Compared to natural environments, the reduced diversity of monoculture agricultural landscapes can lead bacterial plant pathogens to quickly adapt to local biological and ecological conditions. Because of this, accidental introductions of microbial pathogens into naive regions represents a significant economic and environmental threat. Xylella fastidiosa is a plant pathogen with an expanding host and geographic range due to multiple intra- and intercontinental introductions. X. fastidiosa subsp. fastidiosa infects and causes disease in grapevines (Pierce’s disease of grapevines, PD).
KEYWORDS: allopatric, emerging disease, Pierce's disease, Xylella fastidiosa
ABSTRACT
Within the landscape of globally distributed pathogens, populations differentiate via both adaptive and nonadaptive forces. Individual populations are likely to show unique trends of genetic diversity, host-pathogen interaction, and ecological adaptation. In plant pathogens, allopatric divergence may occur particularly rapidly within simplified agricultural monoculture landscapes. As such, the study of plant pathogen populations in monocultures can highlight the distinct evolutionary mechanisms that lead to local genetic differentiation. Xylella fastidiosa is a plant pathogen known to infect and damage multiple monocultures worldwide. One subspecies, Xylella fastidiosa subsp. fastidiosa, was first introduced to the United States ∼150 years ago, where it was found to infect and cause disease in grapevines (Pierce’s disease of grapevines, or PD). Here, we studied PD-causing subsp. fastidiosa populations, with an emphasis on those found in the United States. Our study shows that following their establishment in the United States, PD-causing strains likely split into populations on the East and West Coasts. This diversification has occurred via both changes in gene content (gene gain/loss events) and variations in nucleotide sequence (mutation and recombination). In addition, we reinforce the notion that PD-causing populations within the United States acted as the source for subsequent subsp. fastidiosa outbreaks in Europe and Asia.
IMPORTANCE Compared to natural environments, the reduced diversity of monoculture agricultural landscapes can lead bacterial plant pathogens to quickly adapt to local biological and ecological conditions. Because of this, accidental introductions of microbial pathogens into naive regions represents a significant economic and environmental threat. Xylella fastidiosa is a plant pathogen with an expanding host and geographic range due to multiple intra- and intercontinental introductions. X. fastidiosa subsp. fastidiosa infects and causes disease in grapevines (Pierce’s disease of grapevines [PD]). This study focused on PD-causing X. fastidiosa populations, particularly those found in the United States but also invasions into Taiwan and Spain. The analysis shows that PD-causing X. fastidiosa has diversified via multiple cooccurring evolutionary forces acting at an intra- and interpopulation level. This analysis enables a better understanding of the mechanisms leading to the local adaptation of X. fastidiosa and how a plant pathogen diverges allopatrically after multiple and sequential introduction events.
INTRODUCTION
The worldwide distribution of microbial plant pathogens is constantly shifting. Global trade and the movement of infected plant material enables pathogen introductions from native and endemic areas to naive regions (1, 2). Likewise, the intentional introduction of nonnative plant species of agronomic and ornamental value to novel environments facilitates the host range expansion of endemic pathogens (3, 4). One crucial factor in the formation of novel plant-pathogen associations is the amount of genetic diversity on which natural selection can act, in other words, the adaptive potential (5). Differences in adaptive potential between host and microbial populations can have a significant role in determining the host and geographic range of a pathogen. For instance, in the case of plant pathogens, higher genetic diversity in effector proteins and virulence genes has a positive effect on host range (6–9). Alternatively, multiple studies have highlighted how reduced genetic diversity in plant hosts can enhance the spread of pathogens within a population (10–12).
Factors that influence genetic diversity, whether via the action of distinct evolutionary mechanisms (13, 14) or as a product of ecological and evolutionary history, affect adaptive potential (15). In plant pathogens, geographical and ecological specialization have been frequently described (16, 17). This is partly explained by plant pathogen differentiation and specialization occurring rapidly within agricultural systems (14, 18, 19). Overall, it is expected that in the absence of gene flow, plant pathogens of agricultural crops will rapidly adapt to local environmental, ecological, and biological conditions (20, 21). Therefore, understanding the mechanisms leading to pathogen adaptation, either to a new crop or environmental condition, has great relevance in developing effective management and control strategies (22). This is particularly pertinent in plant pathogens with a proven capacity to adapt to multiple crops as well as having an expanding geographic and host range. This is the case of the emerging pathogen Xylella fastidiosa (23).
The bacterial species X. fastidiosa has been reported to infect 563 plant species from 82 distinct botanical families (23). However, the host range of X. fastidiosa varies among and within described subspecies and phylogenetic clades (24). The geographic distribution of the three main X. fastidiosa subspecies is also unique, with most of them having experienced one or several dispersal and establishment events at the continental scale. For this reason, efficient identification and tracking of X. fastidiosa subspecies has important implications for the development of adequate disease control and mitigation strategies (25, 26). Three X. fastidiosa subspecies have an ancestrally allopatric range that has recently expanded: X. fastidiosa subsp. multiplex is native to temperate and subtropical North America (27, 28) and has been introduced multiple times into Europe (29); X. fastidiosa subsp. pauca is native to South America (28) but was recently reported in the Apulian region in Italy and in Costa Rica (30, 31); and finally, X. fastidiosa subsp. fastidiosa is native to Central America (32, 33) and was introduced to the United States (24, 34) and, subsequently, to Europe (35) and Taiwan (36). Other nonmonophyletic but proposed subspecies include X. fastidiosa subsp. sandyi, found in southern regions of the United States (37, 38) and also introduced into Europe (39), and X. fastidiosa subsp. morus, found only in regions where subsp. multiplex and subsp. fastidiosa cooccur (24, 40).
The hypothesis that subsp. fastidiosa was introduced once to the United States ∼150 years ago, leading to the emergence of Pierce’s disease of grapevines (PD), is well supported (24, 33, 41). PD is a grapevine malady that results in significant economic losses to the wine industry in California (42) and the Southeast United States (43). Current knowledge of the evolution of subsp. fastidiosa suggests that the ability to infect grapevines was acquired after its introduction into the United States (33). Furthermore, there is evidence that local adaptation to environmental factors has occurred in grape-infecting isolates across a latitudinal gradient in California (34). Finally, available genomic and multilocus sequence analysis of environmentally mediated genes data suggest that PD-causing isolates on the West and East Coast of the United States are phylogenetically distinct (34, 44).
These studies are indicative that after its introduction and establishment in the United States, the subsp. fastidiosa clade causing disease in grapevines dispersed to different geographic regions and diversified genetically to adapt to a range of biotic and abiotic conditions. To better understand how X. fastidiosa evolved with the emergence of a novel plant disease (PD) and diversified in allopatry in different regions of the United States, we studied populations of the pathogen from the United States and abroad. We evaluated the evolutionary relationship between U.S. populations and their relationship with recent introduction events derived from them (i.e., introductions to Spain and Taiwan associated with the emergence of PD in those regions). In addition, we identified the evolutionary mechanisms facilitating population diversification by defining intrapopulation patterns of gene gain/loss, intrasubspecific recombination, and nucleotide diversity.
RESULTS
PD isolates are split into regional clades within the United States, with European and Asian introductions originating from these regions.
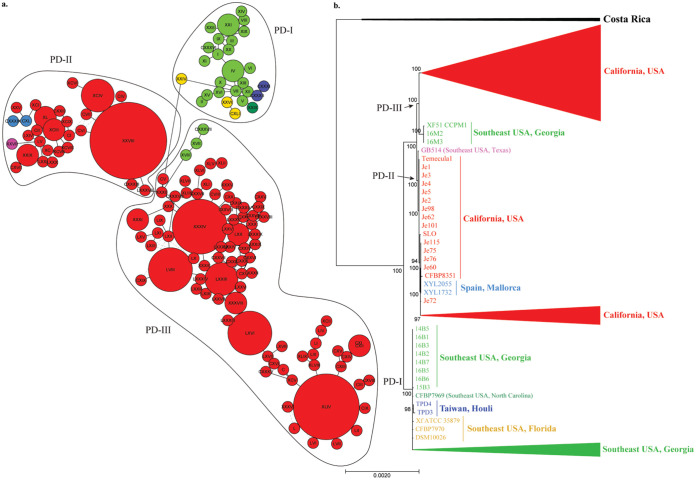
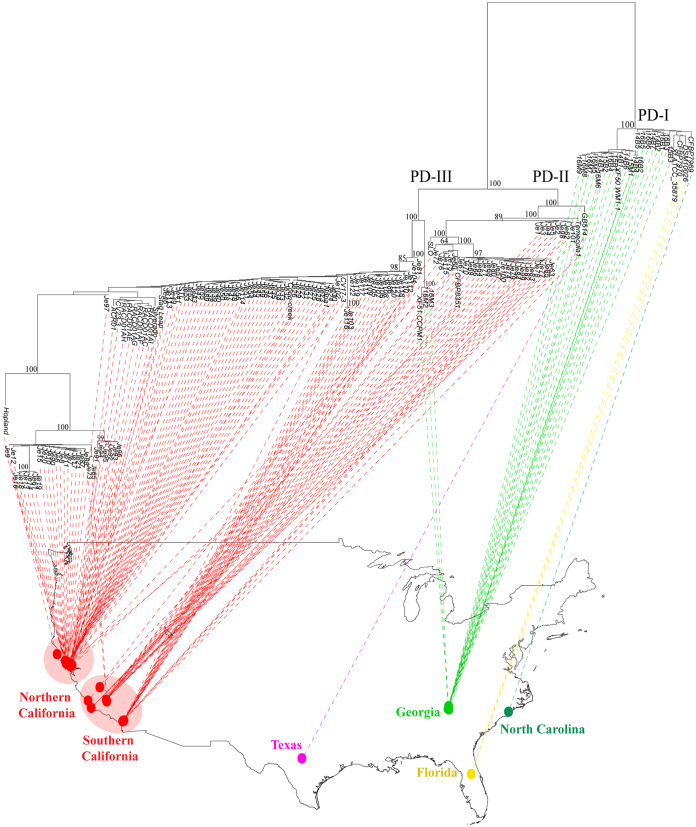
We arbitrarily split grapevine isolates into 3 phylogenetically supported clades, PD-I to -III (Fig. 1). These phylogenetically supported clades were also observed in the nonrecombinant phylogenetic tree (see Fig. S1 in the supplemental material). PD-I only included isolates from the Southeast United States; PD-II and PD-III were dominated by California isolates, but at the base of those clades there was one isolate from Texas (PD-II) and a sister clade from Georgia (PD-III). No isolates from California grapevines clustered within the PD-I clade. From the data available alone, it is not possible to infer the dispersal history of the non-California isolates in PD-II and PD-III (i.e., basal sister clades or introductions to California). Isolates from Taiwan were phylogenetically placed within the PD-I clade, while those from Spain were nested in the PD-II clade. These represent two distinct introductions, originating from different regions in the United States. Isolates from the same geographic region tended to cluster together within each major clade. For instance, in the PD-I clade, most Georgia isolates from Site1 (i.e., 14B1, 14B4, 14B6, 16B2, 15B2, 14B3, and 16B4) and Site2 (i.e., 16M5, 16M6, 16M7, 16M8, and 16M9) clustered together. Isolates from each site formed separate subclades within this group (Fig. 2). Other Georgia isolates from Site1 (i.e., 14B2, 14B5, 14B7, 16B1, 16B3, 16B5, and 16B6) were more closely related to those from Florida and North Carolina. In a similar manner, isolates from the West Coast (i.e., California) tended to group geographically. Specifically, isolates obtained from Southern California (i.e., Je81, Je104, Je112, Je110, etc.) were ancestral to those from Northern California (i.e., Hopland, Stag Leap, Conn-Creek, CV17-3, Je65, Je73, etc.) in the PD-III clade.
FIG 1.
Maximum likelihood (ML) tree and haplotype network showing phylogenetic and geographic diversification of worldwide PD-causing subsp. fastidiosa isolates. Color represents isolates from the same geographical location: California (red), Texas (pink), Georgia (green), North Carolina (dark green), Florida (yellow), Spain (light blue), and Taiwan (dark blue). PD-causing strains have been divided into three phylogenetically supported clades (PD-I, PD-II, and PD-III). (a) Haplotype network of PD-causing subsp. fastidiosa isolates. Haplotypes belonging to each PD-causing clade are shown within black circles. Roman numbers identify detected haplotypes (I-CXLI). The size of the circle indicates the number of isolates belonging to each haplotype. (b) ML tree of PD-causing subsp. fastidiosa isolates. The tree was built using the core genome alignment without removing recombinant segments. Bootstrap values mark branch support. Arrows point toward the base of PD-causing clades (-I to -III).
FIG 2.
Phylogeographic analysis showing diversification of PD-causing isolates within the contiguous United States. Color represents isolates from the same geographical location: California (red), Texas (pink), Georgia (green), North Carolina (dark green), and Florida (yellow). Coordinates were recorded during field sampling. In the absence of this information, coordinates referring to the city or vineyard closest to the sample site were used. Florida coordinates were not available; the location shown on the map represents central Florida. Isolates from Southern and Northern California are shown within pale red circles. PD-causing strains were divided into three phylogenetically supported clades: PD-I (Southeast United States isolates exclusively), PD-II (Southern California and Texas isolates), and PD-III (both Southern and Northern California isolates and three Georgia isolates). The tree was built using the core genome alignment without removing recombinant segments. Bootstrap values mark branch support. The map was generated using the open-source R package phytools (https://www.rdocumentation.org/packages/phytools/versions/0.7-70).
A total of 141 different haplotypes named using roman numerals (I to CXLI) (Fig. 1a) were found in the PD-causing core genome alignment. Haplotypes were structured by geographic location and largely matched the evolutionary relationships observed in phylogenetic analyses (Fig. 1b and 2). Overall, haplotypes were grouped similarly to the phylogenetic analyses. Isolates originating from the West and East Coast were split by 979 mutations. California had the largest number of haplotypes (106) as well as haplotypes with the highest frequency: XXVIII (7), XLIV (6), XXXIV (5), LVIII (4), LXVI (4), LXXIII (3), and XCIV (3). On the other hand, Southeast U.S. haplotypes (31) were generally found in low frequency (i.e., one or two isolates). In addition, Southeast isolates in PD-III formed a distinct group separated from the California group by 243 mutations. Likewise, GB514 (Texas, PD-II) was closely connected to California isolates, from which it differentiated by 159 mutations. Isolates originating from recent introduction events (i.e., Spain and Taiwan) had unique haplotypes. Spain-associated haplotypes were linked to a haplotype originating from California (PD-II) and were differentiated by 61 mutations. Similarly, the Taiwan haplotypes were closely linked to the haplotype group originating from the Southeast United States (PD-I) and differentiated by 13 mutations.
Gene gain and loss events occur following subsp. fastidiosa introduction events.
Estimated rates of gene gain/loss were highest in branches leading to the introduction of subsp. fastidiosa from Central America. Furthermore, a total of 35 core genes were absent from the PD-causing population compared to the Costa Rican isolates, while 49 core genes were present in the PD-causing population but absent from the Costa Rican isolates. In addition, gene gain/loss events also occurred within the U.S. populations. In California (Fig. S2a), gene gain/loss rates were highest in the branches leading to each cluster than within clusters, but PD-III had higher gene gain/loss rates than PD-II. Likewise, two clades were observed within the Southeast U.S. population (Fig. S2b). The first clade was formed by isolates 16M2, 16M3, XF51_CCPM1 (from Georgia, clustering with PD-III), and GB514 (from Texas, clustering with PD-II) and the second by the remaining Southeast U.S. isolates (PD-I).
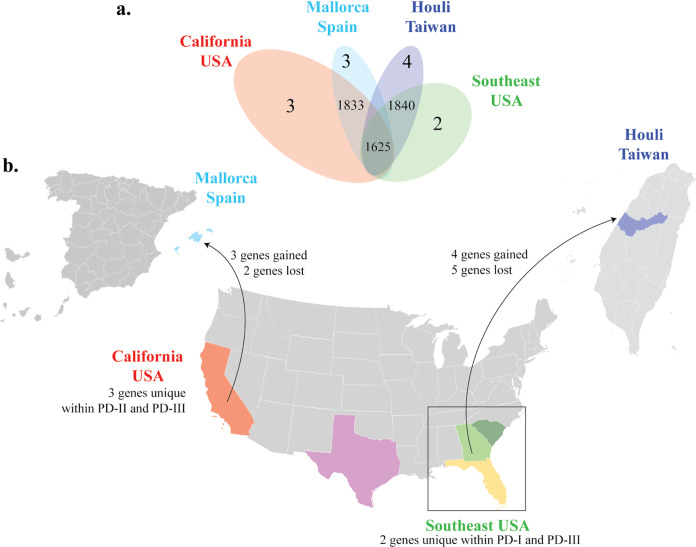
Some unique genes were identified through estimating gene gain/loss rates within each population. We found that, when considering geographical origins of isolates alone, gene presence/absence was similar in PD-II and PD-III isolates regardless of geographical origin (Fig. 3a). In the case of PD-I, PD-II, and PD-III isolates from the Southeast United States, three genes were uniquely found in PD-I and nine in PD-III (Table S3). When gene gain/loss was compared between PD-II and PD-III isolates from California and PD-I, three genes coding for hypothetical proteins were found in PD-II and PD-III isolates from California but absent from PD-I. In addition, two genes were absent from isolates from Spain but present in PD-II and PD-III isolates from California. On the other hand, two genes were found in PD-I but absent from PD-II and PD-III isolates from California (Fig. 3b). In addition, five genes were absent from isolates from Taiwan, which was considered the descendant population of Southeast United States (Table 1).
FIG 3.
Venn diagram and maps showing population-linked gene gain/loss events among PD-causing isolates. Color represents isolates from the same geographical location: California (red), Texas (pink), Georgia (green), North Carolina (dark green), Florida (yellow), Spain (light blue), and Taiwan (dark blue). (a) Venn diagram shows both the number of genes shared between geographic PD-causing populations and genes unique to each population. The size of the oval represents sample size. (b) Estimated number of genes gained and lost between geographical locations and following introduction events. Arrows point from the source population to its descendant following introduction events. California isolates belong to the phylogenetically distinct clades PD-II and PD-III. Included Southeast isolates belong to the phylogenetically distinct PD-I and PD-III clades. All maps were publicly available from Wikimedia commons (Blank US map 1864.svg, Provinces of Spain - blank map.svg, and Blank Taiwan map.svg).
TABLE 1.
List of genes gained/lost among geographic and phylogenetic PD-causing groups
| Category | Annotationa |
|---|---|
| Genes absent from the Taiwan population (found in the contiguous U.S.) | Site-specific DNA-methyltransferase (QIS25725.1); ko K00571, ko K00590, ko K07319 (adenine-specific DNA-methyltransferase); PF01555 |
| Hypothetical protein (QIS26419.1) | |
| Peptidoglycan DD-metalloendopeptidase family protein (QIS26766.1); PF06594, PF00353 (RTX calcium-binding nonapeptide repeat) | |
| Site-specific DNA-methyltransferase (QIS25737.1); ko K00571, ko K00590, ko K07319 (adenine-specific DNA-methyltransferase); PF01555 | |
| Pseudogene | |
| Genes absent from the Spanish population (found in the contiguous U.S.) | CPD*: LacZ, beta-galactosidase/beta-glucuronidase; ko K01192 (beta-mannosidase) |
| Glutamate 5-kinase (AAO28181.1); ko K00931; PF00696 | |
| PD-II and PD-III exclusive genes in the California population | Alpha/beta fold hydrolase (QIS25057.1); ko K02170, ko K07002 (pimeloyl-[acyl-carrier protein] methyl ester esterase) |
| Hypothetical protein (QJP55224.1); PF04014 (antidote-toxin recognition MazE, bacterial antitoxin) | |
| Hypothetical protein (AAO28982.1) | |
| PD-I and PD-III exclusive genes in the Southeast population excluding the Taiwanese clade | Hypothetical protein (QIS26118.1) |
| Phage head morphogenesis protein (QIS26295.1); PF04233 | |
| Genes gained after introduction into Taiwan | CPD*: OM_channels Superfamily, Porin superfamily |
| CPD*: DUF769 superfamily; ko K15125 (filamentous hemagglutinin) | |
| CPD*: entero_EhxA superfamily | |
| Hypothetical protein (QIS25070.1); RTX toxin (QIS25071.1) | |
| Genes gained after introduction into Spain | Pseudogene: glycoside hydrolase family 125 protein; ko K09704 (uncharacterized protein); PF06824 (Metal-independent alpha-mannosidase) |
| DUF596 domain-containing protein (QIS26773.1); PF04591 | |
| Hypothetical protein (QID15519.1); hemagglutinin (QID15518.1); ko K02014 (iron complex outer membrane receptor protein) |
ko, KEGG orthology; PF, Pfam database entry ID; CPD*, conserved protein domain.
These unique genes were annotated using eggNOG-mapper and searched in the GenBank and Pfam databases, using both BLAST and interproscan5 (Table 1, Table S4). Two hypothetical proteins and a gene coding for the HTH-type transcriptional regulator (prtR) were found for PD-I (Table S3); while nine were hypothetical proteins, the protein coding genes traC_2 (DNA primase) and higB_2 (endoribonuclease) were found for the PD-III Southeast U.S. isolates. Two of the three genes found in PD-II and PD-III isolates from California but absent from PD-I coded for hypothetical proteins, and one coded for an alpha/beta fold hydrolase. For the two genes absent from Spain, one of them was listed as glutamate 5-kinase and another had a conserved LacZ, beta-galactosidase/beta-glucuronidase domain. For the two genes found in PD-I but absent from PD-II and PD-III isolates from California, one was annotated as a hypothetical protein and the other one as a phage head morphogenesis protein. For the five genes absent from isolates from Taiwan, two were annotated as site-specific DNA-methyltransferase; another two were annotated as peptidoglycan DD-metalloendopeptidase family protein and hypothetical protein, respectively; the last one could be a pseudogene with unknown function.
Intrasubspecific recombination events are pervasive on both the West and East Coasts.
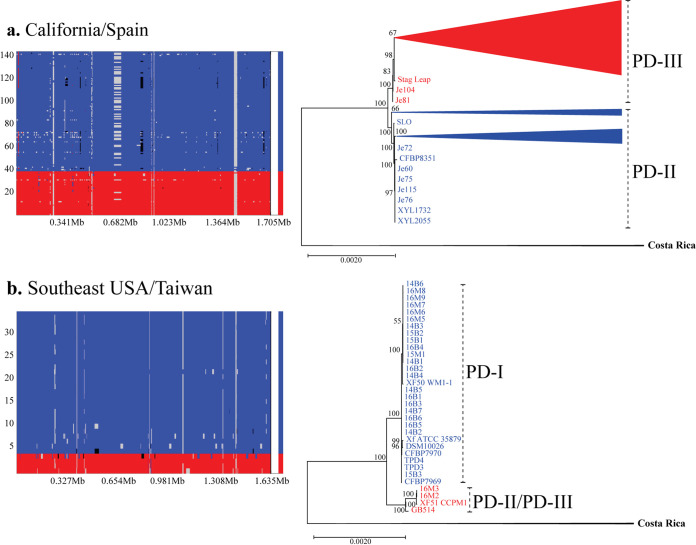
Intrasubspecific recombination was pervasive in both populations (Fig. 4 and Fig. S3 and S4). The r/m estimate (recombination to mutation rates) for the California/Spain core genome alignment was 3.29, while the same estimate for the Southeast U.S./Taiwan core genome alignment was 5.65. In the Southeast United States (Fig. 4b), recombination events were more frequently observed in isolates from the PD-II/PD-III group (recipient) than in isolates from the PD-I group (donor). Within the PD-II/PD-III group, the Texas isolate GB514 (PD-II) was the most frequent recombinant recipient. Donor sequences for the Texas isolate originated from both PD-I and from an “unknown” donor (representing genetic variability present in the population but not characterized in the original sampling). A total of 188 core genes were entirely contained within recombinant regions in the Southeast U.S. population; out of this group, 101 genes were classified as hypothetical proteins. The remaining recombinant core genes belonged to a variety of functions (Table S5). These functions were grouped by their Cluster of Orthologous Groups (COG) class, resulting in 12 genes belonging to the “cellular processes and signaling” class, 5 genes associated with the “information storage and processing” class, 41 genes from the “metabolism” class, and 7 genes belonging to two or more functional classes (“multiple categories”). Based on gene annotation, some coding sequence (CDS) functions are related to virulence and/or host adaptation. These include vitamin B12 import (btuD), ferric uptake regulation protein (fur), response regulator (gacA), virulence protein (PD_1332 in Temecula1 assembly AE009442.1, COG0346), polygalacturonase (pglA), export protein (secB), and ABC transporter (uup).
FIG 4.
Frequency and location of recombination events in fastGEAR identified lineages. Analysis shows results for the California/Spain population (a) and the Southeast United States/Taiwan population (b). FastGEAR’s recombination plots show two distinct lineages on each population (red, PD-III in California/Spain and PD-II/PD-III in Southeast United States/Taiwan; blue, PD-II in California/Spain and PD-I in Southeast United States/Taiwan). The recombination events are shown across the length of the core genome alignment. Larger areas represent recipient sequences, while shorter segments of different color within those areas represent donor sequences from another lineage. Recombinant segments from unidentified lineages are shown in black. Maximum likelihood (ML) trees showing the phylogenetic relationship of isolates within each intrapopulation cluster identified by fastGEAR are also included. Trees were built using the core genome alignment without removing recombinant segments for the California/Spain and Southeast United States/Taiwan populations. Bootstrap values mark branch support.
Likewise, sequence exchange occurred between isolates from the PD-III and the PD-II clusters in California. Recombination events were observed among isolates from the same geographic regions (Fig. 4a). Specifically, recombination was frequent between sequences originating from the Temecula Valley in Southern California (Fig. S3). Sequences in both groups acted as donors and recipients. In addition, Northern California isolates were recipients of recombinant segments from Southern California. This group was also a recipient of unknown sequence fragments. A total of 180 core genes were exclusively contained within these recombinant regions (Table S5). Eighty-five genes were described as hypothetical proteins. The remaining genes were classified by their COG as “cellular processes and signaling” (19 genes), “information storage and processing” (6 genes), “metabolism” (38 genes), and “multiple categories” (6 genes). From these genes, those with annotated function related to host adaptation/virulence include biofilm growth-associated repressor (bigR), periplasmic serine endoprotease (degP) (htrA in Temecula1 assembly AE009442), putative TonB-dependent receptor (phuR in Temecula1 assembly AE009442, COG1629), virulence protein (PD_1332 in Temecula1 assembly AE009442.1, COG0346), sec-independent translocase protein (tatA-D), and PhoH-like protein (ybeZ).
Based on the used genome annotations, a total of 13 recombinant genes were shared in both populations. These genes were glk_1 and glk_2 (glucokinases), glmM_2 (a phosphoglucosamine mutase), glmS_1 and glmS_2 (glutamine–fructose-6-phosphate aminotransferases [isomerizing]), grpE (a GrpE protein), grxD (a glutaredoxin 4), gshB (a glutathione synthetase), gtaB (a UTP–glucose-1-phosphate uridylyltransferase), pepQ (a Xaa-Pro dipeptidase), petA (a ubiquinol-cytochrome c reductase iron-sulfur subunit), petC (an ammonia monooxygenase gamma subunit), an unnamed PKHD-type hydroxylase (COG3128), and a unnamed virulence protein (COG0346).
Grapevine-infecting populations in the East and West United States are largely genetically isolated.
Nucleotide diversity (π) varied within and among populations (Table 2). Overall, nucleotide diversity was higher within the Southeast United States (947 single-nucleotide polymorphisms [SNPs], π = 1.36 × 10e−05) than California (458 SNPs, π = 3.22 × 10e−06). Compared to their corresponding source populations, nucleotide diversity was lower in Spain (2 SNPs, π = 1.38 × 10e−07) and Taiwan (6 SNPs, π = 4.15 × 10e−07). When diversity in phylogenetically distinct clusters was evaluated, PD-I (93 SNPs, π = 7.58 × 10e−07) and PD-II (114 SNPs, π = 9.65 × 10e−07) had lower nucleotide diversity than PD-III (509 SNPs, π = 3.25 × 10e−06).
TABLE 2.
Diversity and neutrality statistics of PD-causing isolates
| Population (n) | Core (nt) | No. of SNPs | π | θ | Tajima's D |
|---|---|---|---|---|---|
| Geographically divided | 14,446,213 | ||||
| California (140) | 458 | 3.22 × 10e−06 | 1.64 × 10e−05 | −1.448 | |
| Southeast U.S. (31) | 947 | 1.36 × 10e−05 | 5.75 × 10e−06 | −0.658 | |
| Spain (2) | 2 | 1.38 × 10e−07 | 1.38 × 10e−07 | —a | |
| Taiwan (2) | 6 | 4.15 × 10e−07 | 4.15 × 10e−07 | —b | |
| Phylogenetically divided | 14,446,213 | ||||
| PD-I (29) | 93 | 7.58 × 10e−07 | 1.64 × 10e−06 | −2.0604 | |
| PD-II (40) | 114 | 9.65 × 10e−07 | 1.87 × 10e−06 | −1.7813 | |
| PD-III (106) | 509 | 3.25 × 10e−06 | 6.72 × 10e−06 | −1.7425 |
Spain isolates were not included.
Taiwan isolates were not included.
The frequency of polymorphism present in the population concerning expectations under neutrality was calculated using Tajima’s D. Briefly, negative Tajima’s D values indicate an excess of rare polymorphisms not expected under neutrality, which can be caused by a selective sweep or a recent population expansion. Positive Tajima’s D values indicate an excess of intermediate frequency polymorphism not expected under neutrality, which suggests balancing selection or a recent population contraction. Tajima’s D in California and the Southeast United States was negative (Table 2); however, the magnitude of the statistic in California was roughly twice that of the Southeast United States (−1.448 and −0.658, respectively). Due to the reduced sample size, it was not possible to estimate Tajima’s D in Spain or Taiwan. When populations were divided phylogenetically, PD-I isolates had a lower Tajima’s D (−2.060) than PD-II (−1.781) and PD-III (−1.743). On the other hand, Watterson’s θ estimates the population mutation rate from the observed nucleotide diversity. This estimator decreases with increased sample size or with recombination rate. Watterson’s θ estimated a higher mutation rate in the Southeast United States (θ = 1.64 × 10e−05) than California (θ = 5.75 × 10e−06). When populations were divided based on phylogeny, the mutation rate was higher in PD-III (θ = 6.72 × 10e−06) than PD-I (θ = 1.64 × 10e−06) or PD-II (θ = 1.87 × 10e−06).
In addition, Fst values were used to measure population differentiation across geographic and phylogenetic groups. Briefly, Fst values compare the amount of genetic variability within and between populations; values of 1 indicate complete population structuring, and values of 0 indicate complete panmixia. Pairwise Fst values (Table S6) for California versus Southeast United States (Fst = 0.814) and California versus Taiwan (Fst = 0.964) were higher than those for California versus Spain (Fst = 0.566). This was also the case for comparisons involving Southeast United States versus Spain (Fst = 0.847) and Southeast United States versus Taiwan (Fst = 0.114). Taiwan versus Spain also showed strong differentiation (Fst = 0.994). Once populations were divided phylogenetically, PD-I was more differentiated from PD-II (Fst = 0.987) and PD-III (Fst = 0.960) than PD-II and PD-III from each other (Fst = 0.541).
A McDonald-Kreitman test (MKT) was used to estimate the rate of synonymous and nonsynonymous polymorphism versus the rate of synonymous and nonsynonymous fixed differences across geographic populations and phylogenetic groups. Under neutrality, it is expected that both rates will be the same (neutrality index [NI] = 1). Therefore, departures from neutrality (NI ≠ 1) will indicate either the action of balancing selection (e.g., maintenance of population polymorphisms; NI > 1) or the action of positive selection (e.g., accumulation of fixed differences between populations; NI < 1). The NI was larger than 1 in all comparisons except for Spain versus Taiwan. NI was significant only for California versus Taiwan (P = 9.87 × 10e−05) (Table S6). Many polymorphisms were observed in Southeast United States and California, while few were observed in Spain or Taiwan. The largest number of fixed differences was observed for Taiwan versus California. When populations were divided phylogenetically, the NI values were larger than 1 only in comparisons between PD-I with PD-II and PD-III. In this instance, the only significant NI was observed for PD-I versus PD-III (P = 6.26 × 10e−05). The number of polymorphisms was larger in PD-III than PD-I and PD-II. The number of fixed differences was similar between PD-I versus PD-II and PD-III but smaller in PD-II versus PD-III.
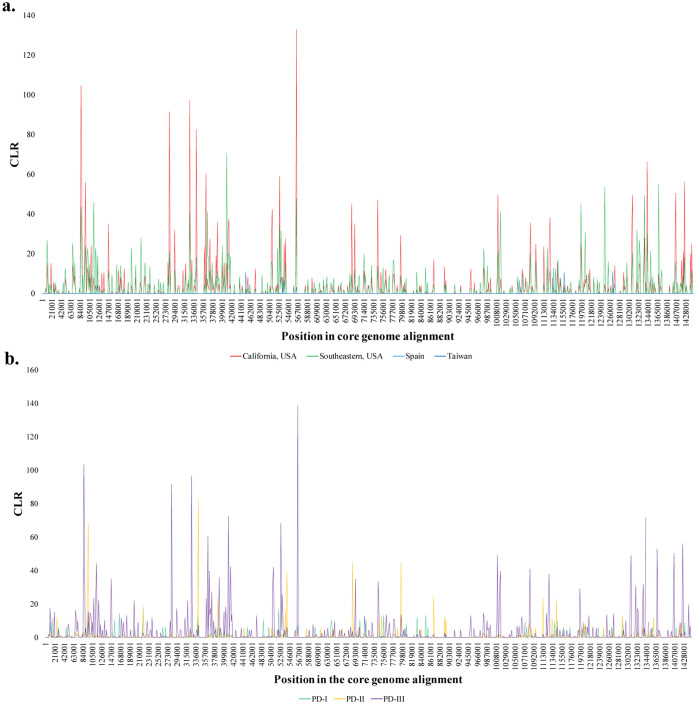
Selective sweep signatures were pervasive in both California and the Southeast United States (Fig. 5a), although the magnitude of the sweep was larger in California. Alternatively, composite likelihood ratio (CLR) peaks were smaller and scattered in Spain and Taiwan. When the populations were split phylogenetically, CLR peaks were more numerous and prominent in PD-III, followed by PD-II and finally PD-I (Fig. 5b). Regardless of whether the populations were subdivided geographically or phylogenetically, some CLR peaks colocated across populations while others were group specific.
FIG 5.
Line plot showing variations in Nielsen’s composite likelihood ratio (CLR) across the length of the core genome alignment (1,500-bp window size). The CLR identifies regions with aberrant allele frequency and determines if their distribution matches those expected from a selective sweep. Peaks represent higher CLR values at that position, which is indicative of a putative selective sweep. Color represents isolates from the same geographical location or phylogenetic cluster. (a) Lines indicate distinct geographic population: California (red), Southeast United States (green), Spain (light blue), and Taiwan (dark blue). (b) Lines indicated distinct phylogenetic clusters: PD-I (teal), PD-II (yellow), and PD-III (purple).
DISCUSSION
Our analyses show that after its introduction from Central America (33, 41), PD-causing subsp. fastidiosa split into two populations: one on the East Coast (31 haplotypes) and one on the West Coast (106 haplotypes). Apart from PD-II/PD-III isolates from the Southeast United States, each population formed a sister monophyletic clade with long basal branch lengths. This indicates that the populations split shortly after introduction to the United States. Moreover, isolates from the same location clustered together, suggesting stronger sequence similarity within than between locations. With the current information available, it is not possible to know if the clustering of PD-II/PD-III isolates from the Southeast United States with the California clades instead of Southeast United States (PD-I) reflects a recent introduction to California or if there is a higher diversity within the Southeast U.S. isolates than currently represented. Alternatively, it is feasible the East and West Coast populations originated via independent introduction events. Previous studies have pointed out the large genetic diversity of subsp. fastidiosa within Central America (33) and the importation of plant material from this region into the United States (45). Our data do not exclude the possibility that additional subsp. fastidiosa strains circulate within Central America and could have been introduced to the United States in relatively simultaneous events. This is a hypothesis that should be evaluated as additional whole genomic data from both native and introduced populations of subsp. fastidiosa become available. However, previously published multilocus sequence typing data (41, 45) and results based on whole-genome sequence analysis (monophyly of the PD-causing population, age and diversity of PD-causing clades, and their evolutionary relationship with the native subsp. fastidiosa population) are indicative of a single introduction event.
Pathogen introductions into Spain and Taiwan were closely related to isolates from California and the Southeast United States, respectively. Although closely related to their source populations, both Spain and Taiwan had unique core haplotypes, which indicates early local adaptation. Nonetheless, we cannot discard the possibility that differences in unique core haplotypes also are the result of a founder effect. Small sample size in both populations does not allow us to test between these two possibilities; however, this should be addressed once additional genomic data become available.
Gene gain/loss events are common between and within populations.
Bacterial gene content is in constant flux (46); in bacteria, evolution via gene gain and loss often precedes evolution at the sequence level (i.e., nucleotide substitutions and indels) (47). Therefore, variations in gene content can act as a source for adaptive differentiation (48). Gene gain and loss rates were highest following introduction to the United States (e.g., 35 genes gained and 49 lost versus 4 genes gained and 5 lost between the East Coast and Taiwan); however, gene content changes were also detected within each geographic population. The higher number of gene gain/loss events observed in basal tree branches can be explained by a founder event. However, they could also be the result of accumulated gene gain/loss events over longer evolutionary time. It is likely that both factors contribute to gene gain/loss between the native and ancestral subsp. fastidiosa populations. The highest intrapopulation gene gain/loss rates were localized in branches following clade splits. Within California, intrapopulation splits were associated with locations along a latitudinal gradient (PD-II in Southern California versus PD-III in Southern and Northern California). In other organisms, selection-driven gene gain/loss has been described in genes involved in environmental interactions (47, 49, 50). Likewise, previous studies have found evidence of local adaptation to environmental conditions within California (34). Thus, it is possible that changes in gene content are adaptive to the local environment. This is further supported by PD-III, which encompasses a larger latitudinal gradient, having four times higher gene gain/loss rates than PD-II.
Alternatively, while gene gain/loss rates were higher in PD-II/PD-III Southeast samples than in PD-I samples, the difference was not as pronounced as that seen in California. Sampling of PD-causing isolates has been more extensive in California; therefore, detecting environmentally linked gene gain/loss might require further sampling in the Southeast United States. Based on the current annotation, it is difficult to interpret the possible benefit or disadvantage of unique genes found in specific X. fastidiosa populations. Functional analysis of these genes will be needed to understand their biological role. Still, a small number of genes involved in transcription regulation (prtR and higB_2) and DNA replication (traC_2) were exclusive to PD-I and PD-III Southeast U.S. isolates. These functions are linked to changes in bacterial transcription and replication in response to environmental cues (51, 52).
However, it should be noted that gene gain/loss events can also be a product of nonadaptive evolution. In bacteria, genetic drift promotes genome reduction, and neutral gene losses are favored by small population size (53, 54). In addition, homologous recombination facilitates core genome homogenization but might not affect accessory genes, leading to gene content divergence and pangenome expansion (47). As such, these gene gain/loss events might not be linked to the adaptive potential of each population. Likewise, this could also be the case of more recent introduction events and smaller population sizes (i.e., Spain and Taiwan populations).
Unequal recombination frequencies drive inter- and intrapopulation differentiation.
r/m estimates showed that recombination contributes more than mutation to genetic diversity. The r/m values for California/Spain (r/m = 3.29) and Southeast United States/Taiwan (r/m = 5.65) were higher than those of previous reports on subsp. fastidiosa (r/m = 2.074 [33]). However, both values were lower than those of reports focused specifically on a California population (r/m = 6.797 [34]). Location-specific core genome analyses can detect nucleotide changes unique to a geographic region. Therefore, the high r/m found here is likely due to location-specific SNPs.
The number of genes located within intrasubspecific recombinations was similar across functional classes, showing that there were no specific gene functions more prone to recombination. These results are like those found in a previous analysis (33). On the other hand, the frequency of recombination varied among phylogenetic clusters. PD-II/PD-III Southeast isolates were recipients of sequence fragments from both PD-I and an unknown group. Similarly, recombination occurred among geographically close isolates from the PD-II and PD-III clusters in California. These results show that genetic exchange is actively occurring on the West and East Coasts. Variations in recombination frequency across isolates have been reported in native subsp. fastidiosa populations (33). Furthermore, recombinant genotypes form distinct phylogenetic groups in subsp. multiplex (55), and in vitro analyses have shown that the natural competency in both subsp. fastidiosa and subsp. multiplex is strain dependent (56, 57). Taken together, this shows that intra- and intersubspecific recombination does not affect all strains equally and that different gene functions, at least within the core genome, are not differentially prone to recombination.
Recombination events also contribute to the differentiation between the East and West Coasts as well as between PD-I, PD-II, and PD-III. Previous studies have shown allele exchange between cooccurring subsp. multiplex and subsp. fastidiosa isolates in the Southeast United States but not in California (40). Therefore, the presence of multiple X. fastidiosa subspecies within the same geographic regions can enable divergence of recombinant prone isolates or clades. Moreover, highly recombinant clades also experienced higher gene gain/loss on the East and West Coasts. Homologous recombination can aid in maintaining core genome cohesiveness while allowing extensive gene gain/loss in the accessory genome (47), and variations in gene content can enable ecological divergence (58). Therefore, intrasubspecific recombination can act as a source of differentiation in PD-causing isolates not only by mediating allelic exchange but also by facilitating gene gain/loss.
From the genes found to recombine in the Southeast U.S. and California populations with putative functions in host adaptation and/or virulence, most have been already identified as recombinants among X. fastidiosa populations (57). Genes with the same annotation found in both studies include btuD, secB, uup, tatD, and ybeZ. In other cases, the identified genes were not exactly the same, but genes with similar functions were found in both studies, including genes related to iron acquisition (fur in the current study), biofilm-associated repressor (bigR in the current study) (59–61) and sulfide sensor (62), other members of the sec pathway (tatA-D) (63, 64), and other serine proteases (degP here) (57, 63, 64). Interestingly, the vitamin B12 transporter BtuD was the single annotated gene with highest inter- and intrasubspecific recombination identified in a previous study (57) and has been described in other bacteria as regulating gene expression, the abundance of microorganisms, and virulence (65, 66), although no functionality has been attributed to X. fastidiosa. Genes like fur and gacA have been identified as transcriptionally regulated by calcium (67), an abundant element inside xylem vessels. Other genes, like the putative TonB-dependent receptor (phuR in the Temecula1 assembly AE009442, COG1629), are involved in twitching motility and biofilm formation (68), and PhoH-like protein (ybeZ) is putatively linked to detection and response to changes of phosphate concentration (69).
West and East Coast populations show unique trends of genetic diversity and mutation rate.
At first glance, isolates originating from the Southeast U.S. population were more genetically diverse than those originating from California. However, this trend was less clear when populations were assigned phylogenetically. PD-II (California plus 1 Texas isolate) had slightly higher genetic diversity than PD-I (exclusively Southeast United States), and PD-III (California plus 3 Georgia isolates) had higher genetic diversity than either PD-I or PD-II.
The negative Tajima’s D values indicate an excess of rare polymorphisms, which can be caused by a selective sweep or a recent population expansion. In the case of subsp. fastidiosa, a population expansion could have occurred following a founder effect. This result, in addition to previously published data (24, 33, 45), supports the hypothesis that subsp. fastidiosa was introduced into the United States. Furthermore, they show that limitation on genetic diversity caused by a founder effect can be long lasting. Tajima’s D values were markedly reduced in PD-I compared to those of the geographic Southeast U.S. population [PD-I + PD-II(Texas)/PD-III(Georgia)]. This indicates that there is more than one phylogenetic cluster circulating on the East Coast. Similarly, Tajima’s D was smaller in PD-II and PD-III than in California isolates, further supporting the idea of ongoing latitudinal distinction on the West Coast.
Watterson’s θ estimates were also affected by grouping criteria. In the case of the Southeast United States compared to PD-I, the Watterson estimator remained roughly unchanged, suggesting that mutation rate in the region is captured by current sampling. Watterson’s θ was larger in California than either PD-II or PD-III and lower in PD-III than PD-II. The values were comparable to those in previous reports in California (34). This indicates that the mutation rate on the West Coast is, to a certain extent, location dependent and that mutation itself contributes less to population differentiation than other evolutionary forces.
PD-causing strains have differentiated phylogenetically and geographically.
The Fst values for different groups of PD-causing isolates were higher than those reported for other global bacterial pathogens (70). These values may reflect the rapid differentiation of PD-causing populations. Pairwise Fst values between PD-I (Southeast only) versus PD-II (California plus 1 Texas) and PD-III (California plus 3 Georgia) were higher than those between Southeast United States and California. These results further support the phylogenetic and geographic separation of the East and West Coasts and the more recent differentiation within California. How much this differentiation can be linked to the Southeast U.S. PD-II/PD-III group needs to be further analyzed. Our Fst analyses indicate a complex phylogeographic history between U.S. populations, yet the effects of sample size on these calculations should not be ignored. For example, recently introduced populations (e.g., Spain and Taiwan) showed even higher population differentiation than comparisons involving their source populations. Whether this suggests higher differentiation as a product of a founder effect remains to be determined.
In general, genetic diversity has a high impact on adaptive potential (71); however, some genetic variants might be considered neutral and can be estimated based on the number of synonymous polymorphisms (72). Variables associated with local adaptation are linked to nonsynonymous polymorphisms. There were more nonsynonymous than synonymous polymorphisms on both the East and West Coasts. This suggests that although the number of polymorphisms is limited due to a recent introduction event, each population maintains a certain level of genetic variation (as evidenced by an NI of >1), which could be a source for local adaptation (73).
When populations were divided according to their phylogenetic relationships, a significant NI of >1 was observed only between PD-I and PD-III. Polymorphism largely accumulated in PD-III compared to PD-I. However, the number of fixed differences was comparable between PD-I versus PD-II and PD-III. This shows that a significant number of intraclade polymorphisms in PD-III have not yet been fixed. Instead, fixed differences seem to mostly reside between PD-I compared to PD-II and PD-III. This further supports the idea that East and West Coast populations split early following introduction to the United States, with local population differentiation within a latitudinal gradient in the West Coast.
Selective sweeps have occurred following the introduction of X. fastidiosa to the United States.
Many CLR peaks were colocalized in the same region, while others were group exclusive. The localized nature of CLR peaks in the core genome alignment suggests that selective sweeps can be detected only on certain genes. The location and intensity of selective sweeps are the product of evolutionary and ecological variables. A founder effect can result in reduced selection strength, but it might not affect recombination potential, particularly in X. fastidiosa (74). Therefore, the CLR patterns observed here likely reflect genes undergoing strong selection, either following subsp. fastidiosa introduction from Central America (colocalized CLR peaks) or via selective pressures associated with a specific environment (group-specific CLR peaks). Strong CLR signals in both PD-II and PD-III indicate that selective sweeps have been more prevalent on the West Coast. Some genes located in the CLR peak include outer membrane protein assembly factors (bamA-B), a beta-barrel assembly-enhancing protease (bepA_4), a ubiquinol cytochrome c oxidoreductase (fbcH), a glycine cleavage system transcriptional repressor (gcvR), a glutamine–fructose-6-phosphate aminotransferase (glmS_2), a proton/glutamate-aspartate symporter (gltP), and a sensor histidine kinase (rcsC). Branch-site analyses aimed to detect signals of positive selection should be performed to further evaluate these results.
Conclusions.
We identified a series of evolutionary mechanisms that led to the diversification of PD-causing subsp. fastidiosa populations. Diversification has occurred in core genome sequences via mutation and recombination and in gene content via gain/loss events. These differences have the potential of facilitating local adaptation to environmental conditions and, in the absence of gene flow, lead to pathogen specialization. The host range and geographic distribution of X. fastidiosa is expanding, and each new introduction can result in significant economic and ecological damage. Understanding the mechanisms and speed of local adaptation in X. fastidiosa is important to manage emerging X. fastidiosa diseases and hopefully limit the number of novel epidemics.
MATERIALS AND METHODS
Sampling, culturing, and isolation.
The following study encompasses 175 X. fastidiosa subsp. fastidiosa isolates obtained from infected PD-symptomatic grapevines from diverse geographic regions. The numbers of isolates from each region were the following: California (n = 140), Southeast United States (n = 31), Spain (n = 2), and Taiwan (n = 2). In addition, three non-grapevine-infecting X. fastidiosa subsp. fastidiosa isolates from Costa Rica were used as an outgroup (33). New subsp. fastidiosa isolates were obtained from infected grapevines in the Southeast United States during 2014 to 2016; these isolates were cultured from symptomatic leaves as previously described (44). Colonies growing after ∼1 to 2 weeks under 28°C incubation were restreaked, cloned, and had identity confirmed with X. fastidiosa-specific PCR primer sets (75). Isolates were obtained from different grapevine varieties. Specifically, the varieties found in Site1 were Merlot (n = 5, years 2014 to 2016), Mourvedre (n = 1, year 2014), Cabernet Sauvignon (n = 1, year 2014), Chardonnay (n = 5, years 2014 to 2016), Viognier (n = 2, years 2014 to 2015), Sangiovese (n = 1, year 2014), and Touriga (n = 1, year 2014). The varieties found in Site2 were Montaluce (n = 1, year 2015), Merlot (n = 3, year 2016), Pinot grigio (n = 3, year 2016), and Vidal (n = 3, year 2016). Except for Site1 (n = 16) and Site2 (n = 8), all data included in the following study were previously made publicly available. Detailed metadata on each assembly are compiled in Table S1 in the supplemental material; assembly statistics for new whole-genome sequences are provided in Table S2.
Sequencing, assembly, and annotation of X. fastidiosa subsp. fastidiosa isolates.
All isolates were sequenced using Illumina HiSeq2000. Samples were sequenced at the University of California, Berkeley, Vincent J. Coates Genomics Sequencing Laboratory (California Institute for Quantitative Biosciences; QB3) and the Center for Genomic Sciences, Allegheny Singer Research Institute, Pittsburgh, PA. All raw reads and information regarding each newly sequenced strain can be accessed under the NCBI BioProject accession number PRJNA655351. The quality of raw paired FASTQ reads was evaluated using FastQC (76) and visualized using MultiQC (77). Low-quality reads and adapter sequences were removed from all paired raw reads using seqtk v1.2 (https://github.com/lh3/seqtk) and cutadapt v1.14 (78) with default parameters. After preprocessing, isolates were assembled de novo with SPAdes v3.13 (79, 80) using the -careful parameter and -k values of 21, 33, 55, and 77. Assembled contigs were reordered using Mauve’s contig mover function (81) with the complete publicly available Temecula1 assembly (GCA_000007245.1) used as the reference. Assembled and reordered genomes were then individually annotated using the Prokka pipeline (82). In addition, published genome sequences were reannotated with Prokka.
Core genome alignments, construction of ML trees, and haplotype network.
Roary v3.11.2 (83) was used to calculate the number of genes in the core (genes shared between 99 and 100% of strains), soft-core (genes shared between 95 and 99% of strains), shell (genes shared between 15 and 95% of strains), and cloud (genes shared between 0 and 15% of strains) genomes of PD-causing isolates (n = 175). A core genome alignment of PD-causing isolates plus the three Costa Rica isolates (non-PD) was created using the -e (codon aware multisequence alignment of core genes) and -n (fast nucleotide alignment) flags in Roary. This core genome alignment was used to build a maximum likelihood (ML) tree with RAxML (84). The GTRCAT substitution model was used on tree construction, while tree topology and branch support were assessed with 1,000 bootstrap replicates. In addition, a nonrecombinant tree was constructed by removing detected recombinant segments from the core genome alignment (described below). The ML nonrecombinant tree was constructed using the same parameters as the recombinant tree. Finally, a haplotype network for PD-causing isolates was built following the removal of the outgroup sequences (non-PD). Core genome haplotypes were calculated based on the number of mutations among the analyzed strains, and the haplotype network was built using the HaploNet function in the R package pegas (85). Haplotypes were then color-coded by geographic location.
Estimation of recombinant segments and gene gain/loss rates within populations.
Isolates were divided based on their geographical origin: California, Southeast United States, Spain, and Taiwan. California and the Southeast United States were the source populations for Spain and Taiwan, respectively. Source and descendant relationships between populations were phylogenetically determined (see Results). A core genome alignment was created for California/Spain (n = 142) and Southeast United States/Taiwan (n = 33). The alignment was used to estimate the frequency and location of recombinant events. FastGEAR (86) was used with default parameters to identify lineage-specific recombinant segments (ancestral) and strain-specific recombinant segments (recent). The size and location of recombinant segments across the length of the core genome alignment were mapped within California/Spain and Southeast United States/Taiwan using the R package circlize (87). Donor/recipient recombinant regions were visualized using fastGEAR’s plotRecombinations script. In addition, the number of substitutions introduced by recombination versus random point mutation (r/m) (88) was estimated for the California/Spain and Southeast United States/Taiwan core genome alignments using ClonalFrameML (89). It should be noted that fastGEAR was designed to test recombination in individual gene alignments instead of core genome alignments; a previous study found that fastGEAR was more conservative than other more appropriate recombination detection methods, such as ClonalFrameML (34). Future research should perform an empirical comparison of recombination detection methods for X. fastidiosa.
Additionally, the stochastic probability of gene gain/loss per tree branch was estimated with GLOOME using default parameters (90). Briefly, RAxML was used to build an ML phylogenetic tree for the California/Spain and Southeast United States/Taiwan core genome alignments. The parameters used were the same as those for the PD-causing ML tree. Roary v3.11.2 was used to calculate a binary gene presence (1)/absence (0) matrix within the California/Spain and the Southeast U.S./Taiwan populations. A binary accessory genome matrix was created by removing core genome genes from the data set. Subsequently, the binary accessory presence/absence matrix was transposed and converted into FASTA format. The binary accessory genome and the ML trees were used as inputs to the GLOOME analysis. Unique genes were identified through estimating gene gain/loss rates within each population. These genes were annotated by eggNOG-mapper v1.0.3 (https://github.com/eggnogdb/eggnog-mapper) and searched in the GenBank and Pfam databases using BLAST and interproscan v5.47 (https://github.com/ebi-pf-team/interproscan).
Population genomics analyses.
Global measures of genetic diversity, population differentiation, and selective sweeps were estimated for the PD-causing data set using the R package PopGenome (91). The data set was subdivided in two ways, (i) based on isolates’ geographical origin (i.e., California, Southeast United States, Spain, and Taiwan) and (ii) based on isolates’ phylogenetic relationships (i.e., PD-I, PD-II, and PD-III; see Results). All calculations described below were performed for both the (i) geographic and (ii) phylogenetic subdivisions.
Genetic diversity was estimated by computing nucleotide diversity (π), Tajima’s D (92), and Watterson’s estimator (θ) (93). Briefly, nucleotide diversity measures the average number of nucleotide differences per site in pairwise comparisons among DNA sequences. Tajima’s D evaluates the frequency of polymorphism present in a population and compares that value to the expectation under neutrality. The Watterson θ estimator measures the mutation rate of a population. Population differentiation was estimated by calculating the Fixation Index (Fst) (94) within (i) geographic and (ii) phylogenetic groups. In addition, the McDonald-Kreitman test (MKT) (95) was used to estimate the rate of synonymous (syn-P) and nonsynonymous (nonsyn-P) polymorphism against the rate of fixed synonymous (syn-F) and nonsynonymous (nonsyn-F) differences. In each instance, the neutrality index (NI) was calculated. An NI of >1 suggests an excess of preserved polymorphism maintained via balancing selection. Alternatively, an NI of <1 suggests population divergence via positive selection. Finally, the location and magnitude of selective sweeps was calculated using Nielsen’s composite likelihood ratio (CLR) (96). This test identifies regions with aberrant allele frequency spectra and estimates if the aberrant allele distribution fits the expectations of a selective sweep. The test was performed on a 1,500-bp sliding window across the length of the PD-causing core genome alignment.
Data availability.
The raw sequence data files for the newly published isolates were submitted to the NCBI Sequence Read Archive under accession numbers SAMN15732826 through SAMN15732849. All other data used were previously published. All accession numbers are listed in Table S1.
Supplementary Material
ACKNOWLEDGMENTS
This work has received funding from the PD/GWSS Research Program, California Department of Food and Agriculture, and the European Union’s Horizon 2020 research and innovation program under grant agreement no. 727987, “Xylella fastidiosa Active Containment Through a multidisciplinary-Oriented Research Strategy XF-ACTORS.” Genome sequencing was performed at the UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory, which is supported by an NIH instrumentation grant (S10 OD018174).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pimentel D, Lach L, Zuniga R, Morrison D. 2000. Environmental and economic costs of nonindigenous species in the United States. Bioscience 50:53. doi: 10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2. [DOI] [Google Scholar]
- 2.Fletcher J, Bender C, Budowle B, Cobb WT, Gold SE, Ishimaru CA, Luster D, Melcher U, Murch R, Scherm H, Seem RC, Sherwood JL, Sobral BW, Tolin SA. 2006. Plant pathogen forensics: capabilities, needs, and recommendations. Microbiol Mol Biol Rev 70:450–471. doi: 10.1128/MMBR.00022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pyšek P, Jarošík V, Pergl J. 2011. Alien plants introduced by different pathways differ in invasion success: unintentional introductions as a threat to natural areas. PLoS One 6:e24890. doi: 10.1371/journal.pone.0024890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early R, Bradley BA, Dukes JS, Lawler JJ, Olden JD, Blumenthal DM, Gonzalez P, Grosholz ED, Ibañez I, Miller LP, Sorte CJB, Tatem AJ. 2016. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat Commun 7:12485. doi: 10.1038/ncomms12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mable BK. 2019. Conservation of adaptive potential and functional diversity: integrating old and new approaches. Conserv Genet 20:89–100. doi: 10.1007/s10592-018-1129-9. [DOI] [Google Scholar]
- 6.Baltrus DA, Nishimura MT, Dougherty KM, Biswas S, Mukhtar MS, Vicente J, Holub EB, Dangl JL. 2012. The molecular basis of host specialization in bean pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 25:877–888. doi: 10.1094/MPMI-08-11-0218. [DOI] [PubMed] [Google Scholar]
- 7.Karasov TL, Horton MW, Bergelson J. 2014. Genomic variability as a driver of plant–pathogen coevolution? Curr Opin Plant Biol 18:24–30. doi: 10.1016/j.pbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plissonneau C, Benevenuto J, Mohd-Assaad N, Fouché S, Hartmann FE, Croll D. 2017. Using population and comparative genomics to understand the genetic basis of effector-driven fungal pathogen evolution. Front Plant Sci 8:119. doi: 10.3389/fpls.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokryakov MV, Abdeev IA, Piruzyan ES, Schaad NW, Ignatov AN. 2010. Diversity of effector genes in plant pathogenic bacteria of genus Xanthomonas. Microbiology 79:58–65. doi: 10.1134/S002626171001008X. [DOI] [Google Scholar]
- 10.Rowntree JK, Cameron DD, Preziosi RF. 2011. Genetic variation changes the interactions between the parasitic plant-ecosystem engineer Rhinanthus and its hosts. Philos Trans R Soc Lond B Biol Sci 366:1380–1388. doi: 10.1098/rstb.2010.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González R, Butković A, Elena SF. 2019. Role of host genetic diversity for susceptibility-to-infection in the evolution of virulence of a plant virus. Virus Evol 5:vez024. doi: 10.1093/ve/vez024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan JX, Yang S, Hu L, Leung H, Mew TW, Teng PS, Wang Z, Mundt CC. 2000. Genetic diversity and disease control in rice. Nature 406:718–722. doi: 10.1038/35021046. [DOI] [PubMed] [Google Scholar]
- 13.Escriu F. 1 March 2012. Diversity of plant virus populations: a valuable tool diversity of plant virus populations: a valuable tool for epidemiological studies. IntechOpen doi: 10.5772/66820. [DOI] [Google Scholar]
- 14.Brown JKM. 2015. Durable resistance of crops to disease: a Darwinian perspective. Annu Rev Phytopathol 53:513–539. doi: 10.1146/annurev-phyto-102313-045914. [DOI] [PubMed] [Google Scholar]
- 15.Zhuh W, Zhan JS. 2016. Population genetics of plant pathogens. Yi Chuan 34:157–166. doi: 10.3724/sp.j.1005.2012.00157. [DOI] [PubMed] [Google Scholar]
- 16.Giraud T, Gladieux P, Gavrilets S. 2010. Linking emergence of fungal plant diseases and ecological speciation. Trends Ecol Evol 25:387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Le Saux MF, Manceau C, Poussier S, Pruvost O, Lemaire C, Jacques MA. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474. doi: 10.1371/journal.pone.0058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan A, Hu J, Hu X, Zhou Z, Hui M, Wang S, Peng W, Wang M, Bao Z. 2009. Fine-scale population genetic structure of zhikong scallop (chlamys farreri): do local marine currents drive geographical differentiation? Mar Biotechnol 11:223–235. doi: 10.1007/s10126-008-9138-1. [DOI] [PubMed] [Google Scholar]
- 19.McDonald BA, Linde C. 2002. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124:163–180. doi: 10.1023/A:1015678432355. [DOI] [Google Scholar]
- 20.Slatkin M. 1985. Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430. doi: 10.1146/annurev.es.16.110185.002141. [DOI] [Google Scholar]
- 21.McDermott JM, McDonald BA. 1993. Gene flow in plant pathosystems. Annu Rev Phytopathol 31:353–373. doi: 10.1146/annurev.py.31.090193.002033. [DOI] [Google Scholar]
- 22.Pruvost O, Boyer K, Ravigné V, Richard D, Vernière C. 2019. Deciphering how plant pathogenic bacteria disperse and meet: molecular epidemiology of Xanthomonas citri pv. citri at microgeographic scales in a tropical area of Asiatic citrus canker endemicity. Evol Appl 12:1523–1538. doi: 10.1111/eva.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.EFSA. 2018. Update of the Xylella spp. host plant database. EFSA J 16:e05408. doi: 10.2903/j.efsa.2018.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanhove M, Retchless AC, Sicard A, Rieux A, Coletta-Filho HD, Fuente LD, La Stenger DC, Almeida PP. 2019. Genomic diversity and recombination among Xylella fastidiosa subspecies. Appl Environ Microbiol 85:e02972-18. doi: 10.1128/AEM.02972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bragard C, Dehnen-Schmutz K, Di Serio F, Gonthier P, Jacques MA, Miret JA, Justesen AF, MacLeod A, Magnusson CS, Milonas P, Navas-Cortés JA, Potting R, Reignault PL, Thulke HH, Van der Werf W, Vicent Civera A, Yuen J, Zappalà L, Makowski D, Delbianco A, Maiorano A, Muñoz Guajardo I, Stancanelli G, Guzzo M, Parnell S. 2019. Effectiveness of in planta control measures for Xylella fastidiosa. EFSA J 17:e05666. doi: 10.2903/j.efsa.2019.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almeida RPP, De La Fuente L, Koebnik R, Lopes JRS, Parnell S, Scherm H. 2019. Addressing the new global threat of Xylella fastidiosa. Phytopathology 109:172–174. doi: 10.1094/PHYTO-12-18-0488-FI. [DOI] [PubMed] [Google Scholar]
- 27.Nunney L, Hopkins DL, Morano LD, Russell SE, Stouthamer R. 2014. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl Environ Microbiol 80:1159–1169. doi: 10.1128/AEM.02920-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunney L, Yuan X, Bromley RE, Stouthamer R. 2012. Detecting genetic introgression: high levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl Environ Microbiol 78:4702–4714. doi: 10.1128/AEM.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landa BB, Castillo AI, Giampetruzzi A, Kahn A, Román-Écija M, Velasco-Amo MP, Navas-Cortés JA, Marco-Noales E, Barbé S, Moralejo E, Coletta-Filho HD, Saldarelli P, Saponari M, Almeida RPP. 2019. Emergence of a plant pathogen in Europe associated with multiple intercontinental introductions. Appl Environ Microbiol 86:e01521-19. doi: 10.1128/AEM.01521-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giampetruzzi A, Saponari M, Loconsole G, Boscia D, Savino VN, Almeida RPP, Zicca S, Landa BB, Chacón-Diaz C, Saldarelli P. 2017. Genome-wide analysis provides evidence on the genetic relatedness of the emergent Xylella fastidiosa genotype in Italy to isolates from Central America. Phytopathology 107:816–827. doi: 10.1094/PHYTO-12-16-0420-R. [DOI] [PubMed] [Google Scholar]
- 31.Saponari M, Giampetruzzi A, Loconsole G, Boscia D, Saldarelli P. 2019. Xylella fastidiosa in olive in Apulia: where we stand. Phytopathology 109:175–186. doi: 10.1094/PHYTO-08-18-0319-FI. [DOI] [PubMed] [Google Scholar]
- 32.Nunney L, Azad H, Stouthamer R. 2019. An experimental test of the host-plant range of nonrecombinant strains of North American Xylella fastidiosa subsp. multiplex. Phytopathology 109:294–300. doi: 10.1094/PHYTO-07-18-0252-FI. [DOI] [PubMed] [Google Scholar]
- 33.Castillo AI, Chacón-Díaz C, Rodríguez-Murillo N, Coletta HD, Almeida RPP, Rica C. 2020. Impacts of local population history and ecology on the evolution of a globally dispersed pathogen. BMC Genomics 21:1–51. doi: 10.1186/s12864-020-06778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanhove M, Sicard A, Ezennia J, Leviten N, Almeida RPP. 2020. Population structure and adaptation of a bacterial pathogen in California grapevines. Environ Microbiol 22:2625–2638. doi: 10.1111/1462-2920.14965. [DOI] [PubMed] [Google Scholar]
- 35.Gomila M, Moralejo E, Busquets A, Segui G, Olmo D, Nieto A, Juan A, Lalucat J. 2019. Draft genome resources of two strains of Xylella fastidiosa XYL1732/17 and XYL2055/17 isolated from Mallorca Vineyards. Phytopathology 109:222–224. doi: 10.1094/PHYTO-08-18-0298-A. [DOI] [PubMed] [Google Scholar]
- 36.Castillo AI, Tuan S-J, Retchless AC, Hu F-T, Chang H-Y, Almeidaa RPP. 2019. Draft whole-genome sequences of Xylella fastidiosa subsp. fastidiosa strains TPD3 and TPD4, isolated from grapevines in Hou-li, Taiwan. Microbiol Resour Announc 8:e00835-19. doi: 10.1128/MRA.00835-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuenzel EL, Scally M, Stouthamer R, Nunney L. 2005. A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 71:3832–3839. doi: 10.1128/AEM.71.7.3832-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X, Morano L, Bromley R, Spring-Pearson S, Stouthamer R, Nunney L. 2010. Multilocus sequence typing of Xylella fastidiosa causing Pierce’s disease and oleander leaf scorch in the United States. Ecol Epidemiol 100:601–611. doi: 10.1094/PHYTO-100-6-0601. [DOI] [PubMed] [Google Scholar]
- 39.Cella E, Angeletti S, Fogolari M, Bazzardi R, De L, Ciccozzi M, Cella E, Angeletti S, Fogolari M, Bazzardi R. 2018. Two different Xylella fastidiosa strains circulating in Italy: phylogenetic and evolutionary analyses. J Plant Interact 13:428–432. doi: 10.1080/17429145.2018.1475022. [DOI] [Google Scholar]
- 40.Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R. 2014. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol 80:3025–3033. doi: 10.1128/AEM.04112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunney L, Ortiz B, Russell SA, Sánchez RR, Stouthamer R. 2014. The complex biogeography of the plant pathogen Xylella fastidiosa: genetic evidence of introductions and subspecific introgression in Central America. PLoS One 9:e112463. doi: 10.1371/journal.pone.0112463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tumber KP, Alston JM, Fuller KB. 2014. Pierce’s disease costs California $104 million per year. Cal Ag 68:20–29. doi: 10.3733/ca.v068n01p20. [DOI] [Google Scholar]
- 43.Hickey C. 2019. Pierce’s disease of grape: identification and management. UGA Coop Ext Bull 1514:1–6. [Google Scholar]
- 44.Parker JK, Havird JC, De La Fuente L. 2012. Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E). Appl Environ Microbiol 78:1385–1396. doi: 10.1128/AEM.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astúa M, Moreira L, Ortiz B, Stouthamer R. 2010. Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce’s disease of grapevine in the U.S. PLoS One 5:e15488. doi: 10.1371/journal.pone.0015488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. 2014. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol 12:66. doi: 10.1186/s12915-014-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iranzo J, Wolf YI, Koonin EV, Sela I. 2019. Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat Commun 10:5376. doi: 10.1038/s41467-019-13429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartmann FE, Croll D. 2017. Distinct trajectories of massive recent gene gains and losses in populations of a microbial eukaryotic pathogen. Mol Biol Evol 34:2808–2822. doi: 10.1093/molbev/msx208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, Steglich C, Church GM, Richardson P, Chisholm SW. 2007. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3:e231–e252. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moulana A, Anderson RE, Fortunato CS, Huber JA. 2020. Selection is a significant driver of gene gain and loss in the pangenome of the bacterial genus Sulfurovum in geographically distinct deep-sea hydrothermal vents. mSystems 5:1–18. doi: 10.1128/mSystems.00673-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frick DN, Richardson CC. 2001. DNA primases. Annu Rev Biochem 70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 52.Browning DF, Busby SJW. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 53.Kuo CH, Moran NA, Ochman H. 2009. The consequences of genetic drift for bacterial genome complexity. Genome Res 19:1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet 17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 55.Nunney L, Vickerman DB, Bromley RE, Russell SA, Hartman JR, Morano LD, Stouthamer R. 2013. Recent evolutionary radiation and host plant specialization in the Xylella fastidiosa subspecies native to the United States. Appl Environ Microbiol 79:2189–2200. doi: 10.1128/AEM.03208-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kandel PP, Almeida RPP, Cobine PA, De La Fuente L. 2017. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol Plant Microbe Interact 30:589–600. doi: 10.1094/MPMI-02-17-0053-R. [DOI] [PubMed] [Google Scholar]
- 57.Potnis N, Kandel PP, Merfa MV, Retchless AC, Parker JK, Stenger DC, Almeida RPP, Bergsma-Vlami M, Westenberg M, Cobine PA, De La Fuente L. 2019. Patterns of inter- and intrasubspecific homologous recombination inform eco-evolutionary dynamics of Xylella fastidiosa. ISME J 13:2319–2333. doi: 10.1038/s41396-019-0423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmutzer M, Barraclough TG. 2019. The role of recombination, niche-specific gene pools and flexible genomes in the ecological speciation of bacteria. Ecol Evol 9:4544–4556. doi: 10.1002/ece3.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbosa RL, Rinaldi FC, Guimarães BG, Benedetti CE. 2007. Crystallization and preliminary X-ray analysis of BigR, a transcription repressor from Xylella fastidiosa involved in biofilm formation. Acta Crystallogr Sect F Struct Biol Cryst Commun 63:596–598. doi: 10.1107/S1744309107028722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbosa RL, Benedetti CE. 2007. BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth. J Bacteriol 189:6185–6194. doi: 10.1128/JB.00331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guimarães BG, Barbosa RL, Soprano AS, Campos BM, De Souza TA, Tonoli CCC, Leme AFP, Murakami MT, Benedetti CE. 2011. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem 286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Lira NPV, Pauletti BA, Marques AC, Perez CA, Caserta R, De Souza AA, Vercesi AE, Paes Leme AF, Benedetti CE. 2018. BigR is a sulfide sensor that regulates a sulfur transferase/dioxygenase required for aerobic respiration of plant bacteria under sulfide stress. Sci Rep 8:1–13. doi: 10.1038/s41598-018-21974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Federici MT, Marcondes JA, Picchi SC, Stuchi ES, Fadel AL, Laia ML, Lemos MVF, Lemos EGM. 2012. Xylella fastidiosa: an in vivo system to study possible survival strategies within citrus xylem vessels based on global gene expression analysis. Electron J Biotechnol 15:717–3458. doi: 10.2225/vol15-issue3-fulltext-4. [DOI] [Google Scholar]
- 64.Da Silva Neto JF, Koide T, Gomes SL, Marques MV. 2007. The single extracytoplasmic-function sigma factor of Xylella fastidiosa is involved in the heat shock response and presents an unusual regulatory mechanism. J Bacteriol 189:551–560. doi: 10.1128/JB.00986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee KM, Go J, Yoon MY, Park Y, Kim SC, Yong DE, Yoon SS. 2012. Vitamin B 12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect Immun 80:1639–1649. doi: 10.1128/IAI.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cordonnier C, Le Bihan G, Emond-Rheault JG, Garrivier A, Harel J, Jubelin G. 2016. Vitamin B12 uptake by the gut commensal bacteria bacteroides thetaiotaomicron limits the production of Shiga toxin by enterohemorrhagic Escherichia coli. Toxins 8:14. doi: 10.3390/toxins8010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H, De La Fuente L. 2020. Calcium transcriptionally regulates movement, recombination and other functions of Xylella fastidiosa under constant flow inside microfluidic chambers. Microb Biotechnol 13:548–561. doi: 10.1111/1751-7915.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cursino L, Li Y, Zaini PA, De La Fuente L, Hoch HC, Burr TJ. 2009. Twitching motility and biofilm formation are associated with tonB1 in Xylella fastidiosa. FEMS Microbiol Lett 299:193–199. doi: 10.1111/j.1574-6968.2009.01747.x. [DOI] [PubMed] [Google Scholar]
- 69.Santos-Beneit F. 2015. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. doi: 10.3389/fmicb.2015.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh J, Khan A. 2019. Distinct patterns of natural selection determine sub-population structure in the fire blight pathogen, Erwinia amylovora. Sci Rep 9:1–13. doi: 10.1038/s41598-019-50589-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ørsted M, Hoffmann AA, Sverrisdóttir E, Nielsen KL, Kristensen TN. 2019. Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet 15:e1008205. doi: 10.1371/journal.pgen.1008205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holderegger R, Kamm U, Gugerli F. 2006. Adaptive vs. neutral genetic diversity: implications for landscape genetics. Landscape Ecol 21:797–807. doi: 10.1007/s10980-005-5245-9. [DOI] [Google Scholar]
- 73.Moutinho AF, Bataillon T, Dutheil JY. 2020. Variation of the adaptive substitution rate between species and within genomes. Evol Ecol 34:315–338. doi: 10.1007/s10682-019-10026-z. [DOI] [Google Scholar]
- 74.Kung SH, Almeida RPP. 2011. Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 77:5278–5284. doi: 10.1128/AEM.00730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francis M, Lin H, Rosa JC, La Doddapaneni H, Civerolo EL. 2006. Genome-based PCR primers for specific and sensitive detection and quantification of Xylella fastidiosa. Eur J Plant Pathol 115:203–213. doi: 10.1007/s10658-006-9009-4. [DOI] [Google Scholar]
- 76.Andrews S, Wingett SW, Hamilton RS. 2018. FastQ Screen: a tool for multi-genome mapping and quality control. F10000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ewels P, Magnusson M, Lundin S, Käller M. 2016. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marcel M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:5–7. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 79.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelski A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel S, Woyke T, McLean J, Lasken R, Tesler G, Alekseyev M, Pevzner P. 2013. Assembly single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 83.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paradis E. 2010. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 86.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. 2017. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol 34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 88.Guttman DS, Dykhuizen DE. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 89.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041-18. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. 2010. GLOOME: gain loss mapping engine. Bioinformatics 26:2914–2915. doi: 10.1093/bioinformatics/btq549. [DOI] [PubMed] [Google Scholar]
- 91.Pfeifer B, Wittelsbu U, Ramos-Onsins SE, Lercher MJ. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genet Soc Am 123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 94.Wright S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420. [Google Scholar]
- 95.McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 96.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. 2005. Genomic scans for selective sweeps using SNP data. Genome Res 15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data files for the newly published isolates were submitted to the NCBI Sequence Read Archive under accession numbers SAMN15732826 through SAMN15732849. All other data used were previously published. All accession numbers are listed in Table S1.