Abstract
A(syn)-T and G(syn)-C+ Hoogsteen base pairs in protein-bound DNA duplexes can be difficult to resolve by X-ray crystallography due to ambiguous electron density and by NMR spectroscopy due to poor chemical shift dispersion and size limitations with solution-state NMR spectroscopy. Here we describe an NMR strategy for characterizing Hoogsteen base pairs in protein-DNA complexes, which relies on site-specifically incorporating 13C/15N labeled nucleotides into DNA duplexes for unambiguous resonance assignment and to improve spectral resolution. The approach was used to resolve the conformation of an A-T base pair in a crystal structure of a ~43 kD complex between a 34-bp duplex DNA and the integration host factor (IHF) protein. In the crystal structure (PDB: 1IHF), this base pair adopts an unusual Hoogsteen conformation with a distorted sugar-backbone that is accommodated by a nearby nick used to aid in crystallization. The NMR chemical shifts and inter-proton NOEs indicate that this base pair predominantly adopts a Watson-Crick conformation in the intact DNA-IHF complex under solution conditions. Consistent with these NMR findings, substitution of 7-deazaadenine at this base pair resulted in only a small (~2-fold) decrease in the IHF-DNA binding affinity. The NMR strategy provides a new approach for resolving crystallographic ambiguity and more generally for studying the structure and dynamics of protein-DNA complexes in solution.
Graphical Abstract
INTRODUCTION
Following the discovery of the DNA double helix1, much effort was directed towards solving single crystal X-ray structures of isolated purine-pyrimidine dimers to test the fine details of Watson-Crick base pairing proposed by Watson and Crick2–8. The first successful attempt was reported in 1959, when Karst Hoogsteen solved the X-ray co-crystal structure of base pair derivatives containing 9-methyladenine and 1-methylthymine2. Rather than observing an A–T Watson-Crick base pair (bp), an alternative pairing mode called ‘Hoogsteen’ was observed (Figure 1A). Relative to a Watson-Crick bp, the adenine base flips 180° to adopt a syn rather than anti conformation (Figure 1A). The base forms a unique set of hydrogen bonds (H-bonds) that require that the two bases come into closer proximity by ~2.0–2.5 Å3, 9 (Figure 1A), which in turn induces a kink in the DNA double helix10, 11. Analogous G–C+ Hoogsteen bps12 form by flipping the guanine base but also require protonation of cytosine N3 to form a second H-bond (Figure 1A).
Figure 1.
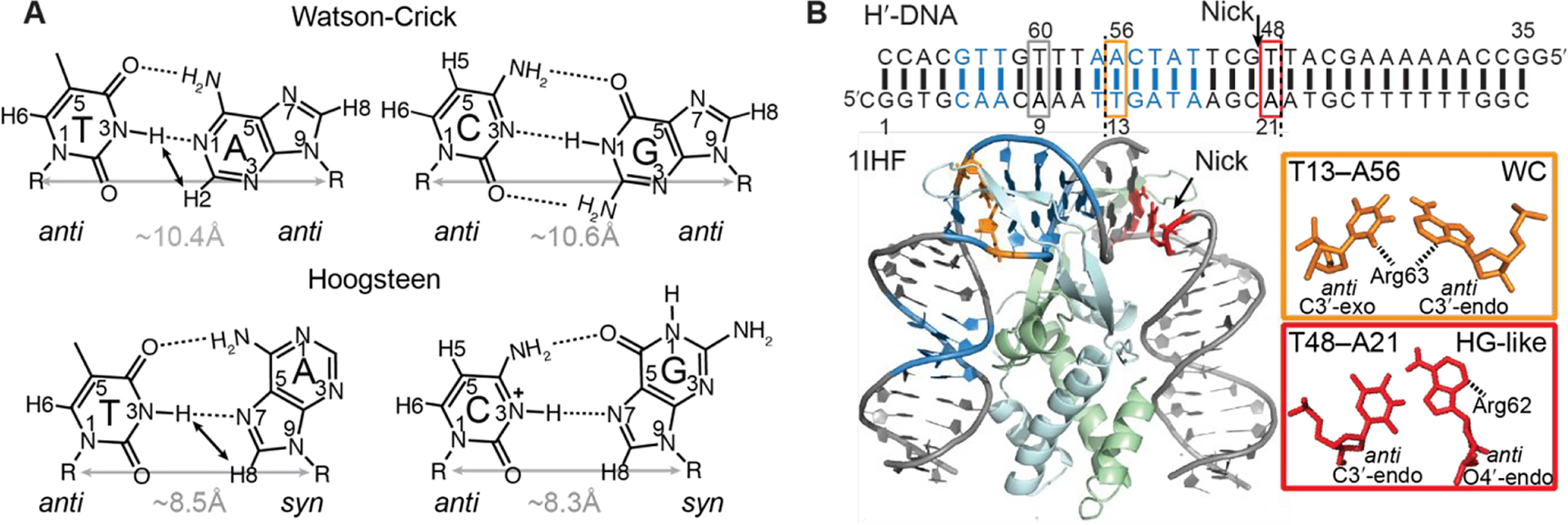
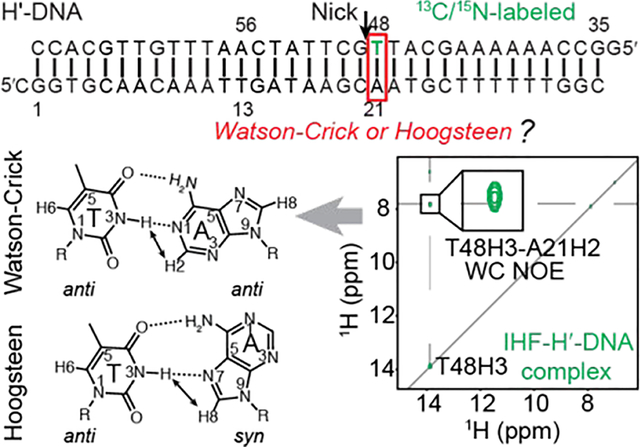
Structures of Watson-Crick and Hoogsteen base pairs and crystal structure of IHF-DNA complex. (A) Chemical structures of Watson-Crick and Hoogsteen base pairs. Differences in hydrogen bonding, cytosine protonation, purine base orientation (anti versus syn) and C1′–C1′ distance are highlighted. Close atomic distances involving imino protons that give rise to characteristic NOEs in A-T Watson-Crick (AH2-TH3) and Hoogsteen (AH8-TH3) bps are indicated using black arrows. (B) X-ray structure of the IHF-DNA complex (PDBID: 1IHF13). The consensus sequence for specific protein recognition is shown in blue. The A21–T48 Hoogsteen-like (HG-like) bp, the A56-T13 Watson-Crick (WC) bp, and the control A9-T60 Watson-Crick bp are shown in red, orange and grey rectangles, respectively. The nick in one DNA strand is indicated with an arrow. Two pseudosymmetry-related large kink sites are marked with dashed lines in the DNA sequence, corresponding to positions of intercalating proline residues.
Since their discovery, Hoogsteen bps have been observed in X-ray structures of AT-rich sequences that form duplexes entirely composed of Hoogsteen bps14–17; in structures of DNA bound to small molecule bis-intercalators12, 18–22; in damaged DNA bases incapable of forming Watson-Crick bps where they are thought to play roles in damage accommodation23–25, recognition26, and repair27; in Y-family ‘low fidelity polymerases’ which replicate DNA using Hoogsteen pairing as a means of bypassing lesions on the Watson-Crick face during replication28–31. More recently, Nuclear Magnetic Resonance (NMR) relaxation dispersion (RD) methods32–34 revealed that in canonical duplex DNA, A–T and G–C Watson-Crick bps exist in dynamic equilibrium with short-lived (lifetimes of 0.2–2.5 ms) and sparsely populated (0.1–5%) Hoogsteen bps11, 35–39.
Hoogsteen bps are also observed in some X-ray structures of DNA-protein complexes where they are thought to play roles in DNA shape recognition13, 40–42. For example, two consecutive G–C+ Hoogsteen bps are observed in the complex between DNA and the TATA box-binding protein41. In one of these Hoogsteen bps, the syn guanine base appears to alleviate a steric clash between the guanine-amino group and a nearby leucine side chain41. In addition, one of the syn guanines partially stacks with the phenylalanine residue that inserts between the two G-C Hoogsteen bps22. In the structure of DNA in complex with the DNA binding domain of p53 tumor suppressor protein, two consecutive A–T Hoogsteen bps in the consensus sequence contribute to a narrowed and more negatively charged minor groove in the flanking regions, which may favor electrostatic interactions with a positively charged arginine residue42. Thus, proteins seem to employ a variety of mechanisms to interact with Hoogsteen bps.
Considering that it is now well established that Hoogsteen bps form robustly in naked duplex DNA23, 35, 40 with a preference for sites of major groove kinking10, 11; it comes as something of a surprise that Hoogsteen bps have not been more widely observed in structures of protein-DNA complexes, where structural distortions could destabilize Watson-Crick bps. Indeed, recent studies on DNA-drug complexes show that recognition of DNA can result in a variety of behaviors; with some bps predominantly (>90% populations) adopting Watson-Crick or Hoogsteen conformations, while others having high (~10%) fractional Hoogsteen populations relative to the naked DNA (<1%)22, 40, 43. Depending on the quality of the electron density, it can be difficult to resolve Hoogsteen bps from Watson-Crick bps when solving X-ray structures of DNA44–46.
For example, Aggarwal and coworkers showed using X-ray crystallography and biochemical experiments that human DNA polymerase ɩ (Polɩ), a member of the Y-family polymerases, employs Hoogsteen base-paring to replicate damaged and undamaged DNA28, 31, 47. The X-ray structure of Polɩ showed a template syn adenine in the active site forming a Hoogsteen bp with an incoming dTTP28. This structure was met with some skepticism when Wang44 argued that the weak electron density of the active site A–T bp makes it is difficult to unambiguously assign a Watson-Crick versus Hoogsteen geometry. Subsequent structural and biochemical studies, including studies examining the impact of single-atom substitutions that destabilize Hoogsteen pairing in the DNA template, confirmed that this polymerase does indeed employ Hoogsteen mechanism to replicate certain base pairs30, 48. Other studies have noted the difficulty in resolving Hoogsteen versus Watson-Crick bps in X-ray structures of protein-DNA complexes including the DNA-homeodomain45 and DNA-p73 complexes46. Thus, it is conceivable that other Hoogsteen bps in crystal structures of DNA have been potentially mis-modeled as pure Watson-Crick bps. Sites that experience significant Hoogsteen breathing can also be difficult to resolve by crystallography, as the elevated local dynamics could contribute to the lack of features in the local electron density49 or be suppressed by cryocooling of the crystals that is routinely performed to improve resolution of crystal structure50. Conversely, there is also a need for methods that can independently verify Hoogsteen bps observed in crystal structures of DNA. For example, AT-rich duplexes composed entirely of Hoogsteen bps based on X-ray crystallography have been shown to form regular Watson-Crick bps when examined by solution NMR15. Here, another potential example is given by the structure of the integration host factor (IHF) protein in complex with DNA13 (Figure 1B).
IHF is a bacterial DNA-binding protein that aids in the compaction of prokaryotic genomes by inducing significant DNA bending51. It plays important roles in site-specific recombination of λ phage into the bacteria host genes52, enhancing bacterial DNA replication53, regulation of gene expression54, 55, and in Cas1-Cas2-mediated spacer integration56. IHF induces a global conformational change in the DNA duplex (i.e. ~160° bending) upon binding, resulting in two sharp major groove directed kinks separated by ~7 Watson-Crick bps in pseudo-symmetry related sites (Figure 1B). In the crystal structure (PDB: 1IHF), a nick between T48 and G49 (Figure 1B) was introduced at a kinked site to aid crystallization13. Next to this site is an unusual Hoogsteen-like A21–T48 bp (Figure 1B). The adenine base adopts an anti, rather than syn conformation that is uncharacteristic of Hoogsteen bps observed so far in duplexes. Additionally, the entire T48 nucleotide is flipped about the sugar-phosphate backbone to adopt a left-handed Z-DNA like conformation. This large sugar-backbone distortion at T48 is likely made possible by the nick between T48 and G4913 (Figure 1B). Indeed, the corresponding A56–T13 bp at the pseudo-symmetry related site adopts a Watson-Crick conformation, though the bp is distorted13 (Figure 1B). Whether or not the A21–T48 bp forms a Hoogsteen bp remains to be established experimentally in the native IHF-DNA complex without nicks.
Thus, there is a need for complementary approaches that can help resolve Watson-Crick versus Hoogsteen conformations and their dynamics in DNA under solution conditions. Watson-Crick and Hoogsteen bps can in principle readily be resolved using solution NMR spectroscopy and this could provide a means to resolve crystallographic ambiguity11, 23, 35. However, application of NMR-based approaches can be difficult for large DNA-protein complexes owing to severe spectral overlap and unfavorable relaxation properties, which leads to significant line-broadening and losses in sensitivity. Here, we propose and employ an approach that relies on using commercially available 13C/15N labeled nucleotides to site-specifically incorporate 13C/15N nucleotides at specific positions in the DNA duplex. Results from the NMR analysis are then verified biochemically by examining the impact of site-specifically incorporating 7-deazapurine (7dA), which destabilizes Hoogsteen bps30, 36, on DNA-protein binding affinity. We apply this approach to resolve the conformation of the A21–T48 bp in the intact IHF-DNA complex with full length DNA under solution conditions. The results indicate that the A21–T48 bp predominantly forms a Watson-Crick bp.
METHODS
Sample Preparation
Preparation of DNA and IHF protein samples for NMR and FP studies:
Single-stranded DNA oligonucleotides with one (A21 or A9) or two (A56 and T48) uniformly 13C/15N-labeled deoxynucleotides were purchased from the Yale Keck Oligo Synthesis Resource with DNA cartridge (Glen-Pak) purification. The only exception was the A21-labeled DNA strand, which was purchased with standard desalting and subjected to in-house purification via 20% denaturing polyacrylamide gel electrophoresis (PAGE). Final yields of purchased site-specifically labeled DNA oligonucleotides were 150 – 200 nmol and ~90 nmol for cartridge-purified and PAGE-purified oligonucleotides, respectively.
5′-Fluorescein-dT labeled oligonucleotides for FP studies were purchased with reverse-phase HPLC purification from IDT (Integrated DNA Technologies, Inc.). DNA oligonucleotides with 7dA21 and 7dA56 modifications were purchased from the Midland Certified Reagent Company and the Yale Keck Oligo Synthesis Resource, respectively, with reverse-phase HPLC purification. Unmodified DNA oligonucleotides were purchased with standard desalting from IDT (Integrated DNA Technologies, Inc.). Complementary DNA strands were annealed at a final duplex concentration of 1 mM. Annealing was performed by heating the mixed single strands at 95 °C for 5 min and slow cooling at room temperature for 20–30 minutes.
Preparation of IHF protein:
IHF protein (UniprotKB: P0A6X7 and P0A6Y1) was expressed in E. coli Rosetta(DE3)plysS cells from a pET21a vector (available from Addgene). Cells from a frozen glycerol stock were streaked onto an agar plate containing LB (Luria-Bertani broth), ampicillin (50 μg/mL) and chloramphenicol (30 μg/mL) and grown overnight at 37 °C. Several single colonies were then resuspended into 20 mL liquid LB with 100 μg/mL ampicillin, which was added to 500 mL of the same medium, grown at 37 °C until A600 reached ~0.7 and kept at 4°C overnight. About 80 mL of that culture was used to inoculate 6 1-liter cultures, which were also grown at 37 °C. Once reached A600=0.7–0.8, IPTG was added to a final concentration of 0.5 mM to induce protein expression. Cells were harvested 2–3 hours later by centrifugation at 8,000 rpm for 7 minutes (Sorvall F10S rotor), and the pellet was stored at −20 °C. Cell pellets were thawed on ice, then resuspended in 200–300 mL 100 mM Tris (pH 8.0), 1 mM EDTA, 10% Surcrose (w/v), 10% (v/v) Glycerol, 1 M NaCl, with 6 tablets of Complete Mini Protease Inhibitor cocktail (Roche). After lysozyme was added to a final concentration of 200 μg/mL, cells were incubated on ice for 10–20 minutes, then sonicated. Cell debris was pelleted by centrifugation for 1 hour at 18,000 rpm in an SS-34 rotor.
The supernatant was collected and subjected to (NH4)2SO4 fractionation. Briefly, IHF remained in the supernatant at 50% saturation and was in the pellet at 80% saturation. Solid (NH4)2SO4 was first added to a final saturation of 50% by gradual addition of solid (NH4)2SO4 with gentle stirring over ice, with an additional for 30 minutes of gentle stirring after all the solid had dissolved. The supernatant was then collected after centrifugation for 30 minutes at 18,000 rpm in an SS-34 rotor. (NH4)2SO4 was gently stirred into the supernatant as above to a final saturation of 80%, followed by a similar centrifugation, and the pellet was stored at 4 °C. The sample was further purified by chromatography on heparin and monoS columns. The (NH4)2SO4 pellet was dissolved in 50 mL or more buffer A (20 mM MES pH 5.5, 0.1 mM EDTA, 5% glycerol) and loaded onto a 20 mL heparin column (Amersham). After washing with 20% buffer B (buffer A with 2 M NaCl), a gradient of 20–80% for buffer B was run in 90 minutes, with IHF eluting at ~1 M NaCl. IHF-containing fractions were pooled, dialyzed into 20% buffer B (80% buffer A), and rechromatographed on the same column. Fractions were then pooled and dialyzed overnight into 2.5% buffer B and loaded onto a 1 ml monoS column (Amersham). The column was washed with 2.5% buffer B, and eluted with a gradient of 2.5% to 47.5% buffer B in 45 minutes. IHF eluted at ~0.35 M NaCl. Pooled fractions were concentrated to about 10 mg/mL, then were dialyzed into 25 mM HEPES pH 7.5, 1 mM EDTA, 20% Glycerol, 0.2 M NaCl, and flash frozen in small aliquots, 100μl each. Samples were tested for endonuclease contamination by incubating 10 μg/μL IHF with 1 μg/μL pUC19 plasmid in the presence of 10 mM MgCl2. Samples were examined by agarose gel electrophoresis after a 2-hour incubation at 37 °C, and no degradation of the plasmid was seen. The concentration of IHF was determined using a calculated absorption coefficient (IHF at 2.95 mg/mL has OD280 of 1).
Preparation of NMR sample of the DNA-IHF complex:
NMR samples of IHF-DNA complex were prepared by slowly titrating and mixing 50 μM duplex DNA into 50 μM IHF protein. The resulting 25 μM DNA-IHF complex was then buffer exchanged by centrifugal concentrators (EMD Millipore with 3 kDa cut-off) to obtain the NMR sample (~250 μL) and then supplied with 10% D2O. Final concentrations of free DNA and DNA-protein complex NMR samples were ~0.3 mM.
NMR buffer:
DNA or DNA-protein complex samples were buffer exchanged into the NMR buffer consisting of 25mM HEPES, 100 mM NaCl, 0.1 mM EDTA with pH 7.0 and 10% D2O three times using a centrifugal concentrator at 4 °C (EMD Millipore with 3 kDa cut-off) until containing >99.9% of the desired buffer.
NMR Experiments
NMR data were collected on a 600 MHz Bruker NMR spectrometer equipped with an HCN cryogenic probe, a 600 MHz Bruker Avance III spectrometer equipped with an HCPN cryogenic probe, a 700 MHz Bruker Avance III spectrometer equipped with a triple-resonance HCN cryogenic probe, and an 800 MHz Varian DirectDrive2 spectrometer equipped with a triple-resonance HCN cryogenic probe and. Data were processed and analyzed using NMRpipe57 and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco), respectively. Chemical shifts data were obtained using both TROSY and conventional 2D HSQC experiments; resonance assignments were analyzed using 15N-edited 2D [1H, 1H] NOESY (mixing time 180 ms), 2D H1′-C1′-AN9/TN1 HCN and conventional HSQC experiments with broadband 15N decoupling and selective decoupling on purine-N9 or pyrimidine-N1. Spectra were recorded at 25 °C unless otherwise stated.
The resonances in the IHF-DNA complex could be unambiguously assigned for single-site labeled samples (H′-DNAA21 and H′-DNAA9). The aromatic resonances (A56-C8 and T48′-C6) in IHF bound H′-DNAA56T48 complex were assigned by turning on and off the selective carbon decoupling on T-C5 (~110 ppm) in the conventional 2D HSQC experiment58 (data not shown). A56-C1′ and T48-C1′ in the free H′-DNAA56T48 were assigned by a 1H-15N 2D version of HCN experiment59 that correlates H1′ to adenine-N9 or thymine-N1 which has distinct chemical shifts (i.e. adenine-N9 ~ 170 ppm while thymine-N1~140 ppm) (Figure S2A). However, the same HCN experiment could not detect signals in the DNA-protein complex due to significant transverse relaxation before acquisition. The A56-C1′ and T48-C1′ resonances in the DNA-protein complex were assigned by selectively decoupling adenine-N9 or thymine-N1, in comparison with the broadband 15N decoupling in a regular 2D HSQC experiment (Figure S2B).
A weak NOE cross peak is observed which can be tentatively assigned to A21H1’ in the 15N-edited NOESY spectra of the complex (see Figure 4B). Such an NOE is not expected for a regular Watson-Crick bp in which the distance between the protons is >6 Å nor is it expected for the distorted Watson-Crick bp in the IHF-protein crystal structure (>6 Å). Such a weak NOE could arise if T48 and A21 came slightly into closer proximity in solution, perhaps even transiently, when bound to IHF protein. This is consistent with observation of ~0.22 ppm downfield shifted T48-H3 in the complex form relative to that in free DNA, which suggests stronger hydrogen bonding. It is also consistent with previously reported (Dhavan et al J Mol Biol 1999) reduced imino proton exchange rates measured in a short 19 bp DNA upon binding to IHF. While a Hoogsteen bp could also bring these protons into close proximity (~5.5 Å), we can rule out this possibility because we do not observe any evidence for the T48H3-A21H8 NOE (Figure S3).
Figure 4.
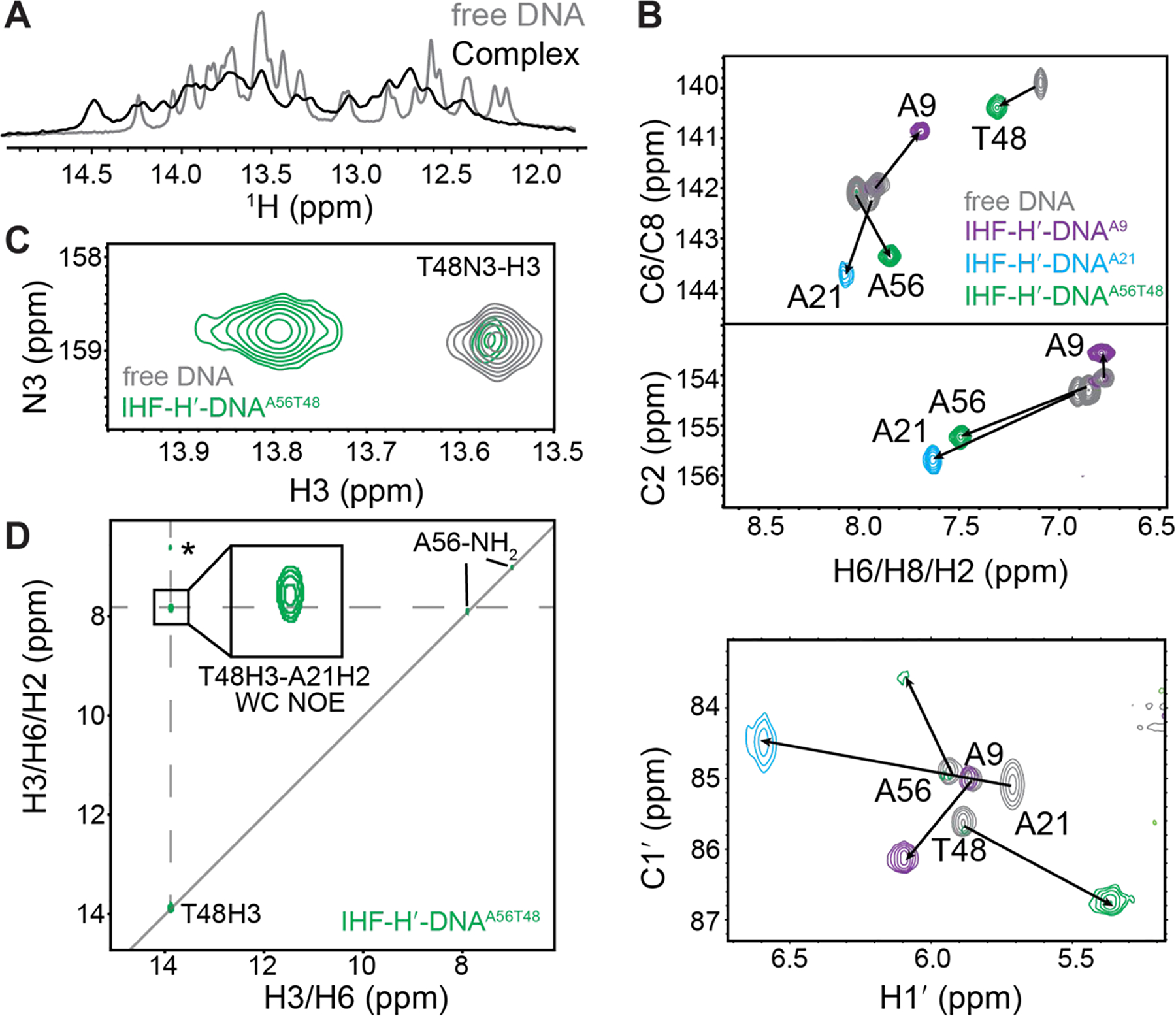
NMR analysis of IHF-DNA complex formation. (A) 1D 1H spectra showing imino proton resonances in the IHF-H′-DNAA56T48 complex overlaid with unbound DNA. (B) 2D [13C, 1H] TROSY HSQC spectra for IHF-DNA complexes (colored) overlaid with unbound DNA (in grey). Chemical shift perturbations are indicated by the black arrows. (C) Overlay of 2D [15N,1H] SOFAST HMQC spectra for IHF-H′-DNAA56T48 complex (in green) and unbound H′-DNAA56T48 (in gray) at 37 °C. Minor unbound DNA population is observed in the spectrum consistent with slow exchange. Complex-only spectra are shown in Figure S3. (D) 2D 15N-edited [1H, 1H] NOESY spectrum for IHF-H′-DNAA56T48 complex shows T48H3-A21H2 NOE cross-peaks consistent with Watson-Crick pairing at A21-T48 bp. The T48H3-A21H1′ NOE assignment (labeled with *) is tentative and we cannot rule out that the resonance assigned to A21H1′ corresponds to other neighboring sugar or amino protons.
R1ρ RD experiments on the free H′-DNAA56T48 duplex were performed on the 700 MHz Bruker spectrometer as previously described35, 61, 62 at 37 °C, with spinlock powers (ωSL 2π−1 Hz) and offset frequencies (Ω 2π−1 Hz) listed in Table S1. Because of the large size of the duplex and low sample concentration (0.3 mM), a large number of scans (ns=512) were used in the RD experiments targeting A56-C1′ and the T48-C1′ to achieve 10–20 signal-to-noise ratio in the absence of the spin-lock. Because of the low sensitivity, only two delay points (0 ms and 20 ms) were used to monitor mono-exponential decay. RD measurements for aromatic spins (adenine-C8/C2) were not performed due to the much lower sensitivity arising due to the larger R261 and potentially larger chemical exchange contributions.
AFQM/MM Calculations
Structures of the IHF-DNA complex for the automated fragmentation quantum mechanics/molecular mechanics (AFQM/MM) calculations were prepared based on the crystal structure (PDBID: 1IHF). Due to the presence of the nick near A21, we focused on computing chemical shifts based on the pseudo-symmetry related kink site without any nick. We directly took the crystal structure (PDB: 1IHF) as the model structure for 100% Watson-Crick conformation for the calculation. For modeling A56 at 100% Hoogsteen conformation, the glycosidic bond at A56 was manually flipped 180°. The structure with flipped A56 was hydrated using SPC/E water, and then neutralized by adding Na+ ions, followed by energy-minimization for 1000 steps and heating gradually from 0 K to 298 K in 20 ps. The system was subsequently equilibrated for 1 ns using Langevin dynamics with the collision frequency 3 ps−1 and NPT ensemble at 298 K. We observed Hoogsteen H-bonds (AN6-H6---TO4 and AN7---TH3-N3) formed in the structure after the equilibration step and the resultant structure was used in the AFQM/MM calculation.
Chemical shift calculations followed the procedure described earlier39, 63. For structures of IHF-DNA complex with either Hoogsteen or Watson-Crick bp modeled at A56-T13, quantum fragments were constructed centered on nucleotides A9 and A56. The effects of atoms in the IHF-DNA complex outside the quantum region (nucleotide A9 or A56), and of water and ions in the solvent, were represented by point charges uniformly distributed on the molecular surface of the quantum region, were resolved by fitting to Poisson–Boltzmann calculations using the “solinprot” program from the MEAD package64. The quantum region (A9 or A56) was assigned a local dielectric of ε = 1 (vacuum), the remaining nucleic acid region had ε = 4, and the solvent region ε = 80. GIAO chemical shift calculations were carried out for each quantum fragment, using version 4.3.6 of the demon-2k program65, using the OLYP functional66 with the TZVP basis set and the GEN-A2* fitting set. Further details are available elsewhere39, 63, 67.
Fluorescence Polarization Assay for Measurement of Binding Affinity
Fluorescence polarization (FP) experiments were performed using a Tecan F500 plate reader using 485/20 nm excitation and 535/25 nm emission polarization filters suitable for fluorescein and a Beacon2000 Fluorescence Polarization System. Experiments were conducted using a black polypropylene 384-well round-bottom plate (Corning Inc.). The concentration of fluorescein-labeled DNA was kept constant at 3 nM, and the IHF protein concentration was varied from 0.1 nM to 20 nM. The FP assay was carried out at 25 °C in the NMR buffer (25mM HEPES, 100 mM NaCl, 0.1 mM EDTA with pH 7.0) and the anisotropy values measured in triplicates or duplicates. Fluorescence anisotropy (A) was calculated based on the parallel and perpendicular emission light intensities:
where A is the measured anisotropy at a given concentration. The normalized anisotropy, Anorm, was obtained using:
in which A0 refers is the anisotropy in absence of protein, Amax is the maximum value of measured anisotropy. Anorm was measured at varying protein concentration (x) and then fitted to an equation describing single-site binding using the Levenberg-Marquardt non-linear curve fitting algorithm implemented in the OriginPro 2016 software (OriginLab Corporation):
where D is the constant fluorescein-labeled DNA concentration, B and C determine the anisotropies for free DNA and bound DNA, respectively, and Kd is the apparent dissociation constant.
Since 7dA56 and 7dA21 are located in different strands, we kept the fluorescein-dT at the 5′-end of the opposing strand to the one containing the 7dA substitution (Figure 6A). Therefore, two control DNAs without 7-deaza substitution were made with the fluorescein label at either of the two strands (Figure 6). Based on the FP assay, the two control DNAs yielded similar Kd values (Kd1 = 1.0 ± 0.3 nM and Kd2 = 1.4 ± 0.4 nM) for binding of unmodified H-DNA duplexes to IHF (Figure S4).
Figure 6.
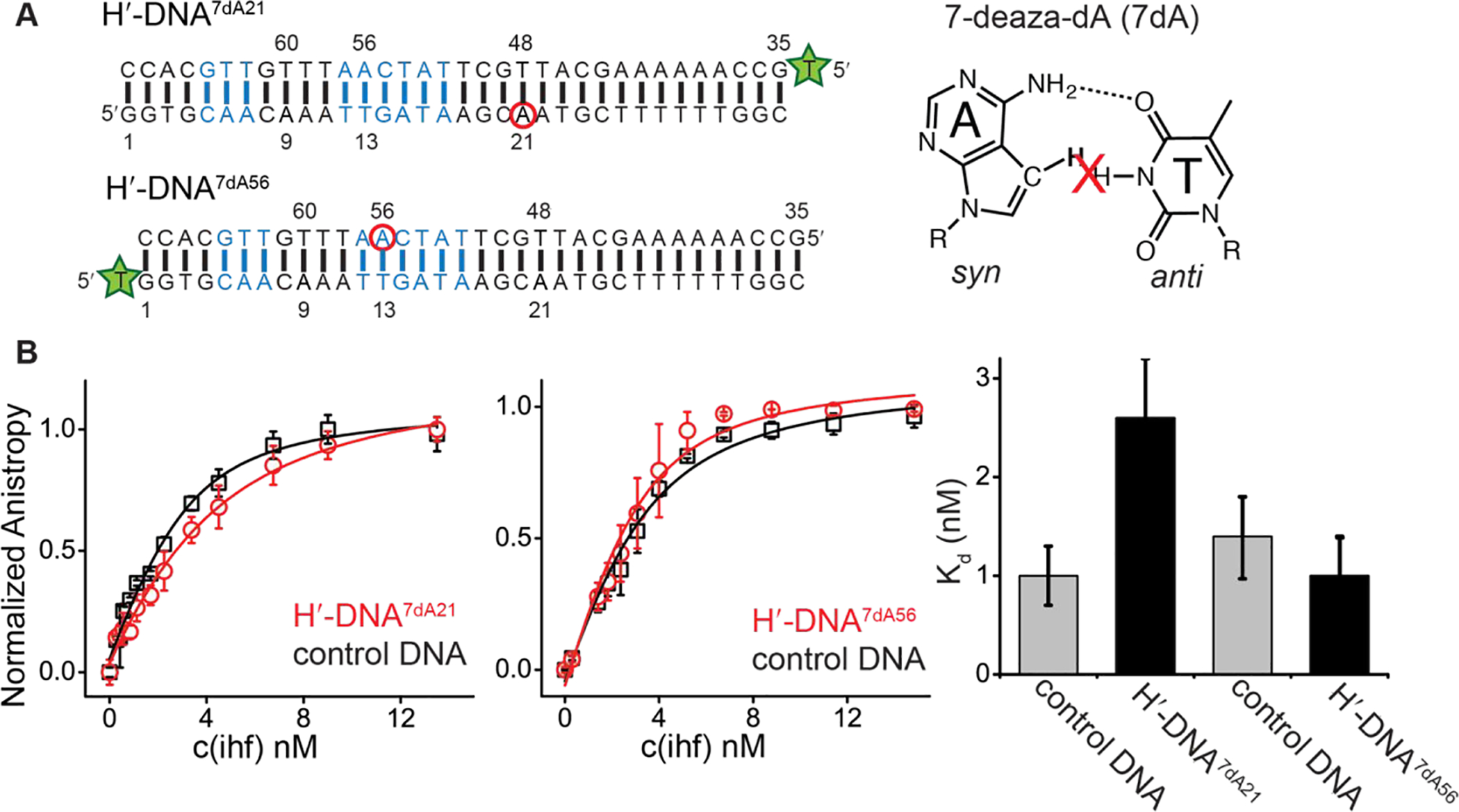
Impact of 7-deazapurine substitutions on IHF-DNA binding affinity. (A) Sequences of DNA duplexes containing 7-deaza-dA modification (red circles) and the fluorescein-dT label (green star) at the 5′-end of the unmodified strand. Also shown is the chemical structure of 7-deaza-dA and its destabilization impact on Hoogsteen H-bonding. (B) Comparison of FP binding curves for 7-deaza-dA modified DNA with its unmodified counterpart (control DNA). Control DNA refers to the unmodified DNA duplex with the fluorescein-dT label at the same position as the modified duplex. Data is fit to single-site binding equation (see methods). Error represents standard deviation from triplicate (left) or duplicate (right) measurements. Fitted Kd and uncertainty values shown on the right.
RESULTS
NMR characterization of site-specifically labeled DNA duplex
Three site-specifically labeled 34 bp DNA duplexes were prepared (see methods) to study the IHF-DNA complex (Figure 1B and Figure S2A). In each case, specific nucleotide positions were uniformly 13C/15N labeled (Figure 1B and Figure S2A). In one sample (H′-DNAA21), the A21 that forms a Hoogsteen bp in the crystal structure was labeled (Figure 1B and Figure S2A). A second sample (H′-DNAA56T48) was prepared by labeling the A21 bp partner T48 as well as A56 at the pseudo-symmetry related site which forms a Watson-Crick bp in the crystal structure (Figure 1B and Figure S2A). A21, A56 and T48 are located at sites in which the DNA kinks sharply toward the major groove (Figure 1B and Figure S1). As a negative control, a third sample (H′-DNAA9) was prepared labeling A9 which is located in a similar CAA sequence context but falls outside the consensus sequence in a region of the DNA structure that has a smaller degree of major groove kinking (Figure 1B and Figure S1). The buffer conditions used for the NMR studies (25 mM HEPES, 100 mM NaCl, 0.1 mM EDTA with pH 7.0) were similar to those used in the crystallization buffer (10 mM HEPES, 100 mM NaCl, 0.1 mM EDTA, 8% glycerol with pH 7.0)68.
We first analyzed the NMR spectra of the unbound DNA duplexes (Figure 2A). As expected, the 1D 1H spectra for all three DNA duplexes in the absence of IHF were essentially identical with well-resolved imino proton resonances that are consistent with a Watson-Crick B-form DNA duplex (Figure 2B). Excellent quality 2D NH and CH HSQC TROSY-based NMR spectra were obtained for all three duplexes clearly showing only a subset of resonances belonging to the labeled nucleotides (Figure 2C). The 1H, 13C, and 15N chemical shifts in the absence of IHF were all consistent with Watson-Crick pairing in B-form DNA. Resonances could readily be assigned in these site-labeled duplexes (see Methods and Figure S2). Assigning large nucleic acids (>20 kDa), such as the 34-bp DNA duplex, with conventional 2D/3D NOESY experiments would be costly (~1 mM for 13C/15N-labeled samples) and challenging owing to low sensitivity and severe spectral overlap69–71. In such cases, having a site-specifically labeled sample even at low concentration (~0.3 mM in this study) provides a convenient methodology in a targeted approach towards identifying Watson-Crick/Hoogsteen bps in large nucleic acids molecules, potentially at lower external magnetic field strengths (say 400 MHz) and non-cryogenic probes.
Figure 2.
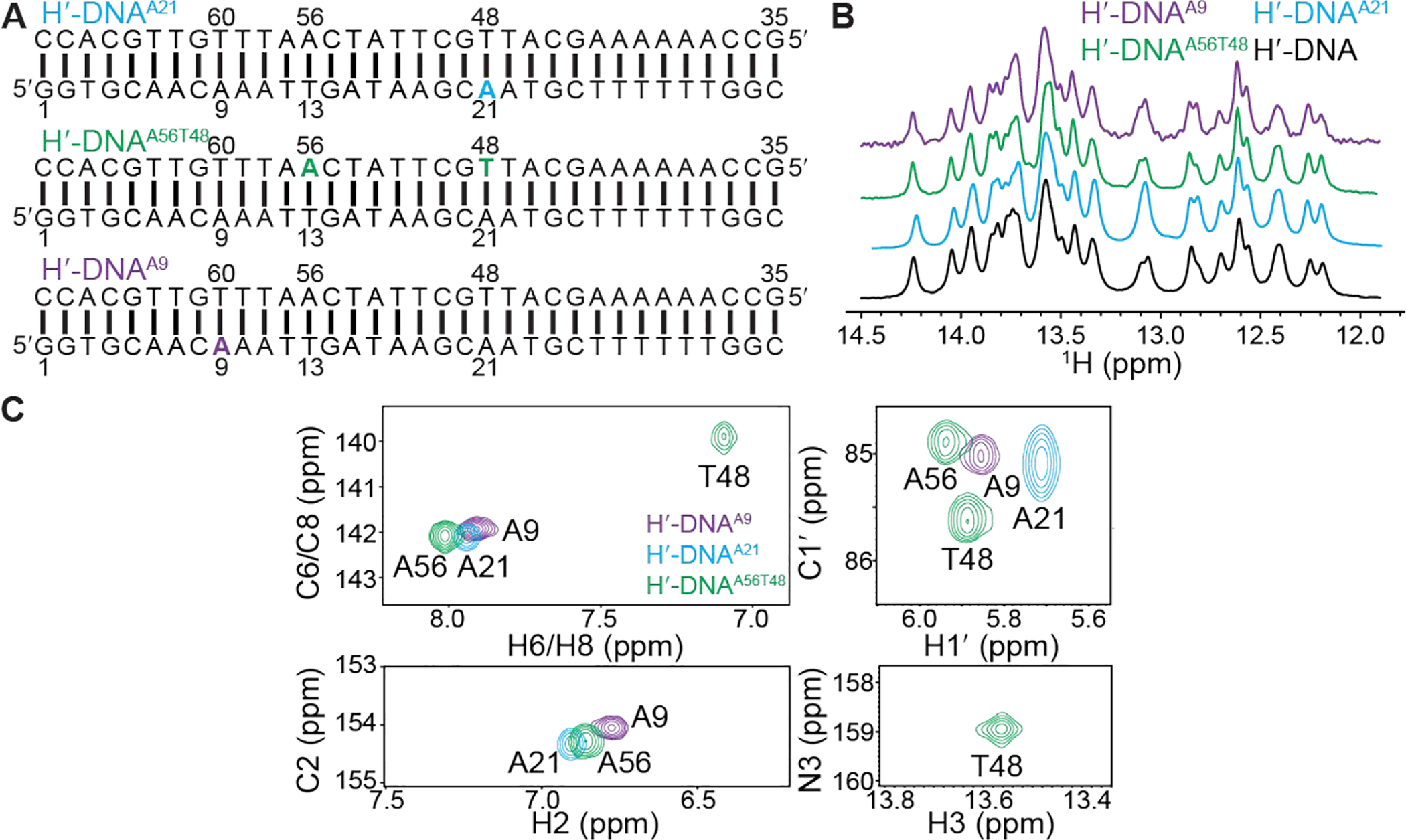
NMR characterization of site-specifically 13C/15N-labeled H′-DNA duplexes. (A) H′-DNAA21, H′-DNAA56T48 and H′-DNAA9 sequences with 13C/15N-labeled nucleotides highlighted in color. (B) 1D 1H NMR spectra showing imino proton resonances in site-labeled DNA overlaid with unlabeled H′-DNA measured at 25 °C. (C) Overlay of 2D [13C,1H] TROSY72 and 2D [15N, 1H] SOFAST HMQC73, 74 NMR spectra measured for the three site-specially labeled unbound H′-DNA sequences at 25 °C. Note that line broadening of A21-C1′H1′ relative to other resonance is not observed in 2D HSQC (not TROSY) spectra (see Figure S3).
Transient Hoogsteen bps in long naked DNA duplexes
Thus far, studies have examined transient Hoogsteen bps in relatively short (<15 bp) duplexes35, 38 using R1ρ relaxation dispersion NMR experiments. To examine whether Watson-Crick-Hoogsteen exchange is detectable in the longer (34 bp) IHF DNA duplexes, we carried out R1ρ RD measurements on A56-C1′ and T48-C1′ in H′-DNAA56T48 in the absence of IHF protein (Table S1). The R1ρ experiment33, 34, 75 measures the chemical exchange contribution (Rex) to intrinsic transverse relaxation rate (R2,int) of NMR resonances during a relaxation period when the spin is irradiated with an applied radiofrequency (RF) pulse with varying powers (ωSL) and frequencies (ωRF). Experiments were performed at T = 37 °C.
Indeed, we observed the expected RD at A56-C1′ (Figure 3). Fitting of the data to 2-state exchange model yields exchange parameters (pB = 0.3±0.1%, kex = 1611±525 s−1 and Δω = 4.2 ± 0.3 ppm) that are consistent with chemical exchange directed to Hoogsteen bps35, 38. The measured pB falls within the range (0.2% - 0.3%) measured in short (~12 bp) DNA duplexes containing the same CAA trinucleotide sequence context at 30.5 °C35. The measured exchange rate (kex = 1611±525 s−1) is only slightly slower (~3000 to 5000 s−1) than values measured in shorter duplexes35. As a negative control, we did not observe any RD at T48-C1′ (Figure 3) at 37 °C as expected for chemical exchange directed to Hoogsteen bps35, 39. These results indicate that the Watson-Crick-Hoogsteen equilibrium is not significantly altered in longer DNA duplexes.
Figure 3.
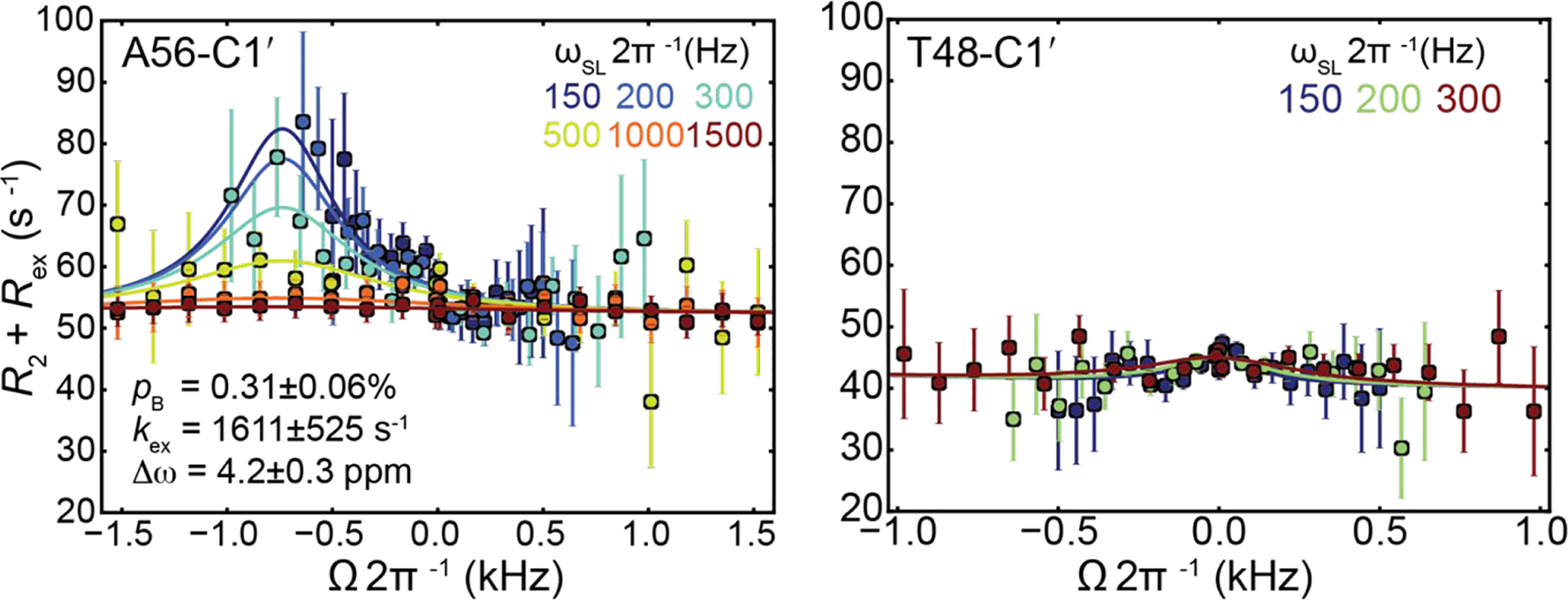
Off-resonance relaxation dispersion profiles for unbound H′-DNAA56T48. Shown are off-resonance profiles as a function of spin-lock offset (Ω 2π−1 kHz, where Ω = Ωobs − ωRF) and power (ωSL 2π−1 Hz, color coded as indicated in the inset) measured at 37 °C. Error bars represent experimental uncertainties estimated from mono-exponential fitting of n = 2 (A56-C1′) and n = 2 (T48-C1′) independently measured peak intensities using a Monte Carlo–based method (Methods). The solid line represents a fit to two-state exchange35.
Characterizing IHF-DNA complex formation
Two assays were used to assess IHF-DNA complex formation prior to NMR analysis. We observed the expected migration retardation in the electrophoretic mobility shift assay (EMSA)76 when incubating the DNA duplexes with increasing IHF concentration (Figure S4). Saturation was observed at 1:1 molar ratio consistent with 1:1 binding stoichiometry (Figure S4A). The apparent dissociation constant (Kdapp = 1.0 ± 0.3 nM) measured using a fluorescence polarization (FP) assay77 (Methods) employing a DNA duplex tagged with fluorescein-dT at the 5′-terminus of one strand (Figure S4B) is in good agreement with values previously reported for the same 34-bp H′-DNA with the IHF protein using gel mobility shift assays with 32P labeling (Kdapp ~ 2.0 ± 0.5 nM)68, and for a shorter (30 bp) H′-DNA (Kdapp ~ 1.65 ± 0.09 nM)78 as well as for a longer (56 bp) H′-DNA duplex (Kdapp ~ 1.5 nM)79 measured by EMSA.
NMR spectra indicate that A21–T48 forms a predominantly Watson-Crick conformation in the DNA-IHF complex.
Next, we characterized IHF-DNA complex formation using NMR. The imino protons belonging to the DNA duplex experienced chemical shift perturbations and line-broadening upon addition of IHF to a 1:1 ratio (Figure 4A). Similar changes were also observed for aromatic and aliphatic resonances consistent with the DNA undergoing changes in structure upon complex formation (Figure 4B). Binding of IHF to DNA is slow on the NMR time scale as evidenced by observation of two sets of resonances that belong to free and bound DNA in spectra with excess DNA. These results establish the utility of using site-specifically labeled DNA to study relatively large DNA-protein complexes by NMR.
Interestingly, the addition of IHF resulted in similar NMR chemical shift perturbations at A56 and A21 (Figure 4B). This indicates that the two adenine nucleotides experience similar changes in their environment upon complex formation in solution, as expected for two pseudo-symmetrically related sites (Figure 1B). Nevertheless, the A56–T13 and A21–T48 chemical shifts in the IHF-DNA complex are not identical, as expected given differences in their sequence and structural contexts (Figure 1B). For example, A56 is located within the consensus sequence while A21 is not, and in the X-ray structure, the A56-T13 uniquely forms H-bonds with a nearby Arg63 residue (Figure 1B).
We observed the T48-H3 imino resonance in the 2D 15N-1H SOFAST HMQC spectra of IHF-DNA complex (Figure 4C) with 1H and 15N chemical shift (T48-H3 13.8 ppm and T48-N3 158.8 ppm) that fall within the region expected for a Watson-Crick bp. We did not observe any evidence for an upfield shifted thymine imino resonance (~1.5–2.0 ppm in 1H and ~2 ppm in 15N) that would be expected for an A-T Hoogsteen bp11, 22, 35. The fact that the T48-H3 imino is observable and not significantly upfield shifted also argues in favor of a stable Watson-Crick conformation and against a high population of transient Hoogsteen conformation that are in fast exchange with the Watson-Crick on the NMR timescale. A predominant A21-T48 Watson-Crick bp is in good agreement with the prior NMR studies on a smaller DNA construct containing the consensus sequence of the H1 binding site60. In contrast, the control site A9 showed distinct and generally smaller chemical shift perturbations (Figure 4B), consistent with the crystal structure showing weaker distortions at this site (Figure S1).
Next, we carried out 15N-edited NOESY experiments (Figure 4D) to directly obtain structural information regarding the nature of base pairing at A21–T48 in the DNA-IHF complex. If A21–T48 adopts a Hoogsteen conformation, we would expect to observe an NOE cross peak between T48-H3 and A21-H811, 23, 35. This NOE cross-peak has previously been reported for a variety of Hoogsteen bps in DNA duplexes, including in modified m1A–T Hoogsteen bp in duplex DNA11, 23, 35 and A–T Hoogsteen bps in DNA-echinomycin complexes22, 80. We did not observe this NOE in the IHF-DNA complex (Figure 4D and Figure S3). Instead, we observed the A21H2-T48H3 NOE as expected for Watson-Crick pairing (Figure 4D).
Using NMR chemical shift fingerprinting to resolve Watson-Crick and Hoogsteen bps
The aromatic and aliphatic 13C/15N/1H chemical shifts of A21 undergo large perturbations on complex formation that are inconsistent with the perturbations expected for formation of Hoogsteen bps in unbound DNA duplexes11, 35, 39. For example, formation of Hoogsteen bps is expected to induce 2–3 ppm downfield shift in A21-C1′35, but a 0.6 ppm upfield shift is observed upon IHF-DNA formation (Figure 4B). Formation of Hoogsteen bps lead to very minor changes in A21-C2 chemical shift35, yet large changes (downfield shifted by 1.4 ppm) are observed on complex formation. Many of the 1H chemical shifts are also inconsistent with expectations of Hoogsteen formation. For example, A-H8 and A-H1′ are typically upfield shifted by ~0.5 and ~ 0.1 ppm, respectively, when forming m1A–T Hoogsteen bps11, 23, 35; but downfield shifts are observed (by 0.14 and 0.88 ppm, respectively) on complex formation (Figure 4B).
The above analysis is however complicated by the fact that chemical shift perturbations likely include other contributions such as bending of the DNA and changes in stacking of bases upon binding resulting in altered ring current effects and possibly interactions with the IHF protein. The Hoogsteen chemical shift signatures may also differ in the highly bent IHF-DNA as compared to relaxed duplex DNA. To further understand the observed chemical shift perturbations on complex formation, we used automated fragmentation quantum mechanics/molecular mechanics (AFQM/MM)63 to compute chemical shifts based on the crystal structure of the IHF- NA complex for models in which the A56 –T13 adopts a Watson-Crick (native) versus Hoogsteen (modeled) conformations. We then compared these predicted values with those measured experimentally using the site-labeled NMR samples. The AFQM/MM approach was recently shown to accurately predict chemical shifts in DNA duplexes with Hoogsteen and Watson-Crick bps39. The chemical shifts measured at the control site A9–T60 was used as an internal reference. The difference in chemical shift between values predicted for a given site (A56 or A21) and for the corresponding nucleus in A9 (Δω = ωA56 (or A21) − ωA9) were predicted computationally and then compared to the experimental data (Figure 5). This approach is clearly limited by the accuracy of the predicted chemical shifts including the modeling of the complex using a single structure rather than an ensemble of conformations as done previously39. Nevertheless, better agreement was observed between measured chemical shifts for both A21 and A56 and values calculated for the distorted Watson-Crick bp as in the X-ray structure as compared to the modeled Hoogsteen bp (Figure 5). While the agreement is somewhat better for the Hoogsteen form for AC8, and no distinctions can be made for AH1′ and AH2, AC1′, AC2 and AH8 resonances suggest better overall agreement with the Watson-Crick conformation The chemical shift data therefore indicate that for the intact IHF-DNA complex in solution, the predominant conformation adopted by both A21–T48 and A56–T13 is similar to that observed for the distorted A56–T13 Watson-Crick bp in the X-ray at the site without the nick. Indeed, this observed result is consistent with the X-ray structure of DNA in complex with Hbb, an IHF-family bacterial DNA-bending protein. Although Hbb recognizes a different DNA sequence from IHF, the two protein-DNA complexes present similar overall highly bent DNA structures upon binding and two distorted A-T Watson-Crick bps are observed at the two sharp kinks in the Hbb-DNA complex determined without any nicks (PDBID: 2NP281).
Figure 5.
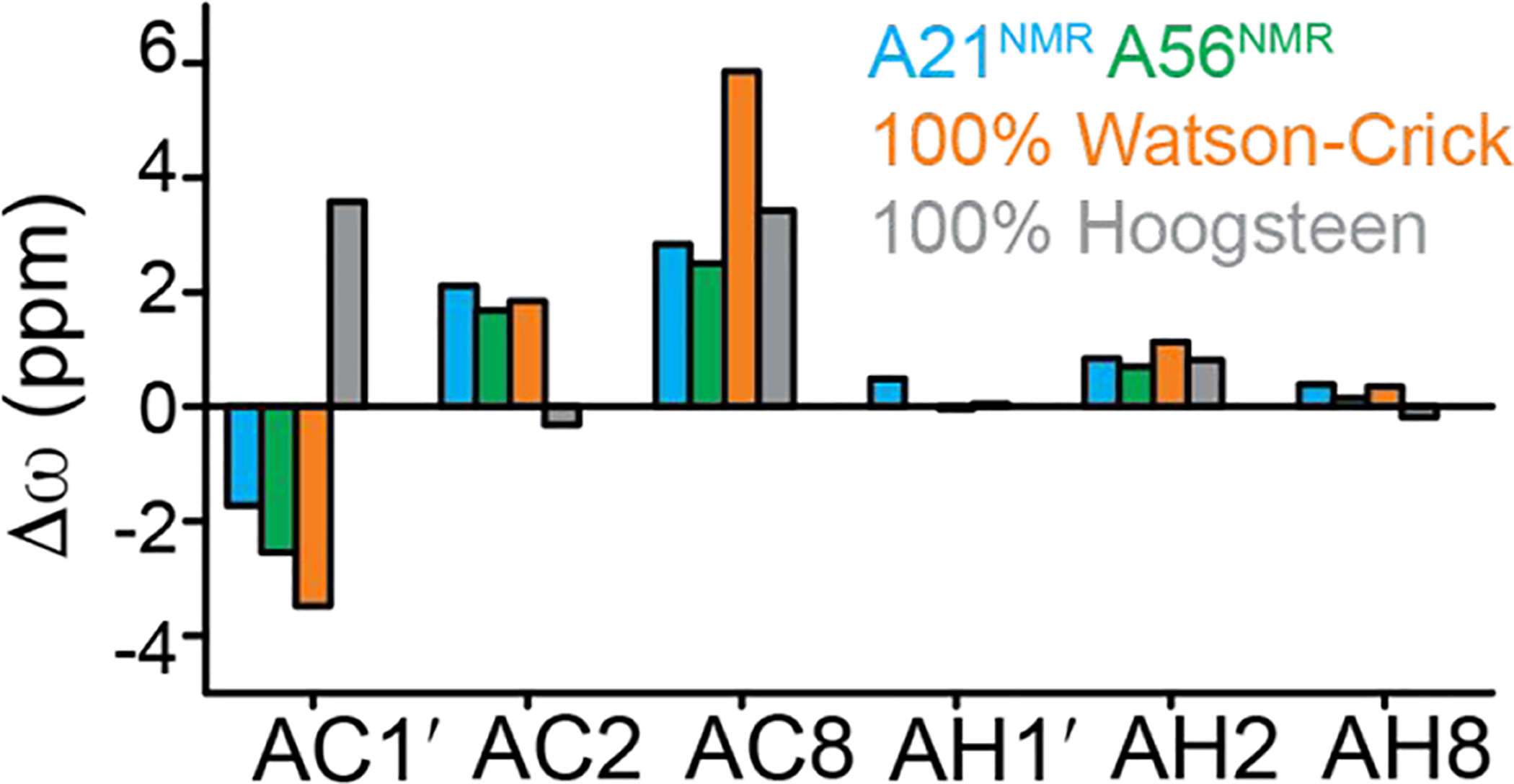
Comparison of measured and calculated chemical shifts for Watson-Crick and Hoogsteen models in the IHF-DNA complex. Shown is the difference between chemical shifts, Δω = ωAix − ωA9x in which i refers to different nucleotides (A21 or A56) and x to different type of nuclei (e.g. C1′, C8, C2). The residue A9 is used as an internal reference. Shown are AFQM/MM predicted values at A56 for 100% Hoogsteen (gray), 100% Watson-Crick (orange), and values measured by NMR for A21(cyan) and A56 (green).
7-deazaadenine substitutions minimally affect the IHF-DNA binding affinity
Our results indicate that predominant conformation for A21-T48 is Watson-Crick. If this were the case, selective destabilization of the Hoogsteen bp relative to the Watson-Crick should minimally affect the IHF-DNA binding affinity. 7-deazapurines have previously been shown to selectively destabilize Hoogsteen bps by knocking out one hydrogen bond (N7---H3-N3) while not affecting Watson-Crick type hydrogen bonding82, 83. The modification destabilizes Hoogsteen bps relative to Watson-Crick bps by at least ~1 kcal/mol (i.e. 10-fold reduction in transient Hoogsteen population), resulting in undetectable RD in naked DNA duplexes36. 7-deazapurine was also shown to significantly inhibit DNA replication by the low-fidelity Y-family polymerase iota (Polɩ) that replicates DNA via Hoogsteen base-pairing by suppressing the formation of Hoogsteen bps30.
We examined the impact of 7-deazaadenine (7dA) substitutions at the kink sites on the IHF-DNA binding affinity. Using the FP assay, the 7dA21 substitution only decreased the binding affinity ~2-fold (Kd = 2.6 ± 0.6 nM for H′-DNA7dA21 compared to 1.0 ± 0.3 nM for the control DNA) (Figure 6). In comparison, the 7dA substitution at the pseudo-symmetry related site (7dA56) had no measurable effect on the binding affinity (Kd = 1.0 ± 0.4 nM). These results argue against a predominantly Hoogsteen conformation in the complex (>99%) at A21 but do not rule out having a small portion of Hoogsteen population in equilibrium with the major Watson-Crick conformation. The small differences observed for the two sites (A21 and A56) are also consistent with small differences in NMR chemical shifts at these two bps in the IHF-DNA complex (Figure 4B).
DISCUSSION
Resolving Hoogsteen versus Watson-Crick bps is not always straightforward by X-ray crystallography. Here, we demonstrated an NMR approach for examining Watson-Crick to Hoogsteen dynamics in large protein-DNA complexes. Application to the IHF-DNA complex indicates that A21-T48 adopts a predominantly Watson-Crick conformation and that the Hoogsteen bp observed in the crystal structure most likely arises due to the introduction of a nick near the kinked site. In this regard, it is interesting to note that Hoogsteen bps have been observed in structural contexts in which the nucleotide is not chemically linked to both neighbors, including bps at duplex termini, and also replication bps in the polymerase active site during replication10, 49. The NMR strategy presented here could facilitate studies of Hoogsteen bps in such contexts and possibly illuminate their biochemical consequences.
In a recent study, it was shown that the binding of the echinomycin antibiotic to DNA increased the Hoogsteen population to 8%22. Our data cannot rule out the possibility that the transient Hoogsteen population for A21-T48 in the IHF-DNA complex is also elevated by comparable amounts. While we observed line broadening for A21-C1′ and A56-C1′ relative to A9 and T48 in the IHF-DNA complex, further studies are needed to resolve whether these differences reflect μs-to-ms chemical exchange at A21-C1′ and A56-C1′ or faster ps-to-ns dynamics that lead to narrowing of A9–C1′ and T48-C1′. TROSY based NMR RD experiments combined with the site-specific labeling strategy used here may help extend the methodology to allow the study of transient Hoogsteen bps in large protein-DNA complexes72, 84–86. These studies could illuminate the DNA bending motions reported previously in the IHF-DNA complex87 as well as other dynamics that could impact the kinetic rates of DNA and IHF assembly87, 88.
While our data suggest that the bps in the IHF-DNA complex form a predominantly Watson-Crick conformation, recent solution state studies on p53-DNA complex provide strong evidence for a predominantly Hoogsteen conformation which was observed in crystallographic structures42, 89. These crystal structures did not feature any nicks in the DNA. In particular, inosine or 2-oxo adenine substitutions that weaken or enhance the Hoogsteen bp formation, as verified by X-ray crystallography, had more dramatic (~6-fold) effects on DNA-protein binding affinities89. This strongly suggests that Hoogsteen bps can form in protein-DNA complexes under solution conditions where they can contribute to shape recognition.
The NMR strategy presented in this work opens the door for characterizing the dynamics of DNA in protein-DNA complexes. Indeed, very few studies60, 90, 91 have examined DNA dynamics at atomic detail when in complex with proteins and the potential role of these fluctuations in the stability and function of the complex. This approach can be applied to systems as large as 50 kDa but could potentially be extended to much larger complexes with the use of solid-state NMR. The site-labeling approach also provides an important avenue for cataloguing valuable chemical shift structure relationships that can be harnessed in the future to aid chemical-shift-based characterization of DNA structure and dynamics.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Evgenia Nikolova, Dr. Janghyun Lee, Dr. Yu Chen, Dr. Isaac Kimsey, Honglue Shi, Atul Rangadurai, Dr. Mary Clay, Dr. Anisha Shakya, and Nicole Orlovsky for assistance and insightful discussions. We acknowledge technical support and resources from the Duke Magnetic Resonance Spectroscopy Center and the Duke Compute Cluster. This work was supported by NIH grants (R01GM089846 to H.M.A).
Footnotes
REFERENCES
- 1.Watson JD, and Crick FHC (1953) Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid, Nature 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 2.Hoogsteen K (1959) The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine, Acta Crystallographica 12, 822–823. [Google Scholar]
- 3.Hoogsteen K (1963) The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine, Acta Crystallographica 16, 907–916. [Google Scholar]
- 4.Pauling L, and Corey RB (1956) Specific hydrogen-bond formation between pyrimidines and purines in deoxyribonucleic acids, Arch. Biochem. Biophys 65, 164–181. [DOI] [PubMed] [Google Scholar]
- 5.Haschemeyer AE, and Sobell HM (1963) THE CRYSTAL STRUCTURE OF AN INTERMOLECULAR NUCLEOSIDE COMPLEX: ADENOSINE AND 5-BROMOURIDINE, Proc. Natl. Acad. Sci. U. S. A 50, 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobell HM, Tomita KI, and Rich A (1963) The crystal structure of an intermolecular complex containing a guanine and a cytosine derivative, Proc. Natl. Acad. Sci. U. S. A 49, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haschemeyer AE, and Sobell HM (1965) The crystal structure of a hydrogen bonded complex of deoxyguanosine and 5-bromodeoxycytidine, Acta Crystallographica 19, 125–130. [DOI] [PubMed] [Google Scholar]
- 8.Haschemeyer AE, and Sobell HM (1965) The crystal structure of a hydrogen bonded complex of adenosine and 5-bromouridine, Acta Crystallographica 18, 525–532. [DOI] [PubMed] [Google Scholar]
- 9.Wang AH, Ughetto G, Quigley GJ, Hakoshima T, van der Marel GA, van Boom JH, and Rich A (1984) The molecular structure of a DNA-triostin A complex, Science 225, 1115–1121. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Hintze BJ, Kimsey IJ, Sathyamoorthy B, Yang S, Richardson JS, and Al-Hashimi HM (2015) New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey, Nucleic Acids Res 43, 3420–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathyamoorthy B, Shi H, Zhou H, Xue Y, Rangadurai A, Merriman DK, and Al-Hashimi HM Insights into Watson–Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A, Nucleic Acids Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quigley GJ, Ughetto G, van der Marel GA, van Boom JH, Wang AH, and Rich A (1986) Non-Watson-Crick G.C and A.T base pairs in a DNA-antibiotic complex, Science 232, 1255–1258. [DOI] [PubMed] [Google Scholar]
- 13.Rice PA, Yang S, Mizuuchi K, and Nash HA (1996) Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn, Cell 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- 14.Abrescia NG, Thompson A, Huynh-Dinh T, and Subirana JA (2002) Crystal structure of an antiparallel DNA fragment with Hoogsteen base pairing, Proc. Natl. Acad. Sci. U. S. A 99, 2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrescia NG, González C, Gouyette C, and Subirana JA (2004) X-ray and NMR studies of the DNA oligomer d(ATATAT): Hoogsteen base pairing in duplex DNA, Biochemistry 43, 4092–4100. [DOI] [PubMed] [Google Scholar]
- 16.De Luchi D, Tereshko V, Gouyette C, and Subirana JA (2006) Structure of the DNA coiled coil formed by d(CGATATATATAT), ChemBioChem 7, 585–587. [DOI] [PubMed] [Google Scholar]
- 17.Pous J, Urpí L, Subirana JA, Gouyette C, Navaza J, and Campos JL (2008) Stabilization by extra-helical thymines of a DNA duplex with Hoogsteen base pairs, J. Am. Chem. Soc 130, 6755–6760. [DOI] [PubMed] [Google Scholar]
- 18.Singh UC, Pattabiraman N, Langridge R, and Kollman PA (1986) Molecular mechanical studies of d(CGTACG)2: complex of triostin A with the middle A - T base pairs in either Hoogsteen or Watson-Crick pairing, Proc. Natl. Acad. Sci. U. S. A 83, 6402–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ughetto G, Wang AHJ, Quigley GJ, van der Marel GA, van Boom JH, and Rich A (1985) A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment, Nucleic Acids Res 13, 2305–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, and Patel DJ (1988) NMR studies of echinomycin bisintercalation complexes with d(A1–C2–G3–T4) and d(T1–C2–G3–A4) duplexes in aqueous solution: sequence-dependent formation of Hoogsteen A1.cntdot.T4 and Watson-Crick T1.cntdot.A4 base pairs flanking the bisintercalation site, Biochemistry 27, 1744–1751. [DOI] [PubMed] [Google Scholar]
- 21.Cuesta-Seijo JA, and Sheldrick GM (2005) Structures of complexes between echinomycin and duplex DNA, Acta Crystallogr D Biol Crystallogr 61, 442–448. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, McSally J, Andricioaei I, and Al-Hashimi HM (2018) Modulation of Hoogsteen dynamics on DNA recognition, Nature Communications 9, 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Zhan Y, Fenn D, Chi LM, and Lam SL (2008) Effect of 1-methyladenine on double-helical DNA structures, FEBS Lett 582, 1629–1633. [DOI] [PubMed] [Google Scholar]
- 24.Lu L, Yi C, Jian X, Zheng G, and He C (2010) Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system, Nucleic Acids Res 38, 4415–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, and Wilson SH (2015) Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide, Nature 517, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, and Lam SL (2009) Effect of 1-methyladenine on thermodynamic stabilities of double-helical DNA structures, FEBS Lett 583, 1548–1553. [DOI] [PubMed] [Google Scholar]
- 27.Ling H, Boudsocq F, Plosky BS, Woodgate R, and Yang W (2003) Replication of a cis-syn thymine dimer at atomic resolution, Nature 424, 1083–1087. [DOI] [PubMed] [Google Scholar]
- 28.Nair DT, Johnson RE, Prakash S, Prakash L, and Aggarwal AK (2004) Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing, Nature 430, 377–380. [DOI] [PubMed] [Google Scholar]
- 29.Nair DT, Johnson RE, Prakash L, Prakash S, and Aggarwal AK (2006) Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase [igr], Nat. Struct. Mol. Biol 13, 619–625. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RE, Prakash L, and Prakash S (2005) Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase iota, Proc. Natl. Acad. Sci. U. S. A 102, 10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova AV, and Kulbachinskiy AV (2012) Structure of human DNA polymerase iota and the mechanism of DNA synthesis, Biochemistry (Mosc) 77, 547–561. [DOI] [PubMed] [Google Scholar]
- 32.Palmer AG III. (2014) Chemical exchange in biomacromolecules: past, present, and future, J. Magn. Reson 241, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekhar A, and Kay LE (2013) NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers, Proc. Natl. Acad. Sci. U. S. A 110, 12867–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y, Kellogg D, Kimsey IJ, Sathyamoorthy B, Stein ZW, McBrairty M, and Al-Hashimi HM (2015) Chapter Two - Characterizing RNA Excited States Using NMR Relaxation Dispersion, In Methods Enzymol (Sarah AW, and Frédéric HTA, Eds.), pp 39–73, Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolova EN, Kim E, Wise AA, O’Brien PJ, Andricioaei I, and Al-Hashimi HM (2011) Transient Hoogsteen base pairs in canonical duplex DNA, Nature 470, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolova EN, Gottardo FL, and Al-Hashimi HM (2012) Probing Transient Hoogsteen Hydrogen Bonds in Canonical Duplex DNA Using NMR Relaxation Dispersion and Single-Atom Substitution, J. Am. Chem. Soc 134, 3667–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolova EN, Goh GB, Brooks CL III, and Al-Hashimi HM (2013) Characterizing the protonation state of cytosine in transient G•C Hoogsteen base pairs in duplex DNA, J. Am. Chem. Soc 135, 6766–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvey HS, Gottardo FL, Nikolova EN, and Al-Hashimi HM (2014) Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics, Nat Commun 5, 4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Clay MC, Rangadurai A, Sathyamoorthy B, Case DA, and Al-Hashimi HM (2018) Atomic structures of excited state A–T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations, J. Biomol. NMR 70, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolova EN, Zhou H, Gottardo FL, Alvey HS, Kimsey IJ, and Al-Hashimi HM (2013) A historical account of hoogsteen base-pairs in duplex DNA, Biopolymers 99, 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, and Burley SK (1999) TATA element recognition by the TATA box-binding protein has been conserved throughout evolution, Genes Dev 13, 3217–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, and Shakked Z (2010) Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs, Nat. Struct. Mol. Biol 17, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfoh R, Cuesta-Seijo JA, and Sheldrick GM (2009) Interaction of an echinomycin-DNA complex with manganese ions, Acta Crystallographica Section F 65, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J (2005) DNA polymerases: Hoogsteen base-pairing in DNA replication?, Nature 437, E6–E7. [DOI] [PubMed] [Google Scholar]
- 45.Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, and Wolberger C (2002) A Hoogsteen base pair embedded in undistorted B-DNA, Nucleic Acids Res 30, 5244–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ethayathulla AS, Tse PW, Monti P, Nguyen S, Inga A, Fronza G, and Viadiu H (2012) Structure of p73 DNA-binding domain tetramer modulates p73 transactivation, Proc. Natl. Acad. Sci. U. S. A 109, 6066–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirouac KN, and Ling H (2011) Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota, Proceedings of the National Academy of Sciences 108, 3210–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair DT, Johnson RE, Prakash L, Prakash S, and Aggarwal AK (2005) Human DNA Polymerase ι Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair, Structure 13, 1569–1577. [DOI] [PubMed] [Google Scholar]
- 49.Hintze BJ, Richardson JS, and Richardson DC (2017) Mismodeled purines: implicit alternates and hidden Hoogsteens, Acta Crystallographica Section D 73, 852–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenwick RB, van den Bedem H, Fraser JS, and Wright PE (2014) Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR, Proceedings of the National Academy of Sciences 111, E445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swinger KK, and Rice PA (2004) IHF and HU: flexible architects of bent DNA, Curr. Opin. Struct. Biol 14, 28–35. [DOI] [PubMed] [Google Scholar]
- 52.Weisberg RA, Enquist LW, Foeller C, and Landy A (1983) Role for DNA homology in site-specific recombination: The isolation and characterization of a site affinity mutant of coliphage λ, J. Mol. Biol 170, 319–342. [DOI] [PubMed] [Google Scholar]
- 53.Gamas P, Burger AC, Churchward G, Caro L, Galas D, and Chandler M (1986) Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF, Molecular and General Genetics MGG 204, 85–89. [DOI] [PubMed] [Google Scholar]
- 54.Goosen N, van Heuvel M, Moolenaar GF, and van de Putte P (1984) Regulation of Mu transposition II. The Escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu, Gene 32, 419–426. [DOI] [PubMed] [Google Scholar]
- 55.Friedman DI (1988) Integration host factor: A protein for all reasons, Cell 55, 545–554. [DOI] [PubMed] [Google Scholar]
- 56.Nuñez James K., Bai L, Harrington Lucas B., Hinder Tracey L., and Doudna Jennifer A. CRISPR Immunological Memory Requires a Host Factor for Specificity, Mol. Cell 62, 824–833. [DOI] [PubMed] [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A NMRPipe: A multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 58.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, and Markley JL (2008) BioMagResBank, Nucleic Acids Res 36, D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sklenář V, Peterson RD, Rejante MR, and Feigon J (1993) Two-and three-dimensional HCN experiments for correlating base and sugar resonances in 15N, 13C-labeled RNA oligonucleotides, J. Biomol. NMR 3, 721–727. [DOI] [PubMed] [Google Scholar]
- 60.Dhavan GM, Lapham J, Yang S, and Crothers DM (1999) Decreased imino proton exchange and base-pair opening in the IHF-DNA complex measured by NMR, J. Mol. Biol 288, 659–671. [DOI] [PubMed] [Google Scholar]
- 61.Hansen AL, Nikolova EN, Casiano-Negroni A, and Al-Hashimi HM (2009) Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R1ρ NMR Spectroscopy, J. Am. Chem. Soc 131, 3818–3819. [DOI] [PubMed] [Google Scholar]
- 62.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, and Al-Hashimi HM (2015) Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes, Nature advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swails J, Zhu T, He X, and Case DA (2015) AFNMR: automated fragmentation quantum mechanical calculation of NMR chemical shifts for biomolecules, J. Biomol. NMR 63, 125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson WH, Peng C, Bashford D, Noodleman L, and Case DA (1997) Incorporating solvation effects into density functional theory: Calculation of absolute acidities, Int. J. Quantum Chem 61, 207–217. [Google Scholar]
- 65.Koster AM, Geudtner G, Calaminici P, Casida ME, Dominguez VD, Flores-Moreno R, Gamboa GU, Goursot AHT, Ipatov AJ,F, del Campo JM, Reveles JU, Vela A, Zuniga-Gutierrez B, and Salahub DR (2011) deMon2k, Version 4, The deMon developers,Cinvestav, Mexico City [Google Scholar]
- 66.Handy NC, and Cohen AJ (2001) Left-right correlation energy, Mol. Phys 99, 403–412. [Google Scholar]
- 67.Case DA (2013) Chemical shifts in biomolecules, Curr. Opin. Struct. Biol 23, 72–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lynch TW, Read EK, Mattis AN, Gardner JF, and Rice PA (2003) Integration Host Factor: Putting a Twist on Protein–DNA Recognition, J. Mol. Biol 330, 493–502. [DOI] [PubMed] [Google Scholar]
- 69.Lukavsky PJ, Kim I, Otto GA, and Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR, Nature Structural Biology 10, 1033. [DOI] [PubMed] [Google Scholar]
- 70.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, and Summers MF (2011) NMR Detection of Structures in the HIV-1 5′-Leader RNA That Regulate Genome Packaging, Science 334, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dieckmann T, and Feigon J (1997) Assignment methodology for larger RNA oligonucleotides: Application to an ATP-binding RNA aptamer, J. Biomol. NMR 9, 259–272. [DOI] [PubMed] [Google Scholar]
- 72.Pervushin K, Riek R, Wider G, and Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution, Proceedings of the National Academy of Sciences 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farjon J, Boisbouvier J, Schanda P, Pardi A, Simorre J-P, and Brutscher B (2009) Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution, J. Am. Chem. Soc 131, 8571–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sathyamoorthy B, Lee J, Kimsey I, Ganser LR, and Al-Hashimi H (2014) Development and application of aromatic [13C, 1H] SOFAST-HMQC NMR experiment for nucleic acids, J. Biomol. NMR 60, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palmer AG III (2016) A dynamic look backward and forward, J. Magn. Reson [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang C-C, and Nash HA (1989) The interaction of E. coli IHF protein with its specific binding sites, Cell 57, 869–880. [DOI] [PubMed] [Google Scholar]
- 77.Ozers MS, Hill JJ, Ervin K, Wood JR, Nardulli AM, Royer CA, and Gorski J (1997) Equilibrium Binding of Estrogen Receptor with DNA Using Fluorescence Anisotropy, J. Biol. Chem 272, 30405–30411. [DOI] [PubMed] [Google Scholar]
- 78.Yang SW, and Nash HA (1995) Comparison of protein binding to DNA in vivo and in vitro: defining an effective intracellular target, The EMBO Journal 14, 6292–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Cosstick R, Gardner JF, and Gumport RI (1995) The specific binding of Escherichia coli integration host factor involves both major and minor grooves of DNA, Biochemistry 34, 13082–13090. [DOI] [PubMed] [Google Scholar]
- 80.Gilbert DE, van der Marel GA, van Boom JH, and Feigon J (1989) Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex, Proceedings of the National Academy of Sciences 86, 3006–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mouw KW, and Rice PA (2007) Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb, Mol. Microbiol 63, 1319–1330. [DOI] [PubMed] [Google Scholar]
- 82.Ganguly M, Wang F, Kaushik M, Stone MP, Marky LA, and Gold B (2007) A study of 7-deaza-2′-deoxyguanosine–2′-deoxycytidine base pairing in DNA, Nucleic Acids Res 35, 6181–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kowal EA, Ganguly M, Pallan PS, Marky LA, Gold B, Egli M, and Stone MP (2011) Altering the Electrostatic Potential in the Major Groove: Thermodynamic and Structural Characterization of 7-Deaza-2′-deoxyadenosine:dT Base Pairing in DNA, The Journal of Physical Chemistry B 115, 13925–13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, and Kay LE (2004) Probing Slow Dynamics in High Molecular Weight Proteins by Methyl-TROSY NMR Spectroscopy: Application to a 723-Residue Enzyme, J. Am. Chem. Soc 126, 3964–3973. [DOI] [PubMed] [Google Scholar]
- 85.Igumenova TI, and Palmer AG (2006) Off-Resonance TROSY-Selected R1ρ Experiment with Improved Sensitivity for Medium- and High-Molecular-Weight Proteins, J. Am. Chem. Soc 128, 8110–8111. [DOI] [PubMed] [Google Scholar]
- 86.Kitevski-LeBlanc JL, Yuwen T, Dyer PN, Rudolph J, Luger K, and Kay LE (2018) Investigating the Dynamics of Destabilized Nucleosomes Using Methyl-TROSY NMR, J. Am. Chem. Soc 140, 4774–4777. [DOI] [PubMed] [Google Scholar]
- 87.Vivas P, Velmurugu Y, Kuznetsov SV, Rice PA, and Ansari A (2012) Mapping the Transition State for DNA Bending by IHF, J. Mol. Biol 418, 300–315. [DOI] [PubMed] [Google Scholar]
- 88.Sugimura S, and Crothers DM (2006) Stepwise binding and bending of DNA by Escherichia coli integration host factor, Proceedings of the National Academy of Sciences 103, 18510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Golovenko D, Bräuning B, Vyas P, Haran TE, Rozenberg H, and Shakked Z (2018) New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins, Structure 26, 1237–1250. e1236. [DOI] [PubMed] [Google Scholar]
- 90.Fernandez C, Szyperski T, Ono A, Iwai H, Tate S, Kainosho M, and Wuthrich K (1998) NMR with 13C, 15N-doubly-labeled DNA: the Antennapedia homeodomain complex with a 14-mer DNA duplex, J. Biomol. NMR 12, 25–37. [PubMed] [Google Scholar]
- 91.Dupureur CM (2006) Unique 31P Spectral Response to the Formation of a Specific Restriction Enzyme–DNA Complex, Nucleosides, Nucleotides Nucleic Acids 25, 747–764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


