Abstract
Auxin influences all aspects of plant growth and development and exerts its function at scales ranging from the subcellular to the whole-organism level. A canonical mechanism for auxin signaling has been elucidated, which is based on derepression of downstream genes via ubiquitin-mediated degradation of transcriptional repressors. While the combinatorial nature of this canonical pathway provides great potential for specificity in the auxin response, alternative noncanonical signaling pathways required to mediate certain processes have been identified. One such pathway affects gene regulation in a manner that is reminiscent of mechanisms employed in animal hormone signaling, while another triggers transcriptional changes through auxin perception at the plasma membrane and the stabilization of transcriptional repressors. In some cases, the exact perception mechanisms and the nature of the receptors involved are yet to be revealed. In this review, we describe and discuss current knowledge on noncanonical auxin signaling and highlight unresolved questions surrounding auxin biology.
The effects of auxin during plant development were first documented in the 1800s, when it was postulated that the movement of so-called “growth regulators” between tissues influenced both morphogenesis and responses to abiotic stimuli, such as gravity and light (for review, see Enders and Strader 2015). However, indole 3-acetic acid (IAA), the natural auxin predominantly responsible for mediating these effects, was not isolated from plants until much later (Haagen-Smit et al. 1941). Auxin has morphogenic properties and is implicated in signaling over both short and long distances, as well as in rapid responses to environmental fluctuations and long-term developmental processes (for review, see Bhalerao and Bennett 2003; Cho et al. 2007; Mroue et al. 2018; Zhao 2018).
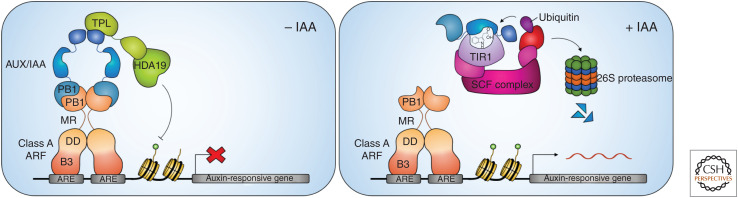
Auxin has a profound effect on gene expression. The exogenous application of auxin to a variety of tissues has demonstrated its genome-wide effects, which include the modulation of hundreds to thousands of genes depending on cellular context (Paponov et al. 2008; Simonini et al. 2017; Kalve et al. 2020). An elegant molecular mechanism for how auxin controls gene expression has been elucidated (for review, see Salehin et al. 2015; Weijers and Wagner 2016; Leyser 2018). Central to this mechanism are proteins of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) family, which act as auxin receptors. In the absence of auxin, repression is maintained at target loci through the Aux/IAA-mediated recruitment of TOPLESS/TOPLESS-RELATED (TPL/TPR) corepressors (Fig. 1; Szemenyei et al. 2008). Upon auxin binding, TIR1/AFBs increase their affinity toward Aux/IAA transcriptional repressors to facilitate their degradation, and thereby relieve the repression of transcription factors belonging to the AUXIN RESPONSE FACTOR (ARF) family (Dharmasiri et al. 2005; Kepinski and Leyser 2005). Plants with mutations in components of the TIR1/AFB pathway have severe developmental defects, demonstrating the prominent role of this pathway in relaying the auxin signal (Berleth and Jürgens 1993; Leyser et al. 1993; Hamann et al. 1999; Prigge et al. 2020). In this review, we will refer to this TIR1/AFB-based mechanism as the canonical auxin signaling pathway or simply the canonical pathway.
Figure 1.
The canonical auxin signaling pathway. In the absence of auxin, AUXIN RESPONSE FACTORS (ARFs) are bound by Aux/IAA repressor proteins, which recruit transcriptional corepressors and prevent the expression of auxin-responsive genes. Auxin increases the affinity between the TIR1/AFB auxin receptor complex and Aux/IAAs, which are subsequently ubiquitinated and degraded. ARFs are then free to activate the expression of auxin-responsive genes. (ARE) Auxin-responsive element, (B3) B3 DNA-binding domain, (DD) dimerization domain, (MR) middle region, (PB1) Phox and Bem1 domain.
An open question in the field of plant biology is how canonical auxin signaling can facilitate the vast array of processes involving auxin. The duplication and neofunctionalization of pathway components are likely to contribute to this versatility. For example, the Arabidopsis genome encodes six TIR1/AFB receptors, 29 Aux/IAAs, and 23 ARFs operating in different combinations in different tissues (Salehin et al. 2015). However, several observations have at some point seemed incompatible with signaling through the canonical pathway either due to their distinct temporal or spatial occurrences, or because they were thought to be regulated by distinct receptors. These observations are (1) acid growth (Hager et al. 1971; for review, see Arsuffi and Braybrook 2018; Gallei et al. 2020), (2) auxin-induced cytoskeletal rearrangements (Bergfeld et al. 1988; for review, see Sauer and Kleine-Vehn 2011; Zhu and Geisler 2015), (3) rapid root-growth inhibition (Fendrych et al. 2018), (4) the rapid influx of Ca2+ during gravitropism (Monshausen et al. 2011), (5) TIR1/AFB2-independent inhibition of PIN cycling (Paciorek et al. 2005; Robert et al. 2010), (6) localization of canonical pathway components to the cytosol (Wang et al. 2016; Powers et al. 2019), and (7) the limited interaction between repressive ARFs and Aux/IAA repressors (Vernoux et al. 2011). While some of these phenomena have since been shown to require canonical pathway components (Dindas et al. 2018; Fendrych et al. 2018; Uchida et al. 2018), or to occur independently of auxin in the case of microtubule rearrangements (Adamowski et al. 2019), the mechanisms underlying others have not yet been elucidated. Evidence for alternative auxin signaling mechanisms has gradually accumulated, and such mechanisms may contribute to the versatility of the auxin response throughout development. Here, we provide a summary of recent findings in this area and aim to highlight critical questions that must be answered to obtain a more comprehensive understanding of auxin biology.
INCOMPATIBILITIES BETWEEN AUXIN EFFECTS AND THE CANONICAL PATHWAY
The acid growth hypothesis was proposed around 50 years ago (Hager et al. 1971) and is one of the many phenomena that prompted the idea of noncanonical auxin signaling. In acid growth, auxin activates the movement of protons into the apoplast through the plasma membrane (PM) proton-pumping ATPase (PM H+-ATPase). The acidification of the apoplast upon proton accumulation leads to alterations in the properties of the cell wall, promoting hyperpolarization of the PM to allow turgor pressure-mediated cell expansion (Kutschera 1994; Hager 2003; Takahashi et al. 2012; Arsuffi and Braybrook 2018). The activity of PM H+-ATPases depends on their phosphorylation status, and phosphorylation was observed in tir1 afb2 double mutants implying that auxin receptors other than TIR1 and AFB2 are responsible for regulating ATPase activity (Takahashi et al. 2012).
Initially, it was proposed that AUXIN-BINDING PROTEIN1 (ABP1) was responsible for mediating nontranscriptional, rapid auxin responses such as cell expansion, cytoskeletal rearrangement, and the inhibition of endocytic PIN cycling (Chen et al. 2001, 2014; Robert et al. 2010) and could therefore be considered a candidate for the non-TIR1/AFB receptor in acid growth (Sauer and Kleine-Vehn 2011). However, it has since been shown that abp1 loss-of-function does not cause developmental defects in Arabidopsis, and its importance in auxin biology is now disputed (Dünser and Kleine-Vehn 2015; Gao et al. 2015; Habets and Offringa 2015).
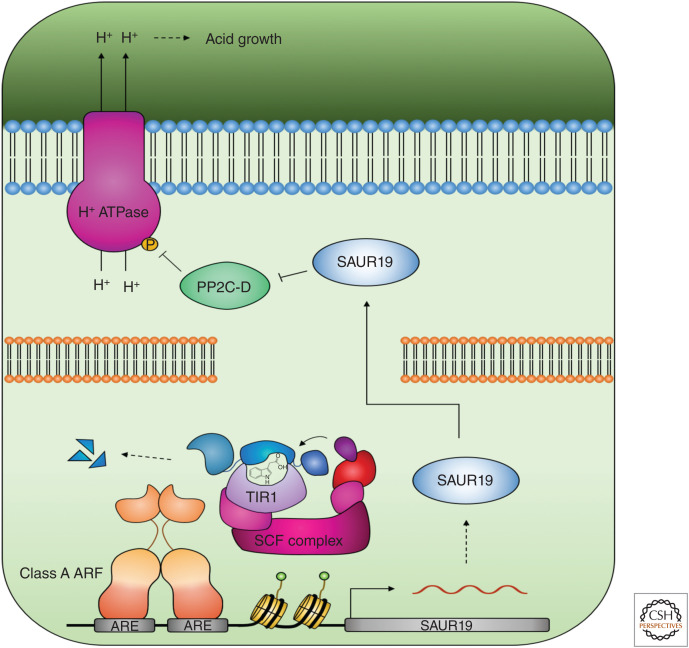
Later, it was discovered that a subset of the SMALL AUXIN UP-RNA (SAUR) gene family are induced through the canonical pathway and inhibit the activity of PP2C-D protein phosphatases to promote PM H+-ATPase phosphorylation, thus directly linking the activity of the TIR1/AFB pathway to acid growth (Fig. 2; Spartz et al. 2014). A synthetic auxin response system has since been developed, using a convex auxin ligand (cvxIAA), which hijacks the downstream auxin signaling machinery in the presence of a concave TIR1 (ccvTIR1) receptor (Uchida et al. 2018). When expressing this synthetic ccvTIR1 receptor in the tir1 afb2 double mutant under control of the endogenous TIR1 promoter, treatment with cvxIAA-induced SAUR19 expression, acid growth, and PM H+-ATPase phosphorylation, providing conclusive evidence that the canonical pathway regulates acid growth (Uchida et al. 2018). Consistent with Takahashi et al. (2012), treatment of the tir1 afb2 mutant with natural IAA also induced H+-ATPase phosphorylation. Moreover, rapid cell elongation in response to natural IAA was still observed in the tir1 afb2 ccvTIR1 line, suggesting that other auxin receptors contribute to this response. Whether these receptors include other members of the AFB family remains to be established (Uchida et al. 2018).
Figure 2.
Auxin triggers the up-regulation of SAUR19 through the canonical signaling pathway. SAUR19 inhibits PP2C-D activity leading to the phosphorylation and activation of H+-ATPases, the acidification of the apoplast, and acid growth.
A second phenomenon that suggested a role for noncanonical signaling during growth and development is auxin-mediated root growth inhibition. Using microfluidics and live imaging, Fendrych et al. (2018) showed that root growth was rapidly inhibited upon the exogenous application of auxin on a time scale that is incompatible with a transcriptional response. Equally, removal of auxin led to the rapid reestablishment of root growth, suggesting a rapid and reversible mechanism of auxin-mediated growth regulation. Interestingly, the response still depends on TIR1/AFB, which suggests the existence of a nontranscriptional output from the canonical pathway (Fendrych et al. 2018; Dubey et al. 2020).
Root growth inhibition on the lower side of the root forms the basis of gravitropism (Shih et al. 2015), and gravistimulation is followed by the redistribution of auxin in the root, PM depolarization, and the influx of Ca2+. Although auxin-mediated calcium signaling was initially thought to occur independently of the canonical receptors (Monshausen et al. 2011), the influx of Ca2+ depends on the intracellular perception of auxin through the canonical pathway and on the CNGC14 Ca2+ channel (Shih et al. 2015; Dindas et al. 2018).
The involvement of the canonical receptors in these rapid responses raises questions on how the nuclear-localized TIR1/AFBs can mediate nontranscriptional signaling and alterations in signaling components at the PM (Retzer et al. 2018). Both Fendrych et al. (2018) and Dindas et al. (2018) speculated that canonical pathway components may be translocated out of the nucleus to mediate their effects at the cell periphery. TIR1/AFBs have been observed in the cytosol, and upon their stabilization, when mutations in the receptors release them from their associated E3 ubiquitin ligases, auxin-resistant phenotypes were observed (Yu et al. 2015; for review, see Kubeš and Napier 2019). Prigge et al. (2020) observed that fluorescently tagged TIR1 exhibited a predominantly nuclear localization, whereas AFB1 was primarily cytosolic and AFB2-5 partitioned between the nucleus and the cytoplasm (Prigge et al. 2020). Furthermore, TIR1/AFBs are chaperoned by cytoplasmic Heat-Shock Protein 90s (HSP90s), which appears to stabilize the receptors to promote their nuclear localization and temperature-dependent auxin responses (Wang et al. 2016). It remains unclear whether localization of the canonical receptors to the cytosol has physiological consequences; however, other canonical pathway components have been observed in the cytosol. ARF7 and ARF19 were found to form cytoplasmic assemblies with physiological consequences on auxin sensitivity (Powers et al. 2019), although this nucleocytoplasmic partitioning has not to date been linked to HSP90 activity.
Another open question relating to ARF function in the canonical pathway is the role of ARF repressors in gene regulation. ARF proteins are B3-type transcription factors that have been divided into specific classes based on phylogeny and ability to activate/repress gene expression (Tiwari et al. 2003; Finet et al. 2013; Mutte et al. 2018). The ARF family in Arabidopsis has 23 members with a general domain structure where the DNA-binding domain is situated in the amino-terminal half followed by a middle region (MR) and a carboxy-terminal Phox and Bem1 (PB1) domain. The amino acid sequence of the MR dictates the function of the ARF as either activating (A-ARFs) or repressive (B-ARFs and C-ARFs) (Tiwari et al. 2003). At the carboxy-terminus, most ARFs contain a PB1 domain that facilitates Aux/IAA interactions, repressing ARF activity in the absence of auxin (Salehin et al. 2015).
The canonical model of auxin signaling proposes that the lifting of Aux/IAA-mediated repression in the presence of auxin leads to the expression of auxin-responsive genes. This model fits well with the activity of A-ARFs, but the role of B-ARFs and C-ARFs in this model was less clear until recently. Through a study in the liverwort, Marchantia polymorpha, which contains one ARF of each class, it was demonstrated that the B-ARFs compete with A-ARFs to bind auxin response elements (AREs) in the promoters of common target genes independently of auxin (Kato et al. 2020). Whether B-ARFs function in a similar way in Arabidopsis where the ARF clade has undergone expansion is unknown; however, it has been shown that B- and C-ARFs have a limited capacity to interact with the Aux/IAA repressors despite the presence of a carboxy-terminal PB1 domain, suggesting that they may also function as auxin-independent transcriptional repressors in Arabidopsis (Vernoux et al. 2011; Piya et al. 2014).
ETT-MEDIATED AUXIN SIGNALING
ETT, ARF13, ARF17, and ARF23 are atypical ARFs that entirely lack the PB1 domain required for interactions with Aux/IAAs (Guilfoyle and Hagen 2007; Lokerse and Weijers 2009). While the carboxyl termini of ARF13, ARF17, and ARF23 are simply truncated, ETT contains a region at its carboxy terminus, which does not share homology with other eukaryotic proteins. This domain was named the ETTIN-specific (ES) domain (Simonini et al. 2016). Given their atypical domain structures, these ARFs do not fit into the canonical model of signaling. So far, no biological function has been assigned to ARF13 or ARF23, but ARF17 is important for pollen development and anther dehiscence (Yang et al. 2013; Wang et al. 2017; Xu et al. 2019), although the molecular mechanism is unknown.
ett loss-of-function mutants have a range of auxin-related phenotypes most prominently during gynoecium development (Sessions and Zambryski 1995; Sessions et al. 1997). In addition, defects in ovule integument development, lateral root emergence, and primary branch formation have been reported (Sessions and Zambryski 1995; Sessions et al. 1997; Garcia et al. 2006; Marin et al. 2010; Kelley et al. 2012; Simonini et al. 2016). The auxin-related developmental defects of ett mutants led to the hypothesis that ETT can mediate auxin signaling independently of the canonical pathway. It was suggested that ETT translates local auxin concentrations to developmental outputs in the gynoecium, but the molecular mechanisms underpinning this were unknown (Nemhauser et al. 2000; Pekker et al. 2005; Simonini et al. 2016). Since then, data supporting a mechanism that is fundamentally different from canonical auxin signaling are emerging. For example, it has been established that several protein–protein interactions involving ETT are sensitive to the natural auxin, IAA, but not other auxinic compounds (Simonini et al. 2016). Several of these interactions are with transcription factors belonging to a range of different families and it has been hypothesized that such interactions are relevant for auxin responsiveness of specific tissues or cell types during development (Simonini et al. 2016). As auxin levels rise, interactions between ETT and its protein partners are broken. Chromatin immunoprecipitation (ChIP) experiments have shown that ETT remains bound to the DNA of its target loci both in the presence and absence of auxin (Simonini et al. 2016). Moreover, a genome-wide analysis of ETT-mediated gene regulation showed that the transcriptome regulated by ETT changes dynamically depending on the auxin levels (Simonini et al. 2016, 2017). For a subset of targets that were analyzed in further detail, ETT-dependent, auxin-responsive gene expression was shown to be independent of the canonical pathway (Kuhn et al. 2020). Biophysical data have shown that ETT is able to directly interact with the auxin molecule via the ES domain suggesting that binding of auxin disrupts the interaction between ETT and its partners (Kuhn et al. 2020).
Like animal genomes, plant genomes encode corepressors of the conserved Tup1/ GROUCHO/TLE family, and their functions have been described in both monocots and dicots (Long et al. 2006; Liu et al. 2019). In Arabidopsis, this family is comprised of TOPLESS (TPL) and TPL-related (TPR) proteins and has been shown to associate with transcription factors affecting most aspects of plant development (Causier et al. 2012). The molecular function of TPL was first described in the context of auxin signaling where TPL was found to interact with Aux/IAAs when auxin levels are low to repress gene expression. As auxin levels increase and Aux/IAAs dissociate from ARFs, TPL also dissociates from the locus to allow derepression (Szemenyei et al. 2008). TPL-mediated repression occurs through the recruitment of histone deacetylases (HDACs) such as HDA19, which maintain a repressed state at target loci that can subsequently be reversed upon Aux/IAA degradation and TPL dissociation (Krogan et al. 2012).
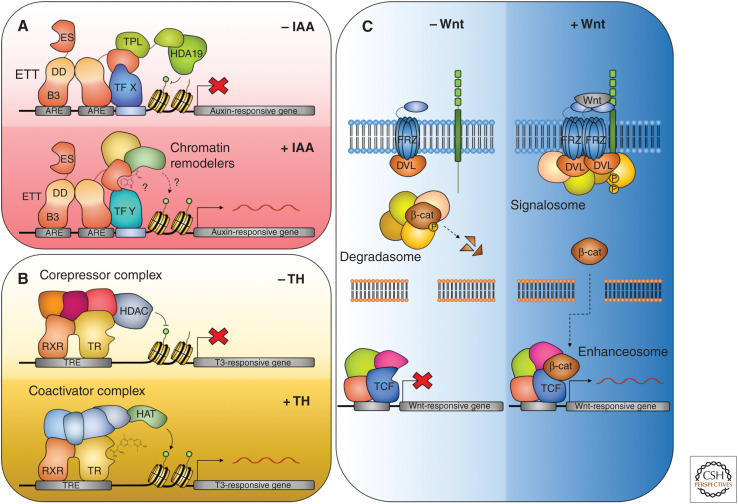
Members of the TPL/TPR family also play a central role in ETT-mediated auxin signaling. Under low auxin conditions, ETT can interact directly with TPL/TPR proteins, but this interaction is disrupted when auxin concentrations rise (Fig. 3A). The interaction occurs via a repressive motif in the ES domain of ETT with the amino acid sequence RLFG found as a common motif in proteins interacting with TPL/TPR (Causier et al. 2012; Liu et al. 2019; Kuhn et al. 2020).
Figure 3.
Ligand-mediated switch in the activity of transcription factor complexes. (A) The ETT-mediated auxin signaling pathway. In the absence of auxin, ETT recruits the corepressor TOPLESS (TPL) to target loci. TPL, in turn, recruits HDA19, which deacetylates histones and represses target gene expression. Auxin binds to ETT directly through the ETT-specific (ES) domain and triggers the dissociation of the repressive complex. Histone acetylation occurs, potentially through the recruitment of coactivators by ETT, and target genes are derepressed. (B) Thyroid hormone (TH) signaling pathway. In the absence of TH, the DNA-bound thyroid hormone receptor (THR) is associated with corepressors and histone deacetylases (HDACs), which repress target gene expression. In the presence of TH, the corepressors are exchanged for coactivators, and histone acetyltransferases (HATs) are recruited, activating target genes. (C) Wnt/β-catenin signaling. In the absence of Wnt, β-catenin is degraded by the degradasome. When the Wnt ligand binds to the Frizzled (FRZ) receptor, degradasome components are recruited to the plasma membrane by the active receptors, leading to the formation of the signalosome, and the accumulation of β-catenin. β-Catenin enters the nucleus and associates with the TCF transcription factor and other components of the enhanceosome, the Groucho corepressors dissociate, and Wnt-responsive genes are expressed.
Generally, hormone signaling in plants involves the ubiquitination and subsequent degradation of transcriptional repressors upon ligand perception and facilitates the activation of gene expression (for review, see Santner and Estelle 2010; Kelley and Estelle 2012). The canonical auxin signaling pathway provides one such example (Salehin et al. 2015; Weijers and Wagner 2016).
In contrast, hormone signaling in animals often involves direct binding of the hormone ligand (as in thyroid hormone [TH] signaling) or an effector protein (as in Wingless [Wnt]/β-catenin signaling) to transcription factors. For instance, in the absence of TH, the thyroid hormone receptor (THR) binds its target genes in complex with corepressors that attract HDACs to the locus to maintain the chromatin in a repressed state. When TH levels increase, TH binds to the DNA-bound THR resulting in a conformational change and the exchange of corepressors for coactivators, triggering the recruitment of histone acetyl transferases (HATs) and the activation of target-gene expression (Fig. 3B; Tsai and O'Malley 1994).
In Wnt/β-catenin signaling the perception of the Wnt ligand at the PM triggers the release of β-catenin from the degradasome (Fig. 3C; Gammons and Bienz 2018). Components of the degradasome are sequestered at the PM forming the signalosome, allowing β-catenin to accumulate and enter the nucleus. Here, β-catenin associates with components of the enhanceosome, including the DNA-bound TCF transcription factor. Wnt association triggers a conformational change and the dissociation of GROUCHO corepressors and activates Wnt-responsive gene expression.
The mechanism by which ETT relays changes in auxin levels is reminiscent of the TH and Wnt/β-catenin pathways in animals. Like THR and TCF, ETT is constitutively bound to at least a subset of its targets; however, it remains to be established whether ETT is involved in the formation of an activating complex upon ligand perception as in the case of THR during TH perception (Fig. 3). A fundamental difference between the ETT-mediated and canonical models of auxin signaling is that the former does not in its core mechanism invoke the requirement for protein degradation to activate gene expression. To reinstate repressive conditions in the canonical pathway, a new pool of Aux/IAA repressors must first be synthesized. In contrast, the ETT-based mechanism is immediately reversible, thus providing an ability to function in specialized processes where a fast and reversible response is important.
The evolutionary origin of the ETT-mediated auxin signaling pathway is so far unknown. While the canonical pathway is conserved at least back to the extant bryophytes such as M. polymorpha (Mutte et al. 2018; Kato et al. 2020), homologs of ETT do not appear outside the angiosperm phylum and ETT-mediated signaling may therefore be absent in nonflowering plants. The domains and residues in ETT-ES known to be important for auxin perception and TPL interaction are highly conserved throughout the angiosperms (Finet et al. 2010; Kuhn et al. 2020). ETT proteins in the basal angiosperms, such as Amborella trichopoda, contain a carboxy-terminal PB1 domain, as well as the conserved RLFG motif required for interaction with TPL/TPR, and this is likely to represent the ancestral state of ETT (Fig. 4; Finet et al. 2010, 2013; Causier et al. 2012; Kuhn et al. 2020). The PB1 domain was then lost independently in several lineages of extant angiosperms, including in the magnoliids and monocots (Finet et al. 2013).
Figure 4.
ETT domains and their conservation across the seed plants. ETT in the eudicot Arabidopsis thaliana and Solanum lycopersicum contains a highly conserved B3 DNA-binding domain, a nuclear localization signal (NLS), two tasi-ARF-binding sites, and an EAR-like TPL-interacting domain. The ARF3/4-like ortholog of the gymnosperm Ginkgo biloba and the ETT of the basal angiosperm Amborella trichopoda share these domains but also contain a full-length PB1 domain. Within the magnoliids, the PB1 domain is retained in the Liriodendron tulipifera ortholog but has been lost in the Persea americana ortholog. The monocot Oryza sativa has two clades of ETT paralogs with either a partial or total loss of the PB1 domain.
The closest homolog of ETT in Arabidopsis is another B-ARF, ARF4 (Pekker et al. 2005). A phylogenetic study strongly suggested that ETT and ARF4 were generated by a duplication event that took place after angiosperms diverged from gymnosperms, but before the last common ancestor of extant angiosperms (Finet et al. 2010). In contrast to ETT, ARF4 contains a carboxy-terminal PB1 domain and was previously shown to interact with Aux/IAA repressors, indicating its regulation via canonical auxin signaling in Arabidopsis (Vernoux et al. 2011). ETT and ARF4 have overlapping functions and arf4 loss-of-function mutants severely enhance defects of ett mutants (Pekker et al. 2005). Intriguingly, ARF4 is also truncated through alternative splicing in several angiosperm lineages, suggesting that the truncation of ETT and ARF4 proteins generally has a functional relevance (Finet et al. 2013). Experiments involving chimeric ETT and ARF4 constructs suggested that the unique carboxy-terminal domain of ETT is important for its function, and that ARF4 function similarly depends on the presence of the PB1 domain (Finet et al. 2010). Further domain swap experiments using ETT and ARF4 domains from species across the angiosperm lineage will be required to fully understand how and when ETT and ARF4 adopted distinct functions, and to reveal the evolutionary origins of ETT-mediated auxin signaling.
Several mutant screens carried out in Arabidopsis to identify genetic factors critical for development or auxin responsiveness led to the discovery of genes encoding components of the canonical pathway (Berleth and Jürgens 1993; Leyser et al. 1993; Hardtke and Berleth 1998; Hamann et al. 1999). This clearly reflects the predominance of this pathway and its importance throughout the plant life cycle. The ETT-based mechanism is not employed to such an extent; however, its immediate reversibility suggests that it is particularly well-suited for certain processes such as the changing of tissue polarity (Simonini et al. 2016). Finally, given the mechanistic conservation between ETT-based auxin signaling and certain animal hormone pathways, it is possible that direct binding of plant hormones or other small molecules to transcription factors is a more common mechanism of regulating gene expression in plants than previously expected. Approaches involving highly sensitive proteomics and biophysics techniques will be essential to elucidate other such mechanisms.
NONCANONICAL SIGNALING THROUGH PROTEIN KINASES
Auxin gradients underpin the formative cell division and expansion patterns that are required for appropriate organ morphogenesis and the formation of the plant body plan (for review, see Bhalerao and Bennett 2003; Friml 2003). While auxin-mediated cell expansion has been widely studied, and many of the mechanisms relating to asymmetric cell growth have been elucidated (for review, see Majda and Robert 2018; Du et al. 2020), processes involved in cell division patterning are more poorly understood. The Trans-Membrane Kinase 1 (TMK1) pathway has been implicated in regulating both differential cell elongation and the establishment of cell division patterns in response to auxin.
There are four kinases in the TMK subfamily in Arabidopsis, all of which contain an extracellular leucine-rich-repeat (LRR) motif linked to an intracellular kinase domain through a single transmembrane region (Dai et al. 2013). The TMK subfamily was first linked to auxin signal transduction when the phenotypes of tmk mutant combinations were analyzed. While single mutants showed no obvious phenotypic abnormalities, double, triple, and quadruple mutants showed cell expansion and proliferation defects, miniaturized organs, infertility, and a reduced sensitivity to exogenously applied auxin (Dai et al. 2013). TMK1 has since been implicated in apical hook maintenance, lateral root development (along with TMK4), and pavement cell morphogenesis (Xu et al. 2014; Cao et al. 2019; Huang et al. 2019). The roles of other TMK family members in auxin signaling have not yet been elucidated.
TMK1 was initially thought to regulate auxin-mediated signaling through its interaction with Auxin-Binding Protein 1 (ABP1) at the PM (Xu et al. 2010, 2014). It was observed that the auxin-mediated activation of the TMK1 pathway triggered Rho-like GTPases from plants (ROPs) at the PM, leading to alterations in the cytoskeleton and the distribution of PINs that seemed to be important for lobe development in the pavement cells of the leaves (Xu et al. 2010, 2014). However, the significance of ABP1 in plant biology has since been put into question (Gao et al. 2015; Habets and Offringa 2015), and so the receptor responsible for extracellular auxin binding and subsequent TMK1 activation remains elusive.
The TMK1 pathway was discovered using the apical hook as a model system (Cao et al. 2019; for review, see Gallei et al. 2020). The apical hook is a structure produced after germination in dicots that protect the shoot apical meristem and cotyledons from damage as the hypocotyl elongates through the soil in search of light. Its formation is facilitated through asymmetric growth on either side and this process is regulated by auxin (for review, see Abbas et al. 2013). The production of the apical hook occurs in three stages: formation, maintenance, and opening. During the formation phase, auxin is concentrated at the concave (inner) side of the apical hook, inhibiting cell elongation, while the convex (outer) side of the hook elongates. This difference in growth leads to hook bending.
Auxin biosynthesis mutants show variable phenotypes relating to the abolition of differential growth in the apical hook (Cao et al. 2019). Ectopic auxin production in the yuc1-D mutant represses growth at the convex side of the apical hook, while auxin deficiency in the wei8-3 tar2-1 mutant promoted growth at the concave side, highlighting that the asymmetric distribution of auxin is vital for differential growth and apical hook formation. The tmk1 mutant also exhibited growth at the concave side of the apical hook. However, the exogenous application of auxin, which rescued defects in the wei8-3 tar2-1 apical hook, did not rescue the tmk1 phenotype, implying that tmk1 was impaired in auxin perception and/or signaling.
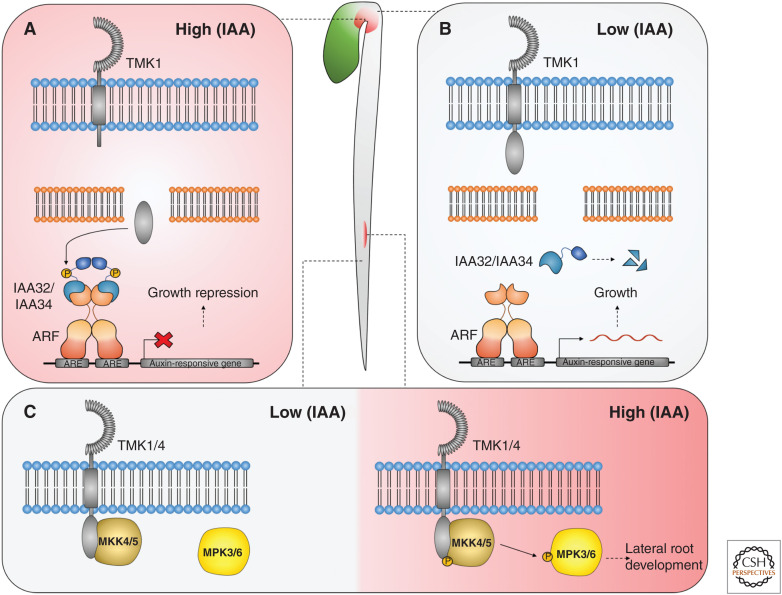
The carboxyl-terminus of TMK1 is cleaved during the maintenance phase at the concave side of the apical hook, in response to the relatively high concentrations of auxin present in this region of the hypocotyl (Fig. 5A; Cao et al. 2019). The carboxy-terminal TMK1 fragment is then translocated to the nucleus, where it interacts with and phosphorylates the Aux/IAA repressors, IAA32 and IAA34, leading to their stabilization and ultimately the repression of growth at the concave side of the apical hook. Unlike Aux/IAA proteins in the canonical pathway, IAA32 and IAA34 lack the degron domain required for interaction with TIR1/AFB receptors and are stabilized in the presence of auxin by TMK1-mediated phosphorylation (Calderon-Villalobos et al. 2010; Cao et al. 2019). Under low auxin concentrations, TMK1 is inactive and IAA32/34 are destabilized, allowing auxin-responsive gene expression and growth (Fig. 5B).
Figure 5.
Trans-Membrane Kinase 1 (TMK1)-mediated auxin signaling during apical hook maintenance and lateral root emergence. (A) Relatively high auxin concentrations at the concave side of the apical hook activate the cleavage of the TMK1 carboxyl terminus. (B) At the convex side of the apical hook, relatively low auxin concentrations promote growth due to the destabilization of IAA32/34. The carboxyl-terminus of TMK1 is translocated to the nucleus, where it phosphorylates and stabilizes IAA32/34 to repress gene expression and inhibit growth. (C) Under high auxin conditions, TMK1 phosphorylates MKK4/5, triggering the downstream phosphorylation of MPK3/6 to regulate lateral root emergence.
The TMK1-mediated auxin signaling pathway has also been implicated in the control of mitogen-activated protein kinase (MAPK) signaling (Fig. 5C; Huang et al. 2019). Lateral root emergence requires tight regulation of cell division and expansion. Auxin maxima form at the locations where lateral roots begin to initiate, and disruption of auxin distribution (De Smet et al. 2007) or of MAPK signaling (Zhu et al. 2019) caused defects in lateral root development. It was previously observed that the application of exogenous auxin triggered the activation of MAPK signaling in the root, although the mechanism by which this occurs is unknown (Mockaitis and Howell 2000).
Huang et al. (2019) showed that auxin triggers the activation of TMK1 and TMK4, which directly interact with and phosphorylate the MAPK-kinases (MAPKKs) 4/5 through their carboxy-terminal intracellular kinase domains. MAPK3/6 were reported to act downstream of MAPKK4/5 in response to several stimuli and are coexpressed with TMK1 during lateral root emergence. The tmk1 tmk4 double mutant and RNAi knockdown lines of mapkk4/5 and mapk3/6 all showed the same defects in lateral root emergence and aberrant cell division patterning in the emerging lateral root primordia, suggesting a shared role in regulating these processes (Huang et al. 2019).
In wild-type plants, the application of auxin triggers lateral root emergence. In auxin-deficient mutants, which produce no lateral roots, lateral root development can be restored by the application of auxin (Huang et al. 2019). The lateral root phenotypes in the tmk1 tmk4 double mutant, and mapkk4/5 and mapk3/6 RNAi knockdown lines, could not be rescued in this manner implying that the TMK1/4-MAPKK4/5-MAPK3/6 module transduces the auxin signal during lateral root development. Furthermore, the level of MAPK3/6 phosphorylation directly correlated with the levels of auxin in the lateral root, and MAPK3/6 phosphorylation was abolished in the tmk1 tmk4 but not in tir1 afb2 afb3 mutant lines, showing that this TMK1/4-dependent pathway is independent of canonical auxin signaling.
Auxin was previously shown to regulate lateral root emergence through the IDA-HAE/HSL2 pathway, which is also upstream of the MAPKK4/5-MAPK3/6 module (Kumpf et al. 2013; Zhu et al. 2019). This raises questions about how peptide and phytohormone signaling modules undergo cross talk to regulate developmental processes, and how specificity in signaling is established through shared signaling modules. The canonical pathway also has important roles during lateral root development, and the exact relationships between all of the auxin-related pathways will now need to be dissected to get a global view of auxin signaling and its regulation of the lateral roots (for review, see He and Meng 2020). Lateral root emergence phenotypes have also been reported in both ett-3 (Simonini et al. 2016), and canonical receptor mutants (Prigge et al. 2020), and lateral roots may therefore provide an excellent model system in which to dissect interactions between all currently known auxin-mediated signaling pathways.
Other noncanonical Aux/IAAs that lack the degron domain have been implicated in growth and development, including IAA20, IAA30, and IAA33. The iaa20 iaa30 double mutant produced ectopic protoxylem implying at least a partially redundant role for these Aux/IAAs in xylem patterning (Müller et al. 2016). The expression of both IAA20 and IAA30 is induced by auxin through the binding of MONOPTEROS(MP)/ARF5 to their promoters, these Aux/IAAs then bind to MP repressing its activity in a feedback loop (Sato and Yamamoto 2008; Krogan et al. 2014; Müller et al. 2016). The expression levels of these noncanonical Aux/IAAs and MP are further modulated by the HD-ZIP transcription factor PHABULOSA (PHB), which stabilizes the auxin response to allow proper xylem development (Müller et al. 2016). It remains unclear whether IAA20 and IAA30 are phosphorylated to modulate their stability, or whether they function downstream of a kinase-mediated auxin signaling pathway such as the TMK1 pathway.
IAA33 maintains stem cell identity in the root by competing with the canonical Aux/IAA IAA5 to bind ARF10 and ARF16, thereby maintaining the repression of these ARFs in the presence of auxin (Lv et al. 2020). IAA33 is stabilized in response to auxin through its phosphorylation by MAPK14, and this stabilization is required for normal stem cell identity specification in the distal root. It is still unclear how auxin triggers the activation of MAPK14, although it is plausible that it could occur through a TMK1-like pathway.
Phosphorylation is a very rapid and reversible change and is an appealing alternative mechanism to explain how the auxin signal can be transduced so swiftly; however, the TMK1 pathway is yet to be directly linked to any of these rapid auxin responses, and many of them have since been shown to be TIR1/AFB-dependent (Fendrych et al. 2018; Uchida et al. 2018; Dubey et al. 2020).
CONCLUDING REMARKS
While the combinatorial nature of the TIR1/AFB-Aux/IAA-ARF pathway of auxin signaling provides great potential to explain diverse effects of auxin on plant development, there is mounting evidence for the existence of alternative noncanonical signaling pathways to mediate certain processes. A common theme in noncanonical auxin signaling is the loss of key domains from canonical pathway components that then allows them to be recruited as novel signaling modules. For instance, the loss of the degron domain from IAA32/34 in the TMK1-mediated pathway and the PB1 domain from ETT have allowed these pathways to regulate specific developmental processes independently of the canonical pathway. Moreover, canonical auxin receptors have now been linked to nontranscriptional mechanisms and it seems likely that canonical and noncanonical pathways are connected into a wider auxin signaling network that is carefully integrated to regulate a wide range of processes.
The existence of different pathways by which auxin relays its effect opens the door for potential interactions between these mechanisms that would increase the potential for specificity in the auxin response. Understanding such interactions will therefore be crucial to fully comprehend the versatility of auxin function in plant biology. Auxin has been intensely studied by plant scientists for decades and there is little doubt that this hugely important plant hormone will continue to provide scientific excitement and fascination for a long time to come.
ACKNOWLEDGMENTS
We are grateful to Yang Dong, André Kuhn, Bhavani Natarajan, and Nicola Stacey for providing critical comments to the manuscript. H.M.M. was funded by the John Innes Foundation, A.C.H.A. by Sainsbury PhD studentship, and L.Ø. by an Institute Strategic Programme Grant from the UKRI Biotechnological and Biological Sciences Research Council (BBSRC) to the John Innes Centre (BB/P013511/1).
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abbas M, Alabadí D, Blázquez MA. 2013. Differential growth at the apical hook: all roads lead to auxin. Front Plant Sci 4: 441. 10.3389/fpls.2013.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Li L, Friml J. 2019. Reorientation of cortical microtubule arrays in the hypocotyl of Arabidopsis thaliana is induced by the cell growth process and independent of auxin signaling. Int J Mol Sci 20: 3337. 10.3390/ijms20133337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsuffi G, Braybrook SA. 2018. Acid growth: an ongoing trip. J Exp Botany 69: 137–146. 10.1093/jxb/erx390 [DOI] [PubMed] [Google Scholar]
- Bergfeld R, Speth V, Schopfer P. 1988. Reorientation of microfibrils and microtubules at the outer epidermal wall of maize coleoptiles during auxin-mediated growth. Bot Acta 101: 57–67. 10.1111/j.1438-8677.1988.tb00012.x [DOI] [Google Scholar]
- Berleth T, Jürgens G. 1993. The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118: 575–587. [Google Scholar]
- Bhalerao RP, Bennett MJ. 2003. The case for morphogens in plants. Nat Cell Biol 5: 939–943. 10.1038/ncb1103-939 [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. 2010. Auxin perception—structural insights. Cold Spring Harb Perspect Biol 2: a005546. 10.1101/cshperspect.a005546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelová Z, et al. 2019. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568: 240–243. 10.1038/s41586-019-1069-7 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438. 10.1104/pp.111.186999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM. 2001. The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J 28: 607–617. 10.1046/j.1365-313x.2001.01152.x [DOI] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, Rakusová H, Benkova E, Perrot-Rechenmann C, Friml J. 2014. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516: 90–93. 10.1038/nature13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Lee OR, Ganguly A, Cho HT. 2007. Auxin-signaling: short and long. J Plant Biol 50: 79–89. 10.1007/BF03030615 [DOI] [Google Scholar]
- Dai N, Wang W, Patterson SE, Bleecker AB. 2013. The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS ONE 8: e60990. 10.1371/journal.pone.0060990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Müller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. 2018. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. 10.1038/s41467-018-03582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Spalding EP, Gray WM. 2020. Rapid auxin-mediated cell expansion. Ann Rev Plant Biol 71: 379–402. 10.1146/annurev-arplant-073019-025907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dubey SM, Serre NBC, Oulehlova D, Vittal P, Fendrych M. 2020. No time for transcription—rapid auxin responses in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünser K, Kleine-Vehn J. 2015. Differential growth regulation in plants—the acid growth balloon theory. Curr Opin Plant Biol 28: 55–59. 10.1016/j.pbi.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Enders TA, Strader LC. 2015. Auxin activity: past, present, and future. Am J Bot 102: 180–196. 10.3732/ajb.1400285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Fourquin C, Vinauger M, Berne-Dedieu A, Chambrier P, Paindavoine S, Scutt CP. 2010. Parallel structural evolution of auxin response factors in the angiosperms. Plant J 63: 952–959. 10.1111/j.1365-313X.2010.04292.x [DOI] [PubMed] [Google Scholar]
- Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F. 2013. Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol 30: 45–56. 10.1093/molbev/mss220 [DOI] [PubMed] [Google Scholar]
- Friml J. 2003. Auxin transport—shaping the plant. Curr Opin Plant Biol 6: 7–12. 10.1016/S1369526602000031 [DOI] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrödinger's cat out of the bag. Curr Opin Plant Biol 22: 43–49. [DOI] [PubMed] [Google Scholar]
- Gammons M, Bienz M. 2018. Multiprotein complexes governing Wnt signal transduction. Curr Opin Cell Biol 51: 42–49. 10.1016/j.ceb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci 112: 2275–2280. 10.1073/pnas.1500365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938. 10.1016/j.cub.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Curr Opin Plant Biol 10: 453–460. 10.1016/j.pbi.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Haagen-Smit AJ, Leech WD, Bergen WR. 1941. Estimation, isolation and identification of auxins in plant material. Science 93: 624–625. 10.1126/science.93.2426.624 [DOI] [PubMed] [Google Scholar]
- Habets ME, Offringa R. 2015. Auxin binding protein 1: a red herring after all? Mol Plant 8: 1131–1134. 10.1016/j.molp.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Hager A. 2003. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res 116: 483–505. 10.1007/s10265-003-0110-x [DOI] [PubMed] [Google Scholar]
- Hager A, Menzel H, Krauss A. 1971. Versuche und hypothese zur primarwirkung des auxins beim Streckungswachstum [Experiments and hypothesis concerning the primary action of auxin in elongation growth]. Planta 100: 47–75. 10.1007/BF00386886 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. 1999. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411. 10.1093/emboj/17.5.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Meng X. 2020. MAPK signaling: emerging roles in lateral root formation. Trends Plant Sci 25: 126–129. 10.1016/j.tplants.2019.11.006 [DOI] [PubMed] [Google Scholar]
- Huang R, Zheng R, He J, Zhou Z, Wang J, Xiong Y, Xu T. 2019. Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc Natl Acad Sci 116: 21285–21290. 10.1073/pnas.1910916116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalve S, Sizani BL, Markakis MN, Helsmoortel C, Vandeweyer G, Laukens K, Sommen M, Naulaerts S, Vissenberg K, Prinsen E, et al. 2020. Osmotic stress inhibits leaf growth of Arabidopsis thaliana by enhancing ARF-mediated auxin responses. New Phytologist 226: 1766–1780. 10.1111/nph.16490 [DOI] [PubMed] [Google Scholar]
- Kato H, Mutte SK, Suzuki H, Crespo I, Das S, Radoeva T, Fontana M, Yoshitake Y, Hainiwa E, van den Berg W, et al. 2020. Design principles of a minimal auxin response system. Nat Plants 6: 473–482. 10.1038/s41477-020-0662-y [DOI] [PubMed] [Google Scholar]
- Kelley DR, Estelle M. 2012. Ubiquitin-mediated control of plant hormone signaling. Plant Physiol 160: 47–55. 10.1104/pp.112.200527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109. 10.1242/dev.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser HMO. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. 2012. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. 10.1242/dev.085407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Yin X, Ckurshumova W, Berleth T. 2014. Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. New Phytol 204: 474–483. 10.1111/nph.12994 [DOI] [PubMed] [Google Scholar]
- Kubeš M, Napier R. 2019. Non-canonical auxin signalling: fast and curious. J Exp Bot 70: 2609–2614. 10.1093/jxb/erz111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Ramans Harborough S, McLaughlin HM, Natarajan B, Verstraeten I, Friml J, Kepinski S, Østergaard L. 2020. Direct ETTIN-auxin interaction controls chromatin states in gynoecium development. eLife 9: e51787. 10.7554/eLife.51787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci 110: 5235–5240. 10.1073/pnas.1210835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U. 1994. The current status of the acid-growth hypothesis. New Phytologist 126: 549–569. 10.1111/j.1469-8137.1994.tb02951.x [DOI] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164. 10.1038/364161a0 [DOI] [PubMed] [Google Scholar]
- Liu X, Galli M, Camehl I, Gallavotti A. 2019. RAMOSA1 ENHANCER LOCUS2-mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiol 179: 348–363. 10.1104/pp.18.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokerse A, Weijers D. 2009. Auxin enters the matrix—assembly of response machineries for specific outputs. Curr Opin Plant Biol 12: 520–526. 10.1016/j.pbi.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. 10.1126/science.1123841 [DOI] [PubMed] [Google Scholar]
- Lv B, Yu Q, Liu J, Wen X, Yan Z, Hu K, Li H, Kong X, Li C, Tian H, et al. 2020. Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J 39: e101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majda M, Robert S. 2018. The role of auxin in cell wall expansion. Int J Mol Sci 19: 951. 10.3390/ijms19040951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. Mir390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. 10.1105/tpc.109.072553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Howell SH. 2000. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J 24: 785–796. 10.1046/j.1365-313x.2000.00921.x [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S. 2011. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318. 10.1111/j.1365-313X.2010.04423.x [DOI] [PubMed] [Google Scholar]
- Mroue S, Simeunovic A, Robert HS. 2018. Auxin production as an integrator of environmental cues for developmental growth regulation. J Exp Bot 69: 201–212. 10.1093/jxb/erx259 [DOI] [PubMed] [Google Scholar]
- Müller CL, Valdés AE, Wang G, Ramachandran P, Beste L, Uddenberg D, Carlsbecker A. 2016. PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in Arabidopsis. Plant Physiol 170: 956–970. 10.1104/pp.15.01204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GK, Weijers D. 2018. Origin and evolution of the nuclear auxin response system. eLife 7: e33399, . 10.7554/elife.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J, Feldmann LJ, Zambryski PC. 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Paciorek T, Zažímalová E, Ruthardt N, Petrášek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K. 2008. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337. 10.1093/mp/ssm021 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910. 10.1105/tpc.105.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T. 2014. Protein–protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744. 10.3389/fpls.2014.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. 2019. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell 76: 177–190.e5. 10.1016/j.molcel.2019.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzer K, Singh G, Napier RM. 2018. It starts with TIRs. Nat Plants 4: 410–411. 10.1038/s41477-018-0196-8 [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. 2010. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121. 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. 10.1105/tpc.114.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. 2010. The ubiquitin–proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040. 10.1111/j.1365-313X.2010.04112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Yamamoto KT. 2008. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 133: 397–405. 10.1111/j.1399-3054.2008.01055.x [DOI] [PubMed] [Google Scholar]
- Sauer M, Kleine-Vehn J. 2011. AUXIN BINDING PROTEIN1: the outsider. Plant Cell 23: 2033–2043. 10.1105/tpc.111.087064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. 1995. Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121: 1519–1532. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. 1997. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Shih HW, DePew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr Biol 25: 3119–3125. 10.1016/j.cub.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. 2016. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev 30: 2286–2296. 10.1101/gad.285361.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S, Bencivenga S, Trick M, Østergaard L. 2017. Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell 29: 1864–1882. 10.1105/tpc.17.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. 10.1105/tpc.114.126037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long J. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641. 10.1104/pp.112.196428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Ann Rev Biochem 63: 451–486. 10.1146/annurev.bi.63.070194.002315 [DOI] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, Endo TA, Kimura S, Zhang H, Nomoto M, Tada Y, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol 14: 299–305. 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Movin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M. 2016. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun 7: 10269. 10.1038/ncomms10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xue JS, Yu YH, Liu SQ, Zhang JX, Yao XZ, Liu ZX, Xu XF, Yang ZN. 2017. Fine regulation of ARF17 for anther development and pollen formation. BMC Plant Biol 17: 243. 10.1186/s12870-017-1185-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Ann Rev Plant Biol 67: 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110. 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. 2014. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028. 10.1126/science.1245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang B, Feng YF, Xue JS, Qian XX, Liu SQ, Zhou J, Yu YH, Yang NY, Xu P, et al. 2019. AUXIN RESPONSE FACTOR17 directly regulates MYB108 for anther dehiscence. Plant Physiol 181: 645–655. 10.1104/pp.19.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tian L, Sun MX, Huang XY, Zhu J, Guan YF, Jia QS, Yang ZN. 2013. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol 162: 720–731. 10.1104/pp.113.214940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhang Y, Moss BL, Bargmann BO, Wang R, Prigge M, Nemhauser JL, Estelle M. 2015. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants 1: 14030. 10.1038/nplants.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. 2018. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Ann Rev Plant Biol 69: 417–435. 10.1146/annurev-arplant-042817-040226 [DOI] [PubMed] [Google Scholar]
- Zhu J, Geisler M. 2015. Keeping it all together: auxin-actin crosstalk in plant development. J Exp Bot 66: 4983–4998. 10.1093/jxb/erv308 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Shao Y, Ge S, Zhang M, Zhang T, Hu X, Liu Y, Walker J, Zhang S, Xu J. 2019. A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat Plants 5: 414–423. 10.1038/s41477-019-0396-x [DOI] [PubMed] [Google Scholar]