Certain methicillin-resistant Staphylococcus aureus (MRSA) strains exhibit β-lactam susceptibility in vitro, ex vivo, and in vivo in the presence of NaHCO3 (NaHCO3-responsive MRSA). Here, we investigate the impact of NaHCO3 on factors required for PBP2a functionality.
KEYWORDS: methicillin-resistant Staphylococcus aureus (MRSA), sodium bicarbonate (NaHCO3), beta-lactam, penicillin-binding proteins (PBP), PBP2a
ABSTRACT
Certain methicillin-resistant Staphylococcus aureus (MRSA) strains exhibit β-lactam susceptibility in vitro, ex vivo, and in vivo in the presence of NaHCO3 (NaHCO3-responsive MRSA). Here, we investigate the impact of NaHCO3 on factors required for PBP2a functionality. Prototype NaHCO3-responsive and -nonresponsive MRSA strains (as defined in vitro) were assessed for the impact of NaHCO3 on the expression of genes involved in PBP2a production-maturation pathways (mecA, blaZ, pbp4, vraSR, prsA, sigB, and floA), membrane PBP2a and PrsA protein content, and membrane carotenoid content. Following NaHCO3 exposure in NaHCO3-responsive (versus nonresponsive) MRSA, there was significantly reduced expression of (i) mecA and blaZ, (ii) the vraSR-prsA gene axis, and (iii) pbp4. Carotenoid production was reduced while floA expression was increased by NaHCO3 exposure in all MRSA strains. This work underscores the distinct regulatory impact of NaHCO3 on a cadre of genes encoding factors required for the maintenance of the MRSA phenotype through PBP2a functionality and maturation.
INTRODUCTION
Staphylococcus aureus is the major causative agent of a number of serious clinical syndromes and a notable public health threat (1–3). Methicillin-resistant S. aureus (MRSA) has been a particular problem due, in part, to its assumed recalcitrance to many first-line therapies useful in the treatment of methicillin-susceptible S. aureus (MSSA) strains, especially β-lactam agents (4). Current anti-MRSA therapies tend to be costlier, less effective, and/or more toxic than those used to treat MSSA infections (e.g., daptomycin, linezolid, and vancomycin [5, 6]); also, patients with MRSA infections are prone to prolonged hospital stays, imposing large economic burdens for their treatment (7).
MRSA strains are preemptively identified in most clinical microbiology laboratories by phenotypic assays (e.g., cefoxitin disk diffusion or PBP2a latex agglutination) (8), as well as by the presence of the mecA gene cassette, encoding the alternative penicillin-binding protein 2a (PBP2a). Additionally, minimum inhibitory concentration (MIC) determinations, as outlined by the Clinical and Laboratory Standards Institute (CLSI), are used to confirm their resistance to standard β-lactam antibiotics (i.e., oxacillin) (9, 10). The CLSI considers S. aureus isolates whose MICs to oxacillin are ≥4 μg/ml in cation-adjusted Mueller-Hinton broth (CA-MHB) to be oxacillin resistant (and resistant to first-generation cephalosporins) (4, 9); the treatment of such strains with all β-lactam agents (excluding ceftaroline and ceftobiprole) is discouraged by published guidelines (4, 9).
We recently discovered that the addition of NaHCO3, the body’s primary biological buffer, to CA-MHB rendered a relatively large proportion of MRSA strains susceptible to two standard β-lactams, oxacillin and cefazolin, by MIC testing; this phenotype has been termed NaHCO3 responsiveness (11, 12). The translational relevance of this NaHCO3-responsive phenotype was verified in a small strain set by successful β-lactam therapy in an ex vivo simulated endocarditis vegetation model as well as in a rabbit model of infective endocarditis (11, 13).
Prior studies suggested that NaHCO3 exerted its impact on antimicrobial susceptibility by targeting and collapsing the proton motive force (PMF) (14); however, this would not adequately explain β-lactam resensitization by NaHCO3. Instead, we recently determined that coexposure of these in vitro-responsive MRSA strains to NaHCO3 plus oxacillin reduced the expression of both mecA and sarA, associated with reduced PBP2a protein production (11). The latter observations were important, as oxacillin binds relatively poorly to PBP2a (encoded by mecA), and deletion of sarA can increase susceptibility to oxacillin, in part, by influencing mecA expression (15).
The above-described finding of NaHCO3-mediated reduction of sarA expression suggested an additional mechanism by which NaHCO3 could influence β-lactam sensitization in MRSA, i.e., via an impact on cell membrane (CM) physiology and PBP2a maturation. The expression of sarA is regulated by the alternative sigma factor, SigB (16), which, in turn, controls the staphyloxanthin biosynthesis operon, crtOPQMN (17). The staphyloxanthin carotenoid pigment, along with flotillin (encoded by floA), are integral parts of functional membrane microdomains (FMMs), which, importantly, provide a scaffolding that anchors PBP2/2a proteins within the MRSA CM (18, 19).
Defining the potential impacts of NaHCO3 on other key aspects of PBP2a regulation (e.g., blaZ expression), maturation (e.g., vraSR and prsA expression), and peptidoglycan cross-linking (e.g., pbp4) is critical to developing a more complete understanding of bicarbonate’s effect on oxacillin susceptibility. Thus, genes within the blaZ operon coregulate mecA expression (20). Of note, VraSR and PrsA have been identified as critical to the see-saw effect, in which daptomycin-nonsusceptible MRSA strains exhibit β-lactam hypersusceptibility (21–23). VraSR is a two-component regulatory system that positively regulates the expression of PrsA, a chaperone required for proper PBP2a maturation, localization, and functionality (24, 25). Moreover, PBP4 is a PBP with transpeptidase activity required to complete peptidoglycan cross-linking in the presence of oxacillin (26, 27).
The current study was designed to adjudicate the above-described preliminary findings in a larger collection of NaHCO3-responsive and -nonresponsive MRSA strains as well as to further define key genetic determinants of these two distinct microbiologic phenotypes.
(These data were presented, in part, at the 4th Annual Texas Medical Center Antimicrobial Resistance and Stewardship Conference [28].)
RESULTS
Impact of NaHCO3 on mecA and blaZ expression.
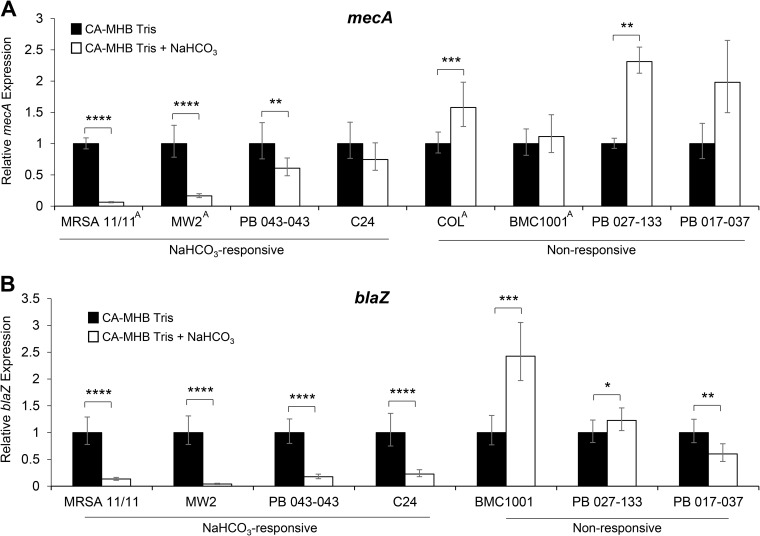
The blaZ operon is important in coregulating mecA expression and PBP2a production (20). As shown in Fig. 1, NaHCO3 exposure substantially decreased mecA gene expression under oxacillin-inducing conditions in all four NaHCO3-responsive strains. In contrast, mecA expression was increased during NaHCO3 exposure in the four nonresponsive strains (Fig. 1A). Similarly, blaZ expression was significantly reduced in all four NaHCO3-responsive strains while being significantly increased in two of the three β-lactamase-positive, NaHCO3-nonresponsive strains (COL is naturally β-lactamase negative) (Fig. 1B).
FIG 1.
Expression of mecA (A) and blaZ (B) in the presence and absence of NaHCO3. qRT-PCR was performed using RNA extracted from stationary-phase cells in the presence of 0.5× MIC of oxacillin. Oxacillin concentrations used are the following: MRSA 11/11, without NaHCO3 (w/o), 16 μg/ml; with NaHCO3 (+ NaHCO3), 0.25 μg/ml; MW2, w/o, 32 μg/ml; + NaHCO3, 1 μg/ml; PB 043-043, w/o, 8 μg/ml; + NaHCO3, 0.5 μg/ml; C24, w/o, 16 μg/ml; + NaHCO3, 0.5 μg/ml; COL, w/o, 256 μg/ml; + NaHCO3, 256 μg/ml; BMC1001, w/o, 128 μg/ml; + NaHCO3, 128 μg/ml; PB 027-133, w/o, 128 μg/ml; + NaHCO3, 128 μg/ml; PB 017-037, w/o, 16 μg/ml; + NaHCO3, 16 μg/ml. Black bars indicate growth in CA-MHB 100 mM Tris, white bars indicate growth in CA-MHB 100 mM Tris plus 44 mM NaHCO3, and error bars indicate the CT standard deviations. No blaZ expression was detected in COL, because the strain lacks the blaZ gene. Student’s t test was used for statistical analyses. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. mecA gene expression data for strains marked with a superscript A were previously published (11).
Influence of NaHCO3 on PBP2a content.
As previously observed in a smaller set of MRSA strains (11), growth in media containing NaHCO3 reduced the amount of PBP2a in NaHCO3-responsive versus nonresponsive strains (Table 1; see also Fig. S2 in the supplemental material).
TABLE 1.
PBP2a agglutination of NaHCO3-responsive and nonresponsive strains grown in medium with or without 44 mM NaHCO3
| PBP2a agglutination levela | ||
|---|---|---|
| Strain | CA-MHB Tris | CA-MHB Tris 44 mM NaHCO3 |
| MRSA 11/11b | ++ | − |
| MW2b | ++ | − |
| PB 043-043 | +++ | + |
| C24 | +++ | + |
| COLb | +++ | +++ |
| BMC1001b | +++ | +++ |
| PB 027-133 | +++ | +++ |
| PB 017-037 | +++ | +++ |
Interpretation of agglutination intensity: +++, high; ++, moderate; +, low; −, none.
Data were previously reported (11).
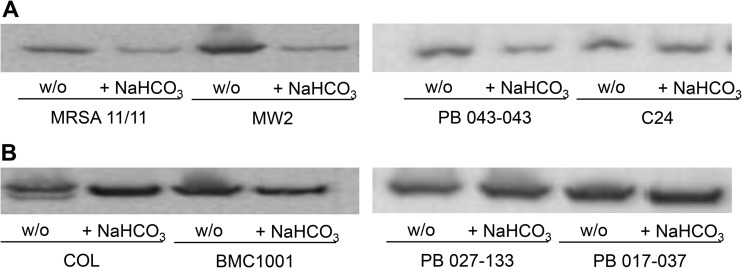
To further verify the impact of NaHCO3 on PBP2a protein production and its CM localization, Western blotting was performed on CM protein extracts from cells grown in the presence of 0.5× MIC of oxacillin (to induce mecA expression), with or without NaHCO3 (Fig. 2). As predicted by the known NaHCO3 repression of the coregulatory mecA-blaZ gene axis expression in NaHCO3-responsive strains, CM-specific PBP2a protein production was visibly decreased in the presence of NaHCO3 in three of four NaHCO3-responsive strains (Fig. 2A). In contrast, NaHCO3 exposure had no impact on PBP2a production in any of the NaHCO3-nonresponsive strains (Fig. 2B). These data indicated that not only was total PBP2a production downregulated in NaHCO3-responsive strains (as per the agglutination assay) but also the amount of protein specifically inserted into the CM was similarly impacted.
FIG 2.
Membrane PBP2a protein content in MRSA strains grown in the presence and absence of NaHCO3. (A) NaHCO3-responsive strains. (B) NaHCO3-nonresponsive strains. Strains were grown in CA-MHB plus 100 mM Tris (w/o) or CA-MHB plus 100 mM Tris plus 44 mM NaHCO3 (+ NaHCO3) in the presence of 0.5× MIC oxacillin. Proteins are the cell membrane fraction extracted from stationary-phase cells.
Effect of NaHCO3 on genes involved in PBP2a maturation and localization.
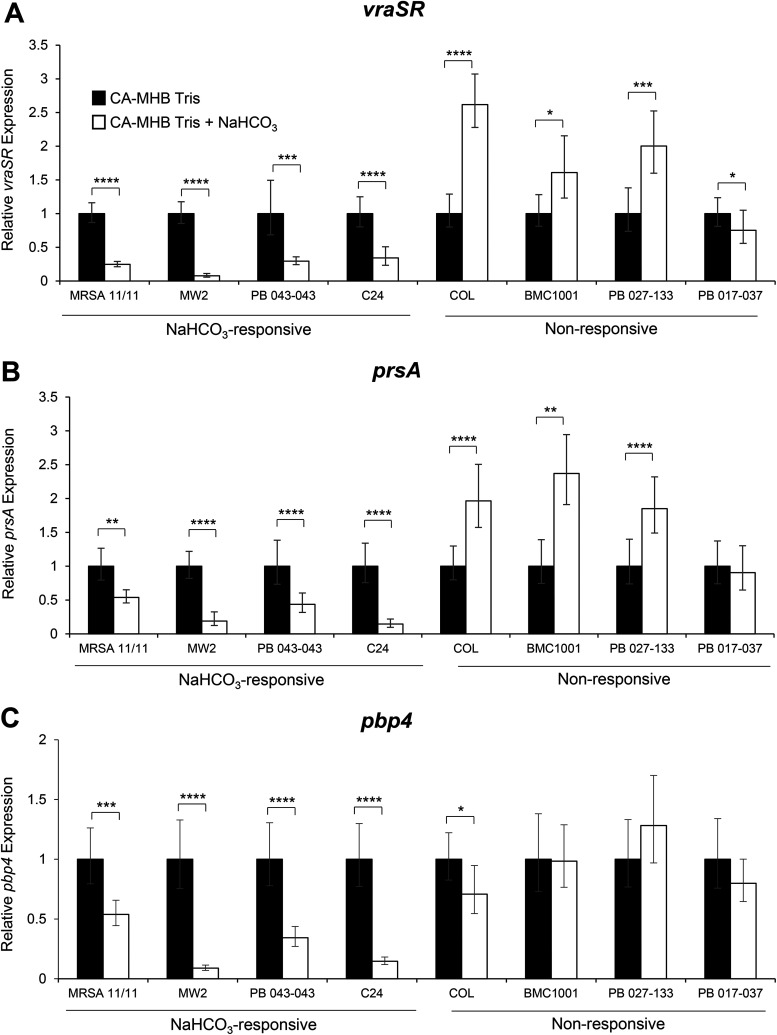
For optimal PBP2a protein functionality, the PrsA chaperone must facilitate posttranslational maturation of PBP2a (21, 25). Regulation of prsA gene expression typically occurs via VraSR, a two-component regulatory system known to be involved in the daptomycin resistance/oxacillin hypersusceptibility see-saw effect (21, 23). Interestingly, NaHCO3 highly repressed expression of both vraSR and prsA only in NaHCO3-responsive strains (Fig. 3A and B).
FIG 3.
Expression of genes related to PBP2a maturation and function in the presence and absence of NaHCO3. (A) vraSR. (B) prsA. (C) pbp4. qRT-PCR was performed using RNA extracted from log-phase cells (OD600, 0.4) in the presence of 0.5× MIC oxacillin for vraSR and prsA or on RNA extracted from stationary-phase cells (pbp4). Black bars indicate growth in CA-MHB with 100 mM Tris, white bars indicate growth in CA-MHB with 100 mM Tris plus 44 mM NaHCO3 for all graphs, and error bars indicate the CT standard deviations. Student’s t test was used for statistical analyses. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
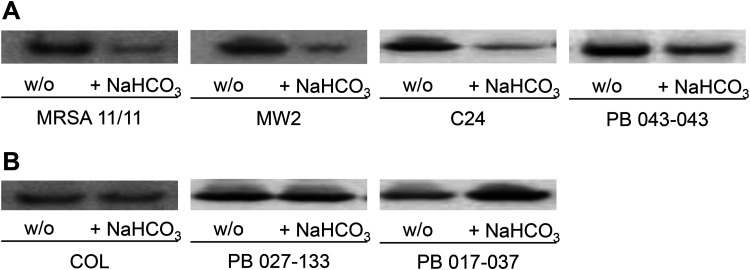
Furthermore, as predicted by the quantitative real-time PCR (qRT-PCR) gene expression data described above, less PrsA protein was present in the CMs of all NaHCO3-responsive MRSA following NaHCO3 exposure versus cells grown in the absence of NaHCO3 (Fig. 4A). In contrast, NaHCO3 had no apparent effect on PrsA CM content in three NaHCO3-nonresponsive strains (Fig. 4B). NaHCO3 did not consistently impact extracellular secretion patterns of PrsA in either NaHCO3-responsive or -nonresponsive MRSA (data not shown). These data support the notion that in the presence of oxacillin induction, NaHCO3 reduces overall PrsA production in NaHCO3-responsive MRSA, resulting in decreased PrsA CM content (as opposed to excess PrsA secretion extracellularly, as seen in the see-saw effect [21]).
FIG 4.
Membrane PrsA protein content in MRSA strains grown in the presence and absence of NaHCO3. (A) NaHCO3-responsive strains. (B) Nonresponsive strains. Strains were grown in CA-MHB plus 100 mM Tris (w/o) or CA-MHB plus 100 mM Tris plus 44 mM NaHCO3 (+ NaHCO3) in the presence of 0.5× MIC oxacillin. Proteins are the membrane fraction extracted from log-phase cells.
pbp4 expression.
In addition to proper protein folding, the PBP2a protein requires PBP4 activity to complete cell wall synthesis (26). Although PBP2a provides the transpeptidase activity involved in peptidoglycan synthesis, highly cross-linked peptidoglycan cannot be produced in many MRSA strains in the presence of oxacillin without PBP4 (18, 26, 29). The qRT-PCR analysis revealed that pbp4 expression was substantially and selectively repressed in the presence of NaHCO3 in NaHCO3-responsive (but not in NaHCO3-nonresponsive) strains (Fig. 3C); this suggested that a less highly cross-linked peptidoglycan is produced in NaHCO3-responsive strains in the presence of NaHCO3.
Influence of NaHCO3 on factors impacting FMM scaffolding for PBP2a.
The alternative sigma factor and stress response regulator, SigB, is involved in regulating genes involved in CM biophysics (e.g., fluidity/rigidity), notably the staphyloxanthin carotenoid biosynthesis operon, crtOPQMN (17). In turn, carotenoids and flotillin (FloA) are critical in providing a key scaffolding upon which PBP2a can oligomerize (19). Additionally, the global regulon sarA is also SigB regulated and is important in maintaining the MRSA phenotype (15, 16, 30). Further, as noted above, we previously determined that NaHCO3 was capable of repressing sarA expression in NaHCO3-responsive strains (11). Thus, we hypothesized that NaHCO3 influences SigB activity, resulting in downstream impacts on SigB-regulated phenotypes important in PBP2a functionality.
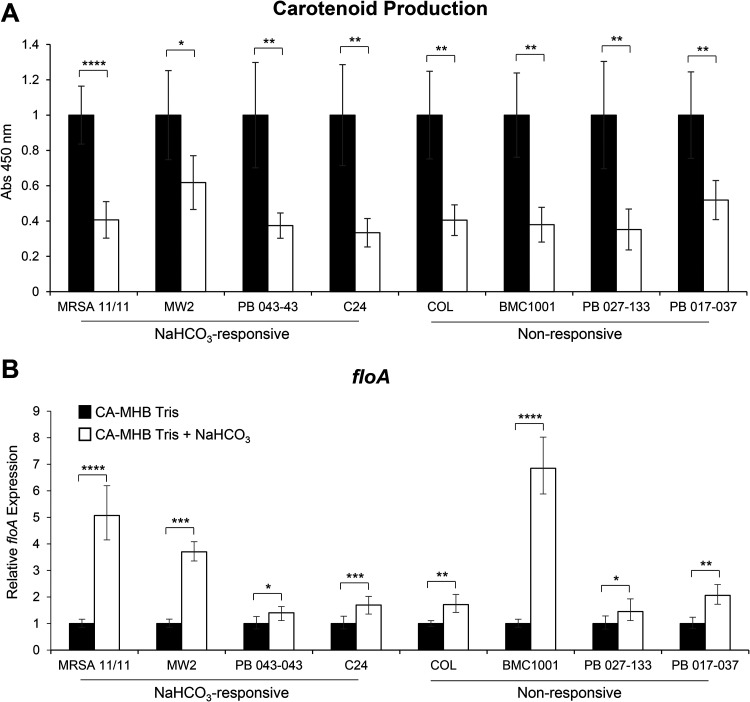
The expression of the constitutively active sigB gene was assessed through expression of asp23, a surrogate reporter for sigB promoter activity (31). Interestingly, in both NaHCO3-responsive and -nonresponsive strains, asp23 expression was substantially and uniformly repressed in the presence of NaHCO3 (data not shown). To confirm the phenotypic outcome of such globally repressed SigB activity, carotenoid content was measured in NaHCO3-responsive and -nonresponsive strains and was found to be universally reduced by NaHCO3 exposures (Fig. 5A).
FIG 5.
Impact of NaHCO3 on FMM-associated factors. (A and B) Carotenoid production (A) and floA gene expression (B) in MRSA strains grown in the presence and absence of NaHCO3. floA gene expression was assessed by qRT-PCR using RNA extracted from log-phase cells (OD600, 0.4) in the presence of 0.5× MIC oxacillin. Black bars indicate growth in CA-MHB with 100 mM Tris, white bars indicate growth in CA-MHB with 100 mM Tris plus 44 mM NaHCO3 for all graphs, and error bars indicate the CT standard deviations. Student’s t test was used for statistical analyses. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Since carotenoids interact with flotillin within the CM to form FMMs (19), we determined whether NaHCO3 also impacts the flotillin component of FMMs by assessing the expression of floA. Interestingly, floA gene expression was increased in all strains in the presence of NaHCO3 (Fig. 5B). Combined with the reduced carotenoid content data described above, the latter data indicate that NaHCO3 is inducing a proportionality disequilibrium in the formation of stable FMMs (i.e, reduced carotenoids plus increased floA expression).
DISCUSSION
Previously, we identified a novel MRSA phenomenon, termed NaHCO3 responsiveness, wherein a relatively large proportion of MRSA strains displayed increased susceptibility to standard β-lactam antibiotics in the presence of NaHCO3 (11, 12). Thus, ∼75% and ∼33% of a collection of 58 recent MRSA bloodstream isolates were significantly more susceptible in vitro to cefazolin and oxacillin, respectively, in the presence versus absence of NaHCO3 (12). We also linked this responsiveness phenomenon to the reduced expression of two genes involved in the maintenance of the MRSA phenotype, mecA and sarA (11). These genetic impacts translated into reduced PBP2a production in such NaHCO3-responsive MRSA in the presence of NaHCO3.
In the present study, we further investigated additional mechanisms potentially involved in NaHCO3–β-lactam responsiveness, in eight representative strains, focusing on the impact of NaHCO3 upon both mecA-blaZ coregulation and key genes involved in the maturation functionality of PBP2a; each of these events is critical in determining ultimate MRSA β-lactam resistance.
Several interesting observations emerged from these investigations. First, in the presence of oxacillin induction, NaHCO3 substantially reduced the expression of both mecA and blaZ only in NaHCO3-responsive strains. Thus, it seems clear that in NaHCO3-responsive strains, NaHCO3 impacts both limbs of the mecA-blaZ coregulatory axis important in the maintenance of the MRSA phenotype (20). Also, our prior studies, utilizing a latex agglutination assay, showed that NaHCO3 could reduce overall production of PBP2a in NaHCO3-responsive versus -nonresponsive MRSA (11). Similar PBP2a production differences were confirmed using this same assay in additional strains as well as with a more precise Western blot assay for CM localization of this PBP.
Second, we established that NaHCO3 exposure in vitro had a rather profound dampening effect on the expression of several key genes involved in PBP2a maturation. For example, the two-component regulatory system, VraSR, and the VraSR-regulated chaperone, PrsA, are known to work in concert; dysfunction in this circuit appears to underlie the see-saw effect, wherein daptomycin-resistant MRSA strains become hypersusceptible to β-lactams (21, 22). Interestingly, such see-saw strains actually demonstrate increased expression of both vraSR and prsA as well as the accumulation of mutations in mprF (21, 23). The collateral effects of the latter genetic changes are the decreased production of PrsA in the CM and concomitant increased extracellular secretion of PrsA, with a resultant decreased localization of PBP2a in the CM (21). In contrast, in NaHCO3-responsive MRSA, there appears to be a totally different mechanism involved in β-lactam hypersusceptibility. Thus, we did not observe excess secretion of PrsA in NaHCO3-responsive strains that could explain the decreased PrsA levels within the CM. These results suggest that NaHCO3 is not directly affecting PrsA functionality but rather more directly affecting overall PrsA protein levels. NaHCO3 reduces the expression of both vraSR and prsA, thereby decreasing the overall production and CM localization of the PrsA chaperone, potentially impairing the ultimate maturation and functionality of PBP2a within the CM.
Third, NaHCO3 reduced pbp4 expression selectively in NaHCO3-responsive strains. Importantly, pbp4 is a nonessential PBP that functions as an auxiliary transpeptidase during cell wall synthesis (32). Of relevance to the present study, deletion of pbp4 in certain community-acquired MRSA strains (e.g., USA300 and MW2, both NaHCO3 responsive) resulted in a 16-fold increased susceptibility to oxacillin, while this deletion had no effect on oxacillin MICs in strain COL (NaHCO3 nonresponsive) (26). Further, in USA300 and MW2, pbp4 mutants exposed to oxacillin exhibited substantially less highly cross-linked cell wall peptidoglycan, indicating a cooperative action between PBP2a and PBP4 in cell wall synthesis during β-lactam exposure. As demonstrated by our data, NaHCO3 reduces both the transcription of pbp4 and overall production of PBP2a, which likely leads to a net decrease in highly cross-linked peptidoglycan in NaHCO3-responsive strains. Although little is known about the regulation of pbp4 expression (33), deletion of pbp4 results in a decrease in pbp2 transcription (26). Recent analyses of the transcriptomes of NaHCO3-responsive strains by RNA sequencing from our laboratory confirm that NaHCO3 downregulates pbp2 expression (unpublished data); this suggests that NaHCO3 is acting on an upstream regulator of both pbp2 and pbp4. Interestingly, PBP2 is one of only three known enzymes with transglycosylase activity in S. aureus (34), indicating that NaHCO3 has additional impacts on peptidoglycan transglycosylation in responsive MRSA strains. Further work is needed to understand the specific impact of NaHCO3 on the regulation of the overall cadre of genes involved in peptidoglycan biosynthesis as well as the impact of NaHCO3 on the abundance of highly cross-linked peptidoglycan muropeptides within the MRSA cell wall. These investigations are ongoing in our laboratories.
Lastly, in addition to strain-specific and selective impacts described above in NaHCO3-responsive versus -nonresponsive MRSA, NaHCO3 also had several, more global effects in all eight prototype strains that could impact CM physiology. For example, NaHCO3 reduced the activity of the global stress response regulator, SigB (35, 36), in all strains studied. In S. aureus, SigB upregulates the expression of the operon crtOPQMN, which is responsible for the biosynthesis of the triterpenoid carotenoid pigment staphyloxanthin (17, 37). Staphyloxanthin, in turn, is a crucial factor in the formation of FMMs, which create the scaffolding for oligomerization of PBP2a within the CM (19). Interestingly, disruption of staphyloxanthin synthesis by clinical statin agents in S. aureus leads to increased susceptibility to β-lactams (19). As expected, based on the NaHCO3 repression of SigB activity, carotenoid production was concomitantly reduced in all strains tested. Conversely, NaHCO3 exposure resulted in increased floA expression in all strains tested, highlighting the possibility that NaHCO3 is disrupting the normal equilibrium of two key factors necessary for stable FMM formation (i.e., relative proportional contents of carotenoids and FloA). This disequilibrium could contribute to the NaHCO3-responsive phenotype when other observed alterations in PBP2a maturation are also present.
There are several limitations to our investigations. For example, we only studied a relatively small number of MRSA strains (n = 8). Additionally, we carefully selected only a specific cadre of genes to study that have been well characterized as to their known impacts on PBP2a regulation, production, and functionality. Further, we have not yet investigated how changes in gene expression impact cell wall morphology and FMM composition/organization. Moreover, the more global genetic impacts of NaHCO3 in MRSA await more extensive analyses, such as whole-genome sequencing and RNA sequencing, studies that are in progress. Finally, genetic swapping studies between NaHCO3-responsive and -nonresponsive strains will be required to more precisely define the role of specific PBP2a genotype species in the β-lactam sensitization phenomenon related to NaHCO3 exposures (38). Importantly, understanding the mechanism(s) of NaHCO3 responsiveness may help guide future clinical microbiologic laboratory screening strategies as well as the repurposing of treatment practices for MRSA infections, resulting in improved patient outcomes using β-lactam-based therapies.
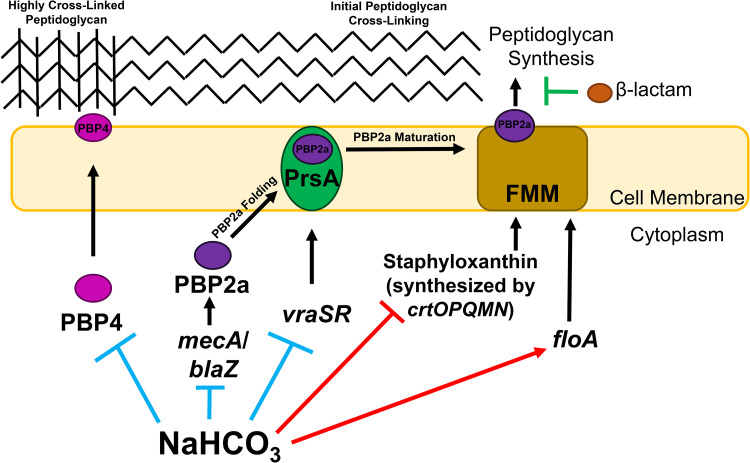
Figure 6 represents one proposed pathway for the overall impact of NaHCO3 on key MRSA genotypes and phenotypes, based on our current investigations.
FIG 6.
Schematic overview of the impact of NaHCO3 on PBP2a maturation/function and peptidoglycan biosynthesis. Impacts specific to NaHCO3-responsive strains (blue inhibition symbols) include the repression of PBP2a protein production, pbp4 gene expression, vraSR gene expression, and PrsA protein production. Impacts of NaHCO3 on all strains tested include repression of carotenoid production (red inhibition symbol) and increased floA gene expression (red enhancement symbol), potentially resulting in less stable functional membrane microdomains (FMMs). Anticipated collateral impact on NaHCO3-responsive strains is an overall decrease in PBP2a mediated peptidoglycan synthesis and the generation of highly cross-linked peptidoglycan, resulting in increased susceptibility to β-lactam antibiotics.
In summary, NaHCO3 was found to have profound effects on PBP2a that were specific to NaHCO3-responsive strains (e.g., mecA and blaZ expression, PBP2a protein production and CM localization, and expression of genes involved in PBP2a maturation and functionality). In contrast, NaHCO3 impacted certain genotypes-phenotypes more globally in both NaHCO3-responsive and -nonresponsive MRSA (i.e., SigB activity, downstream carotenoid production, and floA expression). The latter data indicate that certain NaHCO3-mediated impacts are themselves involved and necessary in the collective NaHCO3-responsive outcome in MRSA, but they are not individually sufficient to cause this overall phenotype without additional perturbations in PBP2a production, maturation, and functionality (Fig. 6).
MATERIALS AND METHODS
Bacterial strains and media.
All MRSA strains used in this study were initially isolated from patients with invasive clinical infections: MRSA 11/11, MW2, PB 043-043, C24, COL, BMC1001, PB 027-133, and PB 017-037 (12, 22, 39–41). Four of these strains (MRSA 11/11, MW2, BMC1001, and COL) were selected based on their inclusion in a prior study on the NaHCO3-responsive phenomenon (11). The other four strains (PB 043-043, C24, PB 027-133, and PB 017-037) were selected from a larger screen of NaHCO3-responsive MRSA (12) based on their oxacillin and cefazolin MICs in the presence/absence of NaHCO3 and their clonal complex (CC) types. The CC types of 7/8 isolates represent genotypes in current widespread clinical circulation (i.e., CC types 5 and 8) (see Table S1 in the supplemental material) (42). MRSA 11/11, MW2, PB 043-043, C24, BMC1001, PB 027-133, and PB 017-037 are all β-lactamase positive; COL is naturally β-lactamase negative. MICs were determined, in the presence or absence of NaHCO3, by standard CLSI methods (Table S1). MICs for MRSA 11/11, MW2, COL, and BMC1001 have been previously reported (11).
For all experiments, strains were grown in ambient air at 37°C in cation-adjusted Mueller-Hinton Broth (CA-MHB; Difco) plus 100 mM Tris (to stabilize the pH at ∼7.3 ± 0.1) with or without 44 mM NaHCO3 (the optimal concentration to disclose the NaHCO3-responsive phenotype in vitro and ex vivo, reflective of deep-tissue HCO3 levels [11–13, 43]). For oxacillin exposure experiments, 0.5× MIC oxacillin was used for a given strain in the specified growth medium, with 2% NaCl incorporated into testing media (Table S1).
MIC assays.
The MICs of oxacillin were determined by broth microdilution according to CLSI guidelines in specified media, as previously described (9, 11, 44). All MICs are the mode of at least six independent determinations.
PBP2a latex agglutination assays.
A semiquantitative, rapid, and reliable latex agglutination method (Seiken, Tokyo, Japan) was used to approximate total PBP2a production, as previously described (11). Agglutination results were scored as high (+++), moderate (++), low (+), or negative (−) based on the presence or absence of an overt agglutination pattern, corresponding to the total amount of PBP2a in the sample. S. aureus ATCC 43300 (MRSA; PBP2a positive) and ATCC 25923 (MSSA; PBP2a negative) were used as positive and negative controls, respectively, in all assays. The grading of agglutination assays was performed blindly as to strain identifications by one of the investigators (S. C. Ersoy).
Membrane (CM) protein extraction.
Cells were grown to stationary phase (PBP2a) or log phase (optical density at 600 nm [OD600], 0.5; for PrsA) in CA-MHB plus 100 mM Tris plus 0.5× MIC oxacillin, with or without 44 mM NaHCO3 (Table S1), and their centrifuged pellets were then resuspended in phosphate-buffered saline (PBS), pH 7.3. Thereafter, 1 ml of resuspended pellets was incubated with 10 μl each of DNase (Ambion, Invitrogen), RNase (Thermo Fisher Scientific), and Halt protease inhibitor cocktail (Thermo Fisher Scientific) for 30 min at 37°C. Cells were then mixed with glass beads and disrupted using a FastPrep cell disrupter. Disrupted cells were centrifuged for 20 min at 4°C and 13,000 rpm, and the supernatant was ultracentrifuged for 2 h at 4°C and 15,000 rpm to collect total CM proteins. The CM protein pellets were resuspended in PBS, and protein concentrations were quantified by Bradford protein assay. Isolated protein extracts were stored at −80°C.
Western blotting for PBP2a and PrsA protein expression.
Forty micrograms of CM protein extract was separated on a 4 to 12% Bis-Tris gel (Invitrogen) and then blotted onto a nitrocellulose membrane (Amersham). Total protein loading was confirmed by staining with 0.25% Ponceau reagent (Fig. S1). The membrane was blocked with 10% dry milk in Tris-buffered saline with Tween (TBST). PBP2a was probed with a chicken anti-PBP2a antibody (RayBiotech) diluted 1:2,500 and detected with an anti-chicken IgY cross-absorbed secondary antibody, horseradish peroxidase (Thermo Fisher Scientific), diluted 1:5,000. In parallel studies, PrsA was probed with a chicken anti-PrsA antibody diluted 1:2,500 (21). Labeled proteins were imaged using a c400 imager (Azure Biosystems). Western blotting for PrsA in strain BMC1001 could not be performed due to a single-nucleotide deletion within its prsA coding region, causing a frameshift mutation resulting in a premature stop codon (prsA g. 796_798del; p. L272X).
Isolation of RNA and qRT-PCR analysis.
RNA was isolated from strains as detailed previously (11). Quantitative real-time PCR (qRT-PCR) was performed using primers for blaZ, prsA, vrsASR, pbp4, floA, mecA, and gyrB as previously described (15, 25, 45–47); these primers are listed in Table S2. The gyrB gene was used as a housekeeping gene for transcript normalization (verification of the adequacy of gyrB as a reproducible control for these assays was independently done using rpoB in parallel experiments [data not shown]) (48). Relative gene expression was calculated using the 2−ΔΔCT method from two independent biological replicates performed in triplicate on at least 2 separate runs for each strain/condition. Data are presented as the relative gene expression in CA-MHB plus 100 mM Tris plus 44 mM NaHCO3 normalized to the relative gene expression in CA-MHB plus 100 mM Tris for each strain, where expression in CA-MHB 100 mM Tris was set to 1.0. A Student’s t test was used for all statistical analyses.
Carotenoid production assay.
To quantify relative levels of staphyloxanthin production, carotenoids were isolated by methanol extraction as previously described (37). Briefly, cells were grown overnight in specified medium, pelleted, and washed twice with PBS. Methanol was added to pellets at a volume that was normalized to total cell weight for each sample (0.0625 g cell weight/ml of methanol). Samples were vortexed vigorously for 10 min to allow carotenoid extraction from the cell pellet into methanol. Methanol absorbance was measured at the OD450. A higher absorbance reading indicates greater carotenoid production. The absorbance for cells grown in CA-MHB plus 100 mM Tris plus 44 mM NaHCO3 was normalized to the absorbance in CA-MHB plus 100 mM Tris for each strain, where CA-MHB plus 100 mM Tris equals 1.0. A Student’s t test was used for all statistical analyses.
Data availability.
The whole-genome shotgun project for strains BMC1001, COL, MRSA 11/11, and MW2 have been deposited at DDBJ/ENA/GenBank as a BioProject (PRJNA697971) under the accession numbers JAFFHU000000000, JAFFHV000000000, JAFFHW000000000, and JAFFHX000000000. The versions described in this paper are versions JAFFHU010000000, JAFFHV010000000, JAFFHW010000000, and JAFFHX010000000.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sabrina Farah for excellent technical assistance.
We have no conflict of interest to declare.
This work was supported by the following grants from the National Institutes of Health: 1RO1-AI146078 (to A.S.B.) and 1R0-1AI139244 (to Y.Q.X.).
All authors have read and approved the final manuscript. Specific contributions were as follows. Concept and design: S.C.E., A.E.R., Y.Q.X., A.S.B., and R.A.P. Collection of data, S.C.E. and A.E.R. Analysis of data, S.C.E., A.E.R., N.N.M., Y.Q.X., and A.S.B. Drafting the manuscript, S.C.E., A.S.B., and R.A.P. Critical revision of the manuscript, S.C.E., A.S.B., Y.Q.X., H.F.C., R.A.P., and N.N.M. Final approval of the manuscript, all authors. All authors agreed to be accountable for all aspects of the work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Murray R. 2005. Staphylococcus aureus infective endocarditis: diagnosis and management guidelines. Intern Med J 35:S25–S44. 10.1111/j.1444-0903.2005.00978.x. [DOI] [PubMed] [Google Scholar]
- 2.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dien Bard J, Hindler JA, Gold HS, Limbago B. 2014. Rationale for eliminating Staphylococcus breakpoints for β-lactam agents other than penicillin, oxacillin or cefoxitin, and ceftaroline. Clin Infect Dis 58:1287–1296. 10.1093/cid/ciu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purrello S, Garau J, Giamarellos E, Mazzei T, Pea F, Soriano A, Stefani S. 2016. Methicillin-resistant Staphylococcus aureus infections: a review of the currently available treatment options. J Glob Antimicrob Resist 7:178–186. 10.1016/j.jgar.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Edwards B, Andini R, Esposito S, Grossi P, Lew D, Mazzei T, Novelli A, Soriano A, Gould I. 2014. Treatment options for methicillin-resistant Staphylococcus aureus (MRSA) infection: where are we now? J Glob Antimicrob Resist 2:133–140. 10.1016/j.jgar.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Goetghebeur M, Landry P-A, Han D, Vicente C. 2007. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can J Infect Dis Med Microbiol 18:27–34. 10.1155/2007/253947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta P, Gulati N, Singla N, Vasdeva HR, Bala K, Chander J, Gupta V. 2011. Evaluation of various methods for the detection of meticillin-resistant Staphylococcus aureus strains and susceptibility patterns. J Med Microbiol 60:1613–1616. 10.1099/jmm.0.032219-0. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein MP, Patel JB, Campeau S, Eliopoulos GM, Galas MF, Humphries RM, Jenkins SG, Li JSL, Limbago B, Mathers AJ, Mazzulli T, Patel R, Richter SS, Satlin M, Swenson JM, Zimmer BL. 2018. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J Clin Microbiol 47:3692–3706. 10.1128/JCM.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ersoy SC, Abdelhady W, Li L, Chambers HF, Xiong YQ, Bayer AS. 2019. Bicarbonate resensitization of methicillin-resistant Staphylococcus aureus to β-Lactam antibiotics. Antimicrob Agents Chemother 63:e00496-19. 10.1128/AAC.00496-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ersoy SC, Otmishi M, Milan VT, Li L, Pak Y, Mediavilla J, Chen L, Kreiswirth B, Chambers HF, Proctor RA, Xiong YQ, Fowler VG, Bayer AS. 2020. Scope and predictive genetic/phenotypic signatures of ‘bicarbonate [NaHCO3]-responsiveness’ and β-Lactam sensitization among methicillin-resistant Staphylococcus aureus (MRSA). Antimicrob Agents Chemother 64:e02445-19. 10.1128/AAC.02445-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose WE, Bienvenida AM, Xiong YQ, Chambers HF, Bayer AS, Ersoy SC. 2019. Ability of bicarbonate supplementation to sensitize selected methicillin-resistant Staphylococcus aureus (MRSA) strains to β-Lactam antibiotics in an ex vivo simulated endocardial vegetation model. Antimicrob Agents Chemother 64:e02072-19. 10.1128/AAC.02072-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farha MA, French S, Stokes JM, Brown ED. 2018. Bicarbonate alters bacterial susceptibility to antibiotics by targeting the proton motive force. ACS Infect Dis 4:382–390. 10.1021/acsinfecdis.7b00194. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Cheung A, Bayer AS, Chen L, Abdelhady W, Kreiswirth BN, Yeaman MR, Xiong YQ. 2016. The global regulon sarA regulates beta-lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J Infect Dis 214:1421–1429. 10.1093/infdis/jiw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bischoff M, Entenza J, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 183:5171–5179. 10.1128/jb.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzif S, Lee E-H, Law AB, Tzeng Y-L, Shafer WM. 2005. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J Bacteriol 187:8181–8184. 10.1128/JB.187.23.8181-8184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster TJ. 2019. Can β-lactam antibiotics be resurrected to combat MRSA? Trends Microbiol 27:26–38. 10.1016/j.tim.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 19.García-Fernández E, Koch G, Wagner RM, Fekete A, Stengel ST, Schneider J, Mielich-Süss B, Geibel S, Markert SM, Stigloher C, Lopez D. 2017. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171:1354–1367. 10.1016/j.cell.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackbarth CJ, Chambers HF. 1993. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 37:1144–1149. 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renzoni A, Kelley WL, Rosato RR, Martinez MP, Roch M, Fatouraei M, Haeusser DP, Margolin W, Fenn S, Turner RD, Foster SJ, Rosato AE. 2017. Molecular bases determining daptomycin resistance-mediated resensitization to β-lactams (seesaw effect) in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 61:e01634-16. 10.1128/AAC.01634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S-J, Xiong YQ, Boyle-Vavra S, Daum R, Jones T, Bayer AS. 2010. Daptomycin-oxacillin combinations in treatment of experimental endocarditis caused by daptomycin-nonsusceptible strains of methicillin-resistant Staphylococcus aureus with evolving oxacillin susceptibility (the “seesaw effect”). Antimicrob Agents Chemother 54:3161–3169. 10.1128/AAC.00487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 56:92–102. 10.1128/AAC.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jousselin A, Renzoni A, Andrey DO, Monod A, Lew DP, Kelley WL. 2012. The post-translocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:3629–3640. 10.1128/AAC.06264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jousselin A, Manzano C, Biette A, Reed P, Pinho M, Rosato A, Kelley WL, Renzoni A. 2015. The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting penicillin-binding protein 2A. Antimicrob Agents Chemother 60:1656–1666. 10.1128/AAC.02333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ersoy SC, Chambers HF, Proctor RA, Rosato AE, Mishra NN, Xiong YQ, Bayer AS. 2021. Impact of bicarbonate on PBP2a production, maturation, and functionality in methicillin-resistant Staphylococcus aureus. 4th Annu Texas Med Center Antimicrob Resist Steward Conf, poster 30. [DOI] [PMC free article] [PubMed]
- 29.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung AL, Zhang G. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front Biosci 7:d1825–d1842. 10.2741/cheung. [DOI] [PubMed] [Google Scholar]
- 31.Giachino P, Engelmann S, Bischoff M. 2001. ςB activity depends on RsbU in Staphylococcus aureus. J Bacteriol 183:1843–1852. 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Costa TM, de Oliveira CR, Chambers HF, Chatterjee SS. 2018. PBP4: a new perspective on Staphylococcus aureus β-lactam resistance. Microorganisms 6:57. 10.3390/microorganisms6030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee SS, Chen L, Gilbert A, Da Costa TM, Nair V, Datta SK, Kreiswirth BN, Chambers HF. 2017. PBP4 mediates β-lactam resistance by altered function. Antimicrob Agents Chemother 61:e00932-17. 10.1128/AAC.00932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. 2011. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol 193:2549–2556. 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan PF, Foster SJ, Ingham E, Clements MO. 1998. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol 180:6082–6089. 10.1128/.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gertz S, Engelmann S, Schmid R, Ziebandt A-K, Tischer K, Scharf C, Hacker J, Hecker M. 2000. Characterization of the ςB regulon in Staphylococcus aureus. J Bacteriol 182:6983–6991. 10.1128/jb.182.24.6983-6991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS. 2011. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother 55:526–531. 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison EM, Ba X, Coll F, Blane B, Restif O, Carvell H, Köser CU, Jamrozy D, Reuter S, Lovering A, Gleadall N, Bellis KL, Uhlemann A-C, Lowy FD, Massey RC, Grilo IR, Sobral R, Larsen J, Rhod Larsen A, Vingsbo Lundberg C, Parkhill J, Paterson GK, Holden MTG, Peacock SJ, Holmes MA. 2019. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat Microbiol 4:1680–1691. 10.1038/s41564-019-0471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol 175:3907–3919. 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 40.Murthy MH, Olson ME, Wickert RW, Fey PD, Jalali Z. 2008. Daptomycin non-susceptible methicillin-resistant Staphylococcus aureus USA 300 isolate. J Med Microbiol 57:1036–1038. 10.1099/jmm.0.2008/000588-0. [DOI] [PubMed] [Google Scholar]
- 41.Bratu S, Eramo A, Kopec R, Coughlin E, Ghitan M, Yost R, Chapnick EK, Landman D, Quale J. 2005. Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units. Emerg Infect Dis 11:808–813. 10.3201/eid1106.040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupper AC, Sullivan MJ, Chacko KI, Mishkin A, Ciferri B, Kumaresh A, Berbel Caban A, Oussenko I, Beckford C, Zeitouni NE. 2020. Blurred molecular epidemiological lines between the two dominant methicillin-resistant Staphylococcus aureus clones p ofz302. Oxford University Press, Oxford, United Kingdom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenn WO. 1928. The carbon dioxide dissociation curve of nerve and muscle. Am J Physiol Legacy Content 85:207–223. 10.1152/ajplegacy.1928.85.2.207. [DOI] [Google Scholar]
- 44.Cockerill FR. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 45.Dumitrescu O, Choudhury P, Boisset S, Badiou C, Bes M, Benito Y, Wolz C, Vandenesch F, Etienne J, Cheung AL, Bowden MG, Lina G. 2011. β-Lactams interfering with PBP1 induce panton-valentine leukocidin expression by triggering sarA and rot global regulators of Staphylococcus aureus. Antimicrob Agents Chemother 55:3261–3271. 10.1128/AAC.01401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. 2011. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:5631–5639. 10.1128/AAC.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mielich-Süss B, Wagner RM, Mietrach N, Hertlein T, Marincola G, Ohlsen K, Geibel S, Lopez D. 2017. Flotillin scaffold activity contributes to type VII secretion system assembly in Staphylococcus aureus. PLoS Pathog 13:e1006728. 10.1371/journal.ppat.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sihto H-M, Tasara T, Stephan R, Johler S. 2014. Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol Lett 356:134–140. 10.1111/1574-6968.12491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome shotgun project for strains BMC1001, COL, MRSA 11/11, and MW2 have been deposited at DDBJ/ENA/GenBank as a BioProject (PRJNA697971) under the accession numbers JAFFHU000000000, JAFFHV000000000, JAFFHW000000000, and JAFFHX000000000. The versions described in this paper are versions JAFFHU010000000, JAFFHV010000000, JAFFHW010000000, and JAFFHX010000000.