Peptidoglycan is assembled on the trans side of the plasma membrane from lipid II precursors into glycan chains that are cross-linked at stem peptides. In S. aureus, SagB, a membrane-associated N-acetylglucosaminidase, cleaves polymerized glycan chains to their physiological length.
KEYWORDS: Staphylococcus aureus, undecaprenol, glycan strand, glucosaminidase, CAAX-like protease, SagB, LyrA, SpdC, peptidoglycan
ABSTRACT
LyrA (SpdC), a homologue of eukaryotic CAAX proteases that act on prenylated substrates, has been implicated in the assembly of several pathways of the envelope of Staphylococcus aureus. We described earlier the lysostaphin resistance (Lyr) and staphylococcal protein A display (Spd) phenotypes associated with loss of the lyrA (spdC) gene. However, a direct contribution to the assembly of pentaglycine cross bridges, the target of lysostaphin cleavage in S. aureus peptidoglycan or of staphylococcal protein A attachment to peptidoglycan, could not be attributed directly to LyrA (SpdC). These two processes are catalyzed by the Fem factors and Sortase A, respectively. To gain insight into the function of LyrA (SpdC), here we use affinity chromatography and LC-MS/MS analysis and report that LyrA interacts with SagB. SagB cleaves glycan strands of peptidoglycan to achieve physiological length. Similar to sagB peptidoglycan, lyrA peptidoglycan contains extended glycan strands. Purified lyrA peptidoglycan can still be cleaved to physiological length by SagB in vitro. LyrA does not modify or cleave peptidoglycan, and it also does not modify or stabilize SagB. The membrane-bound domain of LyrA is sufficient to support SagB activity, but predicted “CAAX enzyme” catalytic residues in this domain are dispensable. We speculate that LyrA exerts its effect on bacterial prenyl substrates, specifically undecaprenol-bound peptidoglycan substrates of SagB, to help control glycan length. Such an activity also explains the Lyr and Spd phenotypes observed earlier.
IMPORTANCE Peptidoglycan is assembled on the trans side of the plasma membrane from lipid II precursors into glycan chains that are cross-linked at stem peptides. In S. aureus, SagB, a membrane-associated N-acetylglucosaminidase, cleaves polymerized glycan chains to their physiological length. Deletion of sagB is associated with longer glycan strands in peptidoglycan, altered protein trafficking and secretion in the envelope, and aberrant excretion of cytosolic proteins. It is not clear whether SagB, with its single transmembrane segment, serves as the molecular ruler of glycan chains or whether other factors modulate its activity. Here, we show that LyrA (SpdC), a protein of the CAAX type II prenyl endopeptidase family, modulates SagB activity via interaction though its transmembrane domain.
INTRODUCTION
Peptidoglycan is a net-like molecule that surrounds bacteria, maintaining the internal turgor of the cell. This large macromolecule is comprised of glycan strands and cross-linked wall peptides. In Staphylococcus aureus, the monomeric unit of the peptidoglycan is the disaccharide [-4(-N-acetylglucosamine-β(1-4)-N-acetylmuramic acid-β)1-]n, abbreviated to GlcNAc-MurNAc, and a wall peptide, l-Ala-d-iGln-l-Lys(Gly5)-d-Ala, that extends from the MurNAc sugar (1). The monomeric unit of peptidoglycan is first assembled in the cytoplasm and linked to the lipid carrier undecaprenyl phosphate, also known as bactoprenol, to form the intermediate lipid II. Lipid II is flipped to the outer face of the membrane, and the GlcNAc-MurNAc disaccharide is polymerized to form glycan chains by penicillin binding protein 2 (PBP2), as well as the monofunctional glycosyltransferases MGT and SgtA (2, 3). New glycans are then incorporated into the peptidoglycan via transpeptidases (PBP1, PBP2, and PBP4) that cross-link the amino group of the pentaglycine cross bridge (Gly5) to the carboxyl group of d-Ala from an available stem peptide within the mature peptidoglycan (4). Glycan strand length varies considerably across different species of bacteria, although the mechanisms governing these lengths are not fully understood. In S. aureus, glycan strands are on average 6 to 8 disaccharides in length (5). The membrane-bound glucosaminidase SagB is a key determinant of glycan chain length (6, 7). A sagB mutant elaborates elongated glycan strands, concomitant with a range of phenotypes including aberrations in protein secretion and changes to antibiotic susceptibility, demonstrating the importance of glycan strand length to S. aureus physiology (6). SagB is also important in relieving cell wall stiffness to facilitate cell enlargement (7).
In this study, we demonstrate that the membrane protein LyrA interacts with SagB. This interaction is necessary for SagB activity. LyrA is classified as a member of the CPBP (CAAX proteases and bacteriocin-processing enzymes) family of intramembrane metalloproteases similar to CAAX type II prenyl endopeptidases of eukaryotes, which proteolytically remove the C-terminal AAX residues of proteins with farnesyl and geranyl modifications at the cysteine (C) of the CAAX motif (C, cysteine; A, aliphatic; X, any amino acid) (8, 9). In eukaryotes, CAAX proteases are located in the membrane of the endoplasmic reticulum and are critical for the membrane localization of proteins with a CAAX motif (8, 10). However, prenylation of proteins is not known to occur in eubacteria. Here, we show that lyrA and sagB mutants closely phenocopy one another. Importantly, we observe elongated glycan chains in the lyrA mutant, indicating reduced SagB activity in this background. We confirm that LyrA does not affect production or processing of SagB, nor does LyrA alter the peptidoglycan substrate of SagB. We find that the transmembrane CPBP domain of LyrA is sufficient to support SagB activity in vivo and discuss possible models for LyrA contribution to peptidoglycan assembly.
RESULTS
The membrane-bound glucosaminidase SagB copurifies with tagged LyrA.
Several phenotypes of lyrA have been reported in the literature, including increased resistance to lysostaphin-mediated lysis (11), decreased display of surface protein A (SpA) (12), and sensitivity to tunicamycin (13). However, the mechanism by which loss of lyrA results in these phenotypes is unknown. To gain insight into the function of LyrA, we devised a pulldown protocol to identify interacting partners. lyrA was cloned with a C-terminal STREP tag for production in S. aureus. Two plasmids carrying untagged and tagged lyrA were transformed into the lyrA mutant, yielding lyrA(plyrA) and lyrA(plyrASTREP) strains, respectively. Both strains displayed lysostaphin susceptibility similar to that of the isogenic wild-type strain Newman, herein referred to as “wild type” (not shown). Next, the two strains were used to purify proteins solubilized in 1% (wt/vol) N-dodecyl β-d-maltoside (DDM) over Strep-Tactin resin. Proteins retained on the column were eluted with desthiobiotin and separated by SDS-PAGE (Fig. 1A). Coomassie staining of gels revealed several bands in the lyrA(plyrASTREP) sample that were absent from the sample with untagged protein. Bands labeled a through e were excised and analyzed after proteolysis by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Bands labeled a and b were identified as LyrA (Fig. 1A). Both these bands had the same peptide coverage; the higher molecular weight species possibly represents dimerization upon oxidation of cysteine residue at position 52. Band c was identified as the membrane-bound glucosaminidase SagB, with all peptides identified by LC-MS/MS shown in bold in Fig. 1B. Band d was identified as a putative amino acid ABC transporter substrate-binding protein encoded by gene nwmn_2313, and band e was identified as the membrane protein insertase YidC (Fig. 1A). A mutant with a transposon insertion in nwmn_2313 was identified from our arranged transposon library (14), crossed back in the wild-type strain Newman, and tested for lysostaphin resistance. The mutant strain exhibited neither Lyr nor Spd phenotypes (not shown), and nwmn_2313 was not further pursued. yidC is believed to be essential to S. aureus and was not further pursued (15). Instead, we sought to validate the SagB-LyrA interaction. The purification was repeated once more over Strep-Tactin resin and aliquots of the flowthrough, wash, and desthiobiotin eluates were analyzed by immunoblot using anti-STREP (αSTREP) and anti-SagB polyclonal serum (αSagB) (Fig. 1C). This analysis revealed the clear enrichment of SagB with LyrASTREP but not with untagged LyrA (Fig. 1C, lanes 3 and 4).
FIG 1.
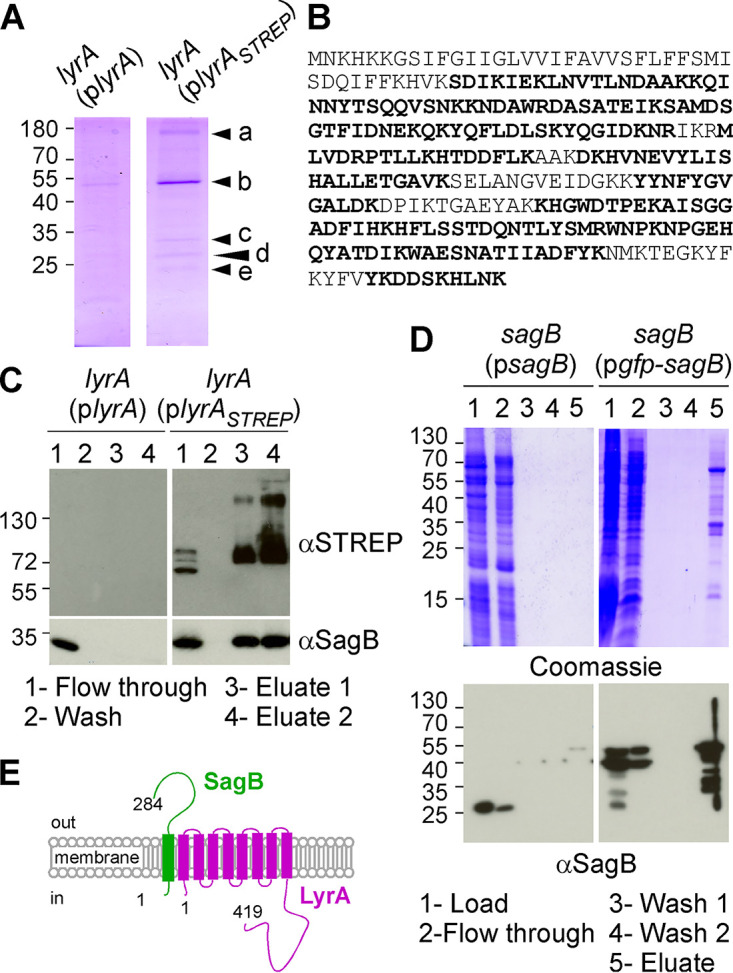
SagB copurifies with LyrA. (A) Coomassie stain of desthiobiotin eluates following affinity purification over Strep-Tactin resin. Samples loaded on the column were enriched for membrane proteins by DDM solubilization of bacterial extracts from lyrA(plyrA) and lyrA(plyrAstrep) cultures, respectively. Arrowheads labeled a through e indicate bands excised for LC-MS/MS analysis. (B) The amino acid sequence of SagB with peptides identified by LC-MS/MS shown in bold. (C) Strep-Tactin purification of samples as described for panel A. The flowthrough, wash, and elution fractions (lanes 1 to 4) were analyzed by immunoblotting (αSagB and αStrep antibodies). (D) DDM fractions prepared from S. aureus overproducing SagB or GFP-SagB were purified over GFP-trap agarose. Aliquots of material loaded on the columns, flowthrough, wash, and eluates (lanes 1 to 5) were separated by SDS-PAGE and stained with Coomassie or analyzed by immunoblotting (αSagB antibodies). (E) Depiction of LyrA and SagB in the membrane. Numbers to the left of gels and blots indicate molecular weight markers in kilodaltons.
Membrane-bound LyrA copurifies with tagged SagB.
To validate the LC-MS/MS data obtained with tagged LyrA, we performed a similar experiment with SagB. First, we asked whether tagged SagB could be purified from DDM extracts. SagB was translationally fused to the C terminus of green fluorescent protein (GFP) to yield GFP-SagB. When expressed from a plasmid (pgfp-sagB), the hybrid rescued all phenotypes of the sagB mutant (not shown). DDM extracts were prepared from strains sagB(pgfp-sagB) and sagB(psagB) as a control, and proteins were purified using GFP-trap agarose (Fig. 1D). SDS-PAGE of eluted proteins revealed several Coomassie-stained bands in the sagB(pgfp-sagB) sample that were absent from the sagB(psagB) purification (Fig. 1D). Western blotting using the SagB polyclonal serum identified SagB in the load and flowthrough of the column loaded with the sagB(psagB) extracts, while the translational hybrid with higher mobility was found in the eluate fraction (Fig. 1D). LC-MS/MS analyses of eluates revealed the specific enrichment for several proteins, including FmhC, LyrA, SecDF, SecA, FemA, and FemB (see Table S1 in the supplemental material). To rule out the possible involvement of these proteins for a LyrA-SagB interaction, C-terminally STREP-tagged LyrA and N-terminally His-tagged SagB (His-SagB) were produced using Escherichia coli BL21(DE3). His-SagB was purified from DDM-solubilized membrane fractions over nickel-nitrilotriacetic acid (Ni-NTA), eluted, and dialyzed. Next, a DDM-solubilized membrane fraction was generated from E. coli overproducing LyrASTREP, split, and mixed with buffer or with purified His-SagB. These suspensions were applied to Strep-Tactin resin via gravity flow. His-SagB preincubated with the LyrAstrep lysate was retained on the Strep-Tactin resin (Fig. S1). As a control, His-SagB alone or an E. coli DDM fraction was not retained on Strep-Tactin resin, suggesting that SagB interacts directly with LyrA independently of any other staphylococcal factor (Fig. S1).
lyrA and sagB mutants phenocopy one another.
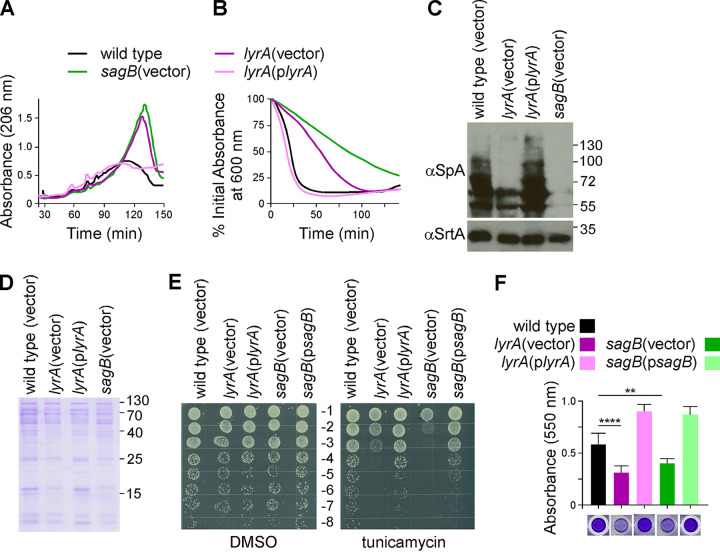
In light of the interaction between LyrA and SagB, a glucosaminidase that cleaves glycan chains, we wondered whether loss of lyrA may also result in aberrant peptidoglycan. Peptidoglycans purified from wild type, sagB(vector), lyrA(vector), and complemented lyrA(plyrA) strains were solubilized with lysostaphin to cleave pentaglycine cross-links while still leaving the glycan chains intact. Reverse-phase high-performance liquid chromatography (RP-HPLC) was used to separate lysostaphin-digested products, and elution was recorded by measuring absorbance at 206 nm. Chromatograms displayed in Fig. 2A show increased amounts of later eluting material for the lyrA(vector) sample relative to those for wild type, indicative of elongated glycan strands (larger and more hydrophobic fragments of peptidoglycan). The elution pattern of lyrA(vector) was strikingly similar to that of sagB(vector) (6, 7). Importantly, complementation of lyrA restored glycan strand length to physiological length, confirming that the elongated glycan strands of the lyrA mutant arise from lack of lyrA.
FIG 2.
The lyrA and sagB mutants phenocopy one another. (A) Peptidoglycan purified from indicated strains was digested with lysostaphin and analyzed by RP-HPLC using a C18 column. (B) Lysostaphin resistance was determined by measuring percentage drop in absorbance of 600 nm over time following addition of lysostaphin. The graph displays the average of three technical repeats per strain and is representative of at least three biological repeats. Error bars have been omitted for clarity. (C) Cell lysates of indicated strains were subjected to immunoblotting with αSpA and αSrtA antibodies. (D) Proteins secreted into the culture medium were concentrated by TCA precipitation, separated by SDS-PAGE. A Coomassie stain of a representative gel is shown. (E) A serial dilution series (−1 through −8) of the strains indicated was plated on tryptic soy agar supplemented with tunicamycin or vehicle only (DMSO) as indicated. The experiment was performed in technical and biological triplicate; a representative image is shown. (F) Biofilm formation after 24 h static growth was measured using a microtiter plate crystal violet assay. Columns represent the mean from 6 technical replicates, and the error bars are the standard deviation. Ordinary one-way ANOVA was used to compare the results with one another. Each comparison was statistically significant, except for lyrA(vector) versus sagB(vector). Statistical significance for lyrA(vector) or sagB(vector) compared to wild type are indicated on the graph: ****, P < 0.0001; **, P < 0.005. Result is representative of 3 biological repeats.
Loss of lyrA has previously been shown to cause increased resistance to lysostaphin and reduced display of staphylococcal protein A (SpA) in the bacterial envelope (6, 11, 12). Resistance to lysostaphin was measured by monitoring the density of cell cultures over time following addition of lysostaphin. As observed earlier, absorbance at 600 nm dropped much more rapidly for wild-type and complement lyrA(plyrA) bacterial cultures compared to that of cultures of the mutant lyrA(vector) (Fig. 2B). Loss of sagB resulted in an even greater increase in resistance toward lysostaphin than did loss of lyrA (Fig. 2B). To compare cell wall anchoring of SpA between these strains, bacterial cultures were spun and washed cells were subjected to prolonged lysostaphin treatment. This process solubilizes proteins, such as SpA, that are covalently anchored to the pentaglycine cross bridges of peptidoglycan by the enzyme sortase A (16). The proteins are released with cell wall fragments of various sizes, causing a banded smear that can be visualized by Western blotting (Fig. 2C). Sample preparations from both lyrA(vector) and sagB(vector) mutants displayed reduced amounts of SpA compared to those of sample preparations from the wild type control. In fact, as with lysostaphin resistance, the defect was exacerbated with the loss of sagB. Both phenotypes could be complemented by plasmid-borne lyrA or sagB, as shown in Fig. 2B and C for lyrA and as reported previously for sagB (6, 11, 17). We previously reported that sagB has a defect in protein secretion that can be complemented with sagB on a plasmid (6). Analysis of lyrA culture supernatant revealed a similar phenotype for lyrA(vector), which could be restored to wild-type levels with a plasmid-borne copy of lyrA (Fig. 2D). lyrA has also been found to confer synthetic lethality when the enzyme TagO is inhibited with the inhibitor tunicamycin (13). An independent study linked lyrA to a defect in biofilm formation (18). TagO encodes the first enzyme in the pathway for wall teichoic acid biosynthesis. Although sagB was not reported as a member of the tagO synthetic lethal network (13), we asked whether sagB might share this phenotype with lyrA. Tenfold serial dilutions of cultures prepared from wild type or lyrA and sagB mutants were spotted on agar supplemented with tunicamycin or mock (dimethyl sulfoxide [DMSO]) (Fig. 2E). The sagB mutant displayed greater susceptibility to tunicamycin than lyrA; in both instances, the growth defect could be reversed by providing the missing genes on a plasmid (Fig. 2E). Next, biofilm formation was assessed in microtiter plates by growing bacteria in tryptic soy broth (TSB) supplemented with 2% (wt/vol) glucose for 24 h. Plates were washed and stained with crystal violet to measure biomass (Fig. 2F). Both lyrA and sagB cultures produced smaller biofilms as judged by reduced absorbance of crystal violet at 550 nm. In both cases, the defect could be complemented by the expression of the respective gene from a plasmid.
LyrA neither modifies peptidoglycan nor perturbs SagB sedimentation with membranes.
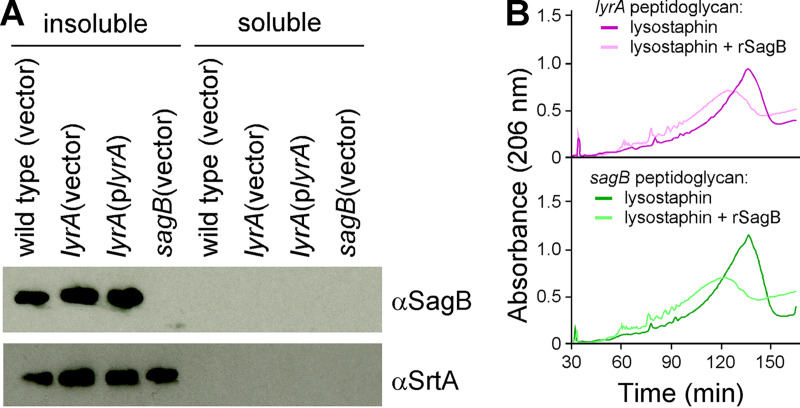
Since eukaryotic CAAX proteases mediate membrane targeting through posttranslational modifications of proteins bearing C-terminal CAAX motif (10), we wondered if SagB insertion in the membrane might be impaired in the lyrA mutant. Washed bacterial cells were lysed and subjected to ultracentrifugation to yield insoluble and soluble fractions enriched for membrane and cytosolic proteins, respectively. Extracts separated by SDS-PAGE for immunoblot revealed the presence of SagB in all insoluble fractions (Fig. 3A). Neither the mobility on SDS-PAGE nor the total amount of SagB was changed in extracts of the lyrA mutant compared to those in extracts of complemented and wild-type strains (Fig. 3A).
FIG 3.
LyrA does not process SagB or modify peptidoglycan in a manner necessary for cleavage by rSagB. (A) Indicated strains were grown to mid-log phase, and cells were sedimented, lysed, and fractionated into soluble or insoluble fractions and analyzed by immunoblotting with αSagB or αSrtA antibodies. (B) Peptidoglycan isolated either from the lyrA (top) or sagB (bottom) mutant was digested either with lysostaphin alone or lysostaphin and purified recombinant SagB (rSagB).
Next, we asked if LyrA may modify glycan repeats for preferential cleavage by SagB. We reasoned that if this were the case, purified peptidoglycan of the lyrA mutant with its extended glycan length would be a poor substrate for recombinant SagB (rSagB). To examine this possibility, purified lyrA peptidoglycan was incubated with rSagB or mock for 8 h followed by lysostaphin overnight to break cross bridges. Purified peptidoglycan from the sagB strain was included as a control since rSagB is active on this substrate (6). Solubilized peptidoglycan samples were separated by RP-HPLC. Chromatograms in Fig. 3B clearly show that rSagB is able to cleave the glycan chains of both lyrA and sagB peptidoglycan as observed by the conversion of fragments with longer to shorter retention times. We conclude that it is unlikely that LyrA modifies the peptidoglycan in a manner that would be necessary for SagB cleavage.
The transmembrane domain but not the three conserved type II CAAX motifs of LyrA is required for optimal SagB activity.
LyrA is a member of the CPBP family of intramembrane metalloproteases (8, 9). Mutational analysis of the yeast CAAX protease Rce1p (19, 20) and biochemical and crystallographic studies of Rce1 from the archaea Methanococcus maripaludis (10) corroborate the notion that the catalytic site of type II CAAX proteases encompasses three conserved motifs: EEXXXR, FXXXH, and HXXXB, where X represents any amino acid and B represents asparagine or aspartate (Fig. S2A) (8, 9). Mutations in any of the underlined residues have been shown to impair the cleavage of a prenylated peptide in vitro (10). In strain Newman, LyrA is 419 amino acids long (Fig. S2B). The N-terminal domain of LyrA (amino acids 1 to 258), consisting of eight transmembrane segments, carries sequences reminiscent of the three motifs: VE135FGFR139, Y163SVFS, and H210ASMT (Fig. S2A and B). These motifs are located between transmembrane segments 4 and 7 of LyrA/SpdC (Fig. S2B). The motifs are better conserved in the related SpdA and SpdB proteins of S. aureus (Fig. S2A). What makes LyrA unique among other CPBP family members is the additional cytosolic domain (amino acids 259 to 419) (Fig. S2C) (12). When aligned with LyrA sequences from a range of staphylococcal species, this C-terminal region varies in both length and sequence (Fig. S2C).
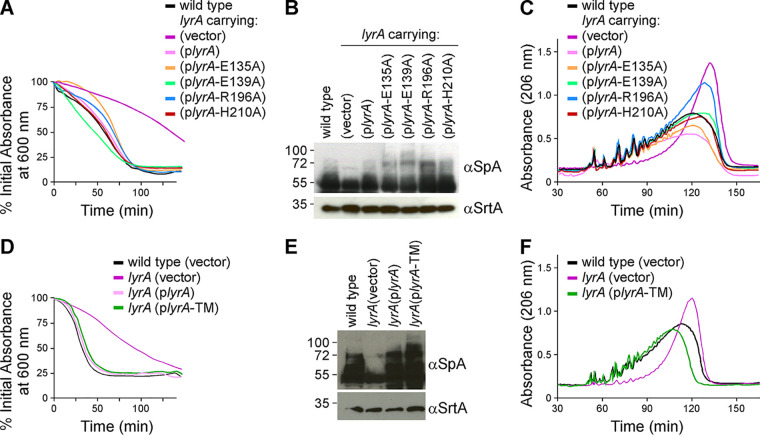
We asked whether the CAAX-like motifs embedded in the N-terminal transmembrane region of LyrA are required for SagB activity in vivo. Alanine substitutions of the nondegenerate amino acids E135, R139, and H210, or residue R196 that is conserved in staphylococcal LyrA sequences, were generated earlier by cloning lyrA variants on a plasmid (11). When transformed in the lyrA mutant, all these plasmids restored susceptibility to lysostaphin, corroborating our earlier findings (Fig. 4A) (11). Similarly, these substitutions did not compromise the activity of LyrA toward SpA production and did not affect the length of glycan strands (Fig. 4B and C), with the exception perhaps of the R196A substitution, which displayed an intermediate phenotype when assessing length of glycan strains (Fig. 4C).
FIG 4.
Neither the CAAX motifs nor the C-terminal domain is required for LyrA function. (A to C) The contribution of the three conserved CAAX protease motif residues (E135, R139, and H210) and the conserved non-CaaX protease R196 residue was tested by expressing alanine (A) substitution variants from a plasmid in the lyrA mutant. The new strains were analyzed for lysostaphin resistance (A), SpA production (B), and length of glycan strands (C). (D to F) The contribution of the C-terminal domain of LyrA was tested by expressing a variant encompassing only the 8 transmembrane segments (lyrA-TM) from a plasmid in the lyrA mutant. The new strains were analyzed for lysostaphin resistance (D), SpA production (E), and length of glycan strands (F).
To determine whether the C terminus of LyrA was necessary for function, a lyrA variant, lyrA-TM, was generated with a stop codon at residue T259 after the eighth transmembrane segment (Fig. S2C). lyrA-TM and full-length lyrA were cloned under the constitutive hprK promoter of plasmid pWWW412 (21). When transformed in the lyrA mutant strain, both plasmids, plyrA-TM and plyrA, restored all major phenotypes associated with loss of lyrA: lysostaphin resistance, SpA display, and glycan strand length (Fig. 4D to F). Therefore, we conclude that the C terminus of LyrA is not necessary for its function. Taken together, these results rule out the contribution of the C-terminal domain as well as the conserved CAAX protease motifs for LyrA function. The results suggest that LyrA is unlikely to contribute a catalytic activity in the SagB-LyrA interaction, yet the membrane-embedded N-terminal domain is sufficient to support SagB activity in vivo.
Overexpression of sagB partially compensates for loss of lyrA.
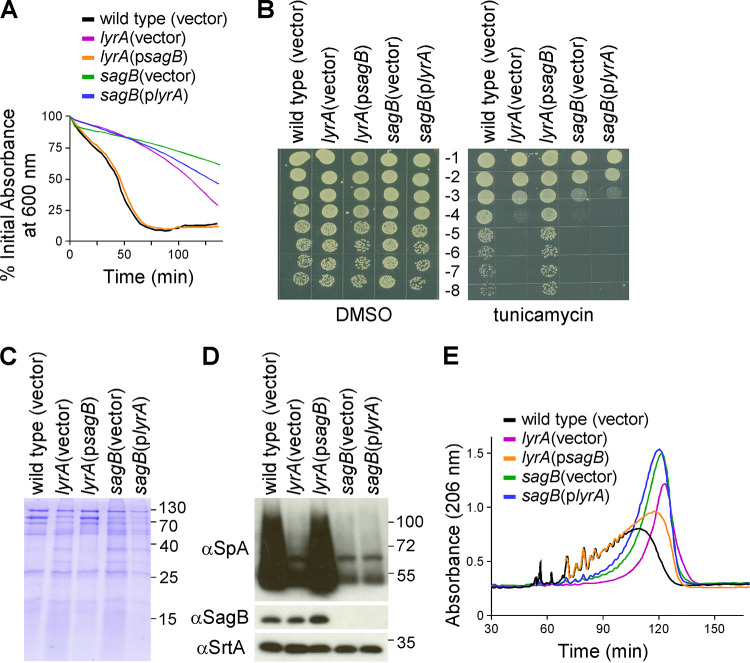
The data so far suggest that LyrA and SagB interact in the cell membrane and that this interaction is necessary for optimum SagB activity. To further explore the dependency of SagB on LyrA, we asked whether the overproduction of SagB could compensate for lack of LyrA and vice versa. Overexpression of lyrA in the sagB mutant reduced lysostaphin resistance only marginally, while overexpression of sagB in the lyrA mutant restored lysostaphin resistance to that of wild-type bacteria (Fig. 5A). Sensitivity toward tunicamycin, defective protein secretion in the growth medium, and reduced SpA production observed in the lyrA background were fully restored upon overexpression of sagB, but the inverse was not observed (Fig. 5B to D). Specifically, overexpression of lyrA in the sagB background did not restore any of these defects, demonstrating that these phenotypes ultimately arise from a lack of SagB activity. As glycan strand length is the direct result of SagB activity, peptidoglycan from these strains was also purified and analyzed following lysostaphin digest and RP-HPLC (Fig. 5E). Overexpression of sagB in the lyrA background resulted in a partial restoration of glycan strand lengths, while overexpression of lyrA in the sagB mutant did not rescue the defect (Fig. 5E).
FIG 5.
sagB overexpression partially compensates for lack of LyrA. The lyrA mutant was transformed with psagB, and the sagB mutant was transformed with plyrA. Strains were analyzed for lysostaphin resistance (A), tunicamycin susceptibility (B), protein secretion (C), SpA production (D), and length of glycan strands (F).
DISCUSSION
Of the six glucosaminidases in S. aureus strain Newman, SagB is one of only two that are membrane bound, the second being SagA (6). This is in line with the “inside-to-outside” model of Gram-positive bacterial growth (22). This model posits that peptidoglycan is synthesized close to the cell membrane and incorporated into the peptidoglycan macromolecule by cross-linking in a loose conformation. As these strands move outwards from the cell during growth, they become extended and taut, lowering the energy of activation for their hydrolysis (22). Limited hydrolysis of membrane-proximal peptidoglycan minimizes the risk of a deleterious breach of cell wall integrity. As such, it is likely that SagB activity is closely regulated. However, regulatory mechanisms for peptidoglycan assembly are not well understood, especially in Gram-positive bacteria.
lyrA was first identified in a screen for mutants resistant to lysostaphin, an endopeptidase that cleaves the pentaglycine cross bridge that cross-links S. aureus peptidoglycan (11). Pentaglycine cross bridges are synthesized on the cis side of the plasma membrane by the Fem factors and LyrA does not play a direct role in this process (11, 23–26). lyrA was later found in a screen for mutants deficient in surface display of SpA, an abundant substrate of sortase A that is covalently attached to pentaglycine cross bridges; the gene was dubbed spdC (12). lyrA/spdC also belongs to a network of genes rendered essential when the first enzyme in the biosynthesis of wall teichoic acids, TagO, is inhibited by tunicamycin treatment (13). These studies all connect LyrA to cell wall processes but via unknown means. More recently, it has been suggested that LyrA interacts with the histidine kinases of 10 two-component systems of S. aureus, including WalK, which is involved in controlling cell wall metabolism, leading to the suggestion that LyrA is a global regulator of two-component system activity (18). Another S. aureus CAAX protease, MroQ, was reported to be a regulator of the Agr quorum sensing system, potentially via interaction with AgrC histidine kinase (27, 28). The related CAAX protease homologue from group B streptococcus, Abx1, has been shown to interact with and regulate the two-component system CovRS (17).
We and others have previously demonstrated that SagB is the key determinant of glycan chain length in S. aureus (6, 7). In this study, we show that LyrA, a member of the CPBP (CAAX proteases and bacteriocin-processing) family, is necessary for SagB activity in vivo. lyrA is conserved across staphylococcal species and correlates with the presence of sagB. Our pulldown experiments using tagged LyrA did not identify WalK or other two-component histidine kinases (18). It is possible that these interactions are not maintained in our experimental conditions. Conversely, SagB pulldown experiments identified LyrA as well as FemA, FemB, FmhC, SecDF, and SecA. FemA and FemB form homodimers that sequentially add two Gly-Gly dipeptides to the peptidoglycan cross bridge, whereas FmhC pairs with FemB to incorporate Gly-Ser dipeptides into cross bridges (23–26). These proteins are enriched at the septum, the site of peptidoglycan assembly. A large amount of SecA/SecDF is also directed to the septum for the secretion of proteins with a YSIRK signal sequence such as SpA (29). LyrA has also been found to localize to the septum (18). The possibility that LyrA interaction with SagB involves additional partners was ruled out by demonstrating the sufficiency of LyrA and SagB interaction in E. coli. This interaction has been corroborated in a recent report that describes the atomic structure of SagB in complex with SpdC (LyrA) lacking its cytoplasmic domain (30). In this study, Schaefer et al. investigated an interaction between SagB and LyrA when transposon reads in the corresponding genes were found to be similarly represented following growth of S. aureus mutant libraries in the presence of cefoxitin, oxacillin, or amdinocillin (30).
While investigating the modulation of SagB activity by LyrA, we did not find a role for LyrA in the synthesis or processing of SagB; this is consistent with both the lack of a CAAX motif in SagB (and bacterial proteins in general) and the fact that predicted catalytic residues of CPBP family proteins are not needed for LyrA to promote SagB activity. Although we cannot exclude the possibility that LyrA acts to hydrolyze an unidentified SagB inhibitor, these findings argue against a catalytic role for LyrA. CAAX protease motifs have been reported to be variably necessary for function in bacterial CPBP proteins (17, 27, 28, 31, 32).
By incubating purified peptidoglycan of a lyrA mutant and recombinant SagB, we determined that LyrA does not modify peptidoglycan for cleavage by SagB, ruling out a possible catalytic activity of LyrA on peptidoglycan. In vitro, recombinant SagB without its transmembrane domain is still able to cleave the glycan chains of lyrA and sagB peptidoglycan preparations to their physiological lengths. While not included here, we failed to observe any enhanced activity with these substrates when using full-length SagB in the presence or absence of full-length LyrA. Nonetheless, when using radiolabeled lipid II bound to non-cross-linked peptidoglycan as a substrate, Schaefer et al. observed that the hydrolase activity of SagB is slightly enhanced when in complex with SpdC (LyrA) (30). However, in vivo we find that the overproduction of SagB compensated partly for lack of lyrA, illustrating that newly polymerized glycan chains remain a substrate of SagB in the absence of LyrA. Based on the data presented here, we can postulate that LyrA influences a rate-limiting step for glycan chain cleavage through direct protein interaction with SagB. Both open and closed structures have been obtained for SagB by X-ray crystallography, suggesting that the enzyme undergoes an active transition to accommodate its substrate (33). The new crystal structure of the SagB-SpdC complex suggests that SpdC could help orient the active site of SagB for cleavage of glycan strands (30). The substrate of SagB is the product of glycosyltransferases, i.e., the extending glycan chain tethered to undecaprenol-PP (Fig. 6) (34–36). Eukaryotic CAAX proteases recognize isoprene modifications on proteins (10). Undecaprenol is an isoprenoid chain. LyrA could bind and present undecaprenol-tethered products of glycosyltransferases to the SagB hydrolase (Fig. 6).
FIG 6.
Model for glycan chain elongation and cleavage in S. aureus. Glycan chain elongation is catalyzed by glycosyltransferase (GT) on the trans side of the plasma membrane. GT initiates the deprotonation of the GlcNAc 4-OH of lipid II via its active site residue. The activated nucleophile attacks the lipid-linked MurNAc carbon C-1 of the growing glycan chain, leading to the formation of a new β-1,4 glycosidic bond and the elongation of the glycan chain (34–36). Cleavage of the extended chain by SagB releases a glycan product that may be incorporated in cell wall peptidoglycan by transpeptidases (6). The second SagB product remains lipid-bound and may serve once more as a GT substrate. In this model, LyrA may help align the substrate into the active site of SagB by binding the lipid moiety of the glycan chain. R: stem peptide with pentaglycine cross bridge. GlcNAc and MurNAc are depicted as blue and red squares or blue and red chemical structures, respectively.
While SagB and LyrA are not essential for growth, the loss of these proteins could impede undecaprenol recycling through the transient accumulation of lipid-linked glycan strands at the membrane. This could indirectly affect the catalytic activity of other enzymes with undecaprenol-bound substrates that are involved in the assembly of peptidoglycan, capsule, wall teichoic acid, or cell wall anchored proteins (37–41). Peptidoglycan with extended glycan strands is also a poor substrate for the incorporation of proteins anchored by sortase A (reduced SpA display) and for lysostaphin (increased lysostaphin resistance). In summary, we have provided a rationale for the previously unexplained phenotypes of lyrA, demonstrating that they are mediated by the lack of SagB activity. We have found that LyrA interacts directly with SagB and is required for SagB activity in vivo, positioning LyrA as a novel regulator of S. aureus peptidoglycan processing by SagB.
MATERIALS AND METHODS
Bacterial strains, bacterial growth, and reagents.
S. aureus strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) supplemented with chloramphenicol 10 μg/ml or kanamycin 50 μg/ml where appropriate. Bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material, and primers are listed in Table S3. Transduction was performed as described earlier (37). E. coli DH5α was used for cloning. E. coli BL21(DE3) was used for purification of rSagB (a variant that lacks the transmembrane segment) (6), as well as for the purification of full-length His-SagB with an N-terminal six-histidine tag and LyrASTREP with a C-terminal STREP tag (this study). Bacterial cultures were grown to absorbance of 0.6 at 600 nm with 100 μg ampicillin/ml Terrific broth at 37°C, and protein production was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 4 h at 37°C. Tunicamycin suspended in DMSO was used at 1 μg/ml.
Preparation of DDM extracts.
DDM was used to enrich for membrane proteins produced in either S. aureus or E. coli. Cultures (2 liters) were grown at 37°C for approximately 4 h before cells were sedimented by centrifugation (10,000 × g, 10 min). Cells were then resuspended in 25 ml 100 mM Tris-HCl (pH 8.0), 150 mM NaCl (buffer A). For staphylococcal preparations, cells were mixed in approximately 1:1 volume to volume with 0.1 mm glass beads (BioSpec) and subjected to lysis by bead beating while encased in an ice-water chamber (BioSpec). For E. coli preparations, cells were lysed by two passages in a French pressure cell at 15,000 lb/in2. All resulting lysates were subjected to ultracentrifugation at 100,000 × g for 1 h. The supernatants were discarded and the pellets were resuspended in 10 ml 100 mM Tris-HCl (pH 8.0), 150 mM NaCl with 1% DDM. The resuspended pellets were left to rotate at room temperature for 1 h. Insoluble material was removed by ultracentrifugation at 100,000 × g for 1 h. The supernatant enriched in membrane proteins was used for further purification.
Purification of proteins.
rSagB was purified as described (6). For purification of membrane proteins, DDM extracts obtained as described above were diluted two times with 100 mM Tris (pH 8.0), 150 mM NaCl and loaded by gravity flow onto 1 ml of preequilibrated Strep-Tactin resin (IBA Lifesciences), Ni-NTA (Qiagen), or GFP-trap agarose (Chromotek). Strep-Tactin resin and Ni-NTA were washed with 20 column volumes of low stringency buffer and eluted with desthiobiotin (2.5 mM) or step gradients of imidazole as previously described (6); purification and elution using GFP-trap agarose were performed according to manufacturer’s recommendations. For identification of proteins, samples were run on SDS-PAGE and gel slices were submitted to the Harvard University Taplin Mass Spectrometry Facility for LC-MS/MS analysis.
Lysostaphin resistance assay.
Cell were washed with 50 mM Tris (pH 7.5) and resuspended in 650 μl 50 mM Tris (pH 7.5). Ninety microliters cells was aliquoted in 2× technical triplicate for each strain into a 96-well plate. To one set of three, 10 μl buffer only was added; to the other, 10 μl of lysostaphin 20 μg/ml was added. Absorbance at 600 nm was monitored every 5 min at 37°C with agitation in a Synergy HT plate reader (BioTek). The change in cell density, expressed as a percentage of the input for each well, was normalized to the negative control and plotted over time. Graphs shown in figures display the average from 3 technical repeats per strain and are representative of at least 3 biological repeats. Error bars were omitted for clarity.
Cellular fractionation and immunoblotting.
S. aureus Newman strains were diluted from overnight cultures and grown to absorbance at 600 nm of 1.0. For analyses of proteins by immunoblotting, bacteria from 2 ml of culture were sedimented, and the supernatant was collected and concentrated by trichloroacetic acid (TCA) precipitation. The sediment was washed with 1 ml Tris-buffered saline (TBS; 50 mM Tris [pH 7.5], 150 mM NaCl), resuspended in the same buffer with 25 μg/ml lysostaphin, and incubated for 30 min at 37°C. To ensure efficient lysis, cells were then exposed to 4× snap-freeze/thaw cycles using a dry ice-ethanol bath and a 60°C water bath. To isolate the insoluble and soluble fractions, lysates were subjected to ultracentrifugation (100,000 × g, 1 h). A total of 800 μl of the resulting supernatant was removed and TCA was precipitated, forming the soluble fraction. The remaining 200 μl was discarded. The pellet was washed with 1 ml TSB and then resuspended in TSB and TCA precipitated, forming the insoluble fraction. TCA-precipitated samples were reconstituted in 50 μl of 0.5 M Tris-HCl (pH 8.0)-4% SDS and heated at 90°C for 10 min. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore) for immunoblot analysis with the relevant rabbit polyclonal antibody. Immunoreactive signals were revealed by horseradish peroxidase-conjugated anti-rabbit antibody and enhanced chemiluminescent substrate (Pierce).
Biofilm assay.
Overnight cultures were standardized to an optical density at 600 nm (OD600) of 3.0 and diluted 1:50 into fresh TSB supplemented with 2% wt/vol glucose into a tissue-culture treated 96-well plate (final volume: 150 μl/well). Six wells were used per strain. The plate was left at 37°C without shaking for 24 h. After 24 h, cultures were tapped out of the wells and the plate was washed by submerging in 1× phosphate-buffered saline (PBS) and discarding the well contents twice. A total of 180 μl of 0.1% (wt/vol) crystal violet was added to each well, and the plate was left at room temperature for 15 min. The crystal violet solution was then discarded, and the plate was washed by submerging the plate in PBS three times. Excess dye was removed with a final vigorous tap of the plate onto a stack of napkin paper. The plate was then left to dry overnight. The dried crystal violet was then resuspended in 180 μl 30% acetic acid and transferred to a new 96-well plate. Absorbance at 550 nm was measured using a Synergy HT plate reader (BioTek). The data were analyzed by ordinary one-way analysis of variance (ANOVA) for statistical significance.
Peptidoglycan purification.
Staphylococci from cultures grown to absorbance at 600 nm between 0.8 and 1.0 were suspended in 50 ml 4% sodium dodecyl sulfate (SDS) buffered with 100 mM Tris (pH 6.8) and boiled for 30 min. Cells were subsequently washed five times in water to remove the detergent and lysed via bead beating (MP Biomedicals). Cellular material was collected by centrifugation (7,500 × g, 10 min), washed two times with water, and suspended in 50 mM Tris-HCl (pH 7.5), 10 mM CaCl2, and 20 mM MgCl2 for digestion with DNase (10 μg/ml) and RNase (50 μg/ml) for 2 h at 37°C and then with trypsin (100 μg/ml) for 16 h at 37°C. The cell wall material was sedimented by centrifugation (3,300 × g, 15 min), suspended in 1% SDS in 100 mM Tris, pH 6.8, boiled for 10 min to inactivate enzymes, and then washed twice with water, once with 8 M LiCl, once with 100 mM EDTA, twice with water, once with acetone, and finally two times with water. Sacculi were suspended in 5 ml 47% hydrofluoric acid for 48 h at 4°C. Peptidoglycan was recovered by centrifugation (33,000 × g for 45 min) and washed twice with water, twice with 100 mM Tris-HCl (pH 7.5), and two further times with water. Sacculi were dried under vacuum, the dry weight was recorded, and they were resuspended to 50 mg/ml with sterile water and stored at −20°C.
Peptidoglycan digestion.
For experiments involving digestion only with lysostaphin, 5 mg of peptidoglycan was resuspended in 1 ml 50 mM Tris (pH 7.5) with 100 μg/ml lysostaphin and left at 37°C overnight. For digestion with rSagB, peptidoglycan was resuspended in 1 ml 20 mM sodium phosphate buffer (pH 5.0) with or without 0.5 mg/ml rSagB, as appropriate. After 8 h of incubation, the pH of the reaction mixture was adjusted to 7 with 1 M NaOH and 100 μg/ml lysostaphin added for overnight digestion. The digestion reaction was ended by heat treatment (95°C, 10 min). Insoluble material was removed by centrifugation at 16,000 × g and the solubilized material was dried under vacuum. The dried fragments were resuspended in 50 μl water and 50 μl 0.5 M sodium borate (pH 9.0), to which 5 mg NaBH4 was added to reduce the peptidoglycan. The reaction was quenched after 15 min with 20% H2PO4. The sample was centrifuged to remove any precipitates before injection onto an HPLC system.
RP-HPLC.
Separation of peptidoglycan digested products by reverse-phase high-pressure liquid chromatography (RP-HPLC) was performed using a Waters 2695 Alliance system. A total of 100 μl of sample was applied to a 250 by 4.6-mm reversed-phase C18 column (ODS-Hypersil, 3 μm; Thermo Scientific) via automated injection. The column was eluted at a flow rate of 0.5 ml/min with a linear gradient starting 5 min after injection of 5% (vol/vol) methanol in 100 mM NaH2P04, (pH 2.5) to 30% (vol/vol) methanol in 100 mM NaH2P04 (pH 2.8) in 150 min. Column temperature was maintained at 52°C. The eluted compounds were detected by absorption at 206 nm.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratory for experimental advice and discussions. We are grateful to Anastasia Tomatsidou, Maksym Bobrovskyy, and Chloe Schneewind for comments and careful reading of the manuscript.
This research was supported by grant AI038897 from the National Institute of Allergy and Infectious Diseases, Infectious Disease Branch.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ghuysen J-M, Strominger JL. 1963. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of the disaccharides. Biochemistry 2:1119–1125. 10.1021/bi00905a036. [DOI] [PubMed] [Google Scholar]
- 2.van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25r–36r. 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- 3.Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. 2011. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol 193:2549–2556. 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffers DJ, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boneca IG, Huang ZH, Gage DA, Tomasz A. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J Biol Chem 275:9910–9918. 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 6.Chan YG, Frankel MB, Missiakas D, Schneewind O. 2016. SagB glucosaminidase is a determinant of Staphylococcus aureus glycan chain length, antibiotic susceptibility, and protein secretion. J Bacteriol 198:1123–1136. 10.1128/JB.00983-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler R, Turner RD, Bailey RG, Salamaga B, Mesnage S, Mohamad SA, Hayhurst EJ, Horsburgh M, Hobbs JK, Foster SJ. 2015. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. mBio 6:e00660. 10.1128/mBio.00660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei J, Grishin NV. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci 26:275–277. 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 9.Pei J, Mitchell DA, Dixon JE, Grishin NV. 2011. Expansion of type II CAAX proteases reveals evolutionary origin of gamma-secretase subunit APH-1. J Mol Biol 410:18–26. 10.1016/j.jmb.2011.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, O’Reilly N, Hanrahan SJ, Thompson AJ, Cronin N, Iwata S, Barford D. 2013. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504:301–305. 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundling A, Missiakas DM, Schneewind O. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol 188:6286–6297. 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. 2010. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol 78:238–252. 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santa Maria JP, Jr, Sadaka A, Moussa SH, Brown S, Zhang YJ, Rubin EJ, Gilmore MS, Walker S. 2014. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc Natl Acad Sci U S A 111:12510–12515. 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101:12312–12317. 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shahmirzadi SV, Nguyen MT, Gotz F. 2016. Evaluation of Staphylococcus aureus lipoproteins: role in nutritional acquisition and pathogenicity. Front Microbiol 7:1404. 10.3389/fmicb.2016.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazmanian SK, Ton-That H, Schneewind O. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol 40:1049–1057. 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 17.Firon A, Tazi A, Da Cunha V, Brinster S, Sauvage E, Dramsi S, Golenbock DT, Glaser P, Poyart C, Trieu-Cuot P. 2013. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog 9:e1003179. 10.1371/journal.ppat.1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poupel O, Proux C, Jagla B, Msadek T, Dubrac S. 2018. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog 14:e1006917. 10.1371/journal.ppat.1006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. 2000. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry 39:4096–4104. 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 20.Plummer LJ, Hildebrandt ER, Porter SB, Rogers VA, McCracken J, Schmidt WK. 2006. Mutational analysis of the ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem 281:4596–4605. 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch AL, Doyle RJ. 1985. Inside-to-outside growth and turnover of the wall of gram-positive rods. J Theor Biol 117:137–157. 10.1016/s0022-5193(85)80169-7. [DOI] [PubMed] [Google Scholar]
- 23.Tschierske M, Mori C, Rohrer S, Ehlert K, Shaw KJ, Berger-Bachi B. 1999. Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol Lett 171:97–102. 10.1111/j.1574-6968.1999.tb13417.x. [DOI] [PubMed] [Google Scholar]
- 24.Sugai M, Fujiwara T, Komatsuzawa H, Suginaka H. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67–75. 10.1016/S0378-1119(98)00508-3. [DOI] [PubMed] [Google Scholar]
- 25.Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bächi B. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc Natl Acad Sci U S A 96:9351–9356. 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willing S, Dyer E, Schneewind O, Missiakas D. 2020. FmhA and FmhC of Staphylococcus aureus incorporate serine residues into peptidoglycan cross-bridges. J Biol Chem 295:13664–13676. 10.1074/jbc.RA120.014371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosgriff CJ, White CR, Teoh WP, Grayczyk JP, Alonzo F. 3rd, 2019. Control of Staphylococcus aureus quorum sensing by a membrane-embedded peptidase. Infect Immun 87. 10.1128/IAI.00019-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marroquin S, Gimza B, Tomlinson B, Stein M, Frey A, Keogh RA, Zapf R, Todd DA, Cech NB, Carroll RK, Shaw LN. 2019. MroQ is a novel Abi-domain protein that influences virulence gene expression in Staphylococcus aureus via modulation of Agr activity. Infect Immun 87. 10.1128/IAI.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu W, Missiakas D, Schneewind O. 2018. Septal secretion of protein A in Staphylococcus aureus requires SecA and lipoteichoic acid synthesis. Elife 7. 10.7554/eLife.34092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer K, Owens TW, Page JE, Santiago M, Kahne D, Walker S. 2021. Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization. Nat Microbiol 6:34–43. 10.1038/s41564-020-00808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas S, Biswas I. 2014. A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J Bacteriol 196:1578–1587. 10.1128/JB.00028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol 192:2068–2076. 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pintar S, Borisek J, Usenik A, Perdih A, Turk D. 2020. Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis. Commun Biol 3:178. 10.1038/s42003-020-0911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derouaux A, Sauvage E, Terrak M. 2013. Peptidoglycan glycosyltransferase substrate mimics as templates for the design of new antibacterial drugs. Front Immunol 4:78. 10.3389/fimmu.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. 2007. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J Am Chem Soc 129:12674–12675. 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Heijenoort J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev 71:620–635. 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan YG, Kim HK, Schneewind O, Missiakas D. 2014. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem 289:15680–15690. 10.1074/jbc.M114.567669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem 277:16241–16248. 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer K, Matano LM, Qiao Y, Kahne D, Walker S. 2017. In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat Chem Biol 13:396–401. 10.1038/nchembio.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caveney NA, Li FK, Strynadka NC. 2018. Enzyme structures of the bacterial peptidoglycan and wall teichoic acid biogenesis pathways. Curr Opin Struct Biol 53:45–58. 10.1016/j.sbi.2018.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.