Abstract
Background
A key clinical feature of COVID-19 is a deep inflammatory state known as “cytokine storm” and characterized by high expression of several cytokines, chemokines and growth factors, including IL-6 and IL-8. A direct consequence of this inflammatory state in the lungs is the Acute Respiratory Distress Syndrome (ARDS), frequently observed in severe COVID-19 patients. The "cytokine storm" is associated with severe forms of COVID-19 and poor prognosis for COVID-19 patients. Sulforaphane (SFN), one of the main components of Brassica oleraceae L. (Brassicaceae or Cruciferae), is known to possess anti-inflammatory effects in tissues from several organs, among which joints, kidneys and lungs.
Purpose
The objective of the present study was to determine whether SFN is able to inhibit IL-6 and IL-8, two key molecules involved in the COVID-19 "cytokine storm".
Methods
The effects of SFN were studied in vitro on bronchial epithelial IB3-1 cells exposed to the SARS-CoV-2 Spike protein (S-protein). The anti-inflammatory activity of SFN on IL-6 and IL-8 expression has been evaluated by RT-qPCR and Bio-Plex analysis.
Results
In our study SFN inhibits, in cultured IB3-1 bronchial cells, the gene expression of IL-6 and IL-8 induced by the S-protein of SARS-CoV-2. This represents the proof-of-principle that SFN may modulate the release of some key proteins of the COVID-19 "cytokine storm".
Conclusion
The control of the cytokine storm is one of the major issues in the management of COVID-19 patients. Our study suggests that SFN can be employed in protocols useful to control hyperinflammatory state associated with SARS-CoV-2 infection.
Keywords: COVID-19, Spike, sulforaphane, biomarkers, inflammation, nutraceuticals
Abbreviations: ARDS, Acute Respiratory Distress Syndrome; COVID-19, coronavirus disease 2019; IL-6, interleukin-6; IL-8, interleukin-8; SFN, sulforaphane; S-protein, Spike-protein; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; RT-qPCR, reverse transcription quantitative polymerase-chain reaction
Graphical abstract
Introduction
The pandemic coronavirus disease 2019 (COVID-19) caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is characterized by two major clinical phases: (a) a SARS-CoV-2 infection of target cells and tissues and (b) a deep inflammatory state, known as “cytokine storm” (Pascarella et al., 2020; Pelaia et al., 2020). A key step for SARS-CoV-2 infection is the binding of the SARS-CoV-2 Spike protein (S-protein) to angiotensin-converting enzyme 2 (ACE2) receptor. This dictates the wide host range and tropism of the infection in human organs and tissues, leading to important clinical manifestations and complications, such as pulmonary failure (Gao et al., 2020; Pascarella et al., 2020). The interaction between the SARS-CoV-2 S-protein and ACE2 is a key step also for inducing the “cytokine storm”. The attachment of the SARS-CoV-2 S-protein to ACE2 is followed by deep cellular changes, among which the “cytokine storm”, including the hyperactivation of Nuclear Factor kappa-B (NF-κB) by IL-6/STATs axis (Ratajczak et al., 2020). This induces in lungs the Acute Respiratory Distress Syndrome (ARDS) that has been frequently observed in COVID-19 patients (Soumagne et al., 2020) and is clearly associated with the severity of the pathology (Grasselli et al., 2020; Matthay et al., 2020). Patients with COVID-19-related ARDS have a form of injury that, in many aspects, is similar to the ARDS unrelated to COVID-19 (Grasselli et al., 2020). Importantly, patients with COVID-19-related ARDS have high mortality rates compared to COVID-19 patients without any ARDS symptoms (Matthay et al., 2020). The impact of anti-inflammatory protocols for anti-SARS-CoV-2 pharmacological strategies is clear, as recently demonstrated by the effective treatments targeting IL-6 (Nasonov et al., 2020) and IL-8 (Andreakos et al., 2021). Notably, the pharmacological approach for treating ARDS steadily needs novel anti-inflammatory reagents as different COVID-19 patients might respond in a different way to these treatments (de Simone et al., 2020).
Several anti-inflammatory strategies to reduce COVID-19 “cytokine storm” and associated ARDS have been proposed using biomolecules derived from herbal medicinal extracts and reviewed by several authors (Khalifa et al., 2020; Matveeva et al., 2020). This was judged to be a key strategy at the beginning of the pandemic, in consideration of the unknown nature of the disease and the lack of effective treatment protocols and approved vaccines (Adem et al., 2020; Ngwa et al., 2020). Repurposing of known plant-derived reagents for anti-inflammatory activity against the COVID-19 “cytokine storm” might be of great interest (Dzobo et al., 2020; Wang et al., 2020). In this respect, sulforaphane (SFN: 1-isothiocyanate-4-methyl sulfonyl butane), one of the main components of Brassica oleraceae L. (Brassicaceae) (common name broccoli) and other spp from the Cruciferae family, deserves consideration (Lattè et al., 2011). SFN has been reported to exhibit anti-inflammatory effects (Sturm et al., 2017; Ruhee et al., 2019) in tissues of several organs, such as joints, kidneys and lungs. In particular, SFN is a potent Nuclear factor erythroid 2-related factor 2 (Nrf2) activator. Interestingly Nrf2 inhibits the recruitment of inflammatory cells and orchestrates the anti-inflammatory process by regulating gene expression through the Antioxidant Response Element (ARE) (Ahmed et al., 2017).
In the context of lung tissue, Qi et al. (2016) reported that SFN dependent Nrf2/ARE activation exerts protective effects against lipopolysaccharide (LPS)-induced acute lung injury (ALI). Thus, SFN may be a potential candidate for use in the treatment of acute lung injury.
The objective of the present study was to determine whether SFN inhibits IL-6 and IL-8, two important molecules involved in the COVID-19 “cytokine storm”. The effects of SFN were studied in an experimental in vitro study using bronchial epithelial IB3-1 cells (Zeitlin et al., 1991) exposed to the SARS-CoV-2 Spike protein (S-protein), following a protocol developed using the S-protein of SARS-CoV-1 (Wang et al., 2007). We first verified whether this novel experimental model employing SARS-CoV-2 S-protein would also induce an increase of proteins characterizing the COVID-19 “cytokine storm”. Then the effects of SFN on the expression of pro-inflammatory genes were analyzed by RT-qPCR and Bio-Plex assay in order to determine mRNA accumulation and extracellular protein release. Both these issues (the development of a SARS-CoV-2 Spike-induced experimental screening system and the effects of SFN on Spike-induced IL-6 and IL-8) are novel and might be considered of some interest for the screnning and characterization of new agents to be proposed as inhibitors of molecules of the COVID-19 “cytokine storm”.
Materials and methods
Materials
All chemicals and reagents were of analytical grade. SFN (D,L-Sulforaphane, 574215-25MG, Merck Millipore, Burlington, MA, USA) (purity > 98% determined by ultra-performance liquid chromatography, UPLC) was diluted in DMSO (D8418, Sigma-Aldrich, St. Louis, MO, USA) at a final stock concentration of 150 mM. SARS-Cov-2 Spike recombinant glycoprotein (ab49046) was purchased by Abcam (Cambridge, UK). The purity was > 90% as determined by SDS-PAGE.
Cell culture conditions
The human bronchial epithelial IB3-1 cell line (Zeitlin et al., 1991) was cultured in humidified atmosphere of 5% CO2/air in LHC-8 medium (Gibco, Thermo Fischer Scientific, Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS, Biowest, Nuaillé, France) in the absence of gentamycin. To verify the effect on proliferation, cell growth was monitored by determining the cell number/ml using a Z2 Coulter Counter (Coulter Electronics, Hialeah, FL, USA).
Stimulation of cells with SARS-CoV-2 Spike protein
SARS-CoV-2 Spike protein (139 KDa; stock concentration = 7.2 μM in 9% urea, 0.32% Tris-HCl pH 7.2, 50% glycerol) was diluted in 200 µl of LHC-8 medium to achieve the final concentrations used to treat IB3-1 cells. Briefly, cells seeded at 50% of confluence, were treated with Spike protein (5–50 nM) and incubated for 30 min at 4 °C, then for 30 min at 37 °C, according with the protocol published by Wang et al. (2007) (this procedure is expected to maximize S-protein interaction with the receptor and the S-protein cellular uptake). After this incubation, LHC-8 medium supplemented with 5% (final concentration) FBS was added to a final 500 μl volume and the cultures were further incubated at 37 °C and for 24 h. Unstimulated cells (treated with DMSO) were used as reference control.
RNA extraction
Cultured cells were trypsinized (0,05% trypsin and 0,02% EDTA; Sigma-Aldrich) and collected by centrifugation at 1,000 × g for 8 min at 4 °C, washed twice with DPBS 1 × (Gibco, Thermo Fischer Scientific) and lysed with Tri-Reagent (Sigma Aldrich), according to the manufacturer's instructions. The isolated RNA was washed once with cold 75% ethanol, dried and dissolved in nuclease-free pure water before use. Obtained RNA was stored at -80 °C until the use (Gasparello et al., 2019).
Quantitative analyses of mRNAs
For ILs mRNA analysis, 500 ng of total RNA were reverse transcribed to complementary DNA (cDNA) using the Taq-Man Reverse Transcription PCR Kit and random hexamers (Applied Biosystems, Thermo Fischer Scientific) in a final reaction volume of 50 µl. Real-time-qPCR experiments were carried out using an assay composed by a primer pair and a fluorescently labeled 5′ nuclease probe purchased from IDT (Integrated DNA Technologies, Coralville, IO, USA; Assays ID: Hs.PT.58.38869678.g for IL-8 and Hs.PT.58.40226675 for IL-6). An amount of 2 µl of cDNA were amplified, in presence of 2 × PrimeTime Gene Expression Master Mix for 40 PCR cycles using CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Relative expression was calculated using the comparative cycle threshold method (ΔΔCT method) and the endogenous control human β-actin was used as normalizer. Negative controls (no template cDNA and RT-minus control) were also run in every experimental plate to assess specificity and to rule out contamination. RT-qPCR reactions were performed in duplicate for both target and normalizer genes (Gasparello et al., 2019).
Analysis of cytokines, chemokines and growth factors
Proteins released into culture supernatants were measured using Bio-Plex Human Cytokine 27-plex Assay (Bio-Rad) as suggested by the manufacturer. The assay allows the multiplexed quantitative measurement of 27 cytokines/chemokines (including FGF basic, Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17A, IP-10, MCP-1 (MCAF), MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, VEGF) in a single well. An amount of 50 μl of cytokine standards or samples (diluted supernatants recovered from IB3-1 cells) was incubated with 50 μl of anti-cytokine conjugated beads in 96-well filter plate for 30 min at room temperature with shaking. The plate was washed by vacuum filtration three times with 100 μl of Bio-Plex Wash Buffer, 25 μl of diluted detection antibody were added, to each wells and plate was incubated for 30 min at room temperature with shaking. After three filter washes, 50 μl of streptavidin-phycoerythrin were added, and plate was incubated for 10 min at room temperature with shaking. Finally, plate was washed by vacuum filtration three times, beads were suspended in Bio-Plex Assay Buffer, plate was read by Bio-Rad 96-well plate reader. Data were collected and analyzed by the Bio-Plex Manager Software (Bio-Rad) (Gasparello et al., 2019).
Effects on cellular viability and apoptosis
Annexin V and Dead Cell assay was performed using Muse Cell Analyzer (Merck Millipore) instrument, according to the instructions supplied by the manufacturer. Cells were washed with sterile DPBS 1 ×, trypsinized, suspended and diluted (1:2) with Muse Annexin V & Dead Cell reagent (Merck Millipore). After an incubation of 15 min at room temperature in the dark, samples were acquired and data were analyzed using Annexin V and Dead Cell Software Module (Merck Millipore) (Gasparello et al., 2020).
Statistics
Results were expressed as mean ± standard error of the mean (SEM) and comparison among groups was made by using analysis of variances (ANOVA).
Results
Induction of IL-6 and IL-8 gene expression in bronchial epithelial IB3-1 cells after exposure to the SARS-CoV-2 Spike protein
We first analyzed the effects of exposure of IB3-1 cells to SARS-CoV-2 Spike protein (S-protein). To this aim, IB3-1 cells were treated with 5, 15 and 50 nM S-protein for 24 h, as indicated in the Materials and methods section and in the experimental flow-chart shown in Fig. 1 A. After the treatment, RNA was purified from the treated cells for RT-qPCR analysis, and cellular supernatants were isolated for the analysis of secreted cytokines, chemokines and growth factors in order to identify SARS-CoV-2 Spike protein-induced alteration of the secretome profile.
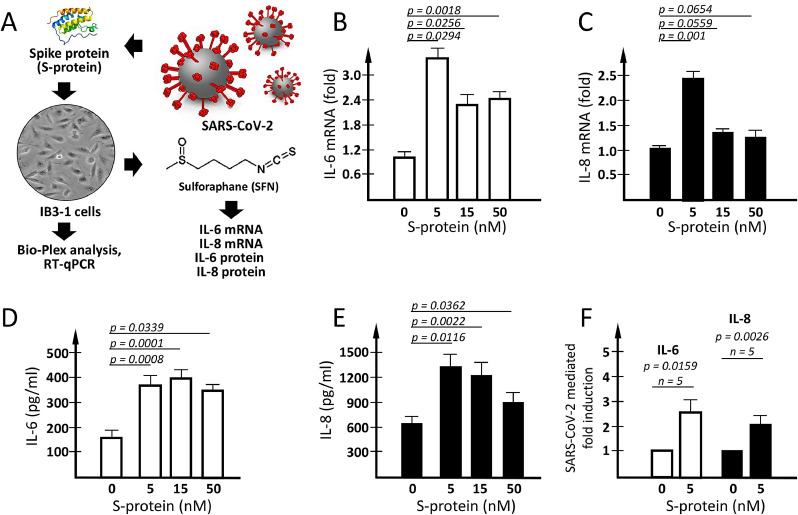
Fig. 1.
IL-6 and IL-8 expression in IB3-1 cells induced by SARS-CoV-2 Spike protein. (A) Flow-chart of the experimental plan. (B-E) Effects of 24 h exposure of IB3-1 cells to increasing amounts of SARS-CoV-2 spike protein (S-protein) on IL-6 (B) and IL-8 (C) mRNA and on IL-6 (D) and IL-8 (E) protein release. (F) Summary of 5 independent experiments performed using 5 nM S-protein to induce IL-6 and IL-8 increased release by treated IB3-1 cells. Results represent the average ± SEM.
Concerning mRNA expression, we have compared the mRNA coding for IL-6 (Fig. 1B) and IL-8 (Fig. 1C). The results obtained show that, after S-protein exposure, IB3-1 cells accumulate larger amounts of IL-6 and IL-8 mRNAs with respect to control untreated IB3-1 cells. This information was derived from RT-qPCR analysis using β-actin as internal control. Similar results were obtained using as internal control GAPDH and RPL13A sequences (data not shown). Interestingly, exposure of IB3-1 cells to 5 nM S-protein was sufficient to have significant differences of mRNA expression (p = 0.0294 for IL-6 and p = 0.001 for IL-8).
The data concerning IL-6 and IL-8 protein release were obtained by Bio-Plex analysis (Fig. 1, panels D-F). Exposure to SARS-CoV-2 Spike protein was associated with a sharp increase of the release of IL-6 (Fig. 1D) and IL-8 (Fig. 1E). In agreement with the RT-qPCR data exposure to 5 nM SARS-CoV-2 S-protein was sufficient to obtain a significant increase (2–3-fold) of the release of IL-6 (p = 0.0008) and IL-8 (p = 0.0116) (see also the summary of 5 independent experiments shown in Fig. 1F).
Data from these experiments showed that the IL-6 and IL-8 gene expression (Fig. 1B and 1C) and the release of their respective proteins (Fig. 1D and Fig. 1F) are operating following treatment of IB3-1 with SARS-CoV-2 Spike protein. As IL-6 and IL-8 gene expression is under transcriptional control of NF-κB (Bezzerri et al., 2011), we also analyzed NF-κB expression by Western blotting and found that exposure of IB3-1 cells to SARS-CoV-2 Spike protein leads to an increase of NF-κB (Supplementary Figure S1). Altogether, these data support the concept that our experimental system recapitulates some of the key step of SARS-CoV-2 Spike-mediated biological changes, including NF-κB stimulation and upregulation of IL-6 and IL-8.
Treatment of IB3-1 cells with SFN reverses IL-6 and IL-8 upregulation induced by SARS-CoV-2 Spike protein
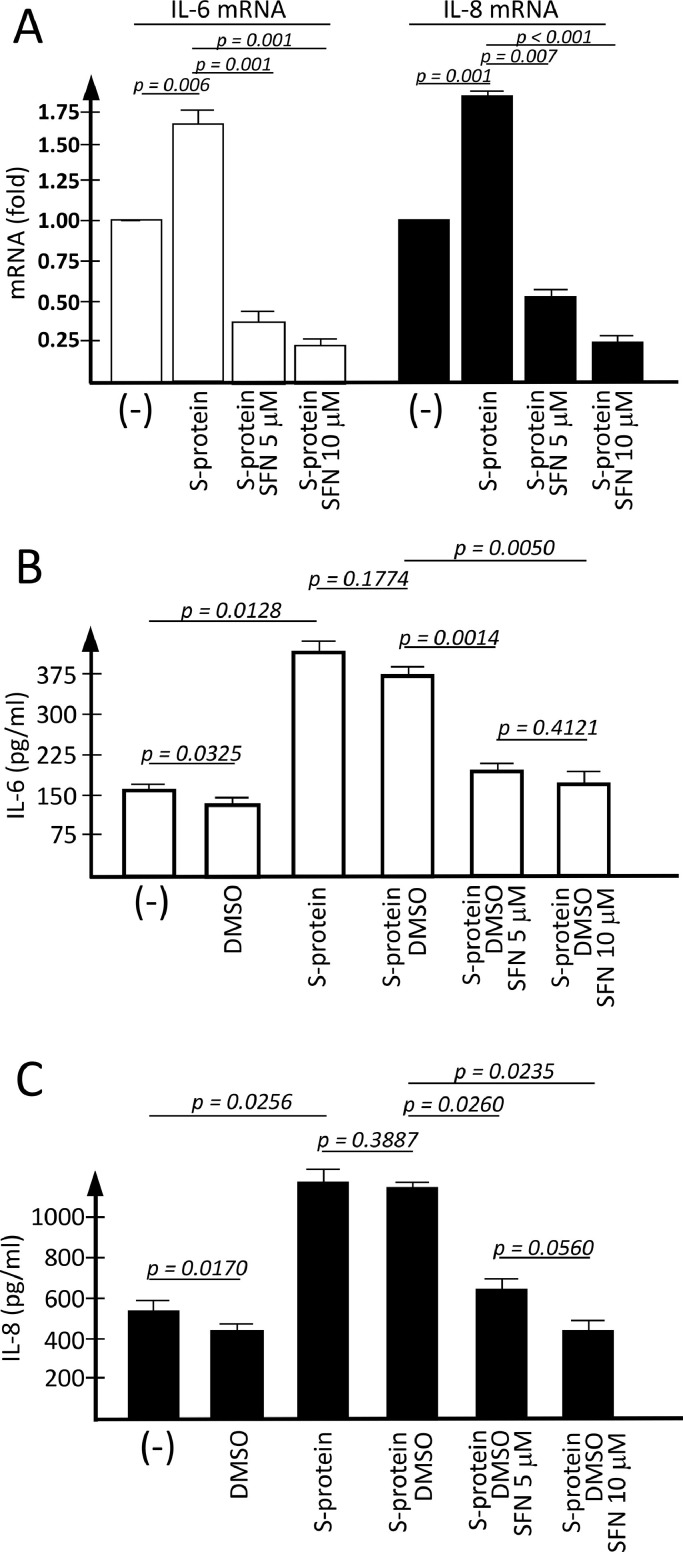
In the experiments shown in Fig. 2 , the effects of SFN on S-protein induced IL-6 and IL-8 gene expression were analyzed. Fig. 2A shows that both 5 and 10 μM of SFN concentrations are potent inhibitors of IL-6 and IL-8 mRNA accumulation. This finding was highly reproducible. We avoided the use of higher concentrations of SFN, since it is known that this molecule is able to induce also pro-apoptotic effects at high concentrations in other types of cells (Gasparello et al., 2020). No major effects of SFN on IB3-1 cell growth were observed when 5 μM concentration was used.
Fig 2.
Effects of SFN on S-protein induced increase of IL-6 and IL-8 gene and protein expression. IB3-1 cells were exposed for 24 h to SARS-CoV-2 Spike protein (S-protein) in the absence (-) or in the presence of DMSO, S-protein and SFN as indicated. (A) Quantification of IL-6 and IL-8 mRNA was performed by RT-qPCR, by comparing SFN-treated cells with untreated cells. (B-C) Extracellular release of IL-6 (B) and IL-8 (C) was studied by Bio-Plex analysis (absolute pg/ml values indicated in the histograms). Results represent the average ± SEM of three independent experiments.
Fig. 2 shows that SFN inhibits the release of IL-6 and IL-8 and that this effect is dose-dependent. In addition, the inhibitory effects on protein release (Fig. 2B and 2C) are, as expected, accompanied by a sharp inhibition of mRNA accumulation (Fig. 2A).
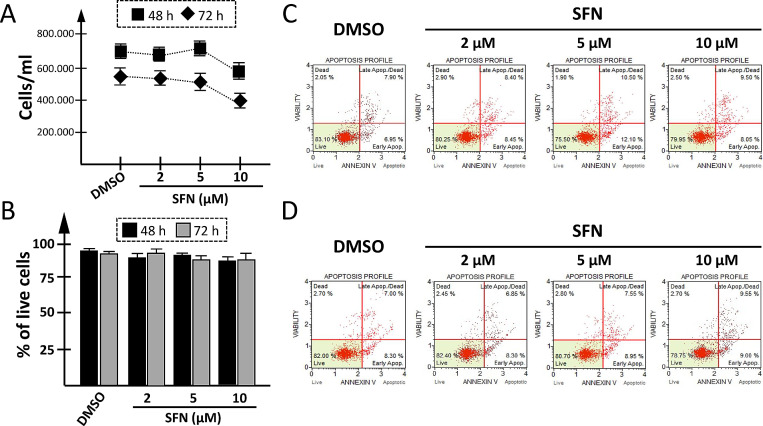
Effects of SFN on toxicity and apoptosis
The experiment shown in Fig. 3 was performed to verify whether SFN treatment of IB3-1 cells was to some extent cytotoxic. In these experiments, IB3-1 cells were cultured for 48 h (Fig. 3C) and 72 h (Fig. 3D) in the presence of the vehicle (DMSO) or in the presence of SFN at the concentrations of 2, 5 and 10 μM. The results obtained demonstrated that SFN, when maintained in contact with IB3-1 cells for 48 or 72 h, had no or very limited effects on cell growth. Moreover, SFN based treatment did not reduce the extent of viable cells (Fig. 3B) and did not induce their apoptosis (Fig. 3C and 3D).
Fig 3.
Effects of SFN on cell growth, viability and apoptosis. IB3-1 cells were cultured in the absence or presence of the indicated concentrations of SFN for 48 h or 72 h and cell growth (A), viability (B) and apoptosis (C,D) were determined. Cell growth (A) was determined after 48 h (black boxes) and 72 h (black rhombuses) treatment with different SFN concentrations (2, 5, 10 μM) or with the DMSO as vehicle. (B) Cell viability was analyzed after 48 h (black boxes) or 72 h (grey boxes) contact with vehicle or SFN. (C-D) Apoptosis was evaluated at 48 h (C) and 72 h (D) using the Muse Annexin V & Dead Cell kit.
Differential effects on secreted proteins in SFN-treated IB3-1 cells exposed to SARS-CoV-2 Spike protein
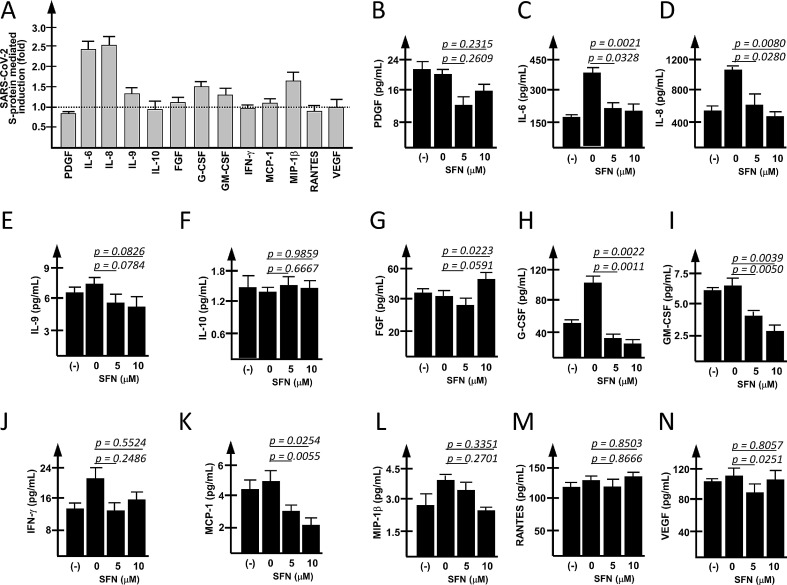
In order to verify the selectivity of the effects of SFN on overall protein released, the Bio-Plex analysis was conducted on secreted proteins exceeding 1 pg/ml (13 proteins) in the extracellular medium of untreated S-protein induced IB3-1 cells. Fig. 4 A shows that 24 h of S-protein treatment induced an increased release of IL-6, IL-8, IL-9, FGF, G-CSF, GM-CSF, MCP-1 and MIP-1β.
Fig. 4.
Effects of SFN on proteins released by S-protein stimulated IB3-1 cells. (A) Effects of the stimulation of IB3-1 cells with SARS-CoV-2 Spike protein on the release of cytokines, chemokines and growth factors by IB3-1 cells. A panel of 27 cytokines/chemokines/growth factors was analyzed in IB3-1 infected cells 24 h after the exposure to the S-protein. Secreted proteins exceeding the concentration of 1 pg/ml in the extracellular medium are reported in the histograms. Data are reported as Fold Change (FC, S-protein infected IB3-1 cells versus control untreated cells). (B-N) Effects of SFN on the inflammation-associated proteins released by S-protein exposed IB3-1 cells. Results represent the average ± SEM of four (A) or three (B-N) independent experiments.
The effects of SFN treatment on the release of the 13 proteins are shown in panels B-N of Fig. 4. These results suggest a differential effect of SFN on the release of cytokines, chemokines and growth factors. SFN-mediated inhibitory effects were evident (in addition to IL-6 and IL-8, as shown in Fig. 2) for PDGF, IL-9, G-CSF, GM-CSF, IFN-γ, MCP-1, MIP-1β. No effect was evident for IL-10, FGF, RANTES and VEGF.
Discussion
One of the clinical features of COVID-19 is the hyperinflammatory activity that is characterized by high expression of IL-6, IL-8 and several other cytokines, chemokines and growth factors (Pelaia et al., 2020). This hyperinflammatory activity is associated with severe forms of COVID-19 and poor prognosis for COVID-19 patients (Zeng et al., 2020). For instance, Del Valle et al. (2020) found that high serum IL-6, IL-8 and TNF-α levels at the time of hospitalization are strong and independent predictors of patient survival. In another study, Burke et al. (2020) found that inflammatory phenotyping (revealing upregulation of IL-6 and IL-8 gene expression) predicts clinical outcome in COVID-19 subjects. Therefore, anti-inflammatory compounds and specific clinical protocols are highly needed.
Concerning this issue, different approaches targeting IL-6 and IL-8 have been proposed in several studies as well as in clinical trials (Ruhee et al., 2019; Sturm et al., 2017). For instance, among these: NCT04381052 (“Study for the Use of the IL-6 Inhibitor Clazakizumab in Patients With Life-threatening COVID-19 Infection”), NCT04343989 (“A Randomized Placebo-controlled Safety and Dose-finding Study for the Use of the IL-6 Inhibitor Clazakizumab in Patients With Life-threatening COVID-19 Infection”) and NCT04363502 (“Use of the Interleukin-6 Inhibitor Clazakizumab in Patients With Life-threatening COVID-19 Infection”) are designed to administer the IL-6 inhibitor clazakizumab to patients with COVID-19 infection characterized by pulmonary failure and a clinical picture consistent with a “cytokine storm” syndrome; NCT04322773 (“Anti-IL6 Treatment of Serious COVID-19 Disease with Threatening Respiratory Failure”), is based on the use of two human monoclonal antibodies against IL-6 receptor, tocilizumab and sarilumab; NCT04486521 (“Clinical Outcome of Anti-IL6 vs Anti-IL6 Corticosteroid Combination in Patients With SARS-CoV-2 Cytokine Release Syndrome”) is based on the use of corticosteroid combinations. As far as IL-8 targeting, NCT04347226 (“Anti-Interleukin-8 for patients with COVID-19”), is designed to administer the IL-8 inhibitor BMS-986253 to COVID-19 patients.
The results presented in our study are a proof-of-principle that the release of two key proteins of the COVID-19 “cytokine storm” (Soy et al., 2020) can be strongly inhibited by sulforaphane (SFN). Since the control of the “cytokine storm” is a major issue in the management of COVID-19 patients (Mustafa et al., 2020), our study could stimulate research activity that can contribute to the development of protocols useful to control hyperinflammatory state associated with SARS-CoV-2 infection. Of particular interest, in addition to IL-6 and IL-8, is the inhibitory effects of SFN on SARS-CoV-2 induced increase of G-CSF, GM-CSF and MCP-1. This paper is expected to sustain research activity on plant extracts and food supplement containing SFN, in order to support the integration of 'phytopreparations' into conventional/official medicine focusing on COVID-19 treatment. SFN is indeed present within several phytomaterials, derived for instance from broccoli sprout, broccoli, cauliflower, kale, Brussels sprout, cabbage, bok choy, collard, rugula, turnips (Farag et al., 2010).
One of the limits of our study is the lack of explanation about the SFN mechanism of action. This should be considered a major issue of the research on this topic in the future, since it could help in finding new targets of possible use in therapeutic protocols. Among the several possibilities (that in any case are not mutually exclusive), SFN can exert its anti-inflammatory activity by JNK/AP-1/NF-κB inhibition and Nrf2/HO-1 activation (Subedi et al., 2019). The SFN-mediated targeting of NRF2 has been described in other studies (Qi et al., 2016). On the other hand, SFN is well known as a potent inducer of endocellular production of hydrogen sulfide (Pei et al., 2011). In consideration of the large variety of anti-inflammatory effects of hydrogen sulfide and hydrogen sulfide-releasing chimeras, this issue should be considered of great interest for future studies.
Conclusion
We have developed a simple experimental system and analytical protocol for the screening of molecules interfering with the expression of proteins known to be involved in the COVID-19 “cytokine storm”. The results here presented demonstrate that exposure of epithelial IB3-1 cells to the SARS-CoV-2 spike protein induces increased expression of NF-κB and increased release particularly of IL-6 and IL-8, but also of IL-9, FGF, G-CSF, GM-CSF, MCP-1 and MIP-1β. This allows the screening of possible inhibitors of these biochemical targets and possible agents to be proposed for the experimental treatment of this clinical phase of the disease.
Treatment with sulforaphane reverses IL-6 and IL-8 upregulation induced by SARS-CoV-2 Spike protein in IB3-1 cells. Furthermore, sulforaphane-mediated inhibitory effects were observed also for PDGF, IL-9, G-CSF, GM-CSF, IFN-γ, MCP-1 and MIP-1β. Therefore, sulforaphane and sulforaphane-containing phytoproducts should be further evaluated as potential inhibitors of the COVID-19 “cytokine storm”.
Further experiments should be programmed to identify other agents able to inhibit changes in gene expression induced by SARS-CoV-2 Spike, not only to find novel molecules to be considered as positive control in this experimental system, but also to verify whether combined therapy with sulforaphane is possible.
Funding Information
This work was funded by Italian Consortium for Biotechnologies (C.I.B), Fondazione Fibrosi Cistica (FFC), Project “Revealing the microRNAs-transcription factors network in cystic fibrosis: from microRNA therapeutics to precision medicine (CF-miRNA-THER)”, FFC#7/2018.
Author Contribution
Jessica Gasparello, investigation, data curation and formal analysis; Elisabetta D'Aversa, data curation; Chiara Papi, data curation; Laura Gambari, resources and methodology; Monica Borgatti, supervision; Alessia Finotti and Brunella Grigolo, supervisors; Roberto Gambari, writing the manuscript and funding acquisition.
Declaration of Competing Interest
No competing financial interests exist. The authors declare the absence of other types of conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phymed.2021.153583.
Appendix. Supplementary materials
References
- Adem Ş., Eyupoglu V., Sarfraz I., Rasul A., Zahoor A.F., Ali M., Abdalla M., Ibrahim I.M., Elfiky A.A. Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine. 2020;153310 doi: 10.1016/j.phymed.2020.153310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.M., Luo L., Namani A., Wang X.J., Tang X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:585–597. doi: 10.1016/j.bbadis.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Andreakos E., Papadaki M., Serhan C.N. Dexamethasone, pro-resolving lipid mediators and resolution of inflammation in COVID-19. Allergy. 2021;76:626–628. doi: 10.1111/all.14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzerri V., Borgatti M., Finotti A., Tamanini A., Gambari R., Cabrini G. Mapping the transcriptional machinery of the IL-8 gene in human bronchial epithelial cells. J. Immunol. 2011;187:6069–6081. doi: 10.4049/jimmunol.1100821. [DOI] [PubMed] [Google Scholar]
- Burke H., Freeman A., Cellura D.C., Stuart B.L., Brendish N.J., Poole S., Borca F., Phan H.T.T., Sheard N., Williams S., Spalluto C.M., Staples K.J., Clark T.W., Wilkinson T.M.A. Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020;21:245. doi: 10.1186/s12931-020-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Simone G., Mancusi C. Finding the right time for anti-inflammatory therapy in COVID-19. Int. J. Infect. Dis. 2020;S1201-9712:32170–32176. doi: 10.1016/j.ijid.2020.09.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzobo K., Chiririwa H., Dandara C., Dzobo W. Coronavirus Disease-2019 treatment strategies targeting interleukin-6 signaling and herbal medicine. OMICS. 2020;25:13–22. doi: 10.1089/omi.2020.0122. [DOI] [PubMed] [Google Scholar]
- Farag M.A., Motaalba A.A., Brown J.G. Sulforaphane composition, cytotoxic andantioxidant activity of crucifer vegetables. J. Adv. Res. 2010;1:65–70. doi: 10.1016/j.jare.2010.02.005. [DOI] [Google Scholar]
- Gao S., Zhang L. ACE2 partially dictates the host range and tropism of SARS-CoV-2. Comput. Struct. Biotechnol. J. 2020;18:4040–4047. doi: 10.1016/j.csbj.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparello J., Gambari L., Papi C., Rozzi A., Manicardi A., Corradini R., Gambari R., Finotti A. High levels of apoptosis are induced in the human colon cancer HT-29 cell line by co-administration of sulforaphane and a Peptide Nucleic Acid targeting miR-15b-5p. Nucleic Acid Ther. 2020;30:164–174. doi: 10.1089/nat.2019.0825. [DOI] [PubMed] [Google Scholar]
- Gasparello J., Lomazzi M., Papi C., D'Aversa E., Sansone F., Casnati A., Donofrio G., Gambari R., Finotti A. Efficient delivery of microRNA and antimiRNA molecules using an argininocalix[4]arene macrocycle. Mol. Ther. Nucleic Acids. 2019;18:748–763. doi: 10.1016/j.omtn.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Tonetti T., Protti A., Langer T., Girardis M., Bellani G., Laffey J., Carrafiello G., Carsana L., Rizzuto C., Zanella A., Scaravilli V., Pizzilli G., Grieco D.L., Di Meglio L., de Pascale G., Lanza E., Monteduro F., Zompatori M., Filippini C., Locatelli F., Cecconi M., Fumagalli R., Nava S., Vincent J.L., Antonelli M., Slutsky A.S., Pesenti A., Ranieri V.M. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir. Med. 2020;8:1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa S.A.M., Yosri N., El-Mallah M.F., Ghonaim R., Guo Z., Musharraf S.G., Du M., Khatib A., Xiao J., Saeed A., El-Seedi H.H.R., Zhao C., Efferth T., El-Seedi H.R. Screening for natural and derived bio-active compounds in preclinical and clinical studies: one of the frontlines of fighting the coronaviruses pandemic. Phytomedicine. 2020;153311 doi: 10.1016/j.phymed.2020.153311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latté K.P., Appel K.E., Lampen A. Health benefits and possible risks of broccoli - an overview. Food Chem. Toxicol. 2011;49:3287–3309. doi: 10.1016/j.fct.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Matthay M.A., Leligdowicz A., Liu K.D. Biological mechanisms of COVID-19 acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2020;202:1489–1491. doi: 10.1164/rccm.202009-3629ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva T., Khafizova G., Sokornova S. In search of herbal anti-SARS-Cov2 compounds. Front. Plant. Sci. 2020;11 doi: 10.3389/fpls.2020.589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa M.I., Abdelmoneim A.H., Mahmoud E.M., Makhawi A.M. Cytokine Storm in COVID-19 patients, its impact on organs and potential treatment by QTY Code-Designed Detergent-Free Chemokine Receptors. Mediators. Inflamm. 2020;2020 doi: 10.1155/2020/8198963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasonov E., Samsonov M. The role of Interleukin 6 inhibitors in therapy of severe COVID-19. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwa W., Kumar R., Thompson D., Lyerly W., Moore R., Reid T.E., Lowe H., Toyang N. Potential of flavonoid-inspired phytomedicines against COVID-19. Molecules. 2020;25:2707. doi: 10.3390/molecules25112707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F., Scarlata S., Agrò F.E. COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Wu B., Cao Q., Wu L., Yang G. Hydrogen sulfide mediates the anti-survival effect of sulforaphane on human prostate cancer cells. Toxicol. Appl. Pharmacol. 2011;257:420–428. doi: 10.1016/j.taap.2011.09.026. [DOI] [PubMed] [Google Scholar]
- Pelaia C., Tinello C., Vatrella A., De Sarro G., Pelaia G. Lung under attack by COVID-19-induced cytokine storm: pathogenic mechanisms and therapeutic implications. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620933508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Xu F., Yan X., Li S., Li H. Sulforaphane exerts anti-inflammatory effects against lipopolysaccharide-induced acute lung injury in mice through the Nrf2/ARE pathway. Int. J. Mol. Med. 2016;37:182–188. doi: 10.3892/ijmm.2015.2396. [DOI] [PubMed] [Google Scholar]
- Ratajczak M.Z., Bujko K., Ciechanowicz A., Sielatycka K., Cymer M., Marlicz W., Kucia M. SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45(-) precursors of hematopoietic and endothelial cells and in response to virus Spike protein activates the Nlrp3 inflammasome. Stem Cell Rev. Rep. 2020:1–12. doi: 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhee R.T., Ma S., Suzuki K. Sulforaphane protects cells against lipopolysaccharide-stimulated inflammation in murine macrophages. Antioxidants. 2019;8:577. doi: 10.3390/antiox8120577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumagne T., Winiszewski H., Besch G., Mahr N., Senot T., Costa P., Grillet F., Behr J., Mouhat B., Mourey G., Fournel A., Meneveau N., Samain E., Capellier G., Piton G., Pili-Floury S. Pulmonary embolism among critically ill patients with ARDS due to COVID-19. Respir. Med. Res. 2020;78 doi: 10.1016/j.resmer.2020.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm C., Wagner A.E. Brassica-derived plant bioactives as modulators of chemopreventive and inflammatory signaling pathways. Int. J. Mol. Sci. 2017;18:1890. doi: 10.3390/ijms18091890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi L., Lee J.H., Yumnam S., Ji E., Kim S.Y. Anti-Inflammatory effect of sulforaphane on LPS-activated microglia potentially through JNK/AP-1/NF-κB inhibition and Nrf2/HO-1 Activation. Cells. 2019;8:194. doi: 10.3390/cells8020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z., She Y. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Vilekar P., Yang S.P., Gupta M., Oh M.I., Meek A., Doyle L., Villar L., Brennecke A., Liyanage I., Reed M., Barden C., Weaver D.F. Small molecule therapeutics for COVID-19: repurposing of inhaled furosemide. PeerJ. 2020;8:e9533. doi: 10.7717/peerj.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin P.L., Lu L., Rhim J.S., Cutting G., Stetten G., Kieffer K.A., Craig R., Guggino W.B. A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am. J. Respir. Cell Mol. Biol. 1991;4:313–319. doi: 10.1165/ajrcmb/4.4.313. [DOI] [PubMed] [Google Scholar]
- Zeng Z., Yu H., Chen H., Qi W., Chen L., Chen G., Yan W., Chen T., Ning Q., Han M., Wu D. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan. China. Crit. Care. 2020;24:525. doi: 10.1186/s13054-020-03255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.