Significance
Siglecs are immunomodulatory receptors that recognize sialic acid–carrying glycans with important functions in immunity. However, many of their natural ligands are poorly defined. We generated a cell-based glycan array comprised of a library of isogenic human cells displaying the greater complexity of sialic acids on diverse glycan structures and glycoconjugates in the natural context of the cell surface. We applied this array to reveal fine binding specificities of Siglecs for sialoglycans, informed of the underlying required glycosyltransferase genes, and provided evidence for selective binding context provided by glycan presentation on distinct protein sequences. Insight into the fine binding specificities of Siglecs will advance understanding their diverse biological functions and benefit therapeutic targeting in autoimmunity, inflammation, cancer, and Alzheimer’s disease.
Keywords: Siglecs, CD33, sialome, sialyltransferase, cell-based glycan array
Abstract
Siglecs are a family of sialic acid–binding receptors expressed by cells of the immune system and a few other cell types capable of modulating immune cell functions upon recognition of sialoglycan ligands. While human Siglecs primarily bind to sialic acid residues on diverse types of glycoproteins and glycolipids that constitute the sialome, their fine binding specificities for elaborated complex glycan structures and the contribution of the glycoconjugate and protein context for recognition of sialoglycans at the cell surface are not fully elucidated. Here, we generated a library of isogenic human HEK293 cells with combinatorial loss/gain of individual sialyltransferase genes and the introduction of sulfotransferases for display of the human sialome and to dissect Siglec interactions in the natural context of glycoconjugates at the cell surface. We found that Siglec-4/7/15 all have distinct binding preferences for sialylated GalNAc-type O-glycans but exhibit selectivity for patterns of O-glycans as presented on distinct protein sequences. We discovered that the sulfotransferase CHST1 drives sialoglycan binding of Siglec-3/8/7/15 and that sulfation can impact the preferences for binding to O-glycan patterns. In particular, the branched Neu5Acα2–3(6-O-sulfo)Galβ1–4GlcNAc (6′-Su-SLacNAc) epitope was discovered as the binding epitope for Siglec-3 (CD33) implicated in late-onset Alzheimer’s disease. The cell-based display of the human sialome provides a versatile discovery platform that enables dissection of the genetic and biosynthetic basis for the Siglec glycan interactome and other sialic acid–binding proteins.
Immune cells are equipped with an array of glycan-binding proteins (GPBs) capable of interpreting the biological information encoded by glycans. Endogenous GBPs recognize host-derived “self” and foreign-derived “nonself” glycans and produce cues that are integrated into the signaling network of immune cells and contribute to immune homeostasis and the immune response (1). Siglecs (sialic acid–binding immunoglobulin-like lectins) serve in self-recognition and transmit immune inhibitory signals upon binding to a select repertoire of sialoglycans expressed by host cells raising the threshold for immune activation (2, 3). The human Siglec family consists of 14 functionally expressed members, and these are composed of an N-terminal V-set immunoglobulin (Ig)-like domain that mediates sialoglycan binding followed by varying numbers of C2-set Ig-like domains. Intracellularly, most Siglecs have immunoreceptor tyrosine-based inhibition motifs, and Siglec-14/15/16 carry immunoreceptor tyrosine-based activation motifs (3–7). Siglecs are broadly expressed throughout the immune system, and several Siglecs are also found outside of the immune system, such as Siglec-4 (MAG), which is expressed by oligodendrocytes and Schwann cells in the nervous system (8). Although the diverse biological functions within and outside of the immune system of Siglecs are not fully understood, Siglecs generally contribute to immune homeostasis by dampening immune activation upon recognition of sialoglycans. For example, Siglec-2 (CD22) can suppress B cell receptor activation (9), and Siglec-9 can dampen neutrophil activation (10). Cancer cells with aberrant sialoglycans and pathogens that express sialic acids can exploit Siglec signaling to modulate immune responses (11, 12). Moreover, Siglec-3 (CD33) is strongly associated with risk for Alzheimer’s disease and expressed on microglia cells (13, 14). Given the potent immune modulatory functions of Siglecs and their wide involvement in autoimmunity, infection, cancer, and neurodegeneration, Siglecs are promising therapeutic targets (7, 15). However, many of the natural ligands of Siglecs have not been fully identified, and endogenous ligands for several Siglecs including Siglec-3/CD33 remain elusive.
Human cells can produce a large diversity of glycans capped with sialic acids (Sia), a family of chemically diverse sugars with N-acetylneuraminic acid (Neu5Ac) being the predominant type in humans. Sialic acids are generally found at the termini of mammalian glycans, and most types of glycoconjugates including N-glycoproteins, multiple types of O-glycoproteins, and glycolipids carry oligosaccharides capped by sialic acids (16, 17). Sialylation is one of the most complex regulated steps in glycosylation with 20 distinct Golgi-located sialyltransferase isoenzymes dedicated to catalyze transfer of sialic acids to galactose (ST3GAL1-6, ST6GAL1 and 2), N-Acetylgalactosamine (Gal-NAc) (ST6GALNAC1-6), or sialic acid (ST8SIA1-6) via α2-3, α2-6, or α2-8 linkages, respectively and with different preferences for the underlying glycan structures and types of glycoconjugate (18–20). The resulting plethora of sialic acid-containing glycans constituting the sialome of cells provides a vast catalog of ligands for Siglecs and potential for distinct instructive cues for the immune response (16). The current insight into the interactome of Siglecs is largely derived from studies with libraries of synthetic and natural glycans printed on glass arrays (21, 22). These glycan arrays have demonstrated distinct structural glycan features that drive selective binding of individual Siglecs, including the linkage type of sialic acids, the core disaccharide carrying sialic acids, and glycan modifications such as sulfation or acetylation (23–27). However, printed glycan arrays may not present glycans in the natural context of the overall glycoconjugate structure and the cell surface with spatial organization and competition dynamics limiting insight into the fine binding specificities of Siglecs and their interactions with the host cell sialome.
Here, we took advantage of our recently developed cell-based glycan array strategy (28–30) and generated an expanded sialome sublibrary with the human embryonic kidney (HEK) 293 for dissection of Siglec binding properties. First, combinatorial gene knockout (KO) was used to delete distinct subsets of sialyltransferase isoenzymes or all endogenous sialylation capacity. Second, using targeted gene knock-in (KI), individual sialyltransferase isoenzymes were introduced in the absence of other isoenzymes. Finally, we introduced selected sulfotransferase isoenzymes to explore cross-talk between sialylation and sulfation. To specifically address the influence of clustered O-glycan presentation for Siglec binding, we introduced a large panel of reporter constructs designed to display human O-glycodomains derived from mucins and mucin-like O-glycoproteins with different densities and patterns of O-glycans. The cell-based sialome array reproduced previous results for binding specificities for Siglec-2 (CD22) and Siglec-9 and led to insight into the binding specificities of Siglec-4/7/15 for distinct GalNAc-type O-glycans and their presentation on O-mucin–like glycoproteins. Finally, we demonstrate that Siglec-3/7/8/15 have preferential binding to sulfated sialoglycans yet have different specificities for underlying glycoconjugate structures. We further discovered the 6′-Su-SLacNAc (Neu5Acα2-3[6-O-sulfo]Galβ1-4GlcNAc) epitope on N-glycans and glycolipids as the ligand for Siglec-3/CD33 as well as Siglec-8. In summary, the cell-based display of the human sialome enables dissection of the Siglec interactome in the natural context of a human cell and provides the biosynthetic and genetic basis for the identified ligands.
Results
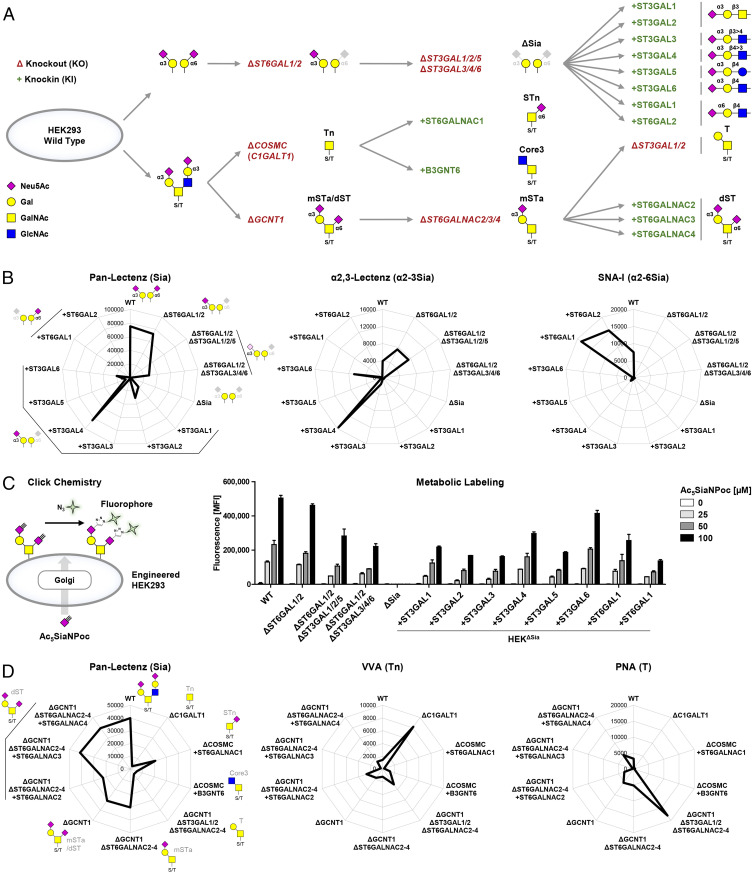
Generation of an Expanded Sialome Cell Library.
We previously built libraries of isogenic HEK cells with selective loss/gain of glycosylation features primarily through KO of endogenously expressed glycosyltransferase genes and with introduction of a limited number of glycosyltransferases not endogenously expressed by site-specific KI (28, 29). These included sublibraries of combinatorial KO of α2-3 and α2-6 sialyltransferase isoenzymes that are useful to display sialoglycans and serve to explore nonredundant functions of sialyltransferases or combinations of these in the presence of other isoenzymes. However, the design does not enable exploration of the full contributions of individual sialyltransferases, including their redundant contributions to the glycome shared by other isoenzymes (30). Here, we expanded the cell-based glycan array platform by eliminating all α2-3/α2-6 sialylation capacities for galactose in HEK wild-type (WT) cells and then reintroducing each of the human ST3GAL1-6 and ST6GAL1-2 sialyltransferases (Fig. 1A). HEKWT cells endogenously express most sialyltransferases for α2-3 and α2-6 sialylation including ST3GAL1-6, ST6GAL1, and ST6GALNAC2-4 and largely lack capacity for α2-8 sialylation (SI Appendix, Table S1) (28). We deleted all α2-3 and α2-6 sialylation capacities for galactose in HEKWT cells through stepwise KO gene targeting of ST3GAL1-6 and ST6GAL1/2 taking advantage of the high efficiency validated gRNAs (guide RNA) assembled in the GlycoCRISPR resource (31). The resulting sialyltransferase empty cells (HEKKO ST6GAL1/2, ST3GAL1-6, designated HEKΔSia) lacked capacities for α2-3, α2-6 sialylation of galactose and α2-8 sialylation, and this was confirmed by flow cytometry analysis showing complete loss of binding to the sialic acid–binding reagents Pan-Lectenz, α2-3-Lectenz, MALII, and SNA-I (Fig. 1B and SI Appendix, Fig. S1A) and gain of binding with the galactose binding lectins PNA and ECL (SI Appendix, Fig. S1A). We further confirmed this by using our previously developed metabolic labeling strategy with alkyne-tagged sialic acid (Ac5SiaNPoc) (32, 33), which showed complete loss of sialylation activity in the HEKΔSia cells (Fig. 1C). Next, the HEKΔSia cells were used to introduce each of the ST3GAL1-6 and ST6GAL1-2 sialyltransferases individually by targeted KI into the human adeno-associated virus “safe harbor” (AAVS1) locus, using a previously established Zinc finger nuclease (ZFN)-based KI approach (34, 35). Successful targeted KI was first confirmed by AAVS1 junction PCR, and activity of the individual sialyltransferase was further evaluated by lectin staining (Fig. 1B and SI Appendix, Fig. S1A), as well as by metabolic labeling that demonstrated induction of efficient sialylation capacity in all the KI cells (Fig. 1C).
Fig. 1.
Generation of the sialoglycan sublibraries. (A) Scheme showing the sublibrary approach for display of α2-3 and α2-6 sialic acid capping of galactose and display of sialylated O-glycans by combinatorial KO (Δ) of endogenous genes (capitalized and italicized) and individual KI (+) of sialyltransferase complementary DNA (capitalized and nonitalicized). Starting from HEKWT cells, ST6GAL1/2 were knocked out to delete α2-6-sialylation of galactose. Next, ST3GAL1/2/5 and ST3GAL3/4/6 were knocked out in sets or combined. ST3GAL1-6 KO was performed, thus deleting α2-3-sialylation capacity. In these empty cells, HEKΔSia, the deleted sialyltransferase genes were knocked in individually. HEKWT cells predictably express a mixture of mSTa (Neu5Acα2-3Galβ1-3GalNAcα1-O-Ser/Thr), dST (Neu5Acα2-3Galβ1-3[Neu5Acα2-6]GalNAcα1-O-Ser/Thr), and sialylated core2 structures. COSMC (or C1GALT1) was knocked out for display of the Tn (GalNAcα-O-Thr/Ser) antigen followed by KI of either ST6GALNAC1 or B3GNT6 yielding STn (Neu5Acα2-6GalNAcα-O-Thr/Ser) or core3 structures (GlcNAcβ1-3GalNAcα1-O-Ser/Thr), respectively. Alternatively, GCNT1 was knocked out resulting in display of a mixture of mSTa/dST and combined KO of ST6GALNAC2/3/4-produced mSTa. Further KO of ST3GAL1/2 produced the T antigen (Galβ1-3GalNAcα1-O-Ser/Thr), and individual KI of ST6GALNAC2/3/4 yielded cells with dST expression. Predicted sialoglycan structures are depicted according to the Symbol Nomenclature for Glycans (92). (B) Radar charts show binding of Pan-Lectenz (Sia), α2-3-Lectenz (α2-3Sia), or SNA-I (α2-6Sia) to the sialic acid capping sublibrary. Lectin binding was quantified by flow cytometry, and data are shown as mean fluorescence intensity (MFI) values from three independent experiments. (C) Scheme illustrating metabolic labeling of cells with Ac5SiaNPoc that is incorporated into cell surface glycans and conjugated to fluorescent azide-biotin using click chemistry. Bar diagram shows metabolic labeling of the capping sublibrary with 0 to 100 µM Ac5SiaNPoc as average MFI ± SEM of three independent experiments. (D) Radar charts show binding of Pan-Lectenz (Sia), VVA (Tn), and PNA (T) to the O-glycan sublibrary as representative MFI values of three independent experiments.
Furthermore, we used combinatorial KO/KI of ST6GALNAC1-4 isoenzymes to display a panel of sialylated O-GalNAc glycans including Tn (GalNAcα1-O-Ser/Thr), sialyl-Tn (STn, Neu5Acα2-6GalNAcα1-O-Ser/Thr), monosialyl-T (mSTa, Neu5Acα2-3Galβ1,3GalNAcα1-O-Ser/Thr), and disialyl-T (dST, Neu5Acα2-3Galβ1-3[Neu5Acα2-6]GalNAcα1-O-Ser/Thr) (Fig. 1A). Deletion of either COSMC or ST6GALNAC2-4 together with introduction of individual genes was probed with Pan-Lectenz, VVA, and PNA (Fig. 1D). STn display by COSMC/C1GALT1 deletion and ST6GALNAC1 KI was confirmed with anti-STn antibody TKH2 (36), and interestingly, we noted that SNA-I recognizes Neu5Acα2-6-Gal generated by ST6GAL1/2, but not STn (SI Appendix, Fig. S1B). Together, these stable isogenic HEK cells form the expanded sialome library (SI Appendix, Table S2). We also included KO cell lines targeting glycoconjugate genes, MGAT1, COSMC, and B4GALT5, which direct the key steps involved in the elaboration of N-glycans, O-glycans, and glycosphingolipids, respectively, to confirm the types of glycoconjugates involved in Siglec binding (28, 29).
Probing the Expanded Sialome Cell-Based Array with Siglecs.
We first probed HEKWT cells with 11 of the 14 human Siglecs by flow cytometry using recombinant Fc chimeras precomplexed with fluorescently labeled anti-human IgG antibody (SI Appendix, Fig. S2A). HEKWT cells do not express Siglecs (SI Appendix, Table S1), and out of the 11 tested Siglec Fc chimera, only Siglec-4/7/9 showed substantial binding to HEKWT, and low Siglec-8 binding was observed (SI Appendix, Fig. S2B). This binding was sialic acid dependent as no binding was observed to sialidase-treated HEKWT cells or HEKΔSia cells. Siglec-2/9 were previously shown to recognize α2-6Sia (Neu5Acα2-6Galβ1-4GlcNAc) and α2-3Sia, respectively, when presented on N-glycans (23, 25, 27, 28, 37, 38). Siglec-2 did not show binding to HEKWT cells, which is in agreement with low expression of α2-6Sia on HEKWT cells (SI Appendix, Table S1) (28). KI of ST6GAL1/2 induced robust binding of Siglec-2 and also greatly enhanced binding of the α2-6Sia-specific lectin SNA-I (Fig. 1B and SI Appendix, Fig. S2C). Siglec-9 binding to HEKWT cells was lost by combined KO of ST3GAL3/4/6 and reinduced in HEKΔSia cells by KI of ST3GAL4, poorly by ST3GAL6, and not by ST3GAL3 or other sialyltransferases including ST6GALNAC1-4 (SI Appendix, Fig. S2C). These findings are in agreement with the reported N-glycan specificity of Siglec-9 (27, 28), and it is noteworthy that the Siglec-9 binding pattern was very similar to α2-3-Lectenz (N-glycan–selective) (Fig. 1B). We did observe very weak binding of other Siglec Fc chimera to the sialyltransferase KI library (e.g., Siglec-3 to ST6GAL1 KI and Siglec-5 to ST3GAL4 KI), but the signals were within baseline variation.
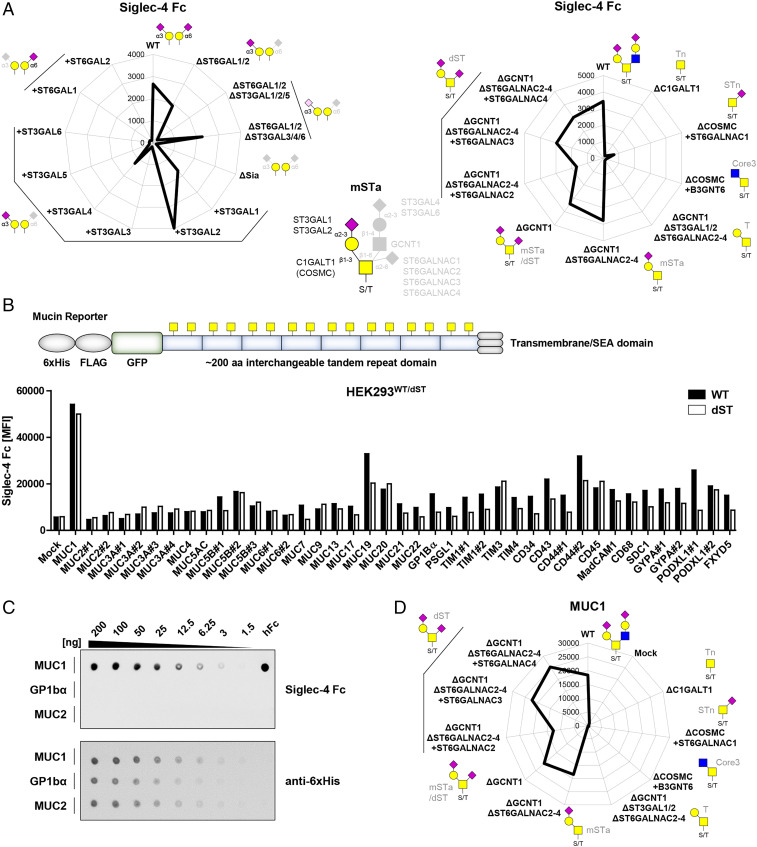
Siglec-4 Binds ST O-Glycans and Preferentially Recognizes MUC1.
Siglec-4/MAG (myelin-associated glycoprotein) showed relatively low binding to HEKWT cells, and the binding was abrogated by KO of ST3GAL1/2/5 and recovered by KI of ST3GAL1/2, but not ST3GAL5 (Fig. 2A). This suggested binding specificity for GalNAc-type O-glycans as ST3GAL1/2 act on those. The biosynthetic pathway and genetic regulation of O-glycosylation in HEKWT cells is illustrated in Fig. 1A, and HEKWT cells express a mixture of core1 (regulated by C1GALT1) and core2 (regulated by GCNT1) O-glycans that are capped by distinct sialyltransferases. KO of C1GALT1, the core1 synthase, resulted in loss of Siglec-4 binding in agreement with the dependency for sialylation by ST3GAL1/2 and GalNAc-type O-glycans (Fig. 2A). The HEKΔSia cells have low endogenous expression of ST6GALNAC2-4, and further dissection with engineering of O-glycosylation showed that Siglec-4 bound to mSTa (HEKKO GCNT1, KO ST6GALNAC2-4) and dST (HEKKO GCNT1 and HEKKO GCNT1, ST6GALNAC2-4, KI ST6GALNAC2/3/4), suggesting that the minimal glycan epitope for Siglec-4 was mSTa (requires C1GalT1/COSMC and ST3GAL1/2), while dST is also bound by Siglec-4 (requires ST6GALNAC2/3/4) (Fig. 2A). These findings support and extend previous results obtained with printed glycan arrays revealing O-glycan and particularly mSTa/b and dST binding preference of Siglec-4 (39) (http://www.functionalglycomics.org/CFGparadigms/index.php/MAG). Recognition of O-glycans may involve more complex features that are induced by the characteristic way O-glycans are often found in dense clusters and distinct patterns in the tandem repeat (TR) regions of mucins and glycoproteins with mucin-like domains (40–44). Selective binding to O-glycan structures on specific O-glycoproteins has also been found for antibodies (45–47) and microbial adhesins (28). Siglec-4 was previously reported to bind MUC1 expressed by cancer cells (48), which prompted us to assess Siglec-4 interactions with heavily O-glycosylated mucin-like O-glycodomains. We utilized a panel of 41 membrane-bound green fluorescent protein (GFP)-tagged reporter constructs containing around 200 amino acids of the TR regions of different human mucins and mucin-like O-glycoproteins for transient expression in HEK293WT and HEKKO GCNT1, ST6GALNAC2-4, KI ST6GALNAC4 cells (Fig. 2B and SI Appendix, Fig. S3 and Table S3) (28, 29). Siglec-4 Fc binding to cells expressing these O-glycodomain reporters was evaluated by flow cytometry gating for GFP-positive–expressing cells with binding to the GFP-negative–nonexpressing cell population serving as base level reference (SI Appendix, Fig. S4). Siglec-4 Fc binding was particularly enhanced with the MUC1 O-glycodomain reporter (ninefold), while MUC19 and CD44 (two- to threefold), as well as most other reporters only induced slightly higher (one- to twofold) binding (Fig. 2B). Selective binding of Siglec-4 to MUC1 was validated by dot blot analysis with secreted and purified MUC1, GP1bα, MUC2, and seven other reporters (Fig. 2C and SI Appendix, Fig. S5A). These reporters were previously validated to have high and homogenous occupancy and carry predominantly the anticipated glycan structures (28, 49). Of note, Siglec-9 was reported to bind sialyl-T (ST) O-glycans on MUC1 (50, 51); however’ we could not observe any binding effect of Siglec-9 toward O-glycoengineered libraries as well as O-glycodomain reporters including MUC1 reporter (SI Appendix, Fig. S5 B and C) (27, 28). The O-glycan dependence of Siglec-4 binding was further dissected by use of O-glycoengineered HEK cells transiently expressing the membrane-tethered MUC1 reporter (Fig. 2D), which demonstrated that Siglec-4 binding to MUC1 was dependent on ST O-glycans (Fig. 2A). These findings were confirmed by dot blot analysis with secreted mucin reporters (SI Appendix, Fig. S5D).
Fig. 2.
Siglec-4 fine binding dissection with cell-based glycan and mucin reporter display. (A) Radar charts show Siglec-4 Fc binding to sialic acid capping (Left) and O-glycan (Right) sublibrary as representative MFI values of three independent experiments. Predicted glycan epitopes and essential glycosyltransferase genes from analysis with GlycoRadar are illustrated. (B) Illustration of the mucin reporter with 6xHis, FLAG tag, and GFP, 200 amino acid–interchangeable mucin TR domain for expression in isogenic cells as membrane-bound or secreted form. Bar diagram shows Siglec-4 binding to HEKWT and HEKdST(KO GCNT1/ST6GALNAC2/3/4, KI ST6GALNAC4) cells transiently transfected with 41 different mucin and mucin-like protein reporters as MFI values from three independent experiments. (C) MUC1, GP1bα, and MUC2 dot blot overlaid with Siglec-4 Fc (above). Equal blotting was confirmed by overlay with anti-6xHis (below). Serial dilutions of the mucin reporters were blotted, and human Fc (hFc) was blotted as positive control. (D) Siglec-4 binding to O-glycoengineered cells expressing membrane MUC1 reporter is shown as radar chart.
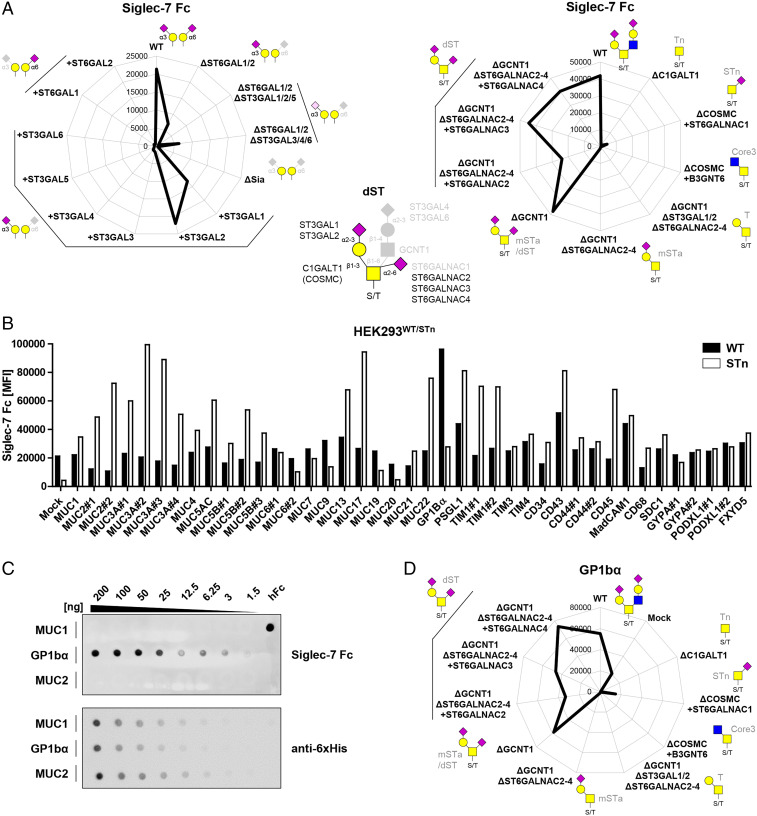
Siglec-7 Binds dST and the Cancer-Associated STn O-Glycan with Distinct Preferences for Mucin O-Glycodomains.
Siglec-7 showed stronger binding to HEKWT than Siglec-4 and a similar binding pattern to the sialome library with the notable exception that all binding was lost by KO of ST6GALNAC2-4 and GCNT1 (Fig. 3A). Thus, Siglec-7 required α2-6 sialylation of the inner GalNAc residue and binds the dST O-glycan without binding to mSTa as for Siglec-4 (Fig. 2A). This is in line with binding to HEKKI ST3GAL1/2 cells that express low levels of ST6GALNAC2-4. Screening HEKWT cells expressing different mucin-like O-glycodomain reporters revealed specific enhancement of binding with O-glycodomains designed from GP1bα (Glycoprotein Ib alpha) (6-fold), PSGL-1 (2.5-fold), and MAdCAM-1 (mucosal addressin cell adhesion molecule 1) (3.5-fold) (Fig. 3B), and this was confirmed by dot blot analysis (Fig. 3C and SI Appendix, Fig. S5A). Remarkably, screening the same reporters expressed in HEKKO COSMC KI ST6GALNAC1 cells, in which they carry only STn O-glycans, we found an entirely different binding pattern with robust Siglec-7 binding induced to several O-glycodomain reporters including those for MUC2, MUC5AC, MUC7, MUC13, and MUC22 (Fig. 3B). Dot blot analysis with some of these reporters confirmed that Siglec-7 bound MUC2 and MUC5AC with STn O-glycans, while binding to the same reporters with core2 O-glycans expressed in HEKWT cells was neglectable (SI Appendix, Fig. S5D). Moreover, the enhanced Siglec-7 binding to the GP1bα reporter expressed in HEKWT cells was strongly reduced when expressed with STn O-glycans. Finally, further dissection of the O-glycan structure dependency of Siglec-7 binding by use of O-glycoengineered HEK cells transiently expressing the membrane tethered GP1bα reporter fully confirmed the requirement for dST O-glycans on GP1bα with no binding to GP1bα with Tn O-glycans and minimal binding with STn O-glycans (Fig. 3D). These findings indicate that Siglec-7 binding is affected partly by the O-glycan structure and partly by the pattern of O-glycan presentation, but perhaps also provides an example illustrating discordance between recognition of structural features of O-glycans and the presentation of these in patterns on proteins. The common structural feature in the dST and STn O-glycans is the sialic acid linked α2-6 to the GalNAc residues attached to the peptide backbone, and it is likely that Siglec-7 recognition involves a clustered patch that is presented in the O-glycodomain of GP1bα with dST O-glycans, while this clustered patch is better and more widely presented by the mucin TR O-glycodomains with STn O-glycans. Regardless, the results clearly demonstrate that Siglecs recognize more complex epitopes than isolated glycans as originally hypothesized (40–44).
Fig. 3.
Siglec-7 fine binding dissection with cell-based glycan and mucin reporter display. (A) Radar charts showing Siglec-7 Fc binding to sialic acid capping (Left) and O-glycan (Right) sublibrary as representative MFI values of three independent experiments. Predicted glycan epitopes and essential glycosyltransferase genes from analysis with GlycoRadar are illustrated. (B) Bar diagram shows Siglec-7 binding to HEKWT and HEKSTn (KO COSMC KI ST6GALNAC1) cells transiently transfected with 41 different mucin reporters as MFI values from three independent experiments. (C) MUC1, GP1bα, and MUC2 dot blot overlaid with Siglec-7 Fc (above) and anti-6xHis (below). Serial dilutions of the mucin reporters were blotted, and human Fc was blotted as positive control. (D) Siglec-7 binding to O-glycoengineered cells expressing membrane GP1bα reporter. MFI from three independent experiments is shown in the radar chart.
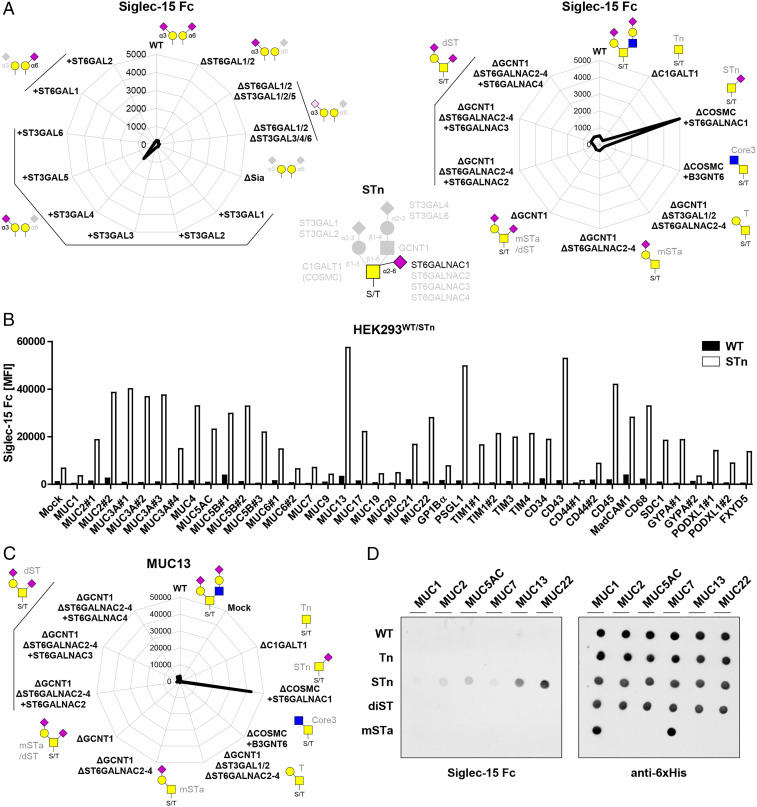
Siglec-15 Binds STn O-Glycans with Distinct Preferences for Patterns.
Siglec-15 did not bind HEKWT cells, and KI of ST3GALs or ST6GALs did not induce significant binding; however, KI of ST6GALNAC1 in cells without core1 O-glycan elongation (HEKKO COSMC KI ST6GALNAC1) to display the STn O-glycan (52) induced relatively strong binding (Fig. 4A). These findings are in line with a previous study suggesting STn recognition by Siglec-15 (53). Screening of Siglec-15 binding to mucin and mucin-like reporters in HEKWT cells showed no significant binding; however’ binding was significantly enhanced for many reporters when expressed with STn O-glycans in HEKKO COSMC KI ST6GALNAC1 cells (Fig. 4B). Interestingly, the pattern of enhancement among the different O-glycodomains were similar to that found for Siglec-7 with the STn glycoforms (Fig. 3B), suggesting that the two Siglecs recognize similar clustered patches of STn O-glycans. Dissection of the O-glycan dependence for Siglec-15 binding to the MUC13 reporter confirmed a restricted specificity for the STn glycoform (Fig. 4C). Dot blot analysis further confirmed selective binding to STn glycoforms of MUC13 and MUC22 (Fig. 4D).
Fig. 4.
Siglec-15 fine binding dissection with cell-based glycan and mucin reporter display. (A) Radar charts showing Siglec-15 Fc binding to sialic acid capping (Left) and O-glycan (Right) sublibrary as representative MFI values of three independent experiments. Predicted glycan epitopes and essential glycosyltransferase genes from analysis with GlycoRadar are illustrated. (B) Bar diagram shows Siglec-15 binding to HEKWT and HEKSTn (KO COSMC KI ST6GALNAC1) cells transiently transfected with 41 different mucin reporters as MFI values from three independent experiments. (C) Radar chart presentation of Siglec-15 binding to membrane MUC13 reporter expressed in O-glycan sublibrary. (D) Secreted mucin reporters (MUC1, MUC2, MUC5AC, MUC7, MUC13, and MUC22) produced in engineered HEK cells with WT, Tn, STn, dST, and mSTa glycosylation were immobilized and overlaid with Siglec-15 or anti-6xHis.
Probing Sulfation—CHST1 Contributes to Siglec-3/7/8/15 Binding.
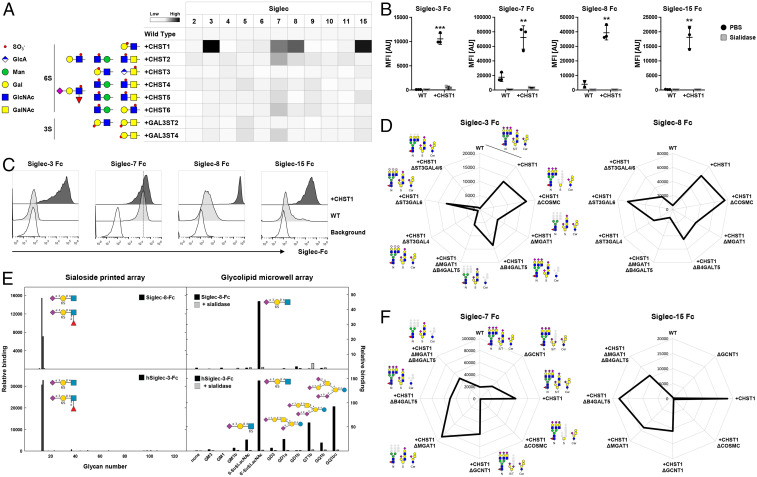
Several Siglecs did not show significant binding to the HEK cell sialome library, and we considered introducing sulfation capacities as previous studies have shown preferred binding of Siglecs (e.g., Siglec-2/CD22, Siglec-8 and its murine paralog Siglec-F or Siglec-9 to sulfated glycans) (25, 26, 54–56). HEKWT cells have no/low expression of nonglycosaminoglycan sulfotransferases (SI Appendix, Table S1), so we introduced the sulfotransferases CHST1/2/3/4/5/6 and Gal3ST2/4 individually into HEKWT cells (Fig. 5A). Introduction of most sulfotransferases had no effect, and we observed two- to fivefold enhancement of Siglec-7 and -9 binding to CHST2-6 and GAL3ST4 KI cells, respectively. Remarkably, CHST1 KI induced robust binding of Siglec-3/8/15 and significantly enhanced (>10-fold) binding of Siglec-7 (Fig. 5A). This binding was completely abolished by sialidase treatment demonstrating the expected requirement for sialic acids (Fig. 5B). CHST1 is predicted to install 6-O-sulfation on the galactose residue of Galβ1-4GlcNAc motifs found in N-glycans, the β6-branch of core2 O-glycans, and lactoseries glycolipids (57, 58). We confirmed these findings with Chinese hamster ovary (CHO) cells that only produce core1 O-glycans, and KI of CHST1 also induced strong binding of Siglec-3/8/15 and enhanced Siglec-7 binding, although to a lesser extent compared to HEK cells (Fig. 5C). Binding competition experiments using a high-affinity Siglec-7 ligand G35 (59) revealed a greater than fivefold increased half-maximal inhibitory concentration when comparing HEKWT cells with HEKKI CHST1 (SI Appendix, Fig. S6). These findings confirm that 6-O-sulfation contributes widely to Siglec recognition of glycans and demonstrates that CHST1 plays a particular role.
Fig. 5.
Probing the contribution of sulfotransferases to Siglec sialoglycan recognition. (A) Sulfotransferases knocked in into HEKWT cells and predicted sulfated glycan structures are shown. Heat map shows Siglec Fc binding to the KI cells as MFI values normalized to HEKWT. (B) Dot plots show Siglec-3/7/8/15 binding to HEKWT and HEKKI CHST1 cells treated with PBS or sialidase. Data of three independent experiments are presented as average MFI ± SEM. (C) Representative histograms show binding of Siglec-3/7/8/15 to CHOWT and CHOKI CHST1 cells. Cells stained only with anti-human IgG AF647 indicate background fluorescence. (D) Radar charts show Siglec-3 (Left) and Siglec-8 (Right) binding to HEKWT and HEKKI CHST1 cells with additional KO of O-GalNAc glycans (KO COSMC), N-glycans (KO MGAT1), glycolipids (KO B4GALT5), N-glycans and glycolipids (KO MGAT1, KO B4GALT5), or α2-3Sia on N-glycans (single or double KO ST3GAL4/6). (E) Siglec binding to glycan arrays. Recombinant Siglec-8 and Siglec-3 Fc chimera were tested for binding to two sialoglycan arrays: a 123-glycan array printed on glass slides (Left) and an 11-glycan sialoglycolipid array on 384-well plates. The glycan structures tested are listed in SI Appendix. Binding is expressed in arbitrary fluorescence units for the printed array and as colorimetric enzyme activity (ΔA405/min × 1,000) for the glycolipid array. Each point is the average of quadruplicate determinations. As indicated, a replicate glycolipid array was pretreated with 50 mU/mL Vibrio cholerae sialidase for 90 min at 37 °C and washed prior to addition of the Siglec Fc chimeras. (F) Radar chart shows Siglec-7 (Left) and Siglec-15 (Right) binding to HEKWT and HEKKI CHST1 cells with additional KO of O-GalNAc glycans (KO COSMC), core2 (KO GCNT1), N-glycans (KO MGAT1), glycolipids (KO B4GALT5), or N-glycans and glycolipids (KO MGAT1, KO B4GALT5). Representative MFI values of three independent experiments are shown, and deleted glycoconjugates are depicted in gray.
The cell-based glycan array enables further dissection of underlying structural requirements for binding using our sublibrary engineering strategy to dissect the glycoconjugates involved (28, 29). Siglec-3 and Siglec-8 binding was strongly reduced when elaboration of both N-glycans (KO MGAT1) and glycolipids (KO B4GALT5) was eliminated, whereas loss of either one only reduced binding (Fig. 5D). This was further corroborated by the finding that KO of ST3GAL4, but not ST3GAL6, which adds α2-3Sia to LacNAc on N-glycans and lactoseries glycolipids (60), resulted in loss of Siglec-3 binding and strongly reduced Siglec-8 binding (Fig. 5D). Conversely, elimination of elaborated O-glycan structures (KO COSMC) did not affect binding of either Siglecs, confirming that O-glycans are not major binding ligands on HEK cells. The results clearly indicated that 6′-Su-SLacNAc is a ligand for Siglec-3 and -8, and this was further confirmed by analysis with printed glycan arrays containing this 6-O-sulfated trisaccharide (61) as well as with a glycolipid array (56, 62, 63) (Fig. 5E,). Both Siglec-3 and -8 recognized almost exclusively 6′-Su-SLacNAc and the fucosylated form (6-sulfo sialyl Lewis X). The glycolipid array further revealed that Siglec-3 also recognized ganglioside glycolipids including GD1a, GT1b, and GQ1bα containing branched sialic acids (Fig. 5E).
In contrast, the effect of CHST1 KI on Siglec-7 and Siglec-15 binding was abolished completely when O-glycan elongation was eliminated (KO COSMC) (Fig. 5F), in agreement with their specificity for O-glycans (Figs. 3 and 4). Since Siglec-7 and Siglec-15 were found to bind simple core1 ST and STn O-glycans without apparent requirement for core2 structures that can contain the Galβ1-4GlcNAc disaccharide predicted to serve as the substrate for CHST1 (64, 65), we sought to further dissect the importance of core2 O-glycans by KO of GCNT1. Interestingly, for both Siglec-7 and Siglec-15, the induced binding to CHST1 KI cells was not significantly altered by elimination of core2 O-glycans (KO GCNT1) but completely abolished by truncation to the Tn structure (KO COSMC) (Fig. 5F). These results suggest that CHST1 sulfates the core1 O-glycan presumably at the terminal galactose residue producing SO3-6Galβ1-3(Neu5Acα2-6)GalNAcα1-O-Ser/Thr with or without α2-3Sia at the terminal Gal residue. Further studies are needed to define the exact structure and interplay between α2-3 and/or α2-6 sialylation of core1 O-glycans and 6-O-sulfation. Siglec-7 binding was dependent on ST3GAL1/2 as well as ST6GALNAC2-4 sialylation of core1 O-glycans (Fig. 3 A and D) but clearly also bound STn in the context of O-glycodomains (Fig. 3B). In contrast, Siglec-15 bound STn and not core1 O-glycans, but with CHST1-induced 6-O-sulfation, binding to core1 O-glycans was found. This appears to provide another scenario to the theme of formation of clustered saccharide patches focused around presentation of the inner Neu5Acα2-6GalNAcα1-O-Ser/Thr epitope. Altogether, our results indicate that 6-O-sulfation is a key determinant for regulating Siglec recognition of sialoglycans.
Discussion
Here, we developed a cell-based array to display the human sialome in the natural context of glycoconjugates at the cell surface and applied this to probe the binding properties of 11 human Siglecs. We were able to detect and dissect binding properties for seven Siglecs (Siglec-2, -3, -4, -7, -8, -9, and -15) (Table 1). In general, the results were in agreement with reported glycan binding specificities, and the analysis of Siglec-4, -7, and -15 that target GalNAc-type O-glycans provided insight into the structural glycan features required and demonstrated evidence for selectivity for O-glycans displayed on distinct mucin-like O-glycodomains. The array provided evidence that sulfation not only augments binding of select Siglecs (Siglec-3, -7, -8, and -15), but sulfation also appears to markedly alter recognition of the overall O-glycan scaffold features. Importantly, we discovered a 6-O-sulfated glycan epitope for Siglec-3 and the enzymes and genes required for the biosynthesis of this epitope in HEK cells. The N-terminal domain of Siglecs contains one variable (V-set) and one or more constant (C-set) Ig domains (4). The V-set domain contains a shallow sialic binding site with a conserved arginine residue that directly interacts with the C-1 carboxyl group of sialic acids, and sequence variation mainly in the V-set domain determines the fine binding specificities (3, 7). While the binding preferences of Siglecs for structural features of sialoglycans such as linkage (α2-3/2-6/2-8Sia), underlying core saccharide, and adjacent sulfation have been elucidated for many Siglecs using traditional glycan arrays (4, 21, 23–27), insight into the natural Siglec ligands is still limited, and the cell-based sialome array presented here clearly demonstrates that Siglec ligand recognition is driven by more complex presentation of clustered glycans orchestrated by the carrier protein (28, 40–44).
Table 1.
Summary of glycosyltransferases essential for Siglec binding to HEK293 cells and contribution of glycoconjugate, mucin reporter, and sulfotransferase context
| Siglec | Sialyltransferase | Linkage | Glycoconjugate | Mucin reporter | Sulfotransferase | Predicted epitope |
| Siglec-2 | ST6GAL1/2 | α2-6 | N-glycan | — | — | Neu5Acα2-6Galβ1-4GlcNAc |
| Siglec-3 | ST3GAL4 | α2-3/6 | N-glycan, GL 6-sulfo-LacNAc | — | CHST1 | Neu5Acα2-3[6-O-sulfo]Galβ1-4GlcNAc |
| Siglec-4 | ST3GAL1/2 | α2-3 | O-glycan, mSTa, dST | MUC1 (MUC19) | — | Neu5Acα2-3Galβ1-3GalNAcα1-O-Ser/Thr |
| Siglec-5/14 | — | — | — | — | — | — |
| Siglec-6 | — | — | — | — | — | — |
| Siglec-7 | ST3GAL1/2, ST6GALNAC2/3/4 | α2-3/6 | O-glycan, dST | GP1βα (CD43, PSGL1, and MADCAM), clustered STn | CHST1, GAL3ST4 | Neu5Acα2-3Galβ1-3[Neu5Acα2-6]GalNAcα1-O-Ser/Thr |
| Siglec-8 | ST3GAL4 | α2-3 | N-glycan, GL 6-sulfo-LacNAc | — | CHST1 | Neu5Acα2-3[6-O-sulfo]Galβ1-4GlcNAc |
| Siglec-9 | ST3GAL4/6 | α2-3 | N-glycan | — | — | Neu5Acα2-3Galβ1-4GlcNAc |
| Siglec-10 | — | — | — | — | — | — |
| Siglec-11/16 | — | — | — | — | — | — |
| Siglec-15 | ST6GALNAC1 | α2-6 | O-glycan, STn | Clustered STn | CHST1 | Neu5Acα2-6GalNAcα1-O-Ser/Thr |
The expanded sialome cell-based library was designed to enable dissection of the nonredundant as well as the redundant contributions of all human α3- and α6-sialyltransferases using combinatorial KO of endogenously expressed sialyltransferase genes (HEKΔSia) as well as site-directed reintroduction of sialyltransferases in these cells (Fig. 1A). It is important to note that the cell-based array was developed in a single human cell line (HEK), and the design of engineering performed was based partly on knowledge of the expression of glycosyltransferases derived transcriptome analysis (28). The cell-based array is therefore not comprehensive with respect to all human enzymes and structural variations of glycans and glycoconjugates possible nor with the expressed proteome and glycosylation variations. Thus, the cell-based array platform can provide positive identification of interactions with the glycome and the underlying biosynthesis but may not necessarily reveal all potential interactions or be representative of the interactions found in vivo. We did not address α2-8 sialylation, and none of the α8-sialyltransferases (ST8SIA1 through 6) are endogenously expressed in HEKWT; however, several Siglecs, including Siglec-7, are reported to bind α2-8Sia (4). Similarly, we did not address fucosylation which has been reported to affect Siglec interactions (66) due to the low expression level of α2/3/4-fucosyltransferases in HEKWT (28, 29). These features can clearly be explored by further expansion of the library. The current design of the cell-based sialome library provides a level of insight into the functions of the many sialyltransferase isoenzymes (18, 19), and we could demonstrate that all serve efficiently to introduce sialylation capacity by metabolic labeling and lectin staining (Fig. 1 B–D). ST3GAL4 was clearly the most efficient isoenzyme in introducing α2-3Sia to LacNAc termini (high α2-3-Lectenz binding), while ST6GAL1 and 2 both efficiently introduced α2-6Sia. Focusing on GalNAc-type O-glycosylation, ST3GAL1 and 2 were both efficient in capping core1 O-glycans with α2-3Sia (Fig. 1D and SI Appendix, Fig. S1A), while we confirmed that only ST6GALNAC1 introduced α2-6Sia to the truncated GalNAc (Tn) O-glycan yielding STn (Fig. 4A). Moreover, the results demonstrate that ST6GALNAC2, 3, and 4 can add α2-6Sia to core1 O-glycans and induce Siglec-7 binding (Fig. 3A). Only ST6GALNAC1 serves in biosynthesis of the prevalent cancer-associated STn biomarker (52, 67, 68) and in the normal intestine in which ST6GALNAC1 is selectively expressed together with 9-O-Acetylated STn O-glycans (69). Interestingly, in contrast to previous reports (70, 71), SNA-I, which is widely used for α2-6Sia detection, only recognized α2-6Sia linked to LacNAc/LacDiNAc but did not recognize STn. Siglec-2 and -15 showed distinct binding specificity to these 2,6Sia structures (SI Appendix, Fig. S1B). Further dissection within this cell-based sialome array now enables wider studies into the human sialyltransferases and their functions in regulation of biological interactions.
We used the expanded sialome array to dissect the binding specificities of the human Siglecs and the biosynthetic pathways needed for binding. Only three Siglecs (4/7/9) showed significant binding to HEKWT cells, and we could demonstrate that Siglec-4 and -7 selectively bound O-glycans (mSTa and dST, respectively), while Siglec-9 binding required ST3GAL4 and to a lower extent ST3GAL6 (Neu5Acα2-3Galβ1-4GlcNAcβ1-R) found on N-glycans, as suggested previously (4, 27, 28, 72). In contrast to previous reports (50, 51), we did not detect Siglec-9 binding to mSTa MUC1. The finding that Siglec-7 required action of ST6GALNAC2-4 (dST) (and ST3GAL1/2 and GCNT1) is supported by a recent genome-wide KO study identifying these particular genes among others (73) and an association study in pancreatic cancer (72). Of note, several reports suggest that Siglec-7 binds to glycolipids such as GD3 (Neu5Acα2-8Neu5Acα2-3Galβ1-4Glcβ1-Cer) (4, 74, 75). We previously found that elimination of elaborated glycolipid glycan structures (KO B4GALT5/6) in HEKWT did not affect Siglec-7 binding (28), but HEK cells may predominantly produce lactoseries glycolipids with little or no ganglioseries glycolipids carrying α2-8Sia, and they do not express α8-sialyltransferases including ST8SIA1 (SI Appendix, Table S1). The HEK cell-based Siglec interactome reported here does not necessarily represent the entirety of Siglec ligands reported so far and to be found in future but offers opportunity to engineer these in a realistic glycan context of the cell.
Interestingly, the endogenous α6-sialylation capacity of HEKWT cells was insufficient to establish robust interactions with Siglecs including Siglec-2 (CD22) with binding specificity for Neu5Acα2-6Galβ1-4GlcNAc on N-glycans (23, 37, 38). HEK cells express ST6GAL1, and KO of ST6GAL1/2 eliminated SNA-I lectin binding (Fig. 1B), but Siglec-2 binding was only found following KI of ST6GAL1/2, which also enhanced SNA-I binding. Presumably, higher levels of α2-6-sialoglycans are required for Siglec-2 to form interaction clusters at the cell membrane (76, 77). KO of ST3GAL3/4/6 did not induce enhanced SNA-I binding (28), suggesting that the low levels of α6-sialylation is not a result of competing α3-sialylation. We noted minor binding of Siglec-3/5/10 to HEKΔSia KI ST6Gal1/2 cells, but it is likely that further engineering of α8-sialylation by instalment of ST8SIAs is required to dissect these Siglecs. It is important to consider that our studies were performed with precomplexed Siglec Fc chimera dimers, which ideally form tetramers. The monomeric Siglecs exhibit low binding affinities (high μM range) (7), and the precomplexes used may bias the binding patterns observed. Furthermore, Siglec interactions may be guided by assembly into microdomains and influenced by other factors (4, 76–78). In this respect, the cell-based sialome array offers a highly flexible platform to address Siglec functions in cell–cell interactions, using the sialome array of cells (with and without expression of specific glycoproteins) as bait in cell–cell assays with the diversity of immune cells naturally expressing Siglecs.
We focused in particular on the O-glycan–binding Siglecs, since O-glycans are often carried on proteins in dense clusters and patterns in regions referred to as mucin-like domains (28, 40, 41, 44–47). Dissection of Siglec-4/7/15 revealed distinct fine binding specificities for mSTa (Siglec-4), dST (Siglec-7), and STn (Siglec-15), and when we included coexpression of a large panel of mucin and mucin-like protein reporters, we discovered selectivity in binding to the O-glycan epitopes presented on different mucin TRs. These mucin and mucin-like protein reporters resemble several aspects of the natural glycoproteins with TR domains and dense O-glycosylation presented on these but may not fully reflect recognition of the natural glycoprotein. Importantly, we previously validated these mucin TR reporters, and for most reporters, we demonstrated homogenous and near-complete site occupancy (28, 49). The majority of the displayed glycans corresponded to the glycan phenotype of the respective isogenic cell and glycosylation, and generally, the glycosylation was comparable between different reporters produced in the same isogenic cells. This supports our notion that the specific peptide sequence or O-glycan pattern provides context for glycan recognition by Siglecs-4/7/15.
Siglec-4 bound selectively to the mSTa glycoforms of MUC1, a biological interaction previously suggested to play a role in pancreatic cancer perineural invasion (48). Similarly, Siglec-7 preferentially bound dST on the major platelet O-glycoprotein GP1bα involved in platelet activation and aggregation (Fig. 3 B–D) (79). Siglec-7 expression on platelets has been reported (80), suggesting that Siglec-7 may engage in cis interactions with platelets through binding to GP1bα. Recently, the dST glycoform of CD43 was identified as a specific target for Siglec-7 (73, 81), and we indeed found that expressing the CD43 membrane reporter in HEK293WT cells induced the second highest increase (approximately threefold) in binding only surpassed by the GP1bα reporter (Fig. 4B). The most remarkable finding though was how the mucin TRs appeared to alter the glycan-dependent binding of Siglec-7. When the mucin-like reporters were expressed in HEKWT cells with the common mST/dST O-glycans, Siglec-7 showed strong preference for GP1bα as well as CD43, but when the same reporters were expressed with the STn glycoform mainly found in cancer, Siglec-7 revealed different preferences for mucin TRs and in particular low binding to STn presented on GP1bα. The molecular basis for these interactions is unclear, but the common structural unit of dST and STn is obviously the Neu5Acα2-6GalNAcα1-O-Ser/Thr core, and we predict that Siglec-7 binding is governed primarily through interactions with the α2-6Sia that may be exposed differently in clusters with the dST (in which loss of the α2-3Sia blocked binding) and the STn O-glycans on different mucin-like O-glycodomains. Siglec-15 bound STn as previously reported (53) and similarly to Siglec-7 showed selectivity for STn O-glycans on distinct mucin TRs with neglectable binding to STn on MUC1 and strong binding to STn on CD43 and CD45 as well as other mucin TRs (Fig. 4B). Recognition of STn displayed on polymers was previously reported for Siglec-2/3/5/6 (66, 82), and density-dependent glycan epitope recognition is a general feature of lectins (40, 41, 44). The molecular basis for the selectivity for mucin TRs is unclear, and a simple correlation between Ser/Thr density and distinct sequence motifs was not found. However, our findings suggest that the protein backbone and glycan density, or secondary features contributed by those, mediate STn recognition by Siglec-7 and -15. STn is a prevalent tumor-associated glycan, and both Siglec-7 and -15 have been associated with tumor immune evasion by inhibiting natural killer cell function (83) and promoting tumor-associated macrophages (84), respectively. Further insight into Siglec recognition of dense STn motifs may help understanding these tumor immune evasion mechanisms (11).
We also started to explore the role of sulfation in determining the Siglec interactome. KI of CHST1 in HEKWT cells, predicted to install capacity for 6-O-sulfation of galactose (57, 58), induced binding of Siglec-3/8/15 (Fig. 5). The binding of Siglec-7 was also enhanced by KI of other sulfotransferases (CHST2/4 to 6 and GAL3ST4). Natural ligands for Siglec-3/CD33 linked to late-onset Alzheimer’s disease have hereto not been reported (13, 14). Siglec-3 is expressed by myeloid cells and microglia cells in the brain, and sialic acid mimetics with high affinity (85) were shown to increase amyloid-β uptake by microglia cells in vitro (86). Previous studies have suggested that Siglec-3 recognizes α2-3/α2-6Sia sialoglycans (66, 87), but here, we identified the branched sialylated and sulfated ligand 6′-Su-SLacNAc, which previously was shown to also serve as a ligand for Siglec-8 (56, 88, 89). In case of Siglec-8, it was shown that 6-O-sulfation of galactose with α2-3Sia mediates specific interactions with three amino acids in the variable C-C′ loop of Siglec-8, creating selective binding (90). It is likely that similar interactions mediate the preferred binding to Siglec-3 and other Siglecs, and these should be further addressed. Both Siglec-3 and -8 were predicted to recognize the ligand epitope primarily on N-glycoproteins and glycolipid, and this glycoconjugate selectivity is likely mediated by interactions in the variable C-C′ loop, too (7). Further introduction of α2-6/8-sialylation capacity in the HEKWT cells in combination with sulfotransferase KI may reveal more sulfated glycan ligands, potentially based on 6-O-sulfo-GlcNAc, as preferred binding to Siglecs.
KI of CHST1 in HEKWT cells also resulted in strong Siglec-7 and Siglec-15 binding, which for both Siglecs was dependent on sialic acid and core1 O-glycans, since sialidase treatment and truncation of O-glycans to Tn (KO COSMC) abrogated binding. Elimination of core2 O-glycan branching (KO GNCT1) did not affect binding.
CHST1 has been reported to sulfate the distal Gal or internal GalNAc residue of mSTa (6-O-sulfo-Galβ1-3GalNAcα1-O-Ser/Thr or Neu5Acα2,3Galβ1-3[6-O-sulfo]GalNAcα1-O-Ser/Thr) (91). We therefore predict that CHST1-mediated 6-sulfation can contribute to both Siglec-7 and -15 binding in a similar manner as the internal α2-6Sia found in dST and STn. Moreover, both Siglecs bind efficiently to the simple STn O-glycans when presented in the context of O-glycodomains. These findings clearly demonstrate the value of a cell-based glycan array for deciphering complex interactions with O-glycans presented in clusters and distinct patterns, and further structural studies are needed to provide insight into how such binding is accommodated. Finally, we note that studies with the cell-based glycan arrays developed here have not necessarily identified the true natural ligands of these Siglecs. The natural Siglec interactome comprises dynamic interactions with specific glycan ligands, their multivalent presentation on specific proteins and lipids, and the organization of these at the cell membrane (4, 20). Although several of these aspects may be captured by the cell-based glycan array, further studies are clearly needed to validate these interactions in more natural systems.
In conclusion, the developed cell-based sialome array enables dissection of the human Siglec interactome and informs of the biosynthetic and genetic regulation of Siglec ligands and structural features of these. Our studies provide evidence that Siglecs recognize the context of O-glycans as presented in mucin-like regions adding a layer of information to the Siglec interactome. Further studies are needed to explore the structural basis, but the cell-based array is clearly useful for uncovering complex glycan epitopes impacted by the carrier protein (30). Dissecting the fine binding specificity of Siglecs in tissues and cell types will advance understanding of their diverse biological roles in the immune system and beyond and may guide the development of glycan-based therapeutics for diseases including immune disorders, Alzheimer’s disease, and cancer.
Materials and Methods
Materials and general laboratory methods, cell culture, CRISPR/Cas9 targeted gene KO in HEK cells, ZFN-mediated gene KI, lectin binding and flow cytometry, metabolic incorporation of Ac5SiaNPoc and click chemistry, transient transfection with mucin reporters, dot blot analysis, printed glycan array and glycolipid microwell arrays, data analysis, and additional necessary information are available in the SI Appendix, SI Materials and Methods.
Genetically Engineered HEK293 Cells.
Stable combinatorial CRISPR/Cas9 targeted KO of glycosyltransferase enzymes was performed with a validated gRNA library (GlycoCRISPR) (31). Stable KI of glycosyltransferases was performed with ZFNs and integration into the human AAVS1 safe harbor site using a modified ObLiGaRe gene KI strategy (35).
Siglec Binding and Flow Cytometry.
Recombinant human Siglec Fc chimera were precomplexed in a 1:2 (wt/wt) ratio for 10 min with Alexa Fluor 647-conjugated goat anti-human IgG and cells were stained with complexes for 1 h at 4 °C followed by flow cytometry analysis.
Expression of Mucin Reporters.
Constructs for transmembrane or secreted expression of 6xHIS and GFP-tagged human mucin tandem repeat domains (150–200 amino acids) are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
This work was supported by the Lundbeck Foundation, the Novo Nordisk Foundation, the Danish National Research Foundation (DNRF107), the European Commission (GlycoImaging H2020-MSCA-ITN-721297, BioCapture H2020-MSCA-ITN-722171), the Mizutani Foundation, the US NIH (U19AI136443), and the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie Grant Agreement No. 787684 (to C.B.).
Footnotes
Competing interest statement: University of Copenhagen has filed a patent application on the cell-based display platform. GlycoDisplay ApS, Copenhagen, Denmark, has obtained a license to the field of the patent application. Y.N. and H.C. are cofounders of GlycoDisplay ApS and hold ownerships in the company.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026102118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Rabinovich G. A., van Kooyk Y., Cobb B. A., Glycobiology of immune responses. Ann. N. Y. Acad. Sci. 1253, 1–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A., Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21, 1121–1124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varki A., Schnaar R. L., Crocker P. R., “I-Type Lectins” in Essentials of Glycobiology, Varki A., et al., Eds. (Cold Spring Harbor, NY, 2015), pp. 453–467. [Google Scholar]
- 4.Duan S., Paulson J. C., Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 38, 365–395 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Macauley M. S., Crocker P. R., Paulson J. C., Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crocker P. R., Paulson J. C., Varki A., Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Büll C., Heise T., Adema G. J., Boltje T. J., Sialic acid mimetics to target the sialic acid-siglec Axis. Trends Biochem. Sci. 41, 519–531 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Figlewicz D. A., Quarles R. H., Johnson D., Barbarash G. R., Sternberger N. H., Biochemical demonstration of the myelin-associated glycoprotein in the peripheral nervous system. J. Neurochem. 37, 749–758 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Collins B. E., Smith B. A., Bengtson P., Paulson J. C., Ablation of CD22 in ligand-deficient mice restores B cell receptor signaling. Nat. Immunol. 7, 199–206 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Lizcano A., et al., Erythrocyte sialoglycoproteins engage Siglec-9 on neutrophils to suppress activation. Blood 129, 3100–3110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Wall S., Santegoets K. C. M., van Houtum E. J. H., Büll C., Adema G. J., Sialoglycans and siglecs can shape the tumor immune microenvironment. Trends Immunol. 41, 274–285 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Chang Y. C., Nizet V., Siglecs at the host-pathogen interface. Adv. Exp. Med. Biol. 1204, 197–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollingworth P.et al.; Alzheimer’s Disease Neuroimaging Initiative; CHARGE consortium; EADI1 consortium , Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naj A. C., et al., Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angata T., Nycholat C. M., Macauley M. S., Therapeutic targeting of siglecs using antibody- and glycan-based approaches. Trends Pharmacol. Sci. 36, 645–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen M., Varki A., The sialome–far more than the sum of its parts. OMICS 14, 455–464 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Varki A., Schnaar R. L., Schauer R., “Sialic acids and other nonulosonic acids” in Essentials of Glycobiology, Varki A., et al., Eds. (Cold Spring Harbor, NY, 2015), pp. 179–195. [Google Scholar]
- 18.Harduin-Lepers A., et al., The human sialyltransferase family. Biochimie 83, 727–737 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Paulson J. C., Rademacher C., Glycan terminator. Nat. Struct. Mol. Biol. 16, 1121–1122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schjoldager K. T., Narimatsu Y., Joshi H. J., Clausen H., Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 21, 729–749 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Gao C., et al., Glycan microarrays as chemical tools for identifying glycan recognition by immune proteins. Front Chem. 7, 833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rillahan C. D., Paulson J. C., Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blixt O., Collins B. E., van den Nieuwenhof I. M., Crocker P. R., Paulson J. C., Sialoside specificity of the siglec family assessed using novel multivalent probes: Identification of potent inhibitors of myelin-associated glycoprotein. J. Biol. Chem. 278, 31007–31019 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Padler-Karavani V., et al., Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J. Biol. Chem. 287, 22593–22608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blixt O., et al., Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campanero-Rhodes M. A., et al., Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem. Biophys. Res. Commun. 344, 1141–1146 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Gao C., et al., Unique binding specificities of proteins toward isomeric asparagine-linked glycans. Cell Chem. Biol. 26, 535–547.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narimatsu Y., et al., An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell 75, 394–407.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Büll C., Joshi H. J., Clausen H., Narimatsu Y., Cell-based glycan arrays-A practical guide to dissect the human glycome. STAR Protoc 1, 100017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narimatsu Y., et al., Genetic glycoengineering in mammalian cells. J. Biol. Chem., 10.1016/j.jbc.2021.100448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narimatsu Y., et al., A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology 28, 295–305 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Büll C., et al., Steering siglec-sialic acid interactions on living cells using bioorthogonal chemistry. Angew. Chem. Int. Ed. Engl. 56, 3309–3313 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Büll C., et al., Sialic acid glycoengineering using an unnatural sialic acid for the detection of sialoglycan biosynthesis defects and on-cell synthesis of siglec ligands. ACS Chem. Biol. 10, 2353–2363 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Lonowski L. A., et al., Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 12, 581–603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto R., et al., Precise integration of inducible transcriptional elements (PrIITE) enables absolute control of gene expression. Nucleic Acids Res. 45, e123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjeldsen T., et al., Preparation and characterization of monoclonal antibodies directed to the tumor-associated O-linked sialosyl-2––6 alpha-N-acetylgalactosaminyl (sialosyl-Tn) epitope. Cancer Res. 48, 2214–2220 (1988). [PubMed] [Google Scholar]
- 37.Powell L. D., Varki A., The oligosaccharide binding specificities of CD22 beta, a sialic acid-specific lectin of B cells. J. Biol. Chem. 269, 10628–10636 (1994). [PubMed] [Google Scholar]
- 38.Powell L. D., Sgroi D., Sjoberg E. R., Stamenkovic I., Varki A., Natural ligands of the B cell adhesion molecule CD22 beta carry N-linked oligosaccharides with alpha-2,6-linked sialic acids that are required for recognition. J. Biol. Chem. 268, 7019–7027 (1993). [PubMed] [Google Scholar]
- 39.Zeng Y., et al., High affinity sialoside ligands of myelin associated glycoprotein. Bioorg. Med. Chem. Lett. 21, 5045–5049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varki A., Selectin ligands. Proc. Natl. Acad. Sci. U.S.A. 91, 7390–7397 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen M., Varki A., Modulation of glycan recognition by clustered saccharide patches. Int. Rev. Cell Mol. Biol. 308, 75–125 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Crottet P., Kim Y. J., Varki A., Subsets of sialylated, sulfated mucins of diverse origins are recognized by L-selectin. Lack of evidence for unique oligosaccharide sequences mediating binding. Glycobiology 6, 191–208 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Cohen M., Hurtado-Ziola N., Varki A., ABO blood group glycans modulate sialic acid recognition on erythrocytes. Blood 114, 3668–3676 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dam T. K., Brewer C. F., Lectins as pattern recognition molecules: The effects of epitope density in innate immunity. Glycobiology 20, 270–279 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Sørensen A. L., et al., Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16, 96–107 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Tarp M. A., et al., Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17, 197–209 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Steentoft C., et al., A strategy for generating cancer-specific monoclonal antibodies to aberrant O-glycoproteins: Identification of a novel dysadherin-Tn antibody. Glycobiology 29, 307–319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson B. J., et al., MUC1 is a counter-receptor for myelin-associated glycoprotein (Siglec-4a) and their interaction contributes to adhesion in pancreatic cancer perineural invasion. Cancer Res. 67, 10222–10229 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Nason R., et al., Display of the human mucinome with defined O-glycans by gene engineered cells. Nat. Commun., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beatson R., et al., The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 17, 1273–1281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beatson R., et al., Cancer-associated hypersialylated MUC1 drives the differentiation of human monocytes into macrophages with a pathogenic phenotype. Commun. Biol. 3, 644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcos N. T., et al., Role of the human ST6GalNAc-I and ST6GalNAc-II in the synthesis of the cancer-associated sialyl-Tn antigen. Cancer Res. 64, 7050–7057 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Takamiya R., Ohtsubo K., Takamatsu S., Taniguchi N., Angata T., The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-β secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology 23, 178–187 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Kumagai T., et al., Airway glycomic and allergic inflammatory consequences resulting from keratan sulfate galactose 6-O-sulfotransferase (CHST1) deficiency. Glycobiology 28, 406–417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macauley M. S., et al., Unmasking of CD22 Co-receptor on germinal center B-cells occurs by alternative mechanisms in mouse and man. J. Biol. Chem. 290, 30066–30077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu H., et al., Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology 27, 657–668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Habuchi O., et al., Purification of chondroitin 6-sulfotransferase secreted from cultured chick embryo chondrocytes. J. Biol. Chem. 268, 21968–21974 (1993). [PubMed] [Google Scholar]
- 58.Torii T., Fukuta M., Habuchi O., Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate Gal-6-sulfotransferase. Glycobiology 10, 203–211 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Rillahan C. D., et al., On-chip synthesis and screening of a sialoside library yields a high affinity ligand for Siglec-7. ACS Chem. Biol. 8, 1417–1422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitagawa H., Paulson J. C., Cloning of a novel alpha 2,3-sialyltransferase that sialylates glycoprotein and glycolipid carbohydrate groups. J. Biol. Chem. 269, 1394–1401 (1994). [PubMed] [Google Scholar]
- 61.Xu R., McBride R., Nycholat C. M., Paulson J. C., Wilson I. A., Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 86, 982–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez P. H., Schnaar R. L., Determination of glycolipid-protein interaction specificity. Methods Enzymol. 417, 205–220 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito H., et al., Systematic synthesis and MAG-binding activity of novel sulfated GM1b analogues as mimics of Chol-1 (alpha-series) gangliosides: Highly active ligands for neural siglecs. Carbohydr. Res. 338, 1621–1639 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Fukuta M., et al., Molecular cloning and expression of chick chondrocyte chondroitin 6-sulfotransferase. J. Biol. Chem. 270, 18575–18580 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Patnode M. L., et al., KSGal6ST generates galactose-6-O-sulfate in high endothelial venules but does not contribute to L-selectin-dependent lymphocyte homing. Glycobiology 23, 381–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinkman-Van der Linden E. C., Varki A., New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope. Sialic acid-binding immunoglobulin superfamily lectins. J. Biol. Chem. 275, 8625–8632 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Kudelka M. R., Ju T., Heimburg-Molinaro J., Cummings R. D., Simple sugars to complex disease–mucin-type O-glycans in cancer. Adv. Cancer Res. 126, 53–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Julien S., Videira P. A., Delannoy P., Sialyl-tn in cancer: (how) did we miss the target? Biomolecules 2, 435–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marcos N. T., et al., ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front. Biosci. (Elite Ed.) 3, 1443–1455 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Silva M. L., Gutiérrez E., Rodríguez J. A., Gomes C., David L., Construction and validation of a Sambucus nigra biosensor for cancer-associated STn antigen. Biosens. Bioelectron. 57, 254–261 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Suzuki H., et al., IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J. Clin. Invest. 118, 629–639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez E., et al., Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 12, 1270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wisnovsky S., et al., Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. U.S.A. 118, e2015024118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hashimoto N., et al., The ceramide moiety of disialoganglioside (GD3) is essential for GD3 recognition by the sialic acid-binding lectin SIGLEC7 on the cell surface. J. Biol. Chem. 294, 10833–10845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamaji T., Teranishi T., Alphey M. S., Crocker P. R., Hashimoto Y., A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues. A comparison with Siglec-9. J. Biol. Chem. 277, 6324–6332 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Gasparrini F., et al., Nanoscale organization and dynamics of the siglec CD22 cooperate with the cytoskeleton in restraining BCR signalling. EMBO J. 35, 258–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ereño-Orbea J., et al., Molecular basis of human CD22 function and therapeutic targeting. Nat. Commun. 8, 764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siddiqui S., et al., Studies on the detection, expression, glycosylation, dimerization, and ligand binding properties of mouse siglec-E. J. Biol. Chem. 292, 1029–1037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du X., Signaling and regulation of the platelet glycoprotein Ib-IX-V complex. Curr. Opin. Hematol. 14, 262–269 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Nguyen K. A., et al., Role of Siglec-7 in apoptosis in human platelets. PLoS One 9, e106239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshimura A., et al., Identification and functional characterization of a Siglec-7 counter-receptor on K562 cells. J. Biol. Chem., 10.1016/j.jbc.2021.100477 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patel N., et al., OB-BP1/Siglec-6. a leptin- and sialic acid-binding protein of the immunoglobulin superfamily. J. Biol. Chem. 274, 22729–22738 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Jandus C., et al., Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 124, 1810–1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J., et al., Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 25, 656–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rillahan C. D., et al., Disubstituted sialic acid ligands targeting siglecs CD33 and CD22 associated with myeloid leukaemias and B cell lymphomas. Chem. Sci. (Camb.) 5, 2398–2406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miles L. A., et al., Small molecule binding to alzheimer risk factor CD33 promotes Aβ phagocytosis. iScience 19, 110–118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodrigues E., et al., A versatile soluble siglec scaffold for sensitive and quantitative detection of glycan ligands. Nat. Commun. 11, 5091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez-Gil A., et al., Sialylated keratan sulfate proteoglycans are Siglec-8 ligands in human airways. Glycobiology 28, 786–801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bochner B. S., et al., Glycan array screening reveals a candidate ligand for Siglec-8. J. Biol. Chem. 280, 4307–4312 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Pröpster J. M., et al., Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8. Proc. Natl. Acad. Sci. U.S.A. 113, E4170–E4179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seko A., Ohkura T., Ideo H., Yamashita K., Novel O-linked glycans containing 6′-sulfo-Gal/GalNAc of MUC1 secreted from human breast cancer YMB-S cells: Possible carbohydrate epitopes of KL-6(MUC1) monoclonal antibody. Glycobiology 22, 181–195 (2012). [DOI] [PubMed] [Google Scholar]
- 92.Varki A., et al., Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.