We characterized a multidrug-resistant (MDR) Enterobacter spp. isolate highlighting the genetic aspects of the antimicrobial resistance genes. An Enterobacter spp. isolate (Ec61) was recovered in 2014 from a transtracheal aspirate sample from a patient admitted to a Brazilian tertiary hospital and submitted to further microbiological and genomic characterization.
KEYWORDS: antimicrobial resistance, carbapenemase, phosphoethanolamine transferase, plasmids
ABSTRACT
We characterized a multidrug-resistant (MDR) Enterobacter spp. isolate highlighting the genetic aspects of the antimicrobial resistance genes. An Enterobacter spp. isolate (Ec61) was recovered in 2014 from a transtracheal aspirate sample from a patient admitted to a Brazilian tertiary hospital and submitted to further microbiological and genomic characterization. Ec61 was identified as Enterobacter hormaechei subsp. xiangfangensis strain ST451, showing an MDR profile and the presence of genes codifying the new β-lactamase variants BKC-2 and ACT-84 and the mobile colistin resistance gene mcr-9.1.
INTRODUCTION
In the past decade, the dramatic increase in the prevalence and clinical impact of infections caused by carbapenemase-producing bacteria became a global health concern. Carbapenemase production is especially problematic when encountered in Enterobacterales members because of their great ability to spread in healthcare settings, resulting in health and economic burdens (1). Brazilian Klebsiella carbapenemase (BKC) is a serine carbapenemase identified for the first time in 2015 from clinical isolates of Klebsiella pneumoniae collected in 2008 (2). BKC-1 is able to hydrolyze penicillins, cephalosporins, monobactams, and carbapenems but not cephamycins and has oxacillin as preferential hydrolytic substrate (2). blaBKC-1 is usually carried by a 10-kb IncQ plasmid and associated with aph(3)-VI and ISKpn23, an insertion sequence belonging to the IS1380 family; however, this resistance gene has been characterized in only a few strains (2–5).
A mobile colistin resistance gene, mcr, which mediates the addition of phosphoethanolamine (pEtN) on bacterial lipopolysaccharide, has been a cause of great concern worldwide (6). So far, 10 variants of mcr genes have been deposited in GenBank. The coproduction of carbapenem and colistin resistance drastically hinders therapeutic options in hospital settings because these mechanisms are frequently associated with non-β-lactam antimicrobial resistance (7).
Herein, we report a new allele of BKC, BKC-2, in MCR-9.1-producing Enterobacter spp., recovered in a Brazilian hospital in São José do Rio Preto, São Paulo, Brazil. The microbiological and molecular properties of BKC-2 have been assessed to evaluate the spectrum of resistance conferred by BKC-2 and the capability of blaBKC-2 to be mobilized and/or transferred.
During a surveillance study conducted from January 2013 to December 2017, a multidrug-resistant (MDR) isolate belonging to the Enterobacter cloacae complex (Ec61) was obtained from a transtracheal aspirate culture that was collected in 2014 from a patient admitted to the general intensive care unit. Due to the MDR phenotype, the isolate was screened for the presence of carbapenemases and other antimicrobial resistance genes (ARGs) by PCR (see Text S1 in the supplemental material). Antimicrobial susceptibility testing was performed by broth microdilution (BMD) and interpreted according to EUCAST v.10.0 clinical breakpoints (8). PCR assay of ARG showed the presence of blaBKC, blaSHV, aac(6′)-Ib, and aph(3′)-IV genes on the Ec61 isolate. Interestingly, to date, only a single isolate of non-Klebsiella pneumoniae carrying blaBKC-1 has been reported (5). Susceptibility profile results confirmed the MDR profile with resistance to most tested antimicrobials (Table 1).
TABLE 1.
Susceptibility profile of Enterobacter hormaechei subsp. xiangfangensis isolates carrying blaBKC-2 (Ec61), transformants, and clones carrying blaBKC-1 and blaBKC-2
| Antimicrobial agent | MIC (mg/liter) |
||||||
|---|---|---|---|---|---|---|---|
| Ec61 (wild type) | TfBKC-1 (E. coli Top10 + p60136 [blaBKC-1]) | TfBKC-2 (E. coli Top10 + pEc61C [blaBKC-2]) | Top 10 (E. coli top 10) | Cl_BKC-1 (E. coli BL21 + pET-26::blaBKC-1)a | Cl_BKC-2 (E. coli BL21 + pET-26::blaBKC-2)a | BL21 + pET26 (E. coli BL21 + pET-26) | |
| Ertapenem | 16 | 0.5 | 0.125 | ≤0.03 | 0.12 | 0.015 | 0.015 |
| Meropenem | 2 | 0.25 | 0.125 | ≤0.03 | 0.12 | 0.06 | 0.06 |
| Imipenem | 4 | 0.5 | 0.125 | ≤0.03 | 0.5 | 0.5 | 0.25 |
| Cefepime | 256 | 16 | 8 | ≤0.03 | 4 | 0.03 | 0.03 |
| Ceftazidime | >256 | 128 | 64 | 0.25 | 8 | 0.125 | 0.06 |
| Aztreonam | >256 | 256 | 64 | 0.06 | 8 | 0.03 | ≤0.015 |
| Amoxicillin | >512 | >512 | 256 | 2 | >256 | 64 | 2 |
| Ampicillin | >512 | >512 | >512 | 1 | >256 | 32 | 1 |
| Cefotaxime | 256 | 256 | 64 | 0.06 | 16 | 0.12 | ≤0.015 |
| Piperacillin-tazobactam | >256/4 | NTb | NT | NT | NT | NT | NT |
| Ceftazidime-avibactam | 2/4 | NT | NT | NT | NT | NT | NT |
| Amikacin | 256 | NT | NT | NT | NT | NT | NT |
| Gentamicin | 64 | NT | NT | NT | NT | NT | NT |
| Tobramycin | 64 | NT | NT | NT | NT | NT | NT |
| Kanamycin | >256 | NT | NT | NT | NT | NT | NT |
| Levofloxacin | 2 | NT | NT | NT | NT | NT | NT |
| Ciprofloxacin | 2 | NT | NT | NT | NT | NT | NT |
| Colistin | 1 | NT | NT | NT | NT | NT | NT |
BMD was performed in the presence of isopropyl-β-d-thiogalactopyranoside 0.2 mM.
NT, not tested.
Molecular techniques were used to better characterize Ec61. Initially, the plasmid was characterized by using S1-pulsed-field gel electrophoresis (S1-PFGE) followed by hybridization to blaBKC (9). Conjugation experiments were performed by using the azide-resistant Escherichia coli strain J53 as the recipient cell at room temperature, 37°C, and 42°C in biological triplicate. Transconjugants were selected on UTI ChromoSelect agar (Merck, NJ, USA) containing 200 mg/liter of ampicillin or 0.5 mg/liter of colistin to screen for blaBKC and mcr, respectively. The S1-PFGE pattern suggested the presence of three plasmids in Ec61, with blaBKC located on an ∼10-kb plasmid (data not shown) according to the hybridization results. The same results were previously observed for BKC-1-producing K. pneumoniae and Citrobacter freundii (2, 3). Despite several attempts, conjugation experiments failed to transfer the plasmid carrying blaBKC.
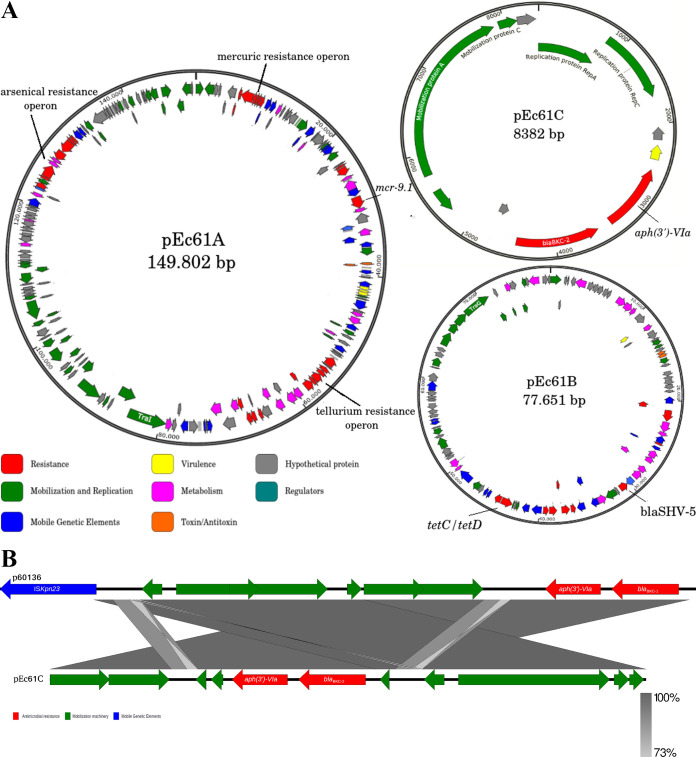
Whole-genome sequencing (WGS) was performed by the Illumina MiSeq platform (MiSeq reagent V3 kit/2 × 300 cycles) and MinION (Oxford Nanopore, UK). Hybrid de novo assembly was performed using Unicycler (v0.4.0) (10), and the automatic annotation was performed using RAST version 2.0 (https://rast.nmpdr.org/). Average nucleotide identity (ANI) was performed to identify the Ec61 bacterial species, using an ANI value of >95% as a threshold for species and >98% for subspecies definition (Text S1) (11). ResFinder v.3.2, MLSTfinder v.2.0.4 based on an E. cloacae scheme, and PlasmidFinder v.2.1 were used for initial characterization of the ARG, MLST, and plasmid replicons present on the Ec61 isolate. ANI analysis confirmed Ec61 to be Enterobacter hormaechei subsp. xiangfangensis, with 99.28% nucleotide identity to the reference sequence NZ_CP017183.1 (see Table S1 in the supplemental material). General genome features revealed the presence of a 4.8-Mb chromosome with 55.3% GC content and three plasmids, i.e., pEc61A (149.8 kb), pEc61B (77.6 kb), and pEc61C (8.3 kb) (Fig. 1A). Ec61 belongs to ST451, a sequence type previously associated with blaKPC-3 in the United States (12).
FIG 1.
(A) Graphic map of the three plasmids detected in E. hormaechei subsp. xiangfangensis carrying blaBKC-2 and mcr-9.1. (B) Colinear analysis of the two different plasmids harboring blaBKC. Arrows, transcription direction of genes and ORFs. Genes were grouped according to their predictive functions, indicated by color.
Point mutations related to antimicrobial resistance, i.e., gyrA, gyrB, parC, parE, ampD, ampR, ompF, and ompC, were investigated by BLASTN using E. hormaechei ATCC 49162 as a reference strain (accession no. MKEQ01000001 to MKEQ01000004), a well-known Ser83Phe substitution in GyrA sequence, causing an increase in the MICs for quinolones. No additional amino acid substitutions were detected in the quinolone resistance-determining region (QRDR) region of GyrB, ParC, and ParE. Analysis of the genes encoding the main outer membrane proteins OmpC and OmpF revealed the presence of mutations on the amino acid chain in the OmpC of the Ec61 isolate and the disruption of the ompF nucleotide sequence by insertion of a new sequence called ISEcl11.
The ARG analysis initially revealed the presence of blaACT-16*, blaBKC-1*, blaSHV-5, sul1, sul2, mcr-9.1, fosA2, aac(6′)-Ib3, aadA1, aph(3′)-VIa, aac(6′)-Ib-cr, and tetD (see Fig. S1 in the supplemental material). However, due to the 97% similarity between genes blaACT-16* and blaBKC-1*, the ResFinder database suggested the presence of new gene variants. The cephalosporinase ACT-16 was aligned with all ACT alleles available in the NCBI database, revealing the presence of a new variant of this chromosomal cephalosporinase named ACT-84 (GenBank accession no. MT136764). ACT-84 differs from ACT-83 by a single amino acid substitution (Pro58Ser). Alignment of the BKC-1 nucleotide sequence (GenBank accession no. NG_048710.1) with that of blaBKC detected on Ec61 showed 20 mismatches leading to 11 amino acid modifications (Fig. S2). Thus, we named the new BKC variant as BKC-2. Like BKC-1, the coding sequence of BKC-2 was located in a small plasmid of the IncQ1 group (pEc61C); however, the plasmid was 1.3 kb smaller than p60136 (GenBank accession no. NG_048710.1). Nucleotide comparison of plasmid-encoding blaBKC-1 with blaBKC-2 revealed that ISKpn23 was absent upstream to blaBKC-2 (Fig. 1B). It has been speculated that the association of ISKpn23 with blaBKC-1 would be responsible for the mobilization and expression of blaBKC-1 (2, 4). Although absent, a nucleotide region of almost 250 bp of ISKpn23 was detected upstream to blaBKC-2, suggesting the exit of ISKpn23 from pEc61C. Nucleotide alignment showed that these 250 nucleotides matched the beginning and the end of the ISKpn23 encoding sequence, where the insertion sequence's promoter is located (see Fig. S3 in the supplemental material) (13).
To verify differences in the susceptibility profile conferred by BKC-1 and BKC-2, plasmids p60136 (carrying blaBKC-1) and pEc61C (carrying blaBKC-2) were transformed into E. coli Top10 by chemical transformation and selected on LB agar plates containing 100 mg/liter of ampicillin, respectively. Additionally, blaBKC-2 was amplified using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) and cloned into pET-26b+ as previously described (2). Transformant BKC-1 and BKC-2 clones were subjected to antimicrobial susceptibility testing by BMD against β-lactam agents. In general, MICs of the BKC-1 transformant were higher than those presented by the BKC-2 transformant. MICs of ertapenem, meropenem, and imipenem were 4, 2, and 2 times higher, respectively, for the BKC-1 transformant than the BKC-2 transformant. Taking into account the BKC clones, MICs of BKC-1 for β-lactams were higher than those of BKC-2, suggesting better activity of BKC-1 against β-lactams. However, although BKC-2 was weaker than BKC-1, in its natural genetic background, BKC-2 showed activities against β-lactam agents similar to those of BKC-1, as suggested by transformant MICs (TfBKC-1 and TfBKC-2) (Table 1). These MIC variations could be explained by differences in the promoter regions between these two carbapenemase-encoding genes. Interestingly, a newly published article suggested that the origin of carbapenemases BKC-1 and GPC-1 related to the Shinella genus (14). Besides, a putative BKC-1 ancestral protein was synthesized in laboratory to antimicrobial susceptibility analysis and named BKC-b (14). BKC-b resulted from a deletion in the duplication segment of amino acids (Ala12 to Ser27) and shares 94% amino acid identity with BKC-2. Our data have been supported by the finding that BKC-b (as well as BKC-2) has weaker hydrolytic activity against the β-lactam agents than BKC-1 (14). Although BKC-2 shows duplication of the peptide Leu24-Arg39, the duplication segment Ala12-Ser27 (present in BKC-1 and responsible for the increase in β-lactamase activity) is absent, explaining the low activity of this new BKC variant.
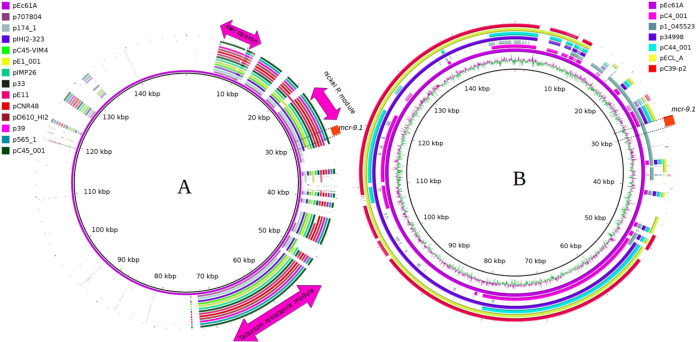
Ec61 harbored colistin resistance gene mcr-9.1, in addition to novel gene blaBKC-2; however, the isolate showed susceptibility to colistin similar to the previously described mcr-9-positive strains (15). mcr-9.1 was carried on a different plasmid, pEc61A (149.8 kb), belonging to IncFIB without other antimicrobial resistance genes; although genes encoding metal resistance, such as arsenic, tellurium, mercuric, and nickel/cobalt resistance, were detected in the plasmid. Since the initial description of mcr-9, its dissemination has been strongly related to IncHI2 plasmids, as supported by our in silico analysis of the GenBank database (16). Only a single plasmid, non-IncHI2, was detected carrying mcr-9.1 (Fig. 2). The mcr-9.1 genetic backbone related to pEc61A contained IS903B::mcr-9.1::wubC::qseC-like::qseB-like::ATPase::integrase::IS26 as previously described in IncHI plasmids (15). Although the mechanism responsible for mcr-9.1 mobilization has not been elucidated, different insertion sequences (IS903-like, IS903B, IS26, and IS1) have been associated with this gene (15, 17).
FIG 2.
(A) Circular comparison between pEc61A and 13 plasmids carrying mcr-9.1 belonging to IncH2 and IncH2a. Pink arrows, conserved regions, including mer operon, nickel resistance module, and tellurium resistance operon. (B) Circular comparison of the six best hit plasmids related to pEc61A on BLASTN analysis of NCBI database. Five of six analyzed plasmids belonged to the IncFIB group, and only p1_045523 also harbored mcr-9. GenBank accession numbers for each plasmid are included in Table S2 in the supplemental material.
IncFIB plasmids are well recognized as conjugative plasmids (18); however, the conjugation assay for the transfer of IncFIB with mcr-9.1 was not successful. The transfer of IncFIB was only found at 42°C, with a low frequency of 10−8, by using a low colistin concentration as the selective agent. Because the MIC for colistin of isolate Ec61 was 1 mg/liter, subinhibitory concentrations (0.25 and 0.5 mg/liter) were used in all attempts and did not provide an ideal selection using the E. coli J53 AziR strain. The findings suggested the limited dissemination of mcr-9.1 by IncFIB. In contrast, IncHI2 plasmids carrying mcr-9.1 have been widely demonstrated disseminating among Enterobacterales and carrying different antimicrobial resistance genes along with mcr-9.1 (15–17).
We need to pay attention to BKC-2 due to its acquisition on distinct bacterial genera, Enterobacter spp., which naturally tend to be microorganisms with high levels of resistance to antimicrobials. Also, the coproduction of a new cephalosporinase, ACT-84, and MCR-9.1 coupled with chromosomal mechanisms revealed a large arsenal of antimicrobial resistance mechanisms in/on Ec61 isolate. Although BKC-2 confers lower β-lactam resistance than BKC-1, its association with chromosomal or other plasmid-acquired mechanisms can lead to carbapenem resistance.
Accession number(s).
The nucleotide sequences have been submitted to the GenBank database under accession no. MT427610 for blaBKC-2, MT136764 for blaACT-84, and CP053103 to CP053106 for the Ec61 genome and its respective plasmids.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kirsty Sands for her contribution on WGS. We are also grateful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for scholarships provided to E.R.M. and L.K.D.A.
The São Paulo Research Foundation (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) provided grants to W.M.B.S.M. (process no. 2018/24431-4) and A.C.G. (process no. 312066/2019-8).
A.C.G. recently received research funding and/or consultation fees from Bayer, Eurofarma, InfectoPharm, MSD, Pfizer, and Zambon. Other authors have nothing to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblad LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicoletti AG, Marcondes MF, Martins WM, Almeida LGP, Nicolás MF, Vasconcelos ATR, Oliveira V, Gales AC. 2015. Characterization of BKC-1 class A carbapenemase from Klebsiella pneumoniae clinical isolates in Brazil. Antimicrob Agents Chemother 59:5159–5164. doi: 10.1128/AAC.00158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins WM, Nicoletti AG, Santos SR, Sampaio JLM, Gales AC. 2016. Frequency of BKC-1-producing Klebsiella species isolates. Antimicrob Agents Chemother 60:5044–5046. doi: 10.1128/AAC.00470-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge SR. 2016. Mobilization of blaBKC-1 by ISKpn23? Antimicrob Agents Chemother 60:5102–5104. doi: 10.1128/AAC.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martins WMBS, Seco BMS, Sampaio JLM, Sands K, Toleman MA, Gales AC. 2020. Detection of BKC-1 in Citrobacter freundii: a clue to mobilisation in an IncQ1 plasmid carrying blaBKC-1. Int J Antimicrob Agents 56:106042. doi: 10.1016/j.ijantimicag.2020.106042. [DOI] [PubMed] [Google Scholar]
- 6.Yang QE, Tansawai U, Andrey DO, Wang S, Wang Y, Sands K, Kiddee A, Assawatheptawee K, Bunchu N, Hassan B, Walsh TR, Niumsup PR. 2019. Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ Int 122:281–290. doi: 10.1016/j.envint.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 7.Huttner B, Jones M, Rubin MA, Neuhauser MN, Gundlapalli A, Samore M. 2012. Drugs of last resort? The use of polymyxins and tigecycline at US Veterans Affairs medical centers, 2005–2010. PLoS One 7:e36649. doi: 10.1371/journal.pone.0036649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Committee on Antimicrobial Susceptibility Testing. 2020. Clinical breakpoints and dosing of antibiotics. http://www.eucast.org/clinical_breakpoints/.
- 9.Toleman MA. 2018. Direct in gel genomic detection of antibiotic resistance genes in S1 Pulsed field electrophoresis gels. Methods Mol Biol 1736:129–136. doi: 10.1007/978-1-4939-7638-6_12. [DOI] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 12.Kanamori H, Parobek CM, Juliano JJ, Van Duin D, Cairns BA, Weber DJ, Rutala WA. 2017. A prolonged outbreak of KPC-3-Producing Enterobacter cloacae and Klebsiella pneumoniae driven by multiple mechanisms of resistance transmission at a large academic burn center. Antimicrob Agents Chemother 61:e01516-16. doi: 10.1128/AAC.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer N, Ebmeyer S, Larsson DGJ. 2020. The class A carbapenemases BKC-1 and GPC-1 both originate from the bacterial genus Shinella. Antimicrob Agents Chemother 64:e01263-20. doi: 10.1128/AAC.01263-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieffer N, Royer G, Decousser JW, Bourrel AN, Palmieri M, Ortiz De La Rosa JM, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. (Erratum, 63:e01866-19, doi:.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Börjesson S, Greko C, Myrenås M, Landén A, Nilsson O, Pedersen K. 2020. A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J Glob Antimicrob Resist 20:285–289. doi: 10.1016/j.jgar.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.