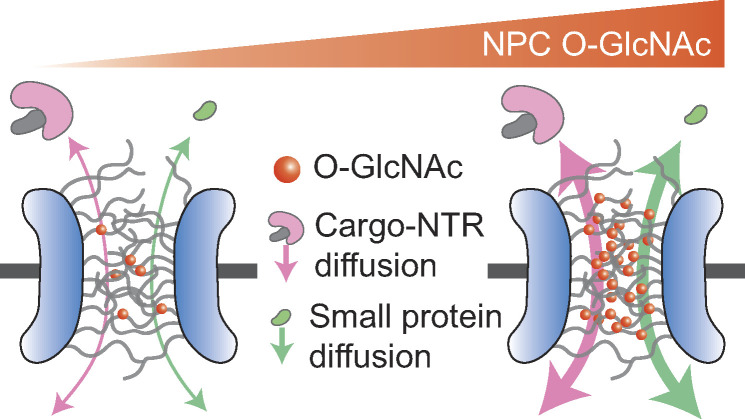
Nuclear pore complexes mediate nuclear transport and are highly modified with O-linked N-acetylglucosamine (O-GlcNAc) on FG repeat domains. Using a new quantitative live-cell imaging assay, Yoo and Mitchison demonstrate acceleration of nuclear import and export by O-GlcNAc modification.
Abstract
Macromolecular transport across the nuclear envelope depends on facilitated diffusion through nuclear pore complexes (NPCs). The interior of NPCs contains a permeability barrier made of phenylalanine-glycine (FG) repeat domains that selectively facilitates the permeation of cargoes bound to nuclear transport receptors (NTRs). FG-repeat domains in NPCs are a major site of O-linked N-acetylglucosamine (O-GlcNAc) modification, but the functional role of this modification in nucleocytoplasmic transport is unclear. We developed high-throughput assays based on optogenetic probes to quantify the kinetics of nuclear import and export in living human cells. We found that increasing O-GlcNAc modification of the NPC accelerated NTR-facilitated transport of proteins in both directions, and decreasing modification slowed transport. Superresolution imaging revealed strong enrichment of O-GlcNAc at the FG-repeat barrier. O-GlcNAc modification also accelerated passive permeation of a small, inert protein through NPCs. We conclude that O-GlcNAc modification accelerates nucleocytoplasmic transport by enhancing the nonspecific permeability of the FG-repeat barrier, perhaps by steric inhibition of interactions between FG repeats.
Graphical Abstract
Introduction
Nucleocytoplasmic transport of macromolecules occurs through thousands of nuclear pore complexes (NPCs) embedded in the nuclear envelope (Watson, 1959). Each NPC consists of ∼30 different nucleoporins (NUPs), many of which are phenylalanine-glycine (FG)-NUPs that project intrinsically disordered regions containing FG repeats into the central channel (Denning et al., 2003; Kim et al., 2018; Lin and Hoelz, 2019). The FG domains constitute a selective permeability barrier that allows the traffic of molecules in a size-dependent manner (Feldherr and Akin, 1997; Keminer and Peters, 1999). Most large molecules are restricted from permeating through the barrier, unless they are complexed with nuclear transport receptors (NTRs), such as importin-βs and exportins (Fried and Kutay, 2003; Ribbeck and Görlich, 2001). The transport of a large NTR–cargo complex is facilitated by specific, transient interactions between FG repeats and hydrophobic pockets of NTR (Hough et al., 2015; Milles et al., 2015). The GTP-bound form of small GTPase Ran (RanGTP) forms a concentration gradient across the nuclear envelope and controls the stability of NTR–cargo interactions, thereby dictating the directionality of NTR-facilitated cargo transport (Christie et al., 2016; Macara, 2001; Matsuura, 2016). In this picture, the overall kinetics of nucleocytoplasmic transport is governed by interactions between FG repeats that determine the passive diffusion properties of NPCs and by interactions between NTRs and FG repeats that determine facilitated diffusion.
FG-NUPs are heavily modified by O-linked β-N-acetylglucosamine (O-GlcNAc), where the monosaccharide is reversibly attached to the hydroxyl oxygen of serine and threonine (Davis and Blobel, 1987; Hanover et al., 1987; Holt et al., 1987; Li and Kohler, 2014; Snow et al., 1987). FG-NUPs were some of the first identified O-GlcNAc–modification substrates and are still among the most heavily modified of all known substrates, yet the functional role of this modification is still unclear. A single pair of enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), regulate O-GlcNAcylation of more than a thousand proteins, making it difficult to determine how changes in cell physiology are caused by modification of specific proteins or assemblies (Chatham et al., 2021; Kreppel et al., 1997; Lubas et al., 1997; Wells et al., 2002). Previous in vitro studies have shown that O-GlcNAcylation alters the structure and permeability of hydrogels derived from FG domains (Labokha et al., 2013), as well as the radius of gyration of FG domains in solution (Tan et al., 2018). These observations made in simplified biochemical model systems predict that O-GlcNAcylation might increase the permeability of NPCs and promote nucleocytoplasmic transport in living cells, but this has never been tested, in large part due to the absence of quantitative tools to measure transport rates in cells. Here, we overcome this limitation by developing optogenetic-based nuclear transport assays and test whether O-GlcNAcylation of the NPC modulates the nuclear transport rate. Alteration of nucleocytoplasmic transport has been implicated in many physiological and pathological processes (Cho and Hetzer, 2020; Grima et al., 2017; Khan et al., 2020; Kim and Taylor, 2017; Lord et al., 2015; Rempel et al., 2019), so tools to measure and modulate it will find multiple uses. In cells, O-GlcNAcylation was proposed to affect NPC integrity and composition (Mizuguchi-Hata et al., 2013; Zhu et al., 2016). This predicts a change in NPC density following O-GlcNAc perturbation, which we also tested.
Results and discussion
Optogenetic-based high-throughput assays are developed to quantify nuclear transport rates in living human cells
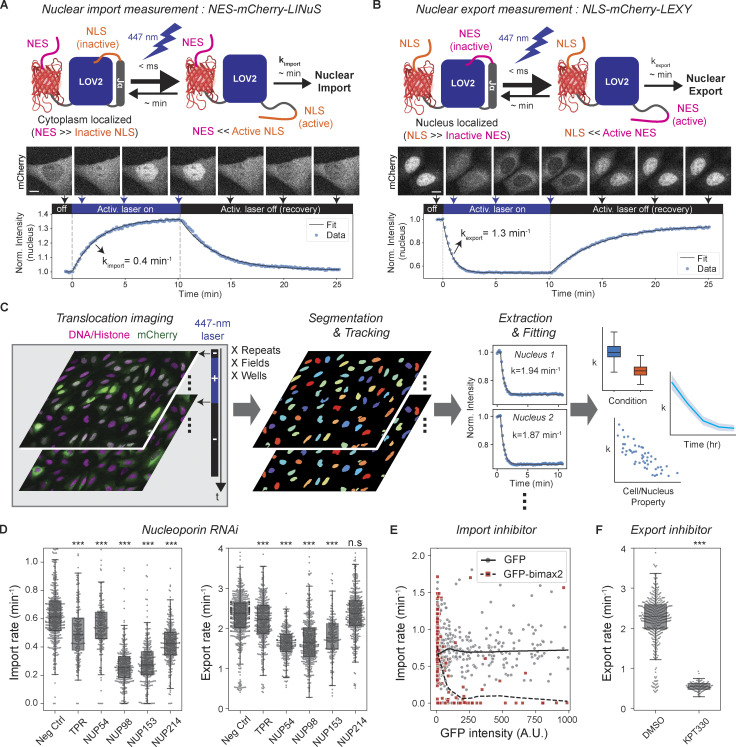
To quantify nuclear import and export kinetics, we repurposed the NES-mCherry-LINuS and NLS-mCherry-LEXY probes previously created for light-controlled gene expression (Niopek et al., 2014; Niopek et al., 2016). These ∼45-kD probes consist of LINuS (LOV2-based photoactivatable nuclear localization sequence [NLS]; Niopek et al., 2014) and LEXY (LOV2-based photoactivatable nuclear export signal [NES]; Niopek et al., 2016) attached to mCherry along with a constitutive NES or NLS that counteracts the photoactivatable signals (Fig. 1, A and B). All the constitutive and photoactivatable NLSs in the probes are classic importin-α/importin-β1–dependent signals, and the NESs are leucine-rich exportin-1–dependent signals. The NES-mCherry-LINuS import probe is mostly localized in the cytoplasm in the dark state but translocates to the nucleus upon blue-light stimulation as the activated LINuS overpowers the NES (Fig. 1 A). Likewise, the NLS-mCherry-LEXY export probe translocates mostly from the nucleus to the cytoplasm upon blue-light stimulation (Fig. 1 B). The nucleus-to-cytoplasm intensity ratio of the photoactivated probes reached 2.21 ± 0.12 (n = 23) for the import probe and 0.28 ± 0.02 (n = 14) for the export probe at equilibrium (mean ± SEM). The kinetics of light-induced changes in the nuclear intensity of the probes was well described by monoexponential decay models. This allowed accurate fitting of kinetic data to output single decay constants that report transport rates. The light-induced conformational change of LOV2 is so fast, within a millisecond (Konold et al., 2016), that it does not contribute to the overall translocation kinetics. In the absence of blue light, the stimulated LOV2 in the probes reverses to the dark state on a minute time scale (Zayner et al., 2012), which enables repeated measurements of the transport rates in individual cells (Fig. 1, A and B). We generated U2OS cell lines stably expressing the transport probes and automated image acquisitions and analysis to allow high-throughput time course measurement and accurate detection of rate changes with high statistical confidence (Fig. 1 C).
Figure 1.
Development and validation of optogenetics-based live-cell nuclear transport assay. (A and B) Measurement of nuclear import and export rates using light-inducible nuclear transport systems, LINuS (import), and LEXY (export). Upon 447-nm laser illumination, NES-mCherry-LINuS and NLS-mCherry-LEXY probes translocate from the cytoplasm to the nucleus and vice versa, respectively. Nuclear transport rates are determined by fitting the light-induced change of the nuclear mCherry intensity to a monoexponential decay model. When the activation radiation ceases, the probes return to their preillumination locations, allowing repeated measurements. Scale bar, 10 µm. Norm., normalized. (C) Automated acquisition and analysis of nuclear transport assay. U2OS cells stably expressing the transport probes and histone marker are imaged in the absence and presence of the 447-nm activation laser illumination. The cycle of activation and recovery is repeated at multiple locations or time points, if necessary. Nuclei are segmented and tracked based on the histone or DNA images. Nuclear import or export rate of each nucleus is determined by monoexponential decay model fitting. Measured transport rates are aggregated for correlation analysis. (D–F) Validation of the nuclear transport assay by measuring the effects of nuclear transport perturbations: NUP RNAi, n > 180 nuclei for each condition (D). Neg Ctrl, negative control. (E) GFP-bimax2 transfection for importin-α sequestration. n = 491 (GFP) and 289 (GFP-bimax2) nuclei. Locally weighted scatterplot smoothing lines (GFP, solid line; GFP-bimax2, dashed line) drawn to show trends. (F) 30-min, 1-µM KPT-330 treatment for export inhibition. n > 90 nuclei. P values were calculated by two-sided Welch’s t test for comparison with negative control. n.s., P > 0.01; ***, P < 1 × 10−4.
To validate the transport assays, we quantified the effects of known perturbations. We first tested the effects of siRNA-mediated depletion of the following FG-repeat–containing NUPs at various locations: NUP54 and NUP98 (inner ring); NUP214 (cytoplasmic filament); and NUP153 and TPR (nuclear basket; Fig. 1 D and Fig. S1). Different NUP depletions differentially affected the transport kinetics of the probes: NUP98 and NUP153 depletion resulted in the largest reduction in the import rate of the LINuS probe (57 and 56%), followed by NUP214 (30%) and TPR (20%). While NUP54 knockdown only slightly reduced the import rate (10%), it greatly affected the export rate of the LEXY probe (31%) to an extent similar to NUP98 and NUP153 depletions (31 and 25%). TPR and NUP214 depletions had a slight (6%) or insignificant influence on the export rate, respectively. These findings demonstrate the capability of the transport assays to separately measure the differential changes in the import and export rates induced by different perturbations. The differential effects of NUP depletions may reflect different roles of individual NUPs in nuclear import and export, which have been studied previously (Ball and Ullman, 2005; Bernad et al., 2006; Frosst et al., 2002; Hutten and Kehlenbach, 2006; Powers et al., 1997; Powers et al., 1995; Ullman et al., 1999; Walther et al., 2001; Wu et al., 2001). However, they could also result from different NUP stability leading to different NUP levels after RNAi or from different long-term translational/transcriptional changes in the nuclear transport machinery, which is to be addressed by future studies.
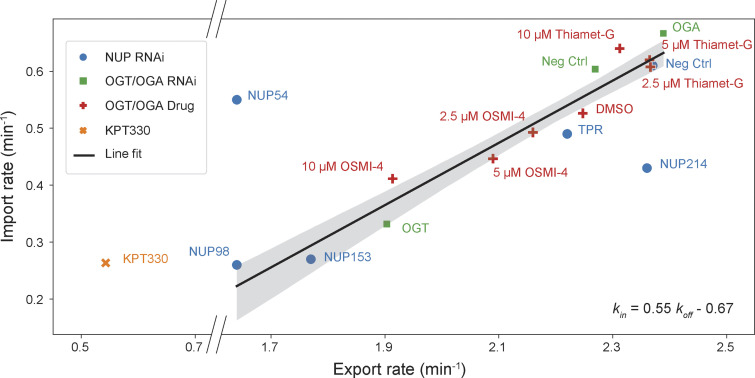
Figure S1.
Nuclear import and export rates measured after NUP depletions (blue dots), OGT/OGA depletions (green squares), OGT/OGA inhibitor treatments (red crosses), and KPT-330 treatment (orange ×). Black line represents the best line fit to the all data points excluding three obvious outliers, NUP54 and NUP214 depletions and KPT-330 treatment, and shaded area represents the 95% confidence interval.
We next tested the effect of nuclear transport inhibitors. We first inhibited nuclear import by expressing bimax2, a peptide that competitively inactivates importin-αs by tightly binding to their NLS-binding domains (Kosugi et al., 2008). We found that the import rate of the LINuS probe sharply decreased with an increasing GFP-bimax2 expression level, while GFP expression alone did not affect the rate (Fig. 1 E). We next inhibited nuclear export using KPT-330, a specific inhibitor of exportin-1 (Lapalombella et al., 2012). After KPT-330 treatment, the export rate of the LEXY probe decreased by 77% (Fig. 1 F). The import rate of the LINuS probe also decreased by 62% after KPT-330 treatment, possibly due to the contribution of the constitutive NES in the probe. The reduction of the export rate relative to the reduction of the import rate nonetheless was much greater for KPT-330 treatment than for other perturbations examined (Fig. S1). These results demonstrate that the LINuS and LEXY probes quantitatively report on the importin-α/β1–dependent import and the exportin-1–dependent export rates, respectively, although they are not fully decoupled.
Increasing O-GlcNAc modification accelerates nuclear import and export
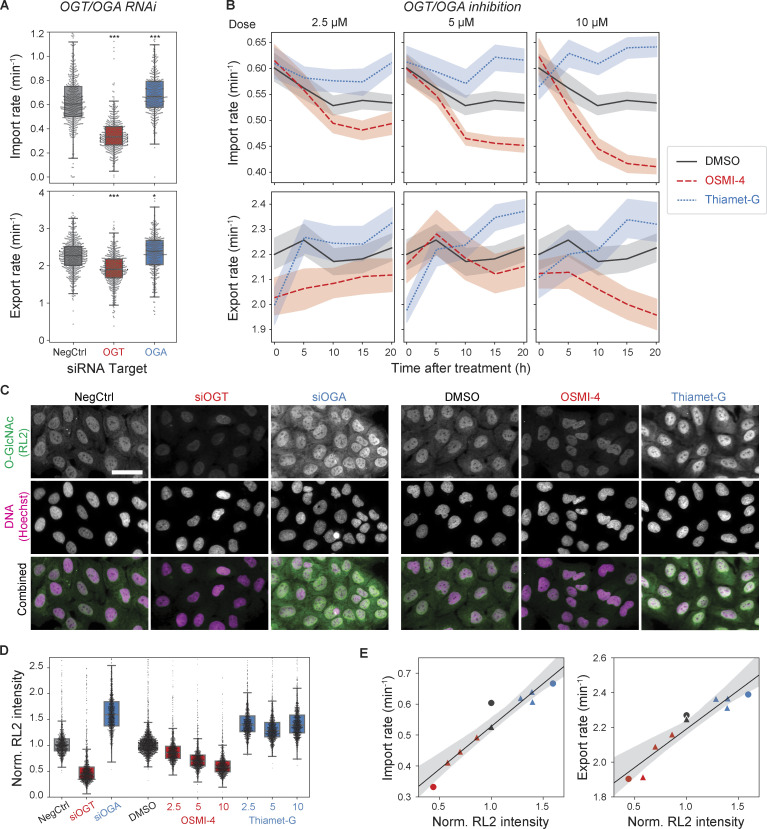
With quantitative assays in hand, we investigated how nuclear transport rates respond to perturbation of O-GlcNAc modifications. We first performed siRNA transfections to deplete OGT or OGA, the sole pair of enzymes that add and remove O-GlcNAc, respectively (Chatham et al., 2021). After 3 d of the siRNA transfections, we found that the import rate was 45% lower in the OGT-depleted cells and 11% higher in the OGA-depleted cells compared with the scrambled-siRNA control (Fig. 2 A). The export rate followed the same trend as the import rate but changed to a smaller extent (Fig. 2 A). Consequently, the import and export rates were ∼100 and 30% higher, respectively, in the OGA-depleted cells than in the OGT-depleted cells.
Figure 2.
Nuclear transport rates are dependent on the O-GlcNAc level. (A) Nuclear import (top) and export (bottom) rates after 3 d of transfection with scrambled (NegCtrl), OGT, or OGA siRNAs. n > 360 nuclei per condition. (B) Import (top) and export (bottom) rates vs. time after treatment with DMSO (black, solid), OSMI-4 (red, dashed), and Thiamet-G (blue, dotted) at 2.5 µM (left), 5 µM (middle), and 10 µM (right). Line and shaded area indicate mean and 95% confidence interval, respectively. n ≥ 120 nuclei per time point per condition. (C) Representative anti-O-GlcNAc (RL2) antibody immunofluorescence images of cells transfected with NegCtrl, OGT, or OGA siRNA for 3 d or treated with DMSO, 10 µM OSMI-4, or 10 µM Thiamet-G for 20 h. Scale bar, 50 µm. (D) Quantification of nuclear RL2 intensity (background corrected and normalized such that the median value of control condition is 1). n > 1,200 nuclei per condition. (E) Median import (left) or export (right) rate (from B) vs. median nuclear RL2 intensity (from D), measured after the knockdowns (circles) or inhibitions (triangles) of OGT (red) or OGA (black). Black line and gray shade are the line fit and its 95% confidence interval. P values were calculated by two-sided Welch’s t test for comparison with negative control. n.s., P > 0.01; *, P < 1 × 10−2; ***, P < 1 × 10−4.
We next used OGT- or OGA-specific small-molecule inhibitors, OSMI-4 (Martin et al., 2018) and Thiamet-G (Yuzwa et al., 2008), to better control the timing and degree of the OGT/OGA inhibitions. Cells were treated with each drug at three different doses and continually assayed for nuclear transport rates over 20 h. The import rate slightly decreased (7%) in the DMSO control over the time of on-stage incubation. It decreased significantly more in the presence of OSMI-4 in a dose-dependent manner, by ≤23% over 20 h (Fig. 2 B). Fitting the OSMI-4 data to a monoexponential decay model, we measured the rate of the decrease to be 0.14 ± 0.02 h−1, which is comparable to the turnover rate of O-GlcNAc at several NUPs measured previously using quantitative mass spectrometry (Wang et al., 2016). On the other hand, the import rate increased after Thiamet-G treatment in a dose-dependent manner, ≤15% over 20 h (Fig. 2 B). Similarly, the nuclear export rate also reached a higher and lower level in the presence of Thiamet-G and OSMI-4, respectively, than in the DMSO control (Fig. 2 B).
To characterize the relationship between the O-GlcNAc level and the transport rates, we performed quantitative immunofluorescence using RL2, anti-O-GlcNAc antibody generated against glycosylated NUPs (Snow et al., 1987). After 3 d of siRNA transfections, the nuclear RL2 signal was 56% lower in the OGT-depleted cells and 59% higher in the OGA-depleted cells, compared with the negative control (Fig. 2, C and D). Compared with the DMSO control, the nuclear RL2 signal decreased after OSMI-4 treatment by ≤42% at the highest concentration tested (10 µM; Fig. 2, C and D). The nuclear RL2 signal increased by ∼39% after Thiamet-G treatments without showing noticeable dose dependence above 2.5 µM, indicating saturation (Fig. 2 D). OSMI-4/Thiamet-G treatments at the highest concentration had smaller influences on the O-GlcNAc level than the OGT/OGA depletions, presumably due to homeostatic mechanisms regulating the expression of OGT/OGA in response to the O-GlcNAc level change (Park et al., 2017; Tan et al., 2020; Zhang et al., 2014). Plotting the transport rates (shown in Fig. 2 B) against the nuclear RL2 signals (shown in Fig. 2 D) revealed that both transport rates increased linearly with increasing nuclear RL2 signal, with the import rate showing a greater dependence (Fig. 2 E). The relationship between the import and export rates measured after the drug-induced OGT/OGA inhibitions was colinear with that after OGT/OGA RNAi, indicating that the O-GlcNAc–dependent change in the rates is independent of the mechanism of perturbation (Fig. S1).
Modification of NPCs drives O-GlcNAc–dependent nuclear transport modulation
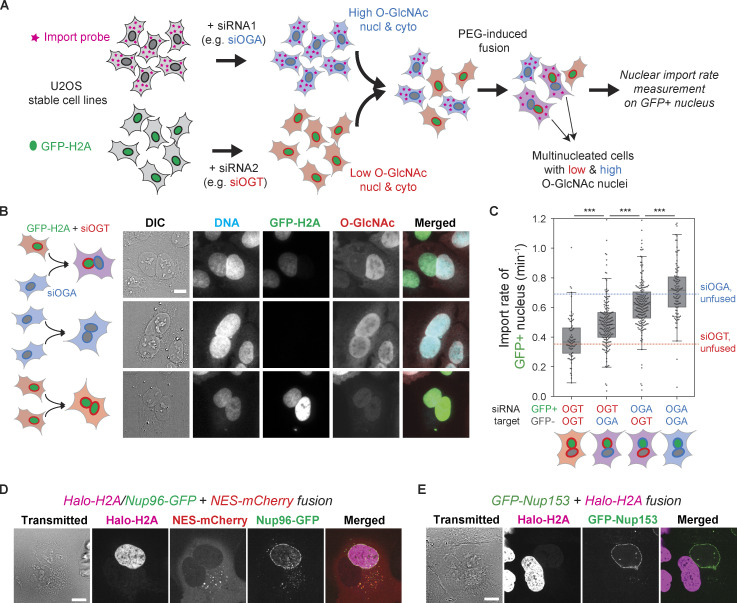
We further asked whether the O-GlcNAc dependence of the transport rates was specifically due to modification of NPCs where O-GlcNAc is abundant (Davis and Blobel, 1987; Hanover et al., 1987; Holt et al., 1987; Snow et al., 1987), as opposed to less direct effects mediated by changes in gene expression (Comer and Hart, 1999) or modification of other factors in the transport machinery. Hypothetically, if the NPCs or other nucleus-confined components were responsible for the O-GlcNAc–dependent transport rates, different nuclei in a multinucleated cell could have different transport rates depending on their O-GlcNAc levels. To test this, we generated heterokaryons using polyethylene glycol (PEG)-mediated cell fusion (Davidson and Gerald, 1976; Fig. 3 A). Briefly, two U2OS stable cell lines, one expressing GFP-labeled histone 2A (H2A) and the other expressing the NES-mCherry-LINuS import probe, were separately transfected with OGT- or OGA-targeting siRNA. Then those two cell populations were combined and treated briefly with PEG to induce cell fusion (Fig. 3 A). The multinucleated cells were assayed after 2–4 h of resting in the presence of both OSMI-4 and Thiamet-G to suppress O-GlcNAc turnover. This maintained prefusion levels of O-GlcNAcylation in each nucleus after fusion (Fig. 3 B).
Figure 3.
Nucleus-confined factors are responsible for the O-GlcNAc dependence of nuclear transport kinetics. (A) Schematics of heterokaryon analysis. Two U2OS stable cell lines, one expressing the NES-mCherry-LINuS import probe and the other expressing GFP-H2A, were separately transfected with OGA- or OGT-targeting siRNA, cocultured, and then fused by brief treatment with PEG. The nuclear import rate was measured on the GFP-positive nuclei in the mCherry-positive fused cells. Cyto., cytoplasm; Nucl., nucleus. (B) RL2 immunofluorescence images of the fused cells, showing that the nuclei largely retain the prefusion O-GlcNAc level. (C) Nuclear import rate of the GFP+ nucleus in four different fusions (from left to right): GFP-H2AsiOGT + import probesiOGT, GFP-H2AsiOGT + import probesiOGA, GFP-H2AsiOGA + import probesiOGT, and GFP-H2AsiOGA + import probesiOGA. Blue and red dotted lines indicate the median nuclear import rates of the unfused cells transfected with siOGA and siOGT, respectively (values from Fig. 2 A). n ≥ 75 nuclei for each condition. P values calculated by two-sided Welch’s t test where ***, P < 1 × 10−4. (D) The fusion of a U2OS cell stably expressing Halo-H2A (magenta) and Nup96-GFP (green) and another U2OS cell expressing NES-mCherry (red). (E) The fusion of U2OS cell stably expressing GFP-Nup153 (green) and another U2OS cell expressing Halo-H2A (magenta). Scale bars, 10 µm.
We performed cell fusions with every combination of the two siRNA transfections (siOGT or siOGA) and the two cell lines (GFP-H2A or NES-mCherry-LINuS) and measured the nuclear import rate of the GFP+ nuclei in mCherry+ cells (Fig. 3, A and C). GFP positivity indicates the origin of the nucleus and therefore its O-GlcNAc state, while mCherry positivity ensures fusion with the other origin. The siOGT/siOGT or siOGA/siOGA fusions displayed import rates similar to those of the OGT- and OGA-depleted unfused cells, respectively, indicating that the cell fusion itself does not alter the transport rates (Fig. 3 C). Interestingly, in the siOGT/siOGA heterokaryons, the import rate was 28% higher at the high–O-GlcNAc nuclei than at the low–O-GlcNAc nuclei (Fig. 3 C), indicating a significant contribution of the nucleus-confined factors to O-GlcNAc–dependent nuclear transport regulation. This difference was smaller than that between siOGA/siOGA and siOGT/siOGT fusions, presumably because of the turnover of O-GlcNAc after the cell fusion or the contribution of the O-GlcNAc-modified diffusive factors.
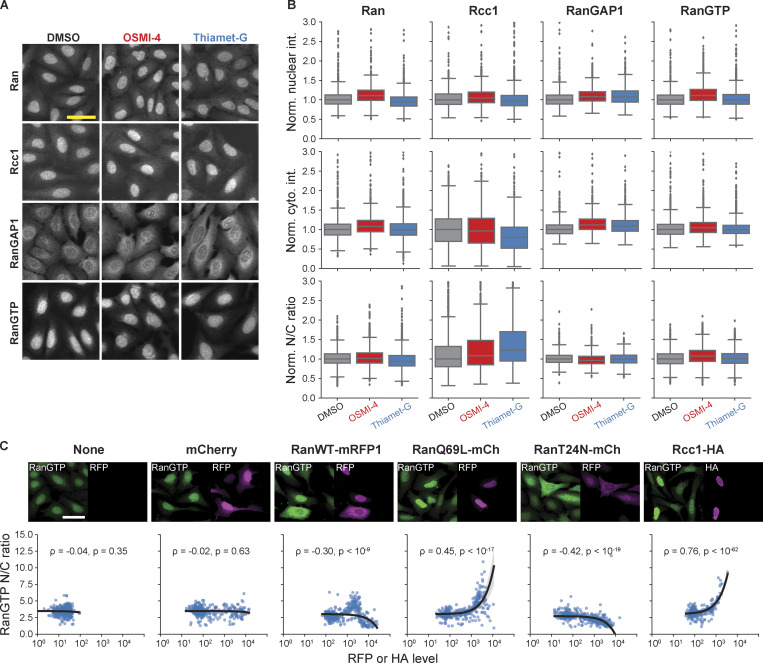
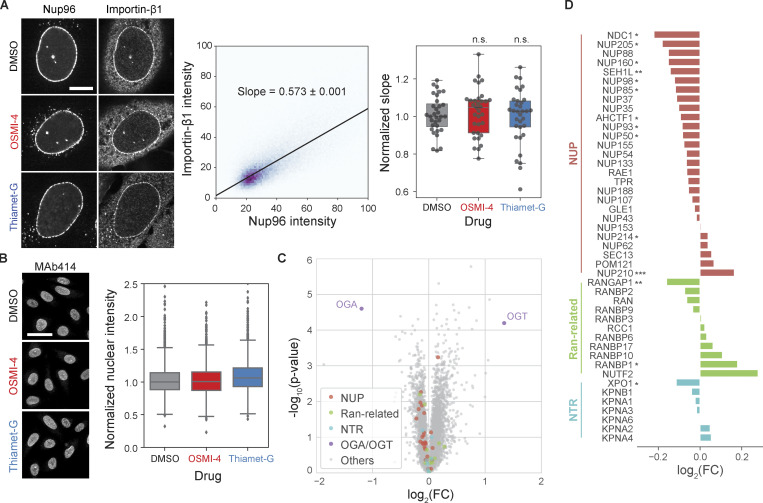
We next asked which nuclear transport machinery components are responsible for the O-GlcNAc dependence of nuclear import observed in the cell fusion assay. The NPC itself is undoubtedly the top qualified candidate not only for the abundance of O-GlcNAc modification but also for its strict confinement to the nucleus (Daigle et al., 2001; Rabut et al., 2004). The strict nuclear confinement was confirmed by heterokaryon fusion analyses: Nup96 was not exchanged between the nuclei in a heterokaryon, and neither was Nup153, despite its dynamic association with the NPC (Daigle et al., 2001; Rabut et al., 2004; Fig. 3, D and E). We also considered the possibility that the Ran pathway plays a role in the O-GlcNAc dependence of the import rate, as Rcc1 and RanGAP1, the two major regulators of Ran (Macara, 2001), are confined to the nucleus at least partially (Mahajan et al., 1997; Matunis et al., 1996; Nemergut and Macara, 2000). However, OSMI-4 or Thiamet-G treatment did not result in any appreciable change in the distributions of Ran, Rcc1, and RanGAP1 (Fig. S2). More importantly, the nuclear/cytoplasmic ratio of RanGTP level, measured by anti-RanGTP antibody (Richards et al., 1995) immunostaining, was not affected by the O-GlcNAc perturbations, but by the overexpression of WT, constitutively active (Q69L) mutant, or dominant-negative (T24N) mutant of Ran (Klebe et al., 1995) or Rcc1, which confirms the validity of the RanGTP immunoassay (Fig. S2). Thus, the NPC, rather than the Ran-regulating system, is the nucleus-confined factor that drives the O-GlcNAc–dependent acceleration of nuclear transport.
Figure S2.
RanGTP gradient is unaltered by O-GlcNAc perturbations. (A) Ran, Rcc1, RanGAP1, and RanGTP immunofluorescence images of cells treated with DMSO, 10 µM OSMI-4, or 10 µM Thiamet-G for 24 h. (B) Mean nuclear and cytoplasmic intensity (cyto. int.) and the nucleus-to-cytoplasm intensity ratio of Ran, Rcc1, RanGAP1, and RanGTP. Data from the same batch were normalized (Norm.) to the median value of the DMSO condition. n > 1,400 cells for each condition. (C) Top: RanGTP immunofluorescence images of cells transfected with mCherry, RanWT-mRFP1, Ran69L-mCherry, RanT24N-mCherry, or Rcc1-HA. Bottom: Scatterplots of RanGTP nucleus-to-cytoplasm intensity ratio vs. RFP or HA level. n > 300 cells for each condition. Black line is the line fit to the data. Shaded gray area represents the 95% confidence interval. Scale bars, 50 µm. ρ, Pearson correlation coefficient; p, P value.
FG-repeat permeability barrier is highly modified with O-GlcNAc
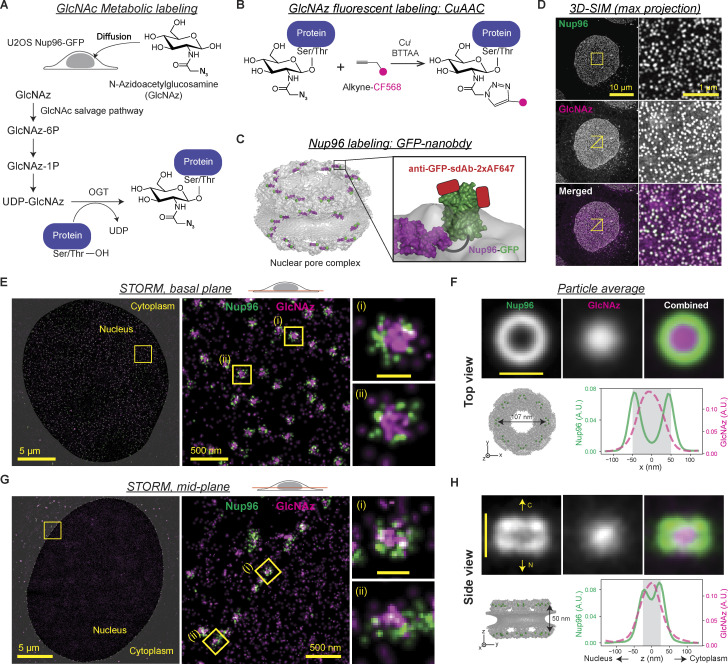
To gain insight into how O-GlcNAcylation alters the NPC, we investigated the distribution of O-GlcNAc within the NPC using metabolic labeling and superresolution microscopy. Previous studies showed that wheat germ agglutinin (WGA), a GlcNAc-binding lectin, is predominantly localized to the center of the NPC (Löschberger et al., 2012; Thevathasan et al., 2019). However, WGA binding may not accurately reflect the O-GlcNAc distribution due to its multiple binding sites and considerable size (36 kD; Nagata and Burger, 1974; Wright and Kellogg, 1996). Moreover, the O-GlcNAc distribution along the transport axis has not been revealed. We therefore employed the combination of metabolic labeling approach (Boyce et al., 2011; Woo et al., 2018; Zaro et al., 2011; Zhu et al., 2015) and copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction to fluorescently label O-GlcNAc sites in situ and performed superresolution microscopy (Fig. 4, A and B). Although per-O-acetylated chemical reporters (e.g., tetra-acetylated N-azidoacetylglucosamine) have been widely used in previous studies for their improved membrane permeability and cellular uptake, they may produce nonspecific background signals by reacting with cysteines via S-glycosylation (Hao et al., 2019; Qin et al., 2018), so we chose to use an unacetylated reporter, N-azidoacetylglucosamine (GlcNAz). To set reference points within the NPC, Nup96 was labeled with Alexa Fluor 647 (AF647) via CRISPR-Cas9–mediated endogenous protein tagging and direct immunostaining using a single-domain antibody (Fig. 4 C). We first imaged the double-stained cells using 3D structured illumination microscopy (SIM) and confirmed a remarkably high level of O-GlcNAc at the NPCs (Fig. 4 D and Video 1). We next used stochastic reconstruction microscopy (STORM) to obtain the detailed O-GlcNAc distribution within the NPC. Viewed from the top, Nup96 delineated the circular periphery of the NPC, whose diameter measured 96 ± 8 nm (± SD; Fig. 4, E and F). O-GlcNAc was concentrated the most at the center and lower at the periphery, resembling the binding pattern of WGA (Löschberger et al., 2012; Thevathasan et al., 2019). In the side view of the NPC, Nup96 was situated along two lines delineating the top and bottom of the symmetric core, separated by ∼50 nm (Fig. 4, G and H). O-GlcNAc was localized mostly between the two lines and symmetrically distributed along the transport axis (Fig. 4, G and H). These results show that O-GlcNAc is particularly abundant within the channel of the symmetric central core of the NPC where the FG repeat permeability barrier is embedded (Lin and Hoelz, 2019).
Figure 4.
O-GlcNAc is highly abundant at the central transport channel of the symmetric core of the NPC. (A–C) In situ fluorescent labeling of Nup96 and O-GlcNAc sites in U2OS cell. (A) Metabolic labeling of O-GlcNAc site. GlcNAz is incorporated into proteins via GlcNAc salvage pathway and OGT activity. (B) The incorporated GlcNAz moieties were labeled with CF568 via CuAAC. (C) Nup96 (purple) was endogenously tagged with GFP (green) at the C-terminus. After fixation, each GFP molecule was labeled with up to two anti-GFP single-domain antibodies conjugated with two AF647 (red). (D) Maximum-intensity projection of 3D SIM image of the double-stained cells. Left column: Full field of view. Right column: Enlarged region of interest (yellow square). (E and G) Representative two-color STORM images of the basal plane (E) and midplane (G) of the nucleus showing the top and side views of the NPCs, respectively. Each localization of molecules is rendered as a normalized gaussian whose SD is set to the localization precision. (F and H) Top: Averaged top (F) or side view (H) STORM image of the NPCs (703 top views from four cells; 122 side views from four cells). Bottom left: Structure (Protein Data Bank accession no. 5A9Q) of the NPC symmetric core, where the C-terminus of Nup96 is marked with a green sphere. Bottom right: Averaged intensity profiles of Nup96-GFP:AF647 (green, solid) and GlcNAz:CF568 (magenta, dashed) along the x axis (F) or z axis (H). Nup96-GFP:AF647 is colored in green and GlcNAz:CF568 in magenta in all merged 3D SIM and STORM images in D–H. Scale bars in individual and averaged pore images in E–H, 100 nm.
Video 1.
3D SIM z-stacks of Nup96-GFP:AF647 (green, left) and GlcNAz:CF568 (magenta, right). Z-spacing, 130 nm. 10 frames per second.
O-GlcNAcylation increases nonspecific permeability of the FG permeability barrier
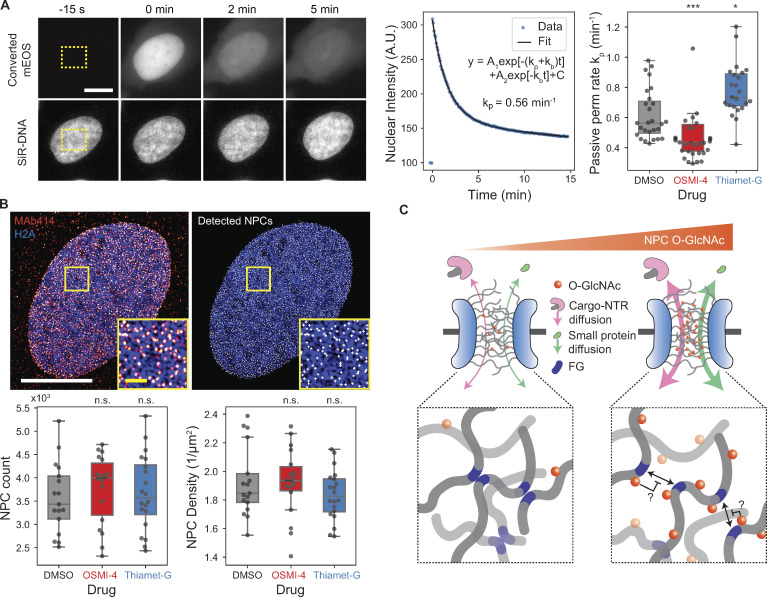
The permeability of the FG repeat barrier to the NTR–cargo complex is determined by two factors: specific binding of NTR to FG repeats and nonspecific interactions between permeants and the FG barrier (Becskei and Mattaj, 2005; Bressloff and Newby, 2013; Stanley et al., 2017). O-GlcNAc modification could alter either. Previous in vitro studies did not report any appreciable change of NTR binding to FG domains following a change in O-GlcNAcylation (Labokha et al., 2013; Tan et al., 2018). Consistent with this observation, we found that the localization of importin-β1 to NPC was unaltered after O-GlcNAc perturbations by OGT/OGA inhibitions (Fig. S3 A). To determine the effect of O-GlcNAc on interactions between permeants and the barrier independent of specific bindings, we measured the transport rate of a protein probe that does not interact specifically with FG repeats. We photoconverted mEOS inside the nucleus and quantified its rate of passive permeation out of the nucleus (Fig. 5 A). We found that the passive permeation rate was 22% lower in the OSMI-4–treated cells and 30% faster in the Thiamet-G–treated cells (Fig. 5 A). These data followed the same trend as NTR-facilitated nuclear transport rates (Fig. 2). Because both facilitated and passive transports are accelerated by O-GlcNAcylation, we conclude that it mainly increases nonspecific permeation rates.
Figure S3.
O-GlcNAc perturbations do not affect importin-β1 localization to NPCs and protein levels of nuclear transport machinery. (A) Left: Importin-β1 and Nup96-GFP immunofluorescence images of cells treated with DMSO, 10 µM OSMI-4, or 10 µM Thiamet-G for 24 h. Middle: For each cell, a linear model (black line) was fitted to the pixel intensities of importin-β1 and Nup96 after background subtraction and thresholding. The slope of the linear model was used as a measure of the localization of importin-β1 at the NPCs. Right: The slopes for each drug condition. n > 31 cells for each condition. n.s., P > 0.5. (B) Left: MAb414 immunofluorescence images of cells treated with DMSO, 10 µM OSMI-4, or 10 µM Thiamet-G for 24 h. Right: Quantified mean nuclear intensity. n > 2,000 cells for each condition. For both A and B, data from the same batch were normalized to the median value of the DMSO condition. (C and D) Proteomics data from Martin et al. (2018) replotted to show the difference in the protein levels of nuclear transport machinery components between HEK293T cells treated with DMSO and those treated with 20 µM OSMI-4 for 24 h. FC, fold-change. *, P < 0.1; **, P < 0.01; ***, P < 0.001.
Figure 5.
O-GlcNAcylation enhances the permeability of the NPC. (A) Passive diffusion rate measurement. Left representative images: mEOS (top row) in the rectangular region (yellow box) inside the nucleus was selectively converted and continually imaged along with DNA (bottom row). Scale bar, 10 µm. Middle plot: A biexponential decay model was fitted to the nuclear intensity of converted mEOS to measure the passive permeation (perm) rate kp. Right box plot: Passive permeation rates for cells treated with DMSO, OSMI-4, or Thiamet-G (10 µM, 24 h). n ≥ 23 cells for each condition. (B) NPC counting. Top images: 3D SIM image of MAb414 immunostaining (red), Halo-H2A:JF646 (blue), and identified NPCs (white). Bottom box plots: Total number (left) and area density (right) of NPCs in cells treated with DMSO, OSMI-4, or Thiamet-G (10 µM, 24 h). n ≥ 17 cells for each condition. Scale bars, 10 µm (white) and 1 µm (yellow; magnified inset). n.s., P > 0.01; *, P < 1 × 10−2; ***, P < 1 × 10−4. (C) Proposed model for O-GlcNAc–dependent modulation of nuclear transport kinetics. Hydrophilic, bulky O-GlcNAc modifications hinder the cohesive interaction between FG repeats, thereby facilitating both passive and NTR-facilitated permeations of molecules through the NPC in both directions.
We next considered the possibility that the change in the rates does not result from the altered per-NPC permeability, but from the change in the number of NPCs, since a previous study suggested that O-GlcNAcylation protects NUPs from proteasomal degradation (Zhu et al., 2016). However, treating cells with 10 µM OSMI-4 or Thiamet-G for 24 h did not significantly affect the number or area density of the NPCs, measured by MAb414 immunostaining and 3D SIM (Fig. 5 B and Fig. S3 B). Moreover, recent proteomics data (Martin et al., 2018) of HEK293T show that the levels of NUPs, as well as other transport machinery components, do not considerably change after 24-h treatment with 20 µM OSMI-4 (Fig. S3, C and D). Therefore, O-GlcNAcylation did not regulate NPC stability in our system. Rather, it increased the nonspecific permeability of the FG barrier, thereby enhancing both facilitated and passive transport in both directions (Fig. 5 C). This confirms predictions made using synthetic FG-repeat–derived hydrogels (Labokha et al., 2013). The enhanced nonspecific permeability might result from the hydrophilic and bulky O-GlcNAc moieties hindering the hydrophobic cohesions of FG domains (Fig. 5 C).
Concluding remarks
In summary, we developed quantitative nuclear transport assays and applied superresolution microscopy to reveal the role of NPC O-GlcNAcylation in modulating transport rates. The transport assays allow high-throughput quantification of the nuclear import and export kinetics in live cells, overcoming the limitations of conventional assays that rely on the measurement of the steady-state nuclear/cytoplasmic localization of cargos or membrane-specific permeabilization of cells. The O-GlcNAc–dependent modulation of the transport rates may underlie the mechanism by which OGA inhibition mitigates the nuclear transport defects in Huntington’s disease (Grima et al., 2017) and may also provide a novel way to treat other neurodegenerative diseases and aging-related stresses compromising the nuclear transport machinery (Cho and Hetzer, 2020; D’Angelo et al., 2009; Hutten and Dormann, 2020; Kim and Taylor, 2017; Rempel et al., 2019). Yeast is the only eukaryote known to lack OGT/OGA, but it uses O-linked mannose to modify NUPs, which may serve the functions equivalent to O-GlcNAcylation in higher eukaryotes (Halim et al., 2015).
Materials and methods
Cell culture
U2OS cell lines (engineered from HTB-96; ATCC) were maintained in complete DMEM (low-glucose [1 g/liter] DMEM [#10567022; Thermo Fisher Scientific] supplemented with 10% FBS [#A31605; Thermo Fisher Scientific], 50 IU/ml penicillin, and 50 µg/ml streptomycin [#15140122; Thermo Fisher Scientific]) at 37°C in a humidified atmosphere with 5% CO2. Cells were validated as mycoplasma free by PCR-based mycoplasma detection kit (#30-1012K; ATCC).
Live-cell nuclear transport assay
Generation of stable cell lines
U2OS cells were engineered to stably express H2A type 1, whose N-terminus was labeled with HaloTag (Halo-H2A) by retroviral transfection and selection with 200 µg/ml hygromycin B (#10687010; Thermo Fisher Scientific). NES-mCherry-LINuS (Niopek et al., 2014) and NLS-mCherry-LEXY (Niopek et al., 2016) coding regions in pcDNA3.1 vectors (#61347 and #72655; Addgene) were cloned into a pBABE-blast packaging vector for retroviral transfections by PCR and Gibson assembly. The Halo-H2A–expressing U2OS cell line was engineered to stably express NES-mCherry-LINuS (import probe) or NLS-mCherry-LEXY (export probe) by retroviral transfection and selection with 2 µg/ml blasticidin (#A1113903; Thermo Fisher Scientific).
Live-cell imaging
Cells were seeded at 10,000–15,000 per well in an eight-well chambered coverslip (#80826; Ibidi) and grown in complete medium for 1–3 d before live-cell imaging. Prior to imaging, the growth medium was replaced with imaging medium: low-glucose (1 g/L) DMEM without phenol red (#11054020; Thermo Fisher Scientific), supplemented with 10% FBS, 50 IU/ml penicillin, 50 µg/ml streptomycin, and GlutaMAX Supplement (#35050061; Thermo Fisher Scientific). For nucleus staining, Halo-H2A cells were incubated in 500 nM JF646-HaloTag ligand (gift from Luke Lavis; Howard Hughes Medical Institute, Janelia Research Campus) in the imaging medium for 3–16 h. Cage microscope incubator (OkoLab) was used to maintain the cells at 37°C in 5% CO2 with high humidity during imaging.
Live-cell imaging was performed on a Nikon Ti motorized inverted microscope with Perfect Focus System, using a spinning disk confocal scanner (CSU-X1; Yokagawa) with Spectral Applied Research Aurora Borealis modification, motorized stage and shutters (Proscan II, Prior), scientific complementary metal–oxide–semiconductor (sCMOS) camera (Flash4.0 V3; Hamamatsu), laser merge module (LMM-5; Spectral Applied Research), CFI Plan Apo 20×/0.75-NA objective lens (Nikon), and Chroma ZT445/514/561/640tpc polychroic mirror. mCherry-labeled import/export probes were imaged using 561-nm laser and ET605/70m emission filter (Chroma), while JF646:Halo-H2A was imaged using 642-nm laser and ET700/75m emission filter (Chroma). In the nuclear transport assays, live-cell time-lapse images were acquired through a ∼25-min imaging cycle (155 frames) that consisted of three acquisition phases: preactivation, activation, and recovery. Throughout the cycle, the mCherry-labeled transport probes and DNA/histone marker were imaged every 10 s with 100-ms exposure time. Each imaging cycle began with a three-frame preactivation phase to determine the baseline nuclear localization of the probe, followed by a 10-min activation phase (61 frames) in which the probes were activated by 200-ms exposure to the activation laser (447 nm) every 10 s. The imaging cycle ended with a 15-min recovery phase (91 frames), during which the probe returned to the prestimulation location in the absence of 447-nm laser stimulation so that the imaging cycle can be repeated on the same field. To increase the throughput, the imaging cycle was executed at three different fields simultaneously. The imaging cycle was repeated at multiple time points and at different positions, which was fully automated by using Journal macro in MetaMorph software. The power of the 447-nm laser was optimized based on the nuclear export rate vs. laser power curve, to the lowest saturation level where a small variation of the power does not influence the transport rate. No sign of photobleaching or photodamaging was noticed when the imaging cycle was repeated every hour for 24 h on the same field.
Image analysis
We created custom Python codes for streamlined computational analysis of the time-lapse images. Briefly, cell nuclei were segmented based on H2A/DNA images using U-Net convolutional neural network trained on manually annotated U2OS nuclei images (Caicedo et al., 2019) and tracked using Trackpy (v0.4.1; https://doi.org/10.5281/zenodo.1226458). Then, for each nucleus, the time trajectory of the mean nuclear mCherry intensity was measured and normalized such that the average intensity during preactivation phase was 1 and the background intensity was 0. The normalized nuclear mCherry intensity trajectory during activation phase was fitted to the following monoexponential decay model to determine the transport rate k:
When the import probe is used, k corresponds to the import rate and b is >1 (i.e., y increases over time), while when the export probe is used, k corresponds to the export rate and b is <1 (i.e., y decreases over time). Trust region reflective least-squares algorithm was used for the nonlinear regression. The fitting result was excluded from further analyses if it met one or more of the following criteria: (1) the fitting algorithm did not converge; (2) there were too many missing time points in the nuclear intensity trajectory (i.e., incomplete nucleus tracking); (3) reduced χ2 statistics were too large; (4) |1 − b| was too small; and (5) k was abnormally large.
siRNA transfections
siRNAs targeting NUP98 (SI00115332), NUP153 (SI03033226), NUP214 (SI04329521), and OGT (SI0266513) and negative control siRNA (1027280) were purchased from Qiagen, OGA siRNA (M-012805-01-0005) and NUP54 siRNA (J-017570-09-0002) from Dharmacon, and TPR siRNA from IDT (synthesized, 5′-UUUAACUGAAGUUCACCCU-3′). Cells were grown to ∼70% confluence in complete DMEM for 1 d in a 6-well plate and transfected with siRNA for each well as follows: 30 pmol of siRNA and 1 µl of Lipofectamine RNAiMAX Transfection Reagent (#13778030; Thermo Fisher Scientific) were diluted separately in two vials of 150 µl Opti-MEM (#31985062; Thermo Fisher Scientific) and kept at RT for 5 min while flicking once every 2 min. Then siRNA solution was added to the reagent solution and incubated for 30 min at RT while flicking once every 10 min. The growth medium was replaced with Opti-MEM + 10% FBS after washing the cells with PBS (both prewarmed to 37°C), and then 300 µl siRNA/reagent cocktail was added and mixed by swirling. The transfected cells were incubated for 24 h at 37°C/5% CO2, before being detached and reattached onto appropriate imaging dishes for nuclear transport, cell fusion, or immunofluorescence assays. The assays were performed 48 h after transfection for NUP RNAi and 60–72 h after transfection for OGT/OGA RNAi. 50–75% depletion of NUP mRNAs and 80–90% depletion of OGT/OGA mRNAs were verified by RT-PCR using QuantiTect Reverse Transcription Kit (#205311; Qiagen), QuantiTect SYBR Green PCR Kit (#204143; Qiagen), and Bio-Rad C1000 (base) with CFX96 (optics).
Drug treatments
Cells were treated with 1 µM KPT-330 (#S7252; Selleckchem) for 30 min for exportin-1 inhibition. OGA inhibitor Thiamet-G was obtained from Cayman Chemicals (#13237), and the second-generation OGT inhibitor OSMI-4 (gift from Suzanne Walker) was obtained as described previously (Martin et al., 2018).
Plasmid transfection
For nuclear import inhibition (Fig. 1 E), psfGFP-bimax2 expression vector was constructed by inserting synthesized DNA fragment (IDT) for bimax2 sequence (RRRRRRKRKREWDDDDDPPKKRRRLD) into XhOI/EcoRI site of psfGFP-C1 vector. Cells were transfected with psfGFP-bimax2 and psfGFP-C1 vectors using TransIT-2020 transfection reagent (#MIR5404; Mirus) and imaged after 6.5 h. For RanGTP immunofluorescence assay validation (Fig. S2 C), mCherry-N1, RanWT-mRFP1 (#59750; Addgene), RanQ69L-mCherry (#30309; Addgene), RanT24N-mCherry (#37396; Addgene), and Rcc1-HA (#106938; Addgene) were transfected using TransIT-2020 24 h before immunostaining.
Cell fusion assay
Two different U2OS stable cell lines, one expressing GFP-H2A and the other expressing the import probe, were grown in a six-well plate and separately transfected with siRNA targeting OGT or OGA as described above. After 24 h, each well was washed with PBS once and treated with 300 µl of trypsin in a tissue culture (TC) incubator for ∼5 min. 700 µl complete DMEM was added to each well and pipetted up and down to detach and dissociate cells. The entire 1-ml cell suspension was transferred to a 1.5-ml tube, and the cell density was measured. ∼105 cells from each cell suspension were combined into another 1.5-ml tube, mixed by brief vortex, and centrifuged at 300 g for 4 min. After gentle removal of the supernatant, the cell pellet was resuspended in 1 ml complete DMEM. The resuspension was filtered through a 35-µm cell strainer and transferred to a 35-mm Petri dish with no. 1.5 poly-d-lysine–coated glass bottom (#P35GC-1.5-14-C; MatTek) with an additional 1 ml complete DMEM. After 2 d, the coculture was washed twice in PBS and treated with ∼300 µl of 50% PEG-1500 (#10783641001; Millipore Sigma) for 2 min at RT after completely aspirating the residual PBS. 3 ml PBS was gently added to the dish, swirled, and aspirated. The cells were gently washed twice more in 3 ml PBS and once in prewarmed imaging medium and placed in 1.5 ml imaging medium containing 1 µM SiR-DNA (CY-SC007; Cytoskeleton) + 10 µM Verapamil (for nucleus staining) and 10 µM OSMI-4 + 10 µM Thiamet-G (to suppress O-GlcNAc turnover). After resting in a TC incubator for 2–4 h, cells were imaged for nuclear import measurement and analyzed as described above or fixed and stained for immunofluorescence analysis.
Superresolution microscopy imaging of O-GlcNAc modification and Nup96
Generation of Nup96-GFP CRISPR-engineered cell line
The C-terminus of endogenous Nup96 in U2OS cells was tagged via homozygous insertion of mEGFP gene into NUP98 gene (encoding Nup98-Nup96 precursor) using CRISPR-Cas9 genome editing as described previously (Koch et al., 2018; Thevathasan et al., 2019). Briefly, pX335 vectors (#42355; Addgene) carrying SpCas9 nickase mutant (D10A) and a pair of sgRNAs, targeting 5′-GTTGGGAGCCTGTGAGCCCC-3′ and 5′-CAGTTCTCGCAGATAGGACT-3′, were used to induce a double-strand break near the C-terminal end of Nup96. Homology-directed repair donor plasmid was prepared by cloning 0.9- and 1-kb homology arms flanking mEGFP gene into pUC19 vector. An eight-residue linker, SACYCELS, was placed between Nup96 and mEGFP. PAM sequences of the sgRNAs in the donor plasmid were muted by site-directed mutagenesis, using QuikChange site-directed mutagenesis kit (#200515; Agilent). U2OS cells were transfected with the plasmids by electroporation, using a Nucleofector 2b device (Lonza) and Ingenio electroporation kit (#50117; Mirus). After 5 d of transfection, individual GFP-positive cells were sorted into 96-well plates and expanded to monoclonal cell lines. Homozygous mEGFP insertion at the C-terminal end of Nup96 was verified by genotyping PCR.
Fluorescent labeling of O-GlcNAc modification and Nup96
All samples for superresolution microscopy (presented in Fig. 4) were prepared in 35-mm Petri dishes with no. 1.5 poly-d-lysine–coated glass bottoms (#P35GC-1.5-14-C; MatTek). For metabolic labeling of O-GlcNAc modifications, Nup96-GFP U2OS cells were grown for 2 d in complete DMEM containing 1 mM N-GlcNAz (#MA30911; Carbosynth), a GlcNAc analogue whose acetamido group has an azide moiety. Cells were fixed with PFA and incubated with AF647-conjugated anti-GFP single-domain antibody (FluoTag-X4, #N0304-AF647; NanoTag Biotech) to fluorescently label the C-terminus of Nup96, as described previously (Thevathasan et al., 2019). Briefly, samples were prefixed for 30 s in TRB (20 mM Hepes, pH 7.5, 110 mM KAc, 1 mM EGTA, and 250 mM sucrose) containing 2.4% PFA; washed twice in TRB; treated with 25 µg/ml digitonin in TRB on ice (membrane-specific permeabilization); washed twice in TBA (1% BSA in TRB); incubated in TBA containing 100 nM antibody for 30 min on ice (first round of staining); washed twice in TBA; further fixed in TBA containing 3% PFA for 10 min; washed twice in TBA; permeabilized by 0.4% Triton X-100 in PBS for 3 min; washed twice in PBS; incubated in 50 nM antibody for 30 min on ice (second round of staining); and washed twice in PBS. Subsequently, incorporated GlcNAz was labeled with alkyne-CF568 (#92088; Biotium) via CuAAC “click” reaction as follows: 100 mM CuSO4 and 50 mM BTTAA (#1236; Click Chemistry Tools) were premixed at a 1:5 ratio. 500 µl of CuAAC reaction cocktail was prepared by sequentially adding 0.5 µl of 10 mM alkyne-CF568, 110 µl of CuSO4:BTTAA premix, and 50 µl of 1 M sodium ascorbate into 340 µl of 100 mM sodium phosphate buffer, pH 7.0, with a brief vortex after each addition. Cells were washed once in PBS with 3% BSA and then incubated in 500 µl of the CuAAC reaction cocktail for 30 min at RT while being protected from light. Samples were then washed twice in PBS with 3% BSA and twice in PBS for 10 min each. GlcNAc/Nup96 double-stained samples were stored in a sealed Petri dish covered with aluminum foil at 4°C for less than a month before being imaged.
3D SIM
Buffer was exchanged for a mounting medium (90% glycerol, 0.5% propyl-gallate, and 20 mM Tris-HCl, pH 8.0) and sealed. 3D SIM images were acquired on a GE DeltaVision OMX Blaze with Olympus 60×/1.42-NA Plan Apo oil-immersion objective and front-illuminated scientific-CMOS camera (PCO). For GlcNAz:CF568 imaging, 568-nm excitation laser, 571/19 (center wavelength/bandwidth) excitation filter, and 609/37 emission filter were used. For Nup96-GFP:AF647 imaging, 642-nm excitation laser, 645.5/15 excitation filter, and 683/40 emission filter were used. 40–60 optical sections were imaged every 125 nm for each channel. 3D SIM reconstruction was performed using a CUDA-accelerated version of the conventional algorithm (Gustafsson et al., 2008), using measured, wavelength-specific optical transfer functions matched to the data (code available at https://github.com/scopetools/cudasirecon). The final reconstructed 3D SIM images have a 40.96 × 40.96–µm (1,024 × 1,024–pixel) field of view and 135–160-nm lateral and 350–380-nm axial resolution. Channel registration was achieved via maximization of cross-correlation.
STORM setup
STORM data were acquired on Nikon Ti motorized inverted microscope with Perfect Focus System, equipped with Nikon total internal reflection fluorescence (TIRF) illuminator and Prior Proscan II motorized stage, filter wheels, and shutters. For illumination, we used the output of Agilent MLC400B monolithic laser combiner with 405-, 561-, and 647-nm laser lines, reflected by a quad-band dichroic beamsplitter (ZT405/488/561/647rpc; Chroma). Emitted fluorescence was collected by a CFI Apo TIRF 100×/1.49-NA oil-immersion objective (Nikon) and filtered by the dichroic beamsplitter, additional quad-band emission filter (ZET405/488/561/647m; Chroma), and an emission filter (ET600/50m or ET700/75m; Chroma) for CF568 and AF647 emissions, respectively. The filtered emission was projected onto electron-multiplying charge-coupled device camera (Ultra DU-897U-CS0-#BV; iXon). Additional 1.5× intermediate magnification was applied, which resulted in a pixel size of 104.3 nm. NIS-Elements software was used to control the hardware.
STORM acquisition
We used GLOX/MEA/COT STORM buffer, which is 86.5 mM Tris-HCl, pH 8, 8.65 mM NaCl, 13.65% (wt/vol) glucose, 560 µg/ml glucose oxidase (#G2133; Millipore Sigma), 40 µg/ml catalase (#C100; Millipore Sigma), 35 mM cysteamine (MEA, #30070; Millipore Sigma), and 2 mM cyclooctatetraene (COT, #138924; Millipore Sigma). The buffer was prepared fresh by mixing stock solutions right before imaging and replaced every 90 min. The sample in a 35-mm glass-bottom dish was briefly washed once in 0.5 ml STORM buffer, placed in 1.5 ml STORM buffer, and clipped down on the microscope stage. Nup96-GFP:AF647 was live-imaged briefly with weak (0.5%) 647-nm illumination to adjust xy position and focus, as well as adjust the TIRF angle, to achieve highly inclined and laminated optical sheet illumination (Tokunaga et al., 2008). The focus was set to either basal plane or midplane of the nucleus so as to acquire top and side views of NPCs, respectively. STORM data for Nup96-GFP:AF647 and GlcNAz:CF568 were acquired sequentially using maximum-power 647- and 568-nm lasers, respectively, while the power of 405-nm stimulation was modulated constantly so as to have 20–40 molecules per frame. ∼6,000 and 40,000 frames were imaged for Nup96-GFP:AF647 and GlcNAz:CF568, respectively, with 30-ms exposure time. The camera was set to a readout rate of 10 MHz, preamplifier setting 1, and EM gain 300× throughout imaging.
STORM postprocessing
STORM image reconstruction was performed using ImageJ plugin ThunderSTORM (Ovesný et al., 2014) as follows. The differences of Gaussian filters with SDs 1 and 1.6 pixels and local maximum detector were used to identify single molecule spots. Each spot was fitted with a symmetric Gaussian PSF model using maximum likelihood estimation. The xy positions were corrected for residual drift by redundant cross-correlation–based drift correction method. Localizations were filtered by the localization precision (<10 nm) and the size of Gaussian PSF (σ < 150 nm). Localizations that appeared in consecutive frames (with a maximum gap of one missing frame) within 30 nm from one another were merged into one localization. Each single-molecule coordinate was rendered as a normalized 2D Gaussian whose SD is equal to the localization precision, where the pixel size of the reconstructed images was set to 1/20 of the raw image pixel size, i.e., 104.3/20 = 5.215 nm. Reconstructed STORM images of Nup96-GFP:AF647 and GlcNAz:CF568 were registered by a custom algorithm written in Python. Briefly, a 2D histogram of single-molecule localizations was constructed for each channel where the bin size was set to 20 × 20 nm. Cross-correlation between two 2D histograms was calculated, and the peak of the cross-correlation was localized to a subbin accuracy by fitting the peak to a symmetric 2D Gaussian using Levenberg–Marquardt least squares optimization. Finally, Nup96-GFP:AF647 STORM image was translated by the offset of the cross-correlation peak from the center.
Particle averaging
The top view average of the NPC was computed using a custom Python code as follows. NPC particles were detected based on the basal nuclear plane STORM image of AF647, by applying Gaussian blurring and detecting the local maxima. Anomalous particles were filtered out based on the peak CF568 and AF647 intensities. xy positions of AF647 within the 240 × 240–nm window were fitted to a circle using robust least squares algorithm, which minimizes
where and r are the center position and radius of the circle, is the localization precision of ith AF647 position , and is the soft l1 loss function. The low-quality NPC particles were further filtered out based on the following exclusion criteria: (1) r < 40 or > 60 nm; (2) standard errors of r, xc, yc > 5 nm; or (3) number of AF647 localizations in the window <15. The 240 × 240-nm STORM images of the remaining NPC particles were translationally aligned based on the center positions of the fitted circles and averaged. The side view average was obtained using the midplane STORM images, by manually determining the center and orientation of the NPCs using Fiji software and translationally and rotationally aligning the NPCs using a custom Python code. No cell-to-cell variation in the averaged images was observed.
Immunofluorescence
Sample preparation
All immunofluorescence samples except those for superresolution microscopy shown in Fig. 4 were prepared as follows. Cells were fixed using 4% PFA/PBS for 10 min, permeabilized using 0.1% Triton X-100/PBS for 10 min, and blocked using 1% BSA/PBS for 1 h. Primary antibodies were diluted using the following ratios in 1% BSA/PBS: anti-O-GlcNAc (RL2, 1:200, #ab2739; Abcam), anti-Rcc1 (1:50, #3589S; Cell Signaling), anti-Ran (1:800, #ab53775; Abcam), anti-RanGAP1 (1F3A4, 1:100, #67146-1-Ig; Proteintech), anti-FG-NUPs (MAb414, 1:1,000, #902901; BioLegend), anti-RanGTP (AR12, 1:200, gift from Ian Macara), anti-importin-β1 (3E9, 1:1,000, #MA3-070; Thermo Fisher Scientific), anti-mCherry (8C5.5, 1:500, #677701; BioLegend), and anti-HA (16B12, 1:1,000, #901516; BioLegend). Cells were incubated with the diluted primary antibodies for 1 h at RT or overnight at 4°C in a humidified chamber. The cells were incubated with goat anti-rabbit or anti-mouse secondary antibody conjugated with AF488 or AF568 (#A11008/#A11001/#A11031; Thermo Fisher Scientific) or donkey anti-mouse secondary antibody conjugated with AF647 (#A13571), diluted (1:1,000 ratio) in 1% BSA/PBS for 1 h at RT. Cells were washed in PBS for 5 min three times after each incubation step. GFP was detected by using AF647-conjugated anti-GFP single-domain antibody (FluoTag-X4, #N0304-AF647; NanoTag Biotech) at 1:500 dilution for 1 h at RT. SiR-DNA (#CY-SC007; Cytoskeleton), JF646-HaloTag ligand (gift from Luke Lavis), or Hoechst 33258 (#09460; Polyscience) was used for nuclei staining.
Image acquisition
Immunofluorescence images shown in Fig. 3 and Fig. S3 were acquired using a Nikon Ti motorized inverted microscope equipped with Perfect Focus System, Yokagawa CSU-X1 spinning, Spectral Applied Research LMM-5 laser merge module, and Hamamatsu ORCA-R2 cooled charge-coupled device camera. CFI Plan Apo 20×/0.75-NA objective lens (Nikon) was used for Fig. 3, while CFI Plan Apo Lambda 100×/1.45-NA oil objective lens (Nikon) was used for NPC localization analysis of importin-β1 (Fig. S3 A). A 488-nm laser and ET525/50m filter (Chroma) were used for AF488 imaging, 561-nm laser and ET620/60m filter (Chroma) for AF568, and 642-nm laser and ET700/75m filter (Chroma) for AF647 and JF646. A Di01-T405/488/568/647 beam splitter (Semrock) was commonly used for all channels. MetaMorph software was used to control the microscope. Immunofluorescence images shown in Fig. 2 and Fig. S2 were acquired using a Nikon Eclipse Ti2 motorized inverted microscope equipped with Perfect Focus System, Lumencor sola light engine, CFI Plan Apochromat Lambda 20×/0.75-NA objective lens (Nikon), and Andor Zyla 4.2 sCMOS. DAPI filter set (ET395/25x excitation, ET460/50m emission, T425lpxr), FITC filter set (ET470/40x excitation, ET525/50m emission, T495lpxr), TRITC filter set (ET545/25x excitation, ET605/70m emission, T565lpxr mirror), and Cy5 filter set (ET640/30x excitation, ET690/50m emission, T660lpxr mirror) were used for Hoechst, AF488, AF568, and AF647 imaging, respectively (all filters from Chroma). NIS-Elements software was used to control the hardware.
Image analysis
U-Net convolutional neural network (described above) or ilastik (Berg et al., 2019) was used for nucleus and cytoplasm segmentations. Importin-β1 localization analysis (Fig. 3 A) was performed as follows. The cytoplasmic background level, determined by finding the mode intensity value of the Gaussian-blurred image (σ = 2 pixels), was subtracted from each Nup96-GFP:AF647 and importin-β1:AF568 image. A nuclear mask was generated based on the Nup96 image by Gaussian blurring, thresholding, and removal of small holes. The background-subtracted pixel intensities of importin-β1 vs. Nup96 within the nuclear mask were fitted to a linear model by robust least squares fitting algorithm using soft l1 loss. The slope of the linear model was used as a measure of the NPC localization of importin-β1.
Photoactivation assay for passive permeation rate measurement
U2OS cells were transfected with mEOS4b-C1 vector (#54812; Addgene) using Nucleofector 2b device (Lonza) and Ingenio electroporation kit (#50117; Mirus) and seeded onto a 35-mm Petri dish with no. 1.5 poly-d-lysine–coated glass bottom (#P35GC-1.5-14-C; MatTek). After resting for 4–6 h in the complete DMEM in the TC incubator, cells were incubated in complete DMEM containing 10 µM OSMI-4, 10 µM Thiamet-G, or DMSO for 22 h. 1–3 h before imaging, the drug-containing complete DMEM was replaced by the imaging medium containing the same drug and 500 nM SiR-DNA (CY-SC007; Cytoskeleton) + 5 µM Verapamil. Photoactivation assay was performed on GE DeltaVision OMX Blaze, the same instrument used for 3D SIM described above, while the sample was maintained at 37°C by stage-top heater and objective heater. 568-nm laser, 571/19 (center wavelength/bandwidth) excitation filter, and 609/37 emission filter were used for imaging converted mEOS, while 642-nm laser, 645.5/15 excitation filter, and 683/40 emission filter were used for imaging SiR-DNA. Images were taken every 5 s for 15 min with 50-ms exposure time. After the third time point, a 0.5-s pulse of 100%-power 405-nm laser was shone onto a rectangular region inside the nucleus by high-speed galvo to selectively convert mEOS in the nucleus. The time course of the nuclear intensity of the converted mEOS was fitted to a biexponential model,
where and are the passive permeation rate and the bleaching rate, respectively. We did not observe any significant difference in the bleaching rate between different O-GlcNAc perturbation conditions (not depicted).
Statistical analysis
All reported point estimations are medians or calculated based on the median values (e.g., percentage change in nuclear transport rates) unless stated otherwise. All reported P values were calculated using two-sided Welch’s t test, after removing outliers higher than Q3 + 1.5 × interquartile range (IQR) or lower than Q1 – 1.5IQR, where Q1 and Q3 are the first and third quartiles and IQR is Q3 – Q1. Data distribution was assumed to be normal but this was not formally tested. Batch effect was examined for all data. For the data showing batch effects (Fig. 2 D, Fig. S2 B, and Fig. S3, A and B), each batch was normalized such that the median of the negative control (e.g., DMSO-treated cells) was equal to 1.
Online supplemental material
Fig. S1 summarizes the results of nuclear import and export rate measurements performed in this study. Fig. S2 shows the nuclear and cytoplasmic localization of Ran, Rcc1, RanGAP1, and RanGTP measured after O-GlcNAc perturbations. Fig. S3 shows importin-β1 localization to NPCs and protein levels of nuclear transport machinery after O-GlcNAc perturbations. Video 1 shows 3D SIM z-stack of Nup96-GFP:AF647 (green, left) and GlcNAz:CF568 (magenta, right), Z-spacing 130 nm.
Acknowledgments
We thank Prof. Suzanne Walker, Dr. Zebulon Levine, and George Fei (all from the Harvard Medical School, Boston, MA) for providing OSMI-4 and RL2 antibody and advice on O-GlcNAc experiments; Prof. Ian Macara (Vanderbilt University, Nashville, TN) for providing anti-RanGTP antibody; Dr. Luke Lavis (Janelia Research Campus, Ashburn, VA) for providing JF dyes; and Dr. Yuyu Song (Massachusetts General Hospital, Boston, MA) for providing anti-Ran antibody. We also thank the Nikon Imaging Center and Cell Biology Microscopy Facility at Harvard Medical School, especially Dr. Talley Lambert, for help with light microscopy and the Methods section; and Dr. Juhyun Oh for helpful discussions.
This study was supported by National Institute of General Medical Sciences (National Institutes of Health) research grant R35GM131753 and postdoctoral fellowship F32GM131585.
The authors declare no competing financial interests.
Author contributions: T. Yoo and T.J. Mitchison conceived the study and wrote the manuscript. T. Yoo designed and performed the experiments and analyzed the data with input from T.J. Mitchison.
References
- Ball, J.R., and Ullman K.S.. 2005. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 114:319–330. 10.1007/s00412-005-0019-3 [DOI] [PubMed] [Google Scholar]
- Becskei, A., and Mattaj I.W.. 2005. Quantitative models of nuclear transport. Curr. Opin. Cell Biol. 17:27–34. 10.1016/j.ceb.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Berg, S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. 2019. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 16:1226–1232. 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- Bernad, R., Engelsma D., Sanderson H., Pickersgill H., and Fornerod M.. 2006. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J. Biol. Chem. 281:19378–19386. 10.1074/jbc.M512585200 [DOI] [PubMed] [Google Scholar]
- Boyce, M., Carrico I.S., Ganguli A.S., Yu S.H., Hangauer M.J., Hubbard S.C., Kohler J.J., and Bertozzi C.R.. 2011. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. USA. 108:3141–3146. 10.1073/pnas.1010045108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressloff, P.C., and Newby J.M.. 2013. Stochastic models of intracellular transport. Rev. Mod. Phys. 85:135–196. 10.1103/RevModPhys.85.135 [DOI] [Google Scholar]
- Caicedo, J.C., Roth J., Goodman A., Becker T., Karhohs K.W., Broisin M., Molnar C., McQuin C., Singh S., Theis F.J., and Carpenter A.E.. 2019. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. Cytometry A. 95:952–965. 10.1002/cyto.a.23863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatham, J.C., Zhang J., and Wende A.R.. 2021. Role of O-Linked N-acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 101:427–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, U.H., and Hetzer M.W.. 2020. Nuclear Periphery Takes Center Stage: The Role of Nuclear Pore Complexes in Cell Identity and Aging. Neuron. 106:899–911. 10.1016/j.neuron.2020.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, M., Chang C.W., Róna G., Smith K.M., Stewart A.G., Takeda A.A., Fontes M.R., Stewart M., Vértessy B.G., Forwood J.K., and Kobe B.. 2016. Structural Biology and Regulation of Protein Import into the Nucleus. J. Mol. Biol. 428(10, 10 Pt A):2060–2090. 10.1016/j.jmb.2015.10.023 [DOI] [PubMed] [Google Scholar]
- Comer, F.I., and Hart G.W.. 1999. O-GlcNAc and the control of gene expression. Biochim. Biophys. Acta. 1473:161–171. 10.1016/S0304-4165(99)00176-2 [DOI] [PubMed] [Google Scholar]
- D’Angelo, M.A., Raices M., Panowski S.H., and Hetzer M.W.. 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 136:284–295. 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., and Ellenberg J.. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, R.L., and Gerald P.S.. 1976. Improved techniques for the induction of mammalian cell hybridization by polyethylene glycol. Somatic Cell Genet. 2:165–176. 10.1007/BF01542629 [DOI] [PubMed] [Google Scholar]
- Davis, L.I., and Blobel G.. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA. 84:7552–7556. 10.1073/pnas.84.21.7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D.P., Patel S.S., Uversky V., Fink A.L., and Rexach M.. 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 100:2450–2455. 10.1073/pnas.0437902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr, C.M., and Akin D.. 1997. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 110:3065–3070. [DOI] [PubMed] [Google Scholar]
- Fried, H., and Kutay U.. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659–1688. 10.1007/s00018-003-3070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst, P., Guan T., Subauste C., Hahn K., and Gerace L.. 2002. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156:617–630. 10.1083/jcb.200106046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima, J.C., Daigle J.G., Arbez N., Cunningham K.C., Zhang K., Ochaba J., Geater C., Morozko E., Stocksdale J., Glatzer J.C., et al. 2017. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron. 94:93–107.e6. 10.1016/j.neuron.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., and Sedat J.W.. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim, A., Larsen I.S., Neubert P., Joshi H.J., Petersen B.L., Vakhrushev S.Y., Strahl S., and Clausen H.. 2015. Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc. Natl. Acad. Sci. USA. 112:15648–15653. 10.1073/pnas.1511743112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover, J.A., Cohen C.K., Willingham M.C., and Park M.K.. 1987. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262:9887–9894. 10.1016/S0021-9258(18)48017-9 [DOI] [PubMed] [Google Scholar]
- Hao, Y., Fan X., Shi Y., Zhang C., Sun D.E., Qin K., Qin W., Zhou W., and Chen X.. 2019. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat. Commun. 10:4065. 10.1038/s41467-019-11942-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, G.D., Snow C.M., Senior A., Haltiwanger R.S., Gerace L., and Hart G.W.. 1987. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J. Cell Biol. 104:1157–1164. 10.1083/jcb.104.5.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, L.E., Dutta K., Sparks S., Temel D.B., Kamal A., Tetenbaum-Novatt J., Rout M.P., and Cowburn D.. 2015. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 4:e10027. 10.7554/eLife.10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten, S., and Dormann D.. 2020. Nucleocytoplasmic transport defects in neurodegeneration - Cause or consequence? Semin. Cell Dev. Biol. 99:151–162. [DOI] [PubMed] [Google Scholar]
- Hutten, S., and Kehlenbach R.H.. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26:6772–6785. 10.1128/MCB.00342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer, O., and Peters R.. 1999. Permeability of single nuclear pores. Biophys. J. 77:217–228. 10.1016/S0006-3495(99)76883-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A.U., Qu R., Ouyang J., and Dai J.. 2020. Role of Nucleoporins and Transport Receptors in Cell Differentiation. Front. Physiol. 11:239. 10.3389/fphys.2020.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J., and Taylor J.P.. 2017. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron. 96:285–297. 10.1016/j.neuron.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J., Fernandez-Martinez J., Nudelman I., Shi Y., Zhang W., Raveh B., Herricks T., Slaughter B.D., Hogan J.A., Upla P., et al. 2018. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 555:475–482. 10.1038/nature26003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe, C., Bischoff F.R., Ponstingl H., and Wittinghofer A.. 1995. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 34:639–647. 10.1021/bi00002a031 [DOI] [PubMed] [Google Scholar]
- Koch, B., Nijmeijer B., Kueblbeck M., Cai Y., Walther N., and Ellenberg J.. 2018. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat. Protoc. 13:1465–1487. 10.1038/nprot.2018.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konold, P.E., Mathes T., Weiβenborn J., Groot M.L., Hegemann P., and Kennis J.T.. 2016. Unfolding of the C-Terminal Jα Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. J. Phys. Chem. Lett. 7:3472–3476. 10.1021/acs.jpclett.6b01484 [DOI] [PubMed] [Google Scholar]
- Kosugi, S., Hasebe M., Entani T., Takayama S., Tomita M., and Yanagawa H.. 2008. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 15:940–949. 10.1016/j.chembiol.2008.07.019 [DOI] [PubMed] [Google Scholar]
- Kreppel, L.K., Blomberg M.A., and Hart G.W.. 1997. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272:9308–9315. 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- Labokha, A.A., Gradmann S., Frey S., Hülsmann B.B., Urlaub H., Baldus M., and Görlich D.. 2013. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32:204–218. 10.1038/emboj.2012.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapalombella, R., Sun Q., Williams K., Tangeman L., Jha S., Zhong Y., Goettl V., Mahoney E., Berglund C., Gupta S., et al. 2012. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 120:4621–4634. 10.1182/blood-2012-05-429506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and Kohler J.J.. 2014. Glycosylation of the nuclear pore. Traffic. 15:347–361. 10.1111/tra.12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.H., and Hoelz A.. 2019. The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 88:725–783. 10.1146/annurev-biochem-062917-011901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.L., Timney B.L., Rout M.P., and Wente S.R.. 2015. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J. Cell Biol. 208:729–744. 10.1083/jcb.201412024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löschberger, A., van de Linde S., Dabauvalle M.C., Rieger B., Heilemann M., Krohne G., and Sauer M.. 2012. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125:570–575. 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- Lubas, W.A., Frank D.W., Krause M., and Hanover J.A.. 1997. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272:9316–9324. 10.1074/jbc.272.14.9316 [DOI] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. 10.1128/MMBR.65.4.570-594.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, R., Delphin C., Guan T., Gerace L., and Melchior F.. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 88:97–107. 10.1016/S0092-8674(00)81862-0 [DOI] [PubMed] [Google Scholar]
- Martin, S.E.S., Tan Z.W., Itkonen H.M., Duveau D.Y., Paulo J.A., Janetzko J., Boutz P.L., Törk L., Moss F.A., Thomas C.J., et al. 2018. Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors. J. Am. Chem. Soc. 140:13542–13545. 10.1021/jacs.8b07328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, Y. 2016. Mechanistic Insights from Structural Analyses of Ran-GTPase-Driven Nuclear Export of Proteins and RNAs. J. Mol. Biol. 428(10, 10 Pt A):2025–2039. 10.1016/j.jmb.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas E., and Blobel G.. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. 10.1083/jcb.135.6.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles, S., Mercadante D., Aramburu I.V., Jensen M.R., Banterle N., Koehler C., Tyagi S., Clarke J., Shammas S.L., Blackledge M., et al. 2015. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 163:734–745. 10.1016/j.cell.2015.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi-Hata, C., Ogawa Y., Oka M., and Yoneda Y.. 2013. Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochim. Biophys. Acta. 1833:2682–2689. 10.1016/j.bbamcr.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Nagata, Y., and Burger M.M.. 1974. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J. Biol. Chem. 249:3116–3122. 10.1016/S0021-9258(19)42646-X [DOI] [PubMed] [Google Scholar]
- Nemergut, M.E., and Macara I.G.. 2000. Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835–850. 10.1083/jcb.149.4.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek, D., Benzinger D., Roensch J., Draebing T., Wehler P., Eils R., and Di Ventura B.. 2014. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun. 5:4404. 10.1038/ncomms5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek, D., Wehler P., Roensch J., Eils R., and Di Ventura B.. 2016. Optogenetic control of nuclear protein export. Nat. Commun. 7:10624. 10.1038/ncomms10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný, M., Křížek P., Borkovec J., Svindrych Z., and Hagen G.M.. 2014. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30:2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.K., Zhou X., Pendleton K.E., Hunter O.V., Kohler J.J., O’Donnell K.A., and Conrad N.K.. 2017. A Conserved Splicing Silencer Dynamically Regulates O-GlcNAc Transferase Intron Retention and O-GlcNAc Homeostasis. Cell Rep. 20:1088–1099. 10.1016/j.celrep.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, M.A., Forbes D.J., Dahlberg J.E., and Lund E.. 1997. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 136:241–250. 10.1083/jcb.136.2.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, M.A., Macaulay C., Masiarz F.R., and Forbes D.J.. 1995. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128:721–736. 10.1083/jcb.128.5.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, W., Qin K., Fan X., Peng L., Hong W., Zhu Y., Lv P., Du Y., Huang R., Han M., et al. 2018. Artificial Cysteine S-Glycosylation Induced by Per-O-Acetylated Unnatural Monosaccharides during Metabolic Glycan Labeling. Angew. Chem. Int. Ed. Engl. 57:1817–1820. 10.1002/anie.201711710 [DOI] [PubMed] [Google Scholar]
- Rabut, G., Doye V., and Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Rempel, I.L., Crane M.M., Thaller D.J., Mishra A., Jansen D.P., Janssens G., Popken P., Akşit A., Kaeberlein M., van der Giessen E., et al. 2019. Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife. 8:e48186. 10.7554/eLife.48186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., and Görlich D.. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S.A., Lounsbury K.M., and Macara I.G.. 1995. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J. Biol. Chem. 270:14405–14411. 10.1074/jbc.270.24.14405 [DOI] [PubMed] [Google Scholar]
- Snow, C.M., Senior A., and Gerace L.. 1987. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 104:1143–1156. 10.1083/jcb.104.5.1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, G.J., Fassati A., and Hoogenboom B.W.. 2017. Biomechanics of the transport barrier in the nuclear pore complex. Semin. Cell Dev. Biol. 68:42–51. 10.1016/j.semcdb.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Tan, P.S., Aramburu I.V., Mercadante D., Tyagi S., Chowdhury A., Spitz D., Shammas S.L., Gräter F., and Lemke E.A.. 2018. Two Differential Binding Mechanisms of FG-Nucleoporins and Nuclear Transport Receptors. Cell Rep. 22:3660–3671. 10.1016/j.celrep.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Z.W., Fei G., Paulo J.A., Bellaousov S., Martin S.E.S., Duveau D.Y., Thomas C.J., Gygi S.P., Boutz P.L., and Walker S.. 2020. O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res. 48:5656–5669. 10.1093/nar/gkaa263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevathasan, J.V., Kahnwald M., Cieśliński K., Hoess P., Peneti S.K., Reitberger M., Heid D., Kasuba K.C., Hoerner S.J., Li Y., et al. 2019. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods. 16:1045–1053. 10.1038/s41592-019-0574-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, M., Imamoto N., and Sakata-Sogawa K.. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161. 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Ullman, K.S., Shah S., Powers M.A., and Forbes D.J.. 1999. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell. 10:649–664. 10.1091/mbc.10.3.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Fornerod M., Pickersgill H., Goldberg M., Allen T.D., and Mattaj I.W.. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703–5714. 10.1093/emboj/20.20.5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Yuan Z.F., Fan J., Karch K.R., Ball L.E., Denu J.M., and Garcia B.A.. 2016. A Novel Quantitative Mass Spectrometry Platform for Determining Protein O-GlcNAcylation Dynamics. Mol. Cell. Proteomics. 15:2462–2475. 10.1074/mcp.O115.049627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, M.L. 1959. Further observations on the nuclear envelope of the animal cell. J. Biophys. Biochem. Cytol. 6:147–156. 10.1083/jcb.6.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, L., Gao Y., Mahoney J.A., Vosseller K., Chen C., Rosen A., and Hart G.W.. 2002. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 277:1755–1761. 10.1074/jbc.M109656200 [DOI] [PubMed] [Google Scholar]
- Woo, C.M., Lund P.J., Huang A.C., Davis M.M., Bertozzi C.R., and Pitteri S.J.. 2018. Mapping and Quantification of Over 2000 O-linked Glycopeptides in Activated Human T Cells with Isotope-Targeted Glycoproteomics (Isotag). Mol. Cell. Proteomics. 17:764–775. 10.1074/mcp.RA117.000261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, C.S., and Kellogg G.E.. 1996. Differences in hydropathic properties of ligand binding at four independent sites in wheat germ agglutinin-oligosaccharide crystal complexes. Protein Sci. 5:1466–1476. 10.1002/pro.5560050803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Kasper L.H., Mantcheva R.T., Mantchev G.T., Springett M.J., and van Deursen J.M.. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA. 98:3191–3196. 10.1073/pnas.051631598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa, S.A., Macauley M.S., Heinonen J.E., Shan X., Dennis R.J., He Y., Whitworth G.E., Stubbs K.A., McEachern E.J., Davies G.J., and Vocadlo D.J.. 2008. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4:483–490. 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- Zaro, B.W., Yang Y.Y., Hang H.C., and Pratt M.R.. 2011. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl. Acad. Sci. USA. 108:8146–8151. 10.1073/pnas.1102458108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayner, J.P., Antoniou C., and Sosnick T.R.. 2012. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J. Mol. Biol. 419:61–74. 10.1016/j.jmb.2012.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Tan E.P., VandenHull N.J., Peterson K.R., and Slawson C.. 2014. O-GlcNAcase Expression is Sensitive to Changes in O-GlcNAc Homeostasis. Front. Endocrinol. (Lausanne). 5:206. 10.3389/fendo.2014.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Liu T.W., Cecioni S., Eskandari R., Zandberg W.F., and Vocadlo D.J.. 2015. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 11:319–325. 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Liu T.W., Madden Z., Yuzwa S.A., Murray K., Cecioni S., Zachara N., and Vocadlo D.J.. 2016. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J. Mol. Cell Biol. 8:2–16. 10.1093/jmcb/mjv033 [DOI] [PMC free article] [PubMed] [Google Scholar]