Burkholderia ubonensis, a nonpathogenic soil bacterium belonging to the Burkholderia cepacia complex (Bcc), is highly resistant to some clinically significant antibiotics. The concern is that B. ubonensis may serve as a resistance reservoir for Bcc or B. pseudomallei complex (Bpc) organisms that are opportunistic human pathogens.
KEYWORDS: Burkholderia, tetracycline, resistance, efflux pump, tetA(64), synergy
ABSTRACT
Burkholderia ubonensis, a nonpathogenic soil bacterium belonging to the Burkholderia cepacia complex (Bcc), is highly resistant to some clinically significant antibiotics. The concern is that B. ubonensis may serve as a resistance reservoir for Bcc or B. pseudomallei complex (Bpc) organisms that are opportunistic human pathogens. Using a B. ubonensis strain highly resistant to tetracycline (MIC, ≥256 µg/ml), we identified and characterized tetA(64) that encodes a novel tetracycline-specific efflux pump of the major facilitator superfamily. TetA(64) and associated TetR(64) regulator expression are induced by tetracyclines. Although TetA(64) is the primary tetracycline and doxycycline resistance determinant, maximum tetracycline and doxycycline resistance requires synergy between TetA(64) and the nonspecific AmrAB-OprA resistance nodulation cell division efflux pump. TetA(64) does not efflux minocycline, tigecycline, and eravacycline. Comprehensive screening of genome sequences showed that TetA(64) is unequally distributed in the Bcc and absent from the Bpc. It is present in some major cystic fibrosis pathogens, like Burkholderia cenocepacia, but absent from others like Burkholderia multivorans. The tetR(64)-tetA(64) genes are located in a region of chromosome 1 that is highly conserved in Burkholderia sp. Because there is no evidence for transposition, the tetR(64)-tetA(64) genes may have been acquired by homologous recombination after horizontal gene transfer. Although Burkholderia species contain a resident multicomponent efflux pump that allows them to respond to tetracyclines up to a certain concentration, the acquisition of the single-component TetA(64) by some species likely provides the synergy that these bacteria need to defend against high tetracycline concentrations in niche environments.
INTRODUCTION
Members of the genus Burkholderia are known to occupy remarkably diverse ecological niches (1). A notable feature of Burkholderia species is that they possess a multipartite genome structure consisting of at least two chromosomes and, in some species, large plasmids (2–5). The ensuing large coding capacity explains at least in part the genus’ metabolic versatility and potential, as well as adaptation to and survival in diverse ecological niches, including adversarial environments (1). A common, yet not very well explored, property is the intrinsic antimicrobial resistance (AMR) of Burkholderia species. This is especially problematic with species that are opportunistic pathogens, which are found in two major Burkholderia complexes, the Burkholderia cepacia complex (Bcc) and the Burkholderia pseudomallei complex (Bpc) (6, 7). Species from both complexes afflict mostly compromised individuals, for instance immunocompromised and cystic fibrosis (CF) patients. Of the opportunistic Bcc bacteria known to be capable of causing serious disease, the two most clinically relevant species are Burkholderia cenocepacia and Burkholderia multivorans, which account for the vast majority of CF infections caused by Bcc bacteria (8). The only known Bpc opportunistic pathogens are B. pseudomallei and Burkholderia mallei, which cause melioidosis and glanders, two diseases with high mortality (9–11). It should be noted that B. mallei is a mammalian host-adapted clone of B. pseudomallei and is incapable of existing outside the host environment for extended periods of time (12). While B. pseudomallei is known to infect individuals with CF (13), the major risk factor for melioidosis is diabetes (11, 14). Bcc and Bpc nonpathogenic and pathogenic bacteria frequently coinhabit soil and aquatic environments in geographic areas where they both exist, mostly in tropical and subtropical regions of the world (15, 16). Burkholderia ubonensis is a nonpathogenic Bcc bacterium that coinhabits soil environments around the globe with B. pseudomallei, e.g., Australia, Thailand, and the Caribbean islands (17). It is highly resistant to some clinically significant antibiotics (17, 18). The concern is that B. ubonensis may serve as a reservoir for horizontal gene transfer of AMR determinants to pathogenic Bcc and Bpc species.
Tetracycline antibiotics have been in clinical use since the first report of chlortetracycline in 1948 (19). In attempts to keep pace with emerging resistance, early naturally occurring tetracyclines, including tetracycline itself that was discovered in 1953, were replaced in time with semisynthetic derivatives (e.g., doxycycline and minocycline, discovered in 1967 and 1972, respectively; the history of discovery of tetracyclines is reviewed in reference 20). After a lengthy pause, efforts were made to identify and develop semisynthetic compounds like tigecycline (discovered in 1993) with efficacy against emerging multidrug-resistant (MDR) organisms (21). The fully synthetic eravacycline is one of the latest derivatives to be introduced into clinical use (22).
For over 70 years, tetracycline antibiotics have been used extensively for management of bacterial infections in human and veterinary medicine and growth promotion in the cattle and poultry industries, as well as treatment of fruit trees for prevention of bacterial diseases (23). The first report of resistance to tetracyclines was in 1953, 5 years after the first clinical deployment of chlortetracycline (24). Resistance mechanisms include tetracycline-specific mechanisms (active efflux from the cell via single-component transporters, production of ribosomal protection proteins, enzymatic degradation, and 16S RNA target mutations) (20, 25) and also nonspecific mechanisms, especially in Gram-negative bacteria, which can include active efflux from the cell via multicomponent transporters and decreased cell-envelope permeability (reviewed in references 20, 25, and 26). The most common tetracycline-specific resistance mechanism is active efflux via single-component pumps belonging to the major facilitator superfamily (MFS) (reviewed in references 20 and 25). At the time of writing the manuscript, there were 35 distinct bacterial tetracycline-specific pumps registered (faculty.washington.edu/marilynr; 20 February 2020 update). Nonspecific multicomponent efflux pumps that actively extrude tetracyclines from Gram-negative bacteria belong to the resistance nodulation cell division (RND) family. Examples are the MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM pumps from Pseudomonas aeruginosa (reviewed in reference 26); the AdeABC pump from Acinetobacter baumannii (27); and the AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC pumps from B. pseudomallei and its closely related near neighbor Burkholderia thailandensis (28–34).
The aggregate of all tetracycline resistance mechanisms discovered and characterized in pathogenic and environmental organisms that are not normally associated with disease is an example of what Gerry Wright and colleagues defined as the resistome for this class of antibiotics (25). According to the resistome concept, it is paramount to investigate resistance in nonpathogenic environmental bacteria since there is ample evidence for them being reservoirs for resistance determinants that can be transmitted to pathogens by horizontal gene transfer (35–37).
We recently defined chromosomally encoded carbapenem resistance mechanisms in the environmental Bcc bacterium B. ubonensis, which has never been associated with animal or human disease (18). During these studies, we noted that the bacterium was highly resistant to tetracycline (TET; MIC ≥256 μg/ml) and doxycycline (DOX; MIC = 32 μg/ml). In this study, we show that the high-level TET resistance is due to synergy between a nonspecific RND efflux pump and a previously unreported tetracycline resistance determinant, tetA(64), that encodes a tetracycline-specific single-component MFS efflux pump, which is transcriptionally regulated by TetR(64). We provide evidence for the presence of tetA(64) in the genomes of some but not all Bcc species and its absence in Bpc species.
RESULTS
Identification of a B. ubonensis mutant with reduced tetracycline susceptibility.
We recently described construction of a random transposon T23 mutant library of B. ubonensis strain Bu278 (B. ubonensis strains used in this study are listed in Table 1) (18). To identify the resistance determinant(s) governing the high (MIC, ≥256 μg/ml) TET resistance (TETr) of strain Bu278, ∼2,000 mutants of this library were screened for increased TET susceptibility as defined by no growth on Lennox broth (LB) plates containing 60 μg/ml TET. We identified one mutant, Bu424, that could no longer grow at this TET concentration, and the MIC of TET for this strain was reduced from ≥256 μg/ml to 16 μg/ml (Table 2). The T23 insertion also caused an 11-fold reduction in the DOX MIC (32 μg/ml for Bu278 versus 3 μg/ml for Bu424), but its effect on the minocycline (MIN) MIC was minimal (3 μg/ml for Bu278 versus 2 μg/ml for Bu424).
TABLE 1.
Burkholderia ubonensis strains used in this study
| Strain | Description | Source |
|---|---|---|
| Bu278a | Wild type; soil isolate, Puerto Rico | 17 |
| Bu333 | Bu278 amrB::T23 | 18 |
| Bu424 | Bu278 tetA(64)::T23 | This study |
| Bu431 | Bu278 ΔtetA(64) | This study |
| Bu436 | Bu431::mini-Tn7T-TMPb | This study |
| Bu434 | Bu431::mini-Tn7T-TMP-PtetA(64)-tetA(64)+ | This study |
| Bu437 | Bu278 ΔamrB | This study |
| Bu441 | Bu278::mini-Tn7T-TMP | This study |
| Bu448 | Bu437::mini-Tn7T-TMP | This study |
| Bu450 | Bu437::mini-Tn7T-TMP-PamrAB-oprA-amrA+B+-oprA+ | This study |
| Bu439 | Bu278 ΔamrB ΔtetA(64) | This study |
| Bu452 | Bu278 ΔtetR(64) | This study |
TABLE 2.
MICs for tetracyclines against B. ubonensis efflux pump mutants
| Strain | Relevant genotype | MIC (µg/ml)a for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TET | DOX | MIN | ERV | TGC | CHL | CIP | GEN | TMP | ||
| Bu278 | Wild type | ≥256 | 32 | 3 | 3 | 6 | 48 | 2 | 128 | 8 |
| Bu424 | Bu278 tetA(64)::T23 | 16 | 3 | 2 | NDb | ND | ND | ND | ND | ND |
| Bu431 | Bu278 ΔtetA(64) | 16 | 3 | 3 | 3 | 6 | 48 | 1.5 | 128 | 8 |
| Bu441 | Bu278::mini-Tn7T-TMP | ≥256 | 32 | 3 | 3 | 6 | ND | ND | ND | ND |
| Bu436 | Bu431::mini-Tn7T-TMP | 12 | 2 | 2 | ND | ND | ND | ND | ND | ND |
| Bu434 | Bu431::mini-Tn7T-TMP-PtetA(64)-tetA(64)+ | ≥256 | 32 | 2 | ND | ND | ND | ND | ND | ND |
| Bu333 | Bu278 amrB::T23 | 96 | 2 | 0.5 | ND | ND | ND | ND | ND | ND |
| Bu437 | Bu278 ΔamrB | 96 | 3 | 0.5 | 0.19 | 0.38 | 24 | 1.5 | 0.5 | 8 |
| Bu448 | Bu437::mini-Tn7T-TMP | 96 | 3 | 0.75 | 0.19 | 0.38 | ND | ND | ND | ND |
| Bu450 | Bu437::mini-Tn7T-TMP-PamrAB-oprA-amrA+B+-oprA+ | ≥256 | 12 | 1.5 | 1.0 | 3 | ND | ND | ND | ND |
| Bu439 | Bu278 ΔtetA(64) ΔamrB | 1 | 0.38 | 0.5 | 0.19 | 0.38 | 24 | 1.5 | 0.5 | 8 |
MIC was determined using the Etest method performed in triplicate on three separate days, and values are reported as the mode of the readings.
ND, not done.
Characterization of the tetracycline resistance locus.
The transposon insertion site in strain Bu424 was mapped to locus CJO66_RS05975 (GenBank assembly accession GCA_002276145.1) (38), which is annotated as a Tet(A)/Tet(B)/Tet(C) family tetracycline MFS transporter (Fig. 1A). The gene encodes a predicted 392-amino acid, 40-kDa protein. BLAST searches showed that the protein was most similar to TetA(41) from environmental Serratia marcescens strain FMC 1-23-O (GenBank accession no. AY264780) (39), exhibiting 70% amino acid identity. Since the amino acid identity of the predicted B. ubonensis protein with any known Tet resistance protein was less than 80%, it was officially assigned the name TetA(64) in accordance with established nomenclature of tetracycline resistance determinants (23).
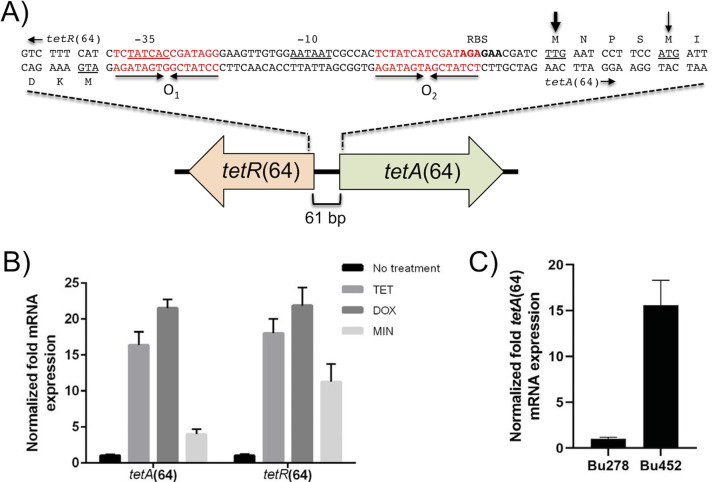
FIG 1.
Genetic organization and functional analysis of B. ubonensis Bu278 tetR(64)-tetA(64) resistance locus. (A) Gene organization and sequence of intergenic region. The tetA(64) gene encoding the TetA(64) MFS efflux pump and the tetR(64) gene encoding the TetR(64) regulatory protein are separated by the 61-bp intergenic region (IR). The thin arrow indicates the TetA(64) ATG start codon (underlined) annotated in the B. ubonensis genome sequences. The more likely TTG start codon (underlined) that is preceded by a putative ribosome-binding site (RBS; bolded) is marked with a thick arrow. The IR contains two regions of imperfect dyad symmetry (inverted arrows), the putative O1 and O2 operator sites, whose sequences are in red type. The predicted −35 and −10 regions of the tetA(64) promoter that share significant homology with bacterial σ70 promoters are underlined. The tetR(64)-tetA(64) region of Bu278 was extracted from GenBank assembly accession no. GCA_002276145.1 and is available via GenBank accession no. MW052058. (B) Transcription of tetA(64) and tetR(64) is induced by tetracyclines. Cells of Bu278 were grown in LB medium in the absence or presence of the indicated tetracyclines, and total RNA was isolated. The tetA(64) and tetR(64) mRNA levels were determined by RT-qPCR. Expression levels are shown relative to uninduced cells. Error bars indicate the standard deviation of comparisons between three biological replicates, of which each was performed in technical triplicate. (C) Transcription of tetA(64) is constitutive in a ΔtetR(64) mutant. Cells of Bu278 and its ΔtetR(64) derivative Bp452 were grown in LB medium, and total RNA was isolated. The tetA(64) mRNA levels were determined by RT-qPCR. Expression levels are shown relative to LB-grown uninduced Bu278 cells. Error bars indicate the standard deviation of comparisons between three biological replicates, of which each was performed in technical triplicate.
A subsequent closer examination of the annotated region upstream of tetA(64) predicts no suitable ribosome-binding site (RBS) ahead of the annotated ATG start codon but instead a putative RBS upstream of a TTG located four codons anterior to the ATG (Fig. 1A). This assignment is supported by the observation that the translation of tetracycline MFS efflux transporters is frequently initiated at a TTG start codon. Examples are S. marcescens TetA(41) and TetA(64) homologs from other Burkholderia species, e.g., B. cenocepacia K56-2 whose TetA(64) is 88% identical to the extended B. ubonensis TetA(64). Other than increasing the tetA(64) gene length from 1,179 to 1,191 bp, shortening of the tetR(64)-tetA(64) intergenic region to 61 bp, and increasing the total amino acid residue count from 392 to 396, the ATG-to-TTG start codon change has no bearing on important TetA(64) parameters, such as percent identity to the nearest tetracycline resistance protein (remains at 70%) and 12 transmembrane α helix domains (see below). In the remainder of the manuscript, we used the TetA(64) gene and protein coordinates that reflect the TTG translation start site.
Mapping of the T32 insertion site in Bu424 showed that the transposon had inserted between nucleotides 287 and 288 in codon 96 of tetA(64). A mutant containing an unmarked 1,179-bp deletion of tetA(64) was constructed, and the resulting ΔtetA(64) strain Bu431 exhibited the same TET, DOX, and MIN susceptibility pattern as the tetA(64)::T23 strain Bu424 (Table 2). Furthermore, TetA(64) did not transport the glycylcycline tigecycline (TGC) and the fluorocycline eravacycline (ERV), of which both were previously shown to not be exported by tetracycline-specific efflux pumps (20). The wild-type Bu278 TET and DOX resistance pattern were restored when ΔtetA(64) was complemented in single copy with wild-type tetA(64) (strain Bu434) but not in strain Bu436 containing the empty mini-Tn7 vector control (Table 2). The results show that TetA(64) is the main TET and DOX resistance determinant of B. ubonensis strain Bu278 but that this MFS transporter does not efflux MIN, ERV, and TGC. TetA(64) is specific for tetracyclines because it does not efflux antibiotics belonging to other classes, including ciprofloxacin (CIP), chloramphenicol (CHL), gentamicin (GEN), and trimethoprim (TMP) (Table 2).
As mentioned above, BLAST searches showed that B. ubonensis TetA(64) is most closely related to S. marcescens TetA(41), which, in turn, was most similar to Acinetobacter sp. Tet(39) (GenBank accession no. AAW66497), a tetracycline efflux pump found in diverse Gram-negative bacteria. Like Tn10 TetA, and the more closely related TetA(41) and Tet(39), TetA(64) belongs to the group 1 family of tetracycline efflux proteins, and it was predicted to contain 12 transmembrane α helices, which is characteristic of this efflux protein family.
Located adjacent to and divergently transcribed from the 1,191-bp tetA(64) gene is locus CJO66_RS05970 (Fig. 1A). This 633-bp gene encodes a predicted TetR family transcriptional regulator, TetR(64) (40). The TetR(64) protein (210 amino acids; 22.8 kDa) is most closely related (62.4% identity) to TetR(41) from S. marcescens strain FMC 1-23-O (GenBank accession no. AY264780) (39) and is predicted to contain a helix-turn-helix DNA-binding domain (amino acids 29 to 50). The genetic tetR-tetA arrangement and the presence of two operator sites in the tetR(64)-tetA(64) intergenic region with strong homology to the TetR consensus-binding sites hints at TetR(64)-mediated transcriptional regulation of tetA(64) and tetR(64) by tetracycline(s). This has been repeatedly observed with other TetR-TetA tetracycline resistance determinants, including the most closely related S. marcescens tetR(41)-tetA(41) (39). Reverse transcription-quantitative PCR (RT-qPCR) analysis confirmed that compared with untreated Bu278, tetA(64) expression is significantly induced by the TetA(64) substrates TET (16-fold) and DOX (22-fold) and to a lesser extent by the nonsubstrate MIN (4-fold) (Fig. 1B). This finding is paralleled by tetR(64) expression, which is significantly induced by TET (18-fold) and DOX (22-fold) and to a lesser extent by the nonsubstrate MIN (11-fold). RT-qPCR analyses also showed that tetA(64) expression is 15.6-fold higher in uninduced strain Bu452 [Bu278 ΔtetR(64)] than baseline levels in uninduced cells of wild-type Bu278 (Fig. 1C). Collectively, the data confirm that TetR(64) is a repressor of tetA(64) expression.
Maximum tetracycline and doxycycline resistance requires TetA(64)-AmrAB-OprA efflux pump synergy.
We previously identified a GEN-susceptible strain of Bu278, Bu333, which had a T23 insertion in the RND transporter component of the AmrAB-OprA efflux pump (18). This pump is ubiquitously present and expressed in Burkholderia species. While best studied in B. pseudomallei, published data suggest that it is responsible for the intrinsic aminoglycoside resistance of Burkholderia species in general (28, 41, 42). Because AmrAB-OprA is known to accommodate tetracycline antibiotics, we also tested the TET, DOX, and MIN susceptibility of the amrB::T23 strain Bu333. The MIC of TET for strain Bu333 was reduced from ≥256 μg/ml to 96 μg/ml (Table 2). The T23 insertion also caused a 16-fold reduction in the DOX MIC (32 μg/ml for Bu278 versus 2 μg/ml for Bu333) and a 6-fold reduction in the MIN MIC (3 μg/ml for Bu278 versus 0.5 μg/ml for Bu333). The same numbers were obtained when MIC determinations were repeated with ΔamrB strain Bu437. Furthermore, use of this strain showed that B. ubonensis AmrAB-OprA also effluxes ERV and TGC; compared with Bu278, the MICs of these antibiotics for Bu437 dropped 16-fold for ERV (from 3 μg/ml to 0.19 μg/ml) and for TGC (from 6 μg/ml to 0.38 μg/ml). The wild-type Bu278 TET MIC was fully restored and DOX, MIN, ERV, and TGC MICs were partially restored when ΔamrB was complemented in single copy with the wild-type amrAB-oprA operon expressed from its native promoter (strain Bu450) but not in strain Bu448 containing the empty mini-Tn7 vector control (Table 2). Deletion of both tetA(64) and amrB had the most profound effect on the resistance to all tetracycline antibiotics tested, especially TET and DOX, whose MICs were lowest in the ΔtetA(64) ΔamrB strain Bu439 (1 μg/ml and 0.38 μg/ml for TET and DOX, respectively). These data are consistent with the synergistic action of the single-component TetA(64) efflux pump and the multicomponent AmrAB-OprA efflux pump to accomplish efflux of TET and DOX from the cytoplasm to the extracellular space (Fig. 2) (43, 44).
FIG 2.
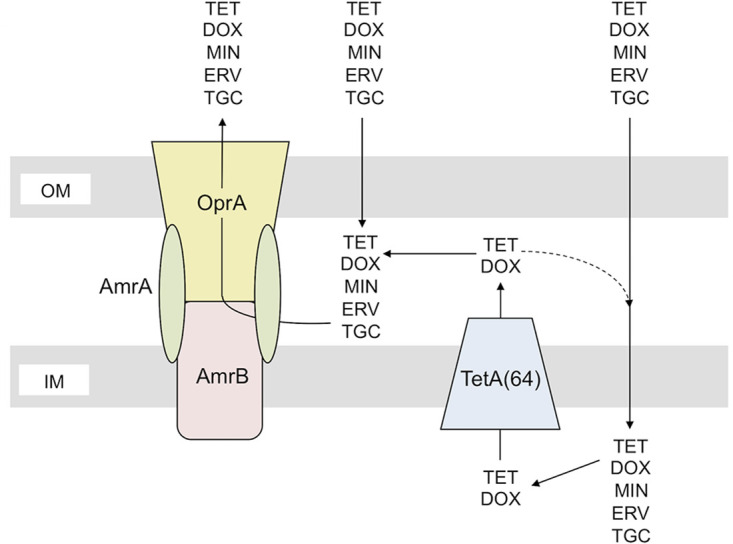
Model of synergistic interaction between single-component and multicomponent efflux pumps in B. ubonensis resistance to tetracycline antibiotics. The single-component MFS transporter TetA(64) removes TET and DOX from the cytoplasm and transports them to the periplasm. The multicomponent AmrAB-OprA RND efflux pump then captures TET and DOX from the periplasm and extrudes the drugs unto the extracellular milieu. A portion of periplasmic TET and DOX may reenter the cytoplasm (dashed line). TetA(64) and AmrAB-OprA efflux pump synergy is required to achieve maximum levels of TET and DOX resistance. In contrast, AmrAB-OprA is an intrinsic resistance determinant for MIN, ERV, and TGC, which are not effluxed by TetA(64).
Distribution of TetA(64)-TetR(64) resistance determinants in Burkholderia species.
To our knowledge, MFS tetracycline-specific efflux transporters and their cognate transcriptional regulators have not yet been definitively demonstrated in Burkholderia species. We therefore used BLAST searches to assess the distribution of TetA(64) and its gene tetA(64), as well as TetR(64) and its gene tetR(64) in the available genomes from over 3,200 Bcc and Bpc bacteria. The sequences were curated to include only those with a full species name and by focusing on nine Bcc species and four Bpc species.
The total number of genome sequences analyzed for the presence of tetA(64) using the 80% identity rule was 2,893, of which 1,600 were B. pseudomallei sequences. We found that tetA(64) is unequally distributed in Bcc bacteria and absent from Bpc bacteria (Table 3). The gene is present in 99.7% of 306 analyzed B. ubonensis genomes and is also present in a few other analyzed Bcc organisms, e.g., 100% Burkholderia stagnalis (100 analyzed), 99.2% B. cepacia (129 analyzed), and 92% B. cenocepacia (300 analyzed). It is absent from other Bcc bacteria, e.g., B. multivorans (n = 211), Burkholderia vietnamiensis (n = 49), Burkholderia diffusa (n = 12), and Burkholderia territorii (n = 34). The tetA(64) gene is also absent from the four Bpc species analyzed, i.e., B. pseudomallei (n = 1,600), B. mallei (n = 82), B. thailandensis (n = 52), and Burkholderia oklahomensis (n = 8) (Table 3).
TABLE 3.
Prevalence of tetA(64) orthologs in the genomes of Bcc and Bpc organismsa
| Presence of tetA(64) | No. of genome sequences analyzed for presence of tetA(64) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Burkholderia cepacia complexb |
Burkholderia pseudomallei complex |
|||||||||
| B. ubonensis (n = 306) | B. cepacia (n = 129) | B. cenocepacia (n = 300) | B. multivorans (n = 211) | B. stagnalis (n = 100) | B. vietnamiensis (n = 49) | B. pseudomallei (n = 1,600) | B. mallei (n = 82) | B. thailandensis (n = 52) | B. oklahomensis (n = 8) | |
| Positive | 305 (99.7%) | 128 (99.2%) | 276 (92%) | None | 100 (100%) | None | None | None | None | None |
| Negative | 1 (0.3%) | 1 (0.8%) | 24 (8%) | 211 (100%) | None | 49 (100%) | 1,600 (100%) | 82 (100%) | 52 (100%) | 8 (100%) |
Species identification was done from a core genome phylogeny.
Other Bcc bacteria analyzed were B. diffusa (n = 12; 100% negative), B. territori (n = 34; 100% negative), and B. ambifaria (n = 10; 9 positive, 1 negative).
Although the TetR proteins are not as conserved as the TetA proteins, the tetA(64) data are mirrored by tetR(64) data. Compared with B. ubonensis TetR(64), TetR proteins are present in TetA(64)-containing Bcc species with identities of 85% to 87% (B. stagnalis), 77% to 78% (B. cepacia), and 76% to 77% (B. cenocepacia). Conversely, TetR proteins are absent from Bcc (e.g., B. multivorans, B. vietnamiensis, B. diffusa, and B. territorii) and Bpc (e.g., B. pseudomallei, B. mallei, B. thailandensis, and B. oklahomensis) species lacking TetA(64).
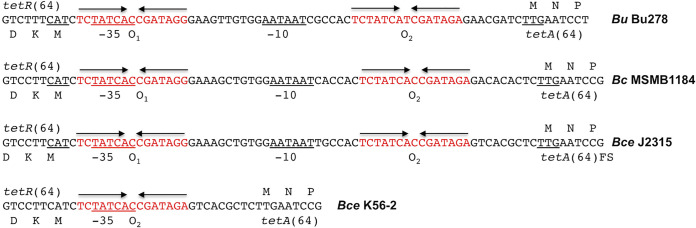
Notable Bcc bacteria containing tetA(64) are K56-2 and J2315, two well-studied B. cenocepacia cystic fibrosis patient isolates of the highly transmissible ET12 lineage (5, 45). It should be noted that tetA(64) is a pseudogene in J2315 due to a deletion of nucleotide T941 (5, 38). This mutation causes a frameshift and production of a 344-residue truncated TetA(64) mutant protein that lacks helices 11 and 12. K56-2 is likely not expressing TetA(64) because it is missing the predicted −10 region of the putative tetA(64) promoter due to a deletion of 38 bp from the tetR(64)-tetA(64) intergenic region caused by a recombination event between the two operator sites (Fig. 3). The latter could also possibly occur in B. ubonensis, B. cepacia, and B. cenocepacia since the intergenic regions are highly conserved in length and sequence across the three species. The B. cepacia MSMB1184WGS and B. cenocepacia J2135 intergenic region (IR) sequences are 94% identical, and both sequences are 92% identical to the IR sequence from B. ubonensis Bu278.
FIG 3.
Conservation of the tetR(64)-tetA(64) intergenic region (IR) in Bcc bacteria. Shown are sequences that are representative of wild-type tetR(64)-tetA(64) IRs (B. ubonensis Bu278, B. cepacia MSMB1184WGS, and B. cenocepacia J2315) and a mutant IR (B. cenocepacia K56-2). Features shown include the predicted amino-terminal amino acid sequence of the tetR(64) and tetA(64) genes on the left and right, respectively, the predicted −35 and −10 regions of the tetA(64) promoter, and the putative O1 and O2 operator sites. The B. cepacia MSMB1184WGS and B. cenocepacia J2315 IRs are identical in length (63 nucleotides), compared with 61 nucleotides for Bu278. The B. cenocepacia K56-2 IR is only 25 nucleotides long and only contains an O2 operator site and a putative −35 sequence but no −10 sequence. FS, frame-shifted and truncated TetA(64) in B. cenocepacia J2315.
Genomic localization of the tetR(64)-tetA(64) locus.
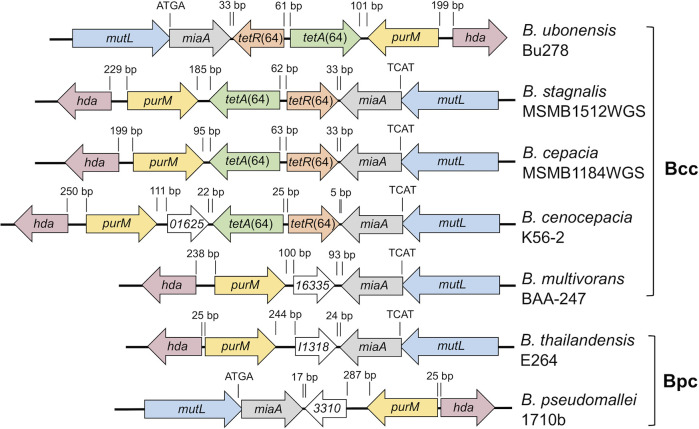
Examination of the genomic location of the tetR(64)-tetA(64) resistance determinants revealed that, when present, they are located on chromosome 1 in a conserved location either upstream or downstream of the miaA gene, dependent on chromosomal orientation (Fig. 4). The tetR(64) gene is always oriented toward miaA and tetA(64) toward purM. In B. ubonensis Bu278, B. stagnalis MSMB1512WGS, and B. cepacia MSMB1184WGS, tetA(64) is located directly adjacent to purM. In B. cenocepacia K56-2, purM and tetA(64) are separated by a 396 bp open reading frame (ORF) that encodes a 131-residue protein. Similar sized ORFs (393 to 402 bp) encoding similar sized proteins (130 to 133 residues) are present in the same location in other Burkholderia species that lack tetR(64) and tetA(64), e.g., B. multivorans BAA-247, B. thailandensis E264, and B. pseudomallei 1710b. The small proteins are very closely related; the K56-2 protein shares 96.6%, 78.5%, and 76.3% identity with the respective proteins from B. multivorans BAA-274, B. thailandensis E264, and B. pseudomallei 1710b. The proteins are either annotated as a ketoisomerase or protein of unknown function. The latter is likely correct since the Burkholderia proteins are missing the active site residues present in other small ketoisomerases, such as the 131-residue ketoisomerase from a Pseudomonas putida species that only shares 22.5% identity with the 131-residue protein from B. cenocepacia K56-2 (46). Except for different chromosomal orientation and some variations due to insertion elements, the genetic makeup of the chromosome 1 regions shown in Fig. 4 for representative strains of the Bcc and Bpc is conserved in the respective species.
FIG 4.
Genetic maps of the chromosome 1 region containing the tetR(64)-tetA(64) resistance determinants in representative Bcc and Bpc strains. In B. ubonensis Bu278, B. stagnalis MSMB1512WGS, and B. cepacia MSMB1184WGS, the tetR(64) and tetA(64) genes are inserted between the purM and miaA genes, with tetR(64) oriented toward miaA and tetA(64) toward purM. B. cenocepacia K56-2 represents a variant where a gene annotated as 01625 (short for locus tag BURCENK562V_RS01625) is inserted between purM and tetA(64). Orthologs of this gene are also present in the same location in Bcc bacteria (e.g., B. multivorans BAA-247) or Bpc bacteria (e.g., B. thailandensis and B. pseudomallei) that do not contain the tetR(64)-tetA(64) locus. Gene annotations are as follows: hda, DNA regulatory inactivator Hda; purM, phosphoribosylformylglycinamidine cyclo-ligase; miaA, tRNA (adenosine[37]-N6)-dimethylallyl transferase; mutL, DNA mismatch repair protein; 01625, 16335 (locus tag NP80_RS16335), I1318 (locus tag BTH_I1318), and 3310 (locus tag BURPS1710b_3310), orthologs that are either annotated as ketosteroid isomerase or hypothetical protein. Distances between genes are indicated in base pairs; ATGA (and reverse) indicates the overlap of the miaA and mutL start and stop codons, respectively.
The intergenic sequences between tetR(64) and tetA(64) and their respective neighboring genes are short, ranging from 22 to 185 bp on the tetA(64) side and 5 to 33 bp on the tetR(64) side in the 4 examples more closely analyzed in this study. These intergenic regions provide no evidence for transposition, i.e., no insertion elements or other significant repeat sequences.
DISCUSSION
Although Burkholderia species are frequently cited as being intrinsically resistant to antibiotics, clinically significant tetracycline resistance in Burkholderia species has to our knowledge been studied and described in any detail only in B. pseudomallei. Historically, TET and more recently DOX have been included in the regimen for oral eradication therapy of B. pseudomallei infections, and DOX remains an important option in instances where other primary (trimethoprim + sulfamethoxazole) or secondary (amoxicillin + clavulanic acid) drugs are contraindicated or ceased because of adverse effects (34, 47). Clinically significant acquired DOX resistance (MIC, ≥16 µg/ml) is rare but has been reported (48), most notably after DOX was used in primary therapy for high-burden B. pseudomallei infection (34). Previous studies with B. pseudomallei and B. thailandensis showed that DOX is exported via the AmrAB-OprA, BpeAB-OprB, and BpeEF-OprA efflux pumps, although tetracyclines are poor RND efflux pump substrates in wild-type strains (28–34). A subsequent study with an isogenic pair of patient isolates of which the second isolate was DOX resistant confirmed an AmrAB-OprA contribution to resistance. However, DOX resistance in this patient isolate was attributed to synergy of loss of methyltransferase function and increased AmrAB-OprA pump efflux activity (34), similar to the synergy noted for TET in this study.
Despite that several Bcc organisms, e.g., B. cenocepacia and B. multivorans, are clinically significant pathogens in CF, very little is known about tetracycline resistance in these species. Tetracyclines such as TET, MIN, and TGC have been included in studies comparing antibiograms for diverse Bcc bacteria, and the studies showed that even newer tetracyclines like TGC are scarcely active (45, 49–51). No tetracyclines were included in recent proposed treatment protocols for Bcc in CF patients (52–54). There are some published data for tetracycline resistance mechanisms in Bcc bacteria (33, 75), and it was shown that the B. cenocepacia RND-4 efflux pump extrudes MIN (55). Based on tetracycline resistance gene amplification by PCR, the Tet(O) ribosomal protection mechanism was proposed for a B. cepacia forest soil isolate (56). The same investigators also proposed a Tet(D) efflux resistance determinant for what is called a B. pseudomallei forest isolate. This is likely a misidentification given the global geographic distribution of B. pseudomallei in subtropical and tropical regions. To our knowledge, neither of these tetracycline resistance determinants have been characterized in detail.
When studying AMR mechanisms in B. ubonensis, we noted that the bacterium exhibited high-levels of resistance to TET (MIC, ≥256 μg/ml) and DOX (MIC, 32 μg/ml) but was considerably more susceptible to MIN (MIC = 3 μg/ml), ERV (MIC = 3 μg/ml), and TGC (MIC = 6 μg/ml). Random transposon mutagenesis of wild-type Bu278 identified a tetracycline-specific efflux pump belonging to the major facilitator superfamily as the primary TET resistance determinant. Although closely related to S. marcescens TetA(41) (70% identity), the B. ubonensis resistance determinant was sufficiently different to warrant assignment of a novel name, TetA(64). Deletion of tetA(64) had a significant effect on TET and DOX resistance, but MIN, ERV, and TGC susceptibility levels were unaffected. Not surprisingly, all tetracyclines tested were substrates of the AmrAB-OprA RND efflux pump, but unlike B. pseudomallei where for instance DOX is a poor AmrAB-OprA substrate, it is a good substrate in B. ubonensis. The Bu439 ΔtetA(64) ΔamrB double mutant was highly susceptible (MICs, 0.19 to 1 µg/ml) to all tetracyclines tested, confirming that tetracycline resistance in B. ubonensis is primarily due to efflux. The double-mutant MIC data also confirmed that TetA(64) pump and AmrAB-OprA efflux pump synergy is required to achieve maximum levels of TET and DOX resistance. This finding is in agreement with that of previous studies with Escherichia coli and Pseudomonas aeruginosa, which showed that synergy between single-component efflux pumps that export antibiotics into the periplasm and multicomponent efflux pumps that then accomplish their efflux into the external medium resulted in resistance levels that were significantly higher than those conferred by each of the pumps operating separately (43, 44). In the present case, the effect for both TET and DOX is multiplicative. While synergistic interactions between pumps of different types have been demonstrated before, the recruitment of a tetracycline-specific resistance determinant in addition to a resident nonspecific tetracycline resistance determinant present in all Burkholderia species to enhance overall resistance levels is a novel observation.
A comprehensive genome analysis from 2,893 Bcc and Bpc organisms using a threshold of 80% amino acid identity over the entire protein length revealed an uneven distribution of the TetA(64) resistance determinant in the Bcc, i.e., either its presence in 92% to 100% of some Bcc species or complete absence in others. The B. stagnalis TetA(64) is most closely related to B. ubonensis TetA(64), which is consistent with the two species being phylogenetically most closely related among the Bcc (57). B. stagnalis has been isolated from the environment and from the respiratory tract of patients (57). Of interest is that of the 24 B. cenocepacia strains whose TetA proteins fell below the 80% identity threshold, 20 were in a cluster with J2315. Eighteen of these strains had identical truncated TetA amino acid sequences as J2315. For two strains, no TetA sequences were available. TetA(64) was found to be absent from the Bpc, including 1,600 B. pseudomallei strains. The presence of TetA(64) in 50% of the Bcc species we examined indicates that its acquisition may be facilitated by these organisms encountering high tetracycline concentrations in the environment.
The data from the present study suggest that it is worth revisiting the sensitivity of individual Bcc pathogens when looking for therapeutic options for infections in CF patients because while resistance to TET and DOX may be present, in some cases the pathogen may be sensitive to MIN, ERV, and TGC.
The synthesis of most Gram-negative Tet efflux pumps is regulated by a repressor protein that is encoded by the tetR gene located adjacent to and transcribed divergently from tetA (58–60). In the presence of tetracycline, the repressor binds the antibiotic and is released from the operators that control tetA and tetR expression (40, 60, 61). B. ubonensis shows a tetR(64)-tetA(64) genetic organization observed in other Gram-negative, including Burkholderia, tetR-tetA systems. The B. ubonensis genes are separated by a 61-bp IR. The IR contains operators that are predicted to govern tetR(64) and tetA(64) expression via TetR(64). In this study, we show that tetA(64) and tetR(64) expression are indeed significantly induced in the presence of the TetA(64) substrates TET and DOX and to a lesser extent also by MIN that is not a substrate of TetA(64). We also showed that TetR(64) is a repressor of tetA(64). Constitutive expression of tetracycline-specific efflux pumps strongly reduces fitness (62). To avoid undue fitness costs, bacteria like B. ubonensis have evolved two mechanisms: (i) tight regulation by TetR and (ii) reduction of translation initiation efficiency by utilization of a TTG start codon for TetA translation. In this context, it is tempting to speculate that the observed mutations in the two B. cenocepacia CF patient isolates J2315 (and at least 18 other strains) and K56-2, which express a truncated TetA(64) or contain a truncated regulatory region, respectively, may reflect tetA(64) purifying selection due to a lack of selective pressure since transitioning from the environment to a human infection.
Most tetracycline-specific MFS efflux pumps are encoded by transposons or plasmids, but some are encoded by chromosomal genes. In B. ubonensis, the tetR(64)-tetA(64) genes are located in a conserved region of chromosome 1 between the miaA (tRNA modification enzyme) and purM (purine metabolism enzyme) genes. Since we could not find any evidence for transposition, the tetR(64)-tetA(64) genes may have been acquired by homologous recombination between conserved flanking housekeeping genes after horizontal gene transfer from a yet unknown source.
In conclusion, we describe the first tetracycline-specific resistance determinant in Burkholderia species TetA(64), which is accompanied by the cognate TetR(64) transcriptional regulator. The tetR(64)-tetA(64) genes are located in the same region on chromosome 1 in all Bcc bacteria that carry them. At present, we do not know where the very closely related, but not identical, tetR(64)-tetA(64) genes observed in various Bcc bacteria originate and how they get integrated into the chromosomes. This begs the question as to whether the resistance genes can be transferred between Burkholderia species. The short tetR(64)-tetA(64)-adjacent gene intergenic regions provide no evidence for transposition, i.e., no insertion elements or other significant repeat sequences. Curiously, in each of the examples examined, the tetR(64)-tetA(64) genes are always oriented with tetA(64) toward purM, which is consistent with site- and orientation-specific transfer of a DNA fragment containing tetR(64)-tetA(64) and neighboring genes. This could happen via homologous recombination between conserved neighboring genes but does not preclude other mechanisms.
Resistance gene transfer between nonpathogenic and pathogenic species is always of concern, especially with organisms like Burkholderia that already contain a formidable armamentarium of AMR determinants (63). Obviously, tetR(46)-tetA(46) have already been acquired by the opportunistic pathogens B. stagnalis, B. cepacia, and B. cenocepacia. The question of whether these resistance genes could be acquired by a high-consequence Bpc pathogen like B. pseudomallei in environments where it frequently coexists with Bcc bacteria like B. ubonensis remains to be answered.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Burkholderia ubonensis strains used in this study are listed in Table 1. Escherichia coli strain DH10B (ThermoFisher Scientific, Waltham, MA) was used for cloning and expression experiments. RHO3 was used for conjugal transfer of plasmids into B. ubonensis (64). Lennox broth (LB) containing 5 g/liter NaCl was used for routine growth of bacteria, and cation-adjusted Mueller-Hinton II agar (MHA; Becton, Dickinson and Company, Sparks, MD) was used for MIC assays. For plasmid selection in E. coli, media were supplemented with 100 µg/ml ampicillin (AMP), 35 µg/ml kanamycin (KAN), or 100 µg/ml trimethoprim (TMP). For use in plasmid maintenance or merodiploid or mini-Tn7 integrant selection in B. ubonensis, KAN and TMP were used at 1,000 µg/ml and 100 µg/ml, respectively.
Antimicrobial susceptibility testing.
MIC assays were performed using Etest strips and following the manufacturer’s (AB bioMérieux, Marcy l’Etoile, France) guidelines.
Deletion mutant construction.
Gene replacement plasmids were constructed by the Gibson assembly method (65). Briefly, overlap sequences for the target plasmid and insert were designed using the NEBuilder assembly tool v.2.2.7 (New England BioLabs, Ipswich, MA). B. ubonensis Bu278 (Bp8955) genomic DNA served a source for PCR-amplified templates and was isolated using the Wizard genomic DNA purification kit (Promega, Madison, WI). Primers were purchased from IDT Technologies (Coralville, IA), and their sequences are provided in Table 4.
TABLE 4.
Primers used in this study
| Primer | Sequence (5′–3′) | Purpose |
|---|---|---|
| P3754 | CCTGTTATCCCTACCCGGGCAATTCCTCGACGGCGCGA | tetA(64) deletion |
| P3755 | CGGATTCCGCGGAAGGATTCAAGATCGTTCTCTATCG | tetA(64) deletion |
| P3756 | GAATCCTTCCGCGGAATCCGGCCGCCCG | tetA(64) deletion |
| P3757 | GGGATAACAGGGTAATCCCGCTCGTGCAGGGCGCCGAG | tetA(64) deletion |
| P3769 | CCGTGCTGCTCGCGTCG | tetA(64) RT-qPCR |
| P3770 | CGTTCGCGCCCGTGATG | tetA(64) RT-qPCR |
| P3773 | CCTGTTATCCCTACCCGGGCCCAGCTCGACTACGCGAC | amrB deletion |
| P3774 | TTCGCAACCTATGAAGATGGCGATCACCC | amrB deletion |
| P3775 | CCATCTTCATAGGTTGCGAAGGATCTCG | amrB deletion |
| P3776 | GGGATAACAGGGTAATCCCGGTCCGACAGGCTCTTCAC | amrB deletion |
| P3777 | CTCGATCATGCATGAGCTCACGCGCCCGTGTCTTTCATC | tetA(64) complementation |
| P3778 | TTCGCGAGGTACCGGGCCCAATTCCGCTTACGCGGCCG | tetA(64) complementation |
| P3853 | CTCGATCATGCATGAGCTCAGTTGCGAGATTCCTTACGTTTTGCTGTC | amrB complementation |
| P3854 | CTGCGGCCGATGTCGCCCACCGTCACGC | amrB complementation |
| P3855 | GTGGGCGACATCGGCCGCAGCGCGGTGC | amrB complementation |
| P3856 | CACGACGAGCACCGACAGCGCGAACAGCATCGG | amrB complementation |
| P3857 | CGCTGTCGGTGCTCGTCGTGTTCCTTGC | amrB complementation |
| P3858 | TTCGCGAGGTACCGGGCCCATCACACGTCGGCTTCCGC | amrB complementation |
| P3887 | ACGAGGTCGGCATCAATG | tetR(64) RT-qPCR |
| P3888 | CATGATCGCTTCCGCCA | tetR(64) RT-qPCR |
| P3889 | CCTGTTATCCCTACCCGGGCCTGTACTACAAGGCGCTG | tetR(64) deletion |
| P3890 | CACGGTCCTGGTGTTCAAGTGCGGCATC | tetR(64) deletion |
| P3891 | ACTTGAACACCAGGACCGTGTCGCGCGT | tetR(64) deletion |
| P3892 | GGGATAACAGGGTAATCCCGTCGACCAGCCGAAATGCGC | tetR(64) deletion |
For construction of a Bu278 derivative containing a 1,179-bp ΔtetA(64), 936 bp of upstream and 854 bp of downstream sequences of the tetA(64) flanking regions were PCR amplified using primer pair 3754 and 3755 and pair 3756 and 3757, respectively. Similarly, for construction of a Bu278 derivative containing a 2,765-bp ΔamrB, 768 bp of upstream and 787 bp of downstream flanking regions of Bu278 ΔamrB were PCR amplified using primer pair 3773 and 3774 and primer pair 3775 and 3776, respectively. For construction of a Bu278 derivative containing a 519-bp ΔtetR(64), 835 bp of upstream and 753 bp of downstream flanking regions of Bu278 ΔtetR(64) were PCR amplified using primer pair 3891 and 3892 and primer pair 3889 and 3890, respectively. DNA fragments were assembled with NotI, and EcoRI linearized pEDL1005 using NEBuilder high-fidelity (hifi) DNA assembly master mix (New England BioLabs), creating pPS3551 [ΔtetA(64)], pPS3556 (ΔamrB), and pPS3596 [ΔtetR(64)] (plasmids used in this study are listed in Table 5).
TABLE 5.
Plasmids used in this study
| Name | Descriptiona | Source |
|---|---|---|
| pEDL1005 | TMPr; allelic exchange vector; sucrose or I-SceI counterselection | 18 |
| pBADSce-Km | KANr; araC-PBAD-I-sceI expression vector with pRO1600(Ts) replicon | Lab collection |
| pPS3551 | TMPr; pEDL1005 with 1,179 bp ΔtetA(64) from ATG start to stop codon | This study |
| pPS3556 | TMPr; pEDL1005 with 2,765 bp ΔamrB (nt 57–2,821 of 3,138-nt ORF) | This study |
| pPS3596 | TMPr; pEDL1005 with 519 bp ΔtetR(64) (nt 43–561 of 633-nt ORF) | This study |
| pPS1897 | TMPr, AMPr; pUC18T-mini-Tn7T-TMP (GenBank accession no. DQ493875) | 18 |
| pPS3553 | TMPr, AMPr; pUC18T-mini-Tn7T-TMP-PtetA-tetA(64) | This study |
| pPS3592 | TMPr, AMPr; pUC18T-mini-Tn7T-TMP-PamrAB-oprA-amrA+B+-oprA+ | This study |
nt, nucleotide(s); Ts, temperature-sensitive.
Plasmid DNA was isolated using the Nucleospin plasmid kit (Macherey-Nagel, Düren, Germany), and plasmid integrity was analyzed by Sanger sequencing. The gene replacement plasmids were transferred to Bu278 by conjugation from E. coli RHO3 (64), and TMP-resistant merodiploids were resolved using I-SceI counter selection as previously described (64). The presence of the desired deletions was verified by PCR amplification and Sanger sequencing.
The resulting mutants are Bu431 [Bu278 ΔtetA(64)], Bu437 (Bu278 ΔamrB), and Bu452 [Bu278 ΔtetR(64)]. The Bu278 ΔtetA(64) ΔamrB mutant Bu452 was obtained by conjugally transferring pPS3556 into strain Bu431 and following the verification steps outlined above for the single mutants.
Deletion mutant complementation.
Mutant complementation was achieved using the mini-Tn7 system, which enables stable and site-specific single-copy insertions into at least two glmS-associated sites in the B. ubonensis genome (18, 66). Delivery plasmids containing mini-Tn7 elements expressing wild-type tetA(64) and amrAB-oprA from their native promoters were constructed by Gibson assembly of PCR fragments amplified from Bu278 genomic DNA.
For pPS3553, a 1,317-bp fragment containing tetA(64) and its promoter was PCR amplified using P3777 and P3778. Purified DNA fragments were assembled with SpeI, and HindIII linearized pPS1897 using NEBuilder hifi DNA assembly master mix (New England BioLabs), creating pPS3553. Plasmid DNA was isolated and its integrity analyzed by Sanger sequencing.
For pPS3592, three DNA fragments with 10- to 20-bp overlaps and the amrAB-oprA operon promoter were PCR amplified using primers P3853 and P3854 (2,058 bp), P3855 and P3856 (1,992 bp), and P3857 and P3858 (1,977 bp), respectively. PCR fragments were assembled with SpeI, and HindIII linearized pPS1897 using NEBuilder hifi DNA assembly master mix, creating pPS3592. Plasmid integrity was analyzed by plasmid next-generation sequencing (MGH CCIB DNA Core, Massachusetts General Hospital, Cambridge, MA).
The mini-Tn7 delivery plasmids pPS3553, pPS3592, and pPS1897 containing the empty mini-Tn7 vector were transferred to Bu278 ΔtetA(64) strain Bu431 (pPS3553 or pPS1897) or the Bu278 ΔamrB strain Bu437 (pPS3592 or pPS1897) by conjugation from E. coli RHO3 (64) or by electroporation (67), along with the transposase-encoding helper pTNS3. Insertions at glmS-associated attTn7 sites were identified using PCR as previously described (18). Complemented deletion mutants are listed in Table 1.
Reverse transcription-quantitative PCR (RT-qPCR).
Expression levels of mRNA levels of target genes were determined in Bu278 grown at 37°C in Lennox Luria Broth medium to mid-log phase (optical density at 600 nm [OD600], 0.4 to 0.6), at which point tetA(64) expression either remained uninduced or was induced for 1 h with 150 µg/ml TET, 16 µg/ml DOX, or 1.5 µg/ml MIN (these concentrations are subinhibitory and correspond to approximately one-half of the respective MIC). Total RNA was extracted using the RNeasy protect bacteria minikit (Qiagen, Valencia, CA), and cDNA synthesis was performed as previously described (30, 68). The primer sets used were Bp23S_F and Bp23S_R previously designed for the B. pseudomallei 23S rRNA housekeeping control (68), P3769 and P3770 for tetA(64), and P3887 and P3888 for tetR(64) (Table 4). Prism (GraphPad Software, La Jolla, CA) was used for data analysis and presentation.
Sequence analysis.
SnapGene software v4.3.9 (GSL Biotech, Chicago, IL) was used to perform general DNA analyses. DNA and protein sequence alignments and similarity predictions were performed using Clustal Omega software (69) on EMBL-EBI (70) (https://www.ebi.ac.uk/Tools/msa/clustalo/). Helix-turn-helix (HTH) DNA-binding domains were predicted by the Rhone-Alpes Bioinformatic Pole Gerland Site (https://npsa-prabi.ibcp.fr) (71). The TMHMM Server 2.0 online at https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 was used for the prediction of transmembrane helices in proteins. For TetA and TetR alignments, the respective sequences for tetA(64) (BW23_977) and tetR(64) (BW23_976) from B. ubonensis were aligned against 3,273 Burkholderia genomes with TBLASTN v2.5.0+ (72) in conjunction with LS-BSR v1.2.0 (73). The blast score ratio (BSR) (74) was calculated by dividing the query bit score by the reference self-alignment bit score. A BSR value of 0.80 is equivalent to 80% peptide identity over 100% of the peptide length.
ACKNOWLEDGMENTS
This work was funded by grant HDTRA1-17-1005-1 from the United States Defense Threat Reduction Agency (DTRA). The contents are solely the responsibility of the authors and do not necessarily represent the official views of DTRA or the Department of Defense.
REFERENCES
- 1.Coenye T, Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 2.Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins TP, Crossman LC, Pitt TL, Churcher C, Mungall KL, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. 2004. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci U S A 101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 5.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahl JW, Vazquez AJ, Hall CM, Busch JD, Tuanyok A, Mayo M, Schupp JM, Lummis M, Pearson T, Shippy K, Colman RE, Allender CJ, Theobald V, Sarovich DS, Price EP, Hutcheson A, Korlach J, LiPuma JJ, Ladner J, Lovett S, Koroleva G, Palacios G, Limmathurotsakul D, Wuthiekanun V, Wongsuwan G, Currie BJ, Keim P, Wagner DM. 2016. The effects of signal erosion and core genome reduction on the identification of diagnostic markers. mBio 7:e00846-16. doi: 10.1128/mBio.00846-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson CHD, Wagner DM, Keim P, Sahl JW. 2018. Developing inclusivity and exclusivity panels for testing diagnostic and detection tools targeting Burkholderia pseudomallei, the causative agent of melioidosis. J AOAC Int 101:1920–1926. doi: 10.5740/jaoacint.18-0014. [DOI] [PubMed] [Google Scholar]
- 8.Jones AM, Dodd ME, Govan JR, Barcus V, Doherty CJ, Morris J, Webb AK. 2004. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59:948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitlock GC, Estes DM, Torres AG. 2007. Glanders: off to the races with Burkholderia mallei. FEMS Microbiol Lett 277:115–122. doi: 10.1111/j.1574-6968.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- 10.Whitmore A. 1913. An account of a glanders-like disease occurring in Rangoon. J Hyg (Lond) 13:1–34. doi: 10.1017/s0022172400005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D. 2018. Melioidosis. Nat Rev Dis Primers 4:17107. doi: 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, Kim HS, Shabalina SA, Pearson TR, Brinkac L, Tan P, Nandi T, Crabtree J, Badger J, Beckstrom-Sternberg S, Saqib M, Schutzer SE, Keim P, Nierman WC. 2010. Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol Evol 2:102–116. doi: 10.1093/gbe/evq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geake JB, Reid DW, Currie BJ, Bell SC, Melioid CFI, Bright-Thomas R, Dewar J, Holden S, Simmonds N, Gyi K, Kenna D, Waters V, Jackson M, O'Sullivan B, Taccetti G, Kolbe J, O'Carroll M, Campbell D, Jaksic M, Radhakrishna N, Kidd TJ, Flight W. 2015. An international, multicentre evaluation and description of Burkholderia pseudomallei infection in cystic fibrosis. BMC Pulm Med 15:116. doi: 10.1186/s12890-015-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis 4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, Wagner DM, Tuanyok A, Wertheim H, Yoke Cheng T, Mukhopadhyay C, Puthucheary S, Day NP, Steinmetz I, Currie BJ, Peacock SJ. 2013. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis 7:e2105. doi: 10.1371/journal.pntd.0002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price EP, Sarovich DS, Webb JR, Ginther JL, Mayo M, Cook JM, Seymour ML, Kaestli M, Theobald V, Hall CM, Busch JD, Foster JT, Keim P, Wagner DM, Tuanyok A, Pearson T, Currie BJ. 2013. Accurate and rapid identification of the Burkholderia pseudomallei near-neighbour, Burkholderia ubonensis, using real-time PCR. PLoS One 8:e71647. doi: 10.1371/journal.pone.0071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price EP, Sarovich DS, Webb JR, Hall CM, Jaramillo SA, Sahl JW, Kaestli M, Mayo M, Harrington G, Baker AL, Sidak-Loftis LC, Settles EW, Lummis M, Schupp JM, Gillece JD, Tuanyok A, Warner J, Busch JD, Keim P, Currie BJ, Wagner DM. 2017. Phylogeographic, genomic, and meropenem susceptibility analysis of Burkholderia ubonensis. PLoS Negl Trop Dis 11:e0005928. doi: 10.1371/journal.pntd.0005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somprasong N, Hall CM, Webb JR, Sahl JW, Wagner DM, Keim P, Currie BJ, Schweizer HP. 2020. Burkholderia ubonensis meropenem resistance: Insights into distinct properties of class A beta-lactamases in Burkholderia cepacia complex and Burkholderia pseudomallei complex bacteria. mBio 11:e00592-20. doi: 10.1128/mBio.00592-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duggar BM. 1948. Aureomycin; a product of the continuing search for new antibiotics. Ann N Y Acad Sci 51:177–181. doi: 10.1111/j.1749-6632.1948.tb27262.x. [DOI] [PubMed] [Google Scholar]
- 20.Grossman TH. 2016. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med 6:a025387. doi: 10.1101/cshperspect.a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sum PE, Petersen P. 1999. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett 9:1459–1462. doi: 10.1016/s0960-894x(99)00216-4. [DOI] [PubMed] [Google Scholar]
- 22.Xiao X-Y, Hunt DK, Zhou J, Clark RB, Dunwoody N, Fyfe C, Grossman TH, O'Brien WJ, Plamondon L, Ronn M, Sun C, Zhang W-Y, Sutcliffe JA. 2012. Fluorocyclines. 1. 7-fluoro-9-pyrrolidinoacetamido-6-demethyl-6-deoxytetracycline: a potent, broad spectrum antibacterial agent. J Med Chem 55:597–605. doi: 10.1021/jm201465w. [DOI] [PubMed] [Google Scholar]
- 23.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts MC. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev 19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 25.Thaker M, Spanogiannopoulos P, Wright GD. 2010. The tetracycline resistome. Cell Mol Life Sci 67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne S, Courvalin P, Périchon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 43:465–470. doi: 10.1128/AAC.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan YY, Tan TMC, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. doi: 10.1128/aac.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mima T, Schweizer HP. 2010. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother 54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507. doi: 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biot FV, Lopez MM, Poyot T, Neulat-Ripoll F, Lignon S, Caclard A, Thibault FM, Peinnequin A, Pagès JM, Valade E. 2013. Interplay between three RND efflux pumps in doxycycline-selected strains of Burkholderia thailandensis. PLoS One 8:e84068. doi: 10.1371/journal.pone.0084068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb JR, Price EP, Currie BJ, Sarovich DS. 2017. Loss of methyltransferase function and increased efflux activity leads to doxycycline resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 61:e00268-17. doi: 10.1128/AAC.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies J, Wright GD. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 36.Marshall CG, Lessard IA, Park I, Wright GD. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother 42:2215–2220. doi: 10.1128/AAC.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liou GF, Yoshizawa S, Courvalin P, Galimand M. 2006. Aminoglycoside resistance by ArmA-mediated ribosomal 16S methylation in human bacterial pathogens. J Mol Biol 359:358–364. doi: 10.1016/j.jmb.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 38.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson SA, Maani EV, Lindell AH, King CJ, McArthur JV. 2007. Novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens. Appl Environ Microbiol 73:2199–2206. doi: 10.1128/AEM.02511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trunck LA, Propst KL, Wuthiekanun V, Tuanyok A, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Peacock SJ, Keim P, Dow SW, Schweizer HP. 2009. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis 3:e0000519. doi: 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Wong J, Tuanyok A, Keim P, Giffard PM, Currie BJ. 2014. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother 58:162–166. doi: 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tal N, Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A 106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varga JJ, Losada L, Zelazny AM, Kim M, McCorrison J, Brinkac L, Sampaio EP, Greenberg DE, Singh I, Heiner C, Ashby M, Nierman WC, Holland SM, Goldberg JB. 2013. Draft genome sequences of Burkholderia cenocepacia ET12 lineage strains K56-2 and BC7. Genome Announc 1:200841-13. doi: 10.1128/genomeA.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somarowthu S, Brodkin HR, D'Aquino JA, Ringe D, Ondrechen MJ, Beuning PJ. 2011. A tale of two isomerases: compact versus extended active sites in ketosteroid isomerase and phosphoglucose isomerase. Biochemistry 50:9283–9295. doi: 10.1021/bi201089v. [DOI] [PubMed] [Google Scholar]
- 47.Chaowagul W, Chierakul W, Simpson AJ, Short JM, Stepniewska K, Maharjan B, Rajchanuvong A, Busarawong D, Limmathurotsakul D, Cheng AC, Wuthiekanun V, Newton PN, White NJ, Day NP, Peacock SJ. 2005. Open-label randomized trial of oral trimethoprim-sulfamethoxazole, doxycycline, and chloramphenicol compared with trimethoprim-sulfamethoxazole and doxycycline for maintenance therapy of melioidosis. Antimicrob Agents Chemother 49:4020–4025. doi: 10.1128/AAC.49.10.4020-4025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenney AW, Lum G, Fisher DA, Currie BJ. 2001. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents 17:109–113. doi: 10.1016/s0924-8579(00)00334-4. [DOI] [PubMed] [Google Scholar]
- 49.Pope CF, Gillespie SH, Moore JE, McHugh TD. 2010. Approaches to measure the fitness of Burkholderia cepacia complex isolates. J Med Microbiol 59:679–686. doi: 10.1099/jmm.0.017830-0. [DOI] [PubMed] [Google Scholar]
- 50.Van Dalem A, Herpol M, Echahidi F, Peeters C, Wybo I, De Wachter E, Vandamme P, Pierard D. 2018. In vitro susceptibility of Burkholderia cepacia complex isolated from cystic fibrosis patients to ceftazidime-avibactam and ceftolozane-tazobactam. Antimicrob Agents Chemother 62:e00590-18. doi: 10.1128/AAC.00590-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodilis J, Denet E, Brothier E, Graindorge A, Favre-Bonte S, Nazaret S. 2018. Comparative genomics of environmental and clinical Burkholderia cenocepacia strains closely related to the highly transmissible epidemic ET12 lineage. Front Microbiol 9:383. doi: 10.3389/fmicb.2018.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El-Halfawy OM, Naguib MM, Valvano MA. 2017. Novel antibiotic combinations proposed for treatment of Burkholderia cepacia complex infections. Antimicrob Resist Infect Control 6:120. doi: 10.1186/s13756-017-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scoffone VC, Chiarelli LR, Trespidi G, Mentasti M, Riccardi G, Buroni S. 2017. Burkholderia cenocepacia infections in cystic fibrosis patients: Drug resistance and therapeutic approaches. Front Microbiol 8:1592. doi: 10.3389/fmicb.2017.01592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia BA, Carden JL, Goodwin DL, Smith TA, Gaggar A, Leon K, Antony VB, Rowe SM, Solomon GM. 2018. Implementation of a successful eradication protocol for Burkholderia cepacia complex in cystic fibrosis patients. BMC Pulm Med 18:35. doi: 10.1186/s12890-018-0594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. 2014. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother 58:7424–7429. doi: 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popowska M, Rzeczycka M, Miernik A, Krawczyk-Balska A, Walsh F, Duffy B. 2012. Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrob Agents Chemother 56:1434–1443. doi: 10.1128/AAC.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Smet B, Mayo M, Peeters C, Zlosnik JEA, Spilker T, Hird TJ, LiPuma JJ, Kidd TJ, Kaestli M, Ginther JL, Wagner DM, Keim P, Bell SC, Jacobs JA, Currie BJ, Vandamme P. 2015. Burkholderia stagnalis sp. nov. and Burkholderia territorii sp. nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int J Syst Evol Microbiol 65:2265–2271. doi: 10.1099/ijs.0.000251. [DOI] [PubMed] [Google Scholar]
- 58.Bertrand KP, Postle K, Wray LV, Jr, Reznikoff WS. 1983. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene 23:149–156. doi: 10.1016/0378-1119(83)90046-X. [DOI] [PubMed] [Google Scholar]
- 59.Chalmers R, Sewitz S, Lipkow K, Crellin P. 2000. Complete nucleotide sequence of Tn10. J Bacteriol 182:2970–2972. doi: 10.1128/jb.182.10.2970-2972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertram R, Hillen W. 2008. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol 1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Møller TS, Overgaard M, Nielsen SS, Bortolaia V, Sommer MO, Guardabassi L, Olsen JE. 2016. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol 16:39. doi: 10.1186/s12866-016-0649-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol 6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 63.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7:1389–1399. doi: 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopez CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 66.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 67.Choi K-H, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select agent compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. 2008. Expression of resistance-nodulation-cell division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from Northern Australia. Trans R Soc Trop Med Hyg 102:S145–S151. doi: 10.1016/S0035-9203(08)70032-4. [DOI] [PubMed] [Google Scholar]
- 69.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dodd IB, Egan JB. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res 18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sahl JW, Caporaso JG, Rasko DA, Keim P. 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasko DA, Myers GS, Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernier SP, Son S, Surette MG. 2018. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200:e00156-18. doi: 10.1128/JB.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]