804391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westergren H, Westergren V, Forsum U. 1997. Propionibacterium acnes in cultures from ventriculo-peritoneal shunts: infection or contamination? Acta Neurochir (Wien) 139:33–36. doi: 10.1007/BF01850865. [DOI] [PubMed] [Google Scholar]
- 30.Forgacs P, Geyer CA, Freidberg SR. 2001. Characterization of chemical meningitis after neurological surgery. Clin Infect Dis 32:179–185. doi: 10.1086/318471. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman BA, Tunkel AR, Pryor JC, Dacey RG, Jr.. 1990. Meningitis in the neurosurgical patient. Infect Dis Clin North Am 4:677–701. [PubMed] [Google Scholar]
- 32.Ross D, Rosegay H, Pons V. 1988. Differentiation of aseptic and bacterial meningitis in postoperative neurosurgical patients. J Neurosurg 69:669–674. doi: 10.3171/jns.1988.69.5.0669. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasan L, Kilpatrick L, Shah SS, Abbasi S, Harris MC. 2016. Cerebrospinal fluid cytokines in the diagnosis of bacterial meningitis in infants. Pediatr Res 80:566–572. doi: 10.1038/pr.2016.117. [DOI] [PubMed] [Google Scholar]
- 34.Lolak S, Bunyaratavej K. 2013. C-reactive protein in prediction of ventriculoperitoneal shunt-related infection in high-risk patients. Surg Infect (Larchmt) 14:192–195. doi: 10.1089/sur.2011.070. [DOI] [PubMed] [Google Scholar]
- 35.Schuhmann MU, Ostrowski KR, Draper EJ, Chu JW, Ham SD, Sood S, McAllister JP. 2005. The value of C-reactive protein in the management of shunt infections. J Neurosurg 103:223–230. doi: 10.3171/ped.2005.103.3.0223. [DOI] [PubMed] [Google Scholar]
- 36.Skar GL, Synhorst D, Beaver M, Snowden JN. 2019. CSF inflammatory markers differ in gram-positive versus Gram-negative shunt infections. J Neuroinflammation 16:7. doi: 10.1186/s12974-019-1395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skar GL, Beaver M, Aldrich A, Lagundzin D, Thapa I, Woods N, Ali H, Snowden J, Kielian T. 2019. Identification of potential cerebrospinal fluid biomarkers to discriminate between infection and sterile inflammation in a rat model of Staphylococcus epidermidis catheter infection. Infect Immun 86:e00311-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snowden JN. 2014. Generation of a central nervous system catheter-associated infection in mice with Staphylococcus epidermidis. Methods Mol Biol 1106:193–198. doi: 10.1007/978-1-62703-736-5_18. [DOI] [PubMed] [Google Scholar]
- 39.Rupp ME, Fey PD. 2001. In vivo models to evaluate adhesion and biofilm formation by Staphylococcus epidermidis. Methods Enzymol 336:206–215. doi: 10.1016/s0076-6879(01)36591-6. [DOI] [PubMed] [Google Scholar]
- 40.Thompson TP, Albright AL. 1998. Propionibacterium acnes infections of cerebrospinal fluid shunts. Childs Nerv Syst 14:378–380. doi: 10.1007/s003810050248. [DOI] [PubMed] [Google Scholar]
- 41.Banks JT, Bharara S, Tubbs RS, Wolff CL, Gillespie GY, Markert JM, Blount JP. 2005. Polymerase chain reaction for the rapid detection of cerebrospinal fluid shunt or ventriculostomy infections. Neurosurgery 57:1237–1243. doi: 10.1227/01.neu.0000186038.98817.72. [DOI] [PubMed] [Google Scholar]
- 42.Simon TD, Van Yserloo B, Nelson K, Gillespie D, Jensen R, McAllister JP, Riva-Cambrin J, Stockmann C, Daly JA, Blaschke AJ. 2014. Use of quantitative 16S rRNA PCR to determine bacterial load does not augment conventional cerebrospinal fluid (CSF) cultures among children undergoing treatment for CSF shunt infection. Diagn Microbiol Infect Dis 78:188–195. doi: 10.1016/j.diagmicrobio.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deutch S, Dahlberg D, Hedegaard J, Schmidt MB, Moller JK, Ostergaard L. 2007. Diagnosis of ventricular drainage-related bacterial meningitis by broad-range real-time polymerase chain reaction. Neurosurgery 61:306–312. doi: 10.1227/01.NEU.0000255526.34956.E4. [DOI] [PubMed] [Google Scholar]
- 44.Fulkerson DH, Sivaganesan A, Hill JD, Edwards JR, Shoja MM, Boaz JC, Jea A. 2011. Progression of cerebrospinal fluid cell count and differential over a treatment course of shunt infection. J Neurosurg Pediatr 8:613–619. doi: 10.3171/2011.8.PEDS11236. [DOI] [PubMed] [Google Scholar]
- 45.Snowden JN, Beaver M, Smeltzer MS, Kielian T. 2012. Biofilm-infected intracerebroventricular shunts elicit inflammation within the central nervous system. Infect Immun 80:3206–3214. doi: 10.1128/IAI.00645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lourbopoulos A, Chatzigeorgiou S, Mavridis T, Kokkinakis I, Tascos N, Simeonidou C. 2012. Stereotactic coordinates for intracerebroventricular infusion after permanent focal cerebral ischemia in Wistar rats. Hippokratia 16:51–56. [PMC free article] [PubMed] [Google Scholar]
- 47.Abbondanzo SJ, Chang SL. 2014. HIV-1 transgenic rats display alterations in immunophenotype and cellular responses associated with aging. PLoS One 9:e105256. doi: 10.1371/journal.pone.0105256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campanella M, Sciorati C, Tarozzo G, Beltramo M. 2002. Flow cytometric analysis of inflammatory cells in ischemic rat brain. Stroke 33:586–592. doi: 10.1161/hs0202.103399. [DOI] [PubMed] [Google Scholar]
- 49.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 50.Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during and/or analyzed during the currents study are available in the EBI PRIDE repository, accession number PXD020527. All other data sets generated or analyzed during this study are included here and in the supplemental material.