Abstract
Liquid-liquid phase separation (LLPS) of proteins is involved in a growing number of cellular processes. Most proteins with LLPS harbor intrinsically disordered regions (IDR), which serve as a guideline to search for cellular proteins that potentially phase separate. Herein, we reveal that oligomerization lowers the barriers for LLPS and could act as a general mechanism to enhance LLPS of proteins domains independent of IDR. Using TDP43 as a model system, we found that deleting its IDR resulted in LLPS that was dependent on the oligomerization of the N-terminal domain (NTD). Replacing TDP43’s NTD with other oligomerization domains enhanced the LLPS proportionately to the state of oligomerization. In addition to TDP43, fusing NTD to other globular proteins without known LLPS behavior also drove their separation in a manner dependent on oligomerization. Finally, we demonstrate that heterooligomers composed of NTD-fused proteins can be driven into droplets through NTD interactions. Our results potentiate a new paradigm for using oligomerization domains as a signature to systematically identify cellular proteins with LLPS behavior, thus broadening the scope of this exciting research field.
Keywords: TDP43, liquid-liquid phase separation, oligomerization, membrane-less organelles, RNA-binding proteins
GRAPHICAL ABSTRACT
INTRODUCTION
In recent years, liquid-liquid phase separation (LLPS) has been shown to play increasingly important role in a number of cellular processes, as represented by transcription regulation, stress response, and metabolite production[1–5]. The determining factors behind what allows a protein to undergo LLPS are only starting to be uncovered, with the roles of intrinsically disorder regions (IDR), electrostatic interactions, cation-π interactions, and complexation identified as potential driving forces[6–10]. In particular, oligomerization has been shown to be critical for LLPS of several protein systems. For instance, the speckle-type POZ protein (SPOP) has been shown to form higher-order homo-oligomers that are able to form nuclear speckles with features of liquid droplets[11]. An additional example is provided by representative IDR sequences that are fused to the Cry2 protein whose dimerization is driven by photo-irradiation. In this case, light-activated dimerization is shown to drive these IDR sequences to form optoDroplets in conditions that monomeric IDR sequences cannot undergo LLPS[12]. While these examples suggest the necessity of oligomerization for some protien systems to undergo LLPS, it remains unclear whether oligomerization could serve as a general mechanism to enhance LLPS of a broader scope of proteins and whether the oligomerization domains could be used to identify cellular proteins with potential LLPS behavior.
To initiate the investigation into these questions, we choose the RNA-binding protein (RBP) TAR-DNA binding Protein of 43 kDa (TDP43) as a model system. TDP43 is a nucleo-cytoplasmic protein whose canonical function in the nucleus is to regulate RNA splicing and mRNA stability[13]. When cells are exposed to stress conditions, TDP43 forms granulated structures including anisosomes (an anisotropic nuclear shell structure), nuclear bodies or stress granules[14–18], potentially via LLPS. TDP43 harbors an N-terminal domain (NTD), a RNA binding domain (RBD) that is composed of two RNA-recognizing motifs, and a C-terminal prion-like domain (PrLD) that is intrinsically disordered[19]. Among these domains, PrLD has been shown to exhibit the property to undergo phase separation as either a purified sequence fragment or as a promoting factor to mediate formation of stress-induced granules[20–24]. Furthermore, PrLD allows TDP43 to rapidly self-associate and initiate protein aggregation, potentially a process that is connected with phase separation[25–30]. Distinct from the intrinsically disordered PrLD, NTD of TDP43 adopts a dynamic, solenoid-like structure, wherein the electrostatic charge is positive on one side and negative on the opposite side[31]. Thus, NTD is able to form head-to-tail oligomers both in vitro and in cells, a process that is important for the functional TDP43 dimerization and/or the pathologic cytoplasmic aggregation[31–33].
In this work, we attempted to study whether the oligomerization of NTD could be a promoting factor to promote the LLPS of TDP43. To our surprise, we found that the truncation of PrLD from TDP43, resulting in a PrLD-less TDP43 that harbors only the NTD and RBD, was capable of phase separation to form liquid droplets. The LLPS of the PrLD-less TDP43 was strictly dependent on the presence of NTD, as the RBD domains of TDP43 could only phase separate at much higher protein concentrations. Furthermore, replacing TDP43’s NTD by other oligomerization domains (represented by the dimeric superoxide dismutase and the tetrameric transthyretin) enhanced LLPS proportionately to the state of oligomerization, suggesting that oligomerization could be a critical factor to promote phase separation. In addition to TDP43, we found that fusing TDP43’s NTD to other proteins (such as the super-folding GFP and the SNAP-tag) also enabled or enhanced their ability to LLPS, despite that both proteins are unable to phase separate even at high protein concentrations. Finally, we demonstrate that oligomerization is critical for RNA-binding deficient TDP43 mutants (R171A and F149L) to form spherical structures in cells, where removing oligomerization completely abrogates these structures. Taken together, our results suggest that oligomerization could be a general mechanism to enhance LLPS and that future research could search for oligomerization domains as a signature to reveal the unexplored LLPS behavior of a wider range of cellular proteins.
RESULTS
The NTD of TDP43 enhances LLPS of the RBDs.
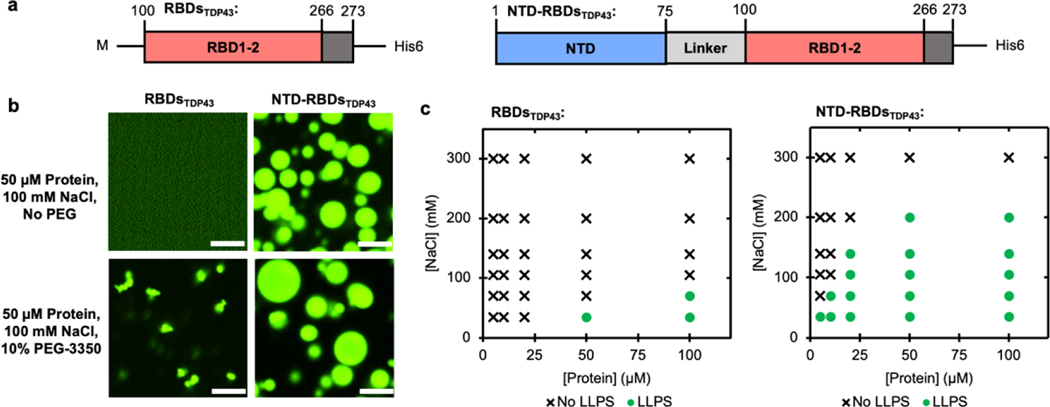
To study the effect of oligomerization on LLPS, we started with a truncation of TDP43 consisting of only the RBD (RBDsTDP43, Figure 1a, left), which was purified from E. coli using a C-terminal His6-tag. To image RBDsTDP43 using fluorescent microscopy, we covalently labeled RBDsTDP43 using a fluorescein-5- maleimide that conjugated to cysteines. The fluorescein labeled RBDsTDP43 was mixed with unlabeled RBDsTDP43 as a ratio of 1:19, thus minimizing the possible interference arising from the fluorescein labels. At 100 mM NaCl, no LLPS is observed (Figure 1b, upper-left). Polyethyleneglycol (PEG) has been previously reported to enhance LLPS of proteins in a manner believed to be dependent on its molecular crowding effect[5, 7, 8, 34, 35]. Therefore, 10% w/v PEG-3350 was used to induce LLPS of RBDsTDP43 at room temperature using 50 μM protein (Figure 1b, lower-left). In this condition, we observed punctate droplets about 1 μm in diameter with a roughly spherical morphology (Supplementary Figure S1a). Because of the size of the droplets, we were unable to examine their liquidity using FRAP. Whereas we found that droplet formation was reversible with the addition of salt or DNA oligos with a sequence previously reported to bind TDP43 (TG8) (Supplementary Figure S1b) [36, 37], suggesting that the observed phenomenon was not aggregation.
Figure 1 |. The NTD of TDP43 enhances LLPS of RBDsTDP43.
(a) The two constructs used in this figure, NTD-RBDsTDP43 (residues 1–273) and RBDsTDP43 (residues 100–273), each with a C-terminal His tag. NTD, N-terminal domain; RBD, RNA-binding domain. (b) Fluorescent confocal microscopy of each construct, with or without crowding agents. Each sample contains 50 μM total protein, consisting of 45 μM unlabeled protein and 5 μM protein labeled with fluorescein via maleimide conjugation. Scale bar is 5 μm. (c) Phase diagrams of RBDsTDP43 (left) and NTD-RBDsTDP43 (right) at varying salt and protein concentrations. LLPS determined by turbidity (Supplemental Figure S1a–b) and defined as absorbance > 0.3 AU.
To study the effect of oligomerizing the RBDsTDP43, we then generated a protein construct adding back TDP43’s homo-oligomerizing NTD connected to the RBDs using the native 25 amino-acid charged linker (NTD-RBDsTDP43, Figure 1a, right). Using a subset of fluorescein-labeled NTD-RBDsTDP43 as described above, we observed significant LLPS of 50 μM protein at 100 mM NaCl, at room temperature in the absence of PEG-3350 (Figure 1b, upper-right). The droplets were distinctly spherical at about 2–5 μm in diameter. To confirm the liquidity of these droplets, we observe partial FRAP recovery at 4 min after inducing LLPS (Supplementary Figure S1c, black). We attribute the lack of full recovery to the rapid hardening of the droplets, verified by a complete lack of FRAP recovery at 11 minutes (Supplementary Figure S1c, gray). Furthermore, droplets exhibited clear spherical morphologies (Supplementary Figure S1d) and droplets formed by NTD-RBDsTDP43 exhibited obvious merging behavior at the time scale of seconds, supporting the liquid material state of these droplets (Supplementary Figure S1e and Supplementary Movie S1). With the addition of 10% w/v PEG-3350, the droplets increase in diameter to a range of 2 μm to 10 μm (Figure 1b, lower-right), demonstrating directly the significant enhancement in phase behavior over RBDsTDP43. Given this enhancement in LLPS by the addition of NTD, we next determined if the NTD itself was able to undergo LLPS in the absence of RBDsTDP43. Consistent with previous reports[31], a construct consisting of the NTD and linker-sequence aggregated into fiber-like structures (Supplementary Figure S2).
To further understand the effect of the NTD on LLPS of the NTD-RBDsTDP43 construct, we used visible light scattering to monitor LLPS in a variety of salt and protein concentrations. At 395 nm, we observed an absorbance value of at least 0.3 AU in samples containing liquid droplets that can be confirmable by microscopy, providing us with a cutoff for declaring LLPS. For the multivalency-driven LLPS process, droplet formation is favored by low ionic strength and high protein concentrations. Consistent to this knowledge, we found that the RBDsTDP43 were capable of LLPS without crowding agents only at low salt concentrations (35–70 mM NaCl) and high protein concentrations (50–100 μM protein, Figure 1c and Supplementary Figure S3a). By contrast, the NTD-RBDsTDP43 were capable of LLPS at NaCl concentrations as high as 200 mM and at protein concentrations as low as 5 μM (Figure 1c and Supplementary Figure S3b). Thus, the NTD is capable of enhancing LLPS of the RBDsTDP43 by both lowering the protein concentration and decreasing the inhibitory effect of NaCl on LLPS.
NTD does not prevent protein-specific inhibition of LLPS.
The observation that NTD does not undergo LLPS indicates that the interactions between the fused proteins is essential for maintaining protein droplets, as opposed to the fibers formed by purified NTD (Supplementary Figure S2). To demonstrate this notion, we next determined if fusion of the NTD altered known inhibition of the protein droplets. DNA binding has previously been shown to inhibit TDP43 aggregation and LLPS[36, 38]. To test if this was true with RBDsTDP43, we added ssDNA either with the TG8 or CA8 sequences that bind with TDP43[39]. Using buffer containing 35 mM NaCl and 10% PEG-3350, we observed complete inhibition of turbidity using 50 μM protein and 50 μM of either ssDNA (Supplemental Figure S3c). We also observed inhibition of LLPS using 8 mM ATP, but interestingly we only found a marginal effect using 5% w/v 1–6 hexanediol, a molecule reported previously to inhibit LLPS[40]. Similar observations were made using NTD-RBDsTDP43 droplets that were formed using 50 μM protein and 100 mM NaCl in the absence of crowding agents (Supplemental Figure S3d), suggesting that the inhibitory effect of RBDsTDP43 interacting with DNA or ATP was not abrogated by fusion with the NTD. Taken together, these results suggest that the RBDsTDP43 intrinsically undergoes LLPS, which is enhanced by the NTD.
NTD enhances LLPS of a variety of globular proteins.
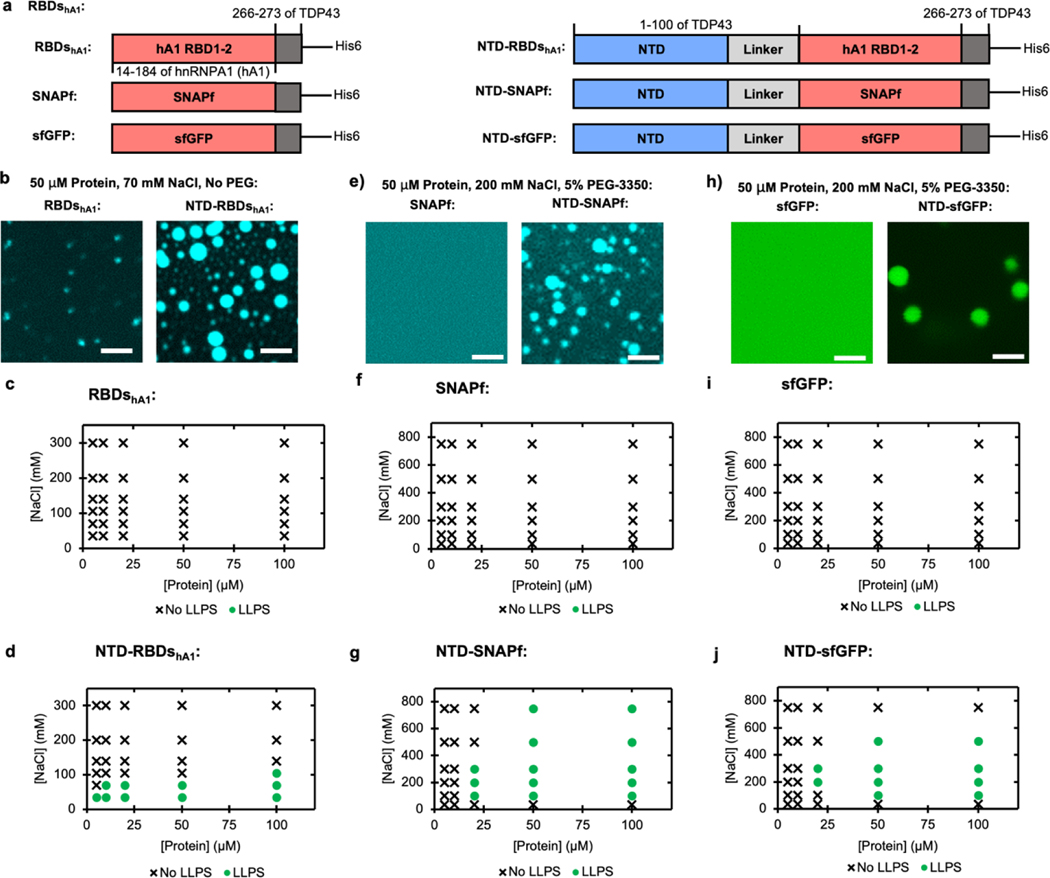
After observing the ability of TDP43’s NTD to enhance LLPS of RBDsTDP43, we sought to understand whether this effect would occur with other proteins as well. We started by studying the RBDs of hnRNPA1 (Figure 2a, top-left). hnRNPA1 is an RNA-binding protein with a similar domain structure to TDP43, having two adjacent RBDs and a prion-like CTD. While similar in function, the RBDs of each protein only contain about 30% sequence homology. A construct comprising the RBD1 and RBD2 of hnRNPA1 (RBDshA1), with a subset of protein covalently labeled with coumarin via maleimide conjugation, was not observed to phase separate at 50 μM protein at 70 mM NaCl without crowding agents (Figure 2b). Only with the addition of 10% w/v PEG-3350 were small punctates observed (Supplementary Figure S4a). To determine if the NTD would affect RBDshA1, we then created a construct that fused the NTD of TDP43 with the RBDs of hnRNPA1 (NTD-RBDshA1) (Figure 2a, top-right), with a subset of protein covalently labeled with coumarin via maleimide conjugation. At 70 mM NaCl, we observed that the NTD-RBDshA1 construct demonstrated droplet formation without the addition of crowding agents, with 50 μM protein forming droplets 2–3 μm in diameter (Figure 2b). In these conditions, the droplets had a morphology consistent with droplets wetting on the coverslip (Supplementary Figure S4b). This enhancement in LLPS is consistent across a range of salt and protein concentrations, where RBDshA1 is not observed to undergo LLPS under any salt condition without crowding agents (Figure 2c and Supplementary Figure S5a). NTD-RBDshA1, however, is capable of phase separation at concentrations as low as 5 μM and salt conditions less than 140 mM NaCl (Figure 2d and Supplementary Figure S5b). The NTD of TDP43 then appears to perform a similar function with RBDshA1 as with RBDsTDP43, lowering the concentration needed to LLPS and enabling LLPS without the addition of any crowding agents.
Figure 2 |. The NTD of TDP43 can be used to enhance LLPS of other proteins.
(a) Schematics of protein fusions, with or without TDP43’s NTD: hnRNPA1 RBDs constructs used in (b - d), SNAPf constructs used in (e - g), and sfGFP constructs used in (h - j). (b) Fluorescent microscopy of hnRNPA1 RBDs without (left) or with (right) fused NTD. Each sample contains 50 μM total protein, including 45 μM unlabeled protein and 5 μM protein labeled with fluorescein via maleimide conjugation. Scale bars are 5 μm. (c-d) Phase diagrams of hnRNPA1 RBDs without (c) or with (d) fused NTD at varying salt and protein concentrations without the addition of PEG. LLPS determined by turbidity values (Supplemental Figure S5a–b) with LLPS defined as absorbance > 0.3 AU. (e) Fluorescent microscopy of SNAPf without (left) or with (right) fused NTD. Each sample contains 50 μM protein with 5 μM coumarin labeled through conjugation to SNAPf’s target substrate, benzyl guanine. Scale bars are 5 μm. (f-g), Phase diagrams of SNAPf without (f) or with (g) fused NTD at varying salt and protein concentrations with the addition of 5% w/v PEG-3350. LLPS determined by turbidity values (Supplemental Figure S5c–d) with LLPS defined as absorbance > 0.3 AU. (h) Fluorescent microscopy of sfGFP without (left) or with (right) fused NTD. Scale bars are 5 μm. (i-j) Phase diagrams of sfGFP without (i) or with (j) fused NTD at varying salt and protein concentrations with the addition of 5% w/v PEG-3350. LLPS determined by turbidity values (Supplemental Figure S5e–f) with LLPS defined as absorbance > 0.3 AU.
We next sought to determine if the LLPS enhancement effect of the NTD could extend to fusions with structurally unrelated proteins. SNAP-tag is a protein capable of covalently binding to a biorthogonal small molecule, and as such is commonly used as a fusion tag to fluorescently label proteins of interest. To determine if the NTD of TDP43 was capable of driving SNAP-tag LLPS, we first worked with a SNAP-tag variant SNAPf[41] (Figure 2a, middle-left), which is unable to undergo LLPS (Figures 2e, 2f and Supplementary Figure S5c). Using an NTD-fused construct (NTD-SNAPf, Figure 2a, middle-right), covalently labeled using sub-saturating amounts of coumarin attached to SNAPf-ligand benzyl-guanine, we were able to observe droplets with 50 μM protein using 200 mM NaCl and 5% PEG-3350, with the droplets mostly appearing about 2 μm in diameter (Figure 2e). These droplets had a spherical morphology (Supplementary Figure S4c). Using turbidity measurements, it appears that in contrast to both NTD-RBDs constructs, NTD-SNAPf favors salt concentrations from 100 to 500 mM NaCl, with no LLPS at 35 mM NaCl (Figure 2g and Supplementary Figure S5d).
Similar observations were made with a super-folding green fluorescent protein[42] (sfGFP; Figure 2a, bottom-left). Using purified sfGFP, LLPS of sfGFP was only observed when at least 1 mM sfGFP was incubated with 10% PEG-3350 (Figure 2h, Supplementary Figures S4d and S5e), and the concentration required to phase separate was inconsistent across protein preps. With a construct with TDP43’s NTD fused to sfGFP (NTD-sfGFP, Figure 2a, bottom-right), we were able to observe droplets from 2 to 5 μm in diameter using 50 μM protein at 200 mM NaCl and 5% PEG-3350 (Figure 2h). NTD-sfGFP droplets were also observed to have a spherical morphology (Supplementary Figure S4e). Turbidity measurements showed NTD-sfGFP favored LLPS between 100 and 500 mM NaCl at most protein concentrations (Figure 2j and Supplementary Figure S5f). Interestingly, no turbidity was observed at 35 mM NaCl, similar to NTD-SNAPf and in contrast to either NTD-RBDs construct. While the reasoning for these differences is not clear, we speculate that it is related to differences in surface charge or conformation of each fused protein. Taken together, our results indicate that NTD is capable of lowering the protein concentration and crowding agents needed to allow various globular, structured proteins to undergo LLPS.
Oligomerization state correlates to LLPS of RBDsTDP43.
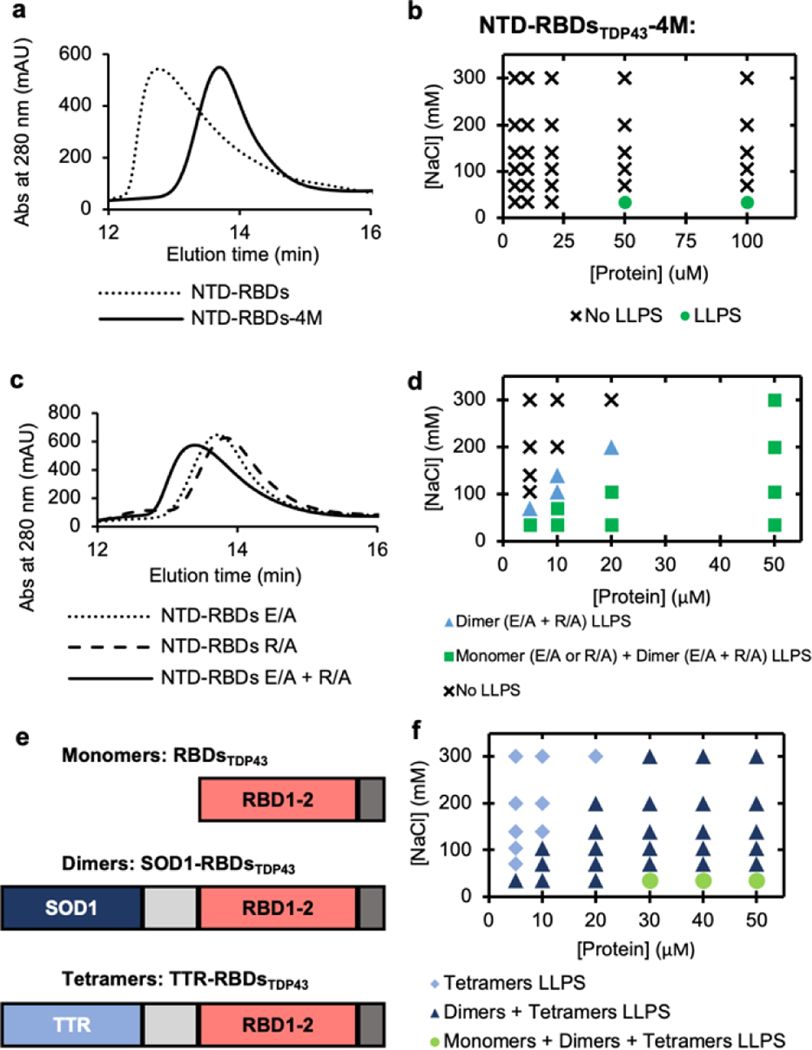
Having established that the NTD of TDP43 was capable of enhancing LLPS of a variety of proteins, we sought to understand how NTD exerts this effect. Previous studies have shown that the NTD is capable of self-oligomerization[43], and this has been regarded as a critical part of aggregation of TDP43 in vivo[33]. To determine if oligomerization was responsible for LLPS of the NTD-RBDsTDP43, we used a set of previously reported mutations (E17A/E31A/R52A/R55A, herein named NTD-RBDsTDP43-4M) that had been shown to reduce oligomerization[31]. Using size-exclusion chromatography (SEC), we were able to show that NTD-RBDsTDP43-4M eluted at a higher retention time, indicating lower apparent molecular weight and decreased oligomerization (Figure 3a). Using turbidity measurements, we found that the ability of the NTD-RBDsTDP43-4M construct to LLPS was comparable to that of RBDsTDP43 (Figure 3b and Supplementary Figure S6a), indicating that oligomerization is primarily responsible for NTD-RBDsTDP43’s enhanced ability to LLPS.
Figure 3 |. Oligomerization as the underlying factor in LLPS enhancement.
(a) Size exclusion chromatography (SEC) elution profile comparing NTD-RBDsTDP43 and oligomerization deficient mutant NTD-RBDsTDP43-4M. (b) Phase diagram of NTD-RBDsTDP43-4M at varying salt and protein concentrations without the addition of PEG. LLPS determined by turbidity values (Supplemental Figure S6a) with LLPS defined as absorbance > 0.3 AU. (c) SEC elution profile comparing monomeric NTD-RBDsTDP43 E/A or R/A to a sample of both monomers incubated together. (d) Phase diagram of salt vs protein concentration for E/A and R/A mutants. Marker denotes if no liquid-liquid phase separation (LLPS) occurs for any mixture (circle), LLPS occurs only for a dimeric mixture of NTD-RBDsTDP43 E/A and R/A (triangle), or if LLPS occurs for E/A, R/A, and a mixture of the two (square). Protein concentration shown as total protein concentration, with dimeric mixture containing 50% E/A and 50% R/A. Samples prepared in 10% w/v PEG-3350. LLPS determined by turbidity values (Supplemental Figure S6c–e) with LLPS defined as absorbance > 0.3 AU. (e) Schematics of protein constructs used to create monomers (RBDsTDP43), dimers (SOD1-RBDsTDP43), and tetramers (TTR-RBDsTDP43). (f) Phase diagram of salt vs protein concentration for oligomeric constructs. Marker denotes if LLPS occurred with only tetramer (diamond), tetramer and dimer (triangle), or tetramer, dimer and monomer (circle). Samples prepared in 10% w/v PEG-3350. LLPS determined by turbidity values (Supplemental Figure S5f–h) with LLPS defined as absorbance > 0.3 AU.
We next explored whether we could control the enhancement to LLPS by tuning the extent of oligomerization. NTD forms oligomers through interactions between two oppositely charged faces[31]. By mutating only one side or the other to alanines, we created two constructs (Supplementary Figure S6b), including one construct that mutated the positive face of the NTD to alanines (R52A/R55A, herein named NTD-RBDsTDP43-R/A) and the other construct that mutated the negative face of the NTD to alanines (E17A/E31A, herein named NTD-RBDsTDP43-E/A), thus reducing the oligomerization of NTD- NTD-RBDsTDP43. As expected, when we used SEC to observe that the NTD-RBDsTDP43-R/A and NTD-RBDsTDP43-E/A exhibited much reduced oligomerization, resulting in retention time that was slightly lower than the monomeric NTD-RBDsTDP43-4M (Figure 3c). Accordingly, both constructs underwent LLPS with 10% PEG-3350 at low salt concentrations (Supplementary Figure S6c–d). When incubated as an equal ratio mixture (Supplementary Figure S6b), we found that NTD-RBDsTDP43-R/A and NTD-RBDsTDP43-E/A likely formed a dimeric complex, whose elution profile shifted to a retention time that was in the middle of NTD-RBDsTDP43 and NTD-RBDsTDP43-4M (Figure 3c). Using the NTD-RBDsTDP43-R/A•NTD-RBDsTDP43-E/A dimer complex, we observed turbidity signals at salt and protein concentration ranges broader than that of the NTD-RBDsTDP43-R/A (or NTD-RBDsTDP43-E/A) monomer (Supplementary Figure S6e), suggesting the dimer’s intermediate LLPS capacity in between the monomeric and oligomeric structures. To better visualize how dimerization affects LLPS, we constructed a diagram of salt vs protein concentration for both the NTD-RBDsTDP43-E/A or R/A monomer (green squares in Figure 3d) and the NTD-RBDsTDP43-R/A•NTD-RBDsTDP43-E/A dimer (blue triangles in Figure 3d). Using turbidity values > 0.3 AU as an indicator of phase separation, dimerization clearly decreased both the protein concentration needed to LLPS and the inhibitory effect of salt on LLPS.
Having established that manipulation of the oligomerization state of the NTD affected LLPS of NTD-RBDsTDP43, we sought to expand this to the other NTD-fused proteins. We applied the 4M mutations to create NTD-RBDshA1-4M, NTD-SNAPf-4M, and NTD-sfGFP-4M. Without the additions of PEG, NTD-RBDshA1-4M only underwent LLPS at much lower salt and higher protein concentrations compared to NTD-RBDshA1 (Supplementary Figure S6f; compare to Figure 2d and Supplementary Figure S5b). Using 5% PEG-3350, we observed that NTD-SNAPf-4M and NTD-sfGFP-4M exhibited significant reductions in overall turbidity compared to NTD-SNAPf and NTD-sfGFP (Supplementary Figures S6g and S6f; compare to Figure 2g and j, and Supplementary Figures S5d and S5f). Thus, abrogation of oligomerization reduces the conditions under which the protein will undergo LLPS.
We next substituted NTD with other oligomerization domains, to determine if the above effect was directly tied to oligomerization or another unidentified behavior of the NTD. To this end, we replaced the NTD of NTD-RBDsTDP43 with either superoxide dismutase (SOD1), a protein known to form stable dimers[44], or transthyretin (TTR), a protein known to form stable tetramers[45] (Figure 3e). We found that purified both SOD1-RBDsTDP43 and TTR-RBDsTDP43 formed droplets with dimensions about 1–2 μm and 1–5 μm, respectively (Supplementary Figure S7). We also measured turbidity at a variety of salt and protein concentrations (Supplementary Figure S8) and plotted the turbidity data in a phase separation diagram using turbidity values of 0.3 AU as a criterion for LLPS (Figure 3f). We found that the critical protein concentration needed to undergo LLPS is lowered proportionately to the degree of oligomerization, with this value being 5 μM, 10 μM and 20 μM for the tetrameric TTR-RBDsTDP43, dimeric SOD1-RBDsTDP43, and monomeric RBDsTDP43, respectively. Thus, these results indicate that oligomerization is a general principle to enhance LLPS of proteins.
NTD drives colocalization of NTD-fusion proteins.
If oligomerization is promoting LLPS, fusing the NTD domain to different proteins should be able to form heterooligomers and drive LLPS of these fusion proteins to colocalize in the same droplets. Therefore, we attempted to test whether NTD-RBDsTDP43 could recruit other NTD-fusion proteins into its droplets using the following two sets of experiments. In the first set of experiments, NTD-RBDsTDP43 was incubated with NTD-sfGFP that was unable to form droplets at the defined conditions. We would expect that the hetero-oligomeric NTD-RBDsTDP43• NTD-sfGFP complex would recruit NTD-sfGFP into the NTD-RBDsTDP43 droplets. In the second set of experiments, NTD-RBDsTDP43 was incubated with NTD-RBDshA1 in conditions that both proteins were able to undergo LLPS separately. We would expect that the NTD-RBDsTDP43•NTD-RBDshA1 complex, either mixed prior to or after LLPS, would colocalize both proteins in the same droplets.
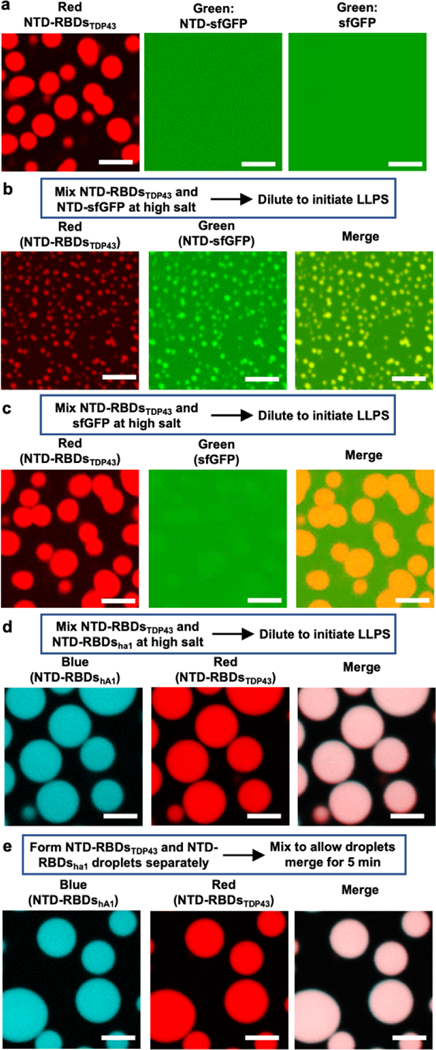
For the first set of experiments, we chose 50 μM protein and 100 mM NaCl, because this condition allows rhodamine-labeled NTD-RBDsTDP43, but not NTD-sfGFP and sfGFP, formed spherical droplets in the absence of crowding agents (Figure 4a). To test if NTD-RBDsTDP43 would colocalize with NTD-sfGFP in droplets, 50 μM of each protein were mixed together and incubated overnight at 4 °C in buffer containing high salt (>300 mM NaCl) to ensure hetero-oligomer formation. After dilution to 100 mM NaCl, we observed droplets that exhibit fluorescent signals from both rhodamine and NTD-sfGFP (Figure 4b), with a visible background of diffusive NTD-sfGFP that may not have formed complexes with NTD-RBDsTDP43. As hypothesized, the hetero-droplets were smaller than the homo-droplets of NTD-RBDsTDP43, suggesting the hetero-oligomers form droplets with a size between either two proteins as homo-oligomers. To verify that hetero-oligomers were responsible for this change in droplet size, this experiment was carried out by incubating sfGFP with NTD-RBDsTDP43. We observed sfGFP preferentially partitioned into the diffusive phase with only slight colocalization to the droplets (Figure 4c). In addition, the morphology and dimension of the droplets were comparable to droplets formed by NTD-RBDsTDP43 homo-oligomers (Figure 4a, left). We found then that NTD-sfGFP was recruited into droplets by the NTD, as sfGFP was not colocalized in droplets. These results suggest that hetero-oligomers undergo phase separation at conditions determined by the proteins fused to the NTD that are driving phase separation.
Figure 4 |. NTD drives colocalization of NTD-fusion proteins.
(a) Fluorescence microscopy of phase-separated NTD-RBDsTDP43 and diffusive NTD-sfGFP or sfGFP (50 μM protein at 100 mM NaCl). (b-c) Colocalization of NTD-RBDsTDP43 with NTD-sfGFP (b) or sfGFP (c). Left, 50 μM protein at 100 mM NaCl. Right, NTD-RBDsTDP43 incubated with either NTD-sfGFP (top row) or sfGFP (bottom row). Channels are Red, showing NTD-RBDsTDP43, Green, showing NTD-sfGFP or sfGFP, and a merge channel. NTD-RBDsTDP43 sample consists of 45 μM unlabeled protein and 5 μM protein labeled with rhodamine via maleimide conjugation. (d) Colocalization of NTD-RBDsTDP43 and NTD-RBDshA1 mixed before inducing LLPS. Left, schematic of experiment. Right, fluorescent confocal microscopy demonstrating colocalization. Channels are Blue, showing NTD-RBDshA1, Red, showing NTD-RBDsTDP43, and a merge channel. NTD-RBDsTDP43 sample prepared as in a. NTD-RBDshA1 sample consists of 45 μM unlabeled protein and 5 μM protein labeled with coumarin via maleimide conjugation. (e) Colocalization of NTD-RBDsTDP43 and NTD-RBDshA1 mixed after inducing LLPS. Left, schematic of experiment. Right, fluorescent confocal microscopy demonstrating colocalization. Channels and sample preparations as in b. All scale bars are 5 μm.
For the second set of experiments, we tested NTD-RBDsTDP43 labeled with rhodamine and NTD-RBDshA1 labeled with coumarin, each of which would separately undergo LLPS at 70 mM NaCl and without crowding agents (Figure 1c and Figure 2b). First, we tested whether the two proteins would colocalize if they were incubated together at high salt before dilution to induce LLPS (Figure 4d). We observed robust droplet formation after dilution to 70 mM NaCl with colocalization of fluorescent signal, and these hetero-droplets exhibited larger sizes than homo-droplets of either proteins (Figure 4d). Next, we tested whether droplets of NTD-RBDsTDP43 and NTD-RBDshA1 could merge after undergoing LLPS. To this end, we prepared droplets of each proteins separately and mixed them 5 minutes after droplet formation. We found droplets that exhibit fluorescent signal from both proteins (Figure 4e), suggesting that droplets created by NTD-fusions can colocalize and merge, as would be expected of liquid droplets.
NTD is required for TDP43 nuclear bodies.
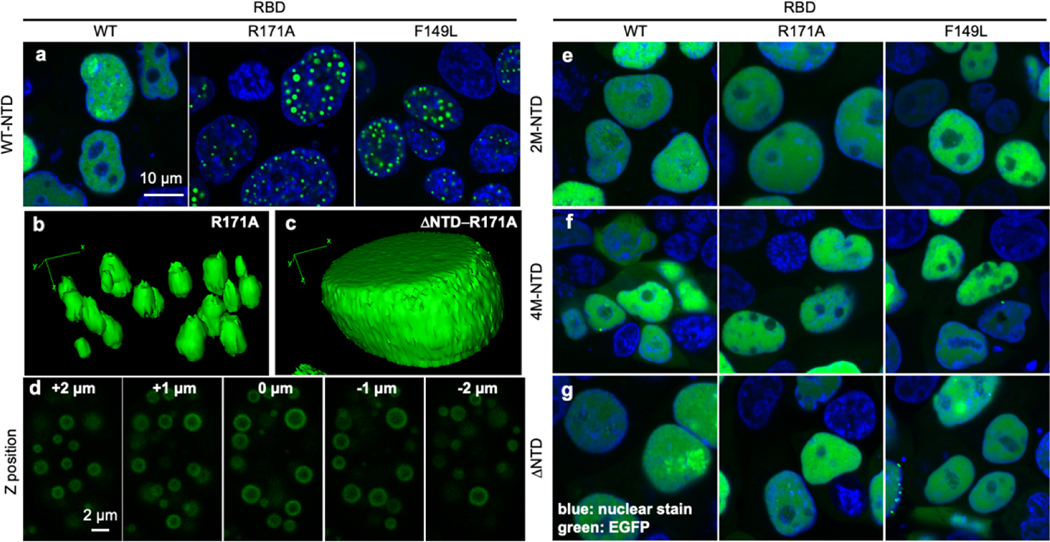
We next asked whether NTD is essential to nuclear bodies formed by TDP43. Previous studies have reported that stress conditions or RNA-binding deficient mutations force TDP43 to go into phase-separated nuclear bodies recently termed “anisosomes” [17, 18, 38, 46]. To this end, we constructed F149L and R171A mutants previously reported to result in decreased RNA-binding affinity (as measured by increased Kd values) from 20.6 nM of the wild-type TDP43 to 86.2 nM and 124.2 nM, respectively[47]. Using enhanced green fluorescence protein (EGFP) fused to the C-terminal of wild-type and mutant TDP43 that were transiently expressed in HEK293T cells, we observed diffusive nuclear fluorescence signal of TDP43-EGFP and granular structures within the nucleus for both TDP43-F149L-EGFP and TDP43-R171A-EGFP (Figure 5a). To ensure the granules were spherical in morphology, the z-stack slices of a selected cell nucleus expressing TDP43-R171A-EGFP was rendered in 3-D space and the morphology of the granule structures were analyzed. The granule structures appeared to be in oval shape in z plane instead of a true spherical shape when presented in 3D rendering (Figure 5b), likely to be an artifact of the diffraction limit of the light microscopy in z plane, where the best resolution between two objects in x-y plane is ~250 nm and ~300 nm in z plane. TDP43-R171A formed spherical and shelled granules that appear to be hollow at the core (Figure 5d), consistent to the structure of recently reported anisosomes that have liquid-like properties [17].
Figure 5 |. Oligomerization is required for LLPS of TDP43 in vivo.
(a) Fluorescence microscopy of transiently transfected GFP-tagged TDP43-WT, TDP43-R171A and TDP43-F149L in HEK293T cells. (b) Z-stack scan of TDP43-R171A-EGFP shows the structure of anisosomes. (c-d) 3D reconstruction of one nucleus containing TDP43-R171A-EGFP (c) and ΔNTD-R171A-EGFP (d). (e-g) Fluorescence microscopy of transiently transfected GFP-tagged TDP43-WT, TDP43-R171A and TDP43-F149L with oligomerization-deficient NTD mutations 2M (e) or 4M (f), or removal of NTD (residues 2–75, g).
To test the role of NTD in formation of nuclear bodies, charged residues within the NTD that are essential for protein oligomerization were mutated to alanine. In particular, a two-amino acid replacement (E17A/E21A, 2M) or a four-amino acid replacement (E17A/E21A/R52A/R55A, 4M), previously reported to abrogate oligomerization[31]. We have chosen not to use the S48E mutation, previously reported to abrogate oligomerization[24] and used in previous studies of TDP43 LLPS[17] to avoid complications that biologically-relevant post-translational modifications could introduce. Furthermore, a ΔNTD construct was generated by deleting the entire NTD (residues 2–75) up to the nuclear localization signal sequence. These mutations (2M, 4M, and ΔNTD) were introduced to TDP43 with both the normal (WT) and deficient RNA-binding constructs (F149L and R171A). As expected, all EGFP-fused NTD mutants exhibited a diffusive structure within the nucleus upon expression (Figures 5e–f), a stark difference from the nuclear granules formed by sequences containing the wild-type NTD sequences (Figure 5a). Three-dimensional distribution of EGFP-ΔNTD-TDP43-R171A expressed in HEK293T cells were also monitored through z-stack. The distribution of EGFP-ΔNTD-TDP43-R171A is evenly throughout the nucleoplasm and distinct from the nuclear bodies formed by EGFP-TDP43-R171A (Figure 5c).
The difference in granular and diffusive structures can be further visualized using intensity profiles of EGFP signal along a cross-section of a nucleus. A diffusive structure, such as EGFP-TDP43 WT, shows a cross-section with medium fluorescent values and broad peaks or troughs representing slight differences in nuclear distribution (Supplementary Figure S9 a+b). Granular structures created by R171A or F149L EGFP-TDP43 constructs, in contrast, exhibit peaks of high brightness separated by low fluorescent values corresponding to almost exclusive distribution within granules (Supplementary Figure S9 c–f). We observe that for all NTD-mutant constructs, cross-section profiles are more representative of diffusive structure (Supplementary Figure S9 g–x). For further quantification, we can compare the area that EGFP-TDP43 fluorescence covers compared to the area of the nucleus, determined by nuclear stain. Diffusive structures would be expected to occupy an area comparable to that of the nucleus, while granular structures occupy a much smaller area than the nucleus. With the wide-field images used to generate Figure 5 a and e–g, we remove nuclei not expressing EGFP-TDP43 and calculate the total area of EGFP-TDP43 (green) and nucleus (blue). We observe that constructs exhibiting diffuse structures, EGFP-TDP43 WT and all NTD mutants, exhibit green area/blue area values around 1 (Supplementary Figure S10, black bars). Only WT NTD constructs R171A and F149L EGFP-TDP43 demonstrate low green area/blue area values of 0.19 to 0.33, consistent with these constructs’ granular structures (Supplementary Figure S10, grey bars). These data suggest that the oligomerization of NTD is essential for the nuclear body formation of TDP43.
DISCUSSION
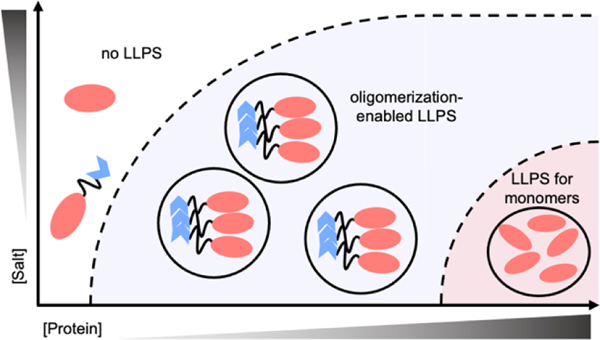
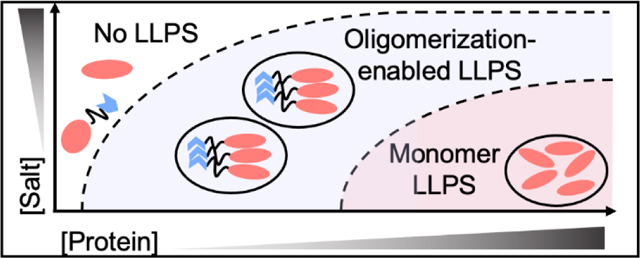
In this work, we demonstrate that the N-terminal domain of TDP43 promotes LLPS of globular protein domains via oligomerization. Our data suggest that the oligomerization could be a general mechanism to enhance LLPS of proteins. Results indicate that oligomerization of NTD lowers critical protein concentrations to undergo LLPS, albeit the actual thermodynamic parameters being modified are as-yet unclear. While not every protein that can undergo LLPS is known to oligomerize, it seems apparent that oligomerization can enhance LLPS when it does occur. We propose that oligomerization is enforcing interactions of proteins in such a way that makes them more readily undergo phase transitions at lower critical protein concentrations and higher salt concentrations (Figure 6), with an additional thermodynamic gain from the entropic considerations of having larger species present in solution.
Figure 6 |. Increasing oligomerization lowers the boundary to LLPS.
A theoretical phase diagram, the dashed-line representing the boundary between 1-phase and 2-phase conditions. As oligomerization increases, the boundaries to LLPS decrease for globular proteins.
Using TDP43 and its homo-oligomerizing NTD as a model system, we report here that NTD-RBDsTDP43 undergoes LLPS in vitro and this is, to our knowledge, the first time that LLPS of TDP43 has been reported without the presence of its PrLD. We note that TDP43 appears to contain two LLPS-capable systems, mediated by RBDs or PrLD. These two systems also appear to be driven by opposite multivalent interactions, with NTD-RBDsTDP43 enhanced at low salt conditions (Figure 1c, right) and the PrLD enhanced at high salt conditions according to previous reports[21–23]. Further, although the PrLD is essential for LLPS in vivo, the few available reports of full-length TDP43 undergoing LLPS in vitro also demonstrate a dependence on low salt conditions[24], consistent with NTD-RBDsTDP43. As such, the role of RBDs in LLPS clearly warrants more study.
NTD-RBDsTDP43 makes for a good example for the following features: it undergoes LLPS in reasonable conditions in vitro, it is composed of well-structured domains that do not aggregate readily, and NTD has a set of mutations that allows for manipulation of oligomerization state[31]. Furthermore, NTD-RBDsTDP43 provides us with a two-part model system to study LLPS by manipulating either the oligomerization domain (NTD) or the fused globular domain (RBDsTDP43). By replacing NTD with other proteins that undergo oligomerization, we are able to demonstrate that the enhancement of LLPS is due to its oligomerization but not due to the presence of just the NTD sequence. Because the NTD has a fairly weak binding affinity (Kd ~2.4 μM) [43], NTD-RBDsTDP43 oligomers are likely in a constant state of exchange, which is a process that we had initially hypothesized to play an important role in enhancing LLPS. By substituting the NTD with SOD1 and TTR with much stronger binding affinities (SOD1: Kd ~0.1 nM [48]; TTR: Kd very low [49]), it becomes apparent that even very stable oligomers exhibit the same LLPS enhancement, indicating that the oligomeric state itself is directly responsible for the LLPS enhancement.
An interesting observation is that the NTD enhances the LLPS of proteins beyond RBDsTDP43. All of the tested fusion proteins have known globular structures, without a reported intrinsically disordered region. While we were able to find LLPS conditions for RBDshA1 and or sfGFP (at >1 mM concentrations), we were not able to find conditions where SNAPf would undergo LLPS. While both proteins have been used extensively as fusion proteins to study LLPS, we have not found reports of either protein undergoing LLPS without the addition of another macromolecule like DNA [50]. This may indicate that there is a variety of proteins without known phase separation behavior that can be induced to undergo LLPS by oligomerization. As the physiological importance of LLPS expands, the search for LLPS-capable proteins is ongoing. There has been heavy focus on finding low-complexity, intrinsically disordered domains like the PrLD in TDP43, as these domains have been demonstrated to drive LLPS in a number of proteins [35, 51]. This work, however, demonstrates the importance of considering both globular and oligomerization domains in the search for LLPS-capable proteins. In addition to searching for disorder and a lack of sequence complexity, we propose that the structural features that govern LLPS be identified and used as well to discover new proteins that undergo LLPS. Within structural database libraries, we expect there will be a number of ordered proteins that can be found to undergo LLPS in specific contexts.
METHODS
Plasmids
EGFP-TDP43: The TDP43 gene was amplified from the wtTDP43tdTOMATOHA (purchased from Addgene, plasmid #28205) and was subcloned into pEGFP_N1 vector (purchased from Addgene, plasmid #32751) using the Polymerase Incomplete Primer Extension (PIPE) PCR method. E. coli DH5a competent cells (NEB, C2987I) were transformed with mixture of PIPE PCR products. A single transformant was collected and inoculated into 7 mL LB medium containing 50 ng/mL of kanamycin, grown for 16 h shaking at 37 °C. Cells were harvested and lysed for DNA extraction using a plasmid purification kit (Omgega Bio-Tek, E.Z.N.A. Plasmid Mini Kit). The successful construction of TDP43-eGFP plasmid was confirmed by sanger sequencing.
EGFP-TDP43 mutants: Various TDP43 mutants were generated using TDP43-eGFP plasmid through Quick Change (QC) PCR. Specific primer sets were created to introduce single or multiple point mutations within NTD or RBD domains of TDP43 gene leading to missense mutation(s) in the protein sequence. QC PCR product was then used to transform DH5α competent cells. A single transformant was collected and inoculated into 7 mL LB medium containing 50 ng/mL of kanamycin, grown for 16 h shaking at 37 °C. Cells were harvested and lysed for DNA extraction using a plasmid purification kit (Omgega Bio-Tek, E.Z.N.A. Plasmid Mini Kit). The mutation was confirmed by sanger sequencing.
NTD-RBDsTDP43: The TDP43 gene was amplified from the wtTDP43tdTOMATOHA (purchased from Addgene, plasmid #28205) and was subcloned into pET29b vector (a gift from the Kelly lab) using the PIPE PCR method, as described above. The region coding for the Prion-Like Domain of TDP43 was removed by QC primers that spanned from the RBDsTDP43 to the HisTag. Sequencing was confirmed by Sanger sequencing.
NTD-RBDsTDP43 Variants: All other constructs for use in vitro were created using the NTD-RBDsTDP43 as a template. Point mutations were created using QC PCR as described above. Domain substitutions were created using PIPE PCR as described above. All constructs were sequenced by Sanger sequencing to confirm final sequence.
Protein expression and purification.
Except where mentioned below, all proteins were expressed in Escherichia coli BL21 (DE3) with C-terminal His tags. Bacterial cells were grown in LB supplemented with 50 mg/L kanamycin at 37 °C until the OD600 reached 0.6 – 0.8. Protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18 °C for 20 – 24 hrs. Cells were harvested and resuspended in Buffer A (20 mM Tris-HCl, pH 8.0, 1 M NaCl, 2 mM imidazole) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). Resuspended cells were frozen to −80 °C and thawed, then sonicated (2 sec on, 8 sec off, 5 min on-time). Lysate was cleared by centrifugation (15k rpm, 60 min) and supernatant was removed and incubated with 6 ml Profinity IMAC resin (BioRad) pre-charged with NiSO4 and equilibrated with Buffer A. After washing with Buffer A, protein was eluted using a gradient of 0–100% Buffer B (20 mM Tris-HCl, pH 8, 500 mM imidazole). Proteins were then buffer-exchanged into Storage Buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 1 mM DTT) using a 60 ml self-packed size-exclusion column with resin recovered from a HiPrep 16/60 Sephacryl S-200 HR (GE Lifesciences). Protein was then flash-frozen in liquid nitrogen and stored at −80 °C.
The following proteins were exceptions to the above: SNAPf, sfGFP, SOD1-RBDsTDP43, and TTR-RBDsTDP43 were expressed in BL21 (DE3) cells containing an ampicillin-resistant, arabinose-inducible plasmid encoding σ32-I54N, as previously reported[52]. Cells were grown in LB supplemented with 50 mg/L kanamycin and 100 mg/L ampicillin. At an OD600 of 0.3–0.4, 2 g/L arabinose was added, and the temperature was lowered to 30 °C until IPTG addition at OD600 of 0.6–0.8, as above. Protein purification continued as above through lysis and Ni-column elution. After Ni-column elution, SNAPf and TTR-RBDsTDP43 were then subjected to size-exclusion chromatography using a HiPrep 16/60 Sephacryl S-200 HR (GE Lifesciences) equilibrated with Storage Buffer, then frozen as above. sfGFP was, after Ni-column elution, dialyzed into Q Buffer A (20 mM Tris-HCl, pH 8), and loaded onto an anion exchange column, a Bio-Scale Mini UNOsphere Q Cartridge (BioRad). sfGFP bound to the column and was eluted with a gradient from 0–100% Q Buffer B (20 mM Tris-HCl, pH 8, 1 M NaCl). The protein was then buffer-exchanged into Storage buffer using the self-packed size-exclusion column as above.
Fluorescent labeling of purified proteins.
To fluorescently label proteins via maleimide conjugation, protein was thawed and a sample removed and exchanged into a buffer of 20 mM HEPES, pH 7.5, and 300 mM NaCl. Maleimide-conjugated dye (Coumarin-maleimide, Thermo Fisher, D10251; Rhodamine-maleimide, AnaSpec, AS-81441–5; Fluorescein-maleimide, Life Tech, 62245) dissolved in DMSO was added in equimolar concentrations to the protein and allowed to incubate for 1 hr at 20 °C. Protein was then exchanged back into Storage Buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 1 mM DTT).
Confocal fluorescent microscopy of in vitro droplets.
Confocal images were acquired at PSU Huck Imaging Core facility on an Olympus FluoView FV1000 confocal microscope equipped with an Olympus PlanApo 60x/1.4 oil immersion objective. Samples were imaged with a 405 nm diode laser for coumarin, 488 nm argon laser for fluorescein and sfGFP, and 543 nm helium-neon laser for rhodamine. Images were processed with Olympus FV10-ASW 4.0 viewer. All samples were prepared and imaged at room temperature, 20 °C. All samples buffered in 20 mM HEPES, pH 7.5, 1 mM DTT, and other conditions as specified.
Negative Staining Electron Microscopy.
A small drop of sample suspension (~ 10 μl) was applied on to a 400-mesh grid supported by carbon-formvar and freshly discharged. The grid was stained with 2% aq. uranyl acetate and then imaged under a FEI BioTwin TEM at 120 kV.
Fluorescence Recovery After Photobleaching (FRAP).
FRAP was conducted on Olympus Fluoview 1000 with a 60x objective and with images processed with Olympus FV10-ASW 4.0 viewer. Samples bleached and imaged with 488 nm laser. All samples were prepared and imaged at room temperature, 20 °C. All samples buffered in 20 mM HEPES, pH 7.5, 1 mM DTT, and other conditions as specified.
3D reconstruction of in vitro samples
3D reconstruction was conducted using fluorescent confocal microscopy on an Olympus Fluoview 1000 with a 60x objective, with images processed with Olympus FV10-ASW 4.0 viewer and 3D reconstruction performed by ImageJ with Fiji plugins. All samples were prepared and imaged at room temperature, 20 °C. All samples buffered in 20 mM HEPES, pH 7.5, 1 mM DTT, and other conditions as specified.
Turbidity assay.
Turbidity was assessed using absorbance readings on a Cary UV-Vis 300 (Agilent), in quartz cuvettes 16.100-Q-10/Z15 (Starna Cells, Inc.). Temperature controlled at room temperature, 20 °C. Absorbance was read at a fixed wavelength of 395 nm for all proteins except sfGFP and NTD-sfGFP, which were read at 600 nm. All samples were assayed in buffers containing 20 mM HEPES, pH 7.5, 1 mM DTT, and other additives as specified.
Size-exclusion chromatography.
Size-exclusion chromatography was performed on an Enrich SEC650 (BioRad). All samples were assayed in buffers containing 20 mM HEPES, pH 7.5, 1 mM DTT, and 300 mM NaCl unless otherwise noted.
Mammalian cell culture.
Mammalian Cell Culture: Human Embryonic Kidney 293T (HEK293T) cells were (purchased from ATCC, CRL-3216) maintained and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10 % fetal bovine serum (FBS) and 1% penicillin/streptomycin/glutamine (PSQ, Gibco) at 37 °C under 5% CO2 in a HERAcell VIOS 160i CO2 incubator (ThermoFisher Scientific). The cells are passaged using trypsin (TrypLE Express, Gibco) when confluency reached 90–95%. A new vial of cells will be revived after the current cells have been passage 20 times.
Transient transfection.
HEK293T cells were plated at 10 % confluency in either a 12-well plate (for epi-fluorescence imaging) or a 35mm Poly-D-Lysine-coated glass-bottomed MatTek dish (for confocal imaging) one day prior to the experiment. The next day (20–25 % confluency), cells were transiently transfected with TDP43 plasmids via lipofection (X-TremeGene 9 DNA Transfection Reagent, Roche). The optimal DNA to Lipofection reagent ratio has been optimized at 1 μg of DNA per 2 μL X-TremeGene 9 reagent in 100 μL Opti-MEM Reduced Serum Medium (Gibco). 1 μg of DNA was used for each single well of the 12-well plate and 3μg of DNA was used for a single 35mm MatTek dish. The transfection mixture was incubated at room temperature for 30 min before added dropwise to the culture medium. Proteins were transiently expressed for 24 h at 37 °C under 5% CO2 before imaging.
Confocal fluorescence microscopy of HEK293T cells.
At 24 hours post-transfection (hpt), the cell nucleus was stained with Hoechst 33342 (0.1 μg/mL) for 30 min at 37 °C in 5% CO2 incubator. Medium containing Hoechst 33342 was replaced with fresh DMEM prior to fluorescence microscopy analysis. Confocal images were acquired at PSU Huck Imaging Core facility on an Olympus FluoView FV1000 confocal microscope equipped with an Olympus PlanApo 60x/1.4 oil immersion objective. The cell nucleus was visualized using A 405 nm diode laser while the eGFP-tagged proteins are visualized using a 488 nm argon laser.
Z-stack: Three-dimensional fluorescence imaging.
The Z-stack fluorescence images were acquired at PSU Huck Imaging Core Facility on a Zeiss LSM 880 confocal microscope equipped with a Zeiss PlanApo 63x/1.4 oil immersion objective. HEK293T cells were maintained at 37 °C and with 5% CO2 throughout the experiment. The cells were seeded and transfected as described in the mammalian cell culture and transient transfection sections. 24 hpt with the desired plasmids, the culture medium was replaced with fresh DMEM prior experiment. Z-stack fluorescence signals of the transfected HEK293T cells were acquired at 250 nm interval in z-axis for total of 6 μm depth. Selected regions of interests were cropped and processed for three-dimensional surface rendering using ImageJ FIJI 3D viewer plugin function.
Analyses of fluorescence images.
Image processing:
All the confocal images were saved in their commercial extension files (.oib for Olympus and .czi for Zeiss). Images from both extension files were processed and exported using NIH ImageJ FIJI. A script was written to automate image processing procedure so that large number of images acquired from the same experiment from the same instrument could be processed in bulk and have consistent adjustments through every image in an efficient manner.
Supplementary Material
HIGHLIGHTS:
The driving forces of liquid-liquid phase separation (LLPS) are poorly understood
TDP43 can undergo LLPS in vitro without its prion-like domain
TDP43 LLPS is dependent on oligomerization of its N-terminal domain (NTD)
Fusion of the NTD to unrelated globular proteins enhances their LLPS
Oligomerization is proposed as a general driving force for LLPS of proteins
ACKNOWLEDGEMENTS
We thank support from the Burroughs Wellcome Fund Career Award at the Scientific Interface (X.Z.), Paul Berg Early Career Professorship (X.Z.), Lloyd and Dottie Huck Early Career Award (X.Z.), Sloan Research Fellowship (X.Z.), PEW Biomedical Scholars Program (X.Z.), National Institute of Health R35 GM133484 (X.Z.). We thank Dr. Gang Ning of the Penn State Microscopy Core Facility for collecting the TEM image.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Adekunle DA, Hubstenberger A. The multiscale and multiphase organization of the transcriptome. Emerg Top Life Sci. 2020;4:265–80. [DOI] [PubMed] [Google Scholar]
- [2].Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, et al. Properties of Stress Granule and P-Body Proteomes. Molecular cell. 2019;76:286–94. [DOI] [PubMed] [Google Scholar]
- [3].Prouteau M, Loewith R. Regulation of Cellular Metabolism through Phase Separation of Enzymes. Biomolecules. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Franzmann TM, Alberti S. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J Biol Chem. 2019;294:7128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–77. [DOI] [PubMed] [Google Scholar]
- [8].Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal Ali R, et al. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell. 2016;63:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brangwynne Clifford P, Tompa P, Pappu Rohit V. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. [Google Scholar]
- [11].Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. Embo J. 2016;35:1254–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shin Y, Berry J, Pannucci N, MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. 2017;168:159–71.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lukavsky PJ, Daujotyte D, Tollervey JR, Ue J, Stuani C, Buratti E, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–9. [DOI] [PubMed] [Google Scholar]
- [14].Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cohen TJ, Hwang AW, Unger T, Trojanowski JQ, Lee VMY. Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. Embo J. 2012;31:1241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–61. [DOI] [PubMed] [Google Scholar]
- [17].Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDR-43 aggregation and cellular toxicity. P Natl Acad Sci USA. 2008;105:6439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Babinchak WM, Haider R, Dumm BK, Sarkar P, Surewicz K, Choi JK, et al. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J Biol Chem. 2019;294:6306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li HR, Chen TC, Hsiao CL, Shi L, Chou CY, Huang JR. The physical forces mediating self-association and phase-separation in the C-terminal domain of TDP-43. Biochim Biophys Acta Proteins Proteom. 2018;1866:214–23. [DOI] [PubMed] [Google Scholar]
- [22].Conicella AE, Dignon GL, Zerze GH, Schmidt HB, D’Ordine AM, Kim YC, et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proceedings of the National Academy of Sciences. 2020;117:5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure. 2016;24:1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. The EMBO journal. 2018;37:e97452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pesiridis GS, Lee VMY, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity (vol 284, pg 20329, 2009). J Biol Chem. 2009;284:25459-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Polymenidou M, Cleveland DW. The Seeds of Neurodegeneration: Prion-like Spreading in ALS. Cell. 2011;147:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lim LZ, Wei YY, Lu YM, Song JX. ALS-Causing Mutations Significantly Perturb the Self-Assembly and Interaction with Nucleic Acid of the Intrinsically Disordered Prion-Like Domain of TDP-43. Plos Biol. 2016;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang LL, Zhao J, Yin XF, He Yang H, Che MX, et al. Two mutations G335D and Q343R within the amyloidogenic core TDP-43 influence its aggregation and inclusion formation. Sci Rep-Uk. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Guo WR, Chen YB, Zhou XH, Kar A, Ray P, Chen XP, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–U102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun. 2017;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chang CK, Wu TH, Wu CY, Chiang MH, Toh EK, Hsu YC, et al. The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem Biophys Res Commun. 2012;425:219–24. [DOI] [PubMed] [Google Scholar]
- [33].Zhang YJ, Caulfield T, Xu YF, Gendron TF, Hubbard J, Stetler C, et al. The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum Mol Genet. 2013;22:3112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McDonald NA, Fetter RD, Shen K. Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature. 2020;588:454–8. [DOI] [PubMed] [Google Scholar]
- [35].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang YC, Lin KF, He RY, Tu PH, Koubek J, Hsu YC, et al. Inhibition of TDP-43 aggregation by nucleic acid binding. PloS one. 2013;8:e64002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun Y, Arslan PE, Won A, Yip CM, Chakrabartty A. Binding of TDP-43 to the 3’UTR of its cognate mRNA enhances its solubility. Biochemistry. 2014;53:5885–94. [DOI] [PubMed] [Google Scholar]
- [38].Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, et al. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron. 2019;102:321–38.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kroschwald S, Maharana S, Simon A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters. 2017. [Google Scholar]
- [41].Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, et al. Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. Chembiochem. 2011;12:2217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. [DOI] [PubMed] [Google Scholar]
- [43].Tsoi PS, Choi KJ, Leonard PG, Sizovs A, Moosa MM, MacKenzie KR, et al. The N-Terminal Domain of ALS-Linked TDP-43 Assembles without Misfolding. Angew Chem Int Ed Engl. 2017;56:12590–3. [DOI] [PubMed] [Google Scholar]
- [44].Parge HE, Hallewell RA, Tainer JA. Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase. Proc Natl Acad Sci U S A. 1992;89:6109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Blake CC, Geisow MJ, Oatley SJ, Rérat B, Rérat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. Journal of molecular biology. 1978;121:339–56. [DOI] [PubMed] [Google Scholar]
- [46].Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet. 2009;18:3353–64. [DOI] [PubMed] [Google Scholar]
- [47].Kuo PH, Chiang CH, Wang Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 2014;42:4712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2004;101:15094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lai Z, Colón W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–82. [DOI] [PubMed] [Google Scholar]
- [50].Cummings CS, Obermeyer AC. Phase Separation Behavior of Supercharged Proteins and Polyelectrolytes. Biochemistry. 2018;57:314–23. [DOI] [PubMed] [Google Scholar]
- [51].Vernon RM, Forman-Kay JD. First-generation predictors of biological protein phase separation. Curr Opin Struct Biol. 2019;58:88–96. [DOI] [PubMed] [Google Scholar]
- [52].Zhang X, Liu Y, Genereux JC, Nolan C, Singh M, Kelly JW. Heat-shock response transcriptional program enables high-yield and high-quality recombinant protein production in Escherichia coli. Acs Chem Biol. 2014;9:1945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.