Zinc is an essential micronutrient for mycobacteria, and its depletion induces multiple adaptive changes in cellular physiology, the most remarkable of which are remodeling and hibernation of ribosomes. Ribosome remodeling, induced upon relatively moderate depletion of zinc, involves replacement of multiple ribosomal proteins containing the zinc-binding CXXC motif (called C+ r proteins) by their motif-free C− paralogs.
KEYWORDS: antibiotic resistance, mycobacterial persistence, ribosome remodeling, tuberculosis, zinc starvation
ABSTRACT
Zinc is an essential micronutrient for mycobacteria, and its depletion induces multiple adaptive changes in cellular physiology, the most remarkable of which are remodeling and hibernation of ribosomes. Ribosome remodeling, induced upon relatively moderate depletion of zinc, involves replacement of multiple ribosomal proteins containing the zinc-binding CXXC motif (called C+ r proteins) by their motif-free C− paralogs. Severe zinc depletion induces binding of mycobacterial protein Y (Mpy) to the 70S C− ribosome, thereby stabilizing the ribosome in an inactive state that is also resistant to kanamycin and streptomycin. Because the Mpy binding region on the ribosome is proximal to the binding pocket of spectinamides (Spa), the preclinical drug candidates for tuberculosis, we addressed the impact of remodeling and hibernation of ribosomes on Spa sensitivity. We report here that while Mpy binding has no significant effect on Spa sensitivity to the ribosome, replacement of S14C+ with its C− counterpart reduces the binding affinity of the drug by ∼2-fold, causing increased Spa tolerance in Mycobacterium smegmatis and Mycobacterium tuberculosis cells harboring the C− ribosome. The altered interaction between Spa and ribosomes likely results from new contact points for D67 and R83 residues of S14C− with U1138 and C1184 of 16S rRNA helix 34, respectively. Given that M. tuberculosis induces ribosome remodeling during progression from the acute to chronic phase of lung infection, our findings highlight new considerations in the development of Spa as effective drugs against tuberculosis.
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis, has surpassed HIV in the annual number of deaths caused by a single infectious agent (1). The World Health Organization estimated 1.5 million TB-related deaths and 10 million new M. tuberculosis infections in 2018 (2), and projections are likely to increase due to the COVID-19-related global crisis. While the frontline treatment regimen comprising rifampin, isoniazid, and pyrazinamide has been crucial in controlling the global TB burden over the last 5 decades, its effectiveness has been progressively diminishing, with increasing frequency of multidrug-resistant TB (MDR-TB) strains (3), raising an urgent need for new TB drugs. The current TB drug pipeline includes multiple ribosome-targeting compounds such as spectinomycin (Spc) analogs, spectinamides (Spa), and sutezolid (4–6), highlighting the mycobacterial ribosome as a key drug target. The mycobacterial ribosome, however, undergoes significant changes in its composition and function in response to a changing growth environment (7–9). Therefore, the question as to how changes in ribosomes impact the activities of the pipeline drugs such as Spa and sutezolid assumes significance. Addressing this question will likely maximize the therapeutic potential of these and future ribosome-targeting antibiotics.
Perhaps the most conspicuous changes in the composition and function of the mycobacterial ribosome have been observed under zinc starvation, which first induces remodeling of the ribosome (8, 9) and then hibernation of the remodeled ribosome (8, 10, 11). Remodeling is associated with replacement of multiple ribosomal (r) proteins containing the CXXC motif (referred to as C+ proteins) with their C− r protein paralogs lacking the motif (8, 9). The replacement is transcriptionally regulated by a zinc-responsive repressor, Zur, which binds to an upstream zur box element of the c− genes under zinc-rich conditions, thereby repressing the transcription of these genes (12, 13). Zinc limitation causes Zur to dissociate from the element, leading to transcription derepression of the c− genes, which are often organized in an operon (8, 9, 13). Expression of the c− genes is sufficient for the incorporation of the corresponding proteins in the 70S ribosome (8), suggesting that C− r proteins kinetically outcompete their C+ counterparts during ribosome assembly. Restoration of zinc homeostasis appears to be an apparent purpose of ribosome remodeling, because mutations in c− genes cause hypersensitivity to zinc starvation (8, 9). In addition, other subtler consequences of ribosome remodeling on the physiology of mycobacteria have been proposed (14, 15), which possibly originate from the differences between the primary sequences of C+ and C− r proteins that may subsequently impact the structure and function of the ribosome.
While the full scope of structural changes from ribosome remodeling are yet to be deciphered from the recently published structures of the C+ (3.3 Å) and C− (3.5 Å) ribosomes (8, 16), early analysis aided by genetic and biochemical evidence identified 70S C− ribosomes as the target for hibernation by mycobacterial protein Y (Mpy) (8, 11). Ribosome hibernation, a highly conserved process induced in growth-arrested bacterial cells, involves binding of specialized proteins to the decoding center of ribosomes, thereby stabilizing the ribosomes as inactive and drug-resistant particles (reviewed in reference 17). These specialized proteins contain a highly conserved double-stranded RNA-binding domain scaffold with βαβββα topology (17). Members of this protein family are broadly divided into two categories based on their ability to hibernate the ribosome either as 100S disomes or 70S monosomes (17). Those forming 100S disomes are generally called hibernation promoting factor (Hpf), which are encoded either as a short variant (Hpfshort) in gamma-proteobacteria, or a long variant (Hpflong) with an extended C-terminal domain (CTD) in other bacterial species (18). While the short variant requires an additional protein called ribosome modulation factor (Rmf) to form a 100S disome, the long variant by itself is sufficient for ribosome dimerization (18–20). The second category of proteins forming hibernating 70S monosomes are usually called protein Y (pY) (21), which, like Hpflong, possess a CTD (17). The nomenclature for Mpy in mycobacteria is derived from the fact that it hibernates the ribosome as a 70S monosome (8). Binding of Mpy to C− ribosomes requires another factor called Mpy recruitment factor (Mrf), which is expressed exclusively under low-zinc conditions (10, 11), thereby implying that zinc starvation is a specific signal for inducing Mpy-dependent ribosome hibernation in mycobacteria. Mpy binds to the 30S subunit of the ribosome, making contacts with helices h31 and h34 (head), h18 (shoulder), h44 (body), h24 (platform), and h28 (neck) and two proteins, S3 and S9 (8). As a result of these contacts, Mpy binding hinders the conformational changes that stabilize the binding of kanamycin (Kan) and streptomycin (Str), conferring resistance against these second-line TB drugs (8). Because Mpy also contacts the 16S rRNA helix h34 near the binding site of Spa (8), we investigated the effect of Mpy on the Spa sensitivity of the C− ribosome.
In this study, we show that while Mpy does not significantly impact Spa sensitivity of the ribosome, replacement of the r protein S14C+ with S14C− reduces Spa binding to the ribosome. The altered Spa sensitivity originates from differences in the contact points of 16S rRNA helix h34 with S14 that arise from C+ to C− substitution. Moreover, the reduced Spa sensitivity of the C− ribosome was also observed in M. tuberculosis, which expresses C− ribosomes during the chronic phase of infection in mouse (8) and likely in human lungs (22).
RESULTS
Reduced Spa sensitivity of C− ribosomes is Mpy independent.
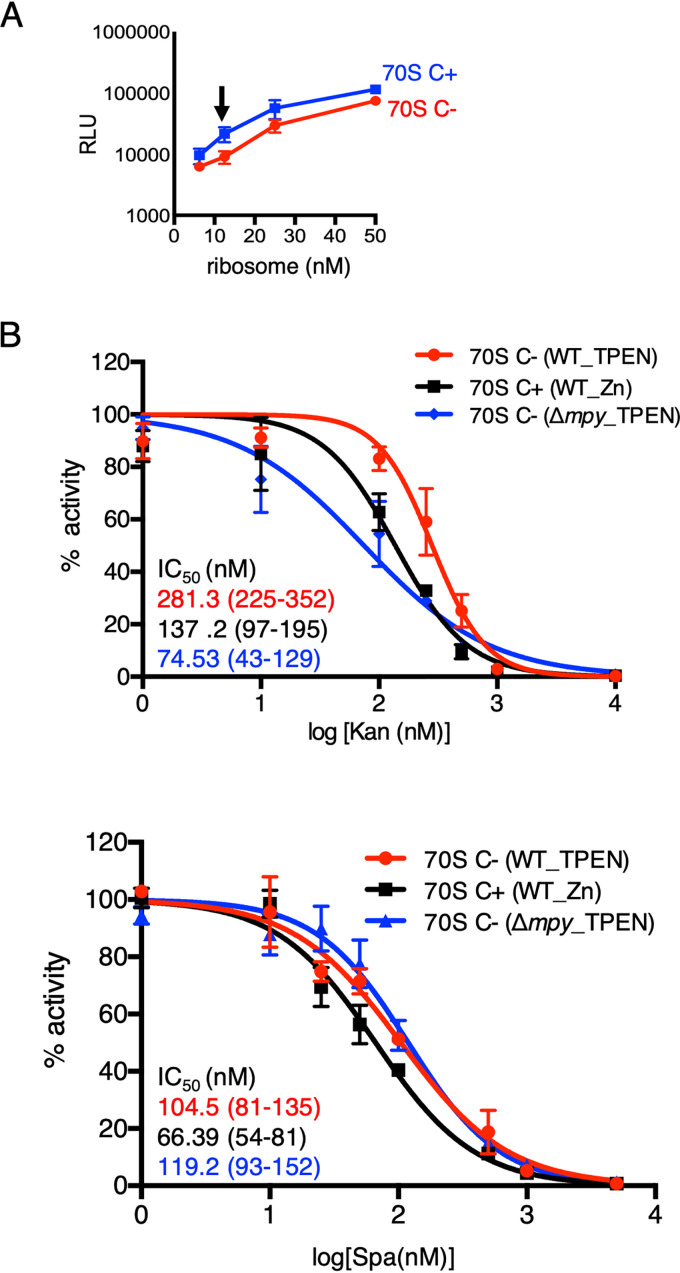
To determine the effect of Mpy on the Spa sensitivity of C− ribosomes, we used a nanoscale luciferase-based in vitro translation assay (11) at a subsaturating ribosome concentration (12.5 nM) (Fig. 1A). We compared the activity of C+ and C− ribosomes from a wild-type strain as well as C− ribosomes from Δmpy strain of Mycobacterium smegmatis in the presence of various concentrations of Spa_1599 (5) (henceforth called Spa). Moreover, activities of these ribosomes in the presence of Kan were also determined as a reference, since Mpy-dependent resistance to Kan was previously demonstrated (8). The 50% inhibitory concentration (IC50) values of both Kan and Spa were higher for C− than for C+ ribosomes, although the extent of difference varied (Fig. 1B). The Kan sensitivity of C− ribosomes from the Δmpy strain was expectedly similar to that of C+ ribosomes, although the Spa sensitivities of C− ribosomes from wild-type and Δmpy strains were indistinguishable (Fig. 1B). We thus conclude that while the C− ribosome is less sensitive to Spa, the effect is independent of its association with Mpy.
FIG 1.
MPY-independent decrease in Spa sensitivity of C− ribosomes of M. smegmatis. (A) Relationship between concentration and activity of C+ ribosomes purified from 96-h-old cultures of M. smegmatis in high-zinc (1 mM ZnSO4) Sauton’s medium (blue) or C− ribosomes purified from 96-h-old cultures of M. smegmatis in low-zinc (1 µM TPEN) Sauton’s medium (red). The activity was measured as relative luminescence units (RLU) of nano-luciferase synthesized in vitro in a transcription-translation coupled reaction. The arrow indicates the concentration (12.5 nM) selected for in vitro translation experiments shown in panel B and in the remainder of this paper. (B) Response of 12.5 nM wild-type (WT) C+, wild-type C−, and Δmpy C− ribosomes to indicated concentrations of Kan and Spa in an in vitro transcription-translation assay utilizing nano-luciferase described for panel A. While WT C+ ribosomes were purified from 96-h-old cultures in high-zinc Sauton’s medium, C− ribosomes from WT and Δmpy strains were purified from 96-h cultures in low-zinc Sauton’s medium. IC50 values (in colors corresponding to the plots) were obtained by GraphPad Prism from the variable slope of the nonlinear dose-response curve. Data represent two biologically independent experiments. Color-coded values in parenthesis denote 95% confidence intervals determined from the standard errors of the corresponding plots.
Reduced Spa sensitivity is intrinsic to C− ribosomes.
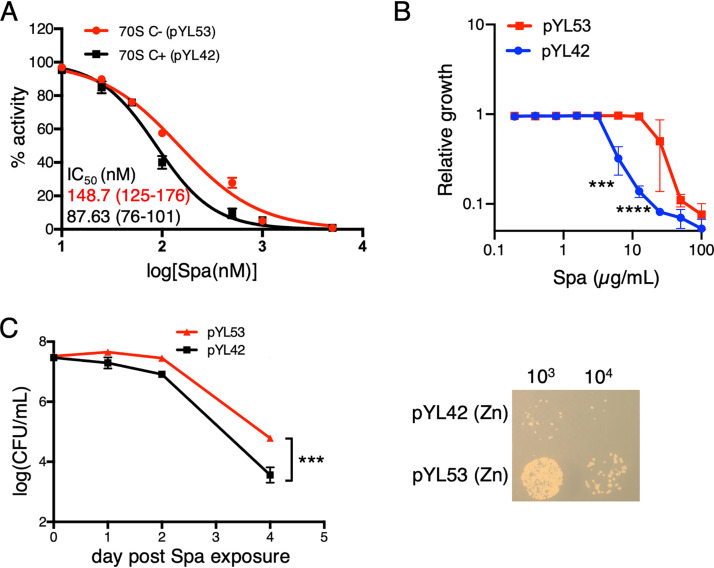
Reduced Spa sensitivity of C− ribosomes can arise either from intrinsic changes in the ribosome structure due to substitution of r proteins or from binding of an unknown extrinsic factor to the C− ribosome under low-zinc conditions. To resolve these possibilities, we utilized a previously constructed recombinant M. smegmatis strain (8), in which the promoter of the c− operon was mutated to abolish its responsiveness to Zur. The mutant promoter cloned upstream of the c− operon on an integrative plasmid (pYL53) rendered constitutive expression of the operon, even in medium with 1 mM ZnSO4 (8). High-zinc cultures of the Δc− strain of M. smegmatis harboring plasmid pYL53 predominantly produced C− ribosomes, while its isogenic counterpart harboring parent plasmid pYL42 produced C+ ribosomes (8) (see Fig. S1 in the supplemental material). Using the luciferase-based in vitro transcription-translation assay, we analyzed the Spa sensitivity of C+ and C− ribosomes purified from high-zinc cultures of pYL42 and pYL53 strains, respectively. Consistent with the property of C− ribosomes from a low-zinc culture of the wild type (Fig. 1B), recombinant C− ribosomes from high-zinc cultures also exhibited reduced sensitivity to Spa compared to that of C+ ribosomes (Fig. 2A). The difference in Spa sensitivity of C+ and C− ribosomes was further recapitulated at the cellular level; the MIC of Spa, measured by resazurin assay (23), was ∼4-fold higher for pYL53 than for pYL42 in high-zinc medium (Fig. 2B), and the bactericidal activity of Spa was at least an order of magnitude lower against pYL53 than against pYL42 strain (Fig. 2C). Taken together, we infer that Spa resistance arises from intrinsic changes in the ribosomes contributed by the C+ to C− r protein replacements but not from secondary effects of zinc depletion.
FIG 2.
Ribosome remodeling is sufficient to reduce Spa sensitivity in M. smegmatis. (A) Spa sensitivity in C+ and C− ribosomes, purified from recombinant M. smegmatis strains pYL53 and pYL42 cultured in high-zinc Sauton’s medium. C+ ribosomes were purified from a high-zinc culture of a Δc− mutant harboring pYL42 (an integrative plasmid containing the wild-type c− operon), whereas C− ribosomes were purified from a high-zinc culture of a Δc− mutant harboring pYL53 (an integrative plasmid constitutively expressing the c− operon). IC50 values were calculated as described for Fig. 1B. Values in parentheses denote 95% confidence intervals determined from the plots of corresponding colors. (B) Growth inhibitory activity of Spa against pYL42 and pYL53 strains in high-zinc Sauton’s medium. Approximately 100 µl of log-phase cultures of each strain from 7H9-ADCTw medium were diluted to an OD600 of 0.025 and mixed with 100 µl 7H9-ADCTw medium containing Spa at the indicated concentrations. Cellular viability was assessed with resazurin dye after 16 h of incubation in the antibiotic containing medium. Data represent averages from three biologically independent experiments. (C) Time-dependent bactericidal activity of Spa against pYL42 and pYL53 strains in high-zinc Sauton’s medium. A saturated culture of each strain was exposed to 50 µg/ml of Spa, and the number of viable cells in the culture was determined at the indicated time points by plating dilutions on 7H10ADC medium and incubating for 3 days. A representative photograph of a plate from a 4-day time point of Spa exposure is shown on the right. Data represent means ± standard deviations (SDs) from three biologically independent experiments. ***, P < 0.001; ****, P < 0.0001 by t test.
Incorporation of S14C− in the ribosome reduces Spa sensitivity.
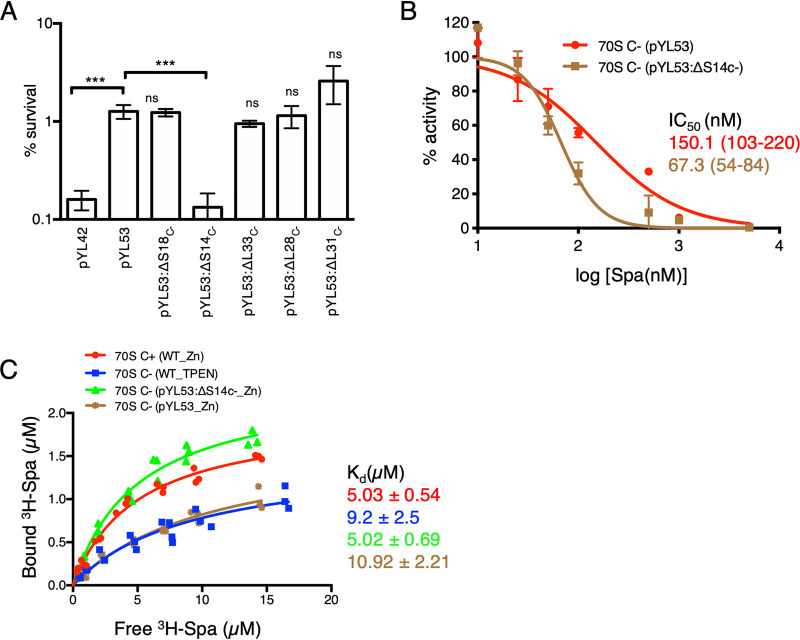
We next asked if the change in Spa sensitivity of C− ribosomes results from C+/C− substitution of any specific r proteins. We constructed in-frame deletions in each of the five genes of the c− operon (encoding S14C−, S18C−, L28C−, L33C−, and L31C−) in the pYL53 template and measured the effect of the mutations on bactericidal activity of Spa. The iTRAQ-MS (isobaric tags for relative and quantitative mass spectrometry) analyses of ribosomes purified from the deletion strains confirmed specific depletion of the corresponding r protein with enrichment of their respective C+ counterparts (8) (see Fig. S2). The only mutation that produced a significant impact on Spa sensitivity was in the gene encoding the r protein S14C− (Fig. 3A), implying that the substitution of S14 is a specific determinant of the altered Spa sensitivity of C+/C− ribosomes. This was further validated by comparing the Spa sensitivity of ribosomes purified from high-zinc cultures of pYL53 (C− ribosomes) and pYL53:ΔS14C− (C− ribosomes with S14C+). The IC50 of Spa for ribosomes from pYL53:ΔS14C− was significantly less than that for ribosomes from pYL53 (Fig. 3B) and closer to that for C+ ribosomes shown in Fig. 2A. Thus, S14 substitution appears to be the singular underlying cause for reduced Spa sensitivity of the C− ribosome.
FIG 3.
S14C− is necessary and sufficient to reduce the Spa sensitivity of C− ribosomes. (A) Survival of the indicated strains of M. smegmatis after 4 days of Spa (50 µg/ml) exposure. The strain harboring pYL53 plasmid constitutively expressed C− ribosomes, while each of the others harboring pYL53-based plasmid with an in-frame deletion in the indicated gene of the c− operon expressed C− ribosomes without the corresponding protein. Protein compositions of 70S ribosomes from pYL53:ΔS14C− and pYL53:ΔS18C− strains were previously analyzed by mass spectrometry (8); those from pYL53:ΔL28C− and pYL53:ΔL33C− are shown in Fig. S2 in the supplemental material, confirming specific loss of the corresponding C− proteins. While incorporation of C+ counterparts of S18, L28, and L33 in the mutant ribosomes was also confirmed by the MS analysis, incorporation of S14C+ is implied from the fact that S14 is an essential protein for ribosome function. The strain harboring pYL42 and expressing C+ ribosomes was used as the control. Data represent means ± SDs from three biologically independent experiments. ***, P < 0.001 by t test; ns, not significant with respect to pYL53. (B) Spa sensitivity in vitro of ribosomes from high-zinc cultures of pYL53 and pYL53:ΔS14C− strains. The translation activity of 12.5 nM ribosomes was measured using a nano-luciferase assay and calculated as percent decrease from the activity measured in the absence of Spa. IC50 of Spa against each of the ribosomes (indicated with the corresponding colors) was determined as described for Fig. 1B. Data represent two biologically independent experiments. The values in parentheses denote 95% confidence intervals determined from the plots of corresponding colors. (C) Saturation binding plot of 3H-Spa to wild-type (WT) C+ and C− 70S ribosomes purified from low- and high-zinc cultures, respectively, as well as recombinant C− ribosomes purified from high-zinc cultures of pYL53 and pYL53:ΔS14C−. Kd values were determined from nonlinear regression curve fit (Y = Bmax × X/Kd + X) using GraphPad Prism. Three biologically independent preparations of each type of ribosomes were used.
The binding pocket of Spa overlaps that of spectinomycin (Spc) (24), which binds to a functionally critical region of h34 within the 30S subunit (25, 26). Given that h34 directly interacts with the S14 protein (26, 27), we predicted that substitution of S14 in the C− ribosomes will directly impact the binding pocket of the drug. The binding affinity of 3H-labeled Spa for C− ribosomes, either from low-zinc wild-type or high-zinc pYL53 cultures, was approximately ∼2-fold lower than that for C+ ribosomes from high-zinc cultures of the wild type (Fig. 3C). Importantly, the Spa affinity for ribosomes from pYL53:ΔS14C− was similar to that for C+ ribosomes, suggesting that C− substitution of S14C+ alters the interaction between the protein and h34.
Structural basis of altered Spa sensitivity by S14 substitution.
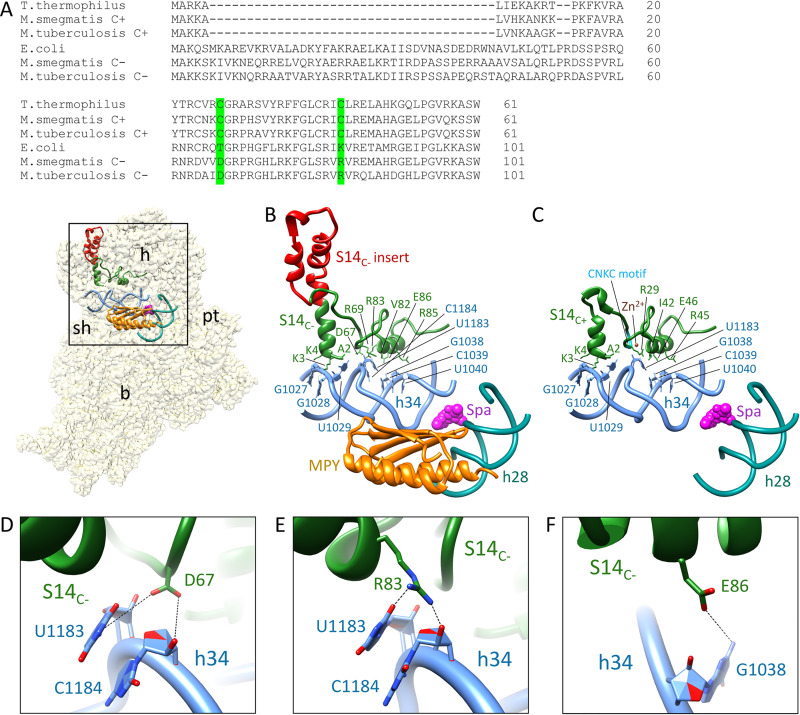
Spectinomycin (or Spa) binds in the minor groove of h34 (contacting G1064, G1068, C1192, and G1193 residues that constitute the crucial hinge 2 region near the h34-h35 junction) in a partially swiveled head of the 30S subunit, thereby inhibiting its rotation during translocation of tRNA and mRNA on the ribosome (25, 28, 29). S14 also contacts h34 near the h34-h35-h38 three-way junction (30). We therefore hypothesized that altered Spa sensitivity originates from changes in the contact points between h34 and the substituted S14C− protein, which substantially differs from S14C+ in its primary sequence (Fig. 4A). In addition to the substitution of cysteines in the two CXXC motifs of S14C+, S14C− has multiple substitutions and a 40-amino acid (aa) insertion in the N-terminal region (Fig. 4A). To decipher the consequence of these differences on the S14-h34 interaction, we analyzed the recently published structures of C+ and C− ribosomes (8, 16). For this purpose, we significantly improved the resolution of our published C− ribosome cryo-electron microscopy (cryo-EM) map (8) to 3.11 Å by implementing the dose weighting during frame alignment (31). In the C+ ribosome, S14 makes seven contacts with h34 (Fig. 4C). In the C− ribosome, S14C− makes two additional contacts with h34, involving D67 and R83, although the 40-aa N-terminal insertion in S14C− does not have any interactions with h34 (Fig. 4B). The residues D67 and R83 contact the bases U1138 and C1184 of h34 through hydrogen bond interactions (Fig. 4D and E).
FIG 4.
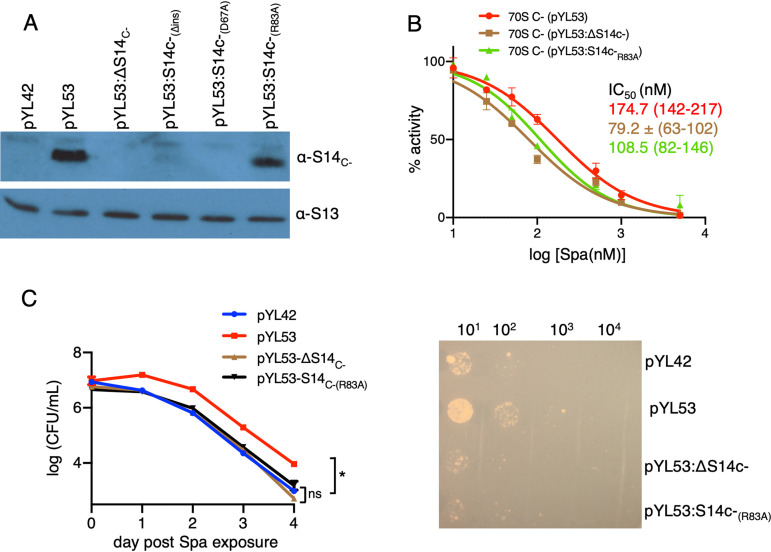
New contact points between S14C− and h34 underlie the reduced Spa sensitivity in C− ribosomes. (A) Sequence alignment of S14 proteins from various bacteria, showing 40-aa residue insertion in the C− paralogs. Residues in S14C− of M. smegmatis making additional contacts with h34 and the corresponding residues in other bacteria are highlighted in green. The alignment was created by Clustal Omega (43). (B) Spc/Spa (magenta)-binding region of the C− ribosome (EMD ID 23076, PDB ID 6DZI) in the context of the interacting residues of the protein S14 (green) with the 16S rRNA helix 34 (light blue). The C−-specific insertion in S14C− is shown in red. The proximity of Mpy (orange) to Spc/Spa binding region on h34 and h28 (dark cyan) are also shown. (C) Spc/Spa-binding region of the C+ ribosome (PDB ID 5O61). The CXXC motif of the S14C+ is shown in cyan, whereas the color of other components is same as in panel B. (D to F) Close-up views of direct interactions of D67, R83, and E86 from S14 with h34 in the C− ribosome. The landmarks of the ribosome labeled in the thumbnails are as follows: h, head; b, body; pt, platform; sh, shoulder.
We predicted that the additional contacts with S14C− are likely to impose rigidity on h34, the flexibility of which is crucial for head rotation and subsequent binding of Spc (25, 28). To test our prediction, we generated Ala substitutions in R83 (S14C−[R83A]) and D67 (S14C−[D67A]). We also constructed in-frame deletion of 40-aa N-terminal insertion (S14C−[Δins]) to determine its contribution to S14C−-dependent Spa sensitivity. Surprisingly, neither S14C−(D67A) nor S14C−(Δins) were incorporated in the C− ribosomes (Fig. 5A), suggesting that these mutations likely affect the assembly of S14C− in the ribosome. The region corresponding to 40-aa N-terminal insertion in S14C− of Escherichia coli was previously implicated in stabilizing the head domain of 16S rRNA or the loose N terminus of S3 (30). In either scenario, incorporation of S14C−(Δins) may result in abortive assembly, which is likely rescued by constitutively expressed S14C+. Similarly, an important role of D67 in incorporation of S14C− suggests that its interaction with U1138 and C1184 of h34 stabilizes the head domain of the 30S ribosome. The lack of these features in S14C+ is perhaps compensated for by the zinc-coordinated CXXC motifs, which are reported to be critical for the function of S14C+ in Bacillus subtilis (32).
FIG 5.
The amino acid residue R83 of S14C− and Spa sensitivity of the C− ribosome. (A) Immunoblot analysis of recombinant C− ribosomes purified from high-zinc cultures of M. smegmatis strains constitutively expressing the c− operon from pYL53 or its derivatives carrying various mutations in S14C−; ΔS14C− denotes in-frame deletion, S14C−(Δins) denotes in-frame removal of the 40-aa-residue insertion, and S14C−(D67A) and S14C−(R83A) denote alanine substitutions at the respective residues. (B) Spa sensitivity in ribosomes from pYL53, pYL53:ΔS14C−, or pYL53:S14C−(R83A). Translation activities of ribosomes shown in panel B were measured using a nano-luciferase assay and calculated as percent decrease from the activity measured in the absence of Spa. IC50 of Spa against each of the ribosomes (indicated with the corresponding colors) was determined as described for Fig. 1B. Data from two biologically independent experiments were plotted. The values in parentheses denote 95% confidence intervals determined from the plots of corresponding colors. (C) Spa sensitivity in cells of the strains described for panel B. High-zinc cultures of these strains were inoculated in high-zinc Sauton’s medium containing 50 µg/ml of Spa and incubated for the indicated time periods prior to plating dilutions and enumerating colonies (after 3 days of incubation). Data represent means ± SDs from three biologically independent experiments. *, P < 0.05 by t test. A representative plate for 4-day time point is shown on the right.
Alanine substitution of R83, however, did not affect the incorporation of S14C−(R83A) in the ribosome (Fig. 5A), although Spa sensitivity of the mutant C− ribosome was substantially increased (Fig. 5B). Consistent with an increased Spa sensitivity in vitro, cells expressing C− ribosomes with S14C−(R83A) exhibited reduced survival against the antibiotic (Fig. 5C). These findings together indicate that additional contact between S14C− and h34 through R83 contributes to reduced Spa sensitivity in the ribosome.
Reduced Spa sensitivity of C− ribosomes in M. tuberculosis.
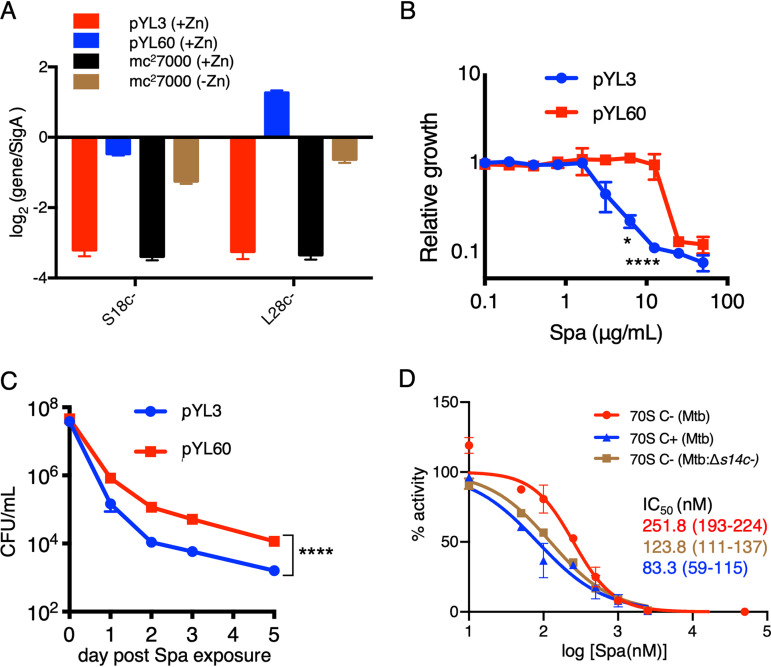
Both D67 and R83 residues are conserved in S14C− of M. tuberculosis (Fig. 4A), suggesting that C+ to C− remodeling of ribosomes, induced in zinc-starved M. tuberculosis during progression from acute to chronic infection of mouse lungs (8), likely reduces the Spa sensitivity in the pathogen. Because this possibility has implications in optimizing treatment efficacy of Spa against TB, we prioritized testing our M. smegmatis findings in M. tuberculosis. To avoid the confounding effects of changing the growth conditions associated with zinc starvation, we chose to compare the Spa sensitivity of recombinant M. tuberculosis cells expressing either C+ or C− ribosomes under the same growth condition, i.e., high-zinc Sauton’s medium. Because transcriptional derepression of the c− operon is sufficient to induce the assembly of C− ribosomes under high-zinc conditions (8), we constructed an M. tuberculosis strain carrying a deletion in the native c− operon while constitutively expressing the operon from an engineered constitutive promoter (Pcosnt) (8) on an integrative plasmid (pYL60) (see Table S1). The quantitative reverse-transcription PCR (qRT-PCR) assay confirmed constitutive expression of the operon in the pYL60 strain, quantitatively comparable to the induced level in the low-zinc culture of the wild type (Fig. 6A). Consistent with the observations in M. smegmatis (Fig. 2A), the MIC50 of Spa was ∼4-fold higher for the pYL60 strain than for the strain carrying empty vector (pYL3) (Fig. 6B). The data were further validated by the significantly decreased bactericidal activity of Spa against the pYL60 strain compared to that against the pYL3 control strain (Fig. 6C). To further confirm that the reduced Spa sensitivity in M. tuberculosis is due to S14C− incorporation in the ribosome, we assessed the Spa IC50 for 70S ribosomes purified from low-zinc cultures of a recombinant strain carrying in-frame deletion in the s14c− open reading frame (ORF) on a plasmid-borne c− operon expressed from the native promoter (pYL97). The genomic copy of the c− operon was deleted in the pYL97 strain. The IC50 of Spa for ribosomes purified from the pYL97 strain was approximately 2-fold less than that for C− ribosomes purified from the wild type (Fig. 6D). In fact, the Spa IC50 for the pYL97 ribosome was closer to that for C+ ribosomes purified from high-zinc cultures of the wild type (Fig. 6D). Taken together, the reduced Spa sensitivity from S14 substitution in the ribosome extends beyond fast-growing mycobacterial species to slow-growing pathogenic mycobacteria, thereby implying a broader implication of our findings.
FIG 6.
Reduced Spa sensitivity in M. tuberculosis (mc27000) expressing C− ribosomes. (A) qRT-PCR-based analysis of the expression of two genes of the c− operon (encoding S18C− and L28C−) in high- (1 mM ZnSO4) and low-zinc (without any zinc supplement) cultures of mc27000 and an isogenic Δc− strain transformed with either pYL60 (expressing c− operon from a constitutive promoter) or an empty vector control (pYL3). Growth inhibitory activity (B) and bactericidal activity (C) of Spa against an mc27000:Δc− strain transformed with either pYL60 or empty vector control (pYL3). Growth inhibition was determined by resazurin assay as described for Fig. 2B. For measuring bactericidal activity of Spa, high-zinc saturated cultures of the indicated strains were inoculated in Sauton’s medium with 100 µg/ml of Spa and incubated for the indicated time periods prior to plating dilutions and enumerating colonies. Data in both panels represent means ± SDs from three biologically independent experiments. *, P < 0.05; ****, P < 0.0001 by t test. (D) Translation activities of 70S ribosomes from the indicated strains in the presence of Spa. 70S C+ and C− ribosomes were purified from high- and low-zinc pellicle cultures of mc27000, while 70S C− ribosomes without S14C− were purified from low-zinc pellicle cultures of an isogenic Δc− strain transformed with pYL97 (a plasmid expressing c− operon without S14c− from the native promoter). IC50 values were calculated as described for Fig. 1B. The values in parentheses indicate 95% confidence intervals determined from the plots of corresponding colors. Data represent two biologically independent experiments.
DISCUSSION
Mycobacteria exhibit multiple responses to zinc starvation, graduated based on the severity of zinc scarcity. While moderate zinc depletion leads to remodeling of the ribosome that involves substitution of multiple ribosomal proteins with CXXC motifs with their motif-free paralogs, severe zinc depletion to a growth prohibitory level induces hibernation of the remodeled 70S ribosome. Hibernation involves binding of Mpy to the decoding center of the 30S subunit. We previously demonstrated that binding of Mpy reduces the sensitivity of ribosomes to kanamycin and streptomycin (8). In this study, we show that substitution of S14 during ribosome remodeling reduces the Spa affinity in the ribosome. The reduced Spa binding to the remodeled ribosome appears to be a consequence of an altered interaction between S14 and h34 of 16S rRNA, which is characterized by two additional contacts between R83 and D67 of S14 and C1184 and G1038 of h34. The corresponding nucleotides (G1058 and C1203) in E. coli 16S rRNA are proximal to the hinge 2 region, which forms a noncovalent connection between the head and the body domains of the 30S subunit (25). It is therefore likely that the additional contact points would increase rigidity of the hinge 2 region, thereby reducing the degree of rotation/swiveling of the 30S C− ribosomes. Moreover, a partially swiveled head of the Spc-bound 30S ribosome suggests that binding of the antibiotic is linked to the degree of swiveling in the head region (28). Thus, it is tempting to speculate that altered head rotation in the 30S C− ribosome caused by a rigid hinge 2 would hinder Spc/Spa access to the binding pocket.
The head rotation is an intrinsic property of the 30S subunit that accompanies ratchet-like (33) and head-swiveling (34) motions during various steps of protein synthesis. A likely restrictive change in head rotation of the 30S subunit from additional contacts of S14C− in the hinge 2 region would negatively impact the rate of protein synthesis in general, and the initiation/elongation steps in particular, by reducing the rate of translocation of tRNAs and mRNA on the C− ribosomes compared to that on the C+ ribosomes. This idea is indeed supported by a recent study that suggests slower translation initiation by C− ribosomes (15).
Given the expression of C− ribosomes during chronic TB in mouse lungs (8) and, possibly, in human lungs (22), our findings highlight new challenges in the development of ribosome-targeting inhibitors as single-agent TB drugs. Consistent with this study, Spa demonstrate lower activity as single agents in mouse infection models of chronic infection than in acute-phase infections when C+ ribosome is expressed (35). Though it should be noted that as combination agents with other TB drugs (rifampin and pyrazinamide), Spa show promising results in chronic infection models, including in C3HeB/FeJ mice that exhibit advanced lung pathology similar to that in human TB infections (36). The divergence of these results may in part be due to secondary pharmacology of Spa, including their ability to avoid intrinsic efflux (24), or the action of partner drugs that interfere with C− ribosome formation. Discovery of novel inhibitors of ribosome remodeling and/or hibernation therefore emerge as a new opportunity in realizing the potential of translation inhibitors as future TB drugs. Meanwhile, the potential of Spa as TB drugs is best realized as combination therapies.
MATERIALS AND METHODS
Bacterial strains and culture.
M. smegmatis wild-type mc2155 and its derivatives were grown either in Middlebrook 7H9 base medium with 10% (vol/vol) albumin-dextrose-catalase (ADC) and 0.05% (vol/vol) Tween 80 or on 7H10-ADC agar plates at 37°C. For purification of ribosomes from high-zinc cultures of wild-type, pYL42, pYL53, or pYL53:ΔS14C− strains, cells were grown in Sauton’s medium containing 1 mM ZnSO4 for 96 h. For purification of ribosomes from low-zinc cultures of wild type and Δmpy strains, cells were grown for 96 h in Sauton’s medium without any supplemental zinc, instead containing 1 µM N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine (TPEN) as the chelator of residual zinc. M. tuberculosis wild type (mc27000) and its derivatives were cultured either in 7H9 medium with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC), 0.05% (vol/vol) Tween 80, and 100 μg/ml pantothenate (pan) or on 7H11-OADCpan agar plates at 37°C. For ribosome purification from pellicle cultures, mc27000 and its derivatives were grown planktonically to an optical density at 600 nm (OD600) of 0.4. Cells were then washed once with detergent-free Sauton’s medium, resuspended at 1:100 dilution in 4 ml of detergent-free Sauton’s medium either with 1 mM ZnSO4 or without the supplemental zinc in 12-well plastic tissue culture plates, and incubated at 37°C for 7 weeks without shaking. Escherichia coli GC5 was grown in LB broth or LB agar at 37°C. For selection of recombinant M. smegmatis, M. tuberculosis, or E. coli colonies, zeocin and kanamycin were used at 25 μg/ml and 20 μg/ml, respectively. Hygromycin was used either at 150 μg/ml (for M. smegmatis and E. coli) or 50 μg/ml for M. tuberculosis.
Construction of recombinant plasmids and strains.
All plasmids and bacterial strains used in this study are listed in Table S1 in the supplemental material. Oligonucleotides used for the construction of recombinant strains are listed in Table S2. Deletion mutations in M. smegmatis (mc2155) and M. tuberculosis (mc27000) were constructed by replacing target genes with a zeor marker using a PCR-based recombineering strategy, followed by unmarking of mutants as previously described (8, 37). In-frame deletion in individual genes of the c− operon was introduced by an overlapping PCR method. Briefly, fragments flanking the deletion region in c− operon from either pYL53 or the M. tuberculosis (mc27000) genome were amplified using primers with 15-bp overhang, followed by a second sowing PCR with distal primers, which joined the flanking fragments. The resulting c− operons with in-frame deletions of genes of interest were cloned into the integrated plasmid pMH94 at SacI and XbaI sites. All recombinant clones were genotyped by PCR and sequencing.
Ribosome purification.
The ribosome samples were purified as previously described (8). Cells from either 500-ml planktonic cultures of M. smegmatis or 7-week pellicles of M. tuberculosis (mc27000) from four 12-well plates were harvested and then flash-frozen in liquid nitrogen. The frozen cells were pulverized for 6 cycles (8 cycles for M. tuberculosis) of 3 min of milling at 15 Hz using a mixer mill (Retsch MM400), followed by 30 s of cooling in liquid nitrogen. The pulverized dried powder was resuspended in 20 ml of low-salt HMA-10 buffer (20 mM HEPES-K [pH 7.5], 30 mM NH4Cl, 10 mM MgCl2, 5 mM β-mercaptoethanol) and centrifuged for 30 min at 30,000 × g in a Thermo Sorvall Lynx 4000 centrifuge at 4°C. The supernatant was transferred to Beckman PC ultracentrifuge tubes and centrifuged for 2 h 15 min at 42,800 rpm in a Beckman rotor type 70Ti at 4°C. The pellet was soaked with 4 ml low-salt HMA-10 buffer overnight and further homogenized in a prechilled glass homogenizer. The homogenate was then treated with 3 units/ml RNase-free Turbo DNase (Invitrogen) for 1 h at 4°C, after which, 4 ml of high-salt HMA-10 buffer (20 mM HEPES-K [pH 7.5], 600 mM NH4Cl, 10 mM MgCl2, 5 mM β-mercaptoethanol) was added and the mixture was incubated at 4°C for another 2 h. The content was centrifuged at 20,000 × g for 15 min in a Thermo Sorvall Lynx 4000 centrifuge, and the supernatant was collected and centrifuged for 2 h 15 min at 42, 800 rpm in a Beckman rotor type 70Ti at 4°C. The crude ribosome pellet was then resuspended with low-salt HMA-10 buffer, layered on a 40-ml 10% to 40% sucrose gradient, and centrifuged for 16 h at 24,000 rpm in a Beckman rotor SW28. The gradient was fractionated by using the density gradient fractionation system (Brandel). The pooled 70S ribosome fractions were further pelleted by ultracentrifugation at 42,800 rpm for 4 h in Beckman rotor Type 70Ti and resuspended in low-salt HMA-10 buffer. The 70S ribosome subunits were quantified by measuring absorbance at 260 nm.
iTRAQ-MS.
Isobaric-tag for relative and quantitative mass spectrometry (iTRAQ-MS) was performed as described previously (8). Briefly, total proteins from 70S particles were extracted at room temperature in 66 mM Tris-HCl (pH 6.8), 5% SDS, and 2% β-mercaptoethanol and precipitated by trichloroacetic acid, followed by acetone wash. The protein pellets were then resuspended in 400 μl of 100 mM Tris-HCl, 0.4% SDS, 5 mM tributylphosphine (pH 8.3), and 7 M urea/2 M thiourea, followed by alkylation with iodoacetamide. Protein concentration was measured using a MicroBCA protein assay kit (Pierce, Rockford, IL) prior to quenching the reactions with 1 mM dithiothreitol. The protein mixtures were then diluted 10-fold in 50 mM Tris-HCl, pH 8.5, and mixed with modified trypsin (Sigma, St. Louis, MO) to a final substrate to enzyme ratio of 30:1. The mixtures were incubated overnight at 37°C. The resulting peptides from trypsin digest were cleaned up using a Discovery DSC-18 cartridge (Sigma, St. Louis, MO). After desalting on a C18 cartridge, 100 μg of the peptide mixtures were lyophilized and resuspended in 30 μl of 0.5 M triethylammonium bicarbonate (TEAB), pH 8.5. The appropriate 4-plex iTRAQ reagent (AB SCIEX, Framingham, MA), dissolved in 70 μl isopropanol, was added, and the reaction mixtures were incubated at room temperature for 2 h, following which they were quenched with 10 μl of 1 M Tris (pH 8.5). The iTRAQ-labeled peptide mixtures were then acidified using 0.8% formic acid to a total volume of 8.0 ml and underwent off-line cation exchange chromatography. A total of 30 fractions were collected, and samples were dried by a speed-vac prior to reversed-phase liquid chromatography-tandem mass spectrometry (RP-LC-MS/MS) analysis by the nanoscale liquid chromatography electrospray ionization quadrupole-quadrupole time of flight (nano-LC ESI QqTOF), as described earlier (38). The data were analyzed using the Paragon algorithm against the M. smegmatis database by ProteinPilot V5.0 software with trypsin as the digest agent, cysteine alkylation, an identification (ID) focus of biological modifications, and other default settings (AB SCIEX). Proteins quantified in at least three spectra (allowing generation of a P value) from two experimental replicates, with a ratio fold change of >1.3 (log10 of 0.11) or <0.7 (log10 of −0.15) and P value of <0.05, were deemed differentially expressed.
In vitro translation assay.
In vitro translation of NanoLuc was performed using a PURExpress ΔRibosome kit (NEB) as previously described (11). Briefly, 2.0 μl of solution A, 0.6 μl of factor mix, and 18 ng of DNA template (containing the T7 promoter, a 5′ untranscribed region [UTR] with a Shine-Dalgarno sequence, and the NanoLuc coding sequence) were mixed with 12.5 nM purified 70S ribosomes in a total volume of 5 μl. After 1 h of incubation at 37°C, relative luminescence units (RLU) produced from the translated luciferase was measured with a Nano-Glo luciferase assay kit (Promega Inc.) according to the manufacturer’s instructions. The inhibition of translation activity of 70S ribosomes purified from designated strains was assayed by adding the designated antibiotic at specified concentrations in the reaction mixture prior to ribosome addition and measuring luciferase activity after 1 h of incubation at 37°C. A reaction mixture without the antibiotic was used as a reference to determine percent inhibition. The IC50 value from a dose-response curve was obtained from nonlinear regression of log10 inhibitor versus activity plot using the variable slope model of GraphPad Prism software. The 95% confidence interval for each best-fit curve, also computed by Graph-Prism software from the standard errors, represents 95% probability that the computed IC50 value is in the given range.
Immunoblotting.
For immunoblotting, 4.8 pmol of 70S ribosome (OD260 of 0.2) purified from designated strains under specified conditions were mixed with 1× SDS loading buffer, resolved in a 15% SDS-PAGE gel, and transferred to a polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was blocked in 5% nonfat milk for 2 h at room temperature on a platform shaker, washed thrice with TBST (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% [vol/vol] Tween 20) for 10 min each, incubated with specified antibodies (S14C− [rabbit], 1:1,000; S13 [DSHB, mouse], 1:100) for 1 h at room temperature, and washed again thrice with TBST for 10 min each. The membrane was subsequently incubated with corresponding horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibodies (1:5,000) for 1 h at room temperature, washed thrice with TBST for 10 min each, and developed with enhanced chemiluminescence (ECL) regents (Thermo Fisher) in a dark room before exposure to BioBlot BXR (Laboratory Product Sales) films.
Antibiotic sensitivity assay.
The indicated strains of M. smegmatis or M. tuberculosis were precultured in 7H9-ADC or 7H9-OADCpan containing 0.05% (vol/vol) Tween 80 and 1 mM ZnSO4 at 37°C to an OD600 of 0.7. Cells were washed three times with phosphate-buffered saline (PBS) containing 0.05% Tween 80, and ∼107 CFU/ml were inoculated into 6 ml of Sauton’s medium containing 0.05% Tween 80, 1 mM ZnSO4, 100 μg/ml pan (for M. tuberculosis strains) and Spa at designated concentrations. At the predetermined time of exposure, an aliquot of cells was diluted and plated on either 7H10-ADC (for M. smegmatis) or 7H11-OADCpan (for M. tuberculosis). While the plates with M. smegmatis were incubated for 3 days, those with M. tuberculosis were incubated for 3 weeks at 37°C prior to imaging and enumerating colonies. Percent survival of population after Spa exposure was determined relative to that of the corresponding Spa-free controls.
Resazurin assay.
The above-indicated strains of M. smegmatis and M. tuberculosis were grown in either 7H9-ADC or 7H9-OADCpan (for M. tuberculosis) containing 0.05% (vol/vol) Tween 80 and 1 mM ZnSO4 at 37°C to an OD600 of 0.7. The cells were then collected, washed three times with PBS containing 0.05% (vol/vol) Tween 80, and resuspended in 7H9-ADC with Tween 80 (7H9-ADCTw) (or 7H9-OADCTw-pan for M. tuberculosis) containing 1 mM ZnSO4 to achieve the OD600 of 0.025 (M. smegmatis) or 0.1 (M. tuberculosis). A 100-μl aliquot of washed cells was placed in each well of a 96-well plate with a 2-fold dilution series of Spa, and the plate was incubated at 37°C without shaking either overnight (for M. smegmatis) or for 7 days (for M. tuberculosis). Wells with cells without Spa were positive controls, whereas wells without any cells were set up as negative (blank) controls. A 30-μl aliquot of 0.01% resazurin was then added to each well, and the color was developed overnight. The absorbance in each well was recorded at 550 nm and 630 nm, and the percentage of growth was calculated using the following formula: [(A550 − A630)sample − (A550 − A630)negative ctrl]/[(A550 − A630)positive ctrl − (A550 − A630)negative ctrl] × 100.
Spa binding assay.
A mixture containing 100 pmol of 70S ribosomes and 3H-spectinamide (1329-T; specific activity, 0.87 mCi/mg) at the designated concentrations in a total volume of 100 μl was incubated for 3 h at 37°C. After incubation, bound and unbound fractions of 3H-spectinamide were separated in 1.2-ml size exclusion columns (Bio-Spin Columns with Bio-Gel P-30; Bio-Rad). The amount of ribosome-bound 3H-spectinamide in the void volume was determined by liquid scintillation analyzer (Tri-Carb 4910TR; PerkinElmer). Dissociation constants (Kd values) were calculated from nonlinear regression plots of bound [3H]Spa versus free [3H]Spa using a binding-saturation model (GraphPad Prism software).
Cryo-electron microscopy and model building.
Cryo-EM data were collected on a FEI Titan Krios electron microscope, equipped with a Gatan K2 Summit direct-electron-detecting camera, at 300 kV and a defocus range of 0.5 to 3 μm at a calibrated magnification of 22,500× of the camera, yielding a pixel size of 1.07 Å on the object scale. A total of 3,589 movie stacks, each containing 40 subframes, were collected. The acquired frames were subjected to mechanical and beam-induced motion correction by implementation of dose weighting performed using MotionCor2 (31), which is known to significantly enhance the high-resolution Thon ring signals that results in better correlation with contrast transfer functions. The subsequent image processing steps were carried out using cryoSPARC (39). A total of 196,786 particle images were picked that yielded a 3.08-Å resolution 70S ribosome map. After three-dimensional (3D) classifications, 125,930 70S particle images were retained and refined to 3.11 Å (EMD ID 23076), as estimated using gold-standard criterion of 0.143 cutoff of Fourier-Shell correlation (40). The structure of S14C− from our previous study (8)was further refined de novo, guided by secondary structural elements and bulky sidechains of amino acids in the cryo-EM density, using UCSF-Chimera 1.14 (41). The model of the 30S head region was finally real-space refined and validated in PHENIX (42).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants to A.K.O. (grants AI132422 and AI144474), R.K.A. (GM061576), and R.E.L. (AI090810).
We thank Patricia Lederman for technical assistance and Qishan Lin from RNA Epitranscriptomics and Proteomics Resource for iTRAQ-MS analysis. Support from the Wadsworth Center Advanced Genomics Technologies and Tissue Culture and Media core facilities is also acknowledged.
Y.L., R.K.K., M.R.S., R.K.A., and A.K.O. designed and performed experiments as well as analyzed data. R.E.L. supplied reagents. Y.L., R.K.K., M.R.S., R.E.L., R.K.A., and A.K.O. wrote the manuscript.
We declare no financial conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ignatius EH, Dooley KE. 2019. New drugs for the treatment of tuberculosis. Clin Chest Med 40:811–827. doi: 10.1016/j.ccm.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2019. Global TB report 2019. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.WHO. 2019. WHO consolidated guidelines on drug-resistant tuberculosis treatment. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 4.Mdluli K, Kaneko T, Upton A. 2015. The tuberculosis drug discovery and development pipeline and emerging drug targets. Cold Spring Harb Perspect Med 5:a021154. doi: 10.1101/cshperspect.a021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RE, Hurdle JG, Liu J, Bruhn DF, Matt T, Scherman MS, Vaddady PK, Zheng Z, Qi J, Akbergenov R, Das S, Madhura DB, Rathi C, Trivedi A, Villellas C, Lee RB, Rakesh Waidyarachchi SL, Sun D, McNeil MR, Ainsa JA, Boshoff HI, Gonzalez-Juarrero M, Meibohm B, Bottger EC, Lenaerts AJ. 2014. Spectinamides: a new class of semisynthetic antituberculosis agents that overcome native drug efflux. Nat Med 20:152–158. doi: 10.1038/nm.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J Libardo MD, Boshoff HI, Barry CE, III. 2018. The present state of the tuberculosis drug development pipeline. Curr Opin Pharmacol 42:81–94. doi: 10.1016/j.coph.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Sun Q, Jiang C, Yang K, Hung LW, Zhang J, Sacchettini JC. 2015. Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure 23:1858–1865. doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Sharma MR, Koripella RK, Yang Y, Kaushal PS, Lin Q, Wade JT, Gray TA, Derbyshire KM, Agrawal RK, Ojha AK. 2018. Zinc depletion induces ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A 115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prisic S, Hwang H, Dow A, Barnaby O, Pan TS, Lonzanida JA, Chazin WJ, Steen H, Husson RN. 2015. Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol Microbiol 97:263–280. doi: 10.1111/mmi.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Sharma MR, Koripella RK, Wade JT, Gray TA, Derbyshire KM, Agrawal RK, Ojha AK. 2019. Reply to Tobiasson et al.: Zinc depletion is a specific signal for induction of ribosome hibernation in mycobacteria. Proc Natl Acad Sci U S A 116:2398–2399. doi: 10.1073/pnas.1821103116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Corro J, Palmer C, Ojha AK. 2020. Progression from remodelling to hibernation of ribosomes in zinc-starved mycobacteria. Proc Natl Acad Sci U S A 117:19528–19537. doi: 10.1073/pnas.2013409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, Kawamura F. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol 52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- 13.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol 189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dow A, Prisic S. 2018. Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc-limiting conditions. PLoS One 13:e0196300. doi: 10.1371/journal.pone.0196300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YX, Xu Z, Wang B, Ge X, Zhu J, Sanyal S, Lu ZJ, Javid B. 2020. Selective translation by alternative bacterial ribosomes. Proc Natl Acad Sci U S A 117:19487–19496. doi: 10.1073/pnas.2009607117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentschel J, Burnside C, Mignot I, Leibundgut M, Boehringer D, Ban N. 2017. The complete structure of the Mycobacterium smegmatis 70S ribosome. Cell Rep 20:149–160. doi: 10.1016/j.celrep.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Prossliner T, Skovbo Winther K, Sorensen MA, Gerdes K. 2018. Ribosome hibernation. Annu Rev Genet 52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 18.Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H, Kato T, Miyata T, Namba K, Wada A. 2013. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 19.Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A 87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueta M, Wada C, Wada A. 2010. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15:43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 21.Agafonov DE, Kolb VA, Nazimov IV, Spirin AS. 1999. A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci U S A 96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai RPJ, Cortes T, Marais S, Rockwood N, Burke ML, Garza-Garcia A, Horswell S, O’Garra A, Young DB, Wilkinson RJ. 9 March 2020. Transcriptomic characterization of tuberculous sputum reveals a host Warburg effect and microbial cholesterol catabolism. bioRxiv doi: 10.1101/2020.03.09.983163. [DOI] [PMC free article] [PubMed]
- 23.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/aac.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Bruhn DF, Lee RB, Zheng Z, Janusic T, Scherbakov D, Scherman MS, Boshoff HI, Das S, Rakesh, Waidyarachchi SL, Brewer TA, Gracia B, Yang L, Bollinger J, Robertson GT, Meibohm B, Lenaerts AJ, Ainsa J, Bottger EC, Lee RE. 2017. Structure-activity relationships of spectinamide antituberculosis agents: a dissection of ribosomal inhibition and native efflux avoidance contributions. ACS Infect Dis 3:72–88. doi: 10.1021/acsinfecdis.6b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan S, Donohue JP, Noller HF. 2014. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A 111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubarenko A, Sergiev P, Wintermeyer W, Dontsova O, Rodnina MV. 2006. Involvement of helix 34 of 16 S rRNA in decoding and translocation on the ribosome. J Biol Chem 281:35235–35244. doi: 10.1074/jbc.M608060200. [DOI] [PubMed] [Google Scholar]
- 27.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 28.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. 2007. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol 2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 30.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16S RNA. J Mol Biol 316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 31.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- 33.Frank J, Agrawal RK. 2001. Ratchet-like movements between the two ribosomal subunits: their implications in elongation factor recognition and tRNA translocation. Cold Spring Harbor Symp Quant Biol 66:67–75. doi: 10.1101/sqb.2001.66.67. [DOI] [PubMed] [Google Scholar]
- 34.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM. 2010. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruhn DF, Scherman MS, Liu J, Scherbakov D, Meibohm B, Bottger EC, Lenaerts AJ, Lee RE. 2015. In vitro and in vivo evaluation of synergism between anti-tubercular spectinamides and non-classical tuberculosis antibiotics. Sci Rep 5:13985. doi: 10.1038/srep13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson GT, Scherman MS, Bruhn DF, Liu J, Hastings C, McNeil MR, Butler MM, Bowlin TL, Lee RB, Lee RE, Lenaerts AJ. 2017. Spectinamides are effective partner agents for the treatment of tuberculosis in multiple mouse infection models. J Antimicrob Chemother 72:770–777. doi: 10.1093/jac/dkw467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Kessel JC, Hatfull GF. 2008. Mycobacterial recombineering. Methods Mol Biol 435:203–215. doi: 10.1007/978-1-59745-232-8_15. [DOI] [PubMed] [Google Scholar]
- 38.Luo J, Ning T, Sun Y, Zhu J, Zhu Y, Lin Q, Yang D. 2009. Proteomic analysis of rice endosperm cells in response to expression of hGM-CSF. J Proteome Res 8:829–837. doi: 10.1021/pr8002968. [DOI] [PubMed] [Google Scholar]
- 39.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 40.Rosenthal PB, Henderson R. 2003. ptimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.