Significance
In organisms that exhibit flexible morphologies, such as plants, the developmental origin of flexibility is an interesting but unsolved mystery. Here, by capturing gene-expression profiles of overlapping cell populations in the Arabidopsis stomatal lineage, we define genetic networks underlying the latent division potential of a multipotent cell type—the stomatal lineage ground cell—and identify roles for specific and general transcriptional regulators in modulating outputs of the stomatal lineage.
Keywords: stomatal lineage ground cell, Arabidopsis, transcriptional profile, MYB16 endoreduplication
Abstract
In many developmental contexts, cell lineages have variable or flexible potency to self-renew. What drives a cell to exit from a proliferative state and begin differentiation, or to retain the capacity to divide days or years later is not clear. Here we exploit the mixed potential of the stomatal lineage ground cell (SLGC) in the Arabidopsis leaf epidermis as a model to explore how cells might balance potential to differentiate with a reentry into proliferation. By generating transcriptomes of fluorescence-activated cell sorting-isolated populations that combinatorically define SLGCs and integrating these data with other stomatal lineage datasets, we find that SLGCs appear poised between proliferation and endoreduplication. Furthermore, we found the RNA polymerase II-related mediator complex interactor DEK and the transcription factor MYB16 accumulate differentially in the stomatal lineage and influence the extent of cell proliferation during leaf development. These findings suggest that SLGC latent potential is maintained by poising of the cell cycle machinery, as well as general and site-specific gene-expression regulators.
The generation and maintenance of diverse cell types in multicellular organisms requires coordination of numerous precursor lineages. These lineages vary in their behavior, with some exhibiting a rigid and predetermined number of progeny and others, such as stem cells, behaving as an immortal pool of continuously replenished cells. Particularly interesting from the perspective of flexibility, are cells whose progeny numbers and final identities vary with different life circumstances: for example, intestinal stem cells that build an organ of different sizes depending on food availability (1), or adult stem-cell lineages where embedded quiescent cells are reactivated in order to maintain and repair tissues. These cells have latent potential, but what is the nature of this state? Does the behavior of these cells reflect their lineage history or their current cellular neighborhood, and how is this information encoded in the transcriptome? The simple, genetically tractable and experimentally accessible stomatal lineage in the Arabidopsis leaf epidermis displays an exemplary latent potential state in the behavior of stomatal lineage ground cells (SLGCs), providing an access point toward identifying mechanisms underlying flexible development.
Upon initiation of the stomatal lineage by physically asymmetric division of a protodermal cell, two distinct daughters are created: a smaller meristemoid (Fig. 1A, red) and a larger SLGC (Fig. 1A, blue or white). Each of these daughters can divide again a small—but variable—number of times. Together, they expand the progenitor pool that ultimately differentiates into either stomatal guard cells or pavement cells (Fig. 1A). The capacity of the meristemoid to continue divisions has been explored in some detail, and it involves nested positive and negative feedback loops between the heterodimeric transcription factor pair SPEECHLESS and SCREAM1/2 and peptide ligand–receptor signaling (2, 3). SLGCs have been more enigmatic. There are no reporters specific to SLGCs, but polarity proteins, such as BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) or BREVIS RADIX LIKE 2 (BRXL2) (Fig. 1B), are preferentially inherited and maintained in SLGCs after asymmetric cell divisions (4–6). Loss of polarity proteins leads to defects in SLGC fate, primarily by failing to make them different from their meristemoid sisters (4, 5). Factors responsible for SLGC identity itself are not known, but SLGC behavior depends on local, systemic, and environmental cues (7–11). For example, cytokinin regulates stomatal production by modulating SLGC divisions (10). How SLGCs lose division potential and become pavement cells is equally unknown. Pavement cells are typically endoreplicated (12), so it is likely that this fate switch is tied to a change in cell cycle programs.
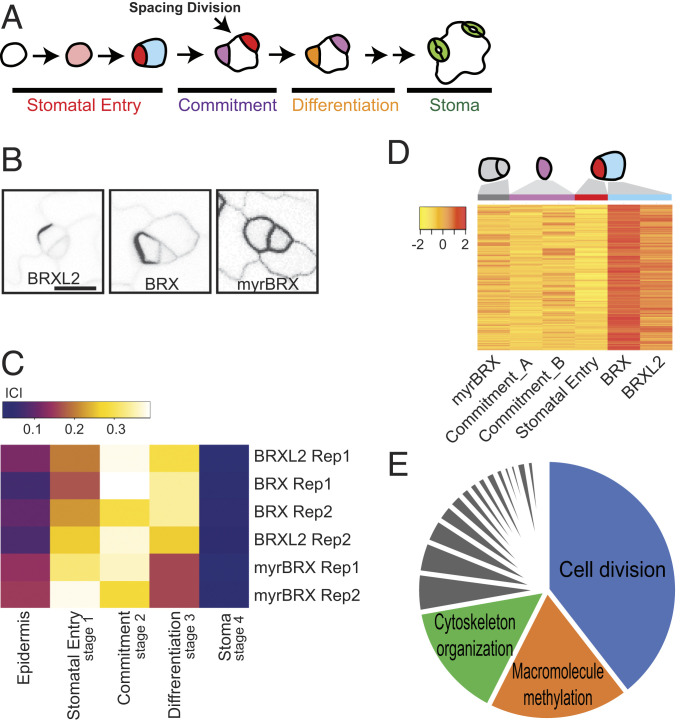
Fig. 1.
Profiling of the SLGC transcriptome. (A) Scheme of Arabidopsis stomatal development. Stomatal entry requires an asymmetric cell division that produces a meristemoid (red) and a SLGC (blue); both have division potential. Meristemoids can divide (not shown) or commit to precursor identity (violet) and subsequent stomatal differentiation (orange), becoming stomata (green). SLGCs are multipotent and can divide to create more meristemoids (spacing division) before differentiating into lobed pavement cells. (B) Confocal images from true leaves of 9-d-old seedlings expressing SLGC-enriched (BRXL2p:BRXL2-YFP and BASLp:BRX-YFP) or SLGC and meristemoid-enriched (BASLp:myrBRX-YFP) markers used for FACS. (Scale bar, 10 μm; all images are at the same scale.) (C) Output of transcriptome-based ICI analysis comparing SLGC-enriched populations to other stomatal lineage cell-type transcriptomes from Adrian et al. (15). BRX and BRXL2 samples are most similar to commitment (stage 2) cells and myrBRX cells resemble stomatal entry (stage 1). (D) Heat map representation of genes highly and differentially expressed in presumptive SLGCs (BRX and BRXL2) (clustering coefficient cutoff 0.6). Mean and median expression values are scaled per gene across samples. Stomatal commitment A and B samples are described in Methods. (E) Enriched GO process terms for SLGC cluster created using REVIGO (48), and represented as a pie chart, with three most significant categories labeled; terms for gray slices are in Dataset S4.
Here we profile fluorescence-activated cell sorting (FACS)-isolated subpopulations of early stomatal lineage cells that, in their overlap, could define SLGCs and use a combinatorial and subtractive computational pipeline to find transcripts enriched in SLGCs. Overall, our results suggest that SLGCs are similar to early stomatal lineage cells, but they express a unique combination of cell cycle regulators, poising them between mitotic and endoreduplication programs. We also identified potential roles for transcriptional and translational regulation of SLGC behavior by functional analysis of the SLGC-enriched genes encoding the MYB16 transcription factor and nucleolar localized DEK family proteins.
Results
Generating an SLGC-Enriched Transcriptome.
The features that make SLGCs biologically interesting—their morphological heterogeneity, their asynchronous production and scattered distribution, and their flexible division patterns—also make them hard to isolate. Thus far, no molecular markers exclusive to this cell type have been reported. To establish transcriptional profiles of the enigmatic SLGCs, therefore, we took a hybrid approach of capturing these cells and their sisters via FACS and then using computational strategies to compare profiles in this mixed population with populations of other epidermal cell types.
Fluorescent reporters based on BASL or the BRX family (6, 13) preferentially, but not exclusively, label SLGCs. Empirically, markers with the greatest specificity for SLGCs were BRXL2p:BRXL2-YFP and BASLp:BRX-YFP that are expressed briefly before asymmetric divisions, but become preferentially localized to the plasma membrane in SLGCs after division (Fig. 1B). Addition of an N-terminal myristoylation site to BASLp:myrBRX-YFP (myrBRX) creates a protein equally distributed at the plasma membrane in both meristemoids and SLGCs, and serves as a comparison to identify SLGC-specific transcripts (Fig. 1B). To obtain cell-type enriched transcriptomes, whole 9-d-old seedling cells expressing BRX, BRXL2, or myrBRX were protoplasted and YFP+ cells were collected by FACS (SI Appendix, Figs. S1 and S2). Two biological replicates per each marker line were used to construct RNA-sequencing (RNA-seq) libraries (schematic of pipeline in SI Appendix, Fig. S3).
Comparison of BRX vs. myrBRX samples yielded ∼1,500 differentially expressed genes with a fold-change cutoff of twofold and adjusted P value of 0.05 (Dataset S1). Pairwise comparison of transcriptomes of cells expressing either BRX or BRXL2, with the same fold-change and adjusted P value criteria resulted in only 301 differentially expressed genes, consistent with live cell imaging that suggested these reporters marked the same epidermal cells (Dataset S2). These results were encouraging that SLGCs and meristemoids had distinct transcriptional identities. To inquire how SLGCs are related to other stomatal lineage cells and what makes them unique, we compared BRX, BRXL2, and myrBRX transcriptomes to those of other stomatal lineage cells using the information-theory based “index of cell identity” (ICI) approach (14) (SI Appendix, Fig. S3 A and B). We first derived sets of informative markers genes for cells at stomatal entry (stage 1), commitment (stage 2), differentiation (stage 3), and maturity (stage 4), as well as the whole epidermis from data in Adrian et al. (15) (Dataset S3). We computed a quantitative score for the similarity between the transcriptomes derived from BRX-, BRXL2-, or myrBRX-sorted populations and cells representing stomatal stages 1 to 4 and the leaf epidermis (Fig. 1C and SI Appendix, Fig. S3). As might be predicted from the appearance of the reporter in both meristemoids and SLGCs, the myrBRX-sorted cell group shows highest similarity to stage 1 (Fig. 1C). BRX- and BRXL2-expressing cells are most similar to stage 2, suggesting SLGCs are at a transition stage beyond meristemoids, but have not gone down the pathway toward stomatal differentiation (stages 3 and 4). The modest degree of similarity to any previously characterized cell types (ICI scores: 0.38, 0.39, 0.29, and 0.37), suggests that SLGCs display unique features at a transcriptional level. Further interrogation of these unique features could therefore identify new regulators of SLGC identity and generate insights into overall cell behaviors.
We made a set of pairwise comparisons (SI Appendix, Fig. S3C) to uncover unique features and dominant expression patterns of BRX and BRXL2 relative to their closest relatives (stages 1 and 2 cells are sisters, mothers, or daughters of SLGCs) (SI Appendix, Fig. S3A). Stage 2 is represented by two independent transcriptomes derived from different but highly overlapping markers, and so is designed as commitment_A and commitment_B (Fig. 1D, purple column headings, and SI Appendix, Fig. S3C). In total, we compared four transcriptome datasets. An unsupervised analysis, fuzzy k-means clustering, was performed to determine dominant expression patterns. To reduce computational time, we removed the genes not differentially expressed in any pairwise comparison, resulting in 19,707 genomic elements for analysis. By fitting clusters from k = 3 to k = 9, distinct patterns emerged at k = 5 for BRX- and BRXL2-sorted cells. This pattern, hereafter called the SLGC cluster, was comprised of 1,016 genes, including the confirmed SLGC-expressed genes ERECTA, SPCH, and BASL (Fig. 1D, blue column heading, and Dataset S4).
Highly enriched processes in the SLGC cluster, as described by gene ontology (GO) terms, were related to cell cycle (39.5%), cytoskeleton organization (14.8%), and macromolecule methylation (18.0%), including histone methylation (Fig. 1E and Dataset S4). These enriched categories are consistent with the previous observation that SLGCs are multipotent and division competent. These main SLGC-enriched GO terms are also found in stomatal lineage cells (15). When the genes within each GO term were compared between the SLGC cluster and the cells at the entry and committed stage, however, only 5 [cluster II in Adrian et al. (15)] (Fig. 1C and Dataset S4) and 27 (cluster III) genes were in common (of 920 and 563 genes, respectively). Thus, it suggests that SLGCs may employ specific subsets of cell cycle proteins or members of gene families during cell division to determine cell fate.
Expression of Cell Cycle-Related Genes in SLGCs Suggests a Poised State between Mitotic and Endoreduplication Potential.
Although we could rationalize the appearance of cell cycle genes in SLGCs, it was surprising to find them enriched relative to stomatal entry (stage 1) and commitment (stage 2) samples, since these stages are also division competent. We therefore analyzed more specific GO terms related to cell cycle, cell division, and cytokinesis and made direct comparisons with lists of core cell cycle genes and individual phases of the cell cycle derived from experimental data (Dataset S4). Two particularly strong signatures were for M-phase regulators (SI Appendix, Fig. S4 A and B) and for genes encoding elements of the mitotic and cytokinetic structures (SI Appendix, Fig. S4C).
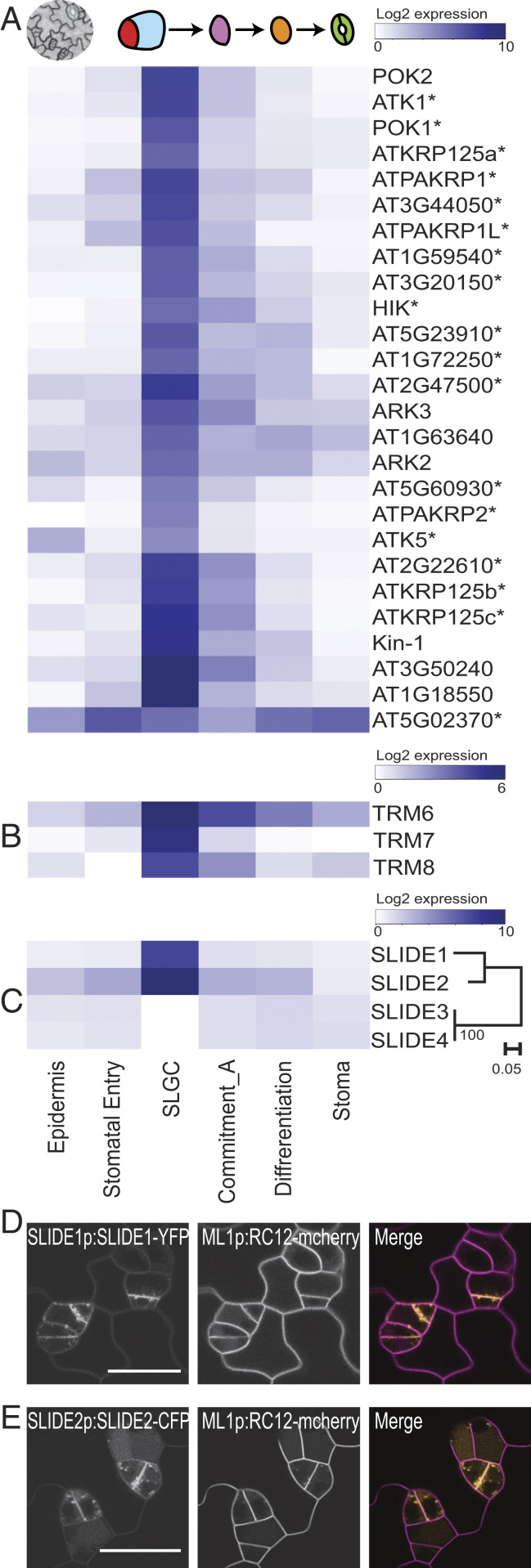
Microtubule rearrangements driven by kinesin motor proteins are essential for mitotic spindle dynamics and phragmoplast formation in cytokinesis. Of 61 Arabidopsis kinesins, 26 were in the SLGC cluster, including 19 of 23 “mitotic” kinesins and the stomatal lineage kinesin, ARK3 (2, 16) (Fig. 2A, SI Appendix, Fig. S4D, and Dataset S5). Asymmetrically dividing cells, such as SLGCs, feature specifically oriented division planes; therefore, it was particularly interesting to see the enriched expression of genes encoding regulators of division plane orientation, such as TANGLED and TON1 RECRUITING MOTIF (TRMs) in the SLGC cluster (Fig. 2B and Dataset S4) (17, 18). We next considered whether the strong cytokinesis signature in SLGCs could be used to identify new potential regulators of cytokinesis and division plane setting. Vesicle trafficking is one activity known to be generally required during cytokinesis, so we looked for SLGC-cluster genes that encoded multipass transmembrane proteins of previously uncharacterized function. Two four-pass transmembrane proteins (AT3G02640 and AT5G16250) were enriched in the SLGC cluster (Fig. 2C) and associated with M-phase in cell culture (19). We named these proteins SLGC IDENTITY ENRICHED (SLIDE), and translational reporters SLIDE1-YFP and SLIDE2-CFP were indeed visible in puncta at the division site, suggesting a role related to vesicle trafficking during stomatal lineage cell division (Fig. 2 D and E).
Fig. 2.
SLGCs profiles are enriched for mitotic kinesins and division plane-associated factors. (A–C) Heatmaps showing genes whose expression is preferentially associated with the SLGC cluster. (A) Twenty-six kinesins (an asterisk indicates mitotic), including POK1, POK2, and ARK3 that associate with the preprophase band (PPB). (B) PPB stabilizing factors, TRM6/7/8. (C) Previously uncharacterized SLIDE family of membrane proteins. Phylogeny indicating closely related SLIDE1 and SLIDE2 are highly expressed in SLGCs. (D and E) Confocal images of leaf epidermal cells showing SLIDE1-YFP and SLIDE2-CFP puncta (yellow in merge) enriched at the cell plate of dividing stomatal lineage cells. Cell outlines marked by ML1p:RC12-mCherry (magenta). (Scale bars, 20 μm.)
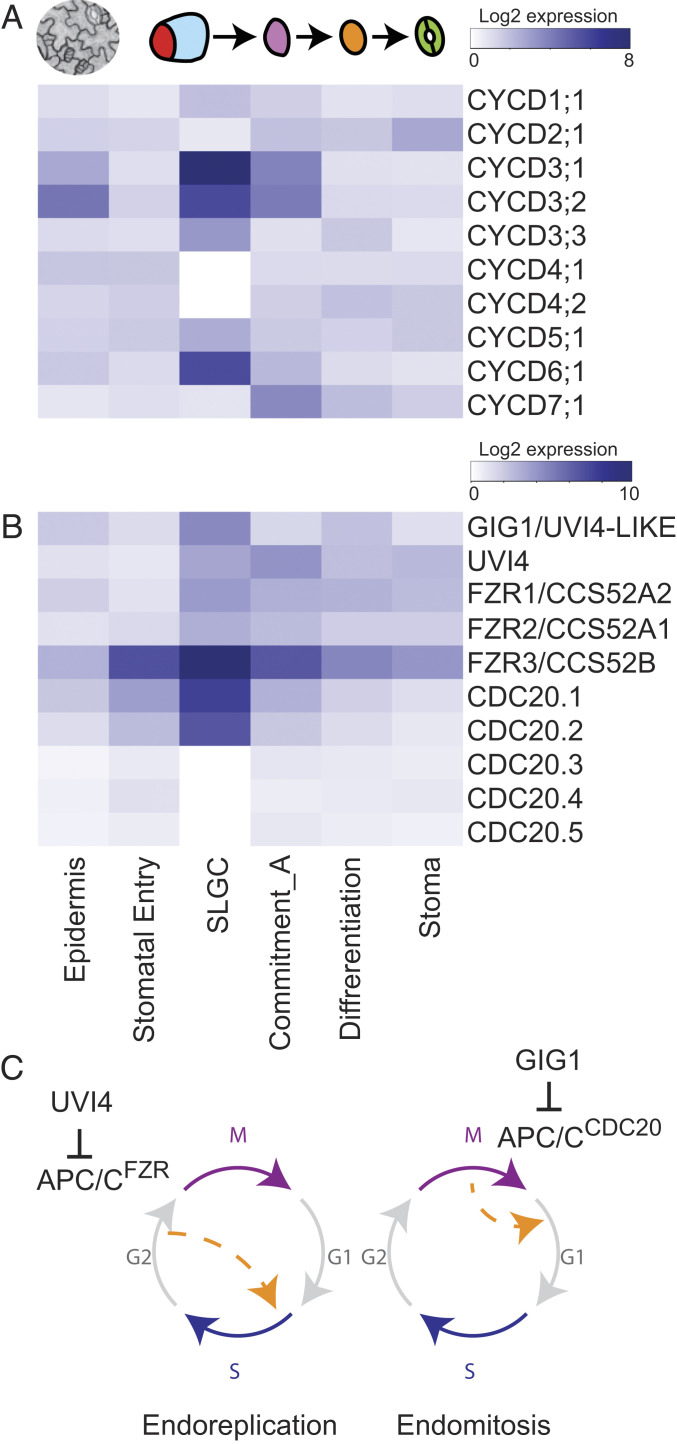
SLGCs are capable of progressing down two different developmental pathways, each of which has been correlated with specific cell-cycle related gene expression. Previous work linked D-type cyclins (CYCDs) to specific cell identities: CYCD6;1 in asymmetric cell division of root stem cells (20) and CYCD7;1 and CYCD5;1 in the stomatal commitment stage (21, 22). SLGCs differed from committed stomatal precursors by expressing CYCD6;1 but not CYCD7;1 and CYCD5;1 (Fig. 3A). SLGCs also expressed all three members of the CYCD3 family (Fig. 3A), previously shown to promote cell proliferation and regulate overall leaf cell number (23, 24).
Fig. 3.
SLGCs preferentially express specific CYCDs and endocycle related components, suggesting a poised division state. (A and B) Heatmaps showing genes whose expression is preferentially associated with the SLGC cluster. (A) Expression of all 10 G1/S regulating CYCD genes. (B) Expression of gene families associated with switch from mitotic to endoreduplication states. SLGCs are enriched for promoters of endoreduplication in the FZR family and CDC20.1/CDC20.2, but also their upstream inhibitors GIG1 and UVI4. (C) Illustration of endoreplication and endomitosis mechanisms for increasing ploidy. Orange dashed arrow indicates where cell cycle stages are bypassed. UVI4 and GIG1 negatively regulate APC/CFZR during endoreplication and APC/CCDC20 during endomitosis, respectively. See also SI Appendix, Fig. S5.
We also investigated genes associated with endocycling. This process of replicating the genome, but not progressing through cytokinesis, is a hallmark of pavement cells, products of the alternative SLGC fate trajectory. The high expression of APC/C coactivators, CELL DIVISION CYCLE20 (CDC20) and CCS52/FZY-RELATED (FZR) families in SLGCs suggests that SLGCs are primed to increase DNA ploidy (Fig. 3B). Yet SLGCs also preferentially express GIG1 and UVI4 (Fig. 3 B and C), which preferentially inhibit APC/CCDC20 and APC/CCCS52A, respectively. GIG1 was previously implicated in stomatal lineage decisions as its loss allows endoreduplicated cells to inappropriately express guard cell identity (25). For a view beyond these well-established regulators, we also profiled the expression of genes corresponding to GO term DNA endoreduplication (GO:0042023) (SI Appendix, Fig. S5 and Dataset S6). Both positive and negative (asterisks in SI Appendix, Fig. S5) endoreplication regulators are present across the stomatal lineage. Of particular interest is the cluster featuring key negative regulators of endoreplication CYCA2;3 (26), KAKTUS (KAK), and ERMO2 (27, 28) that peak in meristemoids and SLGCs. This is accompanied by depletion of positive regulators, such as KRP2 and SIAMESE in SLGCs (SI Appendix, Fig. S5). Collectively, these data suggest SLGCs have mixed potential and are poised between cell cycle progression and endoreduplication.
Regulation of SLGC Behaviors by Chromatin-Associated Gene DEK and Transcription Factor MYB16.
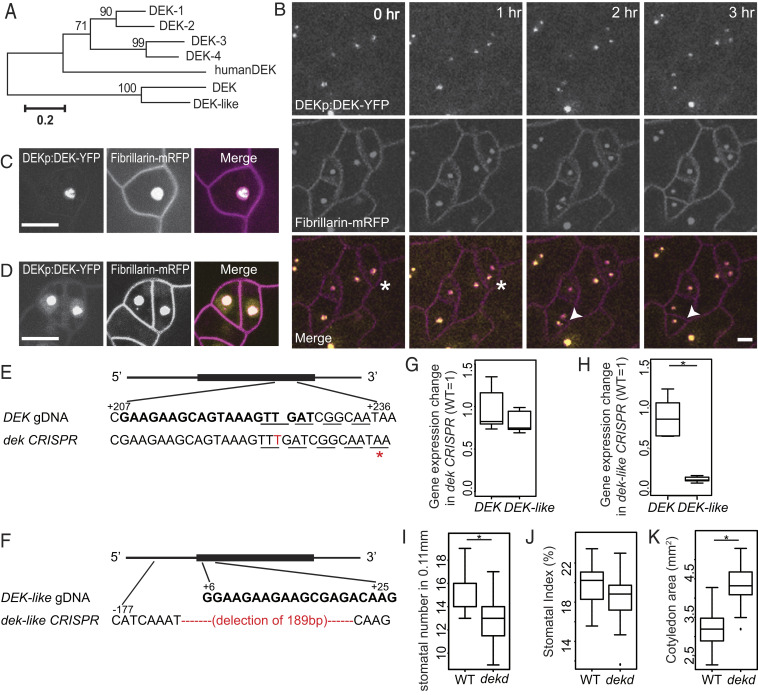
In addition to seeking a broad view of SLGC activities, we also wished to identify key regulators of their identity. We focused on genes predicted to encode proteins involved in gene regulation with the highest SLGC cluster membership rankings. The top candidate was DEK (AT5G42060), a putative chromatin-associated protein that was also verified experimentally to be a mediator complex component (29). DEK and its close homolog, DEK-like (AT1G64490), are outliers in a small family whose core members (AtDEK1-4) encode two canonical DNA binding domains, a C-terminal DEK domain and a Scaffold attachment factor A/B-Acinus-Pias (SAP) domain, defining them as homologs of the human protein DEK (30) (Fig. 4A). Human DEK can act as a transcriptional inhibitor or activator. AtDEK3 regulates nucleosome occupancy and chromatin accessibility in Arabidopsis (31). We found that the translational reporter DEKp:DEK-YFP was expressed in many stomatal lineage cells (Fig. 4B), but appears brighter in SLGCs and dividing guard mother cells (SI Appendix, Fig. S6A). DEK-YFP colocalizes with the nucleolar marker fibrillarin, as seen in time-lapse images of duplicating nucleoli (Fig. 4B). Higher-resolution images show that DEK-YFP does not completely colocalize with fibrillarin, and instead appears to be in a smaller subdomain of the nucleolus (Fig. 4C). Some weak DEK signal also appears in the nucleoplasm (Fig. 4D). DEK and DEK-like reporters also appear in the nucleolus when transiently expressed in Arabidopsis protoplasts or Nicotiana benthamiana leaf epidermal cells (SI Appendix, Fig. S6 B–D)
Fig. 4.
Identification and characterization of nucleolar-associated DEK proteins reveals their role in promoting continued SLGC division. (A) Phylogeny of Arabidopsis DEK-domain containing proteins with DEK and DEK-like as a distinct subgroup. (B) Confocal time-lapse images of dividing stomatal lineage cells from 4-d-old cotyledons coexpressing DEK-YFP (yellow) with a nucleolar marker, fibrillarin (magenta). Asterisks and arrowheads indicate two examples of dividing cells. Time stamps indicate time since start of cell division. Cell outlines marked by ML1p:RC12-mCherry (magenta). (C and D) Still images of DEK (yellow) and fibrillarin (magenta) showing that DEK is restricted to a subdomain of the nucleolus (C), but that also is weakly expressed in the nucleoplasm (D). (Scale bars, 10 μm.) (E and F) Gene-editing design for DEK and DEK-like, guide RNA is shown in bold and codons are denoted by underline. (E) A thymine (T) insertion results in a premature stop codon (red asterisk) in dek CRISPR plants. (F) There is a 189-base pair deletion in dek-like CRISPR plants. (G and H) Quantitative RT-PCR of dek and dek-like CRISPR plants showing unchanged and strongly reduced expression, respectively, of DEK and DEK-like genes. (I–K) Quantification of stomatal numbers and stomatal index in dek and dek-like double CRISPR plants at 7 dpg. The double mutants have lower stomatal density (I) and no difference in stomatal index (J). (K) Quantification of cotyledon size showed the double mutants are larger than WT. Stomatal number in 0.11-mm2 regions (n = 20 seedlings). *P < 0.05 by Wilcoxon rank-sum test. See also SI Appendix, Fig. S6.
We assayed DEK and DEK-like function by examining plants homozygous for T-DNA insertion mutations and generating CRISPR/Cas9 alleles of each gene. No obvious phenotype was observed in plants homozygous for T-DNA insertion alleles DEK (Salk_014079C) or DEK-like (Salk_097030), but T-DNA insertion sites were suboptimal, so we also used CRISPR/Cas9 to create mutant alleles in coding regions. For DEK, an insertion of a single thymine (T) is predicted to result in an early stop (Fig. 4E) and does not change transcript levels (Fig. 4G). For DEK-like, a 189-base pair deletion removes the start of the gene (Fig. 4F) and severely reduces transcript levels (Fig. 4H). In plants bearing mutations in both DEK and DEK-like (dekd), we observed a significant reduction in stomatal density (Fig. 4I) and a trend toward a lower stomatal index (Fig. 4J). Pavement cells were larger (SI Appendix, Fig. S6E), as was the overall organ size (Fig. 4K). Taken together, these phenotypes indicate that DEK and DEK-like promote continued division in SLGCs and delay differentiation into pavement cell fate.
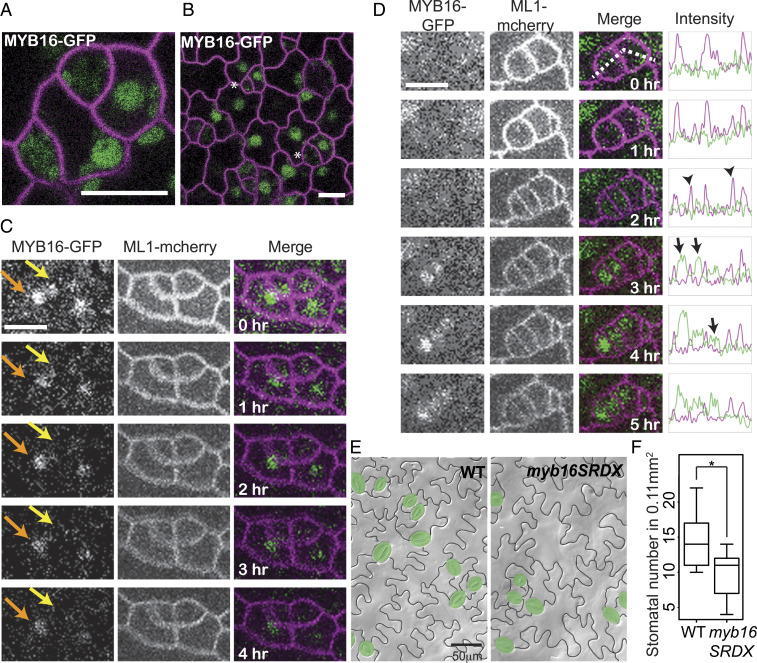
MYB16 was among the top transcription factors in the SLGC cluster (Dataset S4), and was previously connected to two epidermal characters: the promotion of petal cell outgrowth (32) and cuticle formation (33). In leaves, we found MYB16p:MYB16-GFP expressed preferentially in SLGCs (Fig. 5 A and B). The higher expression in SLGCs compared to their sister meristemoids (e.g., Fig. 5B) is opposite that of the meristemoid fate regulator SPEECHLESS (SPCH) (15). Time-lapse imaging showed that SPCH and MYB16 expression asymmetries arise differently. SPCH is expressed in the asymmetrically dividing mother cell, is initially inherited by both daughters, and then disappears from the SLGC (34). In contrast, among meristemoid/SLGC cell pairs that express MYB16 (Fig. 5C, yellow and orange arrows, respectively), the MYB16 meristemoid signal decays more rapidly. In addition, the onset of MYB16 expression often appeared after cell division in both daughter cells (Fig. 5D, 2- to 3-h time points), a pattern particularly easy to see as an increase in nuclear signal while the plasma membrane marker slowly bleaches. MYB16 was shown to be redundant with paralogues in other contexts (33); therefore, to assay for function, we used a dominant-negative form of MYB16 created by fusion to a transcriptional repressor domain (SRDX) (33). As previously reported, myb16SRDX plants showed organ fusion phenotypes in flowers, but rosette leaves were not dramatically affected (SI Appendix, Fig. S7). Pavement cells and stomata are morphologically normal in myb16SRDX but stomatal density (stomata/area) is decreased (Fig. 5 E and F), consistent with MYB16 promoting the division capacity of SLGCs and delaying their differentiation into pavement cells.
Fig. 5.
MYB16 preferentially labels SLGCs after asymmetric cell division and modulates stomatal lineage divisions. (A and B) Confocal image of 4 dpg cotyledons. (A) In meristemoid-SLGC sister pairs, MYB16-GFP (green) expression is higher in SLGCs. (B) In broader view, MYB16-GFP appears absent from meristemoids (asterisks). Cell outlines marked by ML1p:RC12A-mCherry (magenta). (C) Time-lapse confocal images showing that in sister meristemoid/SLGC pairs, MYB16-GFP persists longer in SLGCs (orange arrows) than in meristemoids (yellow arrows). (D) Time-lapse confocal images indicating that MYB16-GFP expression commences several hours after division. Intensity measurements (far right) were made in regions highlighted by a dashed line. In the intensity plot, MYB16-GFP and the cell outline are marked by green and magenta, respectively, two divisions appear at 2 h (arrows). MYB16-GFP (arrows) appears after cytokinesis (arrowheads at 2-h timepoint). (Scale bars, 10 μm.) (E) False-colored DIC images of WT and myb16SRDX epidermis showing morphologically normal stomata and pavement cells. (F) Quantification of changes to stomatal density (*P < 0.05) in 7 dpg WT and myb16SRDX cotyledons. 0.11-mm2 fields were scored (n = 20 seedlings).
Discussion
Plants are characterized by their plastic, adaptive development. Much of this is made possible by variations of cell division behaviors, such as continual divisions of immortal stem cell pools in the shoot and root meristems, or genome amplification through endocycling in differentiated cell types. Focusing on the transcriptional behavior of an epidermal cell type, the SLGC, where it is possible to witness the switch between modes of mitotic proliferation or endocycling, we find that these cells appear “poised” between alternative paths, expressing both activators and repressors of mitosis and endocycles.
DEK, a plant mediator (MED)-interacting protein, was the top SLGC candidate, based on expression enrichment. DEK is nucleolar localized and highly expressed in newly born SLGCs (SI Appendix, Fig. S6) and the phenotypes in dekd plants suggest SLGCs make a premature exit from self-renewal and shift toward differentiation into pavement cells (Fig. 4 I and J). In metazoans, MED may facilitate global down-regulation of transcription to enable a shift toward cell-type–specific transcription (35). In Arabidopsis, AtDEK3 binds histone H3 and H4 and overexpression leads to transcriptional repression of targets, likely through recruitment of histone deacetylases HDA3/HDT1 (31). By analogy, we might expect DEK to repress differentiation genes in SLGCs. DEK, however, localizes to the nucleolus. Our current hypothesis is that DEK promotes ribosome biogenesis by promoting the production of ribosomal RNA (rRNAs). Ribosome biogenesis is highly coordinated with the cell cycle and is associated with cell growth and proliferation (36). High levels of rRNA are found in animal stem cells and are associated with cellular plasticity and dedifferentiation (37). It may be important for SLGC self-renewal, therefore, to possess a high capacity for rRNA expression.
Although our transcriptome approach enabled us to better characterize SLGC activities, we did not identify transcription factors uniquely expressed in these cells, and consider it unlikely that SLGCs will be defined by genes of major impact like FAMA in guard cells (38) or ALTERED PHLOEM DEVELOPMENT (39) and VASCULAR RELATED NAC-PROTEINS (40) in vascular cell types. The best candidate for a transcriptional fate regulator is MYB16, which does differentially accumulate in SLGCs. MYB16 modulates cell division propensity, but the effect is subtle, and MYB16 also affects other epidermal behaviors. In fact, the most highly ranked transcription factors of the SLGC cluster—MYC1, MYC2, and PIF4—have functions in regulating SLGC features (11, 15, 41), but all regulate multiple processes and are expressed in multiple cell types.
For nearly two decades, transcriptional profiling of FACS-isolated pure populations of specific plant cell “types” enabled unprecedented detail in the definition of cell identity and the gene regulatory networks that create and result from that identity. A recent quantum leap in detail accompanied the advent of single-cell RNA-seq (scRNA-seq), where transcriptomes of thousands of individual cells are routinely generated. While there is promise that scRNA-seq can reveal previously unknown cell types through purely computational means, this success has mostly been seen in refining subtypes of terminally differentiated cells like neurons. There is a significant challenge identifying cell types and cell states in multipotent or precursor cell types, especially in plants (42, 43). From a technical standpoint, our combinatorial enrichment approach to identifying SLGCs in the absence of markers that unequivocally mark them, integrated with quantitative cell identity approaches (e.g., ICI) may prove useful for interrogating “unknown” clusters in scRNA-seq. From a conceptual standpoint, themes that emerged from the investigation of SLGCs—that enriched GO categories shared with other cell types, when interrogated more deeply, comprised unique gene sets, that SLGCs simultaneously express antagonistic cell cycle programs, and that gene expression is enriched, but not unique to SLGCs—may find parallels in multipotent precursors of other lineages, and may help us understand the probabilistic nature of such cell states.
Methods
Plant Reporter Lines and Mutants.
Col-0 served as the WT in all experiments and all transgenics were made in this accession. Plant reporters used for cell sorting were: BASLp:BRX-YFP (6), BASLp:myrBRX-YFP (6), and BRXL2p:BRXL2-YFP (6, 13). Details for cloning of new constructs DEKp:DEK-YFP, MYB16pro:MYB16-GFP, and DEK and DEK-like CRISPR alleles are in SI Appendix, Supplementary Methods. Primers are listed in SI Appendix, Table S1. Accession numbers used are as follows: BASL (At5G60880), BRX (At1g31880), BRXL2 (At3g14000), SLIDE1 (At3g02640), SLIDE2 (At5g16250), SLIDE3 (At5g36710), SLIDE4 (At5g36800), DEK1 (At3g48710), DEK2 (At5g63550), DEK3 (At4g26630), DEK4 (At5g55660), DEK (At5g42060), DEK-like (At1g64490), Human DEK (AAH35259), MYB16 (At5g15310).
Generation of SLGC Transcriptomes.
Nine-day postgermination (dpg) seedings of reporter lines were used for protoplast isolation and FACS as described in refs. 15 and 44. FACS was performed on Aria II (BD Biosciences) with an 80-mm nozzle. Gate boundaries and sort scheme are presented in SI Appendix, Supplementary Methods and Fig. S2. Fifty-base pair single-end reads were generated from a HiSeq2000 sequencer (Illumina) in high-output mode. Reads were mapped to TAIR10.18 via Bowtie2 (45) (read statistics are in Dataset S1). Transcripts per million-normalized counts were analyzed by ICI (scheme in SI Appendix, Fig. S3B). Cell commitment stages A and B correspond to MUTEpro:nucYFP and MUTEpro:MUTE-YFP, respectively. To generate an SLGC cluster (SI Appendix, Fig. S3C), counts were normalized via DESeq2 (46) using default settings and differentially expressed genes (19,707 genes) were obtained from all possible pairwise comparisons with a false-discovery rate (FDR) < 0.05, then clustered via FANNY (47) with k = 5 and a probability cutoff of 0.6. RNA-seq datasets are in the Gene Expression Omnibus (GEO) with accession no. GSE129938.
Microscopy.
Seedlings were grown on one-half MS plates at 22 °C with a 16-h light/8-h dark cycle and transferred to a glass slide with water or a custom-built microfluidics chamber (34) filled with one-quarter MS medium for time-lapse. Expression patterns of SLIDES, DEK, and MYB16 were observed in at least five seedlings where cells at appropriate stages exhibited similar localization. Time-lapse images of the abaxial site of 4-dpg seedlings expressing MYB16p:MYB16-YFP and ML1p:RC12A-mCherry were acquired once per hour for 16 h with a 40× objective. At least four to five cells at asymmetric or symmetric cell division events were traced. Z-stacks through the epidermis were generated and analyzed by Leica LAS and by Fiji (ImageJ, NIH). Images of DEK and DEK-like transient expression in Arabidopsis protoplasts were obtained on a Leica Stellaris system with Tau Gating to filter abundant chloroplast signal.
Supplementary Material
Acknowledgments
We thank Jessika Adrian for the initial FACS protocol and Jessica Chang for statistics consulting. Sequencing was performed at the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. This work was initially supported by the Gordon and Betty Moore Foundation. D.C.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021682118/-/DCSupplemental.
Data Availability
The data reported in this paper have been deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE129938). Plant materials are available upon request.
References
- 1.O’Brien L. E., Soliman S. S., Li X., Bilder D., Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603–614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau O. S., et al., Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horst R. J., et al., Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genet. 11, e1005374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J., MacAlister C. A., Bergmann D. C., BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Wang P., Shao W., Zhu J. K., Dong J., The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowe M. H., Dong J., Weimer A. K., Bergmann D. C., A plant-specific polarity module establishes cell fate asymmetry in the Arabidopsis stomatal lineage. bioRxiv [Preprint] (2019). 10.1101/614636 (Accessed 1 January 2020). [DOI]
- 7.Hara K., Kajita R., Torii K. U., Bergmann D. C., Kakimoto T., The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt L., Gray J. E., The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864–869 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Hara K., et al., Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Vatén A., Soyars C. L., Tarr P. T., Nimchuk Z. L., Bergmann D. C., Modulation of asymmetric division diversity through cytokinin and SPEECHLESS regulatory interactions in the Arabidopsis stomatal lineage. Dev. Cell 47, 53–66.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau O. S., et al., Direct control of SPEECHLESS by PIF4 in the high-temperature response of stomatal development. Curr. Biol. 28, 1273–1280.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melaragno J. E., Mehrotra B., Coleman A. W., Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bringmann M., Bergmann D. C., Tissue-wide mechanical forces influence the polarity of stomatal stem cells in Arabidopsis. Curr. Biol. 27, 877–883 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Efroni I., Ip P.-L., Nawy T., Mello A., Birnbaum K. D., Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 16, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adrian J., et al., Transcriptome dynamics of the stomatal lineage: Birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanstraelen M., Inzé D., Geelen D., Mitosis-specific kinesins in Arabidopsis. Trends Plant Sci. 11, 167–175 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Walker K. L., Müller S., Moss D., Ehrhardt D. W., Smith L. G., Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17, 1827–1836 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer E., et al., The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356, 186–189 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Menges M., Hennig L., Gruissem W., Murray J. A., Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Sozzani R., et al., Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weimer A. K., et al., Lineage- and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development 145, dev160671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han S. K., et al., MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev. Cell 45, 303–315.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Dewitte W., et al., Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baekelandt A., et al., Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol. 178, 217–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwata E., et al., GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23, 4382–4393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai K. K., et al., The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18, 382–396 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Refy A., et al., The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol. Genet. Genomics 270, 403–414 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Nakano R. T., et al., GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21, 3672–3685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S., Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Kappes F., et al., Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol. Cell. Biol. 24, 6011–6020 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waidmann S., Kusenda B., Mayerhofer J., Mechtler K., Jonak C., A DEK domain-containing protein modulates chromatin structure and function in Arabidopsis. Plant Cell 26, 4328–4344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann K., et al., Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134, 1691–1701 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Oshima Y., et al., MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25, 1609–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies K. A., Bergmann D. C., Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc. Natl. Acad. Sci. U.S.A. 111, 15585–15590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Alessio J. A., Wright K. J., Tjian R., Shifting players and paradigms in cell-specific transcription. Mol. Cell 36, 924–931 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grummt I., Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Brombin A., Joly J. S., Jamen F., New tricks for an old dog: Ribosome biogenesis contributes to stem cell homeostasis. Curr. Opin. Genet. Dev. 34, 61–70 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Ohashi-Ito K., Bergmann D. C., Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonke M., Thitamadee S., Mähönen A. P., Hauser M. T., Helariutta Y., APL regulates vascular tissue identity in Arabidopsis. Nature 426, 181–186 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Kubo M., et al., Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X., et al., Jasmonate negatively regulates stomatal development in Arabidopsis cotyledons. Plant Physiol. 176, 2871–2885 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Anido C. B., et al., Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev. Cell 56, 1043–1055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satterlee J. W., Strable J., Scanlon M. J., Plant stem-cell organization and differentiation at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 117, 33689–33699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bargmann B. O., Birnbaum K. D., Fluorescence activated cell sorting of plant protoplasts. J. Vis. Exp. 36, 1673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mächler M., Rousseeuw P., Struyf A., Hubert M., Hornik K., Cluster: Cluster Analysis Basics and Extensions (R Package version 1, 2012).
- 48.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE129938). Plant materials are available upon request.