The vaginal microbiota is defined as the community of bacteria residing in the human vaginal tract. Recent studies have demonstrated that the high prevalence of Lactobacillus crispatus species is commonly associated with a healthy vaginal environment.
KEYWORDS: lactobacilli, vaginal microbiota, comparative genomics
ABSTRACT
The vaginal microbiota is defined as the community of bacteria residing in the human vaginal tract. Recent studies have demonstrated that the vaginal microbiota is dominated by members of the Lactobacillus genus, whose relative abundance and microbial taxon composition are dependent on the healthy status of this human body site. Particularly, among members of this genus, the high prevalence of Lactobacillus crispatus is commonly associated with a healthy vaginal environment. In the current study, we assessed the microbial composition of 94 healthy vaginal microbiome samples through shotgun metagenomics analyses. Based on our results, we observed that L. crispatus was the most representative species and correlated negatively with bacteria involved in vaginal infections. Therefore, we isolated 15 L. crispatus strains from different environments in which this species abounds, ranging from vaginal swabs of healthy women to chicken fecal samples. The genomes of these strains were decoded and their genetic content was analyzed and correlated with their physiological features. An extensive comparative genomic analysis encompassing all publicly available genome sequences of L. crispatus and combined with those decoded in this study revealed a genetic adaptation of strains to their respective ecological niche. In addition, in vitro growth experiments involving all isolated L. crispatus strains, together with a synthetic vaginal microbiota, reveal how this species is able to modulate the composition of the vaginal microbial consortia at the strain level. Overall, our findings suggest that L. crispatus plays an important ecological role in reducing the complexity of the vaginal microbiota by depleting pathogenic bacteria.
IMPORTANCE The vaginal microbiota is defined as the community of bacteria residing in the human vaginal tract. Recent studies have demonstrated that the high prevalence of Lactobacillus crispatus strains is commonly associated with a healthy vaginal environment. In the current study, we assessed the microbial composition of 94 public healthy vaginal samples through shotgun metagenomics analyses. Results showed that L. crispatus was the most representative species and correlated negatively with bacteria involved in vaginal infections. Moreover, we isolated and sequenced the genomes of new L. crispatus strains from different environments, and the comparative genomics analysis revealed a genetic adaptation of strains to their ecological niche. In addition, in vitro growth experiments display the capability of this species to modulate the composition of the vaginal microbial consortia. Overall, our findings suggest an ecological role exploited by L. crispatus in reducing the complexity of the vaginal microbiota toward a depletion of pathogenic bacteria.
INTRODUCTION
The human vaginal environment hosts a community of bacteria, also known as the vaginal microbiota (VM), which plays an important role in maintaining vaginal health (1, 2) and in protecting this environment from several urogenital diseases, such as bacterial vaginosis (BV) and viral infections (3–5). In detail, various studies have demonstrated that the presence of members of the Lactobacillus genus in the VM is associated with a positive health status (3, 6–11). The VM has been classified into five distinct community state types (CSTs) obtained through statistical associations and based on specific compositional profiles (6). Four CSTs are characterized by a Lactobacillus-prevalent VM, in which Lactobacillus crispatus, Lactobacillus iners, Lactobacillus gasseri, or Lactobacillus jensenii species dominate the vaginal tract. Furthermore, a fifth CST is composed of different anaerobic bacteria, including Gardnerella vaginalis species and members of the Lachnospiraceae, Leptotrichiaceae, and Prevotellaceae families (6, 12, 13). Notably, the VM is frequently dominated by L. crispatus, commonly associated with a healthy status of the vaginal tract by protecting the host from viral, bacterial, and fungal infections (11, 14–16) due to the production of various antimicrobial compounds, such as hydrogen peroxide, lactic acid, and bacteriocin-like products (17–19). Strains belonging to L. crispatus are commonly isolated from two ecological niches, the human vaginal tract and the gastrointestinal tract (GIT) of certain birds, i.e., chicken and turkey (20, 21), though a small number of strains belonging to this species have been isolated from the human GIT, human oral cavity, and human eyes (22–24). In the latter case, it is not clear if such isolates originated from their true ecological origin or were the consequence of an external contamination during the isolation procedures. Comparative genome analysis of these strains may allow identification of genetic elements that are specifically linked to a particular ecological origin. Furthermore, L. crispatus was reported to be easily replaced by anaerobic bacteria and had shown significant genomic and metabolic differences between strains (25). Nonetheless, several studies have demonstrated that this species regulates the vaginal homeostatic environment and immune barrier functions for the maintenance of a healthy vaginal status (26, 27). For these reasons, L. crispatus is considered to be a probiotic candidate for the prevention of vaginal tract infections and for the reinstatement of a healthy VM environment (28–30).
In this study, we assessed the VM composition of 94 publicly available shotgun metagenomic data sets corresponding to vaginal samples of healthy women. Our findings show that L. crispatus is a highly prevalent bacterial species of the VM. Furthermore, this species negatively correlates with the main bacteria involved in vaginal infections. For this reason, we decided to isolate and characterize novel L. crispatus strains from vaginal swabs of healthy women and also from chicken fecal samples, which represent the two ecological niches for L. crispatus species (20, 21). The genomes of these isolates were decoded and subjected to comparative genome analyses, facilitating the discovery of putative genetic correlations between members of these two distinct ecological origins. Finally, in order to identify novel isolates with high growth capability in an environment resembling vaginal conditions, i.e., mimicking the microbial composition and pH, we performed a comparative in vitro growth experiment among L. crispatus strains isolated from human VM in a simulated vaginal fluid (SVF) (31) with three other typical vaginal Lactobacillus species, L. gasseri, L. iners, and L. jensenii. Our results highlighted the existence of phenotypic strain-specific differences, revealing the superior competitive ability of L. crispatus strain PRL2021 compared to other Lactobacillus species under simulated in vivo conditions employing a simplified human vaginal environment. Moreover, cocultivation experiments using L. crispatus PRL2021 with enteric microbiota revealed that this strain is able to influence the composition of the microbial consortia by provoking a simplification of the overall microbial biodiversity.
RESULTS AND DISCUSSION
Evaluation of vaginal microbiota composition of healthy women.
In 2011, Ravel et al. (6) identified and classified the vaginal microbiota (VM) in five community state types (CSTs). In detail, four CSTs were dominated by different species of Lactobacillus, i.e., L. crispatus (CST 1), L. gasseri (CST 2), L. iners (CST 3), and L. jensenii (CST 5), whereas the fifth (CST 4) possesses a lower proportion of lactic acid bacteria and a higher proportion of strictly anaerobic organisms. Subsequent studies, mostly based on 16S rRNA gene‐based amplicon sequencing, have confirmed the presence of these CSTs (1, 32–35).
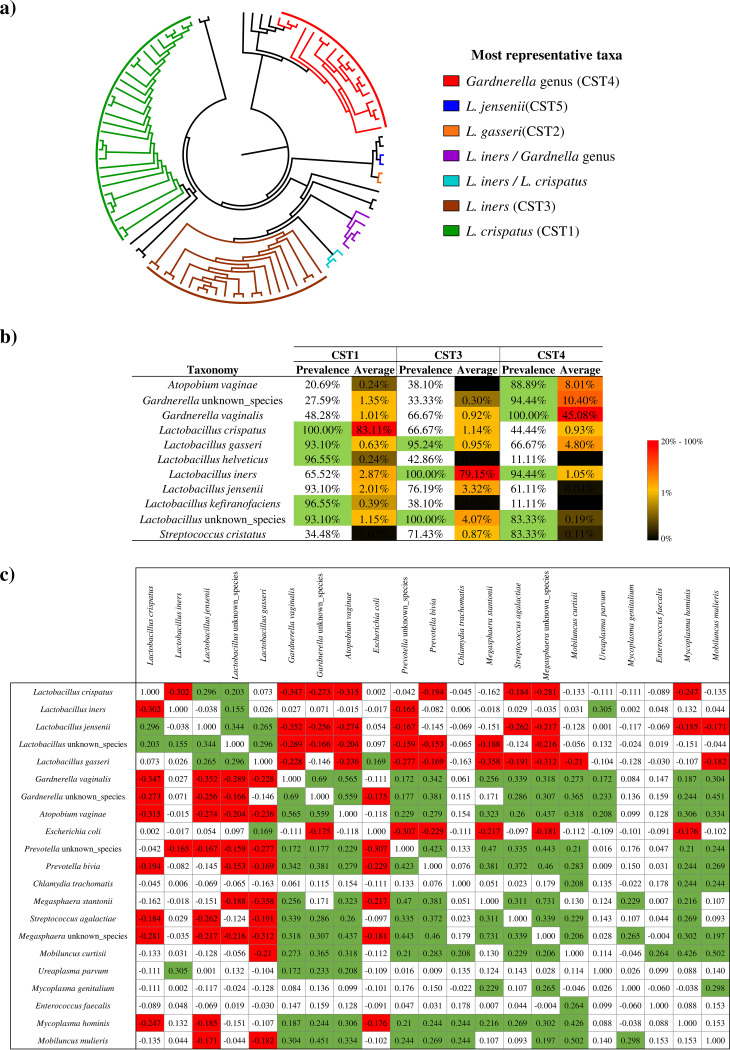
Metagenomic shotgun techniques, i.e., whole-metagenome sequencing (WMS), overcome some of the limitations of 16S rRNA gene‐based amplicon sequencing, such as issues with DNA amplification and primer pair efficiency. Furthermore, WMS allows acquisition of functional information and evaluation of bacterial communities at species level in a more precise manner compared to 16S rRNA gene‐based amplicon sequencing. Consequently, in order to assess the VM composition of healthy women, we decided to perform a comprehensive literature search for vaginal shotgun metagenomic data sets based on Illumina sequencing technology. In detail, we collected publicly available data sets corresponding to metagenomic sequencing of 94 vaginal samples of healthy women based on three studies (36–38) performed in two different countries (Table S1 in the supplemental material). We reanalyzed these data sets using a shallow shotgun metagenomics approach (39), thereby achieving an accurate taxonomic cataloguing of the VM at species level resolution. The metagenomic analysis included a total of 2,361,660 reads with an average of 25,124 ± 23,730 reads per sample (Table S1). Bacterial species analysis showed that 60.64% of the samples represented a vaginal microbiota dominated by Lactobacillus species (Table S2), confirming that in the majority of women a healthy vaginal microbiota is dominated by Lactobacillus species (40). Interestingly, 32.98% and 28.72% of the samples showed L. crispatus and L. iners, respectively, as the most abundant vaginal bacterium. In addition, 29.79% and 23.40% of the samples showed a relative abundance of >40% of L. crispatus and L. iners, respectively. Furthermore, we evaluated the presence of CSTs in the 94 samples collected through hierarchical clustering (HCL) analysis (Fig. 1). The HCL analysis of these publicly available data sets showed the presence of all previously identified CSTs, confirming the predominance of CST1 and CST3, which represented 30.85% and 22.34% of the samples, respectively. Furthermore, 19.15% of the samples displayed a microbial profile corresponding to CST4, characterized by the presence of reads belonging to Gardnerella spp. In order to detect the main bacterial taxa belonging to CST1, CST3, and CST4 identified in this study, taxonomic profiling at species level was assessed to reconstruct the core microbiota members, i.e., core microbiota of the VM of each CST (41). For this purpose, we reconstructed the core microbiota by selecting bacterial species that occur with a prevalence greater than 80% among the collected samples of a particular CST (42, 43) (Fig. 1b). Interestingly, the CST1 and CST3 types showed a core microbiota characterized by bacteria belonging to the Lactobacillus genus, while CST4 showed a more complex core microbiota characterized by the Gardnerella genus, L. iners, Streptococcus cristatus, and potential vaginal pathogens, such as Atopobium vaginae. Furthermore, the identification of the accessory microbiota, i.e., bacterial species different from core microbiota and with prevalence greater than 30% and an average relative abundance greater than 0.5% (44), revealed a lower biodiversity of the CST1 microbiota. In detail, comparison of the accessory microbiota of CST1 with those of CST3 and CST4 showed a 50% and 76.9% decrease in the number of species, respectively. Moreover, the observed concurrence of L. iners and the Gardnerella genus prompted us to investigate possible correlations between various characteristic vaginal bacteria. Consequently, the relative abundances of CST4-associated Lactobacillus species and the most common vaginal pathogens, such as Atopobium vaginae, Streptococcus agalactiae, and Prevotella bivia, found in the 94 samples were employed in a covariance analysis based on Kendall tau rank (Fig. 1c). Interestingly, L. crispatus, L. jensenii, and L. gasseri correlated negatively with at least 50% of the assessed potential vaginal pathogens, suggesting that these microorganisms are able to outcompete and outnumber bacteria involved in vaginal infections. Particularly, these results could reinforce the hypothesis that the L. crispatus species reduces the biodiversity of VM and consequently protects this environment from viral and bacterial infections (11, 14, 15, 28–30).
FIG 1.
Evaluation of vaginal microbiota composition of healthy women. (a) Circular cladogram of the 94 publicly available samples based on hierarchical clustering (HCL) analysis. The samples were subdivided according to the most representative taxa and CST. (b) Prevalence and average abundance of the bacteria that correspond to CST1, CST3, and CST4. The values that represent the core microbiota are highlighted in green. (c) Results of Kendall tau rank covariance analysis based on the fourth CST Lactobacillus species and the most common vaginal pathogens. The significant positive correlations are highlighted in green (P value <0.05), while significant negative correlations are depicted in red (P value <0.05).
Isolation of L. crispatus strains from human and chicken samples.
In order to isolate L. crispatus strains, we collected seven vaginal swab samples from adult Italian women without symptoms of vaginal infections and ten feral chicken fecal samples, following a protocol for the isolation of members of the Lactobacillus genus (see the Materials and Methods section). We decided to screen presumed healthy vaginal swabs and chicken fecal samples based on the previously described high prevalence of L. crispatus strains in these environments (18, 24, 45). Chicken fecal samples were selected to identify possible differences between strains of the same species yet isolated from distinct ecological niches. In total, 18 different strains were isolated, all belonging to the genus Lactobacillus. In this context, 15 strains were determined to represent L. crispatus species, two were shown to be L. jensenii species, and one isolate was demonstrated to belong to the L. gasseri species. Moreover, strain L. crispatus M247 was isolated from a commercial vaginal probiotic product (46) (Table 1). Regarding the novel L. crispatus strains isolated, seven were isolated from poultry fecal samples, whereas eight were isolated from vaginal fluid (Table 1). The relative ease by which such a high number of L. crispatus strains were isolated is consistent with the high abundance level of this species in the human VM and in the gastrointestinal tract of chickens, highlighting its ecological specialization for these environments (18, 24, 45).
TABLE 1.
Lactobacillus strains used in this study and general genome features
| Strain | Size (bp) | No. ORFs | No. rRNA loci | No. tRNA genes | Fold coverage depth | No. contigs | Origin | BioSamplea |
|---|---|---|---|---|---|---|---|---|
| L. crispatus PRL2021 | 2,329,621 | 2,340 | 6 | 64 | 126 | 43 | Vaginal swab | SAMN15357510 |
| L. crispatus LB56 | 2,160,961 | 2,215 | 3 | 55 | 137 | 195 | Vaginal swab | SAMN15357362 |
| L. crispatus LB57 | 2,382,340 | 2,496 | 3 | 65 | 132 | 259 | Vaginal swab | SAMN15357363 |
| L. crispatus LB58 | 2,236,149 | 2,232 | 3 | 65 | 184 | 199 | Vaginal swab | SAMN15357364 |
| L. crispatus LB59 | 2,135,374 | 2,172 | 3 | 66 | 182 | 189 | Vaginal swab | SAMN15357365 |
| L. crispatus LB61 | 2,081,602 | 2,087 | 3 | 66 | 228 | 140 | Vaginal swab | SAMN15357366 |
| L. crispatus LB62 | 2,160,518 | 2,235 | 3 | 63 | 165 | 172 | Vaginal swab | SAMN15357367 |
| L. crispatus LB63 | 2,132,629 | 2,178 | 4 | 60 | 141 | 182 | Vaginal swab | SAMN15357368 |
| L. crispatus M247 | 2,098,939 | 2,083 | 3 | 57 | 256 | 156 | Vaginal probiotic product | Not deposited |
| L. crispatus LB64 | 2,051,142 | 2,040 | 3 | 57 | 259 | 36 | Chicken fecal sample | SAMN15357369 |
| L. crispatus LB65 | 2,061,272 | 2,035 | 3 | 61 | 156 | 44 | Chicken fecal sample | SAMN15357370 |
| L. crispatus LB66 | 2,041,845 | 2,006 | 3 | 56 | 209 | 44 | Chicken fecal sample | SAMN15357371 |
| L. crispatus LB67 | 1,967,651 | 1,912 | 3 | 60 | 193 | 60 | Chicken fecal sample | SAMN15357372 |
| L. crispatus LB68 | 1,966,006 | 1,921 | 2 | 56 | 73 | 111 | Chicken fecal sample | SAMN15357373 |
| L. crispatus LB69 | 1,977,839 | 1,959 | 3 | 55 | 79 | 95 | Chicken fecal sample | SAMN15357374 |
| L. crispatus LB70 | 2,021,157 | 1,988 | 3 | 54 | 76 | 199 | Chicken fecal sample | SAMN15357375 |
| L. gasseri V105C | Vaginal swab | Not sequenced | ||||||
| L. gasseri ATCC 9857 | ATCC collection | Not sequenced | ||||||
| L. jensenii V79H | Vaginal swab | Not sequenced | ||||||
| L. jensenii V94G | Vaginal swab | Not sequenced | ||||||
| L. iners LMG 14328 | LMG collection | Not sequenced |
M247 was not deposited, as it is a commercial strain. V105C, ATCC 9857, V79H, V94G, and LMG 14328 were not sequenced, because they were used only in the physiological experiments and not the genetic experiments.
Evaluation of novel L. crispatus strains to vaginal environment challenges.
In order to evaluate the ecological adaptation of the isolated strains to the vaginal environment, all identified L. crispatus strains were subjected to growth experiments in the presence of environmental stress condition, i.e., acidic stress, simulating those naturally occurring in the vaginal tract (1, 47–51). Interestingly, these isolates do not tolerate harsh acidic conditions, displaying survival rates of <20% when exposed to a pH 2.0 or pH 3.0 for 48 h. Conversely, six strains (PRL2021, LB57, LB61, LB62, LB63, and LB66) reached a survival rate of >85% when the pH of the growth medium was 4.0. Notably, also under this growth condition, strain PRL2021 reached the highest survival rate compared to other assessed L. crispatus isolates (ANOVA P value < 0.05), with a percentage of 94.81% (Fig. 2a). Interestingly, five of the strains mentioned above that were shown to elicit the highest survival rate at pH 4.0 had been isolated from vaginal fluid, suggesting an adaptation of these strains to their natural ecological niche due to the acidic conditions of the vaginal environment (52).
FIG 2.
Physiological features of isolated strains and growth curves of Lactobacillus strains with glycogen as the sole carbon source. (a) Effect of the conditions that simulate the vaginal tract for isolated L. crispatus strains. The heat maps represent the survival rate of each L. crispatus strain to NaCl and acidic conditions. (b) Growth curves of tested strains. On the y axis OD values are reported at a wavelength of 600 nm, whereas the x axis shows times (h). Each Lactobacillus strain is depicted with a different color line as indicated. (c) Box and whiskers plot based on Lactobacillus growth with glycogen as the sole carbon source. The boxes represent the three groups characterized by vaginal L. crispatus isolates, chicken L. crispatus isolates, and the other Lactobacillus species tested. The y axis represents the OD600, while the x axis shows the times (h); the boxes represent 50% of the data set, distributed between the first and the third quartiles. The median divides the boxes into the interquartile range, while the “X” represents the mean. The dots inside the boxes display the data points that lie between the lower whisker line and the upper whisker line, while the dots outside the boxes show the outlier points that are positioned either below the lower whisker line or above the upper whisker line. Statistical analysis between groups were performed through ANOVA analysis. Moreover, we calculated the post hoc analysis LSD (least significant difference) for multiple comparisons.
Different studies have previously shown that the presence of luminal glycogen in the vaginal tract of healthy women is associated with a high prevalence of lactobacilli (53, 54). In order to investigate the utilization of glycogen by Lactobacillus strains, in vitro cultivation assays were performed in growth medium, i.e., semisynthetic MRS without sugar, containing glycogen as the unique carbon source. The control assay was carried out with MRS medium supplemented with lactose and the cultures were monitored for 48 h (see the Materials and Methods). Interestingly, L. crispatus strains isolated from vaginal swabs were shown to grow significantly better compared to strains of poultry origin (t test P value < 0.01), indicating that these strains are adapted to utilize a carbon source naturally present in their ecological niche (Fig. 2). Among strains of vaginal origin, PRL2021 and LB63 showed the highest growth performance, reaching optical density at 600 nm (OD600) values of >0.42 after 12 h of growth (Fig. 2). In contrast, strains of poultry origin were not able to use glycogen as a carbon source, showing low OD600 values (average OD600 0.33 ± 0.15) even after 48 h of growth (Fig. 2b and c). In addition, other tested Lactobacillus strains belonging to L. gasseri, L. jensenii, and L. iners species displayed growth levels 1.5-fold lower compared to L. crispatus strains (t test P value < 0.05) (Fig. 2b and c).
Production of H2O2 by Lactobacillus strains.
Hydrogen peroxide production is generally considered to be a key selective factor allowing Lactobacillus spp. to dominate the vaginal environment (55–57). Moreover, it has been demonstrated that women vaginally colonized by lactobacilli producing hydrogen peroxide were less exposed to bacterial vaginosis compared to women colonized by Lactobacillus strains that do not produce H2O2 (58, 59). For these reasons, in order to investigate the production of hydrogen peroxide, 21 Lactobacillus strains, including L. crispatus, L. iners, L. jensenii, and L. gasseri species (Table 2), were tested by the qualitative tetramethylbenzidine (TMB)-plus peroxidase assay (see the Materials and Methods) (18, 56). Interestingly, all the L. crispatus strains tested, except for L. crispatus LB63, produced variable levels of hydrogen peroxide without any correlation to their ecological origin. The colony-associated blue color intensity of H2O2 producers was different in the examined L. crispatus strains. In particular, four strains, L. crispatus PRL2021, L. crispatus LB58, L. crispatus LB65, and L. crispatus LB66, produced colonies with an intense dark blue color, suggesting strong H2O2 production (Table 2). In contrast, other H2O2-producing strains produced colonies with less blue intensity (Table 2). Furthermore, 60% of the strains of other typical vaginal Lactobacillus species tested did not produce H2O2 (Table 2), reflecting data reported in previous studies and highlighting that L. crispatus species is one of the most potent H2O2-generating species among typical vaginal lactobacilli (18, 60). The H2O2-producing feature is a strain-specific feature, as demonstrated by our results and by previously published studies (18, 60). Strains producing hydrogen peroxide could display a higher ecological fitness (56) and their presence may be important in the modulation of the VM.
TABLE 2.
H2O2 production by lactobacilli isolated in this study
| Strains | H2O2 production | RGB code colora |
|---|---|---|
| L. crispatus PRL2021 | + | 0-0-100 |
| L. crispatus LB56 | + | 0-0-215 |
| L. crispatus LB57 | + | 0-0-160 |
| L. crispatus LB58 | + | 0-0-115 |
| L. crispatus LB59 | + | 0-0-175 |
| L. crispatus M247 | + | 0-0-195 |
| L. crispatus LB61 | + | 0-0-255 |
| L. crispatus LB62 | + | 0-0-200 |
| L. crispatus LB63 | ||
| L. crispatus LB64 | + | 0-0-230 |
| L. crispatus LB65 | + | 0-0-130 |
| L. crispatus LB66 | + | 0-0-150 |
| L. crispatus LB67 | + | 0-0-190 |
| L. crispatus LB68 | + | 0-0-190 |
| L. crispatus LB69 | + | 0-0-245 |
| L. crispatus LB70 | + | 0-0-240 |
| L. jensenii V79H | + | 0-0-105 |
| L. jensenii V94G | + | 0-0-121 |
| L. gasseri ATCC 9857 | ||
| L. gasseri V105C | ||
| L. iners LMG 14328 |
RGB, red, green, and blue color mode.
General genome features of isolated L. crispatus.
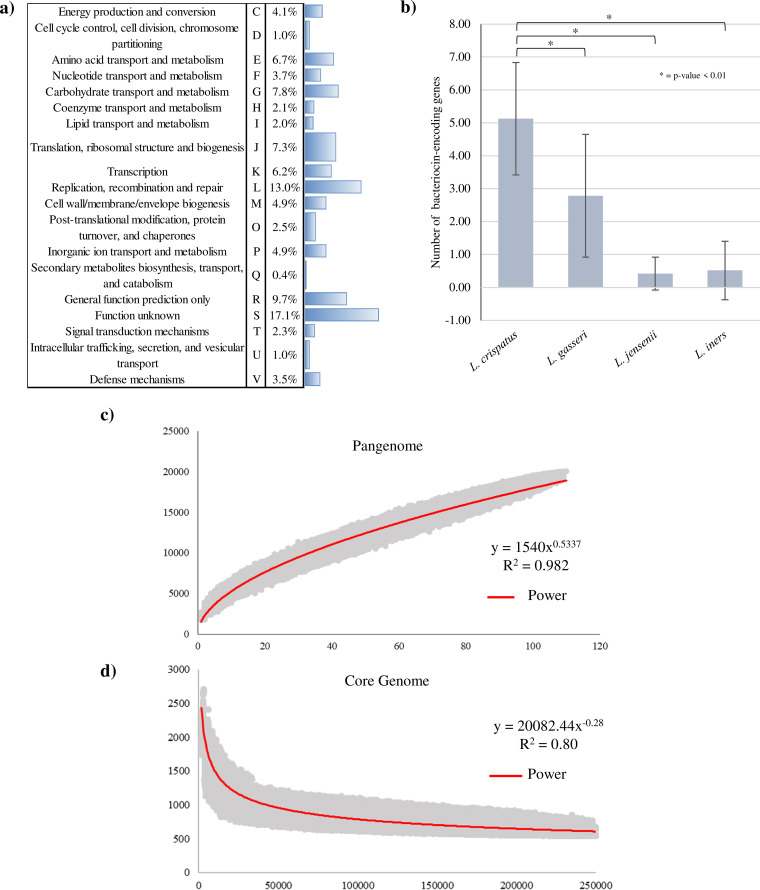
Genome sequences of the 15 newly isolated L. crispatus strains were decoded through whole-genome sequencing employing an Illumina MiSeq platform. Additionally, DNA extracted from L. crispatus PRL2021 was also subjected to whole-genome sequencing using a MinION (Oxford Nanopore, UK). The achieved genome sequences were analyzed to evaluate the genetic content of these isolates. In this context, genome sequences ranged in size from 2,382,340 bp of L. crispatus LB57 to 1,966,006 bp of L. crispatus LB68 (Table 1). Interestingly, the genome sizes of novel isolates from vaginal fluid samples were larger than the genome sizes of the isolated strains from poultry fecal samples (average size 2,190,904 bp ± 104,068 bp and 2,012,416 bp ± 41,196 bp in vaginal and poultry isolates, respectively; t test P value < 0.001), suggesting increased genomic complexity of the strains from human origin. Remarkably, a similar difference in size was also identified in those genomes that are publicly available, confirming that vaginal L. crispatus strains possess an expanded genetic makeup compared to those of poultry origin (average size 2,297,590 bp ± 163,268 bp and 2,056,164 bp ± 7,026 bp in vaginal and poultry origin isolated genomes, respectively; t test P value < 0.001) (Table S3). Moreover, the open reading frame (ORF) prediction revealed that the ORF number ranged from 2,496 in the case of L. crispatus LB57 to 1,912 for L. crispatus LB67 (Table 1). Interestingly, 15 sequenced genomes encompass a similar set of rRNA loci and tRNA genes, which were shown to consist of three or four rRNA operons and between 66 and 54 tRNA genes distributed across the genome (Table 1), except for L. crispatus PRL2021, which harbors six rRNA operons in its genome (Table 1). Notably, functional classification of the L. crispatus ORFeome based on the eggNOG database (61) was possible for the 82.1% of the predicted ORFs. In particular, 15.2% of the predicted proteins are involved in cellular process and signaling (categories D, M, O, T, U, and V), 26.4% referred to proteins related to information storage and processing (categories J, K, and L), and 31.6% described proteins involved in metabolic processes (categories C, E, F, G, H, I, P, and Q) (Fig. 3a). These data underlined the high number of proteins related to carbohydrate and amino acid transport and metabolism, in accordance with the wide range of sugars metabolized by members of this species (31, 62) and with the apparent inability of lactobacilli to synthetize most amino acids (63). Recently, an orthologue of the Escherichia coli glgX gene, which is involved in glycogen metabolism, was identified in L. crispatus genomes (54). Different studies have demonstrated the correlation between high abundance of lactobacilli in the vaginal environment and the presence of high levels of luminal glycogen in the vaginal tract of healthy women (53, 54, 64). Interestingly, we identified three orthologues of this gene in each L. crispatus genome of vaginal origin, whereas genomes of L. crispatus from poultry origin encompassed one or two copies of this gene (Table S4). The presence of glgX orthologues in vaginal isolates suggests the genetic adaptation of these strains to glycogen metabolism.
FIG 3.
Functional classification of the proteome, bacteriocin-encoding genes, and the pangenome and core genome of L. crispatus species. (a) Functional assignment of the isolated L. crispatus proteome based on the eggNOG database. (b) Number of bacteriocin-encoding genes predicted in L. crispatus, L. gasseri, L. jensenii, and L. iners species. (c and d) The pangenome (c) and core genome (d) of the L. crispatus species represented as variation in their gene pool sizes upon sequential addition of the 110 genomes analyzed, respectively.
Prediction of bacteriocin-encoding genes in L. crispatus species.
The production of antimicrobial compounds by lactobacilli, including hydrogen peroxide, organic acids, low-molecular-weight antimicrobial substances, and bacteriocins, is considered an important feature of these microorganisms for counteracting pathogenic bacteria (18, 19). For this reason, in order to predict bacteriocin-encoding genes, the genomes of isolated L. crispatus strains were screened for bacteriocin-encompassing gene clusters by means of the BAGEL3 software and associated database (65). Notably, all tested genomes were predicted to contain bacteriocin-encoding loci, ranging from three genes of L. crispatus LB56 to nine genes of L. crispatus LB57 and L. crispatus LB70 (Table S5). Interestingly, according to BAGEL-mediated classification, all predicted bacteriocin-encoding genes were shown to belong to classes II and III (65) (Table S5). Specifically, all poultry-derived strains, except for LB67 and LB70, only encoded class III bacteriocins (Table S5), whereas vaginal-isolated strains were predicted to produce bacteriocins of both class II and class III (Table S5). The presence of bacteriocin-encoding genes belonging to classes II and III in vaginal-isolated strains suggests a wider range of antimicrobial activity compared to strains of chicken origin. Intriguingly, all isolated strains presented at least one gene predicted to encode a bacteriocin known as helveticin J (Table S5). This bacteriocin was first identified in Lactobacillus helveticus strain 481 (66). Moreover, M23 family metallopeptidase-encoding genes were identified in all sequenced genomes (Table S5). This peptidase family includes proteins that act as bacteriocins that degrade the peptidoglycan of other bacteria (67). Notably, three strains, PRL2021, LB57, and LB70, were shown to contain two adjacent genes in their genome encoding a two-component bacteriocin known as bacteriocin LS2 (68) (Table S5). Interestingly, the genome of strain L. crispatus PRL2021, which exhibits a robust production of other antimicrobial compounds, such as H2O2 (see above), harbors two distinct loci each encoding helveticin J (Table S5), suggesting that this strain produces a high level of antimicrobial activity, which may allow this strain to occupy and be highly competitive in particular ecological niches, such as the human vaginal tract. Moreover, two poultry-isolated strains, LB67 and LB70, harbor a garvicin-encoding gene. Garvicin is a broad spectrum, nonlantibiotic bacteriocin, previously characterized in the Lactococcus garvieae species (69–71). Finally, L. crispatus strain LB70 was the only strain that encodes a lactacin F bacteriocin (Table S5). In order to explore if L. crispatus species exhibit superior antimicrobial activity compared to other Lactobacillus species that are typically present in the human vagina, bacteriocin-encoding genes were predicted also in 94 publicly available L. crispatus genomes and in 116 publicly available genomes belonged to L. gasseri, L. jensenii, and L. iners species (Table S6). This analysis indeed showed that L. crispatus strains are predicted to harbor the highest number of bacteriocin-encoding genes in their genomes, ranging from four genes in five different strains to 10 genes in four different strains (Table S3). Notably, no bacteriocin-encoding genes were detected in 56% and in 74% of L. jensenii and L. iners genomes, respectively (Table S6). Moreover, 67% of the investigated L. gasseri strains encompass one or two bacteriocin-encoding genes in their genomes (Table S6). Interestingly, the number of bacteriocin-encoding genes presented in L. crispatus genomes is significantly higher (t test P value < 0.01) compared to those identified in other analyzed vaginal Lactobacillus species, i.e., L. iners, L. gasseri, and L. jensenii (Fig. 3b, Table S5, and Table S6). The in silico prediction of bacteriocin-encoding genes indicates the production of extensive antimicrobial activity by L. crispatus strains, in particular compared to other vaginal Lactobacillus species, perhaps supporting a superior ecological fitness of this species.
Pangenome and core genome analysis of Lactobacillus crispatus species.
In order to investigate the genomic differences between L. crispatus strains, we performed an extensive comparative genome analysis that not only encompassed the genomes of the 16 isolated L. crispatus strains, i.e., the 15 new isolates and L. crispatus M247, but also the 94 publicly available L. crispatus genomes (Table S3). For this purpose, we performed a reannotation of all publicly available L. crispatus chromosomes using the same bioinformatics pipeline applied for the genomes of the 16 newly isolated strains. The reconstructed genomic data sets of the L. crispatus species, encompassing a total of 110 genomes sequences (i.e., 16 isolated in this study and 94 publicly available), represents the largest reconstructed genetic database for this Lactobacillus species so far. These data were used to predict the pangenome of the L. crispatus species, i.e., the collection of genes of all strains of a species based on the cluster of orthologous groups (COGs) (72). Furthermore, these data were also used to predict the core genome, i.e., the collection of gene families shared between members of a given species, in this case the L. crispatus taxon (73). Plotting on a log-log scale as a function of the number of analyzed genomes, the pangenome size was determined to consist of 20,247 COGs, suggesting that the power trend line has not reached a plateau (Fig. 3c). The number of new genes discovered by sequential addition of genome sequences decreased from 594 COGs for the first two genomes, to 123 COGs for the final addition. These data suggest the existence of an open pangenome within the L. crispatus species (Fig. 3c and d), as already noted for other Lactobacillus species, including L. gasseri, Lactobacillus paragasseri, and L. helveticus (74, 75). Moreover, the 110 L. crispatus genomes were examined to identify shared orthologous genes, as well as unique genes. In silico analyses revealed 452 ORFs shared among all L. crispatus genomes analyzed, representing the core genome of this taxon. In addition, we identified a variable number of truly unique genes (TUGs), which ranged from 233 of L. crispatus LR31 and L. crispatus LR33 to 29 of L. crispatus MGYG-HGUT-02348 and L. crispatus EM-LC1 (Fig. 4 and Table S3). Notably, due to the low quality of some genome sequence data, strains presenting more than 250 TUGs were not considered for this analysis (Table S3). Furthermore, an in silico approach was used to calculate the average nucleotide identity (ANI) values, defined as a measure of nucleotide-level genomic similarity between two genomes (76). This analysis showed a highly syntenic genome structure among members of the L. crispatus species, with associated ANI values ranging from 95.57% to 99.87%. Interestingly, different range of ANI values were recognized between L. crispatus strains isolated from vaginal fluid and from poultry fecal samples. In this context, the lowest ANI value between vaginal isolated strains was 95.57%, while for poultry isolated strains was 97.64%. These data suggest that differences exist between L. crispatus strains isolated from different ecological niches, underlining highly syntenic genome structures among L. crispatus strains of poultry origin.
FIG 4.
Phylogenomic tree of L. crispatus species. A proteomic tree was built on the concatenation of 452 L. crispatus core genes identified in the core genome analysis. This tree was constructed by the neighbor-joining method, and the genome sequence of L. acidophilus DSM 20079 was used as the outgroup. Bootstrap percentages of >70 are shown at node points, based on 1,000 replicates. Circles surrounding the tree represent the isolation origin of the strains and the number of truly unique genes (TUGs) (violet circles).
Phylogenetic analyses of the Lactobacillus crispatus species.
Recently, a taxonomic reclassification of the Lactobacillus genus has resulted in a more robust clade of this bacterial taxon, including microorganisms with shared metabolic and ecological features (77). Notably, these analyses revealed the presence in the Lactobacillus genus of species adapted to vertebrates or invertebrates, including L. crispatus species and typical human vaginal lactobacilli, such as L. iners, L. jensenii, and L. gasseri species (77). The availability of genome sequences of members of L. crispatus species (Table S3) allowed a more robust phylogenetic reconstruction of this taxon. As already mentioned above, in silico analyses identified 452 ORFs shared among analyzed L. crispatus genomes, representing the core genome of this species. A concatenated protein sequence that included the product of each of these core genes was used to build a L. crispatus supertree (Fig. 4). This supertree showed that all 16 L. crispatus strains isolated in this study coclustered with other publicly available L. crispatus genomes. Interestingly, the supertree gave rise to two clades, the first one comprising L. crispatus strains isolated from chicken as well as turkey GIT and the human GIT, while the second one included L. crispatus strains isolated from vaginal swabs and human urine samples (Fig. 4). Interestingly, the L. crispatus DISK12 strain, which was isolated from the human oral cavity, and the type strain L. crispatus DSM20584, which was isolated from human eye (22), coclustered with strains isolated from the gastrointestinal tract of animals and humans (Fig. 4). Notably, strains CIP104459, C037, and OAB24-B, which were isolated from vaginal swabs (78, 79), did not cluster with other vaginal-isolated strains (Fig. 4). Moreover, three strains isolated from human urine samples, L. crispatus UMB0824, L. crispatus UMB0085, and L. crispatus UMB0040 (80), coclustered with strains of animal and human GIT origin (Fig. 4). Despite some exceptions, due to the very low phylogenetic distance among members of L. crispatus species, these analyses showed different evolutionary developments among the strains analyzed, likely reflecting the ecological adaptation of such strains to their ecological niches.
Coculture experiments in simulated vaginal fluid.
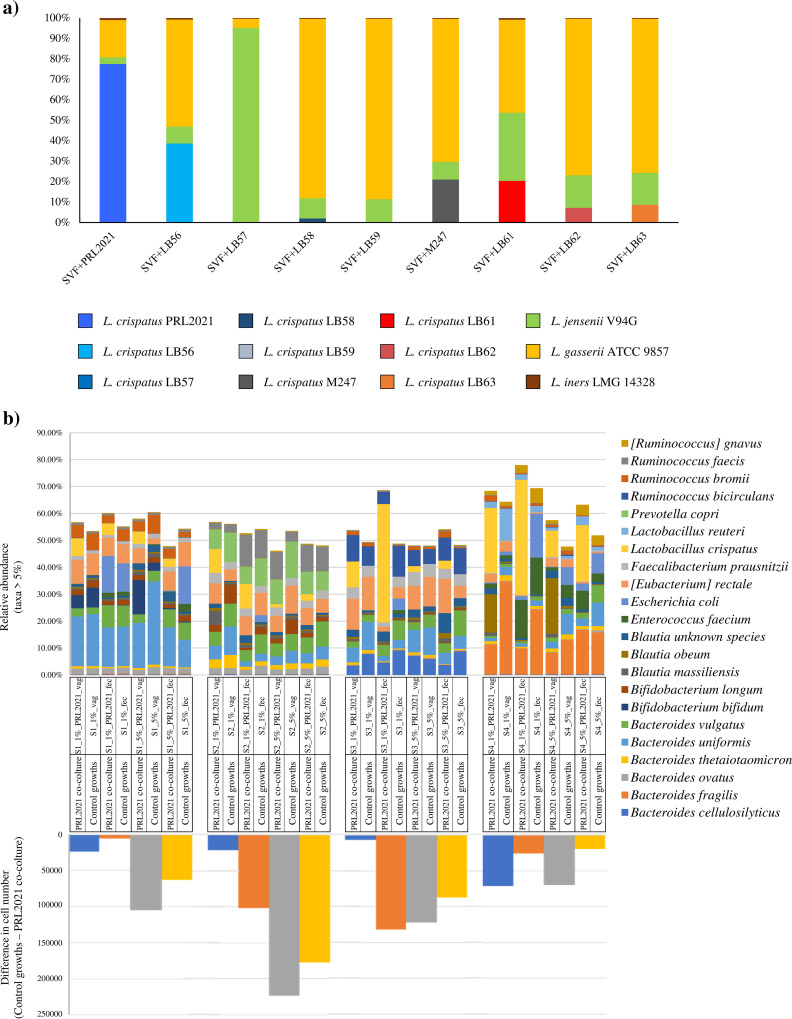
In order to investigate the behavior of vaginal-isolated L. crispatus strains in a simplified human vaginal environment composed of other typical vaginal-Lactobacillus species, we performed growth experiments on simulated vaginal fluid (SVF) that mimics the nutritional and chemical-physical conditions of the vaginal environment, as previously performed by Pan et al. (31). We assessed nine different parallel in vitro experiments for each L. crispatus isolated from human vaginal samples, i.e., PRL2021, LB56, LB57, LB58, LB59, LB61, LB62, and LB63, and a strain originating from a vaginal probiotic product, i.e., M247. In each parallel in vitro experiment, a single vaginal-isolated L. crispatus strain was inoculated (1% vol/vol) with other representative Lactobacillus species of vaginal CSTs exemplified by L. gasseri ATCC 9857, L. iners LMG 14328, and L. jensenii V94G at 1% (vol/vol) each. After 12 h of growth, shallow-shotgun metagenomics was independently performed for each batch culture. Sequencing analyses resulted in a total of 837,443 reads with an average of 93,049 ± 8,520 reads per sample (Table S7).
Notably, after 12 h the average relative abundance of L. iners LMG 14328 corresponded to 0.60% ± 0.26% (Fig. 5a). This low abundance of L. iners LMG 14328 was probably due to the suboptimal growth conditions for this strain, perhaps as a result of its nutritional needs (81). Interestingly, L. gasseri ATCC 9857 seemed to outgrow other strains in seven out of the nine experiments (Fig. 5a) and, in five cases, the relative abundance of this strain was higher than 65% (Fig. 5a). Moreover, in one case L. crispatus PRL2021 was shown to have grown better compared to the other lactobacilli, with a relative abundance of 77.4% (Fig. 5a). Other L. crispatus strains (LB57, LB58, LB59, and M247) were shown to be dramatically outnumbered by other lactobacilli after 12 h of cultivation (Fig. 5a). These data and previous findings (Table 3) suggest that L. crispatus PRL2021 possesses superior ecological fitness compared to the other assessed L. crispatus strains, and its growth performance highlights excellent adaptation to the vaginal environment.
FIG 5.
Growth experiment in simulated vaginal fluid (SVF) and feal medium. (a) The bacterial composition of batch cultures based on shallow shotgun metagenomics profiling. The x axis represents the different analyzed batch-cultures, indicated with the L. crispatus strain name tested in each experiment. The y axis represents the absolute percentage of operational taxonomic units (OTUs) for each sample. (b) The taxonomic absolute abundances and the comparisons between the coculture experiment and its own control.
TABLE 3.
Overview of the overall results regarding L. crispatus strains
| Strain | Origin | Growth performance in 2% NaCl | Growth performance in pH 4 acidic conditions | Utilization of glycogen (OD600 at 12 h) | H2O2 production | No. of predicted bacteriocin-encoding genes | SVF coculture (% relative abundance)a |
|---|---|---|---|---|---|---|---|
| L. crispatus PRL2021 | Vaginal tract | 99.86% | 94.81% | 0.45 | + | 5 | 77.4 |
| L. crispatus LB56 | Vaginal tract | 88.82% | 15.83% | 0.37 | + | 3 | 38.6 |
| L. crispatus LB57 | Vaginal tract | 97.87% | 96.31% | 0.32 | + | 9 | 0.06 |
| L. crispatus LB58 | Vaginal tract | 99.21% | 58.40% | 0.34 | + | 5 | 1.9 |
| L. crispatus LB59 | Vaginal tract | 99.55% | 41.73% | 0.20 | + | 4 | 0.03 |
| L. crispatus LB61 | Vaginal tract | 83.65% | 87.18% | 0.41 | + | 4 | 20.36 |
| L. crispatus LB62 | Vaginal tract | 99.36% | 91.22% | 0.39 | + | 5 | 7.1 |
| L. crispatus LB63 | Vaginal tract | 99.33% | 91.35% | 0.43 | − | 6 | 8.6 |
| L. crispatus M247 | Vaginal probiotic product | 99.46% | 25.58% | 0.40 | + | 5 | 21.0 |
| L. crispatus LB64 | Chicken fecal sample | 60.14% | 38.14% | 0.24 | + | 5 | |
| L. crispatus LB65 | Chicken fecal sample | 99.22% | 34.12% | 0.27 | + | 4 | |
| L. crispatus LB66 | Chicken fecal sample | 99.82% | 88.82% | 0.26 | + | 4 | |
| L. crispatus LB67 | Chicken fecal sample | 10.07% | 11.29% | 0.14 | + | 6 | |
| L. crispatus LB68 | Chicken fecal sample | 67.10% | 74.98% | 0.18 | + | 4 | |
| L. crispatus LB69 | Chicken fecal sample | 37.12% | 12.40% | 0.25 | + | 4 | |
| L. crispatus LB70 | Chicken fecal sample | 67.61% | 9.54% | 0.25 | + | 9 |
SVF, simulated vaginal fluid.
Coculture experiments using L. crispatus PRL2021 and human feces.
The VM composition may be influenced by fecal/rectal bacteria since it is the closest anatomical site characterized by a complex microbiota. In this context, the fecal/rectal microbiota may act as a source or reservoir of microorganisms (82). In fact, transmission of microorganisms from the rectum to the vagina may change the equilibrium of the vaginal microbiota or stimulate local inflammatory responses predisposing women to local infections, such as BV (83). Notably, recent studies have suggested that oral ingestion of Lactobacillus-containing products may play a possible role in the treatment or prevention of genital infections, acting as a gut reservoir for vaginal (re)colonization by lactobacilli (84, 85).
In order to evaluate the ecological effects of L. crispatus PRL2021, which was shown to have properties that were superior among the L. crispatus strains tested here, in shaping the vaginal microbiota, we performed a coculture experiment of PRL2021 with human feces. In detail, we cultivated L. crispatus PRL2021 with four different fecal samples with an inoculum consisting of 1% (wt/vol) or 5% (wt/vol). The growth experiments were performed using both SVF and fecal medium. Control growths were carried out without L. crispatus PRL2021. The ecological effects of strain PRL2021 on the fecal microbiota in the coculture experiments were investigated through a shallow shotgun metagenomics approach followed by an absolute quantification of the identified bacteria through flow cytometry assays. Sequencing analyses resulted in a total of 1,566,052 reads with an average of 43,501 ± 15,086 reads per sample (Table S8). As expected, the metagenomic analysis highlighted the absence of L. crispatus in all four fecal samples and in control growths. In detail, samples grown in SVF and fecal medium with 1% of stool sample showed an average abundance of L. crispatus of 12.33% ± 8.00% and 25.08% ± 21.27%, respectively. Similarly, the samples cultivated with 5% of human stool sample indicated an average abundance of L. crispatus of 4.46% ± 3.83% and 7.40% ± 9.04% in SVF and fecal medium, respectively.
A quantitative microbiome profiling approach, employing flow cytometry enumeration of microbial cells present in each sample, allowed the identification of the microbial load of coculture and control growths (86, 87). The comparison between each coculture experiment with its own control showed a decrease in the number of microbial cells in all samples enriched with L. crispatus PRL2021 (Fig. 5b). More specifically, flow cytometric results showed a drop in cell number in the coculture samples ranging from a thousand- to a hundred thousand-fold (Fig. 4), highlighting the ability of L. crispatus PRL2021 to provoke a simplification of the bacterial community, such as the fecal or vaginal microbiota. Moreover, these results support the notion that L. crispatus PRL2021 is able to grow in a different and more complex environment than that of the vaginal ecosystem, such as the intestinal environment, making this strain an interesting candidate for the development of Lactobacillus-containing probiotic products. Certainly, further investigations based on in vivo preclinical interventions are needed to fully clarify the beneficial role of L. crispatus PRL2021 in women with vaginal dysbiosis.
In conclusion, the human vaginal microbiota (VM) is represented by bacteria that colonize the human vaginal tract and that are assumed to play an important role in supporting a healthy host status. In the current study, we analyzed the VM composition of healthy women and observed that L. crispatus is one of the most representative species and correlates negatively with bacteria involved in vaginal infections. Moreover, this species seems to be able to modulate the VM and appears to play a role in reducing VM biodiversity. However, in this metagenomics analysis, 49% of the samples are of Chinese origin and this could represent a limitation of this study because it can introduce a geographical bias. Certainly, a more in-depth investigation that includes multiple samples of different geographic origins could expand and refine the knowledge on the composition of CST and the healthy vaginal microbiota. Furthermore, the isolation of 15 L. crispatus strains from vaginal swabs and chicken fecal samples confirmed that these two ecological niches are frequently and abundantly colonized by members of this Lactobacillus species. The genome sequencing of the new isolates allowed construction of the largest genomic data set of the L. crispatus taxon, including 94 publicly available L. crispatus genomes. Phylogenomic analyses based on the core genome sequences showed the existence of two different clades in which, with just a few exceptions, L. crispatus strains reflect the ecological niche from which they were isolated, suggesting a genetic adaptation to different environments. Furthermore, VM-simulating in vitro experiments revealed that L. crispatus, and in particular strain PRL2021, is able to outgrow other typical vaginal Lactobacillus species. These experiments also showed how this species exhibits good growth in a fecal environment, thereby highlighting the ability of this microbial taxon to decrease the complexity of a bacterial community. This analysis and in silico prediction of an extensive putative antimicrobial activity of this species suggests that L. crispatus PRL2021 is an interesting strain with an ability to prevent vaginosis and vaginal dysbiosis.
MATERIALS AND METHODS
Selection and metagenomics analyses of public data sets.
In this study, we performed a metagenomics analysis based on three publicly available whole-metagenome sequencing (WMS) data sets based on Illumina sequencing technology, corresponding to a total of 94 vaginal samples (Table S1). Specific metadata regarding age, diet, and any therapies was not available. In order to classify the reads to the lowest possible taxonomic rank, the downloaded fastq files were analyzed with the METAnnotatorX bioinformatics platform (88).
Isolation of new L. crispatus strains.
In order to investigate the occurrence of L. crispatus in vaginal microbiota and in poultry fecal samples, seven human healthy vaginal fluid samples from adult Italian women without symptoms of vaginal infections or local diseases and 10 feral chicken fecal samples were explored. Vaginal samples were collected through vaginal swabs mixed with 4 ml of phosphate-buffered saline (PBS) (pH 6.5), while fecal samples were composed of 10 g of fresh fecal material, which is a sufficient quantity to represent the whole biodiversity of fecal microbiota, as reported in a previous study (89). Serial dilution and subsequent plating were performed using MRS agar supplemented with 1% lactose (wt/vol). Morphologically different colonies that developed on MRS plates were picked and streaked in order to isolate purified bacterial strains.
Taxonomic identification of new isolated strains.
Identification of each isolate was performed by PCR amplification of a portion of the 16S rRNA gene through primer pair P0 (5′-GAAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′). Each 16S rRNA gene thus generated from individual strains was sequenced and was analyzed by BLAST against the GenBank database. Moreover, the genomes of the L. crispatus strains identified were also characterized using L. crispatus species-specific primers: Lbc_fw: 5′-AGGATATGGAGAGCAGGAAT-3′ and Lbc_rv: 5′-CAACTATCTCTTACACACGCC-3′ (90–93). Each sample was subjected to the following thermal cycling conditions: 5 min at 94°C for one cycle, then 20 s at 94°C, 30 s at 57°C, and 40 s at 72°C for 30 cycles, followed by a 5-min elongation period at 72°C. The L. crispatus isolated strains are listed in Table 1.
pH and sodium chloride tolerance tests.
The ability of isolated strains to tolerate various pH levels or NaCl concentrations was evaluated by monitoring optical density (OD) values on 96-well plates. In different wells, MRS medium enhanced with 1% lactose was supplemented with 2%, 6%, or 10% NaCl (wt/vol). In addition, isolated strains were also cultivated in MRS medium supplemented with 1% lactose and set to pH 2.0, pH 3.0, or pH 4.0 by the addition of HCl. Microtiter plates were incubated under aerobic conditions for 48 h at 37°C. Cell density was monitored by OD measurements at OD600 using a plate reader (BioTek, VT, USA). The resistance level to the imposed sodium chloride or pH stress was calculated in each case by comparing the maximum OD600 reached in a particular medium with that of the control medium (MRS + 1% of lactose). Assays were performed in triplicate as independent experiments.
Glycogen growth assays.
Lactobacillus strains were cultivated on semisynthetic MRS medium without sugar supplemented with 1% (wt/vol) of glycogen (Merck, Germany) as the sole carbon source. OD measurements at a wavelength of 600 nm were detected using a plate reader (BioTek, VT, USA). The plate reader was read in intermittent mode, with absorbance readings performed at 3 min intervals for three times after 12, 15, 18, 24, 36, 40, and 48 h of growth, where each reading was ahead of 30 s shaking at medium speed. Cultures were grown in biologically independent triplicates in aerobic conditions at 37°C. The resulting growth data were expressed as the mean of these replicates. Control growth experiments were carried out with strains growth in MRS medium supplemented with 1% lactose (wt/vol).
Detection of hydrogen peroxide production.
Lactobacillus strains were tested for the production of H2O2. Each strain was plated on TMB-plus agar, as previously described (56). In brief, lactobacilli were plated on TMB-plus agar and incubated for 48 h at 37°C in anaerobic conditions (2.99% H2, 17.01% CO2, and 80% N2) in a chamber (Concept 400, Ruskin). After 30 min of exposure to ambient air, the blue color colony morphology of hydrogen peroxide-producing bacteria was evaluated, as previously described (18). Color intensity was defined based on the transposition of the intensity of the colonies blue color on Adobe Photoshop software (Adobe Incorporated, CA, USA) after photography of each plate. The color grade was designated with the combination of the red, green and blue (RGB) color method lights, maintaining the R and the G grade to 0. Each experiment was performed in triplicate.
Genome sequencing and assemblies.
DNA extracted from lactobacillus isolates was subjected to whole-genome sequencing using MiSeq (Illumina, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Illumina, UK). Furthermore, DNA isolated from L. crispatus PRL2021 was also subjected to whole-genome sequencing using a MinION (Oxford Nanopore, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Oxford Nanopore, UK). Fastq files of the paired-end reads obtained from targeted genome sequencing of isolated strains were used as input for genome assemblies through the MEGAnnotator pipeline (94). SPAdes software was used for de novo assembly of each L. crispatus genome sequence (95, 96), while protein-encoding ORFs were predicted using Prodigal (97). The coverage depth of these newly isolated 16 L. crispatus chromosomes ranged from 73- to 259-fold, which upon assembly generated 36 to 259 contigs (Table 1).
Comparative genomics.
A pangenome calculation was performed using the pangenome analysis pipeline PGAP (98), including each L. crispatus genome collected from this study and 94 public available L. crispatus genomes (NCBI database) (Table S1). Each predicted proteome of a given L. crispatus strain was screened for orthologues against the proteome of every collected L. crispatus strain by means of BLAST analysis (99) (cutoff at E value of <1 × 10−4 and 50% identity over at least 80% of both protein sequences). The resulting output was then clustered into protein families by means of MCL (graph theory-based Markov clustering algorithm) (100), using the gene family method. A pangenome profile was built using all possible BLAST combinations for each genome being sequentially added. Using this approach, unique protein families encoded by the analyzed L. crispatus genomes were also identified. Protein families shared between analyzed genomes allowed us to identify the core genome of the L. crispatus species. Each set of orthologous proteins belonging to the core genome was aligned using Mafft software (101) and phylogenetic trees were constructed using ClustalW (102). Based on these comparative analyses, an L. crispatus supertree was visualized using FigTree (https://github.com/rambaut/figtree/releases). The average nucleotide identity (ANI) values among L. crispatus genomes analyzed was calculated with fastANI software (103). Functional annotation of each protein of L. crispatus strains was performed employing the eggNOG database (61).
Prediction of putative bacteriocin-encoding genes.
The genome sequences of the 16 isolated and 94 publicly available L. crispatus genomes were screened for bacteriocin-encoding genes. Moreover, 116 genomes publicly available belonging to L. iners, L. jensenii, and L. gasseri species were explored for the bacteriocin prediction. The screening was carried out using the BAGEL3 database (65) through BLASTP analysis (E value cutoff of 1e−5) (99). Afterward, a manual examination of the sequences predicted to encode a bacteriocin-like protein was performed. The predicted bacteriocin genes were classified according to the bacteriocin classification reported in the BAGEL3 database (65).
Growth experiment of the isolated vaginal L. crispatus in simulated vaginal fluid and shallow shotgun metagenomics.
The simulated vaginal fluid (SVF) was prepared as described by Pan et al. (31). For growth experiments, overnight lactobacillus cultures were diluted to an OD value of 1.0. Each culture was inoculated at 1% (vol/vol) into SVF. We performed nine different experiments in which were inoculated L. gasseri ATCC 9857, L. iners LMG 14328, and L. jensenii V94G (isolated in this study), together with one new vaginal-isolated L. crispatus strain or the vaginal probiotic product strain L. crispatus M247. Batch cultures were incubated under aerobic conditions for 12 h at 37°C. After 12 h of growth, the cultures were centrifuged at 3,000 rpm for 8 min and the pellets were harvested. The pellets were subjected to DNA extraction using the GeneElute bacterial genomic DNA kit (Sigma, Germany) following the manufacturer’s instruction. The extracted DNA was prepared following the Illumina Nextera XT protocol. Briefly, DNA samples were enzymatically fragmented, barcoded, and purified involving magnetic beads. Then, samples were quantified using fluorometric Qubit quantification system (Life Technologies, USA), loaded on a 2200 Tape Station instrument (Agilent Technologies, USA), and normalized to 4 nM. Sequencing was performed by paired-end reads using an Illumina NextSeq 500 sequencer with NextSeq High Output v2 kit chemicals (Illumina Inc., San Diego, USA). The retained reads were analyzed with the METAnnotatorX bioinformatics platform (88).
Growth experiments employing L. crispatus PRL2021 in SVF and fecal medium.
An overnight L. crispatus PRL2021 culture was diluted to reach an OD600 value of 1.0 and was then inoculated at 1% (vol/vol) into SVF and fecal medium. The SVF was prepared as described by Pan et al. (31), whereas the fecal medium was prepared as described by Macfarlane et al. (104). We performed four different experiments for each growth medium in which were inoculated a woman’s fecal sample at 1% (wt/vol) or 5% (wt/vol) together with L. crispatus PRL2021. Control growth experiments were carried out without L. crispatus PRL2021. After 12 h of growth, the cultures were centrifuged at 3,000 rpm for 8 min and the pellets were harvested. The pellets were subjected to DNA extraction using the QIAamp DNA stool minikit (Qiagen) following the manufacturer’s instructions. The extracted DNA was prepared following the Illumina Nextera XT protocol as mentioned previously for coculture experiments.
Evaluation of cell density by flow cytometry assay.
For bacterial cell counting, 500 μl of batch culture was diluted in physiological solution (phosphate-buffered saline, PBS). Subsequently, bacterial cells were stained with 1μl of SYBR Green I and incubated in the dark for at least 15 min before measurement. All count experiments were performed using an Attune NxT Flow flow cytometer (Invitrogen, ThermoFisher Scientific) equipped with a blue laser set at 50 mW and tuned to an excitation wavelength of 488 nm. Multiparametric analyses were performed on both scattering signals (FSC, SSC) and SYBR Green I fluorescence was detected on FL1 channel. Cell debris and eukaryotic cells were excluded from acquisition analysis by a sample-specific FL1 threshold. All data sets were statistically analyzed with Attune NxT flow cytometer software. Utilizing these cell counts to normalize the sequencing data into absolute abundance of each profiled taxon, we were able to perform quantitative microbiome profiling using a previously described method (86, 87).
Statistical analysis.
The hierarchical clustering (HCL) of samples was obtained using bacterial composition at the species level and was calculated through TMeV 4.8.1 software using Pearson correlation as a distance metric based on information at the genus level. The data obtained were represented by a cladogram. SPSS software (IBM, Italy) was used to perform statistical analysis by Student’s t test, ANOVA, and by Kendall tau rank cooccurrence. Moreover, we calculated the post hoc analysis LSD (least significant difference) for multiple comparisons. The figures were generated through Microsoft Excel (Microsoft 365) software.
Data availability.
Raw sequences of the shallow shotgun metagenomics profiling experiments are accessible through SRA study accession number PRJNA641015. Newly isolated Lactobacillus genomes were sequenced and deposited with the BioSample numbers reported in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma. D.v.S. is a member of the APC Microbiome Institute, Ireland, which is funded by SFI through the Irish government’s National Development Plan (grant numbers SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ma B, Forney LJ, Ravel J. 2012. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66:371–389. 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis FM, Bernstein KT, Aral SO. 2017. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 129:643–654. 10.1097/AOG.0000000000001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Łaniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, Herbst-Kralovetz MM. 2018. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep 8:7593. 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamarelle J, Thiebaut ACM, de Barbeyrac B, Bebear C, Ravel J, Delarocque-Astagneau E. 2019. The vaginal microbiota and its association with human papillomavirus, Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect 25:35–47. 10.1016/j.cmi.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stapleton AE. 2016. The vaginal microbiota and urinary tract infection. Microbiol Spectr 4. 10.1128/microbiolspec.UTI-0025-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687. 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. 2012. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res 160:267–282. 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunn KL, Forney LJ. 2016. Unraveling the dynamics of the human vaginal microbiome. Yale J Biol Med 89:331–337. [PMC free article] [PubMed] [Google Scholar]
- 9.Martin DH, Marrazzo JM. 2016. The vaginal microbiome: current understanding and future directions. J Infect Dis 214 Suppl 1:S36–41. 10.1093/infdis/jiw184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onywera H, Williamson AL, Mbulawa ZZA, Coetzee D, Meiring TL. 2019. The cervical microbiota in reproductive-age South African women with and without human papillomavirus infection. Papillomavirus Res 7:154–163. 10.1016/j.pvr.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwasniewski W, Wolun-Cholewa M, Kotarski J, Warchol W, Kuzma D, Kwasniewska A, Gozdzicka-Jozefiak A. 2018. Microbiota dysbiosis is associated with HPV-induced cervical carcinogenesis. Oncol Lett 16:7035–7047. 10.3892/ol.2018.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgdorff H, van der Veer C, van Houdt R, Alberts CJ, de Vries HJ, Bruisten SM, Snijder MB, Prins M, Geerlings SE, Schim van der Loeff MF, van de Wijgert J. 2017. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One 12:e0181135. 10.1371/journal.pone.0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dols JA, Molenaar D, van der Helm JJ, Caspers MP, de Kat Angelino-Bart A, Schuren FH, Speksnijder AG, Westerhoff HV, Richardus JH, Boon ME, Reid G, de Vries HJ, Kort R. 2016. Molecular assessment of bacterial vaginosis by Lactobacillus abundance and species diversity. BMC Infect Dis 16:180. 10.1186/s12879-016-1513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Wang Q, Yang E, Yan L, Li T, Zhuang H. 2017. Antimicrobial compounds produced by vaginal Lactobacillus crispatus are able to strongly inhibit Candida albicans growth, hyphal formation and regulate virulence-related gene expressions. Front Microbiol 8:564. 10.3389/fmicb.2017.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahui Palomino RA, Vanpouille C, Laghi L, Parolin C, Melikov K, Backlund P, Vitali B, Margolis L. 2019. Extracellular vesicles from symbiotic vaginal lactobacilli inhibit HIV-1 infection of human tissues. Nat Commun 10:5656. 10.1038/s41467-019-13468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ. 2014. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J Bacteriol 196:1458–1470. 10.1128/JB.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borges S, Silva J, Teixeira P. 2014. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet 289:479–489. 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 18.Hutt P, Lapp E, Stsepetova J, Smidt I, Taelma H, Borovkova N, Oopkaup H, Ahelik A, Roop T, Hoidmets D, Samuel K, Salumets A, Mandar R. 2016. Characterisation of probiotic properties in human vaginal lactobacilli strains. Microb Ecol Health Dis 27:30484. 10.3402/mehd.v27.30484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prabhurajeshwar C, Chandrakanth RK. 2017. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: an in vitro validation for the production of inhibitory substances. Biomed J 40:270–283. 10.1016/j.bj.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asghar S, Arif M, Nawaz M, Muhammad K, Ali MA, Ahmad MD, Iqbal S, Anjum AA, Khan M, Nazir J. 2016. Selection, characterisation and evaluation of potential probiotic Lactobacillus spp. isolated from poultry droppings. Benef Microbes 7:35–44. 10.3920/BM2015.0020. [DOI] [PubMed] [Google Scholar]
- 21.Dec M, Puchalski A, Nowaczek A, Wernicki A. 2016. Antimicrobial activity of Lactobacillus strains of chicken origin against bacterial pathogens. Int Microbiol 19:57–67. 10.2436/20.1501.01.264. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power SE, Harris HM, Bottacini F, Ross RP, O'Toole PW, Fitzgerald GF. 2013. Draft genome sequence of Lactobacillus crispatus EM-LC1, an isolate with antimicrobial activity cultured from an elderly subject. Genome Announc 1:e01070-13. 10.1128/genomeA.01070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hidalgo-Cantabrana C, Goh YJ, Pan M, Sanozky-Dawes R, Barrangou R. 2019. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc Natl Acad Sci U S A 116:15774–15783. 10.1073/pnas.1905421116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid G. 2019. The need to focus on therapy instead of associations. Front Cell Infect Microbiol 9:327. 10.3389/fcimb.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doerflinger SY, Throop AL, Herbst-Kralovetz MM. 2014. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis 209:1989–1999. 10.1093/infdis/jiu004. [DOI] [PubMed] [Google Scholar]
- 27.Rose WA, 2nd, McGowin CL, Spagnuolo RA, Eaves-Pyles TD, Popov VL, Pyles RB. 2012. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS One 7:e32728. 10.1371/journal.pone.0032728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stapleton AE, Au-Yeung M, Hooton TM, Fredricks DN, Roberts PL, Czaja CA, Yarova-Yarovaya Y, Fiedler T, Cox M, Stamm WE. 2011. Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin Infect Dis 52:1212–1217. 10.1093/cid/cir183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kort R. 2014. Personalized therapy with probiotics from the host by TripleA. Trends Biotechnol 32:291–293. 10.1016/j.tibtech.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Liu Z, Zhang X, Chen X, Wang S. 2019. Local probiotic Lactobacillus crispatus and Lactobacillus delbrueckii exhibit strong antifungal effects against vulvovaginal candidiasis in a rat model. Front Microbiol 10:1033. 10.3389/fmicb.2019.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan M, Hidalgo-Cantabrana C, Goh YJ, Sanozky-Dawes R, Barrangou R. 2019. Comparative analysis of Lactobacillus gasseri and Lactobacillus crispatus isolated from human urogenital and gastrointestinal tracts. Front Microbiol 10:3146. 10.3389/fmicb.2019.03146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DS, Wong RJ, Shaw G, Stevenson DK, Holmes SP, Relman DA. 2015. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A 112:11060–11065. 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Seta F, Campisciano G, Zanotta N, Ricci G, Comar M. 2019. The vaginal community state types microbiome-immune network as key factor for bacterial vaginosis and aerobic vaginitis. Front Microbiol 10:2451. 10.3389/fmicb.2019.02451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma ZS, Li L. 2017. Quantifying the human vaginal community state types (CSTs) with the species specificity index. PeerJ 5:e3366. 10.7717/peerj.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4:132ra52. 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Chen C, Wei W, Wang Z, Dai J, Hao L, Song L, Zhang X, Zeng L, Du H, Tang H, Liu N, Yang H, Wang J, Madsen L, Brix S, Kristiansen K, Xu X, Li J, Wu R, Jia H. 2018. The metagenome of the female upper reproductive tract. Gigascience 7:giy107. 10.1093/gigascience/giy107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Zhang Y, Zhang Q, Chen H, Feng Y. 2018. Characterization of pelvic and cervical microbiotas from patients with pelvic inflammatory disease. J Med Microbiol 67:1519–1526. 10.1099/jmm.0.000821. [DOI] [PubMed] [Google Scholar]
- 38.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, McDonald D, Franzosa EA, Knight R, White O, Huttenhower C. 2017. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550:61–66. 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D. 2018. Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendling W. 2016. Vaginal microbiota. Adv Exp Med Biol 902:83–93. 10.1007/978-3-319-31248-4_6. [DOI] [PubMed] [Google Scholar]
- 41.Salonen A, Salojarvi J, Lahti L, de Vos WM. 2012. The adult intestinal core microbiota is determined by analysis depth and health status. Clin Microbiol Infect 18 Suppl 4:16–20. 10.1111/j.1469-0691.2012.03855.x. [DOI] [PubMed] [Google Scholar]
- 42.Alessandri G, Milani C, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M. 2019. Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ Microbiol 21:1331–1343. 10.1111/1462-2920.14540. [DOI] [PubMed] [Google Scholar]
- 43.Alessandri G, Milani C, Mancabelli L, Longhi G, Anzalone R, Lugli GA, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M. 2020. Deciphering the bifidobacterial populations within the canine and feline gut microbiota. Appl Environ Microbiol 86:e02875-19. 10.1128/AEM.02875-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mancabelli L, Milani C, Lugli GA, Turroni F, Ferrario C, van Sinderen D, Ventura M. 2017. Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Environ Microbiol 19:1379–1390. 10.1111/1462-2920.13692. [DOI] [PubMed] [Google Scholar]
- 45.Adhikari B, Kwon YM. 2017. Characterization of the culturable subpopulations of Lactobacillus in the chicken intestinal tract as a resource for probiotic development. Front Microbiol 8:1389. 10.3389/fmicb.2017.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castagliuolo I, Galeazzi F, Ferrari S, Elli M, Brun P, Cavaggioni A, Tormen D, Sturniolo GC, Morelli L, Palu G. 2005. Beneficial effect of auto-aggregating Lactobacillus crispatus on experimentally induced colitis in mice. FEMS Immunol Med Microbiol 43:197–204. 10.1016/j.femsim.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Linhares IM, Summers PR, Larsen B, Giraldo PC, Witkin SS. 2011. Contemporary perspectives on vaginal pH and lactobacilli. Am J Obstet Gynecol 204:120. 10.1016/j.ajog.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. 2015. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol 6:81. 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SB, Ravel J. 2017. The vaginal microbiota, host defence and reproductive physiology. J Physiol 595:451–463. 10.1113/JP271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rastogi R, Su J, Mahalingam A, Clark J, Sung S, Hope T, Kiser PF. 2016. Engineering and characterization of simplified vaginal and seminal fluid simulants. Contraception 93:337–346. 10.1016/j.contraception.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nami Y, Haghshenas B, Yari Khosroushahi A. 2018. Molecular identification and probiotic potential characterization of lactic acid bacteria isolated from human vaginal microbiota. Adv Pharm Bull 8:683–695. 10.15171/apb.2018.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amabebe E, Anumba DOC. 2018. The vaginal microenvironment: the physiologic role of lactobacilli. Front Med (Lausanne) 5:181. 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirmonsef P, Hotton AL, Gilbert D, Burgad D, Landay A, Weber KM, Cohen M, Ravel J, Spear GT. 2014. Free glycogen in vaginal fluids is associated with Lactobacillus colonization and low vaginal pH. PLoS One 9:e102467. 10.1371/journal.pone.0102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Veer C, Hertzberger RY, Bruisten SM, Tytgat HLP, Swanenburg J, de Kat Angelino-Bart A, Schuren F, Molenaar D, Reid G, de Vries H, Kort R. 2019. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: implications for in vivo dominance of the vaginal microbiota. Microbiome 7:49. 10.1186/s40168-019-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chetwin E, Manhanzva MT, Abrahams AG, Froissart R, Gamieldien H, Jaspan H, Jaumdally SZ, Barnabas SL, Dabee S, Happel AU, Bowers D, Davids L, Passmore JS, Masson L. 2019. Antimicrobial and inflammatory properties of South African clinical Lactobacillus isolates and vaginal probiotics. Sci Rep 9:1917. 10.1038/s41598-018-38253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabe LK, Hillier SL. 2003. Optimization of media for detection of hydrogen peroxide production by Lactobacillus species. J Clin Microbiol 41:3260–3264. 10.1128/jcm.41.7.3260-3264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallor AC, Antonio MA, Hawes SE, Hillier SL. 2001. Factors associated with acquisition of, or persistent colonization by, vaginal lactobacilli: role of hydrogen peroxide production. J Infect Dis 184:1431–1436. 10.1086/324445. [DOI] [PubMed] [Google Scholar]
- 58.Hawes SE, Hillier SL, Benedetti J, Stevens CE, Koutsky LA, Wolner-Hanssen P, Holmes KK. 1996. Hydrogen peroxide-producing lactobacilli and acquisition of vaginal infections. J Infect Dis 174:1058–1063. 10.1093/infdis/174.5.1058. [DOI] [PubMed] [Google Scholar]
- 59.Klebanoff SJ, Hillier SL, Eschenbach DA, Waltersdorph AM. 1991. Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J Infect Dis 164:94–100. 10.1093/infdis/164.1.94. [DOI] [PubMed] [Google Scholar]
- 60.Ocana VS, Pesce de Ruiz Holgado AA, Nader-Macias ME. 1999. Selection of vaginal H2O2-generating Lactobacillus species for probiotic use. Curr Microbiol 38:279–284. 10.1007/pl00006802. [DOI] [PubMed] [Google Scholar]
- 61.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–9. 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q, Zhang L, Ross P, Zhao J, Zhang H, Chen W. 2020. Comparative genomics of Lactobacillus crispatus from the gut and vagina reveals genetic diversity and lifestyle adaptation. Genes (Basel) 11:360. 10.3390/genes11040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koryszewska-Bagińska A, Gawor J, Nowak A, Grynberg M, Aleksandrzak-Piekarczyk T. 2019. Comparative genomics and functional analysis of a highly adhesive dairy Lactobacillus paracasei subsp. paracasei IBB3423 strain. Appl Microbiol Biotechnol 103:7617–7634. 10.1007/s00253-019-10010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. 2013. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev 37:762–792. 10.1111/1574-6976.12029. [DOI] [PubMed] [Google Scholar]
- 65.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. 2013. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–53. 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joerger MC, Klaenhammer TR. 1986. Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J Bacteriol 167:439–446. 10.1128/jb.167.2.439-446.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoo D, Bagon BB, Valeriano VDV, Oh JK, Kim H, Cho S, Kang DK. 2017. Complete genome analysis of Lactobacillus fermentum SK152 from kimchi reveals genes associated with its antimicrobial activity. FEMS Microbiol Lett 364. 10.1093/femsle/fnx185. [DOI] [PubMed] [Google Scholar]
- 68.Busarcevic M, Dalgalarrondo M. 2012. Purification and genetic characterisation of the novel bacteriocin LS2 produced by the human oral strain Lactobacillus salivarius BGHO1. Int J Antimicrob Agents 40:127–134. 10.1016/j.ijantimicag.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 69.Tosukhowong A, Zendo T, Visessanguan W, Roytrakul S, Pumpuang L, Jaresitthikunchai J, Sonomoto K. 2012. Garvieacin Q, a novel class II bacteriocin from Lactococcus garvieae BCC 43578. Appl Environ Microbiol 78:1619–1623. 10.1128/AEM.06891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tymoszewska A, Diep DB, Wirtek P, Aleksandrzak-Piekarczyk T. 2017. The non-lantibiotic bacteriocin garvicin Q targets Man-PTS in a broad spectrum of sensitive bacterial genera. Sci Rep 7:8359. 10.1038/s41598-017-09102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tymoszewska A, Diep DB, Aleksandrzak-Piekarczyk T. 2018. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci Rep 8:15790. 10.1038/s41598-018-34087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102:13950–13955. 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou X, Yang B, Stanton C, Ross RP, Zhao J, Zhang H, Chen W. 2020. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genomics 21:119. 10.1186/s12864-020-6527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fontana A, Falasconi I, Molinari P, Treu L, Basile A, Vezzi A, Campanaro S, Morelli L. 2019. Genomic comparison of Lactobacillus helveticus strains highlights probiotic potential. Front Microbiol 10:1380. 10.3389/fmicb.2019.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Ganzle MG, Lebeer S. 2020. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 78.Clabaut M, Boukerb AM, Racine PJ, Pichon C, Kremser C, Picot JP, Karsybayeva M, Redziniak G, Chevalier S, Feuilloley MGJ. 2020. Draft genome sequence of Lactobacillus crispatus CIP 104459, isolated from a vaginal swab. Microbiol Resour Announc 9:e01373-19. 10.1128/MRA.01373-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Price TK, Shaheen M, Kalesinskas L, Malki K, Hilt EE, Putonti C, Wolfe AJ. 2016. Draft genome sequence of a urinary isolate of Lactobacillus crispatus. Genome Announc 4:e01278-16. 10.1128/genomeA.01278-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas-White KJ, Kliethermes S, Rickey L, Lukacz ES, Richter HE, Moalli P, Zimmern P, Norton P, Kusek JW, Wolfe AJ, Brubaker L, National Institute of Diabetes and Digestive and Kidney Diseases Urinary Incontinence Treatment Network. 2017. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol 216:55.e1–55.e16. 10.1016/j.ajog.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falsen E, Pascual C, Sjoden B, Ohlen M, Collins MD. 1999. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol 49 Pt 1:217–221. 10.1099/00207713-49-1-217. [DOI] [PubMed] [Google Scholar]
- 82.Antonio MA, Rabe LK, Hillier SL. 2005. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J Infect Dis 192:394–398. 10.1086/430926. [DOI] [PubMed] [Google Scholar]
- 83.Brotman RM, Melendez JH, Ghanem KG. 2011. A case control study of anovaginal distance and bacterial vaginosis. Int J STD AIDS 22:231–233. 10.1258/ijsa.2011.010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reid G, Bruce AW, Fraser N, Heinemann C, Owen J, Henning B. 2001. Oral probiotics can resolve urogenital infections. FEMS Immunol Med Microbiol 30:49–52. 10.1111/j.1574-695X.2001.tb01549.x. [DOI] [PubMed] [Google Scholar]
- 85.Hilton E, Isenberg HD, Alperstein P, France K, Borenstein MT. 1992. Ingestion of yogurt containing Lactobacillus acidophilus as prophylaxis for candidal vaginitis. Ann Intern Med 116:353–357. 10.7326/0003-4819-116-5-353. [DOI] [PubMed] [Google Scholar]
- 86.Vandeputte D, Kathagen G, D'Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 87.Mancino W, Duranti S, Mancabelli L, Longhi G, Anzalone R, Milani C, Lugli GA, Carnevali L, Statello R, Sgoifo A, van Sinderen D, Ventura M, Turroni F. 2019. Bifidobacterial transfer from mother to child as examined by an animal model. Microorganisms 7:293. 10.3390/microorganisms7090293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milani C, Casey E, Lugli GA, Moore R, Kaczorowska J, Feehily C, Mangifesta M, Mancabelli L, Duranti S, Turroni F, Bottacini F, Mahony J, Cotter PD, McAuliffe FM, van Sinderen D, Ventura M. 2018. Tracing mother-infant transmission of bacteriophages by means of a novel analytical tool for shotgun metagenomic datasets: METAnnotatorX. Microbiome 6:145. 10.1186/s40168-018-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yeruva T, Rajkumar H, Donugama V. 2017. Vaginal lactobacilli profile in pregnant women with normal & abnormal vaginal flora. Indian J Med Res 146:534–540. 10.4103/ijmr.IJMR_774_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brolazo EM, Leite DS, Tiba MR, Villarroel M, Marconi C, Simoes JA. 2011. Correlation between api 50 ch and multiplex polymerase chain reaction for the identification of vaginal lactobacilli in isolates. Braz J Microbiol 42:225–232. 10.1590/S1517-83822011000100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang R, Daroczy K, Xiao B, Yu L, Chen R, Liao Q. 2012. Qualitative and semiquantitative analysis of Lactobacillus species in the vaginas of healthy fertile and postmenopausal Chinese women. J Med Microbiol 61:729–739. 10.1099/jmm.0.038687-0. [DOI] [PubMed] [Google Scholar]
- 93.Song Y, Kato N, Liu C, Matsumiya Y, Kato H, Watanabe K. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol Lett 187:167–173. 10.1111/j.1574-6968.2000.tb09155.x. [DOI] [PubMed] [Google Scholar]
- 94.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 95.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 100.Vlietstra WJ, Zielman R, van Dongen RM, Schultes EA, Wiesman F, Vos R, van Mulligen EM, Kors JA. 2017. Automated extraction of potential migraine biomarkers using a semantic graph. J Biomed Inform 71:178–189. 10.1016/j.jbi.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 101.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Macfarlane GT, Macfarlane S, Gibson GR. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187. 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences of the shallow shotgun metagenomics profiling experiments are accessible through SRA study accession number PRJNA641015. Newly isolated Lactobacillus genomes were sequenced and deposited with the BioSample numbers reported in Table 1.