Bacterial persisters represent noninheritable drug-tolerant populations that are linked to recalcitrance of infections in health care settings. The rise of antibiotic resistance and the depletion of new antibiotics in the drug discovery pipeline have made the task of persister eradication more daunting.
KEYWORDS: drug tolerance, rifampicin, reactive oxygen species, sodB, drug combination
ABSTRACT
Bacterial persisters represent noninheritable drug-tolerant populations that are linked to recalcitrance of infections in health care settings. The rise of antibiotic resistance and the depletion of new antibiotics in the drug discovery pipeline have made the task of persister eradication more daunting. In the present study, we report that treatment of Acinetobacter baumannii with the last-resort antibiotic polymyxin B yields continuous variations in tolerance among different clinical isolates. Mechanistically, polymyxin B persisters exhibit disruption of proton motive force and delocalization of the cell division protein to attain a growth-arrested phenotype. Tolerance studies with mutant strains revealed that superoxide dismutase (sodB) activity is a major contributor to the tolerance of A. baumannii to polymyxin B. Using a dual fluorescence-based persister detection system, screening of various antibiotics to eradicate polymyxin B-induced A. baumannii persisters was performed. Rifampicin exhibited eradication of polymyxin B-tolerant populations by 6-log-unit reductions in magnitude with different clinical isolates of A. baumannii. We established that enhanced generation of reactive oxygen species (ROS) by rifampicin leads to clearance of these polymyxin B persisters. It was further demonstrated, as a proof of concept, that rifampicin potentiates the killing of polymyxin B persisters in a murine wound infection model. We found that the effects were linked to significant downregulation of sodB by rifampicin, which contributes to greater generation of ROS in polymyxin B-tolerant cells. In view of these results, we propose that the combination of polymyxin B and rifampicin is an effective antipersister strategy in clearing polymyxin B-induced A. baumannii persisters.
INTRODUCTION
Treatment of infectious diseases caused by bacterial pathogens with antibiotics is the hallmark achievement in modern medicine. However, by adopting various ingenious mechanisms, bacteria have been able to evade the action of antibiotics. This has been largely attributed to the emergence and spread of antibiotic resistance in pathogenic bacteria (1, 2). Recently, mechanisms other than resistance have been highlighted as major drivers that contribute to the failure of antibiotics in health care settings. With the introduction of new antibiotics in clinics, it became clear that antibiotic treatment often failed to completely eradicate the bacterial subpopulation termed “persisters” (3). Bacteria are able to survive the action of antibiotics even at lethal concentrations by existing as persisters, which are genetically identical to the normal antibiotic-susceptible cells but exhibit a transient and nonheritable tolerance to bactericidal antibiotics (4). Persisters are widely considered to contribute to recurrent infections and to the development of antibiotic resistance (5). These observations led to avenues of research aiming to elucidate the mechanisms of persister formation and to develop strategies to combat these recalcitrant populations of pathogens.
Acinetobacter baumannii is a Gram-negative, opportunistic pathogen with no specificity for any niche. It is primarily associated with nosocomial infections, mainly hospital-acquired pneumonia, skin and soft tissue infections, urinary tract infections, and bacteremia, although cases of community-acquired infections are exceptional. A. baumannii is considered a major global threat due to rising prevalence of clinical isolates that are multidrug resistant (MDR) (6, 7). The World Health Organization (WHO) has identified A. baumannii as a top-priority pathogen for research and development of new therapeutics, due to limited understanding of its pathophysiology (8). The polymyxin class of antibiotics is considered the last line of defense against MDR Gram-negative “superbugs” (9). Worryingly, a recent report on the association between recalcitrance of A. baumannii infections and polymyxin treatment highlights the urgent need for new strategies to combat polymyxin tolerance in A. baumannii (10).
Several studies with Escherichia coli and Pseudomonas aeruginosa clearly indicate redundancy in persister formation. A remarkable study on E. coli persister formation demonstrated that cells survive by stochastically ceasing growth in the presence of ampicillin and resume growth following antibiotic removal (11). Antibiotic persisters exhibit decreases in metabolic activity, compared to their antibiotic-susceptible counterparts. Several studies suggest that major effectors that contribute to membrane depolarization, ATP efflux, blocked transcription and translation, or inhibition of DNA replication are major contributors to cellular growth arrest (12). However, little is known about the aspects of persister formation in A. baumannii. The slower pace of discovery of new antibiotics reflects both financial and scientific barriers to combating the problem of recurrent infections in clinics (13). However, one strategy that has proved effective is to use existing antibiotics in a wiser manner. Antibiotic combinations have been reported to dramatically enhance the actions of traditional antibiotics against these tolerant/resistant pathogens. The polymyxin class of antibiotics display synergistic effects in combination with many different classes of antibiotics (including rifampicin, meropenem, and aminoglycosides) to overcome resistance in Gram-negative pathogens (14, 15). However, there are only a few reports on the potential of combination therapy to eradicate antibiotic-induced persisters. In this respect, it was shown earlier that antibiotic combinations of polymyxin B with meropenem and colistin with amikacin are able to eradicate persisters in A. baumannii (16, 17). However, increased reports of carbapenem- and amikacin-resistant A. baumannii strains in clinics highlight the need to search for alternative treatment options (18).
Here, we report that, by targeting the reactive oxygen species (ROS) scavenger enzyme SodB, complete eradication of polymyxin B persisters in A. baumannii can be achieved. We screened broad classes of antibiotics with a dual fluorescence system in the A. baumannii AB5075 strain, which is capable of differentiating growth-arrested persisters from normally dividing cells. Among the tested antibiotics, rifampicin demonstrated the greatest decrease in polymyxin B-induced A. baumannii persister levels. Investigating the underlying mechanism behind rifampicin-facilitated killing, we observed enhanced ROS production and DNA damage in polymyxin B-induced persisters. Further, exploiting this persister-suppressing ability of rifampicin treatment demonstrated encouraging efficacy in an in vivo murine wound model of polymyxin B-induced A. baumannii persisters. Additionally, we have highlighted the precise mechanism of polymyxin B persister formation in A. baumannii. With anticipated treatment failure of this last-resort drug, our observations pave the way for further strategies to eradicate polymyxin persisters by targeting the ROS scavenger pathway of this pathogen.
RESULTS
A. baumannii isolates display large variations in survival rates upon treatment with polymyxin B.
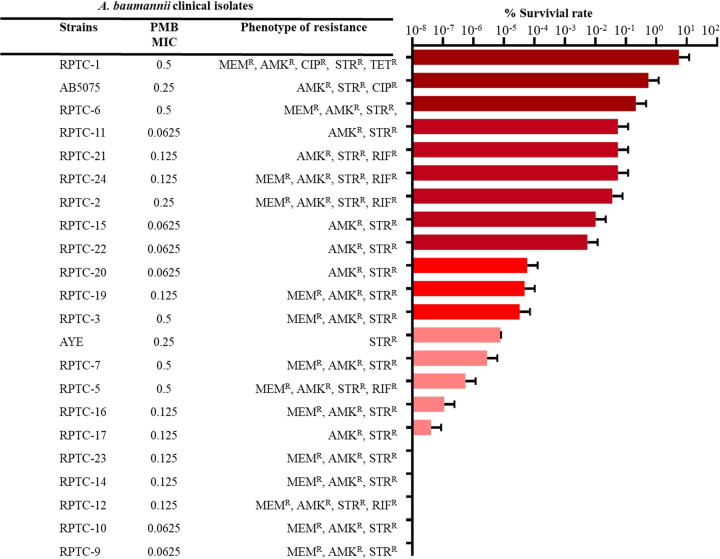
To study the survival rates of polymyxin B-induced persisters, we performed a persister assay with 22 polymyxin B-susceptible clinical isolates of A. baumannii. Most of the isolates displayed high levels of resistance to amikacin, tetracycline, gentamicin, and ciprofloxacin (see Table S1 in the supplemental material). Exponentially growing cells (optical density at 600 nm [OD600] of ∼0.5) of different strains were exposed to a high concentration of polymyxin B (10× MIC) for 3.5 h at 37°C, with shaking. The survival rates for each strain were calculated by determining the pretreatment and posttreatment CFU per milliliter values. Also, isolated colonies from each sample were reinoculated and antimicrobial susceptibility tests were performed to confirm that there were no changes in MIC values.
We observed large variations in survival rates among the clinical isolates. These variations were irrespective of the resistance phenotype of A. baumannii isolates, as strains with the same MIC values for a particular antibiotic displayed differences in persister formation rates (Fig. 1). The highest persister formation rates (0.1 to 1%) upon exposure to polymyxin B were observed for RPTC-1, AB5075, and RPTC-6. However, RPTC-23, RPTC-14, RPTC-12, RPTC-10, and RPTC-9 displayed no persister formation at a high concentration of polymyxin B. RPTC-1 and AB5075, which showed higher persister formation rates upon polymyxin B treatment, were used for persister assays with meropenem, rifampicin, and tigecycline. These strains displayed low persister formation rates with rifampicin and tigecycline (see Fig. S1). This observation may indicate the existence of different mechanisms implied by A. baumannii persister formation upon treatment with different antibiotics. As A. baumannii AB5075 displayed a higher survival rate upon polymyxin B treatment, in addition to the availability of its full genome annotation, it was used for further studies.
FIG 1.
Polymyxin B treatment exhibits variations in persister formation rates in A. baumannii. Persister survival rates, resistance phenotypes, and polymyxin B MICs of 22 clinical isolates of A. baumannii used in this study are shown. MEMR, meropenem resistance; AMKR, amikacin resistance; CIPR, ciprofloxacin resistance; STRR, streptomycin resistance; TETR, tetracycline resistance; RIFR, rifampicin resistance.
Disrupted proton motive force leads to cell division protein delocalization and depletion of intracellular ATP in polymyxin B-induced persisters.
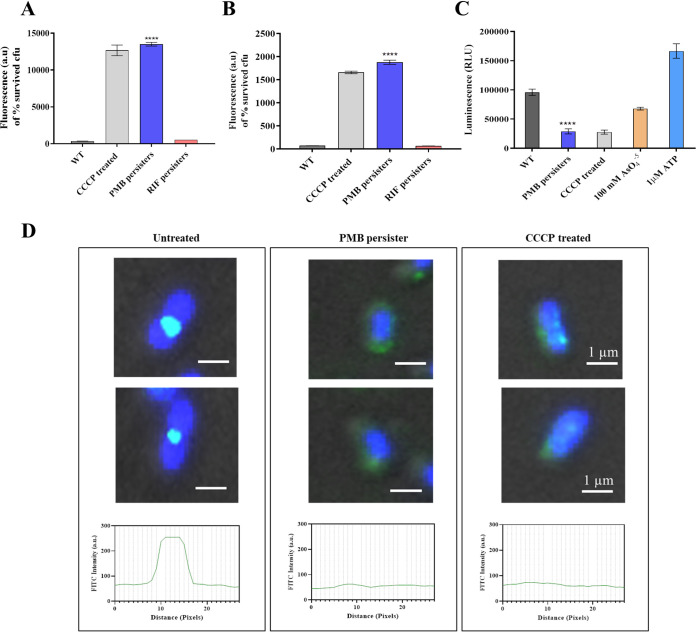
The polymyxin class of antibiotics target the bacterial envelope; therefore, we hypothesized that polymyxin B might alter any membrane-associated process that brings about a quiescent state in A. baumannii cells (19). One such parameter deemed important for cellular growth and division is proton motive force (PMF). In this context, the change of PMF in growth-arrested polymyxin B-induced persisters was studied using bis-(1,3-dibutylbarbituric acid)-trimethine oxonol (DiBAC4) (Δψ sensitive) and 9-amino-6-chloro-2-methoxyacridine (ACMA) (ΔpH sensitive) fluorescent dyes and taking carbonyl cyanide m-chlorophenyl hydrazine (CCCP) (ionophore to disrupt PMF) as the positive control. Treatment with 10× MIC of polymyxin B led to greater membrane distribution of DiBAC4, suggesting disruption of the Δψ component of PMF, comparable to that attained with CCCP. In addition, ACMA exhibited greater fluorescence intensity in polymyxin B-induced persisters, compared to untreated cells of A. baumannii AB5075 (Fig. 2A and B). Overall, polymyxin B-induced persisters exhibited complete disruption of PMF, which would contribute to changes in cellular energy. The intracellular ATP concentration is a direct way to measure the energy state inside a cell. Bacterial cells harness PMF to synthesize ATP by using the F0-F1 ATP synthase complex. To measure the intracellular ATP concentrations in untreated cells and polymyxin B persisters, a luciferase/luciferin assay was performed, taking AsO45− (causes ATP depletion in a PMF-independent manner) and CCCP (causes ATP depletion in a PMF-dependent manner) as positive controls (20, 21). ATP concentrations were significantly lower inside polymyxin B persister cells, compared to untreated cells (Fig. 2C). This decrease is comparable to results with CCCP, indicating a direct relationship with PMF disruption.
FIG 2.
Polymyxin B persisters exhibit membrane depolarization-mediated delocalization of the cell division protein. (A) Measurement of the membrane potential component Δψ using the membrane potential-sensitive fluorescent probe DiBAC4. (B) Measurement of the ΔpH component using the fluorescent probe ACMA. (C) Drop in intracellular ATP levels determined in polymyxin B persisters using CCCP, with AsO45− as a control where indicated. (D) Subcellular localization of the FtsZ ring and nucleoid segregation studied by immunofluorescence microscopy. Cells were immunostained with polyclonal anti-FtsZ antibody followed by FITC-conjugated secondary antibody, and nucleoid visualization was performed by staining the cell with DAPI. The distribution of FtsZ along the cells was analyzed using ImageJ software. Scale bars for all images, 1 μm. WT, wild-type. ****, P < 0.0001.
On the basis of the observation that PMF is hampered in polymyxin B persisters, we hypothesized an efflux-compromised state in these persisters. From the real-time qPCR analysis, we observed significant upregulation of adeB, adeG, adeJ, and tolC components of resistance-nodulation-division (RND) pumps in polymyxin B-induced persisters. However, polymyxin B persisters demonstrated greater ethidium bromide (EtBr) accumulation, compared to untreated cells, due to disruption of PMF, suggesting an efflux-compromised state in A. baumannii (see Fig. S2). Further, to study the effect of PMF disruption and ATP depletion on cell division of polymyxin B persisters, we performed immunofluorescence microscopy of FtsZ protein spatial localization. In comparison with untreated cells, in which we observed an intense and distinct FtsZ ring at the site of cell division, a loss of spatial localization was observed in polymyxin B persisters, which indicated a condition of halted bacterial cell division. We observed the same distribution of FtsZ in cells treated with 5 μM CCCP (Fig. 2D). Treatment with a high concentration of polymyxin B or addition of CCCP resulted in rapid reduction of the fluorescence signal, suggesting that PMF disruption mediates delocalization of the cell division protein in these growth-arrested A. baumannii persisters.
ROS-scavenging assembly in A. baumannii is important for polymyxin B tolerance.
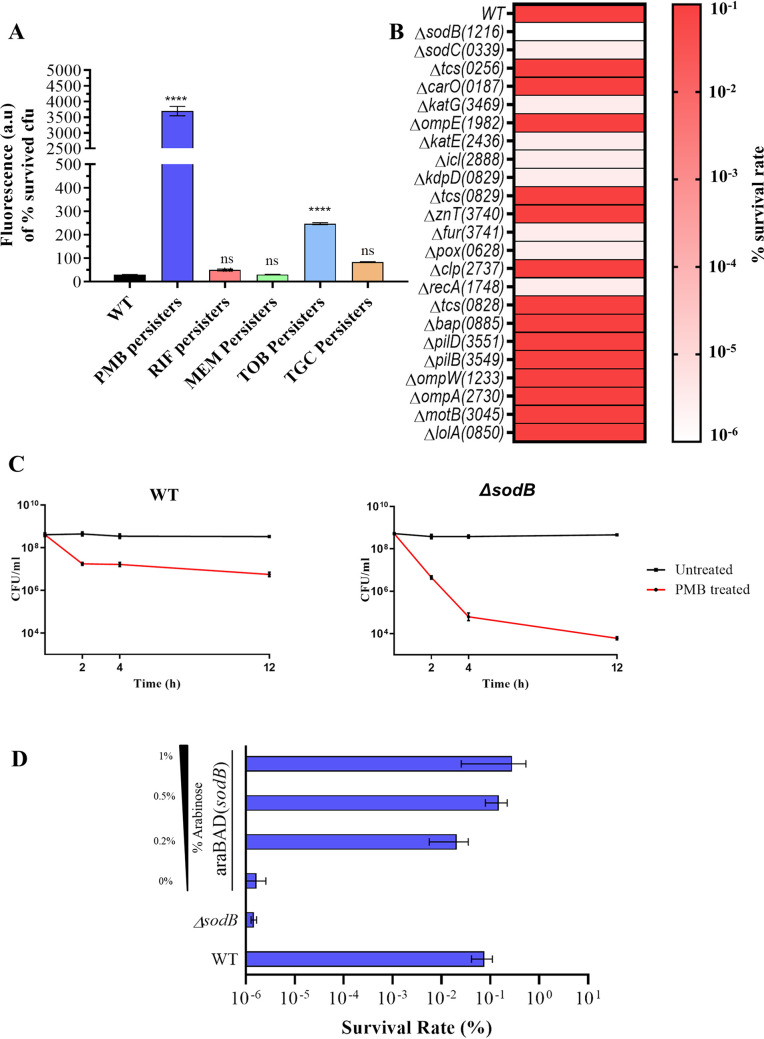
Superoxide free radicals are readily generated as a by-product of aerobic respiration and are the major source of intracellular oxidative stress (22). Additionally, it is widely accepted that, in addition to their mode of action, bactericidal antibiotics mediate killing by generation of ROS (23). Therefore, we sought to determine ROS production in persisters of different antibiotics. We used five different classes of antibiotics to generate A. baumannii persisters. The amount of ROS generated was quantified by using ROS-sensitive dichlorodihydrofluorescein diacetate (DCF-DA), a membrane-permeable fluorescent dye. Interestingly, it was observed that polymyxin B-induced persisters exhibited the greatest ROS generation, followed by tobramycin persisters (Fig. 3A). The amount of ROS generated was lowest for persisters generated upon exposure to meropenem, rifampicin, and tigecycline.
FIG 3.
Identification of the genes responsible for tolerance to polymyxin B in A. baumannii. (A) Measurement of ROS generation in A. baumannii persisters induced upon treatment with five antibiotics by using the ROS-sensitive fluorescent probe DCF-DA. (B) Heat map showing differences in polymyxin B tolerance in A. baumannii AB5075 mutants. Gene names are shown, with ABUW (i.e., gene locus ID of A. baumannii AB5075 strain) numbers in parentheses. The scale shows a gradient from 10−1 (dark orange) to 10−6 (white). (C) Biphasic killing curve showing differences in persister survival rates between wild-type and sodB mutant A. baumannii AB5075 cells upon treatment with 10× MIC of polymyxin B. (D) Complementation of araC-sodB in the A. baumannii AB5075 ΔsodB strain, resulting in restoration of polymyxin B tolerance. WT, wild-type; PMB, polymyxin B; RIF, rifampicin; MEM, meropenem; TOB. tobramycin; TGC, tigecycline. ****, P < 0.0001; ns, not significant.
Further, in order to determine the effector that contributes to polymyxin B tolerance, a simple persister assay was performed with 23 knockout mutants of A. baumannii AB5075. The mutants selected had mutations in genes involved in oxidative stress, persister-associated two-component systems, and polymyxin antibiotic-binding outer membrane proteins (22, 24–27). Strikingly, in groups with lower survival rates for persisters, we found genes involved in neutralization of intracellular ROS. The sodB mutant did not display formation of polymyxin B persisters. Also, the rate of persister formation was greatly reduced in the pox, katE, fur, znuC, and icl knockout strains (Fig. 3B). To further validate this finding, we performed a biphasic killing curve assay with the aforementioned mutants. Again, we observed, on the basis of CFU per milliliter values for the time period of 12 h, that the sodB mutant was highly impaired in tolerance to polymyxin B, with 2 to 2.5 log CFU/ml more killing by polymyxin B, in comparison to wild-type AB5075 cells (Fig. 3C). Next, we constructed a complemented strain in which sodB expression was under the control of an arabinose-inducible promoter, integrated into the chromosome with a method described by Tucker et al. (28). We performed a simple persister assay using polymyxin B against an A. baumannii AB5075 araC-sodB complemented strain with induction with different concentrations of arabinose (0%, 0.2%, 0.5%, and 1%). In contrast to the ΔsodB mutant strain, the complemented strain demonstrated increased polymyxin B tolerance even at 0.2% arabinose induction (Fig. 3D). However, further upregulation of sodB displayed no significant effect on polymyxin B tolerance in A. baumannii AB5075. These results suggested that sodB is important for polymyxin B tolerance in A. baumannii, besides other ROS-scavenging genes.
Rifampicin treatment eradicates polymyxin B persisters.
Persisters represent growth-arrested or reduced-growth phenotypes in a population of physiologically and phenotypically different cells. We sought to develop a dual fluorescence system to identify single cells of persisters from nonpersister populations of the opportunistic pathogen A. baumannii. We designed a dual-fluorescent-protein-based module composed of super folder green fluorescent protein (sfGFP) (green) and tandem dimer Tomato (tdTomato) (red). These proteins differ from one another in having different maturation times; tdTomato, with a longer maturation time (90 min), was transcriptionally fused with sfGFP, which is processed much more quickly (10 min) (29, 30). The expression of the two fluorescent proteins is under the control of a strong constitutive promoter (TTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGC). Next, we incorporated the construct into a neutral chromosomal locus of the pathogenic A. baumannii AB5075 (see Fig. S4). The chosen locus was sulfite reductase (ABUW_0643), which is conserved in most A. baumannii strains and, according to a previous report, is deemed a nonessential gene (31). The construct was integrated by using the double homologous recombination method developed by Tucker et al. (28). Dual fluorescence systems have been widely used to investigate protein distributions in eukaryotes. However, in contrast to eukaryotes, protein degradation in prokaryotes occurs very slowly and the fluorescent protein lifetime depends on the growth rate of the bacteria (32, 33). The doubling time of A. baumannii is ∼43 min; therefore, the maturation of tdTomato and its fluorescence (red) depend on cells with reduced growth rates (34, 35).
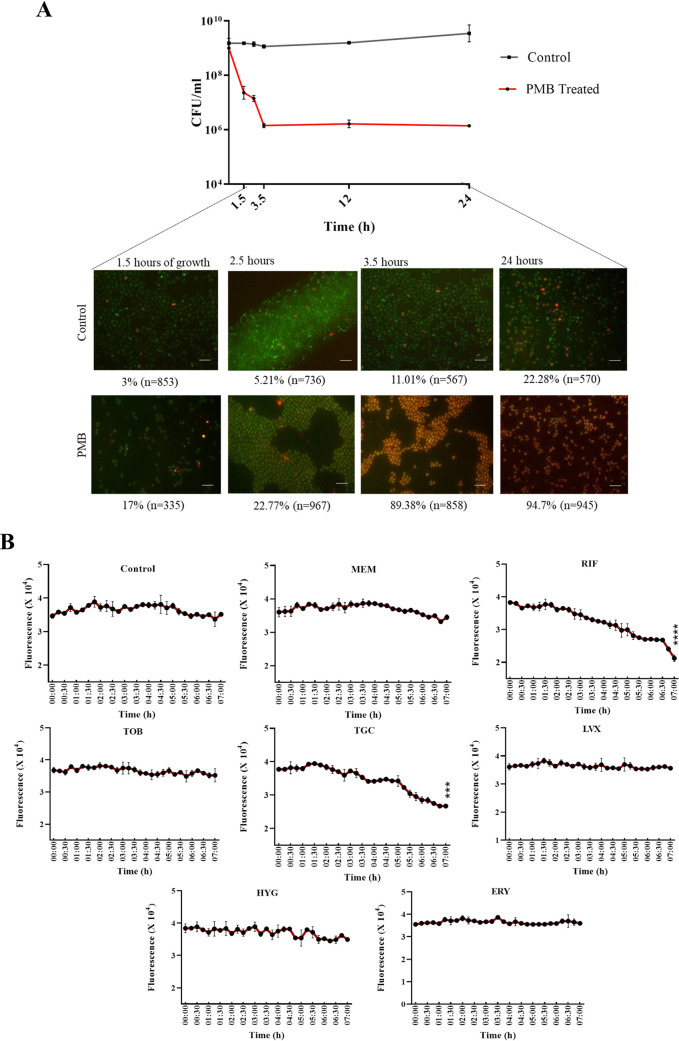
Next, we used this dual fluorescence timer system to detect A. baumannii persisters. Bacterial persisters can be categorized into two types; type 1 persisters represent growth-arrested subpopulations that are triggered under stress conditions, whereas type 2 persisters constitute the subpopulations of slowly growing cells that are spontaneously produced during growth (36). To investigate the detection of type 1 and type 2 persisters using our dual fluorescence system, we collected A. baumannii AB5075(DF) cultures from different growth phases and at different time points after treatment with 10× MIC of polymyxin B. Following quantification of red and green fluorescence of cells from microscopic images, we observed that a small fraction of slowly growing cells were present at mid-exponential phase (3%), which increased in late exponential and stationary phases (11.01% to 22.28%). Further, when a high concentration of polymyxin B was used to treat A. baumannii AB5075(DF), we observed that, following brief exposure (1.5 h) to antibiotic, 17% of total cells attained a growth-arrested state. The total percentage of growth-arrested cells reached 89.38% after 3 h of treatment, which further increased to 94.7% at 12 h (Fig. 4A). The calculated percentages of persisters in each frame represent approximate estimates (as residual dead cells might also have contributed to minor red fluorescence in the treated groups). Therefore, it was evident that our system is capable of differentiating normal dividing cells from nongrowing or slowly growing cells of A. baumannii.
FIG 4.
Dual fluorescent module in A. baumannii for identification of antibiotics to effectively kill polymyxin B-induced A. baumannii persisters. (A) Cells were harvested at different time points, including early exponential phase, late exponential phase, early stationary phase, and late stationary phase. Upon treatment with 10× MIC of polymyxin B, cells were harvested at treatment times ranging from 1.5 h to 24 h. Scale bar for all images, 50 μm. (B) Polymyxin B-induced persisters with the dual fluorescence module of A. baumannii AB5075 were treated with seven antibiotics at concentrations below the clinical breakpoint concentration. Survival kinetics of persisters upon treatment were monitored by taking fluorescence readings of red-fluorescing cells. MICs were as follows: meropenem, 4 μg/ml; rifampicin, 1 μg/ml; tobramycin, 8 μg/ml; tigecycline, 0.5 μg/ml; hygromycin, 16 μg/ml; levofloxacin, 2 μg/ml; erythromycin, 8 μg/ml. PMB, polymyxin B; MEM, meropenem; RIF, rifampicin; TOB. tobramycin; TGC, tigecycline; HYG, hygromycin; LVX, levofloxacin; ERY, erythromycin,. Each treatment group was compared with the control group for statistical significance. ****, P < 0.0001; ***, P < 0.001.
Next, we used our dual timer system to screen for suitable antibiotics that could eradicate the A. baumannii persisters (red fluorescence). Based on the MIC profile of A. baumannii, we used antibiotics of seven different classes (10× MIC) to which A. baumannii AB5075 is sensitive. To ensure that the test bacterial cultures were composed mostly of polymyxin B-induced persisters, the simple persister assay was performed and cells were harvested following 3.5 h of treatment. The harvested cells were concentrated 10-fold in phosphate-buffered saline (PBS), and the final tdTomato red fluorescence of the culture was measured to set a base level of readings. After the addition of antibiotics, the kinetics of persister killing were monitored by measuring decreases in the tdTomato fluorescence intensity for a period of 7 h. We observed that rifampicin and tigecycline could eradicate persisters with significant reductions in fluorescence intensity over the duration of the assay (Fig. 4B). In order to assess whether the drop in red fluorescence signal was due to killing not only of viable but nonculturable cells but also of persisters, we performed persister survival assays with AB5075, RPTC 1, and RPTC 20. We observed that, although tigecycline was effective in reducing the surviving persisters, rifampicin displayed better persister eradication ability than did tigecycline (Fig. S5).
Rifampicin downregulates ROS-scavenging assembly and mediates DNA damage in polymyxin B-induced A. baumannii persisters.
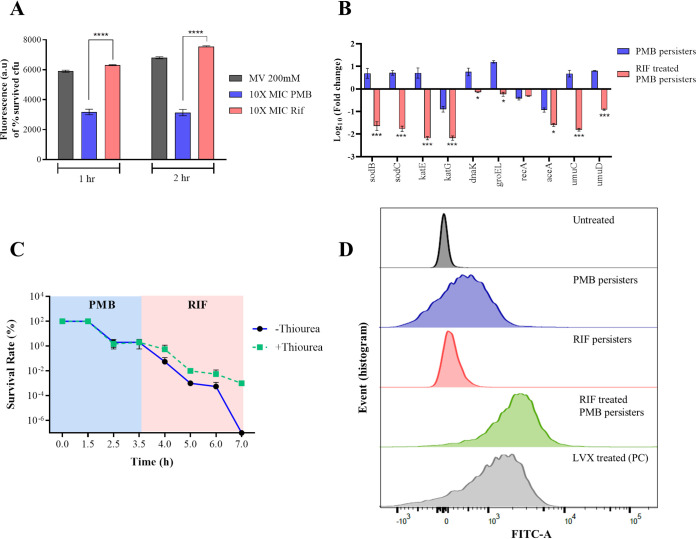
As discussed previously, polymyxin B persisters exhibit high levels of intracellular ROS generation and the ΔsodB mutant displays a defect in polymyxin B persister formation. This warrants investigation of the effect of rifampicin treatment on ROS generation. To this end, we treated A. baumannii AB5075 polymyxin B persisters with rifampicin for 2 h and determined the time-dependent enhancement of ROS production. We observed a time-dependent increase of ROS generation in the two treated groups, which indicates that rifampicin treatment led to an increase in ROS generation in polymyxin B persisters (Fig. 5A). Such time-dependent enhanced production of ROS was not observed in tigecycline-treated polymyxin B persisters (Fig. S5). To investigate the effect of rifampicin on the ROS-scavenging system in persister cells, we performed 16S rRNA-normalized qPCR analysis of genes involved in the liberation of intracellular ROS. It was observed that, following treatment with rifampicin, polymyxin B persister cells underwent 40-fold downregulation of sodB, in addition to other ROS-subsiding genes (Fig. 5B).
FIG 5.
Suppression of sodB leads to enhanced ROS generation and eradication of polymyxin B-induced A. baumannii persisters. (A) Time-dependent increase in ROS generation upon rifampicin treatment measured using the ROS-sensitive fluorescent probe DCF-DA. (B) qRT-PCR analysis of ROS-scavenging genes in A. baumannii. Relative gene expression was calculated by the ΔΔCT method, using 16S rRNA as the reference genes. (C) ROS quenching by using thiourea conferring protection of polymyxin B-induced A. baumannii persisters upon rifampicin treatment, suggesting that ROS-mediated killing is the sole mechanism of persisters eradication. (D) TUNEL assay coupled with flow cytometry to assess rifampicin treatment-induced DNA damage in A. baumannii persisters. The x axis represents the relative FITC fluorescence. MV, methyl viologen; PMB, polymyxin B; Rif, rifampicin; LVX, levofloxacin; PC, positive control; ****, P < 0.0001.
Further, to confirm that ROS generation is the sole mechanism of persister killing, we treated A. baumannii AB5075 cells with polymyxin B followed by rifampicin in the presence and absence of the ROS-quenching agent thiourea. Additionally, to investigate whether rifampicin-induced persisters could be eradicated by polymyxin B treatment, we performed the assay by treating cells with rifampicin first followed by polymyxin B. When polymyxin B was followed by rifampicin treatment in the absence of thiourea, complete eradication of polymyxin B persisters was observed. However, the persister survival rate remained at 0.001% when treatment was performed in the presence of thiourea (Fig. 5C). This strongly suggested that the combination of rifampicin and polymyxin B generated ROS sufficient to kill A. baumannii persisters. Additionally, no such effect was observed when the order of treatment was reversed, suggesting that polymyxin B treatment is ineffective in killing rifampicin persisters in A. baumannii (see Fig. S6).
High intracellular ROS levels in persister cells lead to oxidative damage of DNA and eventually cell death. ROS also oxidize dCTP and dGTP pools, causing misincorporation of bases into DNA, which leads to double-strand breaks (37). This enhanced ROS-mediated DNA damage was evaluated using the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay coupled with flow cytometry, taking DNA-damaging levofloxacin at 16 μg/ml as the positive control. The TUNEL assay revealed that, in the presence of rifampicin, polymyxin B-induced persisters exhibited increased DNA damage, as evident from enhanced incorporation of fluorescein isothiocyanate (FITC)-dUTPs at the site of double-strand breaks (Fig. 5D). Further, fluorescence microscopy was performed to assess the FITC-dUTP incorporation in different treatment groups (see Fig. S7).
Rifampicin treatment facilitates killing of A. baumannii polymyxin B persisters in a murine model.
We used an acute wound infection model to show the in vivo efficacy of rifampicin treatment in persister eradication. To this end, 10-fold concentrated polymyxin B-induced A. baumannii persisters were added to the wound and allowed to dry. Next, we added cell-free medium, cell-free medium with 10× MIC polymyxin B, or cell-free medium with 10× MIC polymyxin and rifampicin to different groups. Cell-free medium supplemented with 10× MIC rifampicin with nonpersister A. baumannii AB5075 was taken as a positive-control group. Quantitative analysis revealed that rifampicin treatment could significantly (P < 0.01) eradicate polymyxin B A. baumannii persisters in the murine acute wound infection model, lowering the persister burden to 3-log-fold (Fig. 6).
FIG 6.
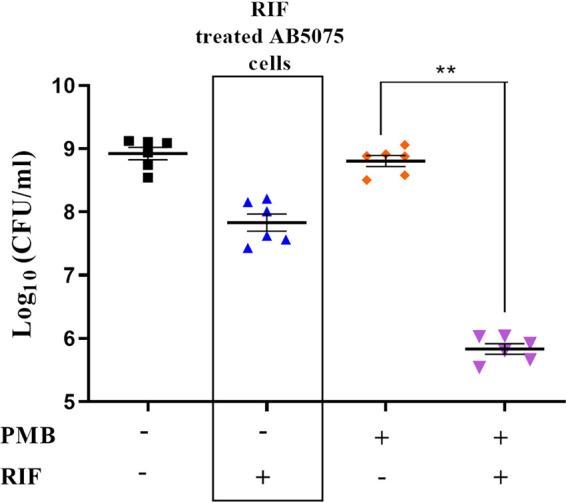
Rifampicin treatment facilitates killing of polymyxin B-induced A. baumannii persisters in a murine model. Survival of polymyxin B-induced A. baumannii persisters was assessed after cells were inoculated in wounds of mice and subjected to rifampicin treatment. Wound lysates were plated to determine the efficacy of rifampicin to eradicate A. baumannii persister cells in vivo. Treatment with rifampicin alone on A. baumannii AB5075 (non-polymyxin B persisters) is represented in the box. PMB, polymyxin B; RIF, rifampicin. **, P < 0.01.
DISCUSSION
Persisters are bacterial subpopulations that exhibit transient tolerance to antibiotics, are ubiquitous among prokaryotes, and have been linked to relapse and recalcitrance of infections in health care settings. Persisters cause high rates of morbidity and death and have been linked to the rise and spread of antibiotic resistance (38, 39). Rapid increases in antibiotic resistance and high rates of exposure of immunocompromised patients to MDR pathogens might contribute to increased numbers of cases of recurrent infections in the near future (40). Therefore, greater understanding of the internal phenomena that might contribute to persister formation will lead to the development of treatment strategies to prevent recalcitrance of bacterial infections.
In this study, the characteristics of persister formation by one of the important ESKAPE pathogens, A. baumannii, were determined following treatment with the drug of last resort, polymyxin B. Polymyxin B-treated A. baumannii isolates exhibit high rates of persister formation, with huge survival rate variations among different clinical isolates. Previous studies of bacterial persisters delineated the involvement of loss of membrane potential, enhanced efflux activity, the toxin/antitoxin system, and stress alarmone ppGpp-mediated regulation (41–45). Membrane potential or PMF, the product of cellular respiration, describes the electrochemical gradient across the cytoplasmic membrane, which is composed of an electrical gradient (Δψ) and a chemical gradient (ΔpH) (46). This electrochemical potential of the cells underpins production of cellular energy for bacterial cells to work in order to maintain a constant PMF (47). Polymyxin B treatment-induced persisters exhibit compromised membrane permeability owing to disruption of Δψ and ΔpH components of PMF. Disrupted PMF results in conversion of the F0-F1 ATP synthase complex to function as an ATP-driven proton pump to drive normal dividing cells into a nongrowing phenotype through eventual depletion of intracellular ATP (48). Also, many multidrug efflux pumps depend on PMF, with energy from the proton gradient being harnessed to facilitate drug efflux from the cell (49). RND pumps are the major class of MDR efflux systems among Gram-negative pathogens (50). RND pumps have been deemed the most important driving force behind efflux of aminoglycosides and other antibiotics (21). Polymyxin B-induced persisters exhibit an efflux-compromised condition in A. baumannii and thus can be eradicated effectively by antibiotics that are otherwise effluxed out. Previously, it was shown that an amikacin-susceptible strain of A. baumannii displayed complete eradication of colistin A. baumannii persisters upon treatment with the combination of colistin and amikacin (17). In this study, however, the combination of colistin and amikacin was not effective in eradicating A. baumannii persisters, due to intrinsic resistance displayed by clinical isolates of A. baumannii. Cellular localization of cell division proteins also depends on cellular PMF. FtsZ forms a ring-like structure (Z-ring) at the site of cytokinesis to establish a scaffold that sequentially recruits the divisome complex to initiate cytokinesis (51). As previously reported, membrane-associated cell division proteins such as MinD and ZipA (which anchor the FtsZ ring in a midcell location) maintain their spatial localization by their membrane-binding amphipathic region. These nonpolar motifs bury themselves in the lipid bilayer, and this interaction is further stabilized by the charged residue with charged lipid head (52). Our results demonstrated that, in growth-arrested polymyxin B-induced persisters, there is a loss of spatial localization of the FtsZ protein, a condition similar to that of cells treated with the PMF disruptor CCCP.
The challenge to discover novel agents that are capable of eradicating bacterial persisters has kept the output of novel antipersister agents to extraordinarily low levels, mainly due to currently available time-consuming antipersister assays. To date, few studies are available in which screening of antipersister agents was performed, due to various CFU per milliliter-based rate-limiting steps, which limits simultaneous screening of large number of agents for antipersister activity. Therefore, we developed a dual fluorescence module in A. baumannii AB5075, in which the killing kinetics of persisters upon exposure to an agent could be monitored in real time, based on decreases in fluorescence reads for an expressed protein with a longer maturation time. This system is proficient in detection of both type 1 and type 2 growth-arrested A. baumannii persisters. Among the screened antibiotics, we found that rifampicin and tigecycline exhibited significant killing of polymyxin B-induced A. baumannii persisters. We further validated this finding using conventional tolerance studies. This is the first report in which a real-time strategy for eradication of A. baumannii persisters has been highlighted.
The generation of ROS and subsequent damage of cellular macromolecules by oxidative stress is a secondary feature shared by many bactericidal antibiotics. We assessed the amount of ROS generated by A. baumannii persisters induced by different classes of antibiotics. Large amounts of ROS are tolerated by the growth-arrested persisters with bacterial ROS scavengers, mainly consisting of superoxide dismutase and catalase. Membrane damage by polymyxin B leads to generation of ROS, most specifically superoxide free radicals (53). Superoxide dismutase (SOD) readily breaks down superoxide free radicals into oxygen and hydroxyl ions; the latter are further neutralized by catalase and peroxidase (54). Earlier report on the treatment of A. baumannii ATCC 17978 with colistin resulted in survival of persister cells via enhanced activity of ROS neutralization assembly and upregulation of the emrAB efflux pump (55). The idea of potentiating the damaging effect of ROS on cell killing and antagonizing the protective effect of the ROS rescue system could implicate enhanced clearance of bacterial persisters. Our study demonstrated that rifampicin, a transcription-inhibiting antibiotic, results in significant downregulation in the expression of various ROS-scavenging genes, thus exceeding the threshold amount of secondary damage by ROS and forcing cell death. ROS also oxidize the dGTP and dCTP pools, causing misincorporation of bases and double-strand breaks in DNA (37). The error-prone polymerase known as UmuD′C2 or DNA polymerase V is used as a last-resort DNA repair system during elevated intracellular ROS-mediated DNA damage and is highly mutagenic (56). A previous study involving antibiotic-induced Escherichia coli persisters demonstrated that the development of the antibiotic-resistant phenotype was greatest in ciprofloxacin-induced persisters due to upregulation of the error-prone DNA repair process facilitated by UmuD′C2 (57). In this study, we observed that elevated expressions of UmuD′C2 in polymyxin B-induced persisters was significantly downregulated upon exposure to rifampicin, which led to diminished repair of double-stranded DNA break-mediated eradication of polymyxin B persisters.
Our results also reveal an unprecedented link between sodB and the tolerance of A. baumannii persisters to polymyxin B. We also demonstrate that deletion of sodB enhances the lethality caused by polymyxin B and abrogates the survival of persister populations. This SOD-dependent pathway is targeted by rifampicin, leading to eradication of growth-arrested A. baumannii persisters by enhanced ROS generation. A. baumannii is a MDR pathogen that is associated with pneumonia, bacteremia, and wound/burn infections. Previously, it was demonstrated that treatment of A. baumannii infections with the polymyxin B class of antibiotics led to high rates of therapeutic failure due to the presence of the persister phenotype. Here, we prepared polymyxin B-induced A. baumannii persisters using the same method as described previously, and we found that rifampicin treatment was able to eradicate the A. baumannii persister burden in a murine wound infection model, an observation similar to that obtained from our in vitro assays.
Rifampicin has been widely used for treatment of infections caused by Staphylococcus aureus and Mycobacterium spp., although its effect in the eradication of the persister phenotype in Gram-negative pathogens has not been explored (58). Here, we report, as a proof of principle, that rifampicin is able to eradicate polymyxin B-induced A. baumannii persisters. Thus, it is convenient to suggest that our study encourages research using a dual timer module to screen for antipersister agents and to target sodB for developing new strategies against polymyxin B-induced A. baumannii persistent infections.
MATERIALS AND METHODS
Strains, culture conditions, and reagents.
Various clinical strains of A. baumannii were isolated from patients attending the Government Medical College and Hospital (Chandigarh, India). A. baumannii AB5075 mutant strains used in this study (ΔsodB, ΔsodC, Δperoxidase, Δcat, Δfur, Δznu, Δicl, Δomps, ΔcarO, Δbap, Δclp, Δpil, and Δtcs) were purchased from the University of Washington (USA). Briefly, an overnight culture of A. baumannii was diluted 1:500 in Muller-Hinton medium and agitated at 37°C for 3.5 h, with or without treatment with antibiotics, for the isolation of cells from the exponential phase of bacterial growth. The antibiotics used in this study, including polymyxin B, rifampicin, tigecycline, meropenem, tobramycin, apramycin, levofloxacin, tobramycin, hygromycin, and erythromycin, were purchased from Sigma-Aldrich. CCCP was purchased from Sigma-Aldrich, and DiBAC4 and ACMA were purchased from Thermo Fisher Scientific. The BacTiter-Glo kit was purchased from Promega. FITC-conjugated goat anti-rabbit IgG secondary antibody was purchased from Thermo Fisher Scientific. All other chemical reagents were of analytical purity.
MIC testing.
The MIC is defined as the minimum concentration of antibiotics at which no visible growth of a microorganism is obtained. The MIC profile for each strain was determined using the broth dilution method. MIC assays were performed with cation-adjusted Muller-Hinton broth in 96-well plates, in a total volume of 200 μl per well. The 2-fold dilution of antibiotics was performed serially across the plate, except that the second last well contained no antibiotics. The bacterial culture was adjusted at 107 CFU/ml per well. The inoculum was added to the plate and incubated at 37°C for 12 to 14 h.
Persister assay.
Persister assays were performed as described previously, with little modification (10, 15). Briefly, different strains of A. baumannii were grown to the exponential phase (OD600 of ∼0.5), followed by exposure to antibiotic (10× MIC) for 3.5 h at 37°C with shaking at 200 rpm. After incubation, cells were harvested, washed three times with PBS, and spread on LB agar plates after serial dilution. To confirm that the surviving colonies were persister cells, two colonies were reinoculated into fresh LB medium and an in vitro susceptibility assay was performed to ensure that the MIC had not changed.
Measurement of PMF.
To determine the Δψ component of PMF, the DiBAC4 fluorescent dye was used. Antibiotic persisters were prepared as described previously. Isolated cells resuspended in PBS were exposed to 30 μM DiBAC4 and incubated for 10 min at room temperature, and fluorescence readings were taken at excitation and emission wavelengths of 485 nm and 528 nm, respectively. Similarly, to determine the ΔpH component of PMF, the ACMA fluorescent dye was used. Normal and antibiotic persisters were exposed to 8 nM ACMA dye and incubated for 37°C for 30 min with shaking at 200 rpm. Fluorescence readings were taken at excitation and emission wavelengths of 419 nm and 483 nm, respectively. Fluorescence was monitored in 96-well black plates using a Synergy H1 plate reader. CCCP was taken as the positive control at a concentration of 5 μM, and cells were treated for 3.5 h. Results are expressed as the percentage of surviving CFU, in comparison to untreated cultures. The untreated group CFU per milliliter was adjusted to match that of the polymyxin B-treated group for comparison.
ATP measurement.
Intracellular ATP levels of polymyxin B persisters and normal cells with addition of 100 mM arsenate and 5 μM CCCP were measured using the BacTiter-Glo kit according to the manufacturer’s instructions. The background ATP level was subtracted from spent medium from each sample, and 3 mM ATP was used as the positive control.
EtBr uptake assay.
The cells of A. baumannii AB5075 were grown to an OD600 of ∼0.5, and polymyxin B-induced persisters were prepared as described previously. The cells were resuspended in PBS supplemented with 0.4% glucose, and the OD600 was adjusted to 0.3. EtBr was added into each sample to yield a final concentration of 10 μg/ml, and intake fluorescence kinetics were measured for 15 min with excitation and emission wavelengths of 480 nm and 610 nm, respectively. Fluorescence was monitored in 96-well black plates using a Synergy H1 plate reader. Results are expressed as the percentage of surviving CFU, in comparison to untreated cultures. The untreated group CFU per milliliter was adjusted to match that of the polymyxin B-treated group for comparison.
Immunofluorescence microscopy of FtsZ.
Normal cells, polymyxin B persisters, and CCCP-treated cells of A. baumannii AB5075 were harvested by centrifugation and washed two times in PBS. The cell pellet was resuspended in 500 μl of PBS, followed by addition of 100 μl of fixative (2.5% glutaraldehyde), and incubated at 4°C for 1 h. Fixed bacterial cells were washed two times with PBS, 10 μl of 2 mg/ml lysozyme was added, and cells were incubated for 20 min at 37°C. After three steps of washing, 2% bovine serum albumin (BSA) was added as a blocking agent. The samples were incubated in a humid chamber for 30 min. Anti-FtsZ antibody (1:100 dilutions in 2% BSA) was added to the samples after thorough washing, and cells were incubated overnight at 4°C in a humidified chamber. The following day, the cells were washed 10 times with PBS, and FITC-conjugated goat anti-rabbit IgG secondary antibody (1:500 dilution in 2% PBS) was added. After incubation for 30 min, the samples were washed three times with PBS. 4′,6-Diamidino-2-phenylindole (DAPI) (4 μg/ml) was used to stain the nucleoid and, after five washing steps in PBS, the samples were added to a 1% agarose pad. After drying, the slides were visualized with a Carl Zeiss fluorescence microscope (×100 magnification) with FITC and DAPI filters.
Detection of intracellular ROS.
DCF-DA was used for detection of intracellular ROS. An overnight culture of A. baumannii AB5075 was diluted 1:500 and allowed to grow to an OD600 of ∼0.5. Antibiotic persisters were washed with PBS and adjusted to an OD600 of ∼0.3. DCF-DA at a working concentration of 100 μM was added and incubated at 37°C for 30 min. The fluorescence of samples was read using a Synergy H1 spectrofluorometer at excitation and emission wavelengths of 485 nm and 528 nm, respectively, using 96-well black plates. Results are expressed as the percentage of surviving CFU, in comparison to untreated cultures. The untreated group CFU per milliliter was adjusted to match that of the polymyxin B-treated group for comparison.
Real-time PCR.
Briefly, 1 μg of total RNA was used to prepare cDNA using the SuperScript III first-strand synthesis kit (Invitrogen). Real-time PCR was performed with the SYBR green master mix (Applied Biosystems) following the manufacturer’s instructions. Measurements were performed using the QuantStudio 6× real-time PCR system (Applied Biosystems) with the following conditions: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min, and a final dissociation cycle of 95°C for 2 min, 60°C for 15 s, and 95°C for 15 s. Relative gene expression was calculated by the ΔΔCT method using 16S rRNA as the reference gene. Experiments were performed in biological duplicates and measured in technical triplicates. Primers used for real-time PCR are given in Table S2 in the supplemental material.
Construction of dual fluorescence module in A. baumannii AB5075 and plasmids.
All oligonucleotides used in this study are listed in Table S2 in the supplemental material. Plasmid pRPT398 was constructed by cloning the PCR product of the upstream and downstream 500 bp of the sulfite reductase gene into the pUC18 vector. A chimeric PCR product consisting of sfGFP (obtained from sfGFP-pBAD) under the control of the constitutive strong promoter J23100 was fused with TdTomato (obtained from TdTomato-pBAD) and subsequently cloned between the upstream and downstream regions of cysI (a linker region was introduced between sfGFP and TdTomato so as not to disturb the functioning of proteins). The apramycin cassette was amplified from the plasmid pMDIAI and cloned into pRPT398 to yield pRPT400. All plasmid sequences are available upon request.
The plasmid pRPT400 was used to obtain a chimeric PCR product consisting of 150-bp upstream and downstream segments of cysI with sfGFP, TdTomato, and apramycin (4.5 kb). The PCRs were performed using high-fidelity HotStar DNA polymerase (Qiagen). Subsequently, each PCR product was purified after running in an agarose gel, and purification was performed by following the manufacturer’s instructions (QIAquick gel extraction kit; Qiagen). The chimeric PCR product was concentrated to 5 μg and transformed into A. baumannii AB5075 electrocompetent cells with pAT02 (induced by 2 mM isopropyl-β-d-thiogalactopyranoside [IPTG]). The transformants were screened for positive knock-in using the primers Up150CysIFP and RPsfxma1. The apramycin cassette was removed from the positive transformants by using pAT03 expressing FLP recombinase. For construction of the complementation strain, intact copies of the araC-BAD promoter and sodB were amplified from the pBAD plasmid and genomic DNA of A. baumannii AB5075 and cloned at BamHI/XmaI and XmaI/KpnI sites of pUC18 (with cloned-500 bp upstream and downstream regions), respectively. The methods involving amplification of the chimeric PCR product and knock-in methods were performed as described above.
Screening of antibiotic treatment against TdTomato-expressing A. baumannii.
Polymyxin B persisters were prepared using the persister assay described previously. Aliquots of 100 μl of polymyxin B persisters suspended in PBS were added to the wells of 96-well black plates. Antibiotics at concentrations of 10× MIC were introduced into the respective wells, and killing kinetics were measured in reference to decreases in the fluorescence intensity of TdTomato, which was read for the period of 7 h with excitation and emission wavelengths of 554 nm and 581 nm, respectively, using a Synergy H1 spectrofluorometer.
Biphasic killing assay.
Exponentially growing A. baumannii cells were challenged with 10× MICs of antibiotics and incubated at 37°C with shaking at 200 rpm. At 1.5, 2.5, 3.5, and 12 h, aliquots of 100 μl were taken out, serially diluted in sterile PBS, and plated on LB agar plates. After overnight incubation at 37°C, colonies were counted to determine the viable cell counts in terms of CFU per milliliter.
ROS-quenching assay.
Intracellular ROS were quantified using DCF-DA dye as described previously. ROS quenching experiments were performed by exposing the polymyxin B persisters to quencher (200 mM thiourea) for 30 min at 37°C with shaking. Following three steps of washing, cells were exposed to a second antibiotic (rifampicin at 10× MIC) and methyl viologen as positive control. To determine the ROS-mediated eradication of polymyxin B persisters by rifampicin, serial exposure to two antibiotics was performed. After 3.5 h of treatment with one antibiotic, cells were harvested and washed three times with PBS, and the other antibiotic was added in the presence or absence of 200 mM thiourea. Aliquots were taken at time points of 1.5, 2.5, 3.5, 4, 5, 6, and 7 h and plated on LB agar to obtain CFU per milliliter.
TUNEL assay.
The TUNEL assay was performed using the BD Pharmingen APO-DIRECT kit (BD Biosciences), as described by the manufacturer. Antibiotic persisters were prepared by the method described previously. The cells were resuspended in 1% paraformaldehyde in PBS (pH 7.4) at a concentration of 106 cells/ml, and this suspension was kept on ice for 60 min. The cells were washed with PBS three times, and cells were resuspended in 70% ice-cold ethanol and allowed to stand for 30 min on ice. For staining, the cell pellet was resuspended in 1 ml wash buffer. After two steps of washing, the pellet was incubated in 50 μl of TUNEL reaction mixture, including FITC-dUTP and deoxynucleotidyl transferase enzyme, at 37°C in the dark for 1 h. After incubation, cells were washed twice with rinse buffer, counterstained with propidium iodide (PI)/RNase staining buffer, and incubated at room temperature for 30 min. Samples were washed and then resuspended in PBS for fluorescence-activated cell sorting (FACS) analysis. The FITC signal was analyzed with an emitting laser at 488 nm and a band pass filter of 525/15 nm using a BD flow cytometer (BD Biosciences) with a 70-μm nozzle. Graphs were generated using FlowJo version 10 software.
Morphometric analysis.
For fluorescence microscopy, untreated and polymyxin B-treated A. baumannii AB5075(DF) cells at different time points were aliquoted and adjusted to an OD of ∼0.3. The samples were washed three times with PBS to remove residual antibiotics, and polymyxin B-treated samples were concentrated 100-fold to achieve comparable cell counts with respect to the untreated group. The samples were loaded on top of a 1% (wt/vol) agarose pad, and sample solutions were allowed to evaporate and to absorb on the pad. A glass coverslip was gently placed to sandwich the cells flat on the image plane. Images were taken using a Carl Zeiss microscope using two emission filters, at 488 nm (sfGFP) and 580 nm (tdTomato). The number of cells in each field was calculated using ImageJ software.
Animal experiments ethics approval.
Animal experiments were performed according to a protocol submitted to the institutional animal ethics committees. Permission was obtained from the IIT Roorkee ethics committee (protocol BT/IAEC/2017/03).
In vivo murine infection.
We used an acute skin wound infection model to determine the efficacy of rifampicin treatment in the eradication of polymyxin B-induced persisters of A. baumannii. Mice were anesthetized (xylazine and ketamine), barbered on the dorsal region, and sterilized and then a whole-skin section (1 cm by 1 cm) was removed using a biopsy punch to make an acute skin wound. Then, 5 μl of 10-fold concentrated polymyxin B-induced persisters of A. baumannii AB5075 (prepared as described previously) was seeded on the wound and allowed to fully dry before addition of 40 μl of cell-free medium without or with antibiotics (with 10× MIC of polymyxin B alone or in combination with rifampicin). After full drying, the wound site was bandaged firmly with medical gauze. After the mice were housed overnight, the scab on the wound was removed and homogenized. The lysates were serially diluted in sterile PBS and plated on LB agar plates for the bacterial survival assay. Normal exponential-phase cells concentrated 10 times and treated with 10× rifampicin were taken as the positive control, and cells with no treatment were taken as the negative control. For quantification, each sample was spread in triplicate.
Statistical analysis.
Statistical analyses were carried out using Prism 8 software (GraphPad, CA USA). The two-tailed Student's t test was used for comparisons between two conditions, while one-way analysis of variance (ANOVA) with Tukey’s multicomparison posttest was used for comparisons between three or more conditions. P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Naveen K Navani, Amit Gaurav, and Mangal Singh for their suggestions. We thank Shahnawaz Ahmad Baba for technical help.
V.D. and R.G. acknowledge independent DBT-JRF fellowships (DBT/2015/IIT-R/301 and DBT/2018/IIT-R/1144, respectively) awarded by the Department of Biotechnology, India.
V.D. and R.P. designed the experiments, V.D. and R.G. performed experimental protocols, V.D. and R.P. wrote the manuscript, and all authors reviewed the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bunker JP. 1999. The rise and fall of modern medicine. BMJ 319:1276. 10.1136/bmj.319.7219.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, Morosini MI. 2011. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991. 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 3.Conlon BP. 2014. Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells: an investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 36:991–996. 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 4.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 5.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 6.Di Venanzio G, Flores-Mireles AL, Calix JJ, Haurat MF, Scott NE, Palmer LD, Potter RF, Hibbing ME, Friedman L, Wang B, Dantas G, Skaar EP, Hultgren SJ, Feldman MF. 2019. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat Commun 10:2763. 10.1038/s41467-019-10706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gootz TD, Marra A. 2008. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther 6:309–325. 10.1586/14787210.6.3.309. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Publication WHO/EMP/IAU/2017.12. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 9.Lowings M, Ehlers MM, Kock MM. 2015. Acinetobacter baumannii: a superbug, p 587–597. In Méndez-Vilas A (ed), The battle against microbial pathogens: basic science, technological advances and educational programs. Formatex Research Center, Badajoz, Spain. [Google Scholar]
- 10.Chung ES, Wi YM, Ko KS. 2017. Variation in formation of persister cells against colistin in Acinetobacter baumannii isolates and its relationship with treatment failure. J Antimicrob Chemother 72:2133–2135. 10.1093/jac/dkx102. [DOI] [PubMed] [Google Scholar]
- 11.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 12.Orman MA, Brynildsen MP. 2013. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57:3230–3239. 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164. 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 14.Yoon J, Urban C, Terzian C, Mariano N, Rahal JJ. 2004. In vitro double and triple synergistic activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 48:753–757. 10.1128/aac.48.3.753-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urban C, Mariano N, Rahal JJ. 2010. In vitro double and triple bactericidal activities of doripenem, polymyxin B, and rifampin against multidrug-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli. Antimicrob Agents Chemother 54:2732–2734. 10.1128/AAC.01768-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo SW, Ferreira CAS, de Oliveira SD. 2017. Combination of polymyxin B and meropenem eradicates persister cells from Acinetobacter baumannii strains in exponential growth. J Med Microbiol 66:1257–1260. 10.1099/jmm.0.000542. [DOI] [PubMed] [Google Scholar]
- 17.Chung ES, Ko KS. 2019. Eradication of persister cells of Acinetobacter baumannii through combination of colistin and amikacin antibiotics. J Antimicrob Chemother 74:1277–1283. 10.1093/jac/dkz034. [DOI] [PubMed] [Google Scholar]
- 18.Ntagiopoulos PG, Paramythiotou E, Antoniadou A, Giamarellou H, Karabinis A. 2007. Impact of an antibiotic restriction policy on the antibiotic resistance patterns of Gram-negative microorganisms in an intensive care unit in Greece. Int J Antimicrob Agents 30:360–365. 10.1016/j.ijantimicag.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother 56:5642–5649. 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 21.Guffanti AA, Clejan S, Falk LH, Hicks DB, Krulwich TA. 1987. Isolation and characterization of uncoupler-resistant mutants of Bacillus subtilis. J Bacteriol 169:4469–4478. 10.1128/jb.169.10.4469-4478.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shvinka JE, Toma MK, Galinina NI, Skards IV, Viesturs UE. 1979. Production of superoxide radicals during bacterial respiration. Microbiology 113:377–382. 10.1099/00221287-113-2-377. [DOI] [Google Scholar]
- 23.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Wang C, Yan B, Lyu L, Takiff HE, Gao Q. 2020. The potassium transporter KdpA affects persister formation by regulating ATP levels in Mycobacterium marinum. Emerg Microbes Infect 9:129–139. 10.1080/22221751.2019.1710090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhagirath AY, Li Y, Patidar R, Yerex K, Ma X, Kumar A, Duan K. 2019. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int J Mol Sci 20:1781. 10.3390/ijms20071781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra-Millán R, Vila-Farrés X, Ayerbe-Algaba R, Varese M, Sánchez-Encinales V, Bayó N, Pachón-Ibáñez ME, Teixidó M, Vila J, Pachón J, Giralt E, Smani Y. 2018. Synergistic activity of an OmpA inhibitor and colistin against colistin-resistant Acinetobacter baumannii: mechanistic analysis and in vivo efficacy. J Antimicrob Chemother 73:3405–3412. 10.1093/jac/dky343. [DOI] [PubMed] [Google Scholar]
- 27.Catel-Ferreira M, Marti S, Guillon L, Jara L, Coadou G, Molle V, Bouffartigues E, Bou G, Shalk I, Jouenne T, Vila-Farrés X, Dé E. 2016. The outer membrane porin OmpW of Acinetobacter baumannii is involved in iron uptake and colistin binding. FEBS Lett 590:224–231. 10.1002/1873-3468.12050. [DOI] [PubMed] [Google Scholar]
- 28.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chishti AA, Hellweg CE, Berger T, Baumstark-Khan C, Feles S, Kätzel T, Reitz G. 2015. Constitutive expression of tdTomato protein as a cytotoxicity and proliferation marker for space radiation biology. Life Sci Space Res (Amst) 4:35–45. 10.1016/j.lssr.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Iizuka R, Yamagishi-Shirasaki M, Funatsu T. 2011. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem 414:173–178. 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch AL, Levy HR. 1955. Protein turnover in growing cultures of Escherichia coli. J Biol Chem 217:947–958. 10.1016/S0021-9258(18)65958-7. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538. 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia A, Han J, Jin Z, Ni L, Yang S, Jin F. 2018. Dual-color fluorescent timer enables detection of growth-arrested pathogenic bacterium. ACS Infect Dis 4:1666–1670. 10.1021/acsinfecdis.8b00129. [DOI] [PubMed] [Google Scholar]
- 36.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higuchi Y. 2003. Chromosomal DNA fragmentation in apoptosis and necrosis induced by oxidative stress. Biochem Pharmacol 66:1527–1535. 10.1016/S0006-2952(03)00508-2. [DOI] [PubMed] [Google Scholar]
- 38.Moreillon P, Tomasz A. 1988. Penicillin resistance and defective lysis in clinical isolates of pneumococci: evidence for two kinds of antibiotic pressure operating in the clinical environment. J Infect Dis 157:1150–1157. 10.1093/infdis/157.6.1150. [DOI] [PubMed] [Google Scholar]
- 39.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Webster TJ. 2018. Bacteria antibiotic resistance: new challenges and opportunities for implant‐associated orthopedic infections. J Orthop Res 36:22–32. 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mordukhova EA, Pan JG. 2014. Stabilization of homoserine-O-succinyltransferase (MetA) decreases the frequency of persisters in Escherichia coli under stressful conditions. PLoS One 9:e110504. 10.1371/journal.pone.0110504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami K, Ono T, Viducic D, Kayama S, Mori M, Hirota K, Nemoto K, Miyake Y. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol Lett 242:161–167. 10.1016/j.femsle.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Vulić M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michiels JE, Van den Bergh B, Verstraeten N, Michiels J. 2016. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist Updat 29:76–89. 10.1016/j.drup.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Korch SB, Hill TM. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol 188:3826–3836. 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell P. 1966. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc 41:445–502. 10.1111/j.1469-185x.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 47.Bakker EP, Mangerich WE. 1981. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol 147:820–826. 10.1128/JB.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hicks DB, Cohen DM, Krulwich TA. 1994. Reconstitution of energy-linked activities of the solubilized F1F0 ATP synthase from Bacillus subtilis. J Bacteriol 176:4192–4195. 10.1128/jb.176.13.4192-4195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farha MA, French S, Stokes JM, Brown ED. 2018. Bicarbonate alters bacterial susceptibility to antibiotics by targeting the proton motive force. ACS Infect Dis 4:382–390. 10.1021/acsinfecdis.7b00194. [DOI] [PubMed] [Google Scholar]
- 50.Blair JM, Richmond GE, Piddock LJ. 2014. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol 9:1165–1177. 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 51.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7:642–653. 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 52.Hu Z, Lutkenhaus J. 2003. A conserved sequence at the C‐terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47:345–355. 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- 53.Edelmann D, Berghoff BA. 2019. Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci Rep 9:14256. 10.1038/s41598-019-50668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ighodaro OM, Akinloye OA. 2018. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): their fundamental role in the entire antioxidant defence grid. Alex J Med 54:287–293. 10.1016/j.ajme.2017.09.001. [DOI] [Google Scholar]
- 55.Kaur A, Sharma P, Capalash N. 2018. Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci Rep 8:11029. 10.1038/s41598-018-29291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. 2010. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol 45:171–184. 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrett TC, Mok WWK, Murawski AM, Brynildsen MP. 2019. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun 10:1177. 10.1038/s41467-019-09058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khameneh B, Bazzaz BSF, Amani A, Rostami J, Vahdati-Mashhadian N. 2016. Combination of anti-tuberculosis drugs with vitamin C or NAC against different Staphylococcus aureus and Mycobacterium tuberculosis strains. Microb Pathog 93:83–87. 10.1016/j.micpath.2015.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.