Abstract
Histone modification critically contributes to the epigenetic control of gene expression by changing the configuration of chromatin and modifying the access of transcription factors to gene promoters. Recently, we observed that histone acetylation and crotonylation mediated the expression of endocytosis-related genes and tumor-related immune checkpoint genes by regulating the enrichment of signal transducer and activator of transcription 3 on these gene promoters in Alzheimer’s disease and tumorigenesis, suggesting that histone modification plays an important role in disease development. Furthermore, studies performed in the past decade revealed that histone modifications affect osteogenic differentiation by regulating the expression of osteogenic marker genes. In this review, we summarize and discuss the histone modification-centric regulation of osteogenic gene expression. This review improves the understanding of the role of histone modifications in osteogenic differentiation and describes its potential as a therapeutic target for osteogenic differentiation-related diseases.
Subject terms: Histone post-translational modifications, Differentiation
Facts
The levels of histone modifiers and their regulators are altered during osteogenic differentiation and the development of osteogenic differentiation-related diseases.
Histone modifications orchestrate osteogenic differentiation.
Histone modifications regulate the expression of osteogenic marker genes by affecting the chromatin structure and transcription factor activity.
Open questions
Which types of histone modifications are mainly responsible for osteogenic differentiation?
How do histone modifications coordinate to regulate the expression of osteogenic marker genes?
Can histone modifications be targeted for treating osteogenic differentiation-related diseases?
Introduction
Histone modification is an important epigenetic process with a key role in diverse biological processes, including transcription, chromosome packaging, and DNA repair1–3. Histone acetylation and methylation are the most widespread and dynamic histone modifications. Histone acetyltransferases and histone deacetylases mediate the addition and removal of acetyl groups at lysine residues in the N-terminal tails of histones, respectively, and histone methyltransferase and histone demethylase mediate the addition and removal of methyl groups at lysine and arginine residues, respectively4,5. Increasing evidence has demonstrated that histone modification is involved in multiple pathological processes, including viral infection, inflammation, neurodegenerative diseases, and tumorigenesis6–10. Our previous study of epigenetic regulation in Alzheimer’s disease and T cell exhaustion showed that histone acetylation is involved in long non-coding RNA NEAT1- and splicing factor SRSF2-mediated expression of endocytosis-related genes and tumor-related immune checkpoint genes, respectively, by recruiting signal transducer and activator of transcription 3 to these gene promoters11,12.
Osteogenic differentiation is a key process in bone formation, and its dysfunction leads to bone metabolism-related diseases13–15. During osteogenic differentiation, some transcription factors and signaling pathways are required for the expression of osteogenic genes, such as runt-related transcription factor 2 (Runx2), bone morphogenetic protein (BMP) signaling, and Wnt signaling16–18. Runx2 is a mammalian homolog of the Drosophila runt19, which functions as a transcription factor required to initiate osteogenic differentiation by regulating the expression of osteogenic marker genes20–23. BMP signaling is a central pathway that induces osteogenic differentiation and bone formation24–27, partly by enhancing the expression of Runx2 by stimulating p300-mediated Runx2 acetylation, inhibiting Smurf1-mediated degradation of Runx2, and promoting the transcription of Runx228. Wnt signaling is another important pathway for osteogenic differentiation and bone formation. The canonical Wnt pathway can activate the expression of Runx2 to promote osteogenic differentiation of bone mesenchymal stem cells (BMSCs)29,30. An increasing number of studies have shown that histone modification is a potential mediator of these regulations by affecting chromatin structure and transcription factor activity31–33.
In this review, we summarize and discuss the regulators and histone modifiers that are altered with modulated levels of histone modification observed during osteogenic differentiation and osteogenic differentiation-related diseases. Additionally, we also discuss the molecular mechanisms by which the regulated histone modification mediates the expression of osteogenic marker genes that affect osteogenic differentiation.
Histone acetylation
Histone acetylation modification, a key mechanism of epigenetic regulation, is closely related to the activation of osteogenic marker gene expression during osteogenic differentiation and bone formation34–36. Furthermore, specific sites of histone acetylation, such as H3K9Ac, have been reported to be involved in this process. In this section, we summarize and discuss the regulators and histone modifiers that modulate H3K9Ac, and their roles in the expression of osteogenic marker genes (Table 1).
Table 1.
Histone acetylation centric-regulation in osteogenic genes expression.
| Regulator/histone modifer | Histone modification | Target gene | Role | Reference |
|---|---|---|---|---|
| PCAF | H3K9Ac | BMP2, BMP4,BMPR1B, Runx2 | Promoted osteogenic differentiation of BMSCs and bone formation | 38 |
| GCN5 | Wnt1, Wnt6, Wnt10a, Wnt10b | Promoted osteogenic differentiation of BMSCs | 43 | |
| HDAC1 | Runx2, OSX, OC, P27 | Inhibited osteogenic differentiation of BMSCs | 44 | |
| NAMPT | Runx2 | Promoted osteogenic differentiation of BMSCs | 47 | |
| MAPK | BGLAP2, IBSP | Promoted osteogenic differentiation of preosteoblast cells | 53 | |
| MAPK | H4K5Ac | Bglap2, LBSP | Promoted osteogenic differentiation of preosteoblast cells | 53 |
H3K9Ac is a hallmark of gene activation and is highly correlated with active promoters37. To date, several regulators- and histone modifiers-mediated H3K9Ac, have been reported to be involved in the regulation of osteogenic marker gene expression during osteogenic differentiation. A study, aimed at understanding the molecular mechanism of epigenetic regulation in the osteogenic differentiation of mesenchymal stem cells (MSCs), showed that the histone acetyltransferase PCAF promoted the expression of BMP pathway genes, including BMP2, BMP4, BMPR1B, and Runx2, by increasing histone H3K9 acetylation in the promoter regions of these genes38. In osteoporosis, a common degenerative bone disease characterized by disrupted osteogenesis and resorption39–41, the expression of GCN5, another histone acetyltransferase that is 73% homologous to PCAF42, was reportedly decreased. This decrease inhibited the osteogenic differentiation of BMSCs by reducing the acetylation of H3K9 on the promoters of Wnt genes such as Wnt1, Wnt6, Wnt10a, and Wnt10b43.
In addition, a study investigating the role of modifications in the chromatin structure in osteogenic differentiation revealed that the expression of histone deacetylase 1 and its enzymatic activity were significantly decreased, resulting in induction of the osteoblast marker genes Runx2, osterix (OSX), and osteocalcin (OC), and cell cycle arrest gene P27, by enhancing H3K9Ac at these gene promoters44. Nicotinamide phosphoribosyltransferase, an enzyme involved in nicotinamide adenine dinucleotide biosynthesis45, was found to be upregulated during the osteogenic differentiation of multi- and omnipotent progenitors46. Investigations of the molecular mechanism revealed that nicotinamide phosphoribosyltransferase promoted osteogenic differentiation by increasing the enrichment of H3K9Ac at the Runx2 promoter, thus enhancing Runx2 transcriptional activity47. The mitogen-activated protein kinase pathway mediates osteogenic differentiation48. Further mechanistic studies revealed that mitogen-activated protein kinase enhances the expression of osteoblast marker genes, such as bone gamma-carboxyglutamate protein 2 and integrin-binding sialoprotein, by elevating H3K9Ac and H4K5Ac, another hallmark of gene activation49,50, which is catalyzed by Tip60 and CBP/p30051,52 at these gene promoters and enhancers by associating with Runx2, as well as by mediating Runx2 S301/S319 phosphorylation53–55.
Histone methylation
Histone methylation is another histone modification involved in regulating osteogenic marker gene expression. It is closely related to the activation or suppression of osteogenic marker gene expression. Methylation of H3K9, H3K27, and H4K20 is often associated with inactive chromatin, whereas methylation of H3K4, H3K36, H3K79, and H3R17 is largely associated with active gene transcription56. In this section, we summarize the regulators and histone modifiers that modulate H3K4, H3K9, and H3K27 methylation, and their roles in the expression of osteogenic marker genes (Table 2).
Table 2.
Histone methylation centric-regulation in osteogenic genes expression.
| Regulator/histone modifer | Histone modification | Target gene | Role | Reference |
|---|---|---|---|---|
| RBP2 | H3K4me3 | OC, OSX, Runx2 | Inhibited osteogenic differentiation of hASCs | 58,59 |
| BCOR | AP-2, EREG | Decreased osteo/dentinogenic potentials of MSCs | 62,63 | |
| Ash1l | OSX, Runx2, HOXA10, Sox9 | Promoted osteogenic differentiation | 65 | |
| NO66 | BSP, OC | Inhibited osteogenic differentiation of osteoprogenitor cells and mineralization | 66 | |
| KDM7A | H3K9me2 | C/ebpα, SFRP1 | Inhibited osteogenic differentiation of osteoprogenitor cells | 73 |
| KDM4B | H3K9me3 | DLX5 | Promoted osteogenic differentiation of MSCs | 69 |
| KDM4A | C/ebpα, SFRP4 | Inhibited osteogenic differentiation of osteoprogenitor cells | 74 | |
| KDM7A | H3K27me2 | C/ebpα, SFRP1 | Inhibited osteogenic differentiation of osteoprogenitor cells | 73 |
| KDM6B | H3K27me3 | HOXC6-1 | Promoted osteogenic differentiation of MSCs | 69 |
| CDK1 | HOXA7, HOXA9,Runx2, TCF7 | Promoted osteogenic differentiation of MSCs | 79 | |
| EZH2 | Runx2, OC, ZBTB16, MX1, FHL1 | Inhibited osteogenic differentiation of MSCs | 76–78 | |
| KDM6A | Runx2, OC | Promoted osteogenic differentiation of MSCs | 76 | |
| TNFα | Runx2 | Inhibited osteogenic differentiation of rMSCs | 80 | |
| DLX3 | DKK4 | Promoted osteogenic differentiation of BMSCs | 82 | |
| BCOR | H3K36me2 | AP-2, EREG | Inhibited osteogenic differentiation of MSCs | 62,63 |
| NO66 | H3K36me3 | BSP, OC | Inhibited osteogenic differentiation of preosteoblasts | 66 |
H3K4 methylation
The first modulator of H3K4 methylation that participates in osteogenic differentiation is retinoblastoma binding protein 2 (RBP2), also known as lysine (K)-specific demethylase 5A, which specifically catalyzes the demethylation of dimethyl or trimethyl histone H3 lysine 4 (H3K4me2 or H3K4me3)57. In a study examining the epigenetic regulation of osteogenic differentiation of human adipose-derived stromal cells, RBP2 was found to negatively regulate the transcription of osteogenic marker genes, OC and OSX, by physically associating with their gene promoters to reduce the level of H3K4me358. In another study of the underlying molecular mechanisms of osteoporosis, RBP2 was shown to be upregulated during osteoporosis. This resulted in inhibition of BMP2-induced osteogenic differentiation. A molecular mechanism study demonstrated that RBP2 repressed osteogenic differentiation by decreasing H3K4me3 levels on the promoters of Runx2, thus inhibiting Runx2 transcription59.
The BCL6 co-repressor (BCOR) represses gene transcription by associating with the transcription repressor BCL-660,61. A study of the roles of BCOR mutations in oculo-facio-cardio-dental syndrome, a rare genetic disorder characterized by canine teeth with extremely long roots, and eye, craniofacial, and cardiac abnormalities, revealed that the BCOR mutation enhanced the osteogenic capacity of MSCs by promoting the expression of AP-2α, a key factor that mediates the osteo/dentinogenic differentiation of MSCs by interfering with the interaction of FBXL10, also known as Jumonji C histone demethylase 1B with the AP-2α promoter, thereby increasing H3K4me3 and H3K36me2 levels at this promoter62. Another study revealed that BCOR associates with FBXL11, a paralog of FBXL10, also known as histone demethylase, K-specific demethylase 2A, to inhibit transcription of the epidermal growth factor EREG by decreasing H3K4me3 and H3K36me2 levels at the EREG promoter. This change results in inhibition of the osteogenic differentiation potential of MSCs63.
Additionally, absent, small, or homeotic disc1-like (Ash1l), a member of the Trx family, was found to promote gene expression via the methyltransferase activity of its SET domain64. In a study investigating the role of Ash1l in the differentiation of MSCs, Ash1l enhanced the osteogenic and chondrogenic differentiation of C3H10T1/2 cells by increasing the enrichment of H3K4me3 at the osteogenic marker gene promoters, including OSX, Runx2, Hoxa10, and Sox965. OSX is an important transcription factor required for osteogenic differentiation and bone formation. In a study performed to understand the regulatory roles of OSX in osteogenic differentiation, the JmjC domain-containing protein NO66 was found to function as a histone demethylase, with reported involvement in osteogenic differentiation and maturation of preosteoblasts by interacting with OSX and regulating OSX target genes, including BSP and OC, by modulating H3K4me3 and H3K36me3 levels at these gene promoters66.
H3K9 methylation
In BMP-stimulated osteogenic differentiation of MSCs, histone K-specific demethylase 4B (KDM4B), also known as JMJD2B, and histone K-specific demethylase 6B (KDM6B), also known as JMJD3, were found to be upregulated and essential for the osteogenic differentiation of MSCs and bone formation. Mechanistically, KDM4B enhanced the expression of DLX5, which mediates osteogenic differentiation in an OSX-dependent manner67,68 by removing H3K9me3 from the gene promoter, whereas KDM6B enhanced the expression of HOXC6-1, a homeodomain-containing transcription factor playing a critical role in osteogenic differentiation by removing H3K27me3 from the gene promoter69.
Furthermore, Wang et al. observed that histone K-specific demethylase 7A (KDM7A) binds to the promoter of CCAAT/enhancer-binding protein α (C/EBPα) and secretes frizzled-related protein 1, which both attenuate the canonical Wnt signaling pathway and play important roles in both adipogenesis and osteogenesis70–72, to promote gene transcription by removing the histone methylation marks H3K9me2 and H3K27me2 at the gene promoter73. They also found that histone K-specific demethylase 4A, also known as JMJD2A, functions to repress Wnt signaling, thereby blocking osteogenic differentiation via enhancing the transcription of C/EBPα and secreted frizzled-related protein 4 by removing H3K9me3 at the gene promoter74.
H3K27 methylation
Enhancer of Zeste homology 1 and 2 (EZH1 and EZH2) are methyltransferases that methylate histone 3 lysine 27 on chromatin to repress target gene expression75. To date, several studies have reported that EZH2 is involved in osteogenic differentiation through the epigenetic regulation of osteogenic marker genes via its methyltransferase activity. They found that EZH2 inhibits expression of the osteogenic marker genes Runx2, OC, ZBTB16, MX1, and FHL1 to inhibit osteogenic differentiation by increasing H3K27me3 levels at these gene promoters76–78, whereas lysine demethylase 6A induces osteogenic differentiation by removing this repressive mark from Runx2 at the OC gene promoter76. Moreover, cyclin-dependent kinase 1 has been found to enhance MSC differentiation into osteoblasts in an EZH2-dependent manner. It was observed that cyclin-dependent kinase 1 inhibited EZH2 methyltransferase activity by promoting the phosphorylation of EZH2 at Thr 487, thus disrupting EZH2 binding with other components of polycomb repressive complex 2. This resulted in upregulation of the osteogenic marker genes HOXA7, HOXA9, Runx2, and TCF7 because of reduction in H3K27Me3 levels at these gene promoters79.
In addition, a study of rat BMSCs revealed that tumor necrosis factor-α inhibited osteogenic differentiation by increasing enrichment of H3K27me3 at the Runx2 gene promoter, thus attenuating Runx2 gene expression80. In another study to clarify the molecular mechanism through which distal-less homeobox 3 (DLX3), a DLX family transcription factor, modulates the osteogenic differentiation of BMSCs, DLX3 was found to promote the osteogenic differentiation of BMSCs by inhibiting the expression of Dickkopf 4, an antagonist of the Wnt/β-catenin pathway by disrupting the interaction of Wnt ligands with LRP5/681, via increasing the enrichment of H3K27me3 at the Dickkopf 4 promoter82.
Histone acetylation and histone methylation
It has been confirmed that different histone modifications can occur simultaneously or sequentially in a combinatorial manner to regulate the expression of osteogenic genes during osteogenic differentiation34,83,84. In this section, we summarize and discuss regulators controlling the expression of osteogenic marker genes by modulating both histone acetylation and histone methylation levels at these gene promoters (Table 3).
Table 3.
Histone acetylation and methylation centric-regulation in osteogenic genes expression.
| Regulator/histone modifer | Histone modification | Target gene | Role | Reference |
|---|---|---|---|---|
| NaBu | H3K9Ac, H3K9Me3 | Runx2 | Promoted osteogenic differentiation of ADSCs | 86 |
| TSA | BMP, ALP | Promoted osteogenic differentiation of nonosteogenic cells | 87 | |
| Wnt/β-catenin | H3K9Ac, H3K14Ac, H4K12Ac, H3K9Me2 | PPARγ2 | Attenuated osteogenic potential of osteoporotic BMSCs | 90 |
| HOXA10 | H4Ac, H3K4Me3 | Runx2, OC, ALP, BSP | Promoted osteogenic differentiation of osteoprogenitor cells | 92 |
| miR-23a | H3K27Ac,H3K27Me3 | OC | Promoted bone formation in mice | 77 |
H3K9 acetylation and methylation
In a study aimed at improving the osteogenic potential of adipose tissue-derived stem cells, sodium butyrate, the sodium salt of the short-chain fatty acid butyric acid that functions as an inhibitor of histone deacetylases85, reportedly increased the osteogenic differentiation capacity of adipose tissue-derived stem cells following stimulation of osteogenic differentiation-specific genes, including Runx2, osteopontin, OC, and alkaline phosphatase (ALP). Investigation of the molecular mechanisms demonstrated that sodium butyrate enhanced Runx2 transcription by increasing the recruitment of transcriptionally permissive histone modification H3K9Ac, and decreasing the recruitment of transcriptionally repressive histone modification H3K9Me3 at the Runx2 promoter86. Furthermore, trichostatin-A, another histone deacetylase inhibitor, was shown to promote the trans-differentiation of non-osteogenic cells (3T3-L1 and NIH3T3) into osteoblasts with enhanced expression of BMP2 and ALP by increasing H3K9Ac and decreasing H3K9Me3 levels at the BMP2 and ALP promoters87.
Moreover, a study of the underlying molecular mechanism through which BMSC adipogenesis overwhelms osteogenesis during osteoporosis, revealed that inhibition of Wnt/β-catenin signaling increased the enrichment of H3K9Ac, H3K14Ac, and H4K12Ac and decreased the levels of H3K9Me2 on the promoter of peroxisome proliferator-activated receptor-gamma isoform 2, a lipid-activated transcription factor required for adipogenesis88,89, thereby elevating its gene expression. This resulted in the inhibition of osteogenic differentiation and promotion of adipogenic differentiation90.
In BMP2-mediated osteogenic differentiation, Hoxa10, an important transcription factor that is essential for bone formation91, is rapidly induced and functions as an activator of Runx2, OC, ALP, and BSP transcription by binding to these gene promoters and altering chromatin modification, for example, by increasing the levels of histone H4 acetylation and H3K4Me3. This resulted in enhanced osteogenic differentiation in a Runx2-dependent or Runx-independent manner92.
H3K27 acetylation and methylation
In a study performed to evaluate the epigenetic mechanisms of miR-23a in regulating bone synthesis and homeostasis, knockdown of the miR-23a cluster was shown to enhance osteogenic differentiation with decreased expression of EZH2 and increased expression of Baf45a, a factor responsible for increasing chromatin accessibility. This resulted in the induction of OC by increasing H3K27ac levels and decreasing H3K27me3 levels at the OC gene promoter77.
Conclusion and perspectives
To date, several regulators and histone modifiers have been reported to affect osteogenic differentiation by regulating histone modification, thus affecting the expression of osteogenic marker genes. In this review, we summarized and discussed the molecules that control histone modification, as well as the molecular mechanism by which histone modification regulates the expression of its target genes. Most of these molecules are deacetylase inhibitors, transcription factors, miRNAs, and protein kinases, which play important roles in modulating the catalytic activity of histone modifiers, including histone acetyltransferases, histone deacetylases, histone methyltransferases, and histone demethylases. Furthermore, histone acetylation and methylation were found to mediate the BMP and Wnt/β-catenin signaling pathways that are required for osteogenic differentiation, as well as control the expression of Runx2 and Runx2-targeted osteogenic marker genes, including OC, OSX, and ALP (Fig. 1). Dysregulation of osteogenic differentiation has been linked to several pathophysiologic processes, such as osteopenia, osteopetrosis, osteogenesis imperfecta, and osteoporosis. Thus, histone modifications may be useful therapeutic targets for treating osteogenic differentiation-related diseases after clarifying the main histone modifications and interplay between these histone modifications during disease development. Overall, this review discusses histone modification-centric regulation of osteogenic marker genes and provides insights into its potential clinical utility in osteogenic differentiation-related diseases.
Fig. 1. Schematic model of the roles of histone modification in osteogenic marker genes expression.
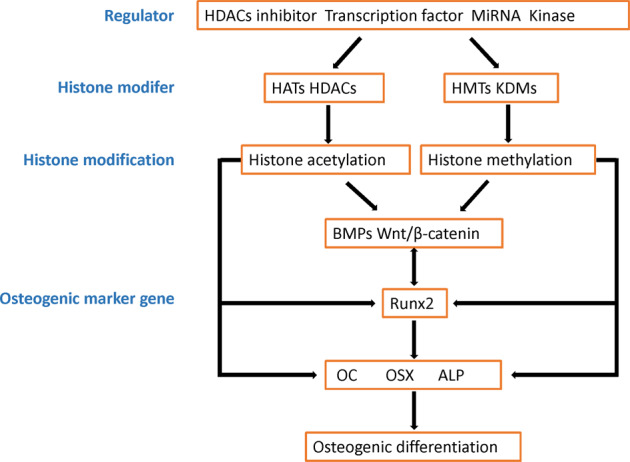
During osteogenic differentiation, numbers of regulators are found to modulate histone acetylation and methylation by affecting the catalytic activity of histone modifiers. The altered histone acetylation and methylation then control expression of osteogenic marker genes, directly or indirectly. HATs histone acetyltransferases, HDACs histone deacetylases, HMTs histone methyltransferases, KDMs histone demethylases, BMPs bone morphogenetic proteins, HOXs homeobox genes, Runx2 runt-related transcription factor 2, OC osteocalcin, OSX osterix, ALP alkaline phosphatase.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32000878), the Natural Science Foundation of Shandong Province (ZR2020LZL008), and the Outstanding University Driven by Talents Program and Academic Promotion Program of Shandong First Medical University (2019LJ001).
Author contributions
K.L. drafted the article; J.H. and Z.W. contributed to conception and design; Z.W. designed the figure and revised the article; All authors read and approved the final paper.
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by Dr. David Michod
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinxiang Han, Email: samsjxh@sina.com.
Ziqiang Wang, Email: yky2009@163.com.
References
- 1.Diehl KL, Muir TW. Chromatin as a key consumer in the metabolite economy. Nat. Chem. Biol. 2020;16:620–629. doi: 10.1038/s41589-020-0517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li K, Wang Z. Histone crotonylation-centric gene regulation. Epigenetics Chromatin. 2021;14:10. doi: 10.1186/s13072-021-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schvartzman JM, Thompson CB, Finley L. Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol. 2018;217:2247–2259. doi: 10.1083/jcb.201803061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Higgins JM. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013;23:175–184. doi: 10.1016/j.tcb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017;18:90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Li K, Wang X, Huang W. MiR-155-5p modulates HSV-1 replication via the epigenetic regulation of SRSF2 gene expression. Epigenetics. 2019;14:494–503. doi: 10.1080/15592294.2019.1600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi Z, et al. Histone deacetylase 3 couples mitochondria to drive IL-1β-dependent inflammation by configuring fatty acid oxidation. Mol. Cell. 2020;80:43–58.e7. doi: 10.1016/j.molcel.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen H, Adlanmerini M, Hauck AK, Lazar MA. Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature. 2020;584:286–290. doi: 10.1038/s41586-020-2576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baksh SC, et al. Extracellular serine controls epidermal stem cell fate and tumour initiation. Nat. Cell Biol. 2020;22:779–790. doi: 10.1038/s41556-020-0525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan K, et al. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci. Adv. 2020;6:eaax0021. doi: 10.1126/sciadv.aax0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, et al. NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell. Mol. Life Sci. 2019;76:3005–3018. doi: 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Modulation of SRSF2 expression reverses the exhaustion of TILs via the epigenetic regulation of immune checkpoint molecules. Cell. Mol. Life Sci. 2020;77:3441–3452. doi: 10.1007/s00018-019-03362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Real A, Riancho-Zarrabeitia L, López-Delgado L, Riancho JA. Epigenetics of skeletal diseases. Curr. Osteoporos. Rep. 2018;16:246–255. doi: 10.1007/s11914-018-0435-y. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li CJ, et al. Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J. Clin. Invest. 2018;128:5251–5266. doi: 10.1172/JCI99044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei J, et al. Glucose uptake and Runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell. 2015;161:1576–1591. doi: 10.1016/j.cell.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauner M, et al. Transferrin receptor 2 controls bone mass and pathological bone formation via BMP and Wnt signaling. Nat. Metab. 2019;1:111–124. doi: 10.1038/s42255-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 20.Franceschi RT, Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J. Cell. Biochem. 2003;88:446–454. doi: 10.1002/jcb.10369. [DOI] [PubMed] [Google Scholar]
- 21.Wagner EF, Karsenty G. Genetic control of skeletal development. Curr. Opin. Genet. Dev. 2001;11:527–532. doi: 10.1016/s0959-437x(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 22.Harada H, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 23.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 24.Bandyopadhyay A, et al. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamiya N, et al. Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J. Bone Miner. Res. 2008;23:2007–2017. doi: 10.1359/JBMR.080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamiya N, et al. Wnt inhibitors Dkk1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J. Bone Miner. Res. 2010;25:200–210. doi: 10.1359/jbmr.090806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D, et al. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J. Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon EJ, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 31.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol. Cell. Biol. 2003;23:8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen J, et al. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol. Endocrinol. 2003;17:743–756. doi: 10.1210/me.2002-0122. [DOI] [PubMed] [Google Scholar]
- 35.Hu B, et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167:1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, et al. SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Wan L, et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 2017;543:265–269. doi: 10.1038/nature21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, et al. Histone H3K9 acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may be involved in osteoporosis. Stem Cells. 2016;34:2332–2341. doi: 10.1002/stem.2424. [DOI] [PubMed] [Google Scholar]
- 39.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook P, Cooper C. Osteoporosis. Lancet. 2006;367:2010–2018. doi: 10.1016/S0140-6736(06)68891-0. [DOI] [PubMed] [Google Scholar]
- 41.Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 42.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 43.Jing H, et al. Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis. 2018;9:176. doi: 10.1038/s41419-017-0231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HW, et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 45.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, He J, He X, Li Y, Lindgren U. Nampt expression increases during osteogenic differentiation of multi- and omnipotent progenitors. Biochem. Biophys. Res. Commun. 2013;434:117–123. doi: 10.1016/j.bbrc.2013.02.132. [DOI] [PubMed] [Google Scholar]
- 47.Ling M, et al. Epigenetic regulation of Runx2 transcription and osteoblast differentiation by nicotinamide phosphoribosyltransferase. Cell Biosci. 2017;7:27. doi: 10.1186/s13578-017-0154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita T, et al. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol. Cell. Biol. 2009;29:5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park CS, Rehrauer H, Mansuy IM. Genome-wide analysis of H4K5 acetylation associated with fear memory in mice. BMC Genomics. 2013;14:539. doi: 10.1186/1471-2164-14-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800. doi: 10.1046/j.1365-2443.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 52.Schiltz RL, et al. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Ge C, Franceschi RT. MAP kinase-dependent RUNX2 phosphorylation is necessary for epigenetic modification of chromatin during osteoblast differentiation. J. Cell. Physiol. 2017;232:2427–2435. doi: 10.1002/jcp.25517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge C, et al. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J. Biol. Chem. 2009;284:32533–32543. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Ge C, Franceschi RT. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J. Bone Miner. Res. 2010;25:154–163. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang C, et al. SUMOylated ORC2 recruits a histone demethylase to regulate centromeric histone modification and genomic stability. Cell Rep. 2016;15:147–157. doi: 10.1016/j.celrep.2016.02.091. [DOI] [PubMed] [Google Scholar]
- 58.Ge W, et al. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells. 2011;29:1112–1125. doi: 10.1002/stem.663. [DOI] [PubMed] [Google Scholar]
- 59.Wang C, et al. KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis. Cell Death Dis. 2016;7:e2335. doi: 10.1038/cddis.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 61.Ghetu AF, et al. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan Z, et al. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat. Cell Biol. 2009;11:1002–1009. doi: 10.1038/ncb1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du J, Ma Y, Ma P, Wang S, Fan Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells. 2013;31:126–136. doi: 10.1002/stem.1255. [DOI] [PubMed] [Google Scholar]
- 64.Gregory GD, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 2007;27:8466–8479. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin B, et al. Epigenetic control of mesenchymal stem cell fate decision via histone methyltransferase Ash1l. Stem Cells. 2019;37:115–127. doi: 10.1002/stem.2918. [DOI] [PubMed] [Google Scholar]
- 66.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hassan MQ, et al. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 69.Ye L, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bafico A, et al. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 1999;274:16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 71.Guo L, Li X, Tang QQ. Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) β. J. Biol. Chem. 2015;290:755–761. doi: 10.1074/jbc.R114.619957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawai M, Rosen CJ. PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nat. Rev. Endocrinol. 2010;6:629–636. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang X, et al. Histone demethylase KDM7A reciprocally regulates adipogenic and osteogenic differentiation via regulation of C/EBPα and canonical Wnt signalling. J. Cell. Mol. Med. 2019;23:2149–2162. doi: 10.1111/jcmm.14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi Q, et al. Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPα and canonical Wnt signaling. Cell. Mol. Life Sci. 2020;77:2407–2421. doi: 10.1007/s00018-019-03289-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemming S, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32:802–815. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 77.Godfrey TC, Wildman BJ, Javed A, Lengner CJ, Hassan MQ. Epigenetic remodeling and modification to preserve skeletogenesis in vivo. Connect. Tissue Res. 2018;59:52–54. doi: 10.1080/03008207.2017.1408599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hemming S, et al. Identification of novel EZH2 targets regulating osteogenic differentiation in mesenchymal stem cells. Stem Cells Dev. 2016;25:909–921. doi: 10.1089/scd.2015.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei Y, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fang B, et al. Involvement of tumor necrosis factor alpha in steroid-associated osteonecrosis of the femoral head: friend or foe? Stem Cell Res. Ther. 2019;10:5. doi: 10.1186/s13287-018-1112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lerner UH, Ohlsson C. The WNT system: background and its role in bone. J. Intern. Med. 2015;277:630–649. doi: 10.1111/joim.12368. [DOI] [PubMed] [Google Scholar]
- 82.Sun S, et al. DLX3 regulates osteogenic differentiation of bone marrow mesenchymal stem cells via Wnt/β-catenin pathway mediated histone methylation of DKK4. Biochem. Biophys. Res. Commun. 2019;516:171–176. doi: 10.1016/j.bbrc.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 83.Tan J, et al. Genome-wide analysis of histone H3 lysine9 modifications in human mesenchymal stem cell osteogenic differentiation. PLoS ONE. 2009;4:e6792. doi: 10.1371/journal.pone.0006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rui Y, et al. Epigenetic memory gained by priming with osteogenic induction medium improves osteogenesis and other properties of mesenchymal stem cells. Sci. Rep. 2015;5:11056. doi: 10.1038/srep11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghosh SK, Perrine SP, Williams RM, Faller DV. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood. 2012;119:1008–1017. doi: 10.1182/blood-2011-06-362434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu X, et al. Histone deacetylase inhibitor sodium butyrate promotes the osteogenic differentiation of rat adipose-derived stem cells. Dev. Growth Differ. 2014;56:206–213. doi: 10.1111/dgd.12119. [DOI] [PubMed] [Google Scholar]
- 87.Cho YD, et al. Epigenetic modifications and canonical wingless/int-1 class (WNT) signaling enable trans-differentiation of nonosteogenic cells into osteoblasts. J. Biol. Chem. 2014;289:20120–20128. doi: 10.1074/jbc.M114.558064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, et al. Epigenetic landscape in PPARγ2 in the enhancement of adipogenesis of mouse osteoporotic bone marrow stromal cell. Biochim. Biophys. Acta. 2015;1852:2504–2516. doi: 10.1016/j.bbadis.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 91.Wahba GM, Hostikka SL, Carpenter EM. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev. Biol. 2001;231:87–102. doi: 10.1006/dbio.2000.0130. [DOI] [PubMed] [Google Scholar]
- 92.Hassan MQ, et al. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


