Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen that causes coronavirus disease 2019 (COVID-19), has significantly impacted the human health and economic status worldwide. There is an urgent need to identify effective prophylactics and therapeutics for the treatment of SARS-CoV-2 infection and associated COVID-19.
KEYWORDS: SARS-CoV-2, coronavirus, fluorescent, luciferase, reporter virus, reverse genetics
ABSTRACT
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen responsible for coronavirus disease 2019 (COVID-19), has devastated public health services and economies worldwide. Despite global efforts to contain the COVID-19 pandemic, SARS-CoV-2 is now found in over 200 countries and has caused a death toll of over 1 million human lives as of November 2020. To date, only one Food and Drug Administration (FDA)-approved therapeutic drug (remdesivir) and a monoclonal antibody (MAb), bamlanivimab, are available for the treatment of SARS-CoV-2. As with other viruses, studying SARS-CoV-2 requires the use of secondary approaches to detect the presence of the virus in infected cells. To overcome this limitation, we have generated replication-competent recombinant SARS-CoV-2 (rSARS-CoV-2) constructs expressing fluorescent (Venus or mCherry) or bioluminescent (Nluc) reporter genes. Vero E6 cells infected with reporter-expressing rSARS-CoV-2 can be easily detected via fluorescence or luciferase expression and display a good correlation between reporter gene expression and viral replication. Moreover, rSARS-CoV-2 expressing reporter genes has plaque sizes and growth kinetics comparable to those of wild-type virus, rSARS-CoV-2/WT. We used these reporter-expressing rSARS-CoV-2 constructs to demonstrate their feasibility to identify neutralizing antibodies (NAbs) or antiviral drugs. Our results demonstrate that reporter-expressing rSARS-CoV-2 represents an excellent option to identify therapeutics for the treatment of SARS-CoV-2, where reporter gene expression can be used as a valid surrogate to track viral infection. Moreover, the ability to manipulate the viral genome opens the feasibility of generating viruses expressing foreign genes for their use as vaccines for the treatment of SARS-CoV-2 infection.
IMPORTANCE Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen that causes coronavirus disease 2019 (COVID-19), has significantly impacted the human health and economic status worldwide. There is an urgent need to identify effective prophylactics and therapeutics for the treatment of SARS-CoV-2 infection and associated COVID-19. The use of fluorescent-protein- or luciferase-expressing reporter viruses has significantly advanced viral research. Here, we generated recombinant SARS-CoV-2 (rSARS-CoV-2) constructs expressing fluorescent (Venus and mCherry) or luciferase (Nluc) reporter genes and demonstrated that these viruses represent an excellent option to track viral infections in vitro. Importantly, reporter-expressing rSARS-CoV-2 constructs display growth kinetics and plaque phenotypes similar to those of their wild-type counterpart (rSARS-CoV-2/WT), demonstrating their usefulness for identifying drugs and/or neutralizing antibodies (NAbs) for the therapeutic treatment of SARS-CoV-2. Henceforth, these reporter-expressing rSARS-CoV-2 constructs can be used to interrogate large libraries of compounds and/or monoclonal antibodies (MAb), in high-throughput screening settings, to identify those with therapeutic potential against SARS-CoV-2.
INTRODUCTION
Late in 2019, a previously unknown coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in Wuhan, China (1). Since then, SARS-CoV-2 has become responsible for the global pandemic of coronavirus disease 2019 (COVID-19) (1). As of November 2020, SARS-CoV-2 has spread worldwide and is responsible for over 40 million confirmed cases and around 1.1 million deaths (2). To date, only one U.S. Food and Drug Administration (FDA)-approved therapeutic antiviral drug, remdesivir, and a monoclonal antibody (MAb), bamlanivimab, are available for the treatment of SARS-CoV-2 infections (3). Recently, two prophylactics (vaccines) against SARS-CoV-2 were FDA approved (4, 5).
SARS-CoV-2 is a single-stranded, positive-sense RNA Betacoronavirus that belongs to the family Coronaviridae. Prior to SARS-CoV-2, only six coronavirus (CoV) species were known to cause disease in humans (6). Of the six, four human CoVs (hCoVs) are prevalent and responsible for causing the common cold in immunocompetent individuals (hCoV-229E, hCoV-OC43, hCoV-NL63, and hCoV-HKU1) (6, 7). The two other CoVs, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have been associated with severe illness and significant morbidity and mortality (8). SARS-CoV was responsible for an outbreak of severe acute respiratory syndrome in 2002-2003 in Guangdong Province, China, with a fatality rate of around 9.5% (9). MERS-CoV was responsible for an outbreak of severe respiratory disease in 2012-2013 in the Middle East, with a fatality rate of around 30% (6, 7, 10). SARS-CoV-2 has a viral genome approximately 30,000 nucleotides in length, with high similarity to that of SARS-CoV (∼79%) and lower similarity to that of MERS-CoV (∼50%). Its overall fatality rate is 3.4%, but it can be as high as 49% in critically ill patients, making the COVID-19 pandemic rival that of the “Spanish flu” in 1918-1919 (11–15).
Studying SARS-CoV-2 in laboratories requires the use of secondary approaches to identify the presence of virus in infected cells. The ability to generate recombinant viruses using reverse genetics approaches represents a powerful tool to answer important questions regarding the biology of viral infections, including mechanisms of viral infection, pathogenesis, and disease. In addition, the use of reverse genetics techniques offers the possibility to generate recombinant viruses expressing reporter genes for their use in cultured cells or in vivo models of infection where reporter gene expression can be used as a valid surrogate the identify the presence of the virus in infected cells (16, 17). Importantly, these reporter-expressing recombinant viruses also represent an excellent tool for the easy and rapid identification of drugs for the prophylactic or therapeutic treatment of viral infections, by allowing high-throughput screening (HTS) approaches to interrogate large libraries of biologicals exhibiting antiviral activity.
Several papers have described the ability to generate recombinant SARS-CoV-2 (rSARS-CoV-2) expressing fluorescent (mNeonGreen and green fluorescent protein [GFP]) or bioluminescent (Nluc) reporter genes (18–20). However, these reverse genetics protocols require laborious in vitro assembly and transcription steps prior to transfecting cells, an inconvenience that should be considered due to the constraints it places on these methods. Here, we describe the generation and characterization of replication-competent rSARS-CoV-2 expressing fluorescent Venus or mCherry or bioluminescent Nluc reporter genes using our recently described bacterial artificial chromosome (BAC)-based reverse genetics approach (21, 22). In Vero E6 cells, rSARS-CoV-2 expressing reporter genes has growth kinetics and plaque phenotype similar to those of wild-type virus (rSARS-CoV-2/WT). Importantly, we have observed a correlation between reporter gene expression and viral replication (21), and infected cells can be easily detected, without the need for secondary approaches, based on reporter gene expression. Using these reporter-expressing rSARS-CoV-2 constructs, we have developed fluorescent-based microneutralization assays that can be used to identify neutralizing antibodies (NAbs) and/or antivirals. The neutralization titers and inhibitory activities of NAbs and antivirals, respectively, obtained in our reporter-based microneutralization assays were similar to those observed in classical microneutralization assays using rSARS-CoV-2/WT (23). These results demonstrate that our reporter-expressing rSARS-CoV-2 represent an excellent tool for studying the biology of the virus and for the identification of therapeutics for the treatment of SARS-CoV-2 infection and also for in vivo studies. Furthermore, because of reporter gene expression, these rSARS-CoV-2 constructs expressing reporter genes represent an ideal option to screen large libraries of biologicals to identify those with antiviral activity. Our results also demonstrate the feasibility of generating rSARS-CoV-2 expressing foreign genes that could be used to generate vaccines for the treatment of SARS-CoV-2 infections and/or associated COVID-19 disease.
RESULTS
Generation of rSARS-CoV-2 expressing reporter genes.
The pBeloBAC11 plasmid carrying the full-length viral genome of SARS-CoV-2 was previously described (21). To generate the reporter-expressing rSARS-CoV-2, open reading frame (ORF) 7a was replaced with a Venus, mCherry, or Nluc gene in the pBeloBAC11 plasmid carrying the remaining viral genome to produce pBeloBAC11-SARS-CoV-2-del7a/Venus, -del7a/mCherry, or -del7a/Nluc plasmids for viral rescue. We then used our previously described BAC-based reverse genetics approach to rescue rSARS-CoV-2-Venus, -mCherry, and -Nluc (Fig. 1A).
FIG 1.
rSARS-CoV-2 expressing reporter genes. (A) Schematic representation of rSARS-CoV-2 expressing Venus (green box), mCherry (red box), and Nluc (blue box) reporter genes instead of the viral ORF 7a. The molecular sizes of the three reporter genes are indicated. The locations of other viral proteins and untranslated regions (UTR) are also shown. (B) Genetic characterization of reporter-expressing rSARS-CoV-2. Vero E6 cells were mock infected or infected (MOI, 0.01) with WT or reporter-expressing rSARS-CoV-2. At 72 h postinfection, total RNA collected from cells was used to amplify, using RT-PCR, the viral NP, the ORF 7a region, and the different reporter genes (Venus, mCherry, or Nluc). Primers used for this RT-PCR analysis are shown on the left. The molecular sizes (base pairs) of the RT-PCR amplified products are shown on the right.
We confirmed the rescue of rSARS-CoV-2 expressing Venus, mCherry, or Nluc reporter genes by reverse-transcription-PCR (RT-PCR) using total RNA from mock-, rSARS-CoV-2/WT-, or rSARS-CoV-2 reporter virus-infected cells with primers specific for the viral NP, ORF 7a, or the individual reporter genes (Fig. 1B). As expected, primers specific for SARS-CoV-2 NP amplified a band of ∼1,260 bp from the RNA extracted from rSARS-CoV-2-infected but not mock-infected cells (Fig. 1B). Bands that were amplified using primers in ORF 7a resulted in the expected ∼566 bp in cells infected with rSARS-CoV-2/WT and ∼920, 911, and 815 bp in the case of cells infected with rSARS-CoV-2-Venus, -mCherry and -Nluc, respectively, based on the different sizes of the reporter genes (Fig. 1B). Primers specific for the reporter genes resulted in RT-PCR amplification of bands only from cells infected with the respective reporter-expressing rSARS-CoV-2 (Fig. 1B). These results demonstrate that substitution of the viral ORF 7a with Venus, mCherry, or Nluc genes results in the successful recovery of rSARS-CoV-2 containing these reporter genes.
Characterization of rSARS-CoV-2 expressing reporter genes.
Next, we characterized the reporter-expressing rSARS-CoV-2 by evaluating the expression levels of Venus, mCherry, or Nluc in cell cultures and compared them to those of cells infected with rSARS-CoV-2/WT (Fig. 2). rSARS-CoV-2 reporter viruses expressing Venus and mCherry were directly visualized under a fluorescence microscope (Fig. 2A). Indirect immunofluorescence microscopy using a MAb against SARS-CoV NP was used to detect rSARS-CoV-2/WT infection (Fig. 2A). As expected, Venus and mCherry expression was observed only in Vero E6 cells infected with rSARS-CoV-2 expressing Venus or mCherry, respectively, but not in cells infected with rSARS-CoV-2/WT (Fig. 2A). Importantly, only cells infected with rSARS-CoV-2-Venus or rSARS-CoV-2-mCherry were detected using green or red filters, respectively (data not shown). As expected, the viral NP was detected in cells infected with rSARS-CoV-2-WT, -Venus, or -mCherry (Fig. 2A). Expression of Nluc in rSARS-CoV-2-Nluc-infected cells was evaluated from tissue culture supernatants at 48 h postinfection (Fig. 2B). High levels of Nluc expression were detected in culture supernatants of cells infected with rSARS-CoV-2-Nluc but not from mock- or rSARS-CoV-2/WT-infected cells (Fig. 2B). These results demonstrate that Vero E6 cells infected with rSARS-CoV-2-Venus, -mCherry, or -Nluc express the corresponding reporter genes and that viral infections can be detected by fluorescence (rSARS-CoV-2-Venus or -mCherry) or luciferase (rSARS-CoV-2-Nluc) without the need for antibodies, which were required for the detection of rSARS-CoV-2/WT.
FIG 2.
Characterization of reporter-expressing rSARS-CoV-2. (A) Fluorescence expression. Vero E6 cells were mock infected or infected (MOI of 0.01) with WT and Venus- or mCherry-expressing rSARS-CoV-2. At 48 h postinfection, cells were fixed and permeabilized, visualized for Venus (left) or mCherry (right) expression, and immunostained with a SARS-CoV NP MAb (1C7). DAPI was used for nuclear staining. Merged images for Venus (left) or mCherry (right), viral NP, and DAPI are presented. Representative images are shown. Magnification, ×20. Bar, 100 μm. (B) Nluc expression. Vero E6 cells were mock infected or infected (MOI of 0.01) with WT and Nluc-expressing rSARS-CoV-2. At 48 h postinfection, Nluc expression in tissue culture supernatants was analyzed using a Synergy LX microplate reader (BioTek). (C) Western blotting. Vero E6 cells were mock infected or infected (MOI 0.01) with WT and Venus (left)-, mCherry (center)-, or Nluc (right)-expressing rSARS-CoV-2. At 48 h postinfection, viral NP and reporter gene protein expression levels were analyzed using specific antibodies. An antibody against beta-actin was used as an internal control. The sizes of molecular markers are shown on the right of each blot.
We next evaluated reporter protein expression levels by Western blotting assay using cell lysates from either mock-, rSARS-CoV-2-WT-, or rSARS-CoV-2-Venus-, -mCherry-, or -Nluc-infected cells using MAbs against the viral NP, the reporter genes, or actin as a loading control (Fig. 2C). As expected, reporter gene expression was detected in lysates of cells infected with the respective reporter-expressing rSARS-CoV-2 but not from mock- or rSARS-CoV-2-WT-infected cells. Viral NP expression was detected in lysates from all virus-infected cells but not mock-infected cells (Fig. 2C).
Next, we assessed reporter gene expression over a period of 96 h in cells that were mock infected (data not shown) or cells infected with WT or reporter-expressing rSARS-CoV-2 (Fig. 3). Venus and mCherry expression levels were determined using fluorescence microscope (Fig. 3A), while Nluc activity in tissue culture supernatants from infected cells was detected using a luminometer (Fig. 3B). Venus and mCherry expression were detected as early as 24 h postinfection, and fluorescent protein expression increased over time until 96 h postinfection, when a decrease in fluorescence was observed because of cytopathic effect (CPE) caused by viral infection (bright-field [BF] microscopy) (Fig. 3A). Similar CPE, but not fluorescence expression, was also observed in cells infected with rSARS-CoV-2/WT (Fig. 3A). Levels of Nluc expression were also detected as early as 24 h postinfection and increased in a time-dependent matter (Fig. 3B).
FIG 3.
Viral growth kinetics and plaque phenotype. (A) Fluorescence expression. Vero E6 cells were infected (MOI of 0.01) with WT (left), Venus-expressing (center), and mCherry-expressing (right) rSARS-CoV-2. At 12, 24, 48, 72, and 96 h postinfection. fluorescence protein expression was determined using a fluorescence microscope. Representative images are shown. Magnification, ×20. Bar, 100 μm. (B) Nluc expression. Vero E6 cells were mock infected or infected (MOI of 0.01) with WT and Nluc-expressing rSARS-CoV-2. At the indicated times postinfection, Nluc expression in the tissue culture supernatants was analyzed using a Synergy LX microplate reader (BioTek). (C) Growth kinetics. Vero E6 cells were infected (MOI of 0.01) with WT or reporter-expressing rSARS-CoV-2. At 12, 24, 48, 72, and 96 h postinfection, the presence of infectious virus in the tissue culture supernatants was determined using a plaque assay. (D) Plaque phenotype. Vero E6 cells were infected with ∼25 PFU of WT (left), Venus-expressing (middle), and mCherry-expressing (right) rSARS-CoV-2. At 72 h postinfection, plaques were observed under a fluorescence microscope to detect Venus or mCherry expression. In the case of rSARS-CoV-2/WT-infected cells, images correspond to fluorescence filters to detect Venus (left) or mCherry (right) expression. Thereafter, viral plaques were detected using the 1C7 SARS-CoV NP MAb. A selected number (n = 6) of plaques were used to determine the percentage of viral plaques expressing fluorescent proteins (Venus or mCherry). Magnification, ×4. Bar, 750 μm.
To assess whether deletion of ORF 7a and insertion of reporter genes compromised viral fitness in cultured cells, we compared growth kinetics of reporter-expressing rSARS-CoV-2 to those of rSARS-CoV-2/WT (Fig. 3C). We found that all the reporter-expressing rSARS-CoV-2 exhibited growth kinetics and peak viral titers of infection similar to those of rSARS-CoV-2/WT (Fig. 3C), suggesting that deletion of ORF 7a and insertion of the reporter genes did not significantly affect viral fitness, at least in cultured cells. These results also support previous findings with SARS-CoV where deletion of ORF 7a and insertion of reporter genes did not impact viral fitness in vitro (24, 25). These results were further confirmed when we evaluated the plaque phenotypes of the rSARS-CoV-2 constructs expressing fluorescent reporter genes and compared them to those of rSARS-CoV-2/WT (Fig. 3D). Similar plaque sizes were observed in Vero E6 cells infected with rSARS-CoV-2/WT and rSARS-CoV-2 expressing Venus or mCherry (Fig. 3D). Notably, Venus-positive or mCherry-positive plaques were detected only in cells infected with rSARS-CoV-2-Venus or -mCherry, respectively, and not in rSARS-CoV-2/WT-infected cells (Fig. 3D). Importantly, fluorescent plaques overlapped those detected by immunostaining using the SARS-CoV NP MAb 1C7. Similar to the growth kinetics data, we found no significant differences between the plaque sizes of reporter-expressing rSARS-CoV-2 and rSARS-CoV-2/WT (Fig. 3D).
Reporter-based microneutralization assay for the identification of antivirals.
To determine the feasibility of using our reporter-expressing rSARS-CoV-2 for the identification of antivirals, we evaluated the ability of remdesivir to inhibit SARS-CoV-2 in reporter-based microneutralization assays (Fig. 4). Remdesivir has been described to inhibit SARS-CoV-2 infection and is the only FDA-approved antiviral for the treatment of SARS-CoV-2 (3, 23, 26). The 50% effective concentrations (EC50s) of remdesivir against rSARS-CoV-2-Venus (1.07 μM) (Fig. 4A and B), -mCherry (1.78 μM) (Fig. 4C and D), and -Nluc (1.79 μM) (Fig. 4E) were similar to those obtained with rSARS-CoV-2/WT (1.51 μM) (Fig. 4F) and to values previously reported in the literature (23). This demonstrates the feasibility of using these reporter-expressing rSARS-CoV-2 constructs and the reporter-based assay to easily identify compounds with antiviral activity based on fluorescent-protein or luciferase expression and without the need for MAbs to detect the presence of the virus in infected cells.
FIG 4.
Reporter-based microneutralization assay for the identification of antivirals. Vero E6 cells (96-well plate format, ∼4 × 104 cells/well, triplicates) were infected with 100 PFU of Venus (A and B), mCherry (C and D), Nluc (E), or WT (F) rSARS-CoV-2. After 1 h viral absorption, postinfection medium containing 3-fold serial dilutions of remdesivir (starting concentration, 50 μM) was added to the cells. At 24 h postinfection, cells were fixed and visualized for Venus (B) and mCherry (D) expression using a fluorescence microscope. In the case of cells infected with rSARS-CoV-2 expressing Nluc, luciferase expression in the tissue culture supernatant was determined at 48 h postinfection using a luciferase assay and a Synergy LX microplate reader (BioTek) (E). For the detection of rSARS-CoV-2/WT, the amount of virus was determined by plaque assay using the SARS-CoV NP MAb 1C7 (F). The amount of viral infection for Venus-expressing, mCherry-expressing, or WT rSARS-CoV-2 (after IFA) was determined using fluorescent images of each well and quantified using Cell Profiler (Broad Institute) cell image analysis software. Nluc activity was quantified using Gen5 data analysis software (BioTek). The 50% effective concentration (EC50) of remdesivir was determined using GraphPad Prism. Dashed lines indicate 50% viral inhibition. Data are means and SD from triplicate wells. Representative images are shown. Magnification, ×10. Bar, 300 μm.
Reporter-based microneutralization assay for the identification of NAbs.
We next evaluated the feasibility of using our reporter-expressing rSARS-CoV in a reporter-based microneutralization assay to identify NAbs against SARS-CoV-2. As proof of concept, we used a human MAb (1212C2) which we recently described to potently bind and neutralize SARS-CoV-2 infection both in vitro and in vivo (27). The half-maximal neutralizing concentrations (NT50s) of 1212C2 against rSARS-CoV-2-Venus (1.94 ng) (Fig. 5A and B), -mCherry (5.02 ng) (Fig. 5C and D), and -Nluc (3.67 ng) (Fig. 5E) were similar to those observed with rSARS-CoV-2/WT (4.88 ng) (Fig. 5F) and to recently reported values (27).
FIG 5.
Reporter-based microneutralization assay for the identification of NAbs: Vero E6 cells (96-well plate format, ∼4 × 104 cells/well, triplicates) were infected with 100 PFU of Venus (A and B), mCherry (C and D), Nluc (E) or WT (F) rSARS-CoV-2. After 1 h viral absorption, postinfection medium containing 3-fold serial dilutions (starting concentration, 500 ng) of a SARS-CoV-2 NAb (1212C2) was added to the cells. At 24 h postinfection, cells were fixed and visualized for Venus (B) and mCherry (D) expression using a fluorescence microscope. In the case of cells infected with rSARS-CoV-2 expressing Nluc, luciferase expression in the tissue culture supernatant was determined at 48 h postinfection using a luciferase assay and a Synergy LX microplate reader (BioTek) (E). For the detection of rSARS-CoV-2/WT, the amount of virus was determined by plaque assay using the SARS-CoV NP MAb 1C7 (F). The amount of viral infection for Venus-expressing, mCherry-expressing, and WT rSARS-CoV-2 (after IFA) was determined using fluorescent images of each well and quantified using Cell Profiler (Broad Institute). Nluc was quantified using Gen5 data analysis software (BioTek). The 50% neutralizing titer (NT50) of 1212C2 was determined using GraphPad Prism. Dashed lines indicate 50% viral neutralization. Data are means and SD from triplicate wells. Representative images are shown. Magnification, ×10. Bar, 300 μm.
Genetic stability of rSARS-CoV-2 in vitro.
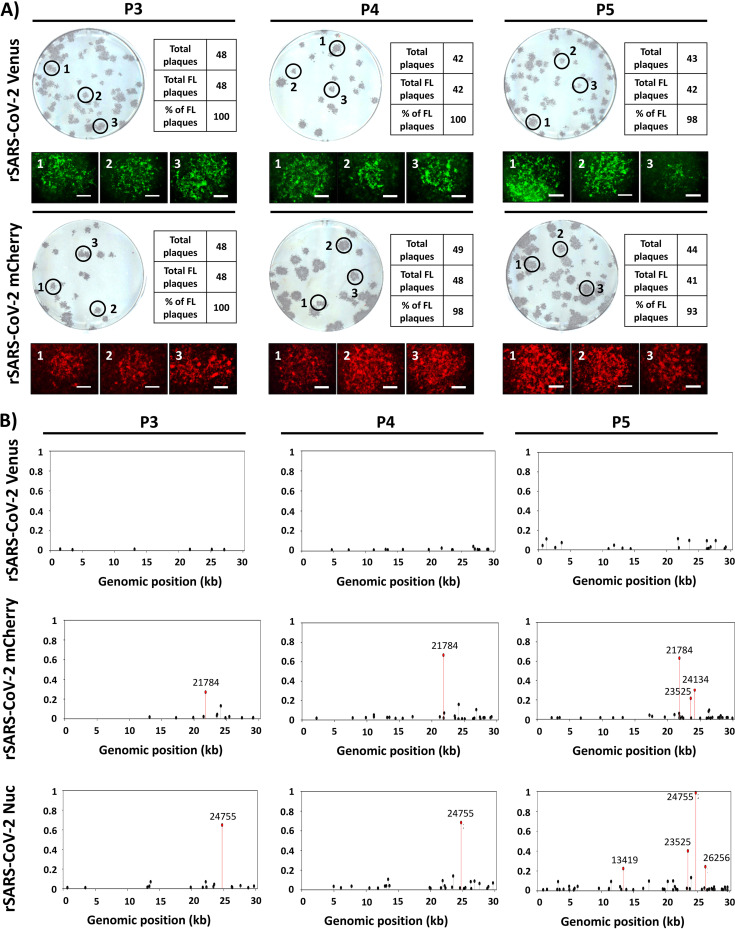
The genetic stability of reporter-expressing recombinant viruses is important to demonstrate their viability in in vitro and/or in vivo studies. To evaluate the ability of our rSARS-CoV-2 constructs to maintain fluorescent reporter gene expression, viruses were consecutively passaged in Vero E6 cells, and Venus and mCherry expression was determined by plaque assay using fluorescence microscopy (Fig. 6A). To that end, we evaluated fluorescence expression of over 40 plaques before immunostaining with anti-SARS-CoV NP MAb 1C7. We found that Venus and mCherry fluorescence expression from our rSARS-CoV-2 reporter viruses was genetically stable with nearly 100% of the plaques analyzed under a fluorescence microscope (Fig. 6A). We also evaluated the complete genome sequences of the reporter-expressing rSARS-CoV-2 constructs used in our studies (passage 3 [P3]) with those of additional passages (P4 and P5) using next-generation sequencing (Fig. 6B). In the case of rSARS-CoV-2-Venus (Fig. 6B, top), few variants were found at low frequencies after two additional passages (P5), indicating no significant changes and/or deletions in the viral genome. However, for rSARS-CoV-2-mCherry (Fig. 6B, middle), variants containing a mutation at position 21784 in the S gene were found in our viral stock (P3), and the frequency of this mutation increased after additional passages (P4 and P5). Two additional mutations at positions 23525 and 24134 (both in the S gene) were also found at P5. In the case of rSARS-CoV-2-Nluc (Fig. 6B, bottom), a mutation at position 24755 (S gene) was found in our viral stock (P3). The frequency of this mutation increased up to 100% after 2 additional passages (P5). Other, less abundant mutations at positions 13419 (nsp12, RNA-dependent RNA polymerase), 23525 (S gene), and 26256 (envelope [E] gene) were also found after 2 additional passages (P5) (Fig. 6B, bottom). Most likely, these mutations arose due to viral adaptation to Vero E6 cells. However, it is also possible that these mutations could be related to the nature of the reporter gene, since they were different between the three reporter-expressing rSARS-CoV-2 constructs.
FIG 6.
Genetic stability of fluorescence-expressing rSARS-CoV-2. (A) Plaque assay. Fluorescence-expressing rSARS-CoV-2 constructs were passaged up to 5 times in Vero E6 cells, and infectious-virus-containing tissue culture supernatants from passages 3 to 5 (P3 to P5) were assessed for Venus or mCherry expression at 72 h postinfection, before immunostaining with the SARS-CoV NP MAb 1C7. The percentage of reporter-expressing viruses was determined from ∼40 to 50 viral plaques per passage. Representative images of immunostaining and fluorescence obtained from each P3 to P5 viral plaque are shown. Magnification, ×4. Bar, 750 μm. (B) Sequence analysis. Reporter-expressing rSARS-CoV-2 nonreference allele frequencies from virus stock (P3) and after two consecutive passages in Vero cells (P4 and P5) were determined using next-generation sequencing, using modified rSARS-CoV-2/WT reference genomes. Nonreference alleles that were below 1% of reads are not shown, and those greater than 20% are indicated in red.
DISCUSSION
Reporter-expressing viruses represent a powerful tool for both basic research and translational studies (16, 17, 28–31). Several research groups, including ours, have described recombinant viruses expressing reporter genes to easily study the biology of viral infections, to evaluate the efficacy of antivirals or NAbs, and for in vivo studies in validated animal models (16, 32–45).
Both fluorescent and luciferase proteins have been used to generate reporter-expressing viruses. However, the innate and differing properties of reporter genes dictate which one might be inserted into a recombinant virus. While fluorescent proteins provide an efficient way to track viral infections using microscopy, luciferase proteins are more readily quantifiable and therefore more amenable to HTS studies (16, 17, 46). For this reason, in this study we generated rSARS-CoV-2 expressing fluorescent proteins (Venus and mCherry) or luciferase (Nluc) (Fig. 1). These reporter genes were selected either because of their distinctive fluorescent properties (Venus and mCherry) or because of their small size, stability, high bioluminescence activity, and ATP independence (Nluc) (47).
Although reporter-expressing rSARS-CoV-2 viruses similar to those reported here were recently described (18–20), this is the first report of a replication-competent rSARS-CoV-2 expressing mCherry. Recombinant viruses expressing a red fluorescent protein represent an advantage over those expressing GFP or mNeonGreen (18–20) in that many genetically modified cell lines and/or animals express green fluorescent proteins. Another limitation of green fluorescent proteins during in vivo imaging is the absorption of the fluorophores’ excitation and emission by hemoglobin and autofluorescence of tissues (48–51). Recombinant viruses expressing red fluorescent proteins represent a better option to combine with genetically modified GFP-expressing cell lines and/or animals and, based on their reduced autofluorescence background, to more accurately capture the dynamics of viral infection and replication.
Reporter-expressing replicating competent viruses can be used to monitor viral infections, assess viral fitness, and evaluate and/or identify antivirals and/or NAbs, where reporter gene expression can be used as a valid surrogate for virus detection in infected cells. Expression of Venus, mCherry, or Nluc from our rSARS-CoV-2 was confirmed by directly visualizing fluorescence expression under a fluorescence microscope (Venus and mCherry) or luciferase activity (Nluc) using a microplate reader (Fig. 2 and 3). Western blot analyses using specific antibodies against each of the reporter genes further confirmed expression from their respective rSARS-CoV-2 constructs (Fig. 2 and 3). Notably, despite deletion of ORF 7a and insertion of a reporter gene, the three reporter-expressing rSARS-CoV-2 constructs displayed growth kinetics and plaque phenotype similar to those of their WT counterpart (Fig. 3). As expected, viral infection was visualized in real time, without the need for secondary approaches (e.g., MAbs) to detect the presence of the virus in infected cells. Overall, reporter gene expression displayed kinetics that correlated with levels of viral replication, further demonstrating the feasibility of using these reporter genes as a valid surrogate to assess viral infection.
Therapeutic treatment of SARS-CoV-2 infection has been limited to the use of remdesivir (3) and the MAb bamlanivimab and, more recently, to two FDA-approved vaccines (4, 5). Notably, there is a possibility, similar to the situation with other respiratory viruses (e.g., influenza virus), of the emergence of drug-resistant SARS-CoV-2 variants that will impose a significant additional challenge in the ongoing COVID-19 pandemic (52). Thus, it is imperative to discover not only new antivirals and other therapeutic approaches but also prophylactics for the treatment of SARS-CoV-2 infections. To that end, rapid and sensitive screening assays to identify compounds with antiviral activity or to assess efficacy of vaccine candidates for the therapeutic and prophylactic treatment of SARS-CoV-2 infections, respectively, are urgently needed. In this study, we demonstrate that reporter-expressing rSARS-CoV-2 constructs represent an excellent option for the rapid identification and characterization of both antivirals (Fig. 4) and NAbs (Fig. 5) for the therapeutic and/or prophylactic treatment of SARS-CoV-2 infections. Importantly, EC50s (antivirals) and NT50s (NAbs) obtained with our reporter-expressing viruses were comparable to those obtained using rSARS-CoV-2/WT or described by others (20, 23, 27), demonstrating the feasibility of using our reporter-based microneutralization assays for the rapid identification of antivirals or NAbs (Fig. 4 and 5, respectively). Furthermore, our results indicate that Venus-, mCherry-, and Nluc-expressing rSARS-CoV-2 constructs are stable up to 5 passages in vitro in Vero E6 cells, including expression of the reporter gene (Fig. 6). We have not yet conducted studies to evaluate the feasibility of using these reporter-expressing rSARS-CoV-2 constructs in vivo. It is possible that, as with other respiratory viruses, rSARS-CoV-2 expressing reporter genes could also be used to study the biology of viral infections in validated animal models of viral infection.
Our SARS-CoV-2 reverse genetics based on the use of a BAC have allowed us to rescue rSARS-CoV-2/WT (21) and rSARS-CoV-2 stably expressing reporter genes. In the case of our reporter-expressing rSARS-CoV-2 constructs, we removed ORF 7a and replaced it with various reporter genes without a significant impact on viral replication. The feasibility of removing viral genes and inserting reporter genes demonstrates the genetic plasticity of the SARS-CoV-2 genome and opens the possibility of generating recombinant viruses expressing other genes of interest for the development of SARS-CoV-2 vaccines that could be used for the control of the ongoing COVID-19 pandemic.
MATERIALS AND METHODS
Biosafety.
All experiments involving infectious SARS-CoV-2 were performed in a biosafety level 3 (BSL3) laboratory at the Texas Biomedical Research Institute. Protocols involving SARS-CoV-2 were approved by the Texas Biomedical Research Institute’s Institutional Biosafety Committee (IBC).
Cell lines.
African green monkey kidney epithelial cells (Vero E6; CRL-1586) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% PSG (100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine) at 37°C with 5% CO2.
Generation of pBeloBAC11-SARS-CoV-2 carrying reporter genes.
The plasmid pBeloBAC11 (New England Biolabs) containing the entire viral genome of SARS-CoV-2 was described previously (21, 53). Briefly, the entire genome sequence of SARS-CoV-2 USA/WA1/2020 (GenBank accession no. MN985325) was chemically synthesized (Bio Basic) in five fragments and cloned into pUC57 plasmids containing unique restriction sites. Silent mutations were introduced to the spike (S) and matrix (M) genes to remove BstBI and Mlul restriction sites, respectively, that were used for the assembly of the entire SARS-CoV-2 genome into the pBeloBAC11 plasmid. These nucleotide changes were also used as genetic markers to distinguish the natural USA/WA1/2020 and the recombinant SARS-CoV-2 (21). The five fragments containing the entire SARS-CoV-2 genome were assembled into pBeloBAC11 using standard molecular biology techniques. To remove the 7a gene and introduce the Venus, mCherry, or Nluc reporter genes, the region flanking the 7a viral gene and each individual reporter gene were amplified by extension and overlapping PCR using specific oligonucleotides in a shuttle plasmid. The modified 7a viral genes were inserted into the pBeloBAC11 plasmid containing the remaining SARS-CoV-2 viral genome using BamHI and RsrII restriction sites to generate pBeloBAC11-SARS-CoV-2-del7a/Venus, pBeloBAC11-SARS-CoV-2-del7a/mCherry, and pBeloBAC11-SARS-CoV-2-del7a/Nluc for the rescue of rSARS-CoV-2-Venus, rSARS-CoV-2-mCherry, and rSARS-CoV-2-Nluc, respectively. Plasmids and pBeloBAC11 constructs were validated by Sanger sequencing (ACGT Inc.).
Rescue of rSARS-CoV-2 expressing reporter genes.
The rSARS-CoV-2/WT and rSARS-CoV-2 expressing reporter genes were rescued as previously described (21, 22). Briefly, confluent monolayers of Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were transfected using Lipofectamine 2000 (LPF2000; Thermo Fisher) with 4 μg/well of pBeloBAC11-SARS-CoV-2/WT or pBeloBAC11-SARS-CoV-2-del7a/Venus, -del7a/mCherry, or -del7a/Nluc. An empty pBeloBAC11 plasmid was included as an internal control. At 14 h, transfection medium was replaced with postinfection medium (DMEM with 2% FBS), and 24 h later, cells were scaled up into T75 flasks. At 72 h, P0 virus-containing tissue culture supernatants were collected and stored at −80°C. Viral rescues were confirmed by infecting fresh Vero E6 cells (1.2 × 106 cells/well, 6-well plates, triplicates) and assessing fluorescence or Nluc expression. P0 viruses were passaged three times, and viral stocks were generated and titrated for in vitro experiments. Viral titers (in PFU per milliliter) were determined by plaque assay in Vero E6 cells (1.2 × 106 cells/well, 6-well plate format).
Sequencing.
Viral RNAs from Vero E6 cells (1.2 × 106 cells/well, 6-well plate format) infected at a multiplicity of infection (MOI) of 0.01 were extracted using TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s specifications. Libraries were generated with a KAPA RNA HyperPrep kit, 100 ng of RNA, and 7 mM adapter. The Illumina HiSeq X was used for sequencing. Raw reads were filtered using Trimmomatic v0.39 (54). SARS-CoV-2 templates were made for each reporter gene by modifying SARS-CoV-2 USA/WA1/2020 (GenBank accession no. MN985325.1). Modifications included deleting ORF 7a, adding T21895C and T26843A mutations, and inserting the appropriate reporter gene (Venus, mCherry, or Nluc) at position 27937. Reads were mapped to the modified SARS-CoV-2 templates with Bowtie v2.4.1 (55), and the total genomic coverage was quantified using MosDepth v0.2.6 (56). Allele frequencies were estimated with LoFreq* v2.1.3.1 (57), and low-frequency variants with less than a 100× read depth or a 1% minor allele frequency were eliminated.
RT-PCR.
Total RNA from mock- or virus-infected (MOI of 0.01) Vero E6 cells (1.2 × 106 cells/well, 6-well plate format) were extracted using TRIzol reagent (Thermo Fisher Scientific). Superscript II reverse transcriptase (Invitrogen) and Expand high-fidelity PCR (Sigma-Aldrich) were used to synthesize and amplify the cDNAs, respectively, using primers specific for the viral nucleoprotein (NP) or ORF 7a region and Venus, mCherry, or Nluc.
Immunofluorescence assays (IFA).
Confluent monolayers of Vero E6 cells (1.2 × 106 cells/well, 6-well format, triplicates) were mock infected or infected (MOI of 0.01) with rSARS-CoV-2 expressing Venus or mCherry or rSARS-CoV-2/WT. At 48 h postinfection, cells were fixed with 10% neutral buffered formalin at 4°C for 16 h for fixation and viral inactivation and permeabilized with phosphate-buffered saline (PBS) containing 0.5% (vol/vol) Triton X-100 for 5 min at room temperature. Cells were washed with PBS and blocked with 2.5% bovine albumin serum (BSA) in PBS for 1 h before incubation with 1 μg/ml of SARS-CoV anti-NP MAb 1C7 in 1% BSA in PBS for 1 h at 37°C. Cells infected with rSARS-CoV-2-Venus or -mCherry were washed with PBS and stained with either Alexa Fluor 594 goat anti-mouse IgG (Invitrogen; 1:1,000) or fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Dako; 1:200), respectively. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Research Organics). Representative images were captured using a fluorescence microscope (EVOS M5000 imaging system) at a magnification of ×20.
Protein gel electrophoresis and Western blots.
Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were mock-infected or infected (MOI of 0.01) with rSARS-CoV-2/WT or rSARS-CoV-2 expressing Venus, mCherry, or Nluc. At 48 h postinfection, cells were lysed with 1× passive lysis buffer (Promega), and proteins were separated by denaturing electrophoresis in 12% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (Bio-Rad) with a Mini-Protean Tetra vertical electrophoresis cell at 100 V for 1 h at 4°C. Membranes were blocked in PBS containing 10% dried skim milk and 0.1% Tween 20 for 1 h and then incubated overnight at 4°C with the following primary MAbs or polyclonal antibodies (PAbs): SARS-CoV NP (mouse MAb 1C7; Thomas Moran, Icahn School of Medicine at Mount Sinai), Venus (rabbit PAb sc-8334; Santa Cruz Biotechnology), mCherry (rabbit PAb; RayBiotech), and Nluc (rabbit PAb; Promega). A MAb against actin (MAb AC-15; Sigma) was included as a loading control. Primary antibodies bound to the membrane were detected using horseradish peroxidase (HRP)-conjugated secondary antibodies against mouse or rabbit (GE Healthcare). Proteins were detected by chemiluminescence using SuperSignal West Femto Maximum Sensitivity substrate (Thermo Scientific) based on the manufacturer’s specifications and imaged in a ChemiDoc imaging system (Bio-Rad).
Plaque assays and immunostaining.
Confluent monolayers of Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were infected with WT or reporter-expressing rSARS-CoV-2 for 1 h at 37°C. After viral absorption, infected cells were overlaid with agar and incubated at 37°C for 72 h. Afterwards, cells were submerged in 10% neutral buffered formalin at 4°C for 16 h for fixation and viral inactivation, and then the agar overlays were gently removed. To observe Venus and mCherry fluorescence expression, PBS was added to each well and plates were imaged under a fluorescence microscope (EVOS M5000 imaging system). For immunostaining, plates were permeabilized with 0.5% Triton X-100–PBS for 10 min at room temperature, blocked with 2.5% BSA–PBS for 1 h at room temperature, and then incubated at 37°C for 1 h with the anti-SARS 2 NP MAb 1C7. Plaques were developed for visualization using the Vectastain ABC kit and DAB HRP substrate kit (Vector Laboratories), in accordance with the manufacturer’s recommendations.
Viral growth kinetics and titrations.
Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were infected (MOI of 0.01) with rSARS-CoV-2/WT or rSARS-CoV-2 expressing Venus, mCherry, or Nluc. After viral adsorption for 1 h at 37°C, cells were washed with PBS, provided with fresh postinfection medium, and then placed in a 37°C incubator with a 5% CO2 atmosphere. At various times postinfection (12, 24, 48, 72, and 96 h), cells were imaged for Venus or mCherry expression under a fluorescence microscope (EVOS M5000 imaging system). Viral titers in the tissue culture supernatants at each time point were determined by titration and immunostaining, as previously described, using the anti-SARS-CoV NP MAb 1C7. Nluc expression in tissue culture supernatants was quantified using Nano-Glo luciferase substrate (Promega) following the manufacturer’s recommendations. Mean values and standard deviations (SD) were determined using GraphPad Prism software (version 8.2).
Reporter-based microneutralization assay for the identification of antivirals.
Vero E6 cells (96-well plate format, 4 × 104 cells/well, quadruplicates) were infected with ∼100 to 200 PFU of rSARS-CoV-2/WT or rSARS-CoV-2 expressing Venus, mCherry, or Nluc for 1 h at 37°C. After viral adsorption, cells were washed and incubated in 100 μl of infection medium (DMEM with 2% FBS) containing 3-fold serial dilutions (starting concentration of 50 μM) of remdesivir or 0.1% dimethyl sulfoxide (DMSO) as a vehicle control and 1% Avicel (Sigma-Aldrich). Cells infected with rSARS-CoV-2/WT or rSARS-CoV-2 expressing fluorescent Venus or mCherry were incubated at 37°C for 24 h, while cells infected with rSARS-CoV-2 expressing Nluc were incubated at 37°C for 48 h. For rSARS-CoV-2/WT and rSARS-CoV-2 expressing fluorescent Venus and mCherry, cells were submerged in 10% neutral buffered formalin at 4°C for 16 h for fixation and viral inactivation. Cells were washed with 100 μl/well of PBS three times, permeabilized with 100 μl/well of 0.5% Triton X-100 in PBS at room temperature for 15 min and blocked with 100 μl/well of 2.5% BSA in PBS at 37°C for 1 h. Next, cells were staining with the anti-NP MAb 1C7 (1 μg/ml) in 1% BSA–PBS at 37°C for 1 h. After incubation with the primary MAb, cells were washed with PBS three times, and a secondary FITC-conjugated goat anti-mouse IgG (Dako; 1:200) in 1% BSA was added to cells for 1 h at 37°C. Cell nuclei were stained with DAPI (Research Organics). Viral infections were determined using fluorescent images of each well and quantified using Cell Profiler (Broad Institute) cell image analysis software. In the case of cells infected with rSARS-CoV-2 expressing Nluc, tissue culture supernatants were collected at 48 h postinfection, and Nluc expression was measured using a luciferase assay and a Synergy LX microplate reader (BioTek).
Reporter-based microneutralization assay for the identification of NAbs.
To test the neutralizing activity of 1212C2, a human MAb recently described to neutralize SARS-CoV-2 (27), confluent monolayers of Vero E6 cells (96-plate format, 4 × 104 cells/well, quadruplicates) were infected with ∼100 to 200 PFU of rSARS-CoV-2/WT or rSARS-CoV-2 expressing Venus, mCherry, or Nluc for 1 h at 37°C. After viral adsorption, cells were washed and incubated with 100 μl of infection medium (DMEM with 2% FBS) containing 3-fold serial dilutions (starting concentration of 500 ng) of 1212C2 or PBS and 1% Avicel (Sigma-Aldrich). Infected cells were incubated at 37°C for 24 h for rSARS-CoV-2/WT and rSARS-CoV expressing Venus or mCherry and for 48 h for rSARS-CoV-2 expressing Nluc. After viral infections, cells infected with rSARS-CoV-2/WT and rSARS-CoV-2 expressing fluorescent Venus and mCherry were submerged in 10% neutral buffered formalin at 4°C for 16 h for fixation and viral inactivation. Cells were washed with 100 μl/well of PBS three times and permeabilized with 100 μl/well of 0.5% Triton X-100 in PBS at room temperature for 15 min. Then, cells were blocked with 100 μl/well of 2.5% BSA in PBS at 37°C for 1 h. Cells were next incubated with the anti-NP MAb 1C7 (1 μg/ml) in 1% BSA–PBS at 37°C for 1 h. Cells were next washed three times with PBS and incubated with a secondary FITC-conjugated goat anti-mouse IgG (Dako; 1:200) in 1% BSA for 1 h at 37°C. Cell nuclei were stained with DAPI (Research Organics). Viral infections were determined using fluorescent images of each well and quantified using Cell Profiler (Broad Institute). In the case of cells infected with rSARS-CoV-2 expressing Nluc, tissue culture supernatants were collected at 48 h postinfection, and Nluc expression was measured using a luciferase assay and a Synergy LX microplate reader (BioTek).
Genetic stability.
Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were infected (MOI of 0.01) with rSARS-CoV-2-Venus or -mCherry P3 stocks and after 1 h viral adsorption, virus inoculum was replaced with infection medium (DMEM with 2% FBS). The cells were incubated at 37°C with 5% CO2 until 70% cytopathic effect (CPE) was observed. Then, tissue culture supernatants were collected and diluted 100-fold in infection medium and used to infect fresh Vero E6 cells (1.2 × 106 cells/well, 6-well format, triplicates) for two additional passages (P5). Venus- and mCherry-expressing plaques (∼50 counted plaques per viral passage) were evaluated by immunostaining and fluorescent-protein expression. Viral plaques were imaged under a fluorescence microscope (EVOS M5000 imaging system) at a magnification of ×4.
Statistical analysis.
Data analysis was conducted using Prism software version 9.0.0 (GraphPad Software, CA, USA). Individual wells from three independent experiments conducted in quadruplicate were used to calculate the average and standard deviation (SD) of viral inhibition using Microsoft Excel software. Nonlinear regression curves, the half-maximal effective concentration (EC50) of remdesivir, and the half-maximal neutralizing concentration (NT50) of 1212C2 were determined using GraphPad Prism software.
Data availability.
All sequence data have been deposited in the NCBI Short Read Archive (BioProject ID PRJNA678001).
ACKNOWLEDGMENTS
We thank Thomas Moran at the Icahn School of Medicine at Mount Sinai for providing us with the SARS-CoV cross-reactive NP MAb 1C7. We also thank BEI Resources for providing the SARS-CoV-2 USA-WA1/2020 isolate (NR-52281) and Marina McDew-White and Robbie Diaz for constructing the NGS libraries. Finally, we also thank members at our institutes for their efforts in keeping them fully operational during the COVID-19 pandemic and the BSC and IACUC for reviewing our protocols in a time-efficient manner.
We dedicate this paper to all COVID-19 victims and to all heroes battling this disease.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh MD, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group Members. 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group. 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, McClung N, Campos-Outcalt D, Morgan RL, Mbaeyi S, Romero JR, Talbot HK, Lee GM, Bell BP, Dooling K. 2021. The Advisory Committee on Immunization Practices' interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep 69:1653–1656. doi: 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig S, Zarbock A. 2020. Coronaviruses and SARS-CoV-2: a brief overview. Anesth Analg 131:93–96. doi: 10.1213/ANE.0000000000004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, Group SW, SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 10.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoye E. 2020. The pandemic in pictures: how coronavirus is changing the world. Nature doi: 10.1038/d41586-020-01048-7. [DOI] [PubMed] [Google Scholar]
- 12.Qian X, Ren R, Wang YF, Guo Y, Fang J, Wu ZD, Liu PL, Han TR, Members of Steering Committee, Society of Global Health, Chinese Preventive Medicine Association. 2020. Fighting against the common enemy of COVID-19: a practice of building a community with a shared future for mankind. Infect Dis Poverty 9:34. doi: 10.1186/s40249-020-00650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vellingiri B, Jayaramayya K, Iyer M, Narayanasamy A, Govindasamy V, Giridharan B, Ganesan S, Venugopal A, Venkatesan D, Ganesan H, Rajagopalan K, Rahman PKSM, Cho SG, Kumar NS, Subramaniam MD. 2020. COVID-19: a promising cure for the global panic. Sci Total Environ 725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones L, Walsh K, Willcox M, Morgan P, Nichols J. 2020. The COVID-19 pandemic: important considerations for contact lens practitioners. Cont Lens Anterior Eye 43:196–203. doi: 10.1016/j.clae.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, Sagnelli C, Bianchi M, Bernardini S, Ciccozzi M. 2019. COVID-19 outbreak: an overview. Chemotherapy 64:215–223. doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogales A, Rodriguez-Sanchez I, Monte K, Lenschow DJ, Perez DR, Martinez-Sobrido L. 2016. Replication-competent fluorescent-expressing influenza B virus. Virus Res 213:69–81. doi: 10.1016/j.virusres.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogales A, Avila-Perez G, Rangel-Moreno J, Chiem K, DeDiego ML, Martinez-Sobrido L. 2019. A novel fluorescent and bioluminescent bireporter influenza A virus to evaluate viral infections. J Virol 93:e00032-19. doi: 10.1128/JVI.00032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, 3rd, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schafer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446.E14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi PY. 2020. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27:841–848.E3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Muruato AE, Zhang X, Lokugamage KG, Fontes-Garfias CR, Zou J, Liu J, Ren P, Balakrishnan M, Cihlar T, Tseng C-TK, Makino S, Menachery VD, Bilello JP, Shi P-Y. 2020. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun 11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye C, Chiem K, Park JG, Oladunni F, Platt RN, 2nd, Anderson T, Almazan F, de la Torre JC, Martinez-Sobrido L. 2020. Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio 11:e02168-20. doi: 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avila-Perez G, Park JG, Nogales A, Almazan F, Martinez-Sobrido L. 2019. Rescue of recombinant Zika virus from a bacterial artificial chromosome cDNA clone. J Vis Exp 2019:e59537. doi: 10.3791/59537. [DOI] [PubMed] [Google Scholar]
- 23.Park JG, Oladunni FS, Chiem K, Ye C, Pipenbrink M, Moran T, Walter MR, Kobie J, Martinez-Sobrido L. 2020. Rapid in vitro assays for screening neutralizing antibodies and antivirals against SARS-CoV-2. J Virol Methods 287:113995. doi: 10.1016/j.jviromet.2020.113995:113995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan K, Huang C, Makino S. 2008. SARS coronavirus accessory proteins. Virus Res 133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piepenbrink MS, Park J-G, Oladunni FS, Deshpande A, Basu M, Sarkar S, Loos A, Woo J, Lovalenti P, Sloan D, Ye C, Chiem K, Erdmann NB, Goepfert PA, Truong VL, Walter MR, Martinez-Sobrido L, Kobie JJ. 2020. Therapeutic activity of an inhaled potent SARS-CoV-2 neutralizing human monoclonal antibody in hamsters. bioRxiv doi: 10.1101/2020.10.14.339150:2020.10.14.339150. [DOI] [PMC free article] [PubMed]
- 28.Li X, Zhang H, Zhang Y, Li J, Wang Z, Deng C, Jardim ACG, Terzian ACB, Nogueira ML, Zhang B. 2019. Development of a rapid antiviral screening assay based on eGFP reporter virus of Mayaro virus. Antiviral Res 168:82–90. doi: 10.1016/j.antiviral.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Kirui J, Freed EO. 2020. Generation and validation of a highly sensitive bioluminescent HIV-1 reporter vector that simplifies measurement of virus release. Retrovirology 17:12. doi: 10.1186/s12977-020-00521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang B, Deng C, Ye H, Xu W, Yuan Z, Shi PY, Zhang B. 2013. Development and characterization of a stable eGFP enterovirus 71 for antiviral screening. Antiviral Res 97:198–205. doi: 10.1016/j.antiviral.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Zou G, Xu HY, Qing M, Wang QY, Shi PY. 2011. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res 91:11–19. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. 2012. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J Virol 86:1433–1448. doi: 10.1128/JVI.05820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breen M, Nogales A, Baker SF, Martínez-Sobrido L. 2016. Replication-competent influenza A viruses expressing reporter genes. Viruses 8:179. doi: 10.3390/v8070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breen M, Nogales A, Baker SF, Perez DR, Martinez-Sobrido L. 2016. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS One 11:e0147723. doi: 10.1371/journal.pone.0147723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czako R, Vogel L, Lamirande EW, Bock KW, Moore IN, Ellebedy AH, Ahmed R, Mehle A, Subbarao K. 2017. In vivo imaging of influenza virus infection in immunized mice. mBio 8:e00714-17. doi: 10.1128/mBio.00714-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckert N, Wrensch F, Gartner S, Palanisamy N, Goedecke U, Jager N, Pohlmann S, Winkler M. 2014. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One 9:e97695. doi: 10.1371/journal.pone.0097695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuyama S, Katsura H, Zhao D, Ozawa M, Ando T, Shoemaker JE, Ishikawa I, Yamada S, Neumann G, Watanabe S, Kitano H, Kawaoka Y. 2015. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat Commun 6:6600. doi: 10.1038/ncomms7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P. 2013. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol 87:8272–8281. doi: 10.1128/JVI.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson EA, Meliopoulos VA, Savage C, Livingston B, Mehle A, Schultz-Cherry S. 2015. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat Commun 6:6378. doi: 10.1038/ncomms7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, Garcia-Sastre A. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J Virol 84:2157–2163. doi: 10.1128/JVI.01433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogales A, Baker SF, Martinez-Sobrido L. 2015. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology 476:206–216. doi: 10.1016/j.virol.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez JT, Garcia-Sastre A, Manicassamy B. 2013. Insertion of a GFP reporter gene in influenza virus. Curr Protoc Microbiol Chapter 15:Unit 15G 4. doi: 10.1002/9780471729259.mc15g04s29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuther P, Gopfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346. doi: 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran V, Poole DS, Jeffery JJ, Sheahan TP, Creech D, Yevtodiyenko A, Peat AJ, Francis KP, You S, Mehle A. 2015. Multi-modal imaging with a toolbox of influenza A reporter viruses. Viruses 7:5319–5327. doi: 10.3390/v7102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsh DK, Noguchi T. 2012. Cellular bioluminescence imaging. Cold Spring Harb Protoc 2012:pdb.top070607. doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- 47.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luker KE, Pata P, Shemiakina II, Pereverzeva A, Stacer AC, Shcherbo DS, Pletnev VZ, Skolnaja M, Lukyanov KA, Luker GD, Pata I, Chudakov DM. 2015. Comparative study reveals better far-red fluorescent protein for whole body imaging. Sci Rep 5:10332. doi: 10.1038/srep10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. 2007. Bright far-red fluorescent protein for whole-body imaging. Nat Methods 4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 50.Yang M, Baranov E, Jiang P, Sun FX, Li XM, Li L, Hasegawa S, Bouvet M, Al-Tuwaijri M, Chishima T, Shimada H, Moossa AR, Penman S, Hoffman RM. 2000. Whole-body optical imaging of green fluorescent protein-expressing tumors and metastases. Proc Natl Acad Sci U S A 97:1206–1211. doi: 10.1073/pnas.97.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam JM, Upadhyay R, Pittet MJ, Weissleder R, Mahmood U. 2007. Improved in vivo whole-animal detection limits of green fluorescent protein-expressing tumor lines by spectral fluorescence imaging. Mol Imaging 6:269–276. doi: 10.2310/7290.2007.00023. [DOI] [PubMed] [Google Scholar]
- 52.Lo MK, Albariño CG, Perry JK, Chang S, Tchesnokov EP, Guerrero L, Chakrabarti A, Shrivastava-Ranjan P, Chatterjee P, McMullan LK, Martin R, Jordan R, Götte M, Montgomery JM, Nichol ST, Flint M, Porter D, Spiropoulou CF. 2020. Remdesivir targets a structurally analogous region of the Ebola virus and SARS-CoV-2 polymerases. Proc Natl Acad Sci U S A 117:26946–26954. doi: 10.1073/pnas.2012294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiem K, Ye C, Martinez-Sobrido L. 2020. Generation of recombinant SARS-CoV-2 using a bacterial artificial chromosome. Curr Protoc Microbiol 59:e126. doi: 10.1002/cpmc.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data have been deposited in the NCBI Short Read Archive (BioProject ID PRJNA678001).