Abstract
The objective of the research article is to propose and validate a combination of machine learning and radiomics features to detect COVID-19 early and rapidly from chest X-ray (CXR) in presence of other viral/bacterial pneumonia and at different severity levels of diseases. It is vital to assess the performance of any diagnosis method on an independent data set and at very early stage of the disease when the disease severity of is very low. In such cases, most of the diagnosis methods fail. A total of 378 CXR images containing both normal lung and pneumonia (both COVID-19 and others lung conditions) were collected from publically available data set. 71 radiomics features for each lung segment were chosen from 100 extracted features based on Z-score heatmap and one way ANOVA test that can detect COVID-19. Three best performing classical machine learning algorithms during the training phase - 1) fine Gaussian support vector machine (SVM), 2) fine k-nearest neighbor (KNN) and 3) ensemble bagged model (EBM) trees were chosen for further evaluation on an independent test data set. The independent test data set consists of 115 COVID-19 CXR images collected from a local hospital and 100 CXR images collected from publically available data set containing normal lung and viral/bacterial pneumonia. Severity was scored between 0 to 4 by two experienced radiologists for each lung with pneumonia (both COVID-19 and non COVID-19) for the test data set. Ensemble Bagging Model Trees (EBM) with the selected radiomics features is the most suitable to distinguish between COVID-19 and other lung infections with an overall sensitivity of 87.8% and specificity of 97% (95.2% accuracy and 0.9228 area under curve) and is robust across severity levels. The method also can detect COVID-19 from CXR when two experienced radiologists were unable to detect any abnormality in the lung CXR (represented by severity score of 0). Once the CXR is acquired and lung is segmented, it takes less than two minutes for extracting radiomics features and providing diagnosis result. Since the proposed method does not require any manual intervention (e.g., sample collection etc.), it can be straightway integrated with standard X-ray reporting system to be used as an efficient, cost-effective and rapid early diagnosis device.
Keywords: COVID-19, Early diagnosis, Chest X-ray, Radiomics, Machine learning
1. Introduction
COVID-19 is currently a major health crisis that the world is experiencing (Chan et al., 2020, Zhu et al., 2020). According to the World Health Organization (WHO) the disease is highly infectious and around 1 out of every 5 people who gets COVID-19 caused by 2019 novel coronavirus (2019-nCoV) needs to be hospitalized due to breathing difficulty (WHO, 2020). To avoid burden on the healthcare system and to reduce the spread of the disease, it is vital to carry out fast and accurate diagnosis of COVID1-9. In clinical practice, real time polymerase chain reaction (RT-PCR) is considered as gold standard in COVID-19 diagnosis. Though the specificity of RT-PCR test is high, the sensitivity varies between 71 to 98% depending on sample collection site and sample quality (Bwire, Majigo, Njiro, & Mawazo, 2020; W. Wang et al., 2020, Watson et al., 2020), stage of disease (Sethuraman, Jeremiah, & Ryo, 2020) and degree of viral multiplication or clearance (Wolfel et al., 2020). Other different types of samples (e.g., blood, urine, stool etc.) were also used for COVID-19 detection with variable results (Xiaolong, 2020).
It has been reported that chest CT demonstrates higher sensitivity for the diagnosis of COVID-19 compared to initial RT-PCR tests of pharyngeal swab samples (Ahmed et al., 2020, Hu et al., 2020). Another study pointed out that chest CT scans suggested presence of COVID-19 related pneumonia for 81% of the patients who were tested negative with RT-PCR. (Ai et al., 2020). According to the Radiological Society of North America (RSNA), the sensitivity of CT to detect COVID-19 infection was 98% compared to RT-PCR sensitivity of 71% (Fang et al., 2020). Thus, CT could be considered as a primary tool for COVID-19 detection, unless the patient cannot be moved (RSNA Press Release, 2020). On the other hand, chest X-ray (CXR) has been considered as an insensitive method specially in the detection of COVID-19 at early disease. Because of lower spatial resolution compared to CT, CXR has a reported low sensitivity of 59% for initial detection and may appear normal until 4-5 days after the start of symptoms (Guan et al., 2020; Y. Wang et al., 2020). Because of availability and portability, it can be utilized as both baseline and follow up imaging method for monitoring disease progression (Simpson et al., 2020).Other than availability and portability, CXR has several advantages over other conventional COVID-19 diagnostic tests, e.g., short examination time, low cost and can be sterilized easily and quickly (Sarkodie, Osei-Poku, & Brakohiapa, 2020).
Several deep neural network based machine learning approaches were proposed to detect COVID-19 directly from CXR. The reported sensitivity and specificity vary from 85.35 to 96.7% and 90 to 100% respectively based on methods, disease types and classification of the diseases (Ahmed et al., 2020, Asif et al., 2020, Basu et al., 2020, Chowdhury et al., 2020, Ozturk et al., 2020). The detail of each method is provided in supplementary Table 1. To obviate the challenges of limited CXR COVID-19 data, transfer learning along with data augmentation was implemented which allows to increase the diversity of the data. However, it is to be noted that data augmentation can only increase the data via geometric transformation of the images and cannot increase the number of images with different COVID-19 conditions.
On the other hand, machine learning techniques that depend on handcrafted features extraction and selection approaches can be trained with smaller data set. In depth analysis of different first, second and third order statistical features known as radiomics have been successfully utilized for decoding the radiographic phenotype in cancer (Aerts et al., 2014, van Griethuysen et al., 2017). Recent studies prove the potential of differentiating glioblastomas (GBM) from metastatic brain tumors (MBTs) on contrast-enhanced T1 weighted imaging with radiomics based machine learning method (Chen, Ou, Wang, Guo, & Ma, 2019). However, only one study so far has reported achieving 93% sensitivity and 90% specificity in detecting COVID-19 by applying different machine learning algorithms on textural features extracted from CXR (Cavallo et al., 2020).
The training and performance evaluation of most of these methods were carried out by randomly splitting the data into training, validation and test sets. The performance was also not assessed on an independent data set. Another vital point to be noted that the performance of any diagnosis method is very much dependent on the level of disease severity. However, the sensitivity and specificity of all previously proposed methods were not separately evaluated for different level of severity.
In this study, we are proposing a radiomics based machine learning approach to detect COVID-19 from CXR. The approach includes selection strategy to pick the most suitable features for binary classification using classical machine learning algorithms. The sensitivity and specificity of the method was verified on a completely independent test data containing both normal, viral and bacterial pneumonia and confirmed COVID-19 with different levels of severity determined by two experienced radiologists.
2. Materials and methods
2.1. Training data set and radiomics features extraction
In the proposed approach, each lung is first manually delineated from training CXR image set. No image smoothing or processing technique was applied before radiomics features extraction. The training dataset was created from three different publically available data repository containing normal and different types of lung conditions. Since the objective is to detect COVID-19 from other lung conditions, all CXR images were grouped into two classes (COVID-19 and non-COVID-19). The details of the training data is provided in Table 1 .
Table 1.
Training data description
| Disease Type | Description | No of CXR | Source |
|---|---|---|---|
| non-COVID-19(Total 152 images) | Normal | 50 | JSRT (Shiraishi et al., 2000) |
| Viral/bacterial pneumonia | 50 | Kaggle (Kaggle, 2020) | |
| Other lung conditions (ARDS (4), SARS (15), Pneumocystis (12), Streptococcus (13), Chlamydophila (1), E. Coli (4), Klebsiella (1) and Legionella (1)) | 52 | GitHub (Github, 2020) | |
| COVID-19 | COVID-19 | 226 | GitHub (Github, 2020) |
To avoid bias in class distribution for training, Adaptive Synthetic (ADASYN) oversampling approach was implemented on non COVID-19 dataset to balance the imbalanced dataset. ADASYN synthetically creates new samples in between difficult-to-classify samples from the minority class (Haibo, Yang, Garcia, & Shutao, 2008). For the manual segmentation of lung on all the CXR images except the normal ones, MATLAB 2019b Image Segmenter app was used (Brown, Wilson, Doust, Gill, & Sun, 1998). For the normal cases, the available lung masks were used [39]. 100 radiomics features were then extracted using the segmented lung and PyRadiomics tool with Python 3.7.6 for each lung separately (van Griethuysen et al., 2017). This yielded 18 first-order statistics, 9 2D shape-based, 22 Gray Level Co-occurrence Matrix (GLCM), 16 Gray Level Run Length Matrix (GLRLM), 16 Gray Level Size Zone Matrix (GLSZM), 5 Neighboring Gray Tone Difference Matrix (NGTDM) and 14 Gray Level Dependence Matrix (GLDM) features.
Among these features, first order features describe the distribution of voxel intensities within the ROI. Shape based features represent 2D size and shape of the ROI, e. g., perimeter, elongation, sphericity etc. The rest five feature matrices represent textural appearance. GLCM records the probability of occurrence of a pixel pair. The number of connected voxels within a distance δ that are dependent on the center voxel is used to generate GLDM. GLRLM is defined as the length in number of pixels having the same gray level value. On the other hand, GLSZM defines a zone of connected voxels with the same gray level intensity. GLSZM quantifies gray level zones within a ROI in an image. The number of connected voxels that share the same gray level intensity defines a gray level zone. NGTDM quantifies the difference between a gray value and the average gray value of its neighbours within distance δ.
2.2. Radiomics features selection
To select the features that have better classification ability between COVID-19 and others, two methods were implemented. The first method is a heatmap of Z-scores to identify features that can classify these two groups. The second method is the one-way ANOVA test to find the features that have a statistically significant difference between the means of the two classes with the criteria p<0.05. It was found that only 71 features out of 100 radiomics features show statistically significant difference and therefore, these features were only used for training the machine learning algorithms. Out of these 71 features 13 are first order, 3 2D shape based, 20 GLCM, 8 GLDM, 10 GLRLM, 13 GLSZM and 4 NGTDM extracted features.
2.3. Training of machine learning algorithms
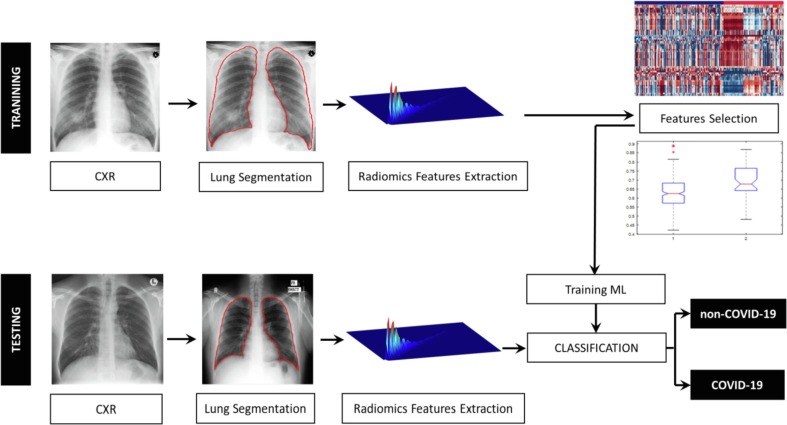
A number of classical supervised and unsupervised machine learning algorithms were evaluated using the Classification Learner App of MATLAB 2019b. To optimize the parameter of the machine learning algorithms to classify between COVID-19 and other cases, 10 fold cross validation approach was implemented (Refaeilzadeh, Tang, & Liu, 2009). The overall process is shown in Fig. 1 .
Fig. 1.
Illustration of the methodology for COVID-19 detection from CXR images.
Area under receiver operating characteristic (AUC-ROC) curve as well as sensitivity, specificity, and accuracy were calculated to evaluate the ability of the classifier to discriminate the COVID-19 CXR cases from the other cases. Sensitivity, specificity and accuracy were calculated as:
where TP, TN, FP and FN refer to true positive, true negative, false positive and false negative respectively.
The three best performing classifiers during the training phase - 1) fine Gaussian support vector machine (SVM), 2) fine k-nearest neighbor (KNN) and 3) ensemble bagged model (EBM) trees were chosen for further evaluation on the test data. Among different classical machine learning algorithms, SVM is suitable for both linear and nonlinear binary classification tasks. It is also one of the mostly used automatic classifiers in healthcare (Cervantes et al., 2020, Huang et al., 2018, Yu et al., 2010). In comparison, KNN is a simpler technique that stores all existing instances and then classifies any new instance based on a user defined similarity measure. However, its performance very much dependent on the size of training examples (Thanh Noi & Kappas, 2017). On the other hand, EBM trees classifier is used due to its stability and found uses in many applications like credit card fraud detection (Zareapoor & Shamsolmoali, 2015).
2.4. Test data set
Test CXR data set used in this study is an independent data set and consists of 165 CXR images (330 lungs) containing 25 normal, 25 viral/bacterial pneumonia and 115 COVID-19 cases. The 115 COVID-19 CXR images of 25 patients were acquired at local hospital. COVID-19 was confirmed with standard RT-PCR test. The details of the test data is shown in Table 2 .
Table 2.
Test data description
| Disease type | Description | No of CXR | Source |
|---|---|---|---|
| non-COVID-19 | Normal | 25 | JSTR (Shiraishi et al., 2000) |
| Viral/bacterial Pneumonia | 25 | Kaggle (2020) | |
| COVID-19 | COVID-19 | 115 (25 patients) | Local Hospital |
Multiple chest X-ray were taken for all the 25 patients. Out of 25 patients 15 patients passed away and 10 patients were released cured from the hospital. Different levels of severity have been observed on each of the different CXR image of the same patient. Even for the same CXR, the severity between the two lungs are sometimes different according to the visual assessment of the radiologists. To evaluate the robustness of machine learning algorithms, the severity of each lung of each CXR for both COVID-19 and viral/bacterial pneumonia was scored. All CXRs were performed in a frontal projection in a posteroanterior view if the patient was able to stand; otherwise, an anteroposterior view in the sitting or supine position was acquired. The CXRs were independently evaluated by two experienced radiologists. In case of discrepant interpretations, the findings were resolved by consensus.
The radiologists rated pulmonary parenchymal involvement on CXR using a semiquantitative severity score (score 0 to 4) depending on the visual assessment on the extent of involvement by ground glass opacities (GGO) (i.e., hazy opacity not obliterating bronchi and vessels) or consolidations (i.e., area of attenuation obscuring airways and vessels). If none of these patterns were seen, then the radiologists would select score 0 (clear lung). Score 1 = <25%, score 2= 25-50%, score 3 = 50-75% and score 4 = >75% involvement. Severity score 0 or 1 corresponds to early stage of both COVID-19 and non-COVID-19 infections.
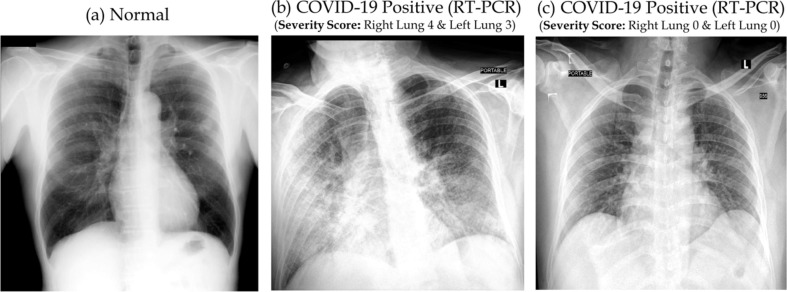
The reason for considering severity is that it is always difficult to detect abnormality at low severity as shown in Fig. 2 . Normal lung and lung with very low severity (scored 0 severity by the radiologists) appear similar though the RT-PCR test confirmed COVID-19 positive. Such low severity makes it difficult for human observer to detect abnormality in the lung in CXR.
Fig. 2.
Normal CXR (a) and RT-PCR test confirmed COVID-19 positive cases with high (b) and low (c) severity score.
Test data set based on severity is shown in Table 3 . This is to be noted that during training severity was not taken in to consideration.
Table 3.
Number of segmented lungs for each severity
| Severity | 0 | 1 | 2 | 3 | 4 | Total |
|---|---|---|---|---|---|---|
| non-COVID-19 | 50 | 17 | 22 | 11 | 0 | 100 |
| COVID-19 | 36 | 88 | 71 | 27 | 8 | 230 |
| Total | 86 | 105 | 93 | 38 | 8 | 330 |
2.5. Validation on test data set
Similar to the training data set, 100 radiomics features were first extracted from each lung segment of each CXR image belonging to the test data set. However, only 71 features that showed statistically significant difference between COVID-19 and other cases during training were used for performance evaluation. The three best performing classifiers during training were then applied using these 71 features to evaluate the classification performance of the machine learning algorithms using sensitivity, specificity and accuracy along with AUC-ROC. The performance was also evaluated on each separate lung to investigate the effects of severity
3. Results
The list of the 71 radiomics features that are statistically significantly different between COVID-19 and non-COVID-19 CXR images are provided in supplementary Table 2 along with the p-value. Fig. 3 shows the Z-score heatmap of the significant radiomics features for each CXR image ordered according to the diagnosis class. The selected features clearly display the difference between the COVID-19 and other cases.
Fig. 3.
Z-score heatmap of 71 radiomics features that yield statistically significant difference between COVID-19 and others. Each row represents one feature and each column represents one CXR image used in the training set.
The performance of the classifiers during training is shown in Table 4 . Each performance value shown in the table is the average of the 10-fold cross-validation results. From the table, we can see that the SVM classifier has the highest average sensitivity and AUC-ROC. But the lowest specificity. The highest specificity is achieved by fine KNN with 97.9% but with the lowest average sensitivity 88.9% and AUC-ROC of 0.9343. The sensitivity, specificity and AUC-ROC for EBM method is always more than 90%.
Table 4.
Performance of the classifiers during training.
| Classifier | Sensitivity | Specificity | Accuracy | AUC-ROC |
|---|---|---|---|---|
| Fine Gaussian SVM | 98.2% | 88.4% | 93.4% | 0.9894 |
| Fine KNN | 88.9% | 97.9% | 93.3% | 0.9343 |
| Ensemble Bagged Model Trees (EBM) | 91.6% | 92.6% | 91.8% | 0.9772 |
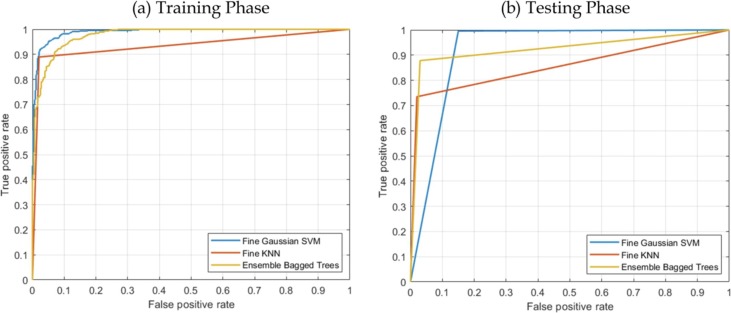
Table 5 shows the performance results of applying the previously trained classifiers to an independent test data set. Fig. 4 shows the AUC of all the three machine learning algorithm during training and testing phases.
Table 5.
Performance of the classifiers during testing.
| Classifier | Sensitivity | Specificity | Accuracy | AUC-ROC |
|---|---|---|---|---|
| Fine Gaussian SVM | 99.6% | 85% | 95.2% | 0.9228 |
| Fine KNN | 73.5% | 98% | 80.9% | 0.8574 |
| Ensemble Bagged Model Trees (EBM) | 87.8% | 97% | 90.6% | 0.9241 |
Fig. 4.
ROC plot of the three best performing classifiers during training (left) and during testing (right).
The SVM classifier was able to correctly predict all the COVID-19 cases except one with 99.6% sensitivity, while EBM correctly predicted COVID-19 with 87.8% sensitivity. The sensitivity of the fine KNN was the lowest with 73.5%. On the other hand, the two highest specificity of 98% and 97% were achieved by KNN and EBM respectively, while the specificity of the SVM method was the lowest with 85%. The highest accuracy and AUC-ROC were achieved by SVM and EBM respectively (95.2% and 0.9241).
The ROC curves of all the classifiers during training and test cases are shown in Fig. 4.
Table 6, Table 7 compare sensitivity and specificity of SVM and EBM classifiers respectively based on the severity. Since the accuracy and AUC-ROC of the fine KNN were less than 90% and 0.9 respectively, it was not considered for further investigation. This is to be noted that for some COVID-19 patients, some of the lung segments were scored as 0 by the radiologists. On the other hand, there were no lung segments with severity score 4 for viral/bacterial pneumonia.
Table 6.
Sensitivity and specificity based on severity for SVM
| Severity | 0 | 1 | 2 | 3 | 4 | Total | |
|---|---|---|---|---|---|---|---|
| COVID-19 | Original | 36 | 88 | 71 | 27 | 8 | 230 |
| Detected | 35 | 88 | 71 | 27 | 8 | 229 | |
| Sensitivity | 97.2% | 100.0% | 100.0% | 100.0% | 100.0% | 99.6% | |
| Non-COVID-19 | Original | 50 | 17 | 22 | 11 | 0 | 100 |
| Detected | 47 | 11 | 20 | 7 | 0 | 85 | |
| Specificity | 94.0% | 64.7% | 90.9% | 63.6% | 0.0% | 85.0% |
Table 7.
Sensitivity and specificity based on severity for EBM.
| Severity | 0 | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|---|
| COVID-19 | Original | 36 | 88 | 71 | 27 | 8 | 230 |
| Detected | 33 | 78 | 64 | 21 | 6 | 202 | |
| Sensitivity | 91.7% | 88.6% | 90.1% | 77.8% | 75.0% | 87.8% | |
| Non-COVID-19 | Original | 50 | 17 | 22 | 11 | 0 | 100 |
| Detected | 50 | 16 | 20 | 11 | 0 | 97 | |
| Specificity | 100.0% | 94.1% | 90.9% | 100.0% | 0.0% | 97.0% |
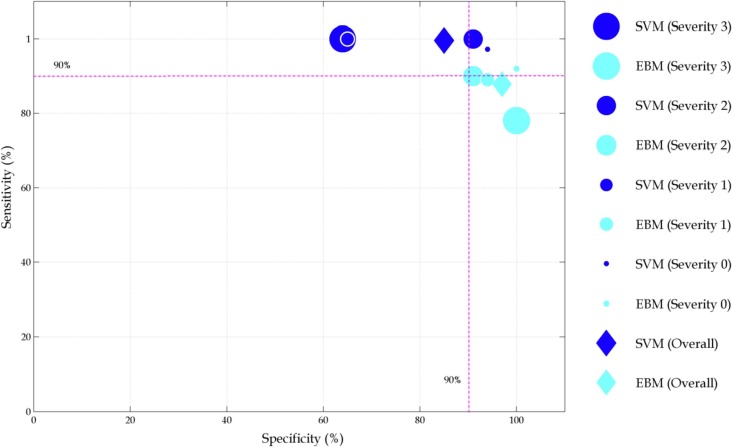
Sensitivity vs. specificity for both SVM and EBM for the whole test data and at each severity level is plotted in Fig. 5 .
Fig. 5.
Sensitivity and specificity for whole test data as well as for each severity level. The size of the filled circles represents severity levels from 0 to 4.
Overall, SVM can detect COVID-19 with 99.6% sensitivity and 85% specificity. On the other hand, the performance of EBM is 87.8% and 97% respectively. For the EBM method, the sensitivity decreases with the increase in severity. On the other hand, the sensitivity of the SVM methods is less dependent on the severity level. However, high level of fluctuations is observed in terms of specificity.
This implies that there are certain radiomics features that are different for COVID-19 patients and these features are dependent on severity level. Appropriate selection of those features also can allow to detect COVID-19 from other diseases.
For the whole test data set, the performance of SVM and EBM was comparable. However, KNN performance was much worse compared to the training phase. The reason behind it could be that during training only average sensitivity and specificity of the 10 fold validation were calculated and due to data augmentation via geometric transformation.
The performance between SVM and EBM methods are more distinguishable if severity is taken into consideration. SVM method shows very good sensitivity (97.2 to 100%) but the specificity in terms of severity is not robust with values ranging from 63.6 to 94%. On the other hand, the sensitivity of EBM decreases with the increase of severity. The specificity of the EBM method is within 10% range for all levels of severity and never falls below 90% (range 90.9 to 100%).
The proposed method can be considered as a rapid diagnosis tool to detect COVID-19 from CXR. Once the CXR is acquired and lung is segmented, it takes less than two minutes for radiomics features extraction on an Intel Core i7 1.5 GHz 4 cores machine with 16 GB RAM and Windows 10 (64-bit) operating system. For classification SVM, KNN and EBM take 1.46, 0.86 and 1.94 seconds respectively on the same machine. Overall time required is less than 2 minutes and no pre-processing steps are involved.
4. Discussion
Several studies have been proposed to use different machine learning algorithms to detect COVID-19 from CXR images (Ahmed et al., 2020, Asif et al., 2020, Ozturk et al., 2020). Most of them have demonstrated good accuracy for disease diagnosis. The performance of those methods vary depending on the definition of the classes used to determine the accuracy. The highest accuracy of 98.08% was achieved by deep learning method (Ozturk et al., 2020). However, it only uses two data classes - COVID-19 and No-Findings. Inclusion of multiple diseases brings the accuracy down to 87.02%. Other studies also reported to achieve similar sensitivity (93 to 96.7%) and specificity (90 to 100%) to classify between COVID-19 and others class, where other class consists of only normal CXR and viral pneumonia. To increase the number of images (sometimes as many as 10 times of the original data), all these algorithm mainly used geometric transformation for data augmentation. Training and test data set were then randomly splited. As a result, it is highly likely that the images of the same patients are present in both the training and test sets resulting in higher accuracies in detection of COVID-19.
A robust method should be able to detect COVID-19 in presence or absence of other possible lung conditions and the performance should not fluctuate considerably for any other independent data set. From this perspective, the proposed method is robust as it not only includes normal and viral pneumonia but also other diseases (e.g., bacterial pneumonia, SARS etc.) in the training data as shown in Table 1. The performance is also tested on a completely independent data set. The other uniqueness of this method is that it can provide similar performance irrespective of the severity levels. The authors are not aware of any such studies to investigate radiomics features to detect COVID-19 at different severity levels.
Performance evaluation based on severity reveals more insight into the radiomics pattern present in CXR images. Visual assessment of 36 lungs by the radiologists reveals no abnormality and were assigned with the severity score of 0. However, RT-PCR test confirmed COVID-19 positive for these lungs and the proposed method also confirmed COVID-19 positive with a sensitivity of 97.2% and 91.7% by SVM and EBM method respectively.
Considering the performance at different severity levels, EBM method proves to be the most robust method. However, the sensitivity of the EBM method decreases with the increase of severity. One of the reasons could be that with the increase of severity, the patterns that serve as radiomics features of COVID-19 vanish and becomes less distinguishable compared to other lung conditions.
Unlike other studies dealing with the diagnosis of COVID-19 from CXR, the performance of the proposed method was validated on a completely independent data set and considering different levels of disease severity. One of the limitations of the study is that the performance of the proposed framework was only implemented for RT-PCR test confirmed COVID-19 cases. It will be really useful to further investigate the performance for cases with RT-PCR being negative at early time point but became positive later.
5. Conclusion
With appropriate selection of radiomics features and machine learning algorithm, it is possible to detect CVOID-19 directly from CXR with a sensitivity and specificity comparable or better than other available techniques, e.g., RT-PCR. The performance of the proposed method with SVM and EBM based machine learning achieved an overall sensitivity of 99.6% and 87.8% and specificity of 85% and 97% respectively. Though the performance are comparable for both the methods, EBM is more robust across severity levels. Since this tool does not require any manual intervention (e.g., sample collection etc.), it can be integrated with any standard X-ray reporting system as an efficient, cost-effective and rapid point-of-care device.
6. Source of Funding
The authors extend his appreciation to the Deputyship for Research & Innovation, Ministry of Saudi Arabia for funding this research work.
CRediT authorship contribution statement
Mahbubunnabi Tamal: Conceptualization, Data curation, Software, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Writing - original draft, Writing - review & editing. Maha Alshammari: Data curation, Software, Formal analysis, Validation, Writing - original draft, Writing - review & editing. Meernah Alabdullah: Data curation, Formal analysis, Validation, Writing - original draft, Writing - review & editing. Rana Hourani: Data curation, Formal analysis, Validation, Writing - original draft, Writing - review & editing. Hossain Abu Alola: Resources, Methodology, Validation, , Writing - review & editing. Tarek M. Hegazi: Resources, Methodology, Validation, , Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eswa.2021.115152.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S.…Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S., Yap, M. H., Tan, M., & Hasan, M. K. (2020). ReCoNet: Multi-level Preprocessing of Chest X-rays for COVID-19 Detection Using Convolutional Neural Networks. Preprint from medRxiv, 2020.2007.2011.20149112.
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W.…Xia L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif, S., Wenhui, Y., Jin, H., Tao, Y., & Jinhai, S. (2020). Classification of COVID-19 from Chest X-ray images using Deep Convolutional Neural Networks. Preprint from medRxiv.
- Basu, S., Mitra, S., & Saha, N. (2020). Deep Learning for Screening COVID-19 using Chest X-Ray Images. Preprint from medRxiv.
- Brown M.S., Wilson L.S., Doust B.D., Gill R.W., Sun C. Knowledge-based method for segmentation and analysis of lung boundaries in chest X-ray images. Computerized Medical Imaging and Graphics. 1998;22:463–477. doi: 10.1016/s0895-6111(98)00051-2. [DOI] [PubMed] [Google Scholar]
- Bwire G.M., Majigo M.V., Njiro B.J., Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol. 2020 doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo, A. U., Troisi, J., Forcina, M., Mari, P., Forte, V., Sperandio, M., Pagano, S., Cavallo, P., Floris, R., & Geraci, F. (2020). Texture Analysis in the Evaluation of Covid-19 Pneumonia in Chest X-Ray Images: a Proof of Concept Study. Preprint from Research Square, 25 Jun 2020. [DOI] [PubMed]
- Cervantes J., Garcia-Lamont F., Rodríguez-Mazahua L., Lopez A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing. 2020;408:189–215. [Google Scholar]
- Chan J.-F.-W., Yuan S., Kok K.-H., To K.-K.-W., Chu H., Yang J.…Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ou X., Wang J., Guo W., Ma X. Radiomics-Based Machine Learning in Differentiation Between Glioblastoma and Metastatic Brain Tumors. Frontiers. Oncology. 2019;9 doi: 10.3389/fonc.2019.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, M. E. H., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M. A., Mahbub, Z., Islam, K. R., Khan, M. S., Al-Emadi, N., & Reaz, M. B. I. (2020). Can AI help in screening Viral and COVID-19 pneumonia? Preprint form arvix.
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Github. (2020). covid-chestxray-dataset. In.
- Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., Liu, L., Shan, H., Lei, C. L., Hui, D. S. C., Du, B., Li, L. J., Zeng, G., Yuen, K. Y., Chen, R. C., Tang, C. L., Wang, T., Chen, P. Y., Xiang, J., Li, S. Y., Wang, J. L., Liang, Z. J., Peng, Y. X., Wei, L., Liu, Y., Hu, Y. H., Peng, P., Wang, J. M., Liu, J. Y., Chen, Z., Li, G., Zheng, Z. J., Qiu, S. Q., Luo, J., Ye, C. J., Zhu, S. Y., Zhong, N. S., & China Medical Treatment Expert Group for, C Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibo H., Yang B., Garcia E.A., Shutao L. In 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence) 2008. ADASYN: Adaptive synthetic sampling approach for imbalanced learning; pp. 1322–1328. [Google Scholar]
- Hu Q., Guan H., Sun Z., Huang L., Chen C., Ai T.…Xia L. Early CT features and temporal lung changes in COVID-19 pneumonia in Wuhan. China. European Journal of Radiology. 2020;128 doi: 10.1016/j.ejrad.2020.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Cai N., Pacheco P.P., Narrandes S., Wang Y., Xu W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics. 2018;15:41–51. doi: 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaggle. (2020). chest-xray-pneumonia. In.
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaeilzadeh, P., Tang, L., & Liu, H. (2009). Cross-Validation. In L. Liu & M. T. ÖZsu (Eds.), Encyclopedia of Database Systems (pp. 532-538). Boston, MA: Springer US.
- RSNA Press Release. (2020). CT Provides Best Diagnosis for COVID-19. In: RSNA.
- Sarkodie, B. D., Osei-Poku, K., & Brakohiapa, E. (2020). Diagnosing COVID-19 from Chest X-ray in Resource-Limited Environment-Case Report,Medical case report. In (pp. 3).
- Sethuraman, N., Jeremiah, S. S., & Ryo, A. (2020). Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. [DOI] [PubMed]
- Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K.-I.…Doi K. Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule. American Journal of Roentgenology. 2000;174:71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- Simpson, S., Kay, F. U., Abbara, S., Bhalla, S., Chung, J. H., Chung, M., Henry, T. S., Kanne, J. P., Kligerman, S., P., K., J.,, & Litt, H. (2020). Radiological Society of North America Expert Consensus Document on Reporting Chest CT Findings Related to COVID-19: Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiology: Cardiothoracic Imaging, 2, e200152. [DOI] [PMC free article] [PubMed]
- Thanh Noi P., Kappas M. Comparison of Random Forest, k-Nearest Neighbor, and Support Vector Machine Classifiers for Land Cover Classification Using Sentinel-2 Imagery. Sensors. 2017;Basel:18. doi: 10.3390/s18010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V.…Aerts H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research. 2017;77 doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X.…Zhou M. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology. 2020;296:E55–E64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369 doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- WHO. (2020). Media Statement: Knowing the risks for COVID-19. In.
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A.…Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xiaolong, C. (2020). Landscape Coronavirus Disease 2019 test (COVID-19 test) in vitro -- A comparison of PCR vs Immunoassay vs Crispr-Based test.
- Yu W., Liu T., Valdez R., Gwinn M., Khoury M.J. Application of support vector machine modeling for prediction of common diseases: The case of diabetes and pre-diabetes. BMC Med Inform Decis Mak. 2010;10:16. doi: 10.1186/1472-6947-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareapoor M., Shamsolmoali P. Application of Credit Card Fraud Detection: Based on Bagging Ensemble Classifier. Procedia Computer Science. 2015;48:679–685. [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J.…Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.