The pathogenic yeast Cryptococcus neoformans presents a worldwide threat to human health, especially in the context of immunocompromise, and current antifungal therapy is hindered by cost, limited availability, and inadequate efficacy. After the infectious particle is inhaled, C. neoformans initiates a complex transcriptional program that integrates cellular responses and enables adaptation to the host lung environment.
KEYWORDS: Cryptococcus neoformans, Pdr802, Titan cells, capsule, fungal pathogens, pathogenesis, pathogenic yeast, quorum sensing, transcription factors
ABSTRACT
Cryptococcus neoformans is a ubiquitous, opportunistic fungal pathogen that kills almost 200,000 people worldwide each year. It is acquired when mammalian hosts inhale the infectious propagules; these are deposited in the lung and, in the context of immunocompromise, may disseminate to the brain and cause lethal meningoencephalitis. Once inside the host, C. neoformans undergoes a variety of adaptive processes, including secretion of virulence factors, expansion of a polysaccharide capsule that impedes phagocytosis, and the production of giant (Titan) cells. The transcription factor Pdr802 is one regulator of these responses to the host environment. Expression of the corresponding gene is highly induced under host-like conditions in vitro and is critical for C. neoformans dissemination and virulence in a mouse model of infection. Direct targets of Pdr802 include the quorum sensing proteins Pqp1, Opt1, and Liv3; the transcription factors Stb4, Zfc3, and Bzp4, which regulate cryptococcal brain infectivity and capsule thickness; the calcineurin targets Had1 and Crz1, important for cell wall remodeling and C. neoformans virulence; and additional genes related to resistance to host temperature and oxidative stress, and to urease activity. Notably, cryptococci engineered to lack Pdr802 showed a dramatic increase in Titan cells, which are not phagocytosed and have diminished ability to directly cross biological barriers. This explains the limited dissemination of pdr802 mutant cells to the central nervous system and the consequently reduced virulence of this strain. The role of Pdr802 as a negative regulator of Titan cell formation is thus critical for cryptococcal pathogenicity.
INTRODUCTION
Cryptococcosis is a fungal infection caused by Cryptococcus neoformans and Cryptococcus gattii. C. neoformans is a ubiquitous opportunistic pathogen that infects mainly immunocompromised patients, while C. gattii is capable of infecting immunocompetent individuals (1). Cryptococcosis causes 180,000 deaths worldwide each year, including roughly 15% of all AIDS-related deaths (2), and is initiated by the inhalation of spores or desiccated yeast cells. In immunocompetent individuals, this typically causes an asymptomatic pulmonary infection that is controlled by the host immune response, although a population of C. neoformans may remain latent for extended periods of time (3–5).
Under conditions of immunocompromise, cryptococci disseminate from the lung to the brain. Mechanisms that have been suggested to mediate fungal crossing of the blood-brain barrier (BBB) include transcellular migration, in which the yeast cells enter and exit vascular endothelial cells (6–9); paracellular movement, in which they cross the BBB at junctions between endothelial cells (10–12); and “Trojan horse” crossing, whereby macrophages harboring C. neoformans enter the brain (13). Cryptococcal meningoencephalitis is difficult to treat and frequently lethal, for reasons that include the availability and cost of therapy (14, 15).
The ability of C. neoformans to survive and proliferate in the lung, and subsequently disseminate to the brain, depends on viability at mammalian body temperature and the expression of multiple virulence traits; these include secreted factors (16, 17), a polysaccharide capsule that surrounds the cell wall (18), and the production of giant (Titan) cells (19, 20). One secreted molecule, the pigment melanin, associates with the cell wall, where its antioxidant properties protect fungal cells from reactive oxygen species produced as a host immune defense (21–25). Urease, a secreted metalloenzyme that converts urea to ammonia and CO2, may affect the course of infection by modulating environmental pH and damaging host tissue structure (11, 12, 26).
The capsule, composed primarily of large polysaccharides (27–29), is a key cryptococcal virulence factor that impairs phagocytosis by immune cells (30–35). This dynamic entity changes its size and structure during interactions with the host or external environment (36–39), contributing to fungal adaptation (40, 41). Capsule polysaccharides that are shed from the cell enable diagnosis of cryptococcal infection and also impede host responses (35, 42).
Titan cells are a cryptococcal morphotype that has been variously characterized as having a cell body diameter (excluding the capsule) greater than 10 or 15 μm or total cell diameter (including the capsule) that exceeds 30 μm (20, 43, 44). These cells are polyploid and produce normal-size cells during infection (19, 45, 46). Titan cell formation is triggered by exposure to the host environment, including nutrient starvation, reduced pH, and hypoxia (47–49), although the extent of induction depends on the host immune response and the duration of infection (45, 50). Titan cell production appears to benefit the development of pulmonary C. neoformans infection, since these large cells are less susceptible to internalization by host phagocytes and more resistant to oxidative stress than normal-size cells (19, 46). Some of these effects may be explained by the highly cross-linked capsule and thickened cell wall of Titan cells (51). In contrast to their success in the lungs, Titan cells show impaired dissemination to the brain (19, 46).
C. neoformans experiences a dramatic change in conditions upon entering a host, including altered nutrient levels and pH. To adapt to the new environment, cryptococci activate a network of transcription factors (TFs) (39, 52). For example, imbalances in ion homeostasis trigger transcriptional changes mediated by the TFs Zap1 (53), Cuf1 (54), Pho4 (55), Cir1 (56), and Crz1 (57). Alkaline pH stimulates expression of the TF Rim101, which enables growth under basic conditions and other stresses, such as high salt and iron limitation; it also promotes the association of capsule polysaccharide with the cell and the formation of Titan cells (47, 58).
Overlapping TF circuits regulate cryptococcal virulence determinants, including polysaccharide capsule production and melanin synthesis. For example, Usv101, an important regulator of capsule thickness and polysaccharide shedding, also regulates three other TFs (Gat201, Crz1, and Rim101) and multiple polysaccharide-related enzymes (59). Gat201 further regulates additional virulence-related transcription factors and the anti-phagocytic protein Blp1 (60), while Crz1 plays a central role in the maintenance of plasma membrane and cell wall stability (57, 61, 62). Crz1 expression is also modulated by the calcineurin signaling pathway, which is required for normal yeast growth at 37°C, virulence, and sexual reproduction (63). A group of TFs, including Usv101, Bzp4, Hob1, and Mbs1 (59, 64), act together to regulate melanin production; deletion of Bzp4 also alters capsule production (52).
In this study, we investigated the TF Pdr802. The corresponding gene has a high rate of nonsynonymous mutations, which suggests that it is evolving rapidly (65). Pdr802 has previously been implicated in C. neoformans virulence (39, 52, 66), but its specific role and targets are not known. We discovered that Pdr802 is induced under host-like conditions, is a negative regulator of Titan cell formation, and influences capsule thickness and phagocytosis by macrophages. It also regulates genes whose products act in cell wall remodeling, virulence factor production, resistance to host temperature and oxidative stress, and quorum sensing. These functions make Pdr802 critical for cryptococcal survival in the lung and dissemination to the brain.
RESULTS
Role of Pdr802 in C. neoformans virulence.
The importance of Pdr802 in C. neoformans virulence has been demonstrated in multiple experimental models. Liu and collaborators first reported in 2008 that partial deletion of PDR802 reduced C. neoformans infectivity in a competition assay of pooled C. neoformans strains (66). In 2015, Maier et al. showed that a pdr802 deletion mutant had reduced virulence when tested individually in a short-term mouse model of infection (39). Later that year, Jung and colleagues reported that Pdr802 was required for full virulence in both wax moth larva and short-term mouse infection using pooled strains (52). Most recently, Lee and collaborators showed that Pdr802 was required for brain infection (67).
To further investigate the role of Pdr802 in pathogenesis, we complemented a complete deletion strain in the KN99α background that we had previously generated (pdr802) (39) with the intact gene at its native locus (PDR802). To examine targets of Pdr802, we also constructed a strain that expresses the protein fused to mCherry at its N terminus (see Fig. S1A in the supplemental material). All of these strains lacked or expressed RNA encoding PDR802 or its modified forms as expected (Fig. S1B), and PDR802 was expressed at wild-type levels in the complemented and modified strains (Fig. S1C).
Mutant strain construction and confirmation. (A) Scheme for generating C. neoformans strains in the KN99α background (middle) that either lack PDR802 (pdr802, top) or encode a tagged copy of the protein (mCherry-PDR802). (B) Qualitative analysis of gene expression in panel A strains and the complemented pdr802 mutant (PDR802). Cryptococcal mRNA isolated from cells grown in DMEM (37°C, 5% CO2, 24 h) was used to generate cDNA; from this, segments of the genes indicated at the left were amplified using the primers listed in Data Set S2, sheet 5, and the products were analyzed by agarose gel electrophoresis. Fragment sizes (in base pairs) are indicated, and the ladder bands shown are 400, 500, 650, 850, and 1,000 bp for the top panel; 200, 300, 400, 500, and 650 bp for the middle panel; and 100, 200, and 300 bp for the bottom panel. (C) Quantitative analysis of PDR802 expression. Samples of RNA isolated as for panel B were analyzed for PDR802 expression by qRT-PCR. All results were normalized to ACT1 expression. Each symbol represents a biological replicate, with the mean and standard deviation also shown. ***, P < 0.001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test. Download FIG S1, PDF file, 0.5 MB (536.1KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
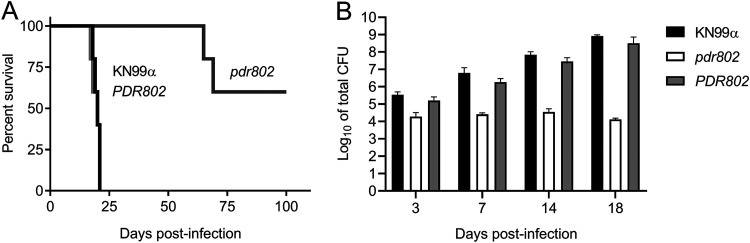
We next assessed the long-term survival of C57BL/6 mice infected with the parental wild-type (WT) strain (KN99α), the deletion mutant (pdr802), or the complemented mutant (PDR802). In this model, mice infected with the parent or complemented strains survived for roughly 3 weeks, while those infected with the deletion mutant showed a striking increase in survival: all animals survived for at least 65 days, and over half survived to the end of the study (100 days) (Fig. 1A). The lung burden measured at the time of death for pdr802-infected mice in this study was approximately 100-fold lower than that of wild-type infections (Fig. S2A), demonstrating the importance of this TF in C. neoformans virulence. Mean brain burden at the time of death was more similar between mutant and wild-type infections (Fig. S2A), although we did note some heterogeneity in this measure for pdr802-infected mice; animals sacrificed at around 2 months of infection (red symbols) showed brain burdens similar to WT levels, while brain burdens in mice sacrificed at day 100 (blue symbols) ranged between zero fungal cells and the WT level.
FIG 1.
The transcription factor Pdr802 influences C. neoformans virulence. (A) Survival of C57BL/6 mice over time after intranasal inoculation with 5 × 104 cryptococci, with sacrifice triggered by weight below 80% of peak. (B) Means and standard deviations (SD) of total colony-forming units (CFU) in lung tissue at various times post-infection. Numbers of CFU inoculated for each strain were 41,400 (KN99α), 42,800 (pdr802 strain) and 26,600 (PDR802 strain). P < 0.05 for the pdr802 strain compared to the other strains at all time points.
Organ burdens. (A) Mean and SD values of total CFU in the indicated tissue of mice from the Fig. 1 survival curve are shown. Each point is the average value for a single animal at the time of death. For pdr802 strain infections, red circles represent mice sacrificed at days 65 and 69, while blue circles represent mice sacrificed at the termination of the study (day 100). (B) Mean and SD of total CFU in the blood and brain at the indicated times post-infection. (C) Mean and SD of total CFU in the lung, blood, and brain 75 days after infection with the pdr802 strain. Each color represents one mouse. Download FIG S2, PDF file, 0.9 MB (948.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next examined the time course of fungal proliferation in the lungs. As expected, the burdens of WT and the complemented mutant strains increased steadily over an 18-day interval (Fig. 1B), eventually reaching roughly 105 times the original inoculum. Towards the end of this period, these cells were also detected in the blood and brain (Fig. S2B). In contrast, the lung burden of pdr802 remained close to the inoculum throughout this period, with no mutant cells detected in the blood or brain. At a late time point of pdr802 infection (75 days), we again noted some heterogeneity of fungal burden: one mouse had high lung burden with no dissemination, another had high lung burden with moderate brain burden, and the third had extremely low lung burden with no dissemination (Fig. S2C). No CFU were detected in the blood of pdr802-infected mice at any point during infection. These results suggest that even though the pdr802 mutant is generally hypovirulent and remains at low levels in the lung, it can occasionally reach the brain and, given enough time, accumulate there (see Discussion).
Given the dramatic effects of Pdr802 on fungal virulence, we wondered about the specific biological processes in which this transcription factor is involved. We first examined the behavior of the pdr802 strain in vitro, including stress conditions that might be encountered in the host. We saw no differences in growth of the mutant compared to WT cells under conditions that challenge cell or cell wall integrity, including the presence of sorbitol, high salt, cell wall dyes, caffeine, sodium dodecyl sulfate (SDS), or ethanol (Fig. S3A to C). The mutant also showed no altered susceptibility to elements of the host response, such as nitrosative or oxidative stresses, or in melanin production. All of these results held when cells were grown on rich medium, whether plates were incubated at 30°C, 37°C, or 37°C in the presence of 5% CO2, which was recently described as an independent stress for C. neoformans (68) (Fig. S3A to C). Finally, the mutant showed no difference from wild-type cells in secretion of urease (Fig. S3D).
Characterization of pdr802 cells. (A to C) Tenfold serial dilutions of WT, pdr802, and PDR802 cells were plated on the media shown and incubated at 30°C (A), 37°C (B), or 37°C in the presence of 5% CO2 (C). Nitrosative (NaNO2) and oxidative (H2O2) stress plates were prepared with YNB medium, and melanization plates containing l-DOPA were prepared as described in Materials and Methods; all other plates were prepared with YPD medium. The lac1 control strain lacks the ability to melanize (88). (D) Urease activity of the indicated strains was evaluated using Christensen’s urea solid medium (see Materials and Methods) at the indicated temperatures. The ure1 control strain does not produce urease (17). Download FIG S3, PDF file, 0.8 MB (795.3KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pdr802 is regulated by “host-like” conditions.
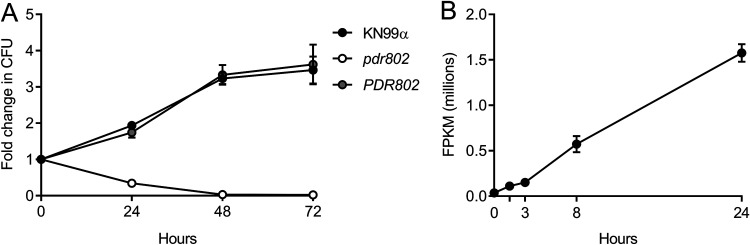
We next tested the growth of the pdr802 mutant under conditions more like those encountered inside the mammalian host, using tissue culture medium (Dulbecco’s modified Eagle medium [DMEM]) at 37°C in the presence of 5% CO2. We found that although the pdr802 mutant grew like WT in rich medium (yeast extract-peptone-dextrose [YPD]), it grew poorly in DMEM (Fig. S4A and B). To test whether the mutant cells were dead or just static after growth in DMEM, we plated aliquots on solid medium to measure CFU over time (Fig. 2A). The pdr802 culture showed a dramatic decrease in viability compared to WT and the complemented strain, which was greatest in the first 24 h. This is the same time frame in which expression of the PDR802 gene shows a striking increase, as measured by transcriptome sequencing (RNA-Seq) (Fig. 2B).
FIG 2.
PDR802 expression is required for cell viability and induced during growth under host-like conditions. (A) Cells grown in DMEM at 37°C and 5% CO2 were sampled at the times indicated and plated on YPD to assess viability (measured as number of CFU and plotted as fold change from time zero). (B) PDR802 expression in KN99α cells grown in DMEM at 37°C and 5% CO2 was assessed by RNA-Seq as in reference 117. FPKM, fragments per kilobase per million. Mean and standard deviation (SD) are shown on both plots.
Growth curves and capsule shedding. (A and B) Growth of the indicated strains in YPD at 30°C (A) or DMEM at 37°C and 5% CO2 (B) was assessed by determining the optical density at 600 nm (OD600) at the times indicated. (C) Conditioned medium from the indicated strains was probed for the presence of GXM after growth in DMEM for 24 or 48 h. Equal volumes of culture supernatant were analyzed without normalization to cell density. Immunoblotting was performed using the anti-GXM monoclonal antibody 302. Download FIG S4, PDF file, 0.6 MB (577.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
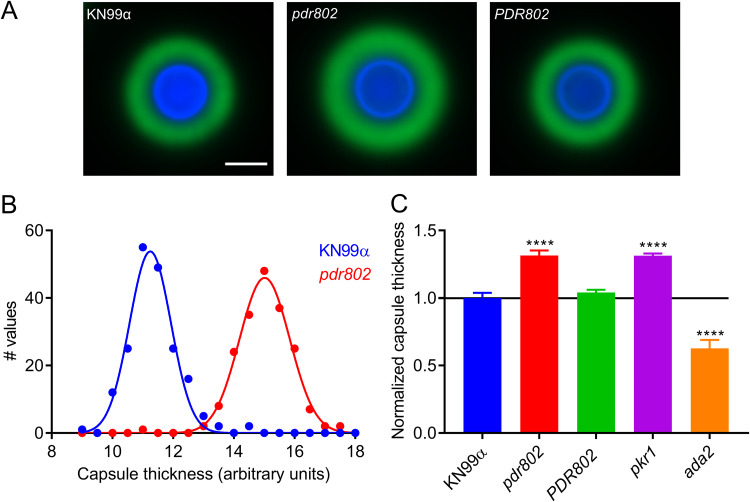
Another important feature that is induced by growth in DMEM at 37°C and 5% CO2 is the polysaccharide capsule, which we previously reported to be regulated by Pdr802, based on negative staining with India ink (39). Fluorescence microscopy confirmed increased capsule thickness of the mutant, which reverted to WT in the complemented strain (Fig. 3A). To quantify this change, we took advantage of a semiautomated assay that we have developed (Fig. S5), which measures capsules on a population scale (Fig. 3B) and is therefore very sensitive. This analysis showed that the capsule thickness of pdr802 cells resembles that of the well-studied hypercapsular mutant pkr1 (39, 69, 70) and is completely restored to WT by complementation at the native locus (Fig. 3C). Previous studies suggest that capsule thickness upon induction reflects the size of the dominant capsule polymer (glucuronoxylomannan [GXM]) (71, 72), which can be analyzed by agarose gel migration and blotting with anticapsule antibodies (71). Consistent with the difference we observed in capsule thickness by imaging, this method showed decreased mobility of GXM from pdr802 as capsule induction progressed (Fig. S4C).
FIG 3.
The pdr802 mutant is hypercapsular. (A) Representative immunofluorescence micrographs of the indicated strains after growth in DMEM (37°C, 5% CO2) for 24 h. The capsule was stained with anti-GXM monoclonal antibody 302 conjugated with Alexa Fluor 488 (green) and the cell wall with calcofluor white (blue). All images are to the same scale; bar, 5 μm. (B) Capsule thickness distribution for the indicated strains. (C) Mean and SD of capsule size, quantified as detailed in Materials and Methods and Fig. S5, with the pkr1 (39) and ada2 (105) strains shown as hypercapsular and hypocapsular controls, respectively. ****, P < 0.0001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test.
Semi-automated assay for cryptococcal capsule imaging. (A) Schematic of applying this method to cryptococcal cells induced to form capsule by growth in DMEM (37°C, 5% CO2) for 24 h, followed by cell wall and capsule staining. Thousands of cells may be imaged per well and analyzed automatically with software that annotates and measures the capsule (annotated in dark blue) and cell wall (annotated in bright green). See Materials and Methods for details. (B) Capsule size distribution of WT cells after induction. Capsule thickness for each cell is the difference between the paired diameters of the cell wall and capsule, which is plotted with reference to the mean value. (C and D) Mean and SD (C) and cumulative percentage (D) analysis of WT compared to hyper- and hypocapsular control strains (here, pkr1 and ada2, respectively). Capsule thickness is in arbitrary units, related to the pixels measured. (E) Time required to analyze the capsule thickness of 1,000 cells by this method compared to manual assessment of India ink images. Download FIG S5, PDF file, 0.7 MB (688.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
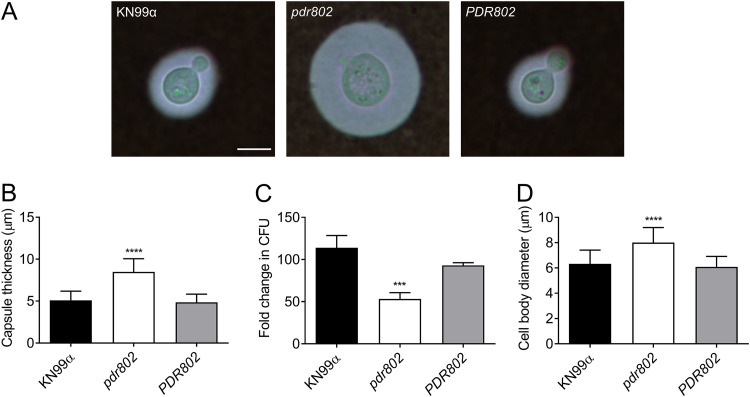
To validate the observations that we had made under standard host-like conditions based on synthetic tissue culture medium, we conducted similar studies in mouse serum at 37°C and 5% CO2. These conditions induced an even more pronounced hypercapsular phenotype of the pdr802 mutant (Fig. 4A and B), as well as reduced cell viability (Fig. 4C) and increased cell body diameter (Fig. 4D).
FIG 4.
Growth in mouse serum elicits increased capsule thickness and cell body diameter in the pdr802 mutant. (A) Light micrographs of the indicated strains after growth in mouse serum (at 37°C, 5% CO2) for 24 h and negative staining with India ink to visualize the capsule. All images are to the same scale; bar, 5 μm. (B) Mean and SD of capsule thickness, assessed by measuring at least 50 cells per strain with ImageJ. (C) Cells grown as described for panel A were plated on YPD to assess CFU. Mean and SD of the fold change compared to 0 h are shown. (D) Mean and SD of cell body diameter, measured as for panel B. ***, P < 0.001, and ****, P < 0.0001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test.
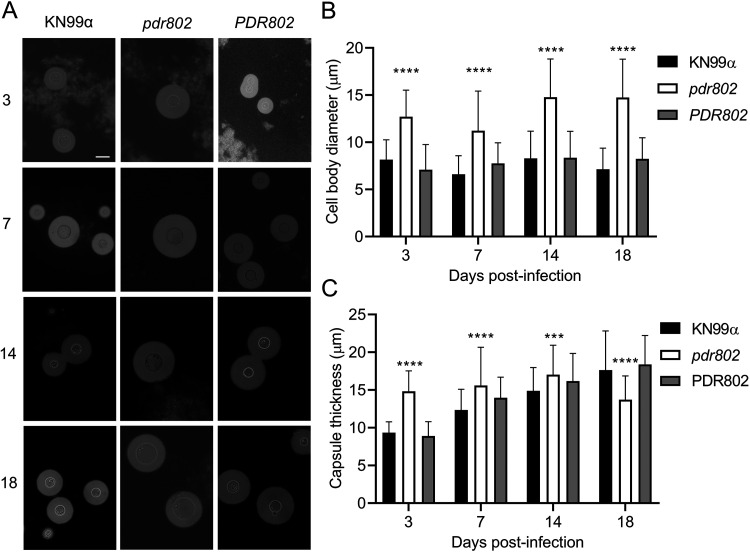
We were intrigued by the enlarged cell body and capsule of the pdr802 mutant cells under host-like conditions in vitro and decided to examine these phenotypes in vivo. For these studies, we isolated fungal cells from the lungs of mice at various times after infection and assessed their morphology by negative staining (Fig. 5A). At each time point, the mean mutant cell body diameter was larger than that of the controls. Additionally, while this parameter was stable for WT and complemented strains throughout the infection period, it trended larger at the end of the infection period for the deletion mutant (Fig. 5B). In contrast, mutant capsule thickness, although initially greater than that of control cells, changed little throughout the period, while capsule thickness of control cells increased to that level or beyond (Fig. 5C). Furthermore, although the total diameter of pdr802 cells consistently exceeded that of WT and complemented cells, their sizes became more comparable late in infection (Fig. S6A). Over time, therefore, the ratio of total cell diameter to cell body diameter for WT and PDR802 cells steadily increased, while it remained roughly constant for the mutant (Fig. S6B).
FIG 5.
Absence of PDR802 yields enlarged cells and loss of capsule induction in the context of animal infection. (A) India ink staining of fungi isolated from the lungs of mice infected with the indicated strains. Numbers at left are days post-infection. All images are to the same scale; bar, 10 μm. (B and C) Means and SD of cell body diameter (B) and capsule thickness (C), assessed by measuring at least 50 cells per strain with ImageJ. ****, P < 0.0001, and ***, P < 0.001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test for each day post-infection.
PDR802 deletion induces the formation of Titan cells. Mean and SD of (A) total cell diameter and (B) ratio of total cell to cell body diameters (diameter ratio), assessed by measuring at least 50 cells per strain with ImageJ. **, P < 0.01, and ****, P < 0.0001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test for each day post-infection. (C) Percent of Titan cells in the indicated strain, evaluated using various published parameters: cell body diameter bigger than 10 or 15 μm (20) or total cell diameter bigger than 30 μm (43). Download FIG S6, PDF file, 0.7 MB (702.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pdr802 negatively regulates Titan cell formation.
We were particularly interested in the cell size phenotype of pdr802 because Titan cells have been strongly implicated in cryptococcal pathogenesis (19). By any definition of this morphotype (cell body diameter greater than 10 or 15 μm or total cell diameter greater than 30 μm), our mutant cell populations were dramatically enriched in Titan cells at every time of infection that we assessed (Fig. S6C).
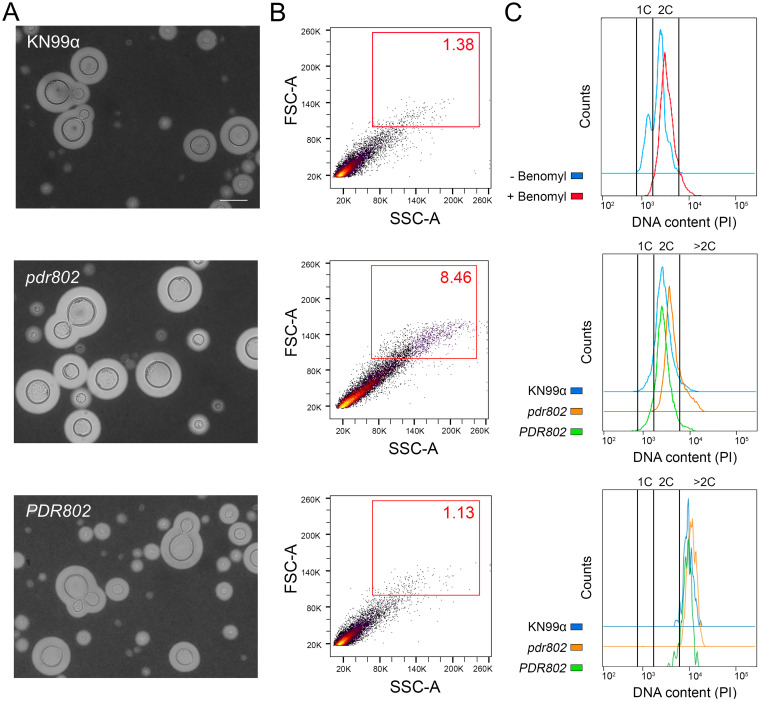
To specifically test Titan cell formation by the pdr802 strain, we subjected mutant cells to in vitro conditions that induce this process (49) (Fig. 6A) and analyzed the resulting population by flow cytometry. Consistent with our in vivo observations, Titan cells constituted a much larger fraction of the population in the mutant culture (8.46%) than in the WT and complemented cultures (1.38% and 1.13%, respectively) (Fig. 6B). As expected for Titan cells, these cells are also polyploid (Fig. 6C).
FIG 6.
Pdr802 is a negative regulator of Titan cell formation. (A) The indicated strains were subjected to in vitro conditions that induce Titan cell formation and imaged with India Ink. All images are to the same scale; bar, 10 μm. Images were selected so that each shows multiple examples of Titan cells, not to reflect abundance of this morphotype. (B) The percentage of Titan cells in each culture was quantified using flow cytometry, gated as indicated by the red square. FSC-A, forward scatter; SSC-A, side scatter. (C) DNA content of cells after staining with propidium iodide (PI) and analysis by flow cytometry. (Top) 1C and 2C gates determined using control cultures of KN99α grown in YNB without or with benomyl, which traps cells at 2C. (Middle) Profiles of the indicated strains after growth under Titan cell-inducing conditions, showing an increased polyploid tail in mutant cultures relative to wild-type cells. (Bottom) DNA content profiles for only the Titan cells (gated as for panel B, red box) for each strain, showing their polyploid nature.
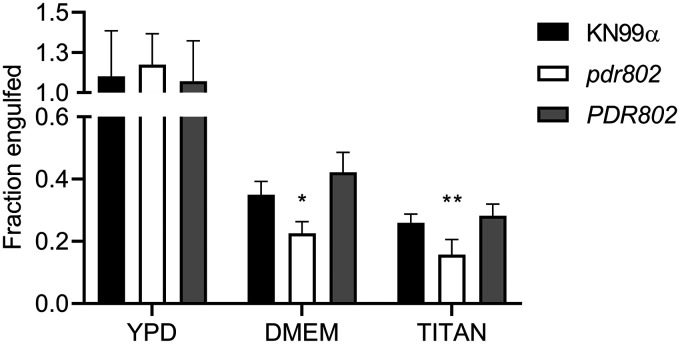
Titan cells are poorly engulfed by host phagocytes (19, 45, 73), which may reflect their increased size as well as alterations in capsule and cell wall (51). We observed this reduced uptake for all strains after growth in conditions that favor Titan cell formation (Fig. 7, Titan versus YPD). Also, all strains showed a reduction in phagocytosis after capsule induction in DMEM (Fig. 7, DMEM versus YPD), which is not surprising, because the capsule is antiphagocytic (31, 73). Notably, the reduction in uptake was greatest for the pdr802 mutant under both of these conditions, even though it showed normal engulfment when all strains were grown under control conditions (YPD). This is likely because the mutant culture is both hypercapsular and enriched in Titan cells.
FIG 7.
Deletion of PDR802 affects phagocytosis after growth under conditions that induce capsule and Titan cell formation. C. neoformans strains were grown in YPD (18 h), DMEM (24 h), or Titan cell induction medium (72 h) and then incubated for 2 h with J774.16 mouse macrophages; host cells were then washed and lysed to assess fungal burden by CFU. Data shown are normalized to the CFU of the initial inoculum. *, P < 0.05, and **, P < 0.01, compared to KN99α by one-way ANOVA with a post hoc Dunnett test.
Identification of direct, functional targets of Pdr802.
To identify direct targets of Pdr802, we next performed chromatin immunoprecipitation followed by sequencing (ChIP-Seq). We compared DNA sequences immunoprecipitated by anti-mCherry monoclonal antibody (MAb) from cells expressing mCherry-Pdr802, which grow similarly to the WT (Fig. S7A), and untagged cells. Both strains were grown for 24 h in DMEM at 37°C and 5% CO2, as these conditions induce PDR802 expression dramatically compared to standard YPD growth conditions (Fig. 2).
Strain viability, Pdr802 interaction with its own promoter, and putative DNA-binding motifs. (A) The indicated strains were grown in DMEM at 37°C and 5% CO2 for the times shown, and samples were tested for their ability to form colonies on YPD medium. Plotted is the fold change in CFU relative to the initial culture. (B) Pdr802 self-regulation. The ratios (log2) of reads from immunoprecipitated (IP) DNA to reads from input DNA were calculated for 1,000 bp upstream of the first coding nucleotide (+1) of PDR802; shown is the difference in these values between tagged and untagged strains. Red triangles, complete Pdr802 DNA-binding motifs (Fig. S7C); blue triangles, partial motifs. (C) Putative Pdr802-binding motifs determined using DREME (76). Primary and secondary hits are shown for analysis of 1,000 bp upstream of the initiating ATG. Download FIG S7, PDF file, 1.0 MB (1.1MB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Using 2-fold-enrichment over control as a cutoff value for peaks with adjusted P values of <0.05, we identified 656 binding sites for mCherry-Pdr802 in genomic DNA. Of these, 540 occurred within 1,000 bp upstream of transcription start sites (Data Set S1, sheets 1 and 2), which we used as an approximation of regulatory regions. Notably, ChIP-Seq data for the region upstream of the PDR802 transcription start site suggested self-regulation, as has been reported for other cryptococcal TFs (74, 75) (Fig. S7B). We further applied discriminative regular expression motif elicitation (DREME) (76) to the set of 540 upstream regions to identify putative Pdr802 binding motifs; these were highly enriched in GA (TC) (Fig. S7C).
Pdr802 ChIP-Seq, RNA-Seq, and dual-threshold optimization (DTO) data. Sheet 1, annotated peaks that occur in gene promoter regions that showed ≥2-fold enrichment when Chip-Seq was performed on strains expressing tagged versus untagged Pdr802. Sheet 2, ChIP-Seq primary data of Pdr802-specific peaks. Sheet 3, RNA-Seq data. Sheet 4, DTO filtered data. Sheet 5, DTO primary data. Sheet 6, ChIP-Seq data of all peaks in mCherry-Pdr802 samples. Sheet 7, ChIP-Seq data of all peaks in untagged (WT) control samples. Download Data Set S1, XLSX file, 2.5 MB (2.5MB, xlsx) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To complement our ChIP studies, we determined the set of genes regulated by Pdr802 under host-like conditions by performing RNA-Seq of WT and pdr802 cells after growth for 24 h in DMEM at 37°C and 5% CO2 (Data Set S1, sheet 3). We then used dual-threshold optimization (DTO) to analyze the RNA-Seq and ChIP-Seq data sets together. This statistical method allowed us to combine the evidence from binding and expression studies to converge on a set of direct and functional TF targets (77); because it uses two independent lines of evidence to suggest regulatory relationships, it increases confidence in highly significant small effects. The Pdr802 target genes yielded by this analysis include key players in multiple processes implicated in cryptococcal virulence, including quorum sensing, Titan cell formation, and stress resistance (Data Set S1, sheets 4 and 5).
Pdr802 regulates the expression of quorum sensing proteins that control Titan cell formation.
The most striking phenotype we observed in cells lacking PDR802 is the marked increase in Titan cell formation. We therefore examined our DTO target list for genes known to influence this phenotype, such as those involved in quorum sensing. Recent studies have shown that the quorum sensing peptide Qsp1 is a negative regulator of Titan cell formation (47, 48); Titan cell formation increases upon deletion of the gene encoding this peptide (QSP1) or proteins that mediate its maturation and import (PQP1 and OPT1, respectively). We found that Pdr802 positively regulates PQP1 and OPT1 gene expression (Table 1), consistent with its repression of Titan cell formation.
TABLE 1.
Pdr802 target genes involved in quorum sensing and Titan cell formation
| Biological process | CNAG IDa | Gene name | Change determined by: |
Description | |
|---|---|---|---|---|---|
| ChIP-Seqb | RNA-Seqc | ||||
| Quorum sensing | 00150 | PQP1 | 1.38 | −0.78 | Peptidase |
| 03013 | OPT1 | 1.25 | −0.55 | OPT small oligopeptide transporter | |
| Titan cell formation | 05835 | LIV3 | 1.52 | −0.84 | Transcription factor |
| 05785 | STB4 | 3.33 | −1.71 | Transcription factor | |
| 05940 | ZFC3/CQS2 | 2.23 | 0.68 | Transcription factor | |
CNAG, Cryptococcus neoformans serotype A genome project gene identifier (118).
Fold change for mCherry-Pdr802 compared to WT.
Log2 fold change for pdr802 compared to WT.
A study of C. neoformans cells exposed to Titan cell-inducing conditions in vitro reported that 562 genes were upregulated under this condition, while 421 genes were downregulated (48). The overlap of these genes with our DTO set of Pdr802-regulated genes included the TF genes LIV3, STB4, and ZFC3 (48) (Data Set S2, sheets 1 and 2). The first two are repressed during Titan cell induction, while ZFC3 (also known as CQS2) is induced. Our analysis showed that Pdr802 positively regulates expression of LIV3 and STB4, while it negatively regulates ZFC3 (Table 1), in concordance with our phenotypic observations of Titan cell formation. Notably, Liv3 and Zfc3 are responsive to the peptide Qsp1 (74, 75) and are important for C. neoformans virulence, while Stb4 influences cryptococcal brain infection (67).
Pdr802 coordinates cryptococcal response to the host environment.
C. neoformans deploys a variety of proteins to resist the many challenges it experiences upon host entry, which include oxidative and temperature stress. Multiple genes that are central to these responses were identified as direct, functional targets of Pdr802 by our DTO analysis (Table 2). For example, Pdr802 induces the expression of genes whose products detoxify reactive oxygen species (ROS), such as CAT1, CAT2, and SOD1 (78, 79), or participate in resistance to these compounds, such as FZC34, MIG1, and CCK1 (52, 80, 81) (Table 2). Both the kinase Cck1 (also known as Yck2) and the TF Fzc34 have been implicated in cryptococcal virulence (80, 82).
TABLE 2.
Genes regulated by Pdr802 involved in adaptation to the host environment
| Biological process | CNAG IDa | Gene name | Change determined by: |
Description | |
|---|---|---|---|---|---|
| ChIP-Seqb | RNA-Seqc | ||||
| Oxidative stress resistance | 04981 | CAT1 | 1.47 | −1.60 | Catalase 1 |
| 05256 | CAT2 | 1.41 | −0.42 | Catalase 2 | |
| 01019 | SOD1 | 2.59 | −0.53 | Superoxide dismutase (Cu-Zn) | |
| 00896 | FZC34 | 2.22 | −0.82 | Transcription factor | |
| 06327 | MIG1 | 1.72 | −0.78 | DNA-binding protein CreA | |
| 00556 | CCK1 | 1.78 | −0.48 | Casein kinase I | |
| Melanin and cell wall formation | 03202 | CAC1 | 1.60 | −0.61 | Adenylate cyclase |
| 01845 | PKC1 | 2.00 | −0.48 | AGC/PKC protein kinase | |
| 07724 | CUF1 | 1.27 | −0.67 | Metal-binding regulatory protein | |
| 00740 | SNF5 | 1.28 | −0.70 | Swi/Snf chromatin-remodeling subunit | |
| 01239 | CDA3 | 2.63 | 1.08 | Chitin deacetylase 3 | |
| 01230 | MP98 | 2.74 | 0.96 | Chitin deacetylase 2 | |
| 00776 | MP88 | 1.86 | 1.23 | Immunoreactive mannoprotein | |
| Growth at 37°C | 00405 | KIC1 | 1.88 | −0.97 | Ste/Ste20/Ysk protein kinase |
| 03670 | IRE1 | 1.66 | −1.14 | IRE protein kinase | |
| Urease activity | 07448 | DUR3 | 2.13 | −3.05 | Urea transporter |
| 01209 | FAB1 | 2.29 | −0.49 | 1-Phosphatidylinositol-3-P 5-kinase | |
| 01938 | KIN1 | 1.61 | −0.34 | CAMK/CAMKL/KIN1 protein kinase | |
| 01155 | GUT1 | 2.96 | 0.62 | Glycerol kinase | |
| 00791 | HLH1 | 1.71 | −1.09 | Transcription factor | |
| Capsule thickness | 02802 | ARG2 | 1.56 | −0.59 | Inositol/phosphatidylinositol kinase |
| 06809 | IKS1 | 2.28 | −0.44 | IKS protein kinase | |
| 01905 | KSP1 | 2.02 | −0.64 | Serine/threonine protein kinase | |
| 02877 | FZC51 | 1.75 | −0.69 | Transcription factor | |
| 07470 | PDE2 | 2.42 | −1.80 | High-affinity phosphodiesterase | |
| 03346 | BZP4 | 3.52 | 1.30 | Transcription factor | |
| Calcineurin signaling | 00156 | CRZ1 | 1.44 | −0.49 | Transcription factor |
| 01744 | HAD1 | 2.26 | 0.47 | Phosphatase | |
CNAG, Cryptococcus neoformans serotype A genome project gene identifier (118).
Fold change for mCherry-Pdr802 compared to WT.
Log2 fold change for pdr802 compared to WT.
As noted above, the pigment melanin has important antioxidant properties that promote cryptococcal survival inside the host (16). Under host-like conditions, Pdr802 regulates genes required for melanization, even though it melanizes normally in vitro. These genes include CAC1, PKC1, CUF1, and SNF5 (Table 2). Cac1 is an adenylyl cyclase responsible for cyclic AMP (cAMP) production in C. neoformans, which plays a central role in melanin synthesis as well as proper capsule production, mating, and virulence (83). The kinase Pkc1 induces production of the laccase (Lac1) that forms melanin and plays a key role in resistance to oxidative and nitrosative stress (84, 85), the TF Cuf1 regulates LAC1 expression and is important for cryptococcal virulence (86, 87), and SNF5 is required for full melanization (88). Melanin occurs in the fungal cell wall, which is another key component in fungal stress resistance. Pdr802 is also a direct, functional regulator of several genes whose products influence cell wall glycan content: two chitin deacetylases (Cda3 and Mp98) and the mannoprotein MP88 (Table 2). This is also consistent with the increases in mannose and chitin that occur in Titan cell walls (51).
Pdr802 positively regulates the expression of several proteins required for yeast survival at 37°C, including the kinases Kic1 and Ire1 (Table 2). Ire1 is a regulator of the cryptococcal unfolded-protein response (UPR) pathway and lack of Ire1 or Kic1 impact C. neoformans virulence (80, 89). Pdr802 also modulates cryptococcal urease activity, which is required for dissemination to the central nervous system (CNS) (11, 12), by regulating the urea transporter Dur3 and other proteins that influence urease activity (e.g., the kinases Fab1, Kin1, and Gut1 and the TF Hlh1) (Table 2). Deletion of FAB1, KIN1, or HLH1 impaired urease activity in C. neoformans, while GUT1 disruption induced it (52, 80).
Above, we document the role of Pdr802 in capsule synthesis, which is dramatically upregulated in the host environment in general and is further increased in cells lacking this TF. We found that Pdr802 is a positive regulator of multiple genes that have been implicated in reducing cryptococcal capsule thickness. These include the kinases Arg2, Iks1, and Ksp1; the TF Fzc51; and the phosphodiesterase Pde2 (Table 2). Notably, null mutants for those genes are hypercapsular, similar to pdr802 cells (52, 80, 90). Pdr802 is a negative regulator of the TF Bzp4, which, as mentioned above, positively regulates capsule (Table 2) (52).
Pdr802 regulates calcineurin target genes.
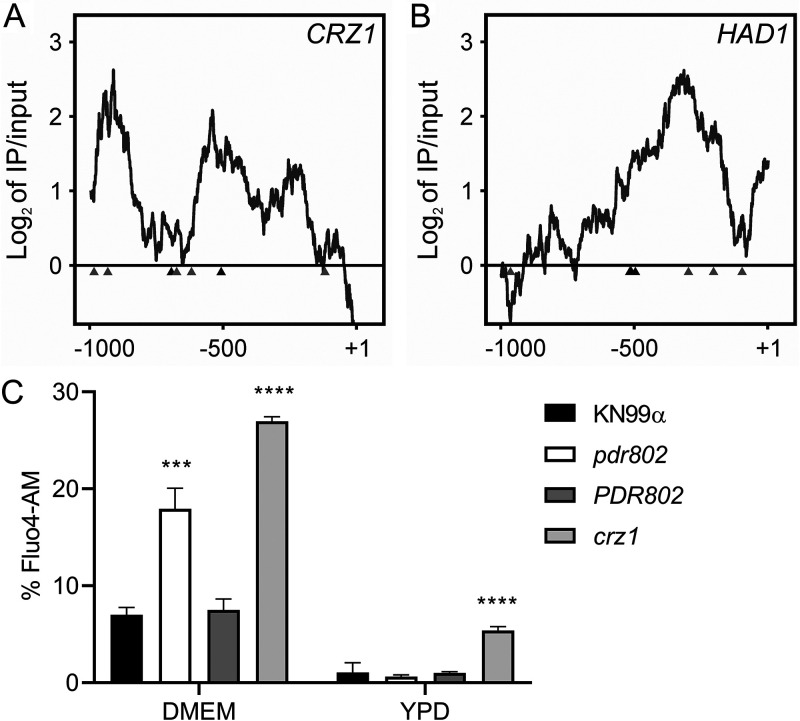
The calcineurin signaling pathway is activated by calcium and governs stress response and virulence in C. neoformans (91–93). One major mediator of calcineurin signaling is the transcription factor Crz1 mentioned above, which is highly responsive to temperature and influences cryptococcal virulence (57, 62). Upon intracellular calcium influx, calcineurin dephosphorylates Crz1, which then translocates to the nucleus and regulates gene expression (57, 63). We found that Pdr802 binds the CRZ1 gene promoter and positively regulates its expression (Fig. 8A, Table 2, and Data Set S2, sheet 3). Pdr802 also binds and regulates five other genes whose products are dephosphorylated by calcineurin; these include the phosphatase Had1, which is important for cryptococcal cell wall remodeling and virulence (Fig. 8B, Table 2, and Data Set S2, sheet 3) (63, 94).
FIG 8.
Pdr802 participates in calcineurin signaling. (A and B) Interactions of Pdr802 with upstream regions of the indicated genes. The ratios (log2) of reads from immunoprecipitated (IP) DNA to input DNA were calculated for 1,000 bp upstream of the first coding nucleotide (+1); shown is the difference in these values between tagged and untagged strains. Black triangles, complete Pdr802 DNA-binding motifs (Fig. S7C); gray triangles, partial motifs. (C) Intracellular calcium measurement by flow cytometry using Fluo-4AM. Each column shows the mean and standard deviation of three biological replicates. ***, P < 0.001, and ****, P < 0.0001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test.
Pdr802 target analysis and primers used in this study. Sheet 1, Pdr802 regulated genes that are down-regulated during Titan cell formation in vitro (48). Sheet 2, Pdr802 regulated genes that are up-regulated during Titan cell formation in vitro (48). Sheet 3, targets regulated by Pdr802 that are also dephosphorylated by calcineurin (63). Sheet 4, the intersection of Pdr802 targets and genes that are Crz1-independent calcineurin targets under conditions thermal stress (55). Sheet 5, primers used in this study. Fold enrichment and adjusted P (q) values throughout are for the DTO results. Download Data Set S2, XLSX file, 0.02 MB (26.6KB, xlsx) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Because Crz1 helps maintain normal cryptococcal Ca2+ concentrations through the regulation of calcium transporters (57), we wondered about the intracellular calcium levels in pdr802 cells. We found that indeed, after 24 h of growth in DMEM, the level of cytosolic calcium in the mutant significantly exceeded that of WT or complemented strains (Fig. 8C). It was still, however, below that of a crz1-null mutant, supporting the idea that Pdr802 is not the sole regulator of CRZ1 expression. Notably, PDR802 deletion had no effect in rich medium (YPD), which reinforces our hypothesis that Pdr802 acts primarily under host-like conditions. To further explore the relationship of Pdr802 and calcineurin, we compared published gene expression profiles of a calcineurin mutant (57) to our DTO data set. Of the 393 genes that are differently expressed in the calcineurin mutant under thermal stress, 26 are regulated by Pdr802 (Data Set S2, sheet 4).
DISCUSSION
We have shown that Pdr802 is a potent regulator of cryptococcal responses to the host environment. In this context, it influences the formation of capsule and Titan cells as well as cellular responses to temperature and oxidative stress, acts as a downstream effector of calcineurin, and modulates calcium availability. The last function is likely achieved through its positive regulation of the transcription factor Crz1, which in turn modulates the calcium transporters Pmc1 and Vcx1 (57). Since calcium ion is a major second messenger in eukaryotic cells, its accumulation in pdr802 cells affects multiple processes central to host interactions, including stress responses, cell wall integrity, and capsule size (61, 62, 92, 95, 96).
C. neoformans dissemination to the brain is the main driver of patient mortality (2). We found that dissemination of pdr802 cells is significantly impaired, although they do occasionally reach the brain. These observations can be explained by a combination of factors. First, the limited accumulation of the pdr802 mutant in the lungs, due to factors summarized above, may directly affect dissemination (97). Second, this strain survives poorly in mouse serum, as demonstrated directly by our culture experiments and indirectly by our inability to detect it in the blood of infected mice, even 75 days after infection. The latter might occur because the cells do not reach the blood or because they are rapidly eliminated, consistent with previous observations (98). Third, the thick capsules of the pdr802 mutant reduce its ability to reach the brain. This is true whether fungal entry occurs directly, by the movement of free fungi across the BBB, or indirectly, via a Trojan horse mechanism that requires macrophage uptake (99); such uptake is impeded by enlarged capsules, independent of cell size (31). Fourth, calcium imbalance directly affects cryptococcal transmigration (100). Finally, pdr802 cells show reduced expression of genes required for urease activity, which promotes C. neoformans dissemination to the CNS (11, 12, 100). Interestingly, despite all of these obstacles to dissemination, mutant cells that do reach the brain are able to proliferate to wild-type levels.
Titan cells are a robust and persistent morphotype of C. neoformans that contribute to yeast virulence (45). We showed that cells lacking Pdr802 demonstrate increased formation of Titan cells in vivo and in vitro, suggesting that this TF is a novel repressor of this process. Although Titan cells enhance aspects of cryptococcal pathogenesis (19, 101), their overproduction negatively impacts dissemination to the brain due to their resistance to phagocytosis by macrophages (19, 45) and decreased penetration of biological barriers (19).
Our combined analysis of DNA binding and gene expression data allows us to understand the increase in Titan cell formation that occurs upon deletion of PDR802. Under host-like conditions, Pdr802 positively regulates Pqp1, Opt1, and Liv3, all key proteins in the cryptococcal quorum sensing pathway, which represses Titan cell formation (47, 48). In the absence of this TF, quorum sensing is impaired, a situation known to increase Titan cell formation. Pdr802 may also indirectly regulate Titan cell formation by regulating other TFs that impact this process, such as Zfc3 (Cqs2) and Stb4.
We know that capsule, a key virulence factor, is typically highly induced in the host or under host-like conditions (102). Our studies in vitro, ex vivo, and in vivo show that Pdr802 normally reins in this process. This likely occurs via a combination of Pdr802’s repression of the TF Bzp4, which positively regulates capsule size, and the induction of other factors (e.g., the TF Fzc1, the phosphodiesterase Pde2, and the kinases Ksp1, Arg2, and Iks1) that negatively regulate capsule size (52, 80, 90).
Overall, we found that Pdr802 influences key cryptococcal phenotypes that influence virulence, including quorum sensing, stress responses, Titan cell formation, and capsule production (Fig. 9). We have further identified multiple genes that are central in these processes and are directly regulated by Pdr802. Some of these targets are also regulated by calcineurin (e.g., Had1 and Crz1) or by another important TF, Gat201 (e.g., Opt1, Liv3, and Zfc3) (60, 74, 75). Finally, the expression of PDR802 itself is regulated by the TFs Gat201 and Hob1 (67, 75). The cross talk between all of these regulatory mechanisms remains to be dissected. Nonetheless, it is evident that Pdr802 is critical for both survival in the lung and dissemination to the brain, thus explaining its role in cryptococcal virulence.
FIG 9.
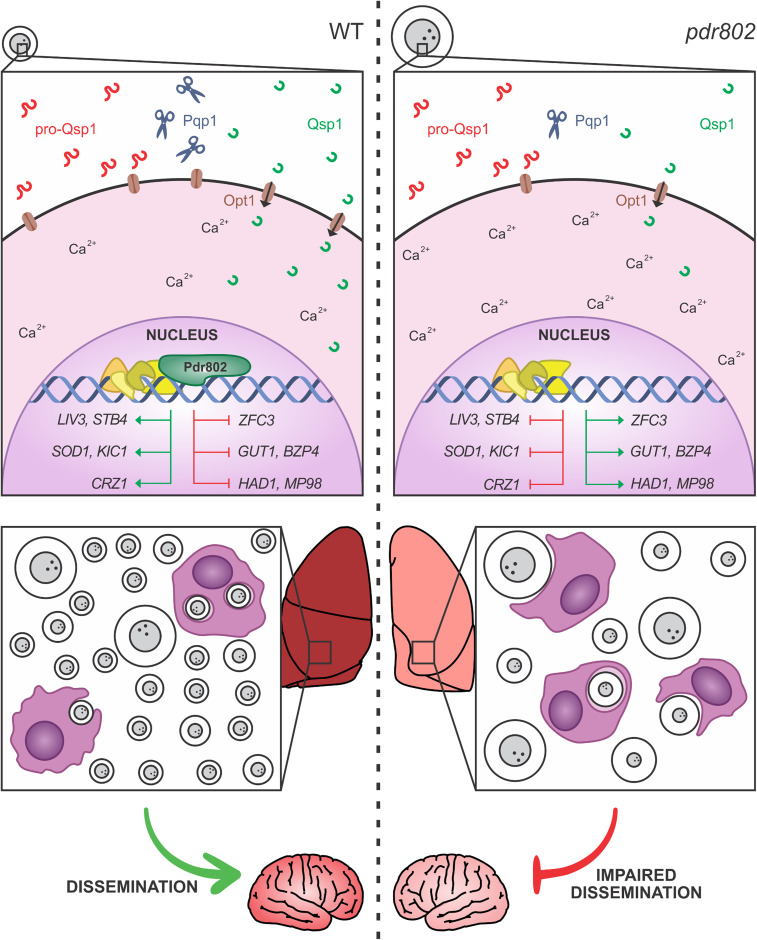
Pdr802 mode of action. (Left) When wild-type C. neoformans enters a host, PDR802 expression is induced and Pdr802 positively regulates elements of the quorum sensing pathway (described in the text) as well as expression of TFs implicated in this pathway (LIV3), Titan cell production (ZFC3), and brain infectivity (STB4). At the same time, Pdr802 regulates two calcineurin targets (CRZ1 and HAD1) and a variety of other genes (see the text). Shown are a few examples of genes involved in the response to oxidative stress (SOD1), growth at 37°C (KIC1), urease activity (GUT1), capsule production (BZP4), and cell wall remodeling (MP98). (Right) In the absence of these regulatory changes, pdr802 cells are poorly equipped to survive the stress of the host environment and are subject to increased intracellular calcium levels, dysregulation of capsule production, and impaired stress resistance. As a result, the cryptococcal population in the lung is smaller and is enriched in Titan cells and hypercapsular cells of normal size, both of which demonstrate reduced phagocytosis by host cells and impaired ability to cross biological barriers; these defects reduce dissemination to the central nervous system.
MATERIALS AND METHODS
Strain construction and cell growth.
We previously reported the PDR802 deletion mutant (pdr802) in the KN99α strain background (103) that was used in this work (39). Complementation of this mutant with the wild-type gene at the native locus (PDR802) and construction of a strain that expresses Pdr802 with N-terminal mCherry (mCherry-Pdr802) are detailed in the supplemental methods. For all studies, C. neoformans strains were inoculated from single colonies into YPD medium (2% [wt/vol] dextrose, 2% [wt/vol] Bacto peptone, and 1% [wt/vol] yeast extract in double-distilled water [ddH2O]) and grown overnight at 30°C with shaking at 230 rpm before further handling as detailed below. To assess viability during growth in tissue culture medium, overnight cultures were washed with phosphate-buffered saline (PBS), diluted to 106 cells/ml in DMEM (Sigma; D6429), plated (1 ml/well) in triplicate in 24-well plates, and incubated at 37°C and 5% CO2. At the indicated times, cells were mixed thoroughly, diluted in PBS, and plated on YPD agar (YPD medium, 2% agar [wt/vol]) for assessment of CFU. To assess viability during growth in mouse serum, mouse blood was collected as described below in “Animal experiments”; YPD-grown cryptococcal cells were incubated in 100 μl of serum in 96-well plates for 24 h at 37°C and 5% CO2, and CFU were counted as described above.
Animal experiments.
All animal protocols were approved by the Washington University Institutional Animal Care and Use Committee (reference 20170131) or Comissão de Ética no Uso de Animais (CEUA) (reference 30936), and care was taken to minimize handling and discomfort. For survival studies, groups of five 4- to 6-week-old female C57BL/6 mice (The Jackson Laboratory) were anesthetized by subcutaneous injection of 1.20 mg ketamine and 0.24 mg xylazine in 120 μl sterile water and intranasally infected with 5 × 104 cryptococcal cells. The mice were monitored and humanely sacrificed when their weight decreased to below 80% of initial weight or if they showed signs of disease, at which point organ burden was assessed. The lungs and brains were harvested, homogenized, diluted, and plated on YPD agar. The resulting CFU were enumerated, and survival differences were assessed by Kaplan-Meier analysis.
For timed organ burden studies, C. neoformans overnight cultures were centrifuged (1,000 × g for 3 min), washed with sterile PBS, and resuspended in PBS to 1 × 106 cells/ml. Groups of three 4- to 6-week-old female C57BL/6 mice (Centro Multidisciplinar para Investigação Biológica na Área da Ciência em Animais de Laboratório [CEMIB]) were anesthetized as described above, intranasally infected with 5 × 104 cryptococcal cells, and monitored as described above. At set time points post-infection (see above), mice were sacrificed and fungal burden was assessed from organs (as described above) or blood (obtained by cardiac puncture). Organ burden was analyzed by Kruskal-Wallis test with Dunn’s multiple comparison post hoc test for each day post-infection.
To assess cryptococcal viability in mouse serum, 6 BALB/c mice were anesthetized with isoflurane and blood was collected from the retro-orbital space using a sterile capillary tube. Collected blood was incubated at 37°C for 30 min, and serum was isolated by centrifugation at 1,000 × g for 15 min and then heat inactivated at 56°C for 30 min.
Capsule analysis.
To qualitatively assess capsule thickness, strains were grown on YPD medium for 16 h and washed with PBS, and 103 cells were incubated in mouse serum for 24 h at 37°C and 5% CO2. After incubation, cells were fixed in 4% paraformaldehyde and washed three times with PBS. C. neoformans cells were placed on glass slides and mixed with similar volumes of India ink, and the capsule was measured as previously described (104).
For population-level capsule measurement, C. neoformans strains were grown overnight in YPD, washed with PBS, and diluted to 106 cells/ml in DMEM. Aliquots (150 μl) were then plated in quadruplicate in the middle 32 wells of a poly-l-lysine-coated 96-well plate (Fisher; 655936) and incubated at 37°C and 5% CO2. After 24 h, the cells were washed with PBS and incubated with 150 μl of a staining mixture (100 μg/ml calcofluor white to stain cell walls, 50 μg/ml of the anticapsular monoclonal antibody 302 conjugated to Alexa Fluor 488 [Molecular Probes], and 1.5% goat serum in PBS) for 30 min at room temperature in the dark. The cells were washed again with PBS, fixed with 4% formaldehyde for 10 min at room temperature, and washed with PBS, and each well was refilled with 150 μl PBS. The cells were imaged using a BioTek Cytation 3 imager, which automatically collected 100 images per well in a grid pattern at the well center. Image files were prepared for analysis with GE Healthcare IN Cell Translator and assembled into .xdce image stacks for analysis with GE Healthcare IN Cell Developer Toolbox 1.9. Cell wall and capsule images were first filtered to remove background noise and border objects, and then cells were identified using shape-based object segmentation (3-pixel kernel, 50% sensitivity) followed by watershed clump breaking to prevent apparent connectivity caused by incomplete segmentation. Target linking was performed to assign each cell wall object to one capsule object based on known 1:1 pairing and location, generating a target set. Capsule and cell wall object diameters were calculated for each target set (hundreds to thousands per well), and the difference between measurements in each pair was defined as the capsule thickness. Data were normalized by the difference in capsule thickness between uninduced and induced WT cells, which were included in each experiment, and compared to data for hypercapsular (pkr1) (39) and hypocapsular (ada2) (105) control strains in each experiment. Capsule sizes were compared by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison post hoc test.
To measure capsule thickness of cryptococcal cells grown in the lungs of infected mice, lung homogenates were filtered through a cell strainer with 40-μm pores using a syringe plunger, fixed in 3.7% formaldehyde, and used for India ink staining and measurement as described above. For the visualization of KN99α and PDR802 cells from mouse lungs after 18 days of infection, the tissue was treated with 50 μg/ml DNase I for 30 min at 37°C.
GXM immunoblotting was conducted as previously described (71). Briefly, 106 cells/ml were grown in DMEM for 24 and 48 h. Culture supernatant fractions were then resolved by gel electrophoresis on 0.6% agarose, transferred onto nylon membranes, and probed with 1 μg/ml anti-GXM antibody 302.
Phenotypic assays.
For stress plates, cryptococcal cells were grown overnight in YPD, washed with PBS, and diluted to 107 cells/ml in PBS. Aliquots (3 μl) of 10-fold serial dilutions were spotted on YPD or YNB agar supplemented with various stressors (sorbitol, NaCl, CaCl2, LiCl, Congo red, calcofluor white, caffeine, SDS, NaNO2, H2O2, and ethanol) at the concentrations indicated in the figures. Melanization was tested on plates made by mixing 10 ml of 2× minimal medium (2 g/liter l-asparagine, 1 g/liter MgSO4 ⋅ 7H2O, 6 g/liter KH2PO4, 2 g/liter thiamine, 2 mM l-3,4-dihydroxyphenylalanine [l-DOPA]; 0.1% dextrose was added for melanization induction, and 0.5% was added for melanization inhibition) with 10 ml of 2% agar-water per plate. A control strain lacking the ability to melanize was used as a control (lac1) (88). For the solid urease assay, 10 μl of a 107 cells/ml suspension in water was plated on Christensen’s urea solid medium (1 g/liter peptone, 1 g/liter dextrose, 5 g/liter NaCl, 0.8 g/liter KH2PO4, 1.2 g/liter Na2HPO4, 0.012 g/liter phenol red and 15 g/liter agar; pH 6.8). Plates were incubated at 30°C or 37°C.
Titan cells.
Titan cell induction was performed in 1× PBS supplemented with 10% heat-inactivated fetal calf serum (FCS) for 72 h at 37°C and 5% CO2 as recently described (49) and quantified by flow cytometry as previously reported (47, 48). For analysis of DNA content, cells were treated as previously described (47) with a few modifications. Briefly, cultures were fixed with 70% ethanol for 1 h at 24°C and then overnight at 4°C, washed twice with PBS, and then treated with propidium iodide (50 μg/ml) and RNase A (0.5 mg/ml) in PBS for 2 h at 30°C with agitation. Quantification was performed using a FACS ARIA III (BD) cytometer with analysis on the Cytobank platform. A total of 20,000 events were recorded, and doublets were filtered out using comparisons of forward-scatter width (FSC-W) versus forward-scatter height (FSC-H) to exclude events with high FSC-W and FSC-H. To define the 1C and 2C gates, cells were grown in yeast nitrogen base (YNB) (24 h, 30°C, 150 rpm) without or with 80 μg/ml benomyl, which traps cells in 2C, as recently reported (106).
Phagocytosis.
J774.16 cells were prepared for uptake experiments by seeding (105 cells/well) in a 96-well plate and incubating in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2 for 24 h. C. neoformans cells were prepared for uptake experiments by inoculating an overnight culture in YPD into either DMEM or Titan cell induction medium (49) and growing at 37°C and 5% CO2 for 24 or 72 h, respectively. To initiate the study, cryptococcal cells were washed with PBS and opsonized with anti-capsular antibody 18B7 (1 μg/ml) for 1 h at 37°C, while macrophages were activated with 50 nM phorbol myristate acetate (PMA) for 1 h at 37°C and 5% CO2; 106 cryptococcal cells were then incubated with the macrophages for 2 h at 37°C and 5% CO2. The wells were then washed three times with warm PBS, and the macrophages were lysed with 0.1% Triton in PBS and plated for CFU enumeration as described above. Fold change in numbers of CFU was assessed by comparison to the CFU of opsonized cells. One-way ANOVA with Dunnett’s multiple-comparison post hoc test was used to compare phagocytosis of pdr802 and PDR802 strains with that of KN99α.
Chromatin immunoprecipitation.
ChIP studies were performed as previously described (39, 105). Briefly, wild type and N-terminal-mCherry-Pdr802 strains were cultivated in DMEM for 24 h at 37°C and 5% CO2. The cells were then fixed with formaldehyde and lysed by mechanical bead-beating, and the cell debris was removed by centrifugation. The supernatant fraction was sheared by sonication and centrifuged, and an aliquot was reserved as “input.” The remaining material was incubated with rabbit IgG anti-mCherry antibody (Abcam; ab213511) tethered to protein A Sepharose (IP) or Sepharose alone overnight at 4°C. The beads were then washed, incubated at 65°C to reverse DNA-DNA and DNA-protein cross-links, and the DNA was recovered by phenol-chloroform-isoamyl alcohol (25:24:1) extraction, ethanol precipitation, and resuspension in nuclease-free water.
Samples were submitted to the Washington University Genome Technology Access Center for library preparation, and DNA samples were sequenced using the Illumina NextSeq platform. The first replicate was sequenced using paired-end 2 × 75-bp reads, and replicates 2 and 3 were sequenced using single-end 75-bp reads; the minimum coverage obtained was ∼16×. The quality of the reads was evaluated by FastQC (107). Fastq files were aligned to the KN99 genome (108) using NextGenMap 0.5.3 (109). SAM files were converted to bam, reads were sorted and indexed, and read duplicates were removed from the final bam files using SAMtools (110). SAMtools was also used to filter out reads with a mapping quality of less than 20 phreds to guarantee single alignment of the reads. Peaks were called using MACS2 (2.1.1.20160309) (111), filtered by size (maximum threshold of 5 kb and no minimum), and annotated using Homer 4.8 (112). The significant peaks were chosen using the cutoff of fold enrichment above 2 and an adjusted P value of <0.05, and read coverage of each peak was obtained using SAMtools (110). Pdr802 binding motifs were identified using DREME (76); partial motifs were defined as at least 5 consecutive base pairs of the motif.
RNA-Seq and DTO.
RNA from wild-type and pdr802 cells grown for 24 h in DMEM (37°C, 5% CO2) was isolated and sequenced as previously described (39). Briefly, cDNA samples were sequenced using the Illumina NextSeq platform for paired-end 2 × 75 bp reads and read quality was evaluated by FastQC (107). Fastq files were aligned to the KN99 genome (108) using NovoAlign (113), SAM files were converted to bam, reads were sorted and indexed, and read duplicates were removed from the final bam files using SAMtools (110). The number of reads mapped per gene was calculated using HTSeq (114), and differential gene expression was analyzed with DESeq2 (115), using the independent hypothesis weighting (IHW) package to calculate the adjusted P values (116). Dual-threshold optimization (DTO) analysis was performed as recently described (77). This is a method for simultaneously finding the best thresholds for significance in a TF binding location data set (e.g., ChIP) and a TF perturbation response data set (e.g., RNA-Seq of a TF mutant). It works by trying out all pairs of thresholds for the two data sets, picking the pair that minimizes the probability of the overlap between the bound and responsive gene sets occurring by chance under a null model, and testing the significance of the overlap by comparison to randomly permuted data. Our application of DTO to our ChIP and RNA-Seq data yielded 1,455 bound genes, 5,186 responsive genes, and 1,167 genes that were both bound and responsive. Based on DTO, Pdr802 has an acceptable convergence from binding and perturbation, with a P value of <0.01 from the random permutation test and minimum expected false discovery rate (FDR) less than or equal to 20% at 80% sensitivity. In addition to requiring a statistically significant overlap between the ChIP-Seq and RNA-Seq gene sets, we filtered out any genes for which traditional differential expression analysis yielded an adjusted P value of ≤0.15 or absolute fold change of ≥0.3, leaving 380 bound targets.
Intracellular calcium measurement.
To measure intracellular free Ca2+, yeast cells were cultured overnight in YPD at 30°C with shaking, washed three times with deionized water, diluted to 106 cells/ml in DMEM (Sigma; D6429), plated (1 ml/well) in triplicate in 24-well plates, and incubated at 37°C and 5% CO2 for 24 h. At the indicated times, cells were mixed thoroughly, diluted in PBS containing 2 μM Fluo4-AM (Thermo Fisher), incubated at 30°C for 30 min, and analyzed using flow cytometry. The overnight culture was used as a control and treated as above.
Data availability.
ChIP-Seq and RNA-Seq data files are available at the NCBI Gene Expression Omnibus under accession numbers GSE153134 and GSE162851, respectively.
Supplemental methods. Download Text S1, PDF file, 0.04 MB (37.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We appreciate helpful discussions with Charley Christian Staats, Augusto Schrank, and members of the Doering, Kmetzsch and Brent labs. We are grateful for assistance from Thomas Hurtaux, Eamim Squizani, and Julia Sperotto with mouse experiments; Guohua Chen with spotting assays; Jessica Plaggenberg with library preparation and sequencing; Chase Mateusiak with RNA-Seq data analysis; and Sandeep Acharya for performing the DTO analysis. We thank Marilene Henning Vainstein and her lab for providing support with in vivo experiments. We also thank Liza Miller for comments on the manuscript, Arturo Casadevall for providing the antibody anti-GXM (18B7), and Pedro Brum for the assistance with the DNA analysis of Titan cells on the FACS ARIA III (BD) cytometer from the Department of Genetics of the Universidade Federal do Rio Grande do Sul.
These studies were supported by National Institutes of Health grant AI087794 to T.L.D. and M.R.B.; National Institutes of Health grants AI136688 and AI140979 to T.L.D.; and grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil; grant number 310510/2018-0), and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS, Brazil) to L.K. CAPES fully supported J.C.V.R. during her studies in Brazil; her studies in the United States were partially supported by this source (Advanced Network of Computational Biology, RABICÓ, Biocomputational Grant 23038.010041/2013-13).
Conceived and designed experiments: J.C.V.R., D.P.A., A.L.C., M.R.B., L.K., and T.L.D.; Performed experiments: J.C.V.R., D.P.A., H.M., H.B., and A.L.C; Analyzed data: J.C.V.R., D.P.A., A.L.C., M.R.B., L.K., and T.L.D.; Contributed reagents and materials: M.R.B., L.K., and T.L.D.; Drafted the paper: J.C.V.R, L.K., and T.L.D.; Revised the paper: J.C.V.R., D.P.A., M.R.B., L.K., and T.L.D.
We declare no competing financial interests.
Footnotes
Citation Reuwsaat JCV, Agustinho DP, Motta H, Chang AL, Brown H, Brent MR, Kmetzsch L, Doering TL. 2021. The transcription factor Pdr802 regulates Titan cell formation and pathogenicity of Cryptococcus neoformans. mBio 12:e03457-20. https://doi.org/10.1128/mBio.03457-20.
REFERENCES
- 1.Kwon-Chung KJ, Fraser JA, Doering TL, Wang ZA, Janbon G, Idnurm A, Bahn YS. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med 4:a019760. 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballou ER, Johnston SA. 2017. The cause and effect of Cryptococcus interactions with the host. Curr Opin Microbiol 40:88–94. 10.1016/j.mib.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Sabiiti W, May RC. 2012. Mechanisms of infection by the human fungal pathogen Cryptococcus neoformans. Future Microbiol 7:1297–1313. 10.2217/fmb.12.102. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans Infection. J Clin Microbiol 37:3204–3209. 10.1128/JCM.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YC, Stins MF, McCaffery MJ, Miller GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS, Kwon-Chung KJ, Paul-Satyasee M. 2004. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect Immun 72:4985–4995. 10.1128/IAI.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S-H, Long M, Wu C-H, Kwon-Chung KJ, Chang YC, Chi F, Lee S, Jong A. 2011. Invasion of Cryptococcus neoformans into human brain microvascular endothelial cells is mediated through the lipid rafts-endocytic pathway via the dual specificity tyrosine phosphorylation-regulated kinase 3 (DYRK3). J Biol Chem 286:34761–34769. 10.1074/jbc.M111.219378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jong A, Wu C-H, Shackleford GM, Kwon-Chung KJ, Chang YC, Chen H-M, Ouyang Y, Huang S-H. 2008. Involvement of human CD44 during Cryptococcus neoformans infection of brain microvascular endothelial cells. Cell Microbiol 10:1313–1326. 10.1111/j.1462-5822.2008.01128.x. [DOI] [PubMed] [Google Scholar]
- 9.Maruvada R, Zhu L, Pearce D, Zheng Y, Perfect J, Kwon-Chung KJ, Kim KS. 2012. Cryptococcus neoformans phospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cell Microbiol 14:1544–1553. 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen SHM, Stins MF, Huang S-H, Chen YH, Kwon-Chung KJ, Chang Y, Kim KS, Suzuki K, Jong AY. 2003. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J Med Microbiol 52:961–970. 10.1099/jmm.0.05230-0. [DOI] [PubMed] [Google Scholar]
- 11.Olszewski MA, Noverr MC, Chen G-H, Toews GB, Cox GM, Perfect JR, Huffnagle GB. 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol 164:1761–1771. 10.1016/S0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J Clin Invest 120:1683–1693. 10.1172/JCI41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago-Tirado FH, Onken MD, Cooper JA, Klein RS, Doering TL. 2017. Trojan horse transit contributes to blood-brain barrier crossing of a eukaryotic pathogen. mBio 8:e02183-16. 10.1128/mBio.02183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis JN, Bicanic T, Loyse A, Namarika D, Jackson A, Nussbaum JC, Longley N, Muzoora C, Phulusa J, Taseera K, Kanyembe C, Wilson D, Hosseinipour MC, Brouwer AE, Limmathurotsakul D, White N, van der Horst C, Wood R, Meintjes G, Bradley J, Jaffar S, Harrison T. 2014. Determinants of mortality in a combined cohort of 501 patients with HIV-associated cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis 58:736–745. 10.1093/cid/cit794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen M-H, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC, TCS. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 50:291–322. 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nosanchuk JD, Casadevall A. 2003. The contribution of melanin to microbial pathogenesis. Cell Microbiol 5:203–223. 10.1046/j.1462-5814.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- 17.Singh A, Panting RJ, Varma A, Saijo T, Waldron KJ, Jong A, Ngamskulrungroj P, Chang YC, Rutherford JC, Kwon-Chung KJ. 2013. Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. mBio 4:e00220-13. 10.1128/mBio.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang YC, Kwon-Chung KJ. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol 14:4912–4919. 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6:e1000953. 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaragoza O, Nielsen K. 2013. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr Opin Microbiol 16:409–413. 10.1016/j.mib.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Aisen P, Casadevall A. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun 63:3131–3136. 10.1128/IAI.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mednick AJ, Nosanchuk JD, Casadevall A. 2005. Melanization of Cryptococcus neoformans affects lung inflammatory responses during cryptococcal infection. Infect Immun 73:2012–2019. 10.1128/IAI.73.4.2012-2019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Casadevall A. 1994. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun 62:3004–3007. 10.1128/IAI.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doering TL, Nosanchuk JD, Roberts WK, Casadevall A. 1999. Melanin as a potential cryptococcal defence against microbicidal proteins. Med Mycol 37:175–181. 10.1046/j.1365-280X.1999.00218.x. [DOI] [PubMed] [Google Scholar]
- 25.Agustinho DP, Nosanchuk JD. 2017. Functions of fungal melanins: reference module in life sciences. Elsevier, New York, NY. [Google Scholar]
- 26.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot Cell 2:655–663. 10.1128/ec.2.4.655-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZA, Li LX, Doering TL. 2018. Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 28:719–730. 10.1093/glycob/cwy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agustinho DP, Miller LC, Li LX, Doering TL. 2018. Peeling the onion: the outer layers of Cryptococcus neoformans. Mem Inst Oswaldo Cruz 113:e180040. 10.1590/0074-02760180040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulmer GS, Sans MD. 1967. Cryptococcus neoformans. II. Phagocytosis by human leukocytes. J Bacteriol 94:1480–1483. 10.1128/JB.94.5.1480-1483.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulmer GS, Sans MD. 1968. Cryptococcus neoformans. J Bacteriol 95:5–8. 10.1128/JB.95.1.5-8.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tacker JR, Farhi F, Bulmer GS. 1972. Intracellular fate of Cryptococcus neoformans. Infect Immun 6:162–167. 10.1128/IAI.6.2.162-167.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monari C, Bistoni F, Vecchiarelli A. 2006. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res 6:537–542. 10.1111/j.1567-1364.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- 34.Hayes JB, Sircy LM, Heusinkveld LE, Ding W, Leander RN, McClelland EE, Nelson DE. 2016. Modulation of macrophage inflammatory nuclear factor κB (NF-κB) signaling by intracellular Cryptococcus neoformans. J Biol Chem 291:15614–15627. 10.1074/jbc.M116.738187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fonseca FL, Nohara LL, Cordero RJB, Frases S, Casadevall A, Almeida IC, Nimrichter L, Rodrigues ML. 2010. Immunomodulatory effects of serotype B glucuronoxylomannan from Cryptococcus gattii correlate with polysaccharide diameter. Infect Immun 78:3861–3870. 10.1128/IAI.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Hermoso D, Dromer F, Janbon G. 2004. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect Immun 72:3359–3365. 10.1128/IAI.72.6.3359-3365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guimarães AJ, Frases S, Cordero RJB, Nimrichter L, Casadevall A, Nosanchuk JD. 2010. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol 12:740–753. 10.1111/j.1462-5822.2010.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordero RJB, Pontes B, Guimarães AJ, Martinez LR, Rivera J, Fries BC, Nimrichter L, Rodrigues ML, Viana NB, Casadevall A. 2011. Chronological aging is associated with biophysical and chemical changes in the capsule of Cryptococcus neoformans. Infect Immun 79:4990–5000. 10.1128/IAI.05789-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maier EJ, Haynes BC, Gish SR, Wang ZA, Skowyra ML, Marulli AL, Doering TL, Brent MR. 2015. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res 25:690–700. 10.1101/gr.184101.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos JRA, Holanda RA, Frases S, Bravim M, Araujo GDS, Santos PC, Costa MC, Ribeiro MJA, Ferreira GF, Baltazar LM, Miranda AS, Oliveira DB, Santos CMA, Fontes ACL, Gouveia LF, Resende-Stoianoff MA, Abrahão JS, Teixeira AL, Paixão TA, Souza DG, Santos DA. 2014. Fluconazole alters the polysaccharide capsule of Cryptococcus gattii and leads to distinct behaviors in murine cryptococcosis. PLoS One 9:e112669. 10.1371/journal.pone.0112669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silveira CP, Piffer AC, Kmetzsch L, Fonseca FL, Soares DA, Staats CC, Rodrigues ML, Schrank A, Vainstein MH. 2013. The heat shock protein (Hsp) 70 of Cryptococcus neoformans is associated with the fungal cell surface and influences the interaction between yeast and host cells. Fungal Genet Biol 60:53–63. 10.1016/j.fgb.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Alspaugh JA. 2015. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol 78:55–58. 10.1016/j.fgb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García-Rodas R, de Oliveira H, Trevijano-Contador N, Zaragoza O. 2018. Cryptococcal Titan cells: when yeast cells are all grown up. Curr Top Microbiol Immunol 422:101–120. 10.1007/82_2018_145. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, Ballou ER. 2018. The Cryptococcus neoformans Titan cell: from in vivo phenomenon to in vitro model. Curr Clin Micro Rpt 5:252–260. 10.1007/s40588-018-0107-9. [DOI] [Google Scholar]
- 45.Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6:e1000945. 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, Fraser JA, Berman J, Nielsen K. 2015. Polyploid Titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6:e01340-15. 10.1128/mBio.01340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclère A, Janbon G, Perfect JR, Fraser JA, Casadevall A, Cuomo CA, Dromer F, Nielsen K, Alanio A. 2018. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog 14:e1006982. 10.1371/journal.ppat.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trevijano-Contador N, de Oliveira HC, García-Rodas R, Rossi SA, Llorente I, Zaballos Á, Janbon G, Ariño J, Zaragoza Ó. 2018. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog 14:e1007007. 10.1371/journal.ppat.1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, Harrison TS, Jaspars M, May RC, Brown GD, Yuecel R, MacCallum DM, Ballou ER. 2018. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog 14:e1006978. 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Barbazán I, Trevijano-Contador N, Rueda C, de Andrés B, Pérez-Tavárez R, Herrero-Fernández I, Gaspar ML, Zaragoza O. 2016. The formation of titan cells in Cryptococcus neoformans depends on the mouse strain and correlates with induction of Th2-type responses. Cell Microbiol 18:111–124. 10.1111/cmi.12488. [DOI] [PubMed] [Google Scholar]
- 51.Mukaremera L, Lee KK, Wagener J, Wiesner DL, Gow NAR, Nielsen K. 2018. Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf 1:15–24. 10.1016/j.tcsw.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung K-W, Yang D-H, Maeng S, Lee K-T, So Y-S, Hong J, Choi J, Byun H-J, Kim H, Bang S, Song M-H, Lee J-W, Kim MS, Kim S-Y, Ji J-H, Park G, Kwon H, Cha S, Meyers GL, Wang LL, Jang J, Janbon G, Adedoyin G, Kim T, Averette AK, Heitman J, Cheong E, Lee Y-H, Lee Y-W, Bahn Y-S. 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun 6:6757. 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider R, de O, Fogaça N, de SS, Kmetzsch L, Schrank A, Vainstein MH, Staats CC. 2012. Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii. PLoS One 7:e43773. 10.1371/journal.pone.0043773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Santamarina S, Festa RA, Smith AD, Yu C-H, Probst C, Ding C, Homer CM, Yin J, Noonan JP, Madhani H, Perfect JR, Thiele DJ. 2018. Genome-wide analysis of the regulation of Cu metabolism in Cryptococcus neoformans. Mol Microbiol 108:473–494. 10.1111/mmi.13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lev S, Kaufman-Francis K, Desmarini D, Juillard PG, Li C, Stifter SA, Feng CG, Sorrell TC, Grau GER, Bahn Y-S, Djordjevic JT. 2017. Pho4 is essential for dissemination of Cryptococcus neoformans to the host brain by promoting phosphate uptake and growth at alkaline pH. mSphere 2:e00381-16. 10.1128/mSphere.00381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung WH, Sham A, White R, Kronstad JW. 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4:e410. 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow EWL, Clancey SA, Billmyre RB, Averette AF, Granek JA, Mieczkowski P, Cardenas ME, Heitman J. 2017. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet 13:e1006667. 10.1371/journal.pgen.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6:e1000776. 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gish SR, Maier EJ, Haynes BC, Santiago-Tirado FH, Srikanta DL, Ma CZ, Li LX, Williams M, Crouch EC, Khader SA, Brent MR, Doering TL. 2016. Computational analysis reveals a key regulator of cryptococcal virulence and determinant of host response. mBio 7:e00313-16. 10.1128/mBio.00313-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chun CD, Brown JCS, Madhani HD. 2011. A major role for capsule-independent phagocytosis-inhibitory mechanisms in mammalian infection by Cryptococcus neoformans. Cell Host Microbe 9:243–251. 10.1016/j.chom.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moranova Z, Virtudazo E, Hricova K, Ohkusu M, Kawamoto S, Husickova V, Raclavsky V. 2014. The CRZ1/SP1-like gene links survival under limited aeration, cell integrity and biofilm formation in the pathogenic yeast Cryptococcus neoformans. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 158:212–220. 10.5507/bp.2013.024. [DOI] [PubMed] [Google Scholar]
- 62.Lev S, Desmarini D, Chayakulkeeree M, Sorrell TC, Djordjevic JT. 2012. The Crz1/Sp1 transcription factor of Cryptococcus neoformans is activated by calcineurin and regulates cell wall integrity. PLoS One 7:e51403. 10.1371/journal.pone.0051403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park HS, Chow EWL, Fu C, Soderblom EJ, Moseley MA, Heitman J, Cardenas ME. 2016. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog 12:e1005873. 10.1371/journal.ppat.1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee D, Jang E-H, Lee M, Kim S-W, Lee Y, Lee K-T, Bahn Y-S. 2019. Unraveling melanin biosynthesis and signaling networks in Cryptococcus neoformans. mBio 10:e02267-19. 10.1128/mBio.02267-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhodes J, Desjardins CA, Sykes SM, Beale MA, Vanhove M, Sakthikumar S, Chen Y, Gujja S, Saif S, Chowdhary A, Lawson DJ, Ponzio V, Colombo AL, Meyer W, Engelthaler DM, Hagen F, Illnait-Zaragozi MT, Alanio A, Vreulink J-M, Heitman J, Perfect JR, Litvintseva AP, Bicanic T, Harrison TS, Fisher MC, Cuomo CA. 2017. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics 207:327–346. 10.1534/genetics.117.203836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee K-T, Hong J, Lee D-G, Lee M, Cha S, Lim Y-G, Jung K-W, Hwangbo A, Lee Y, Yu S-J, Chen Y-L, Lee J-S, Cheong E, Bahn Y-S. 2020. Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat Commun 11:1521. 10.1038/s41467-020-15329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krysan DJ, Zhai B, Beattie SR, Misel KM, Wellington M, Lin X. 2019. Host carbon dioxide concentration is an independent stress for Cryptococcus neoformans that affects virulence and antifungal Susceptibility. mBio 10:e01410-19. 10.1128/mBio.01410-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 21:3179–3191. 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu G, Steen BR, Lian T, Sham AP, Tam N, Tangen KL, Kronstad JW. 2007. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog 3:e42. 10.1371/journal.ppat.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoneda A, Doering TL. 2008. Regulation of Cryptococcus neoformans capsule size is mediated at the polymer level. Eukaryot Cell 7:546–549. 10.1128/EC.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frases S, Pontes B, Nimrichter L, Viana NB, Rodrigues ML, Casadevall A. 2009. Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc Natl Acad Sci U S A 106:1228–1233. 10.1073/pnas.0808995106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaylord EA, Choy HL, Doering TL. 2020. Dangerous liaisons: interactions of Cryptococcus neoformans with host phagocytes. Pathogens 9:891. 10.3390/pathogens9110891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Summers DK, Perry DS, Rao B, Madhani HD. 2020. Coordinate genomic association of transcription factors controlled by an imported quorum sensing peptide in Cryptococcus neoformans. PLoS Genet 16:e1008744. 10.1371/journal.pgen.1008744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Homer CM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, Moresco JJ, Toffaletti D, Upadhya R, Caradonna I, Petnic S, Pessino V, Cuomo CA, Lodge JK, Perfect J, Yates JR, III, Nielsen K, Craik CS, Madhani HD. 2016. Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe 19:849–864. 10.1016/j.chom.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27:1653–1659. 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang Y, Patel NR, Shively C, Recio PS, Chen X, Wranik BJ, Kim G, Scott McIsaac R, Mitra R, Brent MR. 2020. Dual threshold optimization and network inference reveal convergent evidence from TF binding locations and TF perturbation responses. Genome Res 30:459–471. 10.1101/gr.259655.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giles SS, Stajich JE, Nichols C, Gerrald QD, Alspaugh JA, Dietrich F, Perfect JR. 2006. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell 5:1447–1459. 10.1128/EC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun 71:173–180. 10.1128/iai.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee K-T, So Y-S, Yang D-H, Jung K-W, Choi J, Lee D-G, Kwon H, Jang J, Wang LL, Cha S, Meyers GL, Jeong E, Jin J-H, Lee Y, Hong J, Bang S, Ji J-H, Park G, Byun H-J, Park SW, Park Y-M, Adedoyin G, Kim T, Averette AF, Choi J-S, Heitman J, Cheong E, Lee Y-H, Bahn Y-S. 2016. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun 7:12766. 10.1038/ncomms12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caza M, Hu G, Price M, Perfect JR, Kronstad JW. 2016. The zinc finger protein Mig1 regulates mitochondrial function and azole drug susceptibility in the pathogenic fungus Cryptococcus neoformans. mSphere 1:e00080-15. 10.1128/mSphere.00080-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y, Toffaletti DL, Tenor JL, Litvintseva AP, Fang C, Mitchell TG, McDonald TR, Nielsen K, Boulware DR, Bicanic T, Perfect JR. 2014. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 5:e01087-13. 10.1128/mBio.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 1:75–84. 10.1128/ec.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heung LJ, Kaiser AE, Luberto C, Del Poeta M. 2005. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J Biol Chem 280:28547–28555. 10.1074/jbc.M503404200. [DOI] [PubMed] [Google Scholar]
- 85.Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. 2008. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 7:1685–1698. 10.1128/EC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Waterman SR, Hacham M, Hu G, Zhu X, Park Y-D, Shin S, Panepinto J, Valyi-Nagy T, Beam C, Husain S, Singh N, Williamson PR. 2007. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest 117:794–802. 10.1172/JCI30006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang N, Sun N, Xiao D, Pan J, Wang Y, Zhu X. 2009. A copper-responsive factor gene CUF1 required for copper induction of laccase in Cryptococcus neoformans. FEMS Microbiol Lett 296:84–90. 10.1111/j.1574-6968.2009.01619.x. [DOI] [PubMed] [Google Scholar]
- 88.Walton FJ, Idnurm A, Heitman J. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 57:1381–1396. 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- 89.Cheon SA, Jung K-W, Chen Y-L, Heitman J, Bahn Y-S, Kang HA. 2011. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog 7:e1002177. 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hicks JK, Bahn YS, Heitman J. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot Cell 4:1971–1981. 10.1128/EC.4.12.1971-1981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kozubowski L, Lee SC, Heitman J. 2009. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol 11:370–380. 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fox DS, Cruz MC, Sia RAL, Ke H, Cox GM, Cardenas ME, Heitman J. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol Microbiol 39:835–849. 10.1046/j.1365-2958.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 93.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16:2576–2589. 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jung W-H, Son Y-E, Oh S-H, Fu C, Kim HS, Kwak J-H, Cardenas ME, Heitman J, Park HS. 2018. Had1 is required for cell wall integrity and fungal virulence in Cryptococcus neoformans. G3 (Bethesda) 8:643–652. 10.1534/g3.117.300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kmetzsch L, Staats CC, Cupertino JB, Fonseca FL, Rodrigues ML, Schrank A, Vainstein MH. 2013. The calcium transporter Pmc1 provides Ca2+ tolerance and influences the progression of murine cryptococcal infection. FEBS J 280:4853–4864. 10.1111/febs.12458. [DOI] [PubMed] [Google Scholar]
- 96.Kmetzsch L, Staats CC, Simon E, Fonseca FL, de Oliveira DL, Sobrino L, Rodrigues J, Leal AL, Nimrichter L, Rodrigues ML, Schrank A, Vainstein MH. 2010. The vacuolar Ca2+ exchanger Vcx1 is involved in calcineurin-dependent Ca2+ tolerance and virulence in Cryptococcus neoformans. Eukaryot Cell 9:1798–1805. 10.1128/EC.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Denham S, Brown J. 2018. Mechanisms of pulmonary escape and dissemination by Cryptococcus neoformans. J Fungi (Basel) 4:25. 10.3390/jof4010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun D, Sun P, Li H, Zhang M, Liu G, Strickland AB, Chen Y, Fu Y, Xu J, Yosri M, Nan Y, Zhou H, Zhang X, Shi M. 2019. Fungal dissemination is limited by liver macrophage filtration of the blood. Nat Commun 10:4566. 10.1038/s41467-019-12381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Santiago-Tirado FH, Doering TL. 2017. False friends: phagocytes as Trojan horses in microbial brain infections. PLoS Pathog 13:e1006680. 10.1371/journal.ppat.1006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Squizani ED, Oliveira NK, Reuwsaat JCV, Marques BM, Lopes W, Gerber AL, de Vasconcelos ATR, Lev S, Djordjevic JT, Schrank A, Vainstein MH, Staats CC, Kmetzsch L. 2018. Cryptococcal dissemination to the central nervous system requires the vacuolar calcium transporter Pmc1. Cell Microbiol 20:e12803. 10.1111/cmi.12803. [DOI] [PubMed] [Google Scholar]
- 101.Okagaki LH, Nielsen K. 2012. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 11:820–826. 10.1128/EC.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Srikanta D, Santiago-Tirado FH, Doering TL. 2014. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast 31:47–60. 10.1002/yea.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71:4831–4841. 10.1128/iai.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reuwsaat JCV, Motta H, Garcia AWA, Vasconcelos CB, Marques BM, Oliveira NK, Rodrigues J, Ferrareze PAG, Frases S, Lopes W, Barcellos VA, Squizani ED, Horta JA, Schrank A, Rodrigues ML, Staats CC, Vainstein MH, Kmetzsch L. 2018. A predicted mannoprotein participates in Cryptococcus gattii capsular structure. mSphere 3:e00023-18. 10.1128/mSphere.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, Brent MR, Doering TL. 2011. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog 7:e1002411. 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Denham ST, Brammer B, Chung KY, Wambaugh MA, Bednarek J, Guo L, Brown JCS. 2020. Phosphate induces a morphological shift that enhances vascular dissemination of Cryptococcus neoformans. bioRxiv 2020.08.27.270843.
- 107.Babraham Bioinformatics. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 24 June 2019.
- 108.Friedman RZ, Gish SR, Brown H, Brier L, Howard N, Doering TL, Brent MR. 2018. Unintended side effects of transformation are very rare in Cryptococcus neoformans. G3 (Bethesda) 8:815–822. 10.1534/g3.117.300357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sedlazeck FJ, Rescheneder P, von Haeseler A. 2013. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29:2790–2791. 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- 110.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Novocraft Technologies. 2014. NovoAlign. http://www.novocraft.com/. Accessed 25 November 2020.
- 114.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ignatiadis N, Klaus B, Zaugg JB, Huber W. 2016. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat Methods 13:577–580. 10.1038/nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li LX, Rautengarten C, Heazlewood JL, Doering TL. 2018. Xylose donor transport is critical for fungal virulence. PLoS Pathog 14:e1006765. 10.1371/journal.ppat.1006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, Bahn Y-S, Chen W, Chen Y, Chow EWL, Coppée J-Y, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh Y-P, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, Cuomo CA, Dietrich FS. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutant strain construction and confirmation. (A) Scheme for generating C. neoformans strains in the KN99α background (middle) that either lack PDR802 (pdr802, top) or encode a tagged copy of the protein (mCherry-PDR802). (B) Qualitative analysis of gene expression in panel A strains and the complemented pdr802 mutant (PDR802). Cryptococcal mRNA isolated from cells grown in DMEM (37°C, 5% CO2, 24 h) was used to generate cDNA; from this, segments of the genes indicated at the left were amplified using the primers listed in Data Set S2, sheet 5, and the products were analyzed by agarose gel electrophoresis. Fragment sizes (in base pairs) are indicated, and the ladder bands shown are 400, 500, 650, 850, and 1,000 bp for the top panel; 200, 300, 400, 500, and 650 bp for the middle panel; and 100, 200, and 300 bp for the bottom panel. (C) Quantitative analysis of PDR802 expression. Samples of RNA isolated as for panel B were analyzed for PDR802 expression by qRT-PCR. All results were normalized to ACT1 expression. Each symbol represents a biological replicate, with the mean and standard deviation also shown. ***, P < 0.001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test. Download FIG S1, PDF file, 0.5 MB (536.1KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Organ burdens. (A) Mean and SD values of total CFU in the indicated tissue of mice from the Fig. 1 survival curve are shown. Each point is the average value for a single animal at the time of death. For pdr802 strain infections, red circles represent mice sacrificed at days 65 and 69, while blue circles represent mice sacrificed at the termination of the study (day 100). (B) Mean and SD of total CFU in the blood and brain at the indicated times post-infection. (C) Mean and SD of total CFU in the lung, blood, and brain 75 days after infection with the pdr802 strain. Each color represents one mouse. Download FIG S2, PDF file, 0.9 MB (948.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of pdr802 cells. (A to C) Tenfold serial dilutions of WT, pdr802, and PDR802 cells were plated on the media shown and incubated at 30°C (A), 37°C (B), or 37°C in the presence of 5% CO2 (C). Nitrosative (NaNO2) and oxidative (H2O2) stress plates were prepared with YNB medium, and melanization plates containing l-DOPA were prepared as described in Materials and Methods; all other plates were prepared with YPD medium. The lac1 control strain lacks the ability to melanize (88). (D) Urease activity of the indicated strains was evaluated using Christensen’s urea solid medium (see Materials and Methods) at the indicated temperatures. The ure1 control strain does not produce urease (17). Download FIG S3, PDF file, 0.8 MB (795.3KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth curves and capsule shedding. (A and B) Growth of the indicated strains in YPD at 30°C (A) or DMEM at 37°C and 5% CO2 (B) was assessed by determining the optical density at 600 nm (OD600) at the times indicated. (C) Conditioned medium from the indicated strains was probed for the presence of GXM after growth in DMEM for 24 or 48 h. Equal volumes of culture supernatant were analyzed without normalization to cell density. Immunoblotting was performed using the anti-GXM monoclonal antibody 302. Download FIG S4, PDF file, 0.6 MB (577.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Semi-automated assay for cryptococcal capsule imaging. (A) Schematic of applying this method to cryptococcal cells induced to form capsule by growth in DMEM (37°C, 5% CO2) for 24 h, followed by cell wall and capsule staining. Thousands of cells may be imaged per well and analyzed automatically with software that annotates and measures the capsule (annotated in dark blue) and cell wall (annotated in bright green). See Materials and Methods for details. (B) Capsule size distribution of WT cells after induction. Capsule thickness for each cell is the difference between the paired diameters of the cell wall and capsule, which is plotted with reference to the mean value. (C and D) Mean and SD (C) and cumulative percentage (D) analysis of WT compared to hyper- and hypocapsular control strains (here, pkr1 and ada2, respectively). Capsule thickness is in arbitrary units, related to the pixels measured. (E) Time required to analyze the capsule thickness of 1,000 cells by this method compared to manual assessment of India ink images. Download FIG S5, PDF file, 0.7 MB (688.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PDR802 deletion induces the formation of Titan cells. Mean and SD of (A) total cell diameter and (B) ratio of total cell to cell body diameters (diameter ratio), assessed by measuring at least 50 cells per strain with ImageJ. **, P < 0.01, and ****, P < 0.0001, compared to KN99α by one-way ANOVA with a post hoc Dunnett test for each day post-infection. (C) Percent of Titan cells in the indicated strain, evaluated using various published parameters: cell body diameter bigger than 10 or 15 μm (20) or total cell diameter bigger than 30 μm (43). Download FIG S6, PDF file, 0.7 MB (702.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain viability, Pdr802 interaction with its own promoter, and putative DNA-binding motifs. (A) The indicated strains were grown in DMEM at 37°C and 5% CO2 for the times shown, and samples were tested for their ability to form colonies on YPD medium. Plotted is the fold change in CFU relative to the initial culture. (B) Pdr802 self-regulation. The ratios (log2) of reads from immunoprecipitated (IP) DNA to reads from input DNA were calculated for 1,000 bp upstream of the first coding nucleotide (+1) of PDR802; shown is the difference in these values between tagged and untagged strains. Red triangles, complete Pdr802 DNA-binding motifs (Fig. S7C); blue triangles, partial motifs. (C) Putative Pdr802-binding motifs determined using DREME (76). Primary and secondary hits are shown for analysis of 1,000 bp upstream of the initiating ATG. Download FIG S7, PDF file, 1.0 MB (1.1MB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pdr802 ChIP-Seq, RNA-Seq, and dual-threshold optimization (DTO) data. Sheet 1, annotated peaks that occur in gene promoter regions that showed ≥2-fold enrichment when Chip-Seq was performed on strains expressing tagged versus untagged Pdr802. Sheet 2, ChIP-Seq primary data of Pdr802-specific peaks. Sheet 3, RNA-Seq data. Sheet 4, DTO filtered data. Sheet 5, DTO primary data. Sheet 6, ChIP-Seq data of all peaks in mCherry-Pdr802 samples. Sheet 7, ChIP-Seq data of all peaks in untagged (WT) control samples. Download Data Set S1, XLSX file, 2.5 MB (2.5MB, xlsx) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pdr802 target analysis and primers used in this study. Sheet 1, Pdr802 regulated genes that are down-regulated during Titan cell formation in vitro (48). Sheet 2, Pdr802 regulated genes that are up-regulated during Titan cell formation in vitro (48). Sheet 3, targets regulated by Pdr802 that are also dephosphorylated by calcineurin (63). Sheet 4, the intersection of Pdr802 targets and genes that are Crz1-independent calcineurin targets under conditions thermal stress (55). Sheet 5, primers used in this study. Fold enrichment and adjusted P (q) values throughout are for the DTO results. Download Data Set S2, XLSX file, 0.02 MB (26.6KB, xlsx) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods. Download Text S1, PDF file, 0.04 MB (37.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
ChIP-Seq and RNA-Seq data files are available at the NCBI Gene Expression Omnibus under accession numbers GSE153134 and GSE162851, respectively.
Supplemental methods. Download Text S1, PDF file, 0.04 MB (37.8KB, pdf) .
Copyright © 2021 Reuwsaat et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.