Abstract
Epigenetic profiling by chromatin immunoprecipitation followed by sequencing (ChIP-seq) has become a powerful tool for genome-wide identification of regulatory elements, for defining transcriptional regulatory networks, and for screening for biomarkers. However, the ChIP-seq protocol for low-input samples is laborious and time-consuming and suffers from experimental variation, resulting in poor reproducibility and low throughput. Although prototypic microfluidic ChIP-seq platforms have been developed, these are poorly transferable as they require sophisticated custom-made equipment and in-depth microfluidic and ChIP expertise, while lacking parallelization. To enable standardized, automated ChIP-seq profiling of low-input samples, we constructed microfluidic PDMS-based plates capable of performing 24 sensitive ChIP reactions within 30 min of hands-on time and 4.5 h of machine-running time. These disposable plates can be conveniently loaded into a widely available controller for pneumatics and thermocycling. In light of the plug and play (PnP) ChIP plates and workflow, we named our procedure PnP-ChIP-seq. We show high-quality ChIP-seq on hundreds to a few thousand of cells for all six post-translational histone modifications that are included in the International Human Epigenome Consortium set of reference epigenomes. PnP-ChIP-seq robustly detects epigenetic differences on promoters and enhancers between naive and more primed mouse embryonic stem cells (mESCs). Furthermore, we used our platform to generate epigenetic profiles of rare subpopulations of mESCs that resemble the two-cell stage of embryonic development. PnP-ChIP-seq allows nonexpert laboratories worldwide to conveniently run robust, standardized ChIP-seq, whereas its high throughput, consistency, and sensitivity pave the way toward large-scale profiling of precious sample types such as rare subpopulations of cells or biopsies.
To allow proper organization and function, genomes contain regulatory layers of information generally referred to as the epigenome. The epigenome includes the binding of transcription factors (TFs) and the presence of a wide range of chemical modifications that can be deposited on DNA and histones, such as methylation of DNA or acetylation on histone tails (Kouzarides 2007). During embryonic and fetal development of mammalian organisms, establishment and maintenance of cellular identity are regulated through these modifications (Berger 2007). Furthermore, a myriad of diseases is caused or characterized by alteration of epigenetic patterns (Portela and Esteller 2010). Therefore, epigenetic changes represent a highly interesting layer of information for disease stratification and for personalized medicine (Heyn and Esteller 2012; Dirks et al. 2016). A plethora of studies have highlighted the role of various histone post-translational modifications (hPTMs) in the regulation of chromatin structure that are necessary for DNA accessibility during gene expression (Jenuwein and Allis 2001; Barski et al. 2007; Berger 2007; Kouzarides 2007; Dekker 2008). For example, the presence of trimethylation of lysine 4 on histone 3 (H3K4me3) at genomic loci is commonly associated with active promoters (Barski et al. 2007), whereas a combination of H3K27 acetylation (H3K27ac) and H3K4me1 is typical for active enhancers (Creyghton et al. 2010). H3K36me3 generally covers gene bodies of active genes (Barski et al. 2007). Conversely, the hPTMs H3K27me3 and H3K9me3 are associated with repressed genes and/or heterochromatin (Boyer et al. 2006; Barski et al. 2007; Martens et al. 2010). As such, it has become clear that epigenetic profiling of hPTMs allows for the identification of regulatory elements in the genome.
During the past 10 years, chromatin immunoprecipitation followed by sequencing (ChIP-seq) has become the method of choice for genome-wide profiling of TFs and hPTMs (Park 2009; Welboren et al. 2009; Collas 2010; Furey 2012). The ChIP-seq protocol relies on the affinity purification of a DNA-binding protein by the use of antibodies. Characterization of the DNA associated with the protein of interest by high-throughput sequencing allows for identification of the protein binding sites at a genome-wide scale. However, the ChIP-seq workflow requires large amounts of material, is labor intensive, and lacks robustness owing to experimental variation (Ho et al. 2011; Chen et al. 2012; Landt et al. 2012). These drawbacks make the application of ChIP-seq challenging, in particular in settings in which material is limited (Dirks et al. 2016).
To facilitate ChIP-seq profiling of low-input samples, a range of strategies have been developed (O'Neill et al. 2006; Dahl and Collas 2007, 2008a,b; Adli and Bernstein 2011; Brind'Amour et al. 2015; Rotem et al. 2015; Schmidl et al. 2015; Dahl et al. 2016; van Galen et al. 2016; Weiner et al. 2016; Zhang et al. 2016; Skene et al. 2018; Ai et al. 2019; Grosselin et al. 2019; Hainer et al. 2019; Kaya-Okur et al. 2019; Ku et al. 2019). Methods that have been applied include barcoding and pooling of multiple samples in the ChIP reaction (Rotem et al. 2015; van Galen et al. 2016; Weiner et al. 2016), small volume sonication (Adli and Bernstein 2011), substitution of sonication by a native MNase digestion approach (Brind'Amour et al. 2015), the use of carrier material (mainly used for ChIP-qPCR) (O'Neill et al. 2006), and the application of a transposase for DNA cleavage and library generation (Schmidl et al. 2015; Ai et al. 2019; Kaya-Okur et al. 2019). Each of the various ChIP-seq methodologies yield incremental benefits but suffer from (a combination of) low read complexity, a lack of robustness, suboptimal throughput, and lengthy and/or laborious protocols. On the other hand, semiautomated workflows have been developed to increase reproducibility of ChIP-seq and reduce the workload of the laborious protocol (Aldridge et al. 2013; Berguet et al. 2014; Gasper et al. 2014; Wallerman et al. 2015), but these generally require high quantities of input material. Recent studies have shown the feasibility of combining low-input samples with automated workflows using microfluidic devices (Cao et al. 2015; Shen et al. 2015; Murphy et al. 2018). However, these prototypic platforms require dedicated, custom-made sophisticated laboratory equipment; have low throughput owing to the limited number of samples that can be run in parallel (one sample [Cao et al. 2015], four samples [Murphy et al. 2018], and four samples [Shen et al. 2015] in parallel, respectively); and are mainly focused on a few or a single histone modification (H3K4me3). Therefore, despite showing proof of principle, further maturation of these platforms in terms of throughput, flexibility, and standardization of the microfluidic platform is required to allow integration in workflows of major epigenetic profiling endeavors, such as the International Human Epigenome Consortium (IHEC), but also to allow implementation of these platforms in nonexpert laboratories (Bujold et al. 2016; Fernández et al. 2016; Stunnenberg et al. 2016). Similarly, throughput and standardization of ChIP-seq are key for implementation in clinical applications of epigenetic biomarkers.
Here, we aimed to develop a fully automated, integrated, and standardized plug and play (PnP) ChIP-seq microfluidic platform for low-input samples that can easily be implemented, which we call PnP-ChIP-seq. We set the following requirements to be fulfilled by PnP-ChIP-seq: (1) generation of high-quality data to enable genome-wide ChIP profiling of low-input biological samples; (2) a standardized ChIP-seq procedure that is robust and easy to implement in laboratories and facilities; (3) compatibility with ChIP-seq profiling of the hPTMs H3K4me3, H3K36me3, H3K27ac, and H3K4me1, associated with gene activity, and H3K9me3 and H3K27me3, associated with repression of genes (together, this meets the IHEC requirements for generating reference epigenomes from biological samples) (Bujold et al. 2016; Fernández et al. 2016; Stunnenberg et al. 2016); (4) allowing high throughput by processing a large number of samples in parallel; and (5) the ability to perform the complete ChIP-seq workflow within a day.
Results
Automated part of ChIP-seq in microfluidic ChIP workflows
In recent years, a large range of low-input ChIP technologies have been pioneered (Supplemental Fig. S1). Although elegant, these approaches are generally very laborious and prone to multiple sources of noise owing to the large number of handling steps. To increase precision, some low-input workflows have been automated (Cao et al. 2015; Shen et al. 2015; Murphy et al. 2018), but these studies use custom-made devices requiring extensive microfluidics and/or ChIP-seq expertise (Supplemental Fig. S1). Therefore, we set out to develop an automated miniaturized low-input ChIP-seq workflow that can conveniently be adopted by users world-wide. As such, we developed our workflow on a widely available commercial controller for pneumatics and thermocycling, the Fluidigm C1 Controller. We designed disposable polydimethyl siloxane (PDMS)–based integrated fluidic circuit (IFC) devices (Fig. 1), on which 24 parallel ChIPs are performed fully automated after loading into the controller. By facilitating the construction of bead columns, these newly designed PDMS-based IFC devices for microfluidic ChIP are very different from the PDMS devices that have been developed for single-cell captures and single-cell RNA-seq workflows (Frederickson 2002; Durruthy-Durruthy and Ray 2018).
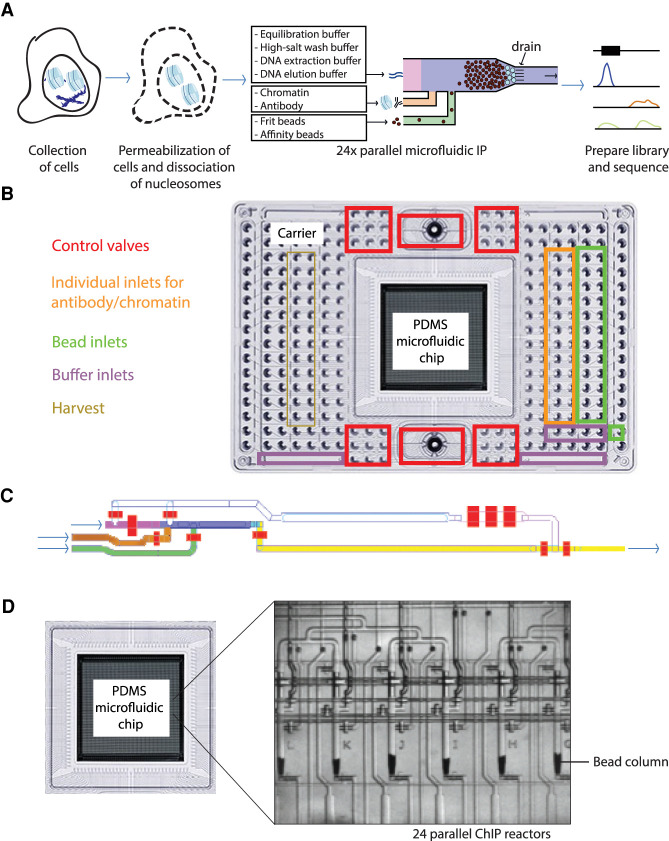
Figure 1.
Overview of the microfluidic chip design for automated microfluidic chromatin immunoprecipitation. (A) Workflow of automated microfluidic ChIP-seq. (B) Overview of the interface plate. At the sides are the inlets, whereas the PDMS microfluidic chip containing the microreactors is located in the center. (C) Architecture of PDMS microfluidic chip, also referred to as integrated fluidic circuit (IFC). The bead inlet is in green; the antibody and chromatin inlets, orange; the channel in which the bead column is constructed, blue; the inlet for various buffers needed in the workflow, pink; and the waste and harvest outlet, yellow. The control valves are colored red. (D) Phase contrast image of six out of 24 parallel microfluidic bead columns on every chip.
The conventional ChIP workflow (Supplemental Fig. S2) starts with the collection of chromatin from cells, after which the chromatin is sheared either by enzymatic digestion (e.g., by the use of MNase) or by ultrasonication. In the case of ultrasonication, the chromatin is usually cross-linked before harvesting to stabilize protein–protein and protein–DNA interactions. Next, the isolated chromatin fragments are probed for proteins of interest by antibodies. The antibodies and associated chromatin fragments are captured, for example, by a mix of Protein A and Protein G antibody binding beads (Prot A/G beads). After washings to remove nonspecific fragments from the scaffold, the DNA fragments are eluted and sequenced to determine the binding sites of the protein of interest at a genome-wide scale. For the microfluidic workflow, we set out to automate the labor-intensive process of (1) coupling the antibody to the beads, (2) binding of the chromatin to the antibodies, (3) washing of the antibody–protein complexes that are bound to the beads to remove nonbound background, and (4) performing elution of the DNA (Fig. 1A; Supplemental Fig. S2). We designed the workflow such that the DNA that is harvested after our ChIP workflow (3 µL in the standard protocol) can be directly used as input for DNA library construction required for sequencing, without the need to perform DNA purification.
Microfluidic disposable plates used for miniaturized ChIP-seq
For the development of the IFC devices used as hardware for our workflow, we designed PDMS valve-operated fluidic circuits produced using multilayer soft lithography (Fig. 1B; Unger et al. 2000). The PDMS chip is mounted to a plastic carrier that forms the pneumatic and thermodynamic operation interface with the controller (the chip together with the plastic carrier will be called plate from here on) and contains 25-µL-volume inlets and four larger reservoirs for reagent loading. The samples, beads, and control line fluids, as well as wash, harvesting, and elution buffers, can be conveniently loaded in the appropriate wells of the plate (Fig. 1B; Supplemental Fig. S3A). Each IFC plate consists of 24 nanoliter-scale reactors that facilitate the parallelization of experiments, whereas in each reactor, a single ChIP experiment is performed (Fig. 1C; Supplemental Fig. S3B). The 24 reactors each have individual inputs for (1) antibody-binding beads and (2) chromatin and antibody, the main reagents used for an immunoprecipitation reaction. Each common wash and elution reagent is prefilled into a single inlet of the microfluidic plate, which serves all 24 reactors (Supplemental Fig. S3A). The plate facilitates loading of up to four of such buffers. To allow maximum flexibility, all control valves can be individually pressurized (Fig. 1C; red valves).
At the start of the procedure, all reagents and the dissociated chromatin suspension are loaded into the inlets (Supplemental Fig. S3A), after which the entire circuitry is loaded onto the controller to start the ChIP protocol. All reagents are dead-end filled at the start of a microfluidic run in order to remove any air bubbles present in the system while operating. We constructed the procedure such that a tight column of micron-sized monodisperse antibody-binding beads is packed (Fig. 1A; loading through the green inlet and blue column), on which the immunoprecipitation is performed (Fig. 1D).
This column is built upon a frit layer of inert beads, which are larger in size compared with the beads used for the column (Fig. 1A; frit layer schematically represented in cyan at the bottom of the column) and function to prevent leaking of relatively small 2.8-µm-diameter beads through the 5-µm-spaced drain at the bottom. The use of 30% glycerol solution as a carrier keeps the beads in suspension during the process of building the separation columns. After packing the beads, the column is washed using an equilibration buffer to remove any remaining glycerol (Fig. 1C, flowing through the pink channel). The chromatin sample, up to 8 µL in volume, is flushed across the antibody binding column (Fig. 1C, flowing through the orange channel). The antibodies used can be loaded together with either the beads or the chromatin. After binding of the specific chromatin fragments to the antibodies on the bead column, the column is washed using an equilibration buffer followed by a high-salt wash buffer (Fig. 1C, flowing through the pink channel). The specific DNA fragments associated with the protein of interest are eluted using a DNA extraction buffer incubated for 20 min at 55°C followed by 1 h at 65°C (which de-cross-links when using fixed chromatin and degrades the Proteinase K) (Fig. 1C, flowing through the pink channel). DNA elution buffer is used to push the eluted DNA fragments to the outlet wells to a final volume of 3 µL. This DNA can directly be used for further processing (no clean-up step is needed) because the DNA extraction buffer containing the DNA fragments (∼10 nL) is highly diluted by the DNA elution buffer. During elution, the resulting DNA fragments are collected into individual outlets (Fig. 1C, via the yellow channel) and can be used for qPCR or sequencing.
Optimization of microfluidic ChIP-qPCR
For optimization of the microfluidic ChIP procedure, we tested a range of variables using ChIP-qPCR on H3K4me3 in mouse embryonic stem cells (mESCs), using a well-known positive locus of a highly active gene (Actb) and a negative locus in a gene desert for background control as read-out. H3K4me3 is mainly present at promoters of active genes (Barski et al. 2007). For testing, the main variables included (1) the composition of the frit layer, (2) the size of the column used for immunoprecipitation, (3) the type of beads, and (4) the pressure used to load the sample on the columns (Fig. 2A–D). For these tests, we used a small quantity of bulk-isolated chromatin to load on each bead column: an amount of chromatin equivalent to 3000 mESCs. We aimed at using small beads as it allows for a large surface area in the columns to capture chromatin fragments. As such, the small columns have an more than 100-fold excess of H3K4me3 binding sites relative to the amount of H3K4me3 present in 3000 mESCs, whereas for the large columns, the H3K4me3 binding sites are more than 1000-fold in excess. The results showed that a frit layer composed of a mixture of 4.5-µm and 6-µm inert beads (Fig. 2A), combined with large columns (Fig. 2B; Supplemental Fig. S4A) composed of an equal mix of 2.8-µm ProtA and ProtG beads (Fig. 2C), resulted in optimal recoveries. The pressure used to load the samples was less critical (Fig. 2D). Altogether, these tests resulted in a significant improvement of ChIP-qPCR recoveries compared with an initial, default workflow that we applied (Fig. 2E). The hands-on time for the optimized microfluidic protocol is very limited, in total ∼30 min (Supplemental Fig. S5A): Preparation including pipetting of the plate takes at maximum 20 min, whereas harvesting of the 24 DNA samples of the ChIP takes another 10 min of hands-on time. The hands-free parallelized immunoprecipitation process that is performed on the bead columns takes ∼ 4.5 h (Supplemental Fig. S5).
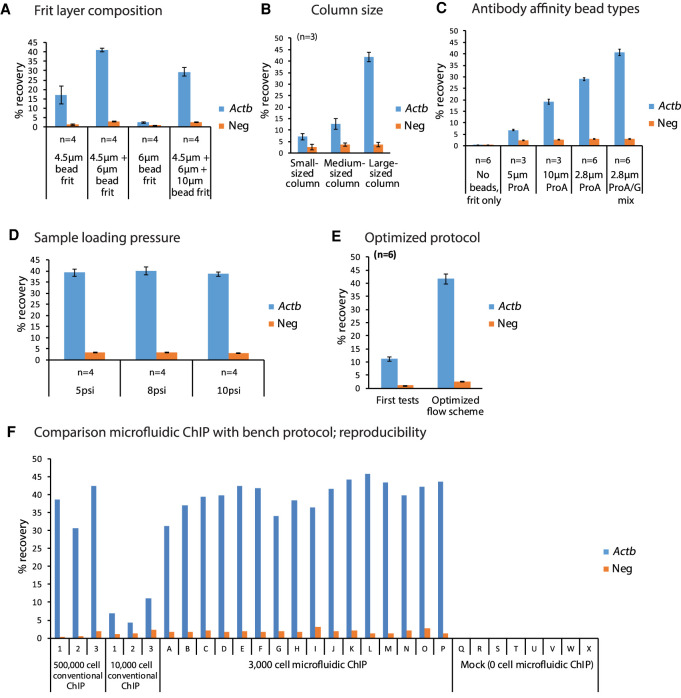
Figure 2.
Optimization of parameters for the automated microfluidic ChIP protocol. ChIP-qPCR are depicted on a positive (Actb) and a negative locus, with H3K4me3 recoveries plotted with ±SEM. (A) Recoveries using various types of frit layer composition that allow packing of antibody binding beads in the microfluidic reactors. (B) Recoveries using various column sizes (Supplemental Fig. S4A) built using different amounts of antibody binding beads. (C) Recoveries of various types of beads used to construct the antibody binding column. (D) Recoveries using varying chromatin loading pressures. (E) Final recoveries with optimized parameters compared with initial testing. (F) Recoveries of conventional versus low-input automated microfluidic ChIP-qPCR, showing high yields and reproducibility of microfluidic ChIP compared with conventional chip.
Microfluidic ChIP is sensitive and robust
We next evaluated the performance of the optimized workflow over the 24 individual reactors of a microfluidic chip by routinely constructing 24 separate, parallel antibody affinity bead columns on each microfluidic chip (Supplemental Fig. S4B). For ChIP-qPCR, the results obtained for individual columns of a single microfluidic chip were highly consistent, with 40.10% ± 0.15% recovery of H3K4me3 over the Actb promoter (Fig. 2F, columns named A–P). This shows that using the optimized workflow, all individual columns are above a critical size, resulting in optimal recoveries. The mock controls, in which no chromatin was present, did not show any recovery for either the positive or negative locus, indicating there was no cross-contamination during our procedures or on the PDMS chip (Fig. 2F, columns named Q–X). Furthermore, we observed very high consistency between ChIP-qPCRs performed on separate microfluidic plates (Supplemental Fig. S4C) run on different days.
To evaluate the results of the optimized microfluidic ChIP procedure, we compared our results to conventional ChIP-qPCRs using the equivalent of 500,000 or 10,000 mESCs from a bulk mESC sonicated sample as input. In line with the fact that lower input quantities affect the efficiency of ChIP (Kidder et al. 2011; Hainer et al. 2019; Ku et al. 2019), we observed a fivefold reduction in recovery in conventional bench ChIP-qPCRs performed using 10,000 mESCs compared with the ChIP-qPCRs performed using 500,000 mESCs (Fig. 2F, left part labeled 1–3). The relative recoveries obtained using the microfluidic ChIP-qPCR procedure using 3000 mESCs (40.10% ± 0.15%) were much higher compared with conventional ChIP-qPCRs using chromatin of 10,000 mESCs (7.44% ± 0.60%) and also slightly higher than the recoveries obtained for conventional ChIP-qPCRs using chromatin of 500,000 mESCs (37.22% ± 0.46%) (Fig. 2F). Along with greatly enhanced Actb recoveries for the optimized and automated low-cell chromatin immunoprecipitations (ChIPs), we consistently observed a slightly increased recovery for the negative control in the microfluidic ChIP-qPCR compared with conventional ChIP-qPCR (Fig. 2F). This might be caused by the fact that the antibody beads are present in a column in our workflow, which does not allow us to resuspend the beads during washing as in regular ChIP. Altogether, this shows that the miniaturized platform is superior over conventional low-input bench ChIP protocols and that the microfluidic platform efficiently generates highly reproducible ChIPs on very small quantities of cells.
Microfluidic PnP-ChIP-seq for epigenetic profiles of histone modifications
Next, we used our optimized automated ChIP workflow for ChIP-seq, a procedure that we named PnP-ChIP-seq (Supplemental Methods). To optimize the PnP-ChIP-seq, we used cross-linked and sonicated chromatin obtained from “bulk” (multimillion) mESC chromatin preparations. We loaded the chromatin equivalent of 3000, 1000, and 500 mESCs on the microfluidic platform to generate H3K4me3 ChIP-seq profiles, with replicate experiments performed on separate microfluidic chips to probe for consistency between runs. Visual inspection shows a high overlap of enriched sites of the low-input PnP-ChIP-seq profiles compared with the bulk reference track (Fig. 3A; Supplemental Fig. S6A), albeit at lower signal intensities. We next performed peak calling and plotted the ChIP-seq signals over the merged peak set. These plots further confirmed the reduction in H3K4me3 signal intensities when using a lower number of cells as input, as reflected in the heatmaps (Fig. 3B) and average plots (Supplemental Fig. S6B). However, the Pearson's correlation of the intensities of the joint peaks between all different H3K4me3 profiles was very high (r > 0.87), both between bulk ChIP-seq and PnP-ChIP-seq as well as between profiles generated by PnP-ChIP-seq using different input quantities (Fig. 3C), confirming the high quality of profiles generated using the microfluidic platform. De novo peak calls on the 3000-cell microfluidic ChIP showed that we detected 85% of the bulk reference peak set (Fig. 3D), with hardly any peaks being detected outside the bulk reference peak set, whereas the profiles generated using the chromatin equivalent of 3000 or 1000 mESCs show a high overlap (Fig. 3E). Furthermore, the ChIP-seq profiles generated using the microfluidic platform are highly reproducible, as shown by the Pearson's correlations (Fig. 3C) and by the peak overlap of the replicate H3K4me3 profiles using the chromatin equivalent of 3000 mESCs (Fig. 3F).
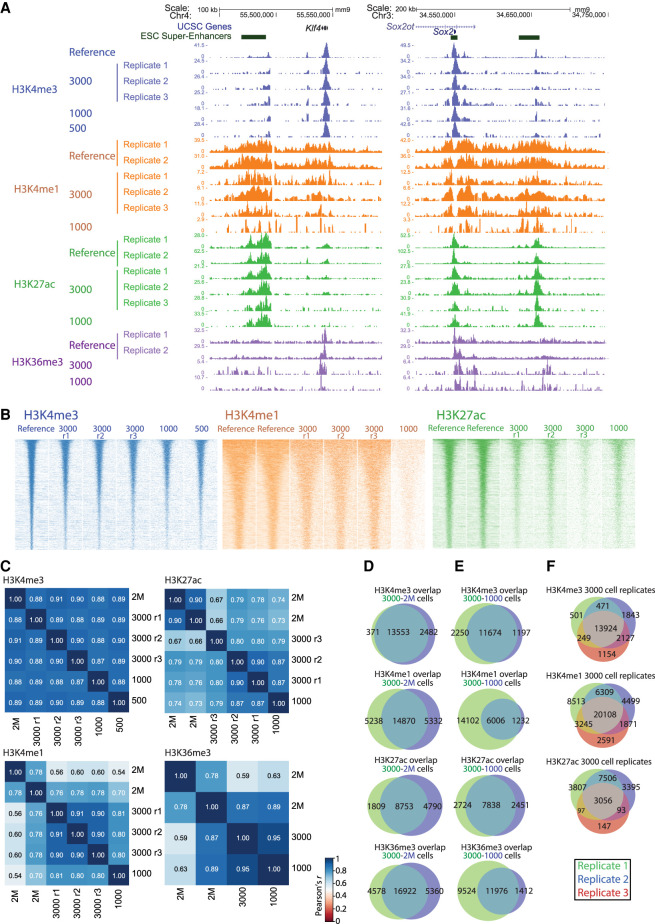
Figure 3.
PnP-ChIP-seq using small quantites of bulk-sonicated cross-linked chromatin. (A) Gene-centered genome browser view for PnP-ChIP-seq of H3K4me3, H3K4me1, H3K27ac, and H3K36me3. (B) Heatmap of merged peak set for various starting amount of sonicated chromatin for PnP-ChIP-seq of H3K4me3, H3K4me1, H3K27ac, and H3K36me3. (r) Replicate. (C) Cross-correlations of PnP-ChIP-seq using tag counts of merged peak set. (D) Overlap between de novo peak calls of PnP-ChIP-seq and bulk ChIP-seq. (E,F) Overlap between de novo peak calls of PnP-ChIP-seq.
In addition to H3K4me3, we set out to use our platform for profiling of the additional hPTMs H3K4me1 and H3K27ac, which together allow to define active and poised enhancers (Creyghton et al. 2010), and H3K36me3, which covers the gene body of active genes (Barski et al. 2007). For the three additional hPTMs, we used the chromatin equivalent of 3000 and 1000 mESCs for PnP-ChIP-seq. Visual inspection of the profiles generated confirmed the anticipated location of enhancers and active gene bodies, respectively, and also showed the similarity between the bulk reference track and the PnP-ChIP-seq tracks (Fig. 3A; Supplemental Fig. S6A). Similar to H3K4me3, the H3K4me1 and H3K27ac profiles show a reduction in signal associated with the number of cells used as input for the PnP-ChIP-seq (Fig. 3B). Further analysis using correlograms showed the PnP-ChIP-seq tracks of H3K4me1, H3K27ac, and H3K36me3 were well in concordance with ChIP-seq tracks using bulk material, albeit the Pearson's correlations were somewhat lower compared with the profiles generated for H3K4me3 (Fig. 3C). The majority of peaks called for H3K4me1, H3K27ac, and H3K36me3 were also present in the bulk reference set, with between 65% and 76% of the bulk peaks being called (Fig. 3D). The use of 1000 mESCs resulted in a clear drop in signals: Although for H3K27ac we were still able to call most of the peaks as present in the bulk reference set, this number dropped to 30% and 56% for H3K36me3 and H3K4me1, respectively (Fig. 3E). Altogether, these analyses show the compatibility of our microfluidic platform to comprehensively profile the main epigenetic hPTM marks associated with gene activity using very low sample quantities of 3000 mESCs, whereas the use of even lower numbers of mESCs generally results in a reduction in sensitivity.
PnP-ChIP-seq is compatible with low-abundance populations of cells
Having established the sensitivity of our platform on small quantities of chromatin prepared from bulk collections, we set out to make the microfluidic platform compatible with ChIP-seq profiling of low-abundance populations of cells that are not easily collected in large amounts. The preparation of chromatin from a low number of cells is challenging, in particular when using sonication for chromatin shearing. We extensively tested sonication on low quantities of cells, but this resulted in a gradual loss of ChIP-seq signal when reducing the amount of input chromatin used for shearing (Supplemental Fig. S7A,B). Therefore, we switched to low-input MNase digestion for shearing of native (non-cross-linked) chromatin. We took a fixed number of 7500 or 15,000 mESCs for MNase digestions and subsequently used the chromatin equivalent of 3000, 1000, 500, and 100 mESCs for H3K4me3 ChIP-seq as input for the microfluidic platform (Fig. 4A; Supplemental Fig. S8A). Visual inspection showed the H3K4me3 PnP-ChIP-seq profiles were very similar to the bulk reference profiles generated by conventional ChIP-seq (using 2 million mESCs), independent of the number of mESCs loaded on the platform. Peak calling on the individual profiles showed a high overlap of peak calls between the bulk reference set and the H3K4me3 PnP-ChIP-seq profiles generated using the 3000 mESC chromatin equivalent (Fig. 4B). The use of a smaller number of mESCs resulted in a concentration-dependent decrease of H3K4me3 peaks and signals (Fig. 4B,C; Supplemental Fig. S8B), which is known for low-input ChIP-seq (Kidder et al. 2011; Hainer and Fazzio 2019; Ku et al. 2019). However, even with as few as 100 mESC chromatin equivalents, we were still able to call 53% of the peaks as present in bulk H3K4me3 ChIP-seq (Fig. 4B). The results between the different starting amounts, 7500 or 15,000 mESCs, were similar (Fig. 4A). Even with these low starting amounts, the H3K4me3 PnP-ChIP-seq profiles generated using the MNase-based protocol showed higher signal-to-noise ratios compared with the H3K4me3 PnP-ChIP-seq profiles generated using chromatin that was sonicated in bulk (Supplemental Fig. S8C). Quantification of the merged H3K4me3 peak set of the MNase-based profiles showed a very high correlation (Fig. 4D), with cross-correlations between low-cell-input experiments and the bulk reference of r > 0.82 (Pearson's correlation) and high consistency between technical replicates separated before MNase treatment (r > 0.88). Also, the majority of peaks were consistently detected in all technical replicates irrespective of the number of mESCs that was used as input for the PnP-ChIP-seq (Supplemental Fig. S8D), further underlining the high quality of the ChIP-seq profiles generated by the microfluidic platform. To further evaluate the performance of the PnP-ChIP-seq, we performed comparative analysis to other low-input ChIP-seq technologies that have been developed and have included H3K4me3 ChIP-seq on mESCs in their studies, in particular ChIP-seq profiles of 1000 mESCs on a previously developed prototype microfluidic platform (Shen et al. 2015) and low-input native ChIP-seq profiles generated using the ULI-NChIP-seq bench protocol (Brind'Amour et al. 2015). Our microfluidic ChIP-seq generated more peaks than ULI-NChIP-seq and a comparable number of peaks as the prototypic microfluids platform (Supplemental Fig. S9A). However, in terms of signal-to-noise ratio (Supplemental Fig. S9B) and similarity to the bulk reference (Supplemental Fig. S9C), our PnP-ChIP-seq seemingly performs better than both other methods that were previously developed.
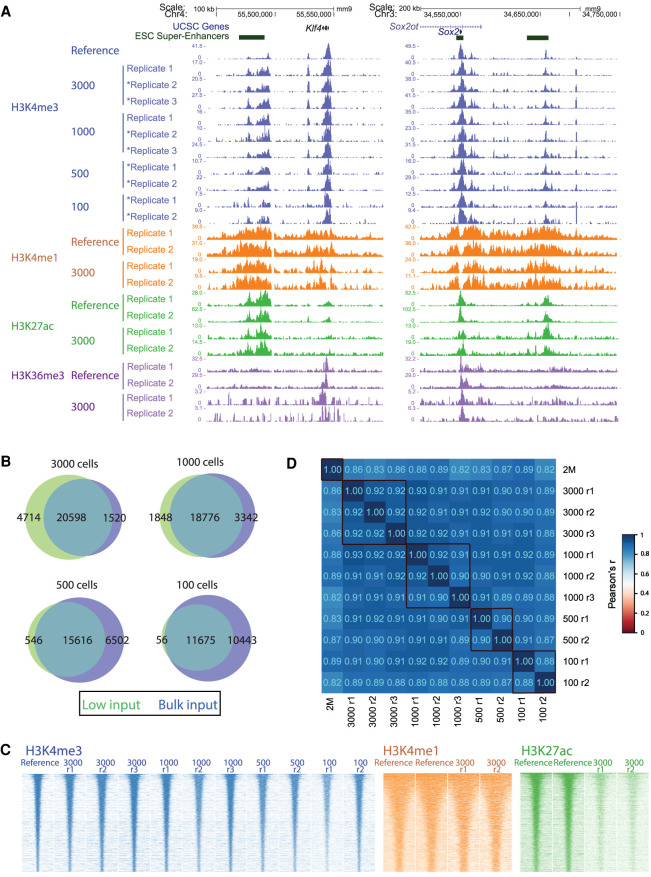
Figure 4.
PnP-ChIP-seq using small cell quantities of mESCs by the use of MNase. (A) Gene-centered genome browser view for PnP-ChIP-seq of H3K4me3, H3K4me1, H3K27ac, and H3K36me3. Profiles labeled with an asterisk were generated from a starting amount of 7500 mESCs; otherwise, 15,000 mESCs were used. (B) Overlap between de novo peak calls of PnP-ChIP-seq and bulk ChIP-seq. (C) Heatmap of merged peak set for various starting amount of sonicated chromatin for PnP-ChIP-seq of H3K4me3, H3K4me1, H3K27ac, and H3K36me3. (D) Cross-correlations of H3K4me3 PnP-ChIP-seq using tag counts of merged peak set.
PnP-ChIP-seq is compatible with profiling of all six reference epigenomes
In view of the high sensitivity of the MNase-based native PnP-ChIP-seq H3K4me3 profiles, we included H3K4me1, H3K27ac, and H3K36me3 for further profiling of the mESCs, using the same protocol with a chromatin equivalent of 3000 mESCs (obtained from a sample of 15,000 mESCs). Visual inspection of the profiles generated using the microfluidic platform confirmed the anticipated location and also showed the similarity between the bulk reference track and the PnP-ChIP-seq profiles (Fig. 4A; Supplemental Fig. S8A). Although the similarity to bulk ChIP-seq for these hPTMs was somewhat lower compared with H3K4me3, the Pearson's correlation of r > 0.58 (Supplemental Fig. S10A), the heatmap over the peaks (Fig. 4C), and the overlap of peaks compared with bulk ChIP-seq (Supplemental Fig. S10B) showed that the PnP-ChIP-seq profiles were of very good quality. Despite the signal intensities of H3K27ac of the 3000 mESCs being reduced compared with the bulk (Fig. 4C), peak calling identified around half of the H3K27ac enriched sites (Supplemental Fig. S10B). Also, the 3000 mESC profiles of H3K4me1, H3K27ac, and H3K36me3 showed high reproducibility (Fig. 4C,D; Supplemental Fig. S10C).
To explore the compatibility of our platform with hPTMs associated with gene silencing, we performed PnP-ChIP-seq for H3K27me3 and H3K9me3. H3K27me3 is typically present at silent genes, in particular for developmental genes in mESCs (Boyer et al. 2006). Accordingly, we observed strong enrichment of H3K27me3 by PnP-ChIP-seq for the developmental regulators Gata6 and Lhx1 and over the Hoxb cluster (Supplemental Fig. S11A), both for the sonication-based and for the MNase based workflows that we developed. The average profile over the merged H3K27me3 peak set as detected in mESCs shows a clear enrichment for the PnP-ChIP-seq profiles, similar to H3K27me3 generated by the low-input STAR ChIP-seq strategy (Supplemental Fig. S11B; Zhang et al. 2016). In mESCs, H3K9me3 is mainly present over major satellite repeats, both at pericentric heterochromatin and intergenic, but also over various other type of repetitive regions such as LINEs/ERVs (Martens et al. 2005; Bulut-Karslioglu et al. 2014). By using PnP-ChIP-seq, we observed a H3K9me3 pattern similar to H3K9me3 mESC bulk ChIP-seq profiling (Supplemental Fig. S11C). As expected, we observed a very high enrichment of major satellite sequences in the PnP-ChIP-seq, whereas it was depleted over repeats such as SINEB (Alu), which is located in gene-rich regions (Deininger 2011) and associated with hPTMs associated with gene activity (Supplemental Fig. S11D). In conclusion, these experiments show that by the use of very small cell quantities, we were able to perform comprehensive epigenetic profiling of all six hPTMs required by IHEC for generating reference epigenomes from biological samples (H3K4me3, H3K36me3, H3K27ac, H3K4me1, H3K9me3, and H3K27me3) using PnP-ChIP-seq in an automated fashion.
PnP-ChIP-seq robustly detects epigenetic differences between two types of mESCs
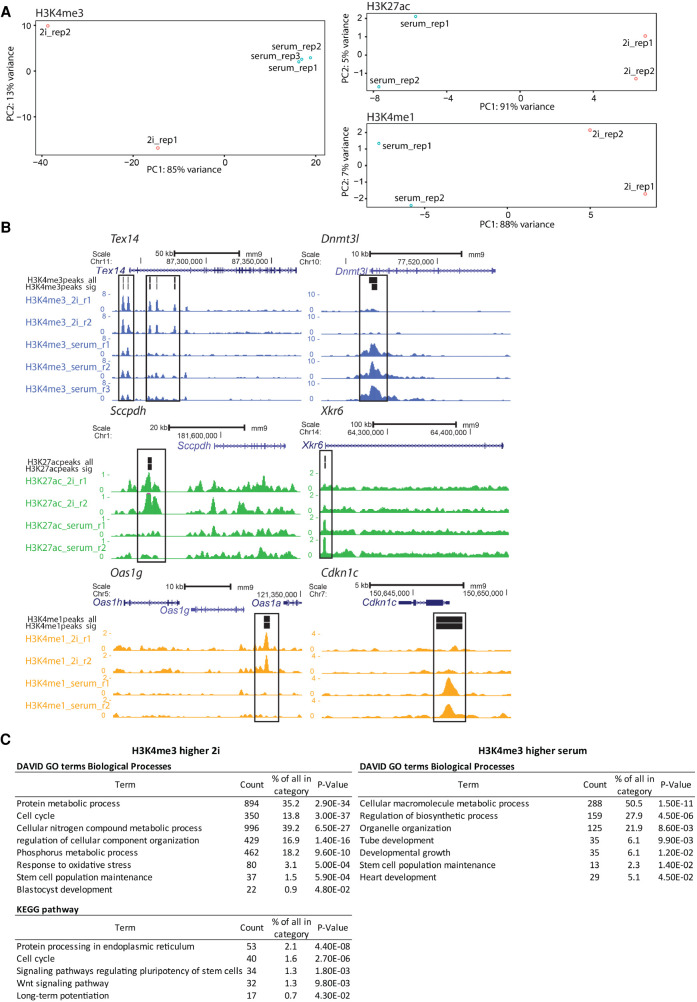
Having shown the compatibility of MNase PnP-Chip-Seq for multiple hPTMs, we next asked whether the workflow we developed would have enough sensitivity to detect differences between two cell populations. Therefore, we performed low-input MNase PnP-ChIP-seq for H3K4me3, H3K27ac, and H3K4me1 on two types of mESCs: ground-state pluripotent ES cells grown under serum-free conditions using two kinase inhibitors (“2i”) (Ying et al. 2008) and metastable serum-grown ES cells (“serum”), which contain features of primed pluripotency (Habibi et al. 2013). Despite the fact that these two cell types are relatively similar (Marks et al. 2012), PnP-ChIP-seq profiling readily picked up differences between these cell types. Using principal component analysis (PCA) on each of the three hPTMs that we profiled, the 2i mESC replicas clearly separated from the serum mESC replicas along the main principal component 1 (PC1), explaining 85%–91% of the variation between the samples (Fig. 5A). For H3K4me3, we detected in total 25,617 H3K4me3 peaks, of which 3459 peaks were significantly higher in either 2i mESCs or serum mESCs (Supplemental Fig. S12; Supplemental Table S1). These included differential peaks that are associated with well-known genes that are significantly higher expressed in 2i mESCs (Tex14 and Ubc) or serum mESCs (Lin28b, Dnmt3l, and Cdk12) (Fig. 5B; Supplemental Fig. S13). Similarly, we were able to call significant differences between 2i and serum mESCs for H3K27ac and H3K4me1 (Fig. 5B; Supplemental Figs. S12, S14, S15; Supplemental Table S1). Functional annotation clustering of differential H3K4me3 by Gene Ontology (GO) and pathway analysis (Fig. 5C) revealed that genes associated with increased H3K4me3 in 2i are significantly enriched for terms associated with metabolic processes, cell cycle, early embryonic development, and Wnt signaling. Genes associated with increased H3K4me3 in serum are significantly linked to the GO terms related to metabolic processes and postimplantation germ layer specification. As similar terms were identified using differential gene expression between 2i and serum mESCs (Marks et al. 2012; Marks and Stunnenberg 2014), changes in H3K4me3 between 2i and serum mESCs are very well reflected in the transcription in these mESCs. Altogether, these experiments show that PnP-ChIP-seq robustly picks up relevant significant epigenetic differences between two closely related cell types.
Figure 5.
PnP-ChIP-seq allows detection of significant differences between 2i and serum mESCs. (A) PCA on peaks of H3K4me3, H3K27ac, and H3K4me1 PnP-ChIP-seq in 2i and serum mESCs. (rep) Replicate. (B) Genome browser views for PnP-ChIP-seq of loci showing a significant difference in H3K4me3, H3K27ac, or H3K4me1 between 2i and serum mESCs (boxed). (H3*peaks_all) All peaks of a hPTM detected in 2i and serum mESCs; (H3*peaks_sig) all peaks of a hPTM that are significantly increased in either 2i or serum mESCs. (C) GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of genes associated with differential H3K4me3 between 2i and serum mESCs.
The H3K4me3 landscape of two-cell-stage-like mESCs is similar to regular mESCs
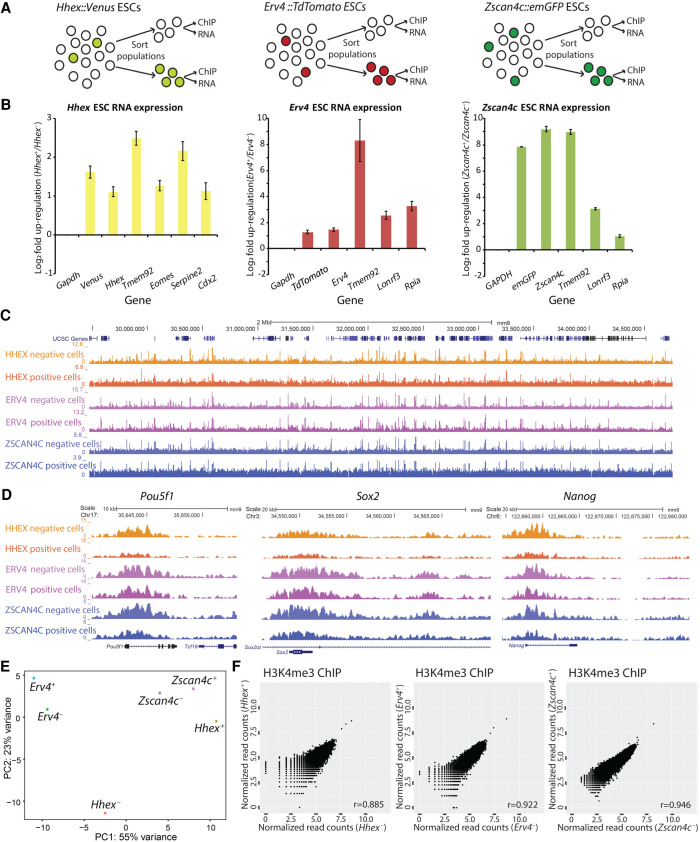
mESC cultures are heterogeneous (Kolodziejczyk et al. 2015), and gene expression analysis showed that a small number of mESCs within the total mESC population represent the two-cell stage of embryonic development (“two cell-stage like” [2C-like] cells) (Morgani and Brickman 2014; Fu et al. 2020). Previous research suggested that enhancers (as profiled by H3K27ac and H3K4me1 ChIP-seq) are similar between 2C-like cells and regular mESCs, whereas differences in the localization of H3K27me3 are minimal (Hayashi et al. 2016; Zhang et al. 2019). In line, we did not find significant differences between 2C-like cells and regular mESCs in the H3K27ac, H3K4me1, and H3K27me3 ChIP-seq profiles generated in these studies (Supplemental Fig. S17A). This might indicate that the 2C-like cells occur in the mESC cultures by stochastic changes in gene expression rather than by epigenetic changes. To discriminate between these two scenarios, we used PnP-ChIP-seq to profile H3K4me3, an epigenetic mark that has a very high correlation with gene expression (Barski et al. 2007). We performed fluorescent-activated cell sorting (FACS) for distinct low-abundant subpopulations of mESCs, as previously reported, based on promoter activity of Hhex (Morgani et al. 2013; Morgani and Brickman 2014), Zscan4c (Falco et al. 2007; Macfarlan et al. 2012; Eckersley-Maslin et al. 2016; Ishiguro et al. 2017), and Erv4 (also known as MuERV-L) (Macfarlan et al. 2012; Eckersley-Maslin et al. 2016). We made use of fluorescent reporters in three different mESC lines: (1) a Venus-positive subpopulation of mESCs sorted using a Hhex::Venus reporter, which has been shown to be totipotent-like (Morgani et al. 2013; Morgani and Brickman 2014); (2) an Emerald(Em)-GFP-positive subpopulation of mESCs sorted using a Zscan4c::Emerald-GFP reporter, which has been reported to be 2C-like cells (Falco et al. 2007; Macfarlan et al. 2012; Eckersley-Maslin et al. 2016; Ishiguro et al. 2017); and (3) a TdTomato-positive population of mESCs sorted using a Erv4::TdTomato reporter, which is a subselection of the Zscan4c-positive mESC population (Fig. 6A; Macfarlan et al. 2012; Eckersley-Maslin et al. 2016). The FACS profiles showed that we were able to collect discrete subpopulations of mESCs based on their fluorescent markers (Supplemental Fig. S16). We validated the sorting by comparing expression of the marker-positive populations versus expression of the marker-negative populations using RT-qPCR. We detected increased RNA expression of the sorted subpopulation marker as well as the corresponding fluorescent transcript and multiple other specific markers for the subpopulations as reported in the original studies (Fig. 6B; Morgani et al. 2013; Eckersley-Maslin et al. 2016), confirming that we obtained the anticipated subpopulations of 2C-like cells. Next, we used PnP-ChIP-seq to profile H3K4me3 for the mESCs populations showing Hhex, Zscan4c, and Erv4 promoter activity by means of positive marker expression, as well as for the populations of mESCs that were negative for the markers (Fig. 6C,D). Visual inspection of the H3K4me3 profiles showed that the Venus-, Emerald-GFP-, and TdTomato-positive mESCs were similar to their negative counterparts (Fig. 6C), including the H3K4me3 signals at the promoters of the core pluripotency factors Nanog, Pou5f1, and Sox2 (Fig. 6D). Next, we quantified genome-wide enrichment of H3K4me3 at 1-kb regions around all promoters. Subsequent PCA showed that none of the main PCs consistently separated the 2C-like cells from the remainder of the mESCs (Fig. 6E). We observed a very high correlation in H3K4me3 intensities between the Venus-, Emerald-GFP-, and TdTomato-positive mESC subpopulations compared with their respective negative mESC subpopulations (Fig. 6F). Statistical analysis for differential H3K4me3 sites using DESeq2 (Love et al. 2014) showed that none of the H3K4me3 enriched loci in the three 2C-like cell populations were significantly different from the remainder of the populations (Fig. 6F), whereas also an overall comparison of three marker-negative versus three marker-positive cell populations did not yield any significant differences (FDR-adjusted P-value <0.05) (Supplemental Table S2). Specifically, when we focused on genes that are changing in 2C-like cells compared with regular mESCs (Fu et al. 2020), the group of up-regulated genes in 2C-like cells did not show a significant difference in H3K4me3, whereas the group of down-regulated genes shows a small but significant decrease in H3K4me3 in the 2C-like cells for all three mESC lines (Supplemental Fig. S17B). Altogether, this shows that the transcriptional changes associated with the 2C-like state (Falco et al. 2007; Macfarlan et al. 2012; Morgani et al. 2013; Eckersley-Maslin et al. 2016) are largely uncoupled from the H3K4me3 epigenetic landscape. This suggests that propagation of the expanded potential of mESCs in the 2C-like or totipotent-like state might occur by a stochastic increase in transcriptional activity of genes associated with these states rather than by stable epigenetic (H3K4me3-associated) alterations.
Figure 6.
Epigenomic analysis of various totipotent-like cells or “two-cell-stage-like” (2C-like) mESC subpopulations. (A) Experimental outline for sorting and analysis of 2C-like mESC subpopulations. (B) RT-qPCR on mESC subpopulations to validate successful FACS sorting. (C,D) A genome browser view depicting a broad genomic region (4Mb; C) and a zoom in D of the H3K4me3 profiles generated for the 2C-like mESC subpopulations and their controls. (E) PCA on the promoter-associated H3K4me3 signals of the various populations of cells. (+) Positive for marker; (−) negative for marker. (F) Correlation of H3K4me3 signal in promoters between Hhex, Erv4, or Zscan4c marker-positive mESCs (2C-like mESC subpopulations) and marker-negative mESCs; no differential sites were detected (FDR-adjusted P-value <0.05).
Discussion
Determining protein binding sites on DNA by means of ChIP-seq is key to our understanding of gene regulation (Jenuwein and Allis 2001; Barski et al. 2007; Berger 2007; Kouzarides 2007; Dekker 2008; Park 2009; Portela and Esteller 2010). Furthermore, it has potential for identification of epigenetic biomarkers for disease stratification and personalized medicine (Heyn and Esteller 2012; Dirks et al. 2016). To facilitate such studies, the compatibility of ChIP-seq with low-cell-quantity input is highly beneficial to enable the use of relevant biological specimens, for example, mouse early embryonic tissues or human biopsies. With respect to epigenetic biomarker discovery and screening, it is essential that the ChIP-seq protocol is sensitive, robust, and high throughput with little hands-on time. For large-scale studies and routine clinical use, it is critical to minimize variation among users and between laboratories. With the development of PnP-ChIP-seq, we achieved reproducible, robust low-input ChIP reactions for 24 samples in parallel with only 30 min of hands-on time and 4.5 h of machine-running time. This uniquely allows us to perform the full ChIP-seq procedure from harvesting of the cells up to loading of the ChIP-seq library on a sequencer in a single day. As the procedure that we pioneered is automated and standardized, PnP-ChIP-seq can conveniently be applied in nonexpert laboratories, provided that these have access to the Fluidigm C1 or a similar type of thermodynamic and pneumatic controller, for example, the Juno system. The fact that such controllers are now commonly available, including at regular core facilities world-wide, makes the PnP-ChIP-seq workflow that we developed widely accessible. As such, PnP-ChIP-seq is unique compared with previously engineered automated ChIP-seq workflows (Cao et al. 2015; Shen et al. 2015; Murphy et al. 2018), which require custom-designed and nontransferable equipment. Furthermore, PnP-ChIP-seq allows us to run more samples in parallel (24 compared with four or less in previous studies), automates a larger part of the workflow, and has lower handling and running times, while we show compatibility with all main hPTMs. Therefore, the automation and parallelization of the low-input ChIP protocol as reported here paves the way toward large-scale ChIP-seq profiling of precious sample types. The standardized procedure of PnP-ChIP-seq will facilitate consistent and reproducible results between laboratories, thereby allowing direct comparisons between ChIP-seq profiles generated in separate laboratories, which have been challenging thus far (Landt et al. 2012).
Because traditional ChIP-seq approaches require large amounts of material (Ho et al. 2011; Chen et al. 2012; Landt et al. 2012), a range of previous studies have worked toward procedures to downscale the ChIP procedure (Supplemental Fig. S1). These include barcoding and pooling of multiple samples in the ChIP reaction (Rotem et al. 2015; van Galen et al. 2016; Weiner et al. 2016; Grosselin et al. 2019), the use of carrier material (O'Neill et al. 2006), and application of a transposase for DNA cleavage and library generation (Schmidl et al. 2015; Ai et al. 2019). Furthermore, single-cell ChIP-seq approaches have been developed, such as single-cell CUT&RUN (Hainer et al. 2019; Hainer and Fazzio 2019) or scChIC-seq (Ku et al. 2019), both of which depend on antibody-fused MNase, and scCUT&Tag (Kaya-Okur et al. 2019), which depends on an antibody-based tethering of a transposase and single-cell ChIP-seq based on droplet technology (Rotem et al. 2015; Grosselin et al. 2019). Within the single-cell approaches, cells are pooled before ChIP. Although single-cell ChIP-seq approaches are very powerful to dissect cellular heterogeneity, they often require a large amount of starting material, whereas also they show very low coverage per cell. In alternative approaches, on-bead ligation of adaptors has recently been pioneered by lobChIP (Wallerman et al. 2015), SLIM-ChIP (Gutin et al. 2018), and iChIP (Lara-Astiaso et al. 2014; Sadeh et al. 2016), during which the DNA is prepared for sequencing while bound to the beads used in IP, thereby alleviating the necessity for further sample preparation after ChIP. As ChIP procedures are dependent on immunoprecipitation, all of these are, in principle, compatible with our PnP-ChIP-seq workflow. The flexibility of our platform—in terms of (1) the reagents to be loaded, (2) the flexible circulation schemes of reagents owing to the large number of independent control valves, and (3) the control over the temperature—will further facilitate automation of the alternative ChIP approaches using our PnP-ChIP-seq. In view of the better performance of our microfluidic platform compared with low-input bench ChIP (Fig. 2F) and previously developed microfluidic platforms from which comparable data were available (Supplemental Figs. S9, S11), the use of the PnP-ChIP-seq might further increase the sensitivity of these procedures.
Although previous studies that engineered miniaturization of ChIP-seq mainly focused on H3K4me3 (Cao et al. 2015; Shen et al. 2015; Murphy et al. 2018), we aimed to develop an automated ChIP-seq workflow for the complete set of six hPTMs, which together comprise the IHEC reference epigenomes (Bujold et al. 2016; Fernández et al. 2016). By using as few as 15,000 cells as input material and 3000 cells per ChIP reaction, PnP-ChIP-seq allows for profiling of these hPTMs, thereby obtaining a comprehensive epigenetic blueprint of cells. In terms of sensitivity, a comparison between 2i and serum mESCs (Fig. 5) shows that this PnP-ChIP-seq workflow robustly detects differences in promoter (H3K4me3) and enhancer (H3K27ac and H3K4me1) hPTMs in two closely related cell types. We were able to generate high-quality H3K4me3 PnP-ChIP-seq profiles by either starting with lower input amounts (7500 mESCs) (Fig. 4A) or loading lower amounts of chromatin per ChIP reaction (equivalent to 100 mESCs) (Fig. 4A). However, such low amounts were not compatible with reproducible high-quality profiles for most of the other hPTMs. Therefore, we advise the use of 15,000 cells as an optimal starting amount.
Low-input cell numbers affect sensitivity of ChIPs (Kidder et al. 2011; Hainer et al. 2019; Ku et al. 2019), which is clear in the current study from the H3K4me3 average profiles (Supplemental Fig. S8). However, the use of 3000 mESCs allowed for the detection of the majority of enriched sites for H3K4me3, H3K4me1, and H3K36me3. Although profiling of H3K27ac appeared to be more challenging, similar to previous observations (Murphy et al. 2018), the H3K27ac PnP-ChIP-seq still showed a clear correlation with bulk ChIP-seq (Supplemental Fig. S10A) and allowed for detection of around half of the total number of enriched sites (Supplemental Fig. S10B). Antibodies against hPTMs such as H3K4me3 are known for their very low dissociation constant (Kd)/high affinity (Hattori et al. 2013), and as such, the lower performance for H3K27ac is likely related to the affinity of the current antibodies against H3K27ac. However, also other features, including the availability of the H3K27ac epitope in mESCs, might cause the lower performance of PnP-ChIP-seq for H3K27ac. Nevertheless, given the high correlation between H3K27ac PnP-ChIP-seq replicates, our platform is highly compatible with H3K27ac ChIP-seq profiling, albeit at a reduced sensitivity compared with bulk ChIP-seq. Altogether, we anticipate that our platform is likely to be compatible with profiling of other hPTMs that we did not include in this study, depending on the affinity of the hPTM antibody used and the availability of the hPTM epitope but likely also on the distribution of the hPTM over the genome and total levels of the hPTM. Profiling of TFs is known to be generally more delicate than profiling of hPTMs, requiring large amounts of cells (Park 2009; Furey 2012). TFs are generally profiled using cross-linked chromatin to stably capture the binding event of TFs to chromatin or DNA. In view of the fact that we make use of MNase for shearing of the chromatin, which is not easily compatible with cross-linked chromatin, profiling of TFs by the use of PnP-ChIP-seq is likely to be challenging. However, it has been shown that ChIP-seq profiling of TFs on non-cross-linked chromatin by the use of MNase is feasible using a method called ORGANIC (Kasinathan et al. 2014). Therefore PnP-ChIP-seq may also be useful for analysis of TFs and other nonhistone proteins.
To gain mechanistic insight, we apply PnP-ChIP-seq to study totipotent-like or 2C-like cells that are present within mESC cell cultures. By comparison of H3K4me3 ChIP-seq of the 2C-like cell population versus the remainder of the pluripotent (non-2C-like) mESC population, we set out to investigate whether the 2C-like cell population arises owing to stochastic gene activation in mESCs or owing to epigenetic activation of genes by means of deposition of H3K4me3. As we found very little significant changes in H3K4me3 between 2C-like cells and the remainder of the mESC population, using either Zscan4c, Erv4, or Hhex promoter activity as a marker for 2C-like or totipotent-like cells, we tentatively conclude that the 2C-like cells likely arise in the mESC population owing to stochastic gene activation. Our findings do not exclude the possibility that the 2C-like state contains unique chromatin features other than those related to H3K4me3, for example, at the level of DNA methylation (Macfarlan et al. 2012; Morgani and Brickman 2014; Eckersley-Maslin et al. 2016). Taken together, our results provide a solid rationale for the observations that mESCs rapidly cycle in and out of the totipotent-like or 2C-like state (Macfarlan et al. 2012; Morgani and Brickman 2014). The absence of a robust epigenetic program of transcription in the transient 2C-like cells likely facilitates the rapid down-regulation of the 2C or totipotency genes after their activation in mESCs.
Altogether, the universal ChIP device as pioneered in the current study will facilitate implementation of the labor-intensive and highly sensitive low-input ChIP procedure in regular laboratories with no expertise in the ChIP procedure. Moreover, given the highly parallelized, automated workflow, the PnP-ChIP workflow will find its way to specialized epigenetic laboratories and core facilities, enabling large-scale projects and consortia. In view of the reproducibility and sensitivity, the robustness of the procedure, and the low-input requirements, we anticipate that the PnP-ChIP-seq will be a first step to discovery and screening of hPTM-based biomarkers in the clinic (Martens et al. 2010; Ross-Innes et al. 2012; Saeed et al. 2012; Jansen et al. 2013; Stelloo et al. 2015; Cejas et al. 2016; Dirks et al. 2016). Whether in a research setting or in the clinic, implementation of PnP-ChIP-seq will benefit from the fact that our workflow is based on a commercially available microfluidic platform. In addition, we foresee that the concept presented here can also be easily adapted to other programmable microfluidic platforms with a similar design, namely, nanoliter-sized affinity purification columns targeting chromatin-associated proteins with pressure-driven laminar flow of buffers and lysates.
Methods
Cell culture and FACS
E14 mESCs (129/Ola background; also referred to as “serum” mESCs) and the reporter mESCs were maintained without feeders in Dulbecco's Modified Eagle Medium (Thermo Fisher Scientific) containing 15% fetal bovine serum (Cell Signaling Technologies), 1000 U/mL leukemia inhibitory factor (LIF; Millipore), 5 µM beta-mercaptoethanol (Sigma-Aldrich), and 1 mM sodium pyruvate (Thermo Fisher Scientific). The so-called “2i” mESCs were ES cells that were cultured without feeders in the presence of 1000 U/mL LIF in serum-free N2B27 supplemented with MEK inhibitor PD0325901 (1 mM) and GSK3 inhibitor CH99021 (3 mM), together known as 2i. Generation of Hhex::Venus reporter mESCs (Morgani et al. 2013), Zscan4c::Emerald(Em)-GFP reporter mESCs (Eckersley-Maslin et al. 2016), and Erv4::TdTomato reporter mESCs (Eckersley-Maslin et al. 2016) have been described previously. Further details about FACS are described in the Supplemental Methods.
ChIP, library preparation for ChIP-seq, and RT-qPCR
Conventional cross-linked ChIP and library preparation for ChIP-seq was performed as described previously (van Mierlo et al. 2019); further details are present in the Supplemental Methods. RT-qPCR was performed according to standard protocols, as described in the Supplemental Methods.
Low-input microfluidic ChIP
Cross-linked chromatin was prepared from a cell suspension according to the conventional protocol as described above, with volumes downscaled to match the concentrations of cells used. For low-volume sonication, we used the Diagenode One sonication device according to the instructions of the manufacturer. For native ChIP-seq, non-cross-linked chromatin of 7500 or 15,000 mESCs was digested using MNase (NEB M0247) for 5–15 min at 20°C, after which the quality of the digestion was checked on a Bioanalyzer (Agilent). After digestion, the chromatin was diluted twofold in 60 mM Tris (pH 8.0), 300 mM NaCl, 1 µg/µL antibody, and 2× protease inhibitor cocktail (freshly prepared). For microfluidic ChIP, final volumes were kept below 20 µL to ensure short loading times across the prepacked antibody affinity bead column. Both cross-linked and native chromatin was snap-frozen in liquid nitrogen and stored at −80°C for later use. The microfluidic ChIP operation protocol is outlined in the Results section and in the Supplemental Figures. The various buffers used are as follows: control valve fluids (0.05% Tween 20), harvesting buffer (30mM Tris at pH 8.5), equilibration buffer (2 mM EDTA, 20 mM Tris at pH 8.0, 1% Triton X-100, 0.1% SDS, 150 mM NaCl), high-salt wash buffer (2 mM EDTA, 20 mM Tris at pH 8.0, 1% Triton X-100, 0.1% SDS, 500 mM NaCl), DNA extraction buffer (150 mM NaCl, 30 mM Tris at pH 8.0, 0.1 µg/µL Proteinase K; Sigma-Aldrich), and DNA elution buffer (10 mM Tris at pH 8.5). Both wash buffers included addition of freshly dissolved EDTA-free complete protease inhibitors (Roche). Microfluidic ChIP-seq libraries were constructed using Rubicon ThruPLEX library preparation kits according to the protocol of the manufacturer using 10 cycles of amplification. Ampure XP beads were used to select for DNA fragments of 300 bp in size (120-bp adaptor and 180-bp insert). Quality control for size and concentration was performed using the Agilent Bioanalyzer. The antibodies used for ChIP are as follows: H3K4me3, Diagenode C1540003; H3K4me1, Diagenode C1540194; H3K27ac, Diagenode C15410196 (lot #A1723-0041d); H3K36me3, Diagenode pAb-192-050; H3K27me3, Millipore 04-779; and H3K9me3, Diagenode C15410193, pAb-193-050 (lot #A1671-001P).
Sequencing and data analysis
Samples were sequenced paired-end, sequencing 42 nucleotides from both ends, using Illumina NextSeq 500. Bowtie 2 (version 2.0.2) (Langmead and Salzberg 2012) was used to map using the mm9 genome. We used mm9 to allow easier comparisons with previous data, but the use of mm10 would not affect our conclusions as the reference genomes mm9 and mm10 are very similar. Unmapped, duplicate, and low-quality (mapq < 15) reads were removed. SICER was used for peak calling (window size 200, gap size 200 for H3K4me3; window size 200, gap size 600 for H3K36me3, H3K4me1, H3K27ac; E-value 0.1). Empirically determined artificially enriched signal was excluded (ENCODE mm9 blacklist) (The ENCODE Project Consortium 2012; Amemiya et al. 2019). BEDTools v2.20.1 and pybedtools were used for peak call intersections and tag counting on peaks or promoter regions. DESeq2 (Love et al. 2014) was used for calling of differential PnP-ChIP-seq loci (FDR-adjusted P-value <0.05 as cut-off for calling significant difference) and PCA analysis. For direct comparison of loci between marker-positive (2C-like cells) versus marker-negative mESCs for either Hhex, Erv4, or Zscan4c, we used a cutoff of more than 10 read-normalized tags per H3K4me3 promoter. Heatmaps and average profiles were created using ngs.plot v2.61 (Shen et al. 2014). GO and KEGG analysis was performed using DAVID v6.8 (Dennis et al. 2003). In the 2i and serum mESC analysis, we linked the H3K4me3 peaks to the closest gene. ChIP-seq repeat analysis was performed as described in the Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE120673.
Supplementary Material
Acknowledgments
We thank Melanie Eckersley-Maslin for sharing of the Erv4::TdTomato (also known as MERVL::TdTomato) and Zscan4c::emGFP reporter mESC lines and Joshua Brickman for sharing of the Hhex::Venus reporter mESC line. We thank Eva Janssen-Megens for assistance with sequencing and Rob Woestenenk for help with FACS. We thank Mark Lynch and Jing Wang (Fluidigm) for helpful discussions about IFC architecture and scripts. This work was supported by the European Union's Seventh Framework Programme (FP7/2007-2013; grant number 282510-BLUEPRINT to H.G.S.) and the Netherlands Organization for Scientific Research (grant number NWO-VIDI 864.12.007 to H.M.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.260745.120.
Competing interest statement
P.C.T. and R.C.J. are former employees of Fluidigm Corporation and may still hold stock in the company.
References
- Adli M, Bernstein BE. 2011. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc 6: 1656–1668. 10.1038/nprot.2011.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai S, Xiong H, Li CC, Luo Y, Shi Q, Liu Y, Yu X, Li C, He A. 2019. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol 21: 1164–1172. 10.1038/s41556-019-0383-5 [DOI] [PubMed] [Google Scholar]
- Aldridge S, Watt S, Quail MA, Rayner T, Lukk M, Bimson MF, Gaffney D, Odom DT. 2013. AHT-ChIP-seq: a completely automated robotic protocol for high-throughput chromatin immunoprecipitation. Genome Biol 14: R124. 10.1186/gb-2013-14-11-r124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9: 9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Berguet G, Hendrickx J, Sabatel C, Laczik M, Squazzo S, Mazon Pelaez I, Saxena R, Pendeville H, Poncelet D. 2014. Automating ChIP-seq experiments to generate epigenetic profiles on 10,000 HeLa cells. J Vis Exp Dec 10: 52150. 10.3791/52150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. 2015. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6: 6033. 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- Bujold D, Morais DAL, Gauthier C, Côté C, Caron M, Kwan T, Chen KC, Laperle J, Markovits AN, Pastinen T et al. 2016. The International Human Epigenome Consortium data portal. Cell Syst 3: 496-499.e2. 10.1016/j.cels.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B et al. 2014. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell 55: 277-290. 10.1016/j.molcel.2014.05.029 [DOI] [PubMed] [Google Scholar]
- Cao Z, Chen C, He B, Tan K, Lu C. 2015. A microfluidic device for epigenomic profiling using 100 cells. Nat Methods 12: 959–962. 10.1038/nmeth.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, Li L, O'Neill NK, Duarte M, Rao P, Bowden M, Zhou CW, Mendiola M, Burgos E, Feliu J, et al. 2016. Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med 22: 685–691. 10.1038/nm.4085 [DOI] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, et al. 2012. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods 9: 609–614. 10.1038/nmeth.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P. 2010. The current state of chromatin immunoprecipitation. Mol Biotechnol 45: 87–100. 10.1007/s12033-009-9239-8 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. 2007. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells 25: 1037–1046. 10.1634/stemcells.2006-0430 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. 2008a. µChIP: a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res 36: e15. 10.1093/nar/gkm1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. 2008b. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat Protoc 3: 1032–1045. 10.1038/nprot.2008.68 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol 12: 236. 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. 2008. Gene regulation in the third dimension. Science 319: 1793–1794. 10.1126/science.1152850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Dirks RA, Stunnenberg HG, Marks H. 2016. Genome-wide epigenomic profiling for biomarker discovery. Clin Epigenetics 8: 122. 10.1186/s13148-016-0284-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy-Durruthy R, Ray M. 2018. Using fluidigm C1 to generate single-cell full-length cDNA libraries for mRNA sequencing. Methods Mol Biol 1706: 199–221. 10.1007/978-1-4939-7471-9_11 [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Svensson V, Krueger C, Stubbs TM, Giehr P, Krueger F, Miragaia RJ, Kyriakopoulos C, Berrens RV, Milagre I, et al. 2016. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep 17: 179–192. 10.1016/j.celrep.2016.08.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MSH. 2007. Zscan4: a novel gene expressed exclusively in late two-cell embryos and embryonic stem cells. Dev Biol 307: 539–550. 10.1016/j.ydbio.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández JM, de la Torre V, Richardson D, Royo R, Puiggròs M, Moncunill V, Fragkogianni S, Clarke L, Flicek P, Rico D, et al. 2016. The BLUEPRINT data analysis portal. Cell Syst 3: 491–495.e5. 10.1016/j.cels.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson RM. 2002. Fluidigm. Biochips get indoor plumbing. Chem Biol 9: 1161–1162. 10.1016/S1074-5521(02)00266-1 [DOI] [PubMed] [Google Scholar]
- Fu X, Djekidel MN, Zhang Y. 2020. A transcriptional roadmap for 2C-like-to-pluripotent state transition. Sci Adv 6: eaay5181. 10.1126/sciadv.aay5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey TS. 2012. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat Rev Genet 13: 840–852. 10.1038/nrg3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper WC, Marinov GK, Pauli-Behn F, Scott MT, Newberry K, DeSalvo G, Ou S, Myers RM, Vielmetter J, Wold BJ. 2014. Fully automated high-throughput chromatin immunoprecipitation for ChIP-seq: identifying ChIP-quality p300 monoclonal antibodies. Sci Rep 4: 5152. 10.1038/srep05152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, et al. 2019. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 51: 1060–1066. 10.1038/s41588-019-0424-9 [DOI] [PubMed] [Google Scholar]
- Gutin J, Sadeh R, Bodenheimer N, Joseph-Strauss D, Klein-Brill A, Alajem A, Ram O, Friedman N. 2018. Fine-Resolution mapping of TF binding and chromatin interactions. Cell Rep 22: 2797–2807. 10.1016/j.celrep.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. 2013. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13: 360–369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hainer SJ, Fazzio TG. 2019. High-resolution chromatin profiling using CUT&RUN. Curr Protoc Mol Biol 126: e85. 10.1002/cpmb.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Bošković A, McCannell KN, Rando OJ, Fazzio TG. 2019. Profiling of pluripotency factors in single cells and early embryos. Cell 177: 1319–1329.e11. 10.1016/j.cell.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Taft JM, Swist KM, Luo H, Witt H, Slattery M, Koide A, Ruthenburg AJ, Krajewski K, Strahl BD, et al. 2013. Recombinant antibodies to histone post-translational modifications. Nat Methods 10: 992–995. 10.1038/nmeth.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Maehara K, Harada A, Semba Y, Kudo K, Takahashi H, Oki S, Meno C, Ichiyanagi K, Akashi K, et al. 2016. Chd5 regulates MuERV-L/MERVL expression in mouse embryonic stem cells via H3K27me3 modification and histone H3.1/H3.2. J Cell Biochem 117: 780–792. 10.1002/jcb.25368 [DOI] [PubMed] [Google Scholar]
- Heyn H, Esteller M. 2012. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13: 679–692. 10.1038/nrg3270 [DOI] [PubMed] [Google Scholar]
- Ho JW, Bishop E, Karchenko PV, Nègre N, White KP, Park PJ. 2011. ChIP-chip versus ChIP-seq: lessons for experimental design and data analysis. BMC Genomics 12: 134. 10.1186/1471-2164-12-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro KI, Nakatake Y, Chikazawa-Nohtomi N, Kimura H, Akiyama T, Oda M, Ko SBH, Ko MSH. 2017. Expression analysis of the endogenous Zscan4 locus and its coding proteins in mouse ES cells and preimplantation embryos. In Vitro Cell Dev Biol Anim 53: 179–190. 10.1007/s11626-016-0097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen MP, Knijnenburg T, Reijm EA, Simon I, Kerkhoven R, Droog M, Velds A, van Laere S, Dirix L, Alexi X, et al. 2013. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res 73: 6632–6641. 10.1158/0008-5472.CAN-13-0704 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S. 2014. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods 11: 203–209. 10.1038/nmeth.2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. CUT&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Zhao K. 2011. ChIP-Seq: technical considerations for obtaining high-quality data. Nat Immunol 12: 918–922. 10.1038/ni.2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, et al. 2015. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17: 471–485. 10.1016/j.stem.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Ku WL, Nakamura K, Gao W, Cui K, Hu G, Tang Q, Ni B, Zhao K. 2019. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat Methods 16: 323–325. 10.1038/s41592-019-0361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science 345: 943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Stunnenberg HG. 2014. Transcription regulation and chromatin structure in the pluripotent ground state. Biochim Biophys Acta 1839: 129–137. 10.1016/j.bbagrm.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812. 10.1038/sj.emboj.7600545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, Altucci L, Stunnenberg HG. 2010. PML-RARα/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 17: 173–185. 10.1016/j.ccr.2009.12.042 [DOI] [PubMed] [Google Scholar]
- Morgani SM, Brickman JM. 2014. The molecular underpinnings of totipotency. Philos Trans R Soc Lond B Biol Sci 369: 20130549. 10.1098/rstb.2013.0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles R, Ko MSH, Brickman JM. 2013. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep 3: 1945–1957. 10.1016/j.celrep.2013.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TW, Hsieh YP, Ma S, Zhu Y, Lu C. 2018. Microfluidic low-input fluidized-bed enabled ChIP-seq device for automated and parallel analysis of histone modifications. Anal Chem 90: 7666–7674. 10.1021/acs.analchem.8b01541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM. 2006. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 38: 835–841. 10.1038/ng1820 [DOI] [PubMed] [Google Scholar]
- Park PJ. 2009. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680. 10.1038/nrg2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. 2010. Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068. 10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 33: 1165–1172. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh R, Launer-Wachs R, Wandel H, Rahat A, Friedman N. 2016. Elucidating combinatorial chromatin states at single-nucleosome resolution. Mol Cell 63: 1080–1088. 10.1016/j.molcel.2016.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S, Logie C, Francoijs KJ, Frige G, Romanenghi M, Nielsen FG, Raats L, Shahhoseini M, Huynen M, Altucci L, et al. 2012. Chromatin accessibility, p300, and histone acetylation define PML-RARα and AML1-ETO binding sites in acute myeloid leukemia. Blood 120: 3058–3068. 10.1182/blood-2011-10-386086 [DOI] [PubMed] [Google Scholar]
- Schmidl C, Rendeiro AF, Sheffield NC, Bock C. 2015. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 12: 963–965. 10.1038/nmeth.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Jiang D, Fu Y, Wu X, Guo H, Feng B, Pang Y, Streets AM, Tang F, Huang Y. 2015. H3k4me3 epigenomic landscape derived from ChIP-Seq of 1000 mouse early embryonic cells. Cell Res 25: 143–147. 10.1038/cr.2014.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13: 1006–1019. 10.1038/nprot.2018.015 [DOI] [PubMed] [Google Scholar]
- Stelloo S, Nevedomskaya E, van der Poel HG, de Jong J, van Leenders GJ, Jenster G, Wessels LF, Bergman AM, Zwart W. 2015. Androgen receptor profiling predicts prostate cancer outcome. EMBO Mol Med 7: 1450–1464. 10.15252/emmm.201505424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg HG, The International Human Epigenome Consortium, Hirst M. 2016. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 167: 1145–1149. 10.1016/j.cell.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. 2000. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288: 113–116. 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- van Galen P, Viny AD, Ram O, Ryan RJ, Cotton MJ, Donohue L, Sievers C, Drier Y, Liau BB, Gillespie SM, et al. 2016. A multiplexed system for quantitative comparisons of chromatin landscapes. Mol Cell 61: 170–180. 10.1016/j.molcel.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo G, Dirks RAM, De Clerck L, Brinkman AB, Huth M, Kloet SL, Saksouk N, Kroeze LI, Willems S, Farlik M, et al. 2019. Integrative proteomic profiling reveals PRC2-dependent epigenetic crosstalk maintains ground-state pluripotency. Cell Stem Cell 24: 123–137.e8. 10.1016/j.stem.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Wallerman O, Nord H, Bysani M, Borghini L, Wadelius C. 2015. lobChIP: from cells to sequencing ready ChIP libraries in a single day. Epigenetics Chromatin 8: 25. 10.1186/s13072-015-0017-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Lara-Astiaso D, Krupalnik V, Gafni O, David E, Winter DR, Hanna JH, Amit I. 2016. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat Biotechnol 34: 953–961. 10.1038/nbt.3652 [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428. 10.1038/emboj.2009.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. 2016. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537: 553–557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen F, Chen R, Xie D, Yang J, Zhao X, Guo R, Zhang Y, Shen Y, Göke J, et al. 2019. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res 47: 8485–8501. 10.1093/nar/gkz594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.