We assessed the performance, stability, and user acceptability of swab-independent self-collected saliva and saline mouth rinse/gargle sample types for the molecular detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in adults and school-aged children. Outpatients who had recently been diagnosed with COVID-19 or were presenting with suspected COVID-19 were asked to have a nasopharyngeal (NP) swab collected and provide at least one self-collected sample type.
KEYWORDS: COVID-19, SARS-CoV-2, gargle, saliva, PCR
ABSTRACT
We assessed the performance, stability, and user acceptability of swab-independent self-collected saliva and saline mouth rinse/gargle sample types for the molecular detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in adults and school-aged children. Outpatients who had recently been diagnosed with COVID-19 or were presenting with suspected COVID-19 were asked to have a nasopharyngeal (NP) swab collected and provide at least one self-collected sample type. Participants were also asked about sample acceptability using a five-point Likert scale. For those previously diagnosed with COVID-19, all samples underwent real-time PCR testing using a lab-developed assay, and the majority were also tested using an FDA-authorized assay. For those presenting with suspected COVID-19, only those with a positive nasopharyngeal swab sample went on to have other samples tested. Saline mouth rinse/gargle and saliva samples were tested daily at time zero, day 1, and day 2 to assess nucleic acid stability at room temperature. Fifty participants (aged 4 to 71 years) were included; of these, 40 had at least one positive sample and were included in the primary sample yield analysis. Saline mouth rinse/gargle samples had a sensitivity of 98% (39/40), while saliva samples had a sensitivity of 79% (26/33). Both saline mouth rinse/gargle and saliva samples showed stable viral RNA detection after 2 days of room temperature storage. Mouth rinse/gargle samples had the highest (mean, 4.9) and health care worker (HCW)-collected NP swabs had the lowest acceptability scores (mean, 3.1). In conclusion, saline mouth rinse/gargle samples demonstrated higher combined user acceptability ratings and analytical performance than saliva and HCW-collected NP swabs. This sample type is a promising swab-independent option, particularly for outpatient self-collection in adults and school-aged children.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic that has shown the potential to overwhelm the capacity of local and national health care systems (1). Rapid scale-up of molecular testing for SARS-CoV-2 has occurred on an unprecedented scale and has presented numerous challenges, including deployment of assays with variable performance, lack of availability of high-throughput testing platforms, and bottlenecks in supply chain procurement of preanalytical materials and testing reagents (2). One of the key weaknesses in the diagnostic cycle has been the challenge of sample acquisition. While nasopharyngeal flocked swab (NPFS) collection has been the gold standard for diagnosis of active SARS-CoV-2 infections using nucleic acid amplification test (NAT) methods (3), such collection is difficult due to the associated discomfort and is resource intensive, requiring health care workers (HCWs), and procurement of the collection devices has proved to be a challenge in many jurisdictions. This quality initiative was carried out in order to meet anticipated increases in outpatient testing demands, the expected challenges associated with the global NPFS supply chain, the difficult nature of NPFS sample collection, and complex handling of highly viscose saliva samples in the laboratory. We compared the analytical performance, sample stability, and user acceptability of saline gargle and saliva samples against health care provider-collected NPFSs from outpatients with COVID-19 infections.
MATERIALS AND METHODS
Individuals from the Vancouver Greater Metropolitan Area of British Columbia were approached for participation in this quality improvement (QI) initiative if they had SARS-CoV-2 detected in any clinical sample collected at an outpatient testing center or if they were identified as a symptomatic household contact of a confirmed COVID-19 case. After 28 August 2020, symptomatic children 4 to 12 years of age presenting to the BC Children’s and Women’s Hospital Campus COVID-19 Collection Centre were also asked to provide a saline mouth rinse/gargle sample in addition to an NPFS. Verbal consent was obtained from all participants, and for those who provided both saliva and saline mouth rinse/gargle samples, the order of sample collection was alternated sequentially (i.e., saliva first versus mouth rinse/gargle sample first). Demographic and clinical information was collected, including age, whether they were a health care worker (HCW), if they had been previously admitted to hospital, the date of onset of their symptoms, and the date of initial molecular diagnosis of COVID-19 (if applicable). Sample collection occurred either at the participant’s residence or at the BC Children’s and Women’s Hospital Campus COVID-19 Collection Centre, and instruction sheets (see the supplemental material) with supporting verbal instructions were provided. Participants were asked to not eat, drink, smoke, brush their teeth, or chew gum 1 h prior to collection.
Self-collection procedures.
Nasopharyngeal flocked swabs (flexible flocked swabs with 3 ml of universal viral transport system medium; Beckon Dickinson, Sparks, MD) were collected via the left naris unless preference was stated for the right naris. NPFSs were inserted the distance from the naris to the external ear canal and then rotated 5 times and left in place for 5 to 10 s prior to being removed as per Infectious Diseases Society of America-recommended instructions for collection (4). Mouth rinse/gargle specimens were self-collected by instructing users to open 5-ml vials of sterile 0.9% saline (AddiPak; Teleflex Medical, Research Triangle Park, NC) and squeeze the contents into their or their child’s open mouth. They were then asked to swish the contents for 5 s followed by tilting their heads back and gargling for 5 s. This swish/gargle cycle was repeated 2 more times and then the saline was expelled into a wide-mouthed sterile empty polypropylene container (Leakbuster 90-ml container; Starplex Scientific, Etobicoke, ON, Canada). For saliva sample collection, participants were asked to pool and spit saliva repeatedly into a wide-mouthed sterile empty polypropylene container (Leakbuster 90-ml container, Starplex Scientific) until at least 5 to 10 ml was collected if possible. All study samples were collected at the same time. Samples were immediately brought to the laboratory and were processed within 12 h of collection. All samples were vortexed for at least 10 s prior to aliquoting for testing.
Laboratory testing.
All samples were first extracted with the QiaSymphony automated extractor using the DSP virus/pathogen minikit (Qiagen, Germantown, MD) and subsequently tested with a laboratory-developed test (LDT) within 24 h of collection on the Applied Biosystems 7500 fast real-time PCR system (Life Technologies, Carlsbad, CA). Saliva samples that were deemed to have too much mucus by the processing technologist were diluted 1:1 in Tris-EDTA (TE) buffer. Mouth rinse/gargle and saliva samples were also stored in the laboratory at room temperature (21°C) and extracted and tested again on day 1 and day 2 after collection. The previously validated LDT is a triplex reverse transcription-PCR (RT-PCR) assay that targets both the pan-sarbecovirus E gene and the RNA-dependent RNA polymerase (RdRp) gene, as well as the human RNase P gene as a sample positive control, and was previously determined to have a limit of detection of 3.901 log copies/ml (95% confidence interval [CI], 3.048 to 4.755) (5). A portion of the NPFS and mouth rinse gargle specimens was also tested using the Cepheid Xpert Xpress SARS-CoV-2 assay on the GeneXpert system (Cepheid, Sunnyvale, CA) as per manufacturer instructions.
User acceptability of sampling.
Participants who had all three samples collected were asked about the acceptability of each self-collected sample type they had obtained as well as about the HCW-collected NPFS using a 5-point Likert scale with 1 being “least acceptable” and 5 being “most acceptable.”
This project was reviewed by the BC Children’s and Women’s Research Ethics Board and deemed a QI/quality assurance (QA) activity.
Statistical analysis.
For the purpose of the analysis of sensitivity for each sample type, individuals were considered positive cases if they had at least one sample test positive for both targets (E gene and RdRp gene) on the LDT. Samples were considered positive with a threshold cycle (CT) value of less than 40 on the LDT and as reported by the Xpert assay (presumptive positive results were considered positive). Using this reference standard for a SARS-CoV-2-positive participant, the sensitivity results of mouth rinse/gargle sampling was compared to those of NPFS and saliva sampling (LDT assay) using the test for comparing two independent proportions (STATA command prtesti); the McNemar exact test was also used to determine the comparability of mouth rinse/gargle sample testing to NPFS or saliva sample testing. The CT values obtained with baseline testing using all sample types were compared using repeated-measures analysis of variance (ANOVA) using Box’s conservative correction factor, with post hoc significant pairwise differences determined using Tukey’s honestly significant difference (HSD); this methodology was also used to compare acceptability ratings of all sample types. For comparison of baseline CT values for viral targets in matched samples, only samples in which the target was detected were included in the analysis. For assessment of stability of SARS-CoV-2 RNA between gargle samples and saliva samples (transport medium-free methods), individuals were included only if they had samples tested at each of those three time points (time zero, day 1, and day 2). Statistical significance was set as a P value of <0.05. All testing was done using STATA v16.1 (StataCorp, College Station, TX).
RESULTS
A total of 50 participants (34 known positives and 16 household contacts/symptomatic children) provided validation samples between 8 May 8 and 11 September 2020. All of the known positive patients had been initially diagnosed via testing with NPFS samples. Of all participants, 40 were found to be positive according to the reference definition at the time of study sample collection. There were 28 (56%) participants that were female, with a median age of 25.1 years (25th to 75th percentile, 13.6 to 35.9 years), and 10 were <18 years of age. Data on symptoms were available for 40 participants; of these, 17 (42%) had fever and 25 (62%) had cough. Among participants found to be positive, the median time from infection confirmation to retesting was 3 days (25th to 75th percentile, 2 to 5 days) and the median time from symptoms to retesting was 7 days (25th to 75th percentile, 4 to 9 days).
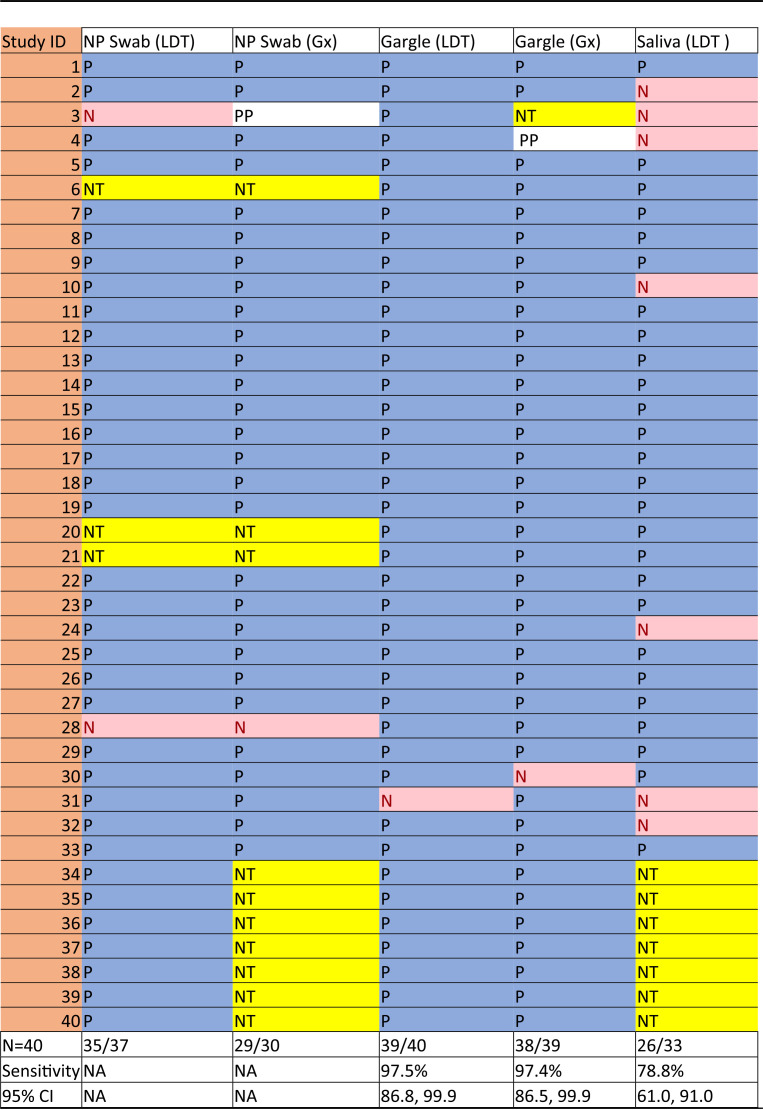
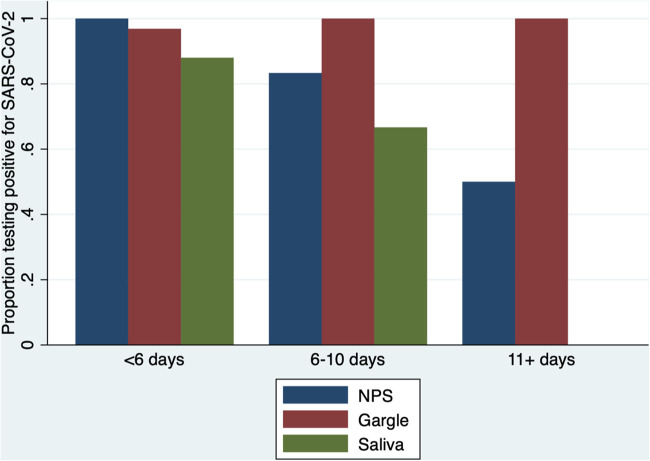
The results of testing for each of the sample types for all positive participants are shown in Table 1. Two of the 33 saliva samples required 1:1 dilution with TE buffer, and both of these tested positive. Mouth rinse/gargle samples were significantly more likely to be positive than saliva samples (difference of 18.7% [95% CI, 3.9 to 33.5%]; P = 0.01). When matched samples were compared, mouth rinse/gargle sample testing results were found to differ from saliva samples (26 positive by both, 6 rinse/gargle positive but saliva negative, and 1 negative by both; McNemar P = 0.03). Positivity proportions for NPFS, mouth rinse/gargle, and saliva sample types based on time from COVID-19 diagnosis are shown in Fig. 1.
TABLE 1.
Performance of sample types in 40 COVID-19-positive participantsa
NP, nasopharyngeal; LDT, laboratory-developed test; Gx, GeneXpert assay; NA, not applicable; P, positive test; N, negative test; PP, presumptive positive; NT, not tested; CI, confidence interval.
FIG 1.
Proportion of samples testing positive based on time from initial COVID-19 diagnosis. NPS, nasopharyngeal swab.
Participant-rated acceptability performance of sample types is shown in Table 2. There were clear differences between the acceptability of the sample types; the mouth rinse/gargle sample had the highest mean acceptability (4.95) and was significantly more acceptable than HCW-collected NPFS (mean acceptability, 3.17) or saliva sampling (mean acceptability, 4.44).
TABLE 2.
Acceptability of sample types as rated on a 5-point Likert scalea
| Sample | Mean acceptability | Acceptability difference | 95% CI of difference | Tukey P value for difference | |
|---|---|---|---|---|---|
| Mouth rinse/gargle | 4.95 | vs saliva | 0.50 | 0.0074–0.99 | 0.046 |
| vs NPFS | 1.78 | 1.28–2.27 | <0.001 | ||
| Saliva | 4.45 | vs NPFS | 1.28 | 0.78–1.77 | <0.001 |
| NPFS | 3.17 | ||||
On the Likert scale, 1 = lowest acceptability and 5 = highest acceptability. NPFS, nasopharyngeal flocked swab.
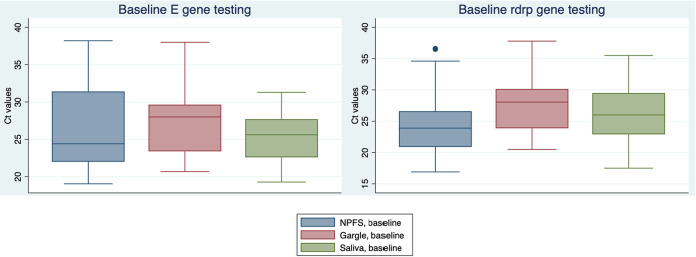
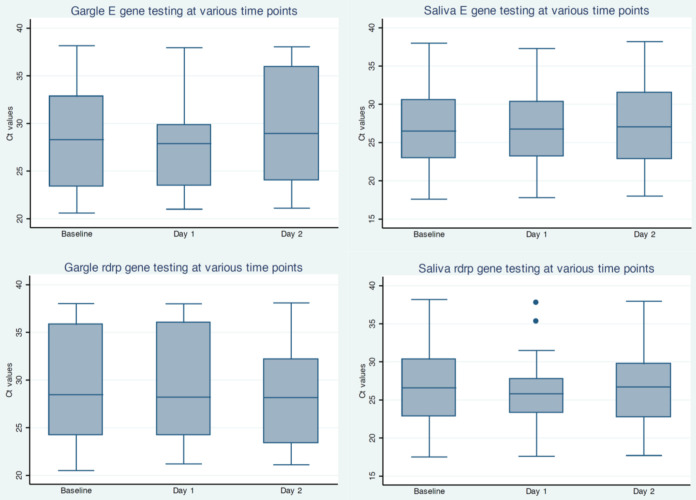
Mean CT values at baseline (day 0) were somewhat higher for saline gargle samples than for NPFS samples (for E gene, saline gargle sample mean CT value, +2.8 [P = 0.01] and for RdRp gene, saline gargle sample mean CT value, +3.6 [P < 0.001]); box plots of these data are shown in Fig. 2. Mean CT data at day zero, day 1, and day 2 for mouth rinse/gargle and saliva samples are shown in Fig. 3. There was no significant loss of RNA recovery over all time points. Also among positive participants with mouth rinse/gargle (n = 30) and saliva (n = 28) samples that were tested on all 3 days, there were similar numbers of negative results (for both targets) on each day (saliva, day 0 = 6, day 1 = 4, and day 2 = 3; mouth rinse/gargle, day 0 = 1, day 1 = 5, and day 2 = 2).
FIG 2.
Threshold cycle (CT) values across different sample types at baseline. NPFS, nasopharyngeal flocked swab.
FIG 3.
Stability of SARS-CoV-2 RNA detection. Shown are threshold cycle values at baseline, day 1, and day 2 for saline gargle and saliva samples with positive values.
DISCUSSION
Laboratory diagnostics are critical for any attempt to contain the COVID-19 pandemic. Our study demonstrates that self-collected mouth rinse/gargle samples are noninferior to HCW-collected NPFS for the detection of SARS-CoV-2 and, at the same time, are significantly more acceptable to patients. Mouth rinse/gargle specimens, in particular, demonstrated the highest sensitivity and were preferred by those undergoing sampling. These data should be considered by all those planning laboratory testing strategies and algorithms; self-collected mouth rinse/gargle samples may obviate the deployment of significant numbers of HCWs trained in sample collection and the consumption of large amounts of personal protective equipment and swabs. It should be emphasized that testing volumes in school-aged children have generally increased with return to in-person schooling, and nasopharyngeal sampling may be more challenging in this age group.
Saline mouth rinse/gargle samples (also known as mouth throat washes) have previously been evaluated for detection of influenza and other respiratory viruses (6, 7), showing promising performance compared to throat swab and other sample types. Interestingly, although there have been numerous evaluations of multiple self-collected sample types, including mid-turbinate swabs, throat swabs, and saliva samples, there have been very few reports to date describing mouth rinse/gargle samples for COVID-19 diagnosis (8, 9). In one study in Germany (10), 5 individuals were evaluated who had both saline gargle samples and throat swab samples, and all 5 gargle samples were positive, whereas only 4 individuals had a positive throat swab sample. The technique we used for acquisition of saline mouth rinse/gargle samples (three cycles for mouth rinse followed by gargle) may allow for improved recovery of viral RNA given that it would effectively sample the entire oropharynx.
Self-collected mouth rinse/gargle samples could also potentially result in a significant amount of savings due to the lack of need for personal protective equipment or trained HCWs for sample acquisition. Its utility might be even higher in low- and middle-income-country settings or remote regions where access to testing clinics would be another barrier to sample acquisition. The stability of RNA recovery in this sample type was preserved for at least 2 days at room temperature, making later drop-off delivery of samples a feasible option.
In our study, saliva samples were both significantly less sensitive and less acceptable than mouth rinse/gargle samples. We do note, however, that a sizable proportion of SARS-CoV-2-positive participants whose matching saliva samples were negative were sampled >5 days from initial diagnosis. Several recent reports have found favorable detection rates for saliva samples; however, most of these have been in inpatient populations (11), in whom viral load levels tend to be higher, and/or additional manual saliva processing steps were utilized (12) that are less amenable to high-throughput processing. In our evaluation, processing of saliva samples was intermittently observed to be impeded by variable sample viscosity; additional manual processing of two saliva samples was required due to excessive amounts of mucus present. For these reasons, among the swab- and transport medium-free options, the mouth rinse/gargle samples appeared more attractive.
Our study design had several strengths. In addition to having a number of participants across the pediatric and adult age ranges, we also enrolled solely individuals presenting with outpatient illness, in whom the largest burden of testing is performed. This is particularly the case for testing surges that are expected with a return to school for children in the fall. Our evaluation also includes the assessment of performance across multiple extraction and PCR platforms, including a Health Canada- and FDA-authorized commercial assay. A main weakness of the study is that a number of the patients enrolled did not have samples collected until >5 days into their illness, and therefore, we had a number of samples with smaller amounts of viral RNA detected. Also, as mentioned above, the population sampled included only relatively well outpatients, and it is unclear if similar performance would be found in those with more severe inpatient disease. Participants were also all initially identified as being positive through testing of NPFS samples, which would bias in favor of this sample type.
Given the very high user acceptability rating, lack of need for swabs and/or transport media, and excellent diagnostic yield, saline mouth rinse/gargle samples appear to be a promising sample type for testing of outpatients with suspected COVID-19.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participants who agreed to contribute to this validation project and also the nursing staff at the BC Children’s and Women’s Campus COVID-19 Collection Centre and the molecular microbiology technologists at the BC Children’s Hospital and the BC Centre for Disease Control for their contributions to this work.
No external funding was provided for this project.
We have no conflicts of interest to disclose.
REFERENCES
- 1.Cesari M, Montero-Odasso M. 2020. COVID-19 and older adults. Lessons learned from the Italian epicenter. Can Geriatr J 23:155–159. doi: 10.5770/cgj.23.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callahan CJ, Lee R, Zulauf KE, Tamburello L, Smith KP, Previtera J, Cheng A, Green A, Abdul Azim A, Yano A, Doraiswami N, Kirby JE, Arnaout RA. 2020. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. J Clin Microbiol 58:e00876-20. doi: 10.1128/JCM.00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Respiratory Virus Infections Working Group. 2020. Canadian Public Health Laboratory Network best practices for COVID-19. Can Commun Dis Rep 46:112–118. doi: 10.14745/ccdr.v46i05a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson KE, Caliendo AM, Arias CA, Englund JA, Lee MJ, Loeb M, Patel R, El Alayli A, Kalot MA, Falck-Ytter Y, Lavergne V, Morgan RL, Murad MH, Sultan S, Bhimraj A, Mustafa RA. 2020. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19. Clin Infect Dis 16:ciaa760. doi: 10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeBlanc JJ, Gubbay JB, Li Y, Needle R, Arneson SR, Marcino D, Charest H, Desnoyers G, Dust K, Fattouh R, Garceau R, German G, Hatchette TF, Kozak RA, Krajden M, Kuschak T, Lang ALS, Levett P, Mazzulli T, McDonald R, Mubareka S, Prystajecky N, Rutherford C, Smieja M, Yu Y, Zahariadis G, Zelyas N, Bastien N, COVID-19 Pandemic Diagnostics Investigation Team of the Canadian Public Health Laboratory Network (CPHLN) Respiratory Virus Working Group. 2020. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol 128:104433. doi: 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett S, Davidson RS, Gunson RN. 2017. Comparison of gargle samples and throat swab samples for the detection of respiratory pathogens. J Virol Methods 248:83–86. doi: 10.1016/j.jviromet.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Bennett S, MacLean A, Gunson R. 2019. Verification of Cepheid Xpert Xpress Flu/RSV assay for use with gargle samples, sputa and endotracheal secretions. J Hosp Infect 101:114–115. doi: 10.1016/j.jhin.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Saito M, Adachi E, Yamayoshi S, Koga M, Iwatsuki-Horimoto K, Kawaoka Y, Yotsuyanagi H. 2020. Gargle lavage as a safe and sensitive alternative to swab samples to diagnose COVID 19: a case report in Japan. Clin Infect Dis 71:893–894. doi: 10.1093/cid/ciaa377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal A, Gupta A, Kumar S, Surjit M, Singh B, Soneja M, Soni KD, Khan AR, Singh K, Naik S, Kumar A, Aggarwal R, Nischal N, Sinha S, Trikha A, Wig N. 2020. Gargle lavage as a viable alternative to swab for detection of SARS-CoV-2. Indian J Med Res 152:77–81. doi: 10.4103/ijmr.IJMR_2987_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malecki M, Lüsebrink J, Teves S, Wendel AF. 11 May 2020. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol doi: 10.1017/ice.2020.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman OE, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. 2020. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams E, Bond K, Zhang B, Putland M, Williamson DA. 2020. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 58:e00776-20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.