Abstract
The tragic deaths of three patients in a recent AAV-based X-linked myotubular myopathy clinical trial highlight once again the pressing need for safe and reliable gene delivery vectors. Non-viral minimized DNA vectors offer one possible way to meet this need. Recent pre-clinical results with minimized DNA vectors have yielded promising outcomes in cancer therapy, stem cell therapy, stem cell reprograming, and other uses. Broad clinical use of these vectors, however, remains to be realized. Further advances in vector design and production are ongoing. An intriguing and promising potential development results from manipulation of the specific shape of non-viral minimized DNA vectors. By improving cellular uptake and biodistribution specificity, this approach could impact gene therapy, DNA nanotechnology, and personalized medicine.
INTRODUCTION
In 2017, we wrote a comprehensive review of the history, key developments, specialized uses, and broad outlook for non-viral minimized DNA vectors as therapeutics, and, in some cases, as critical enablers of other cell-based therapies (e.g., stem cell reprogramming) [1]. We described in detail the many advantages minimized DNA vectors offer. In brief, removal of immunogenic bacterial sequences and antibiotic resistance genes from plasmids allowed for a dramatic reduction in vector length and led to the emergence of a new generation of non-viral gene delivery vectors (minimized DNA vectors). Minimized DNA vectors do not integrate into the genome and encode only therapeutic sequences. Reduced vector length is one of many factors that is likely to account for the observed increased levels and duration of gene expression compared to other non-viral vectors, particularly plasmids (some comparisons of vector systems are summarized in Table 1) [2–6].
TABLE 1.
Outcomes of studies comparing non-viral minimized DNA vectors to other vector systems.
| Vectors used | Vector length | Sequence encoded | Transfection method | Outcomes | Ref. |
|---|---|---|---|---|---|
| Minicircle Plasmid |
3,881 6,233 |
Firefly luciferase | Sequence-defined oligoamino amides/cationic polymer | Compact, rod-shaped polyplexes were 65–100 nm using plasmid and 35–40 nm using minicircle; all formulations of minicircle polyplexes lacked cell cycle dependence. Mini-circle transfected ~3-fold more than equal moles of plasmid. Combined, tyrosine trimer integration, combination polyplexes, and use of minicircle increased gene expression ~200-fold over an equal mass of plasmid. | [2] |
| Minicircle Plasmid |
4,573 8,147 |
Mesothelin CAR | Electroporation | CAR expression, IFNγ and granzyme B secretion, and specific lysis of pancreatic cancer cell lines was significantly increased in NK cells electroporated with minicircle over plasmid. Use of minicircle resulted in increased NK cell viability after electroporation. | [3] |
| Minicircle Plasmid Minicircle Plasmid |
3,700* 7,700* 4,000* 7,900* |
Firefly luciferase TK/NTR |
Microvesicles/cationic lipid | Equal moles of minicircle resulted in prolonged transgene expression in breast cancer cells. Minicircles loaded into microvesicles twice as efficiently as equal moles of plasmid but resulted in a peak bioluminescent signal 14 times higher than in cells treated with microvesicles containing plasmid. Microvesicles loaded with minicircles encoding TK/NTR led to greater activity of prodrug converting enzymes over microvesicles with equal mass of plasmid. | [4] |
| Minicircle Plasmid |
364 8,318 |
Guide RNA (to inhibit PLK1) | LHNPs | Cas9 protein co-delivered in LHNPs with minicircles decreased PLK1 expression more than Cas9 protein co-delivered with plasmid or minicircle co-delivered with Cas9 DNA in vitro. | [5] |
| Minicircle Plasmid Minicircle Lentivirus |
Unknown Unknown Unknown NA |
eGFP 2nd gen. anti-CD19 CAR |
Electroporation | CD34+, H9 hESCs, and T cells electroporated with minicircle encoding eGFP resulted in more and brighter eGFP+ cells, increased cell viability, and increased CFUs compared to equal mass of plasmid. T cells electroporated with CAR minicircle killed tumor cells in vitro and in mice comparably to T cells transduced with lentiviral vector. | [6] |
| Minivector Plasmid siRNA |
400* 3,900* NA |
shRNA/siRNA against GFPor ALK | Cationic lipid | Minivector and siRNA, but not plasmid, decreased GFP expression in difficult-to-transfect Jurkat cells and decreased expression of ALK in Karpas 299 cells; the three vectors were comparable in easy-to-transfect 293 FT cells. Minivector and siRNA, but not plasmid, arrested growth of ALCL cells. Minivector DNA survived human serum > 10-fold longer than plasmid or siRNA. | [9] |
| Minivector Plasmid |
281–2,679 1,711–5,302 |
Multiple different1 | NA | Minivectors ≤ 1,200 bp survived nebulization while longer vectors sheared faster as a function of increasing length. Negative supercoiling afforded up to 2-fold additional protection from nebulization and sonication shear forces. | [10] |
| Minicircle Plasmid Plasmid |
2,257 3,487 5,541 |
GFP | Cationic lipid (niosomes) | Minicircle transfected twice as efficiently as an equal mass of plasmid. Minicircle had higher capacity to deliver to primary retinal cells and rat retinas than equal mass of plasmid. | [28] |
| Minicircle Plasmid Minicircle AAV |
Unknown Unknown 2,500* NA |
GFP Rhodopsin |
Cationic lipid | Minicircle GFP expression in retinal cells was maintained for 7 days while GFP expression from an equal mass of plasmid was lost before 7 days. Gene delivery to retinal cells in vitro using AAV or minicircles encoding rhodopsin was comparable in efficiency. Cells modified ex vivo with AAV or minicircles encoding rhodopsin reconstructed functional retinal tissue and supported vision function in blind mice. | [29] |
| Plasmid Plasmid2 Plasmid Plasmid2 |
7,722 9,668 8,738 10,684 |
eGFP eGFP/Cre recombinase HN HNHis/Cre recombinase |
Cationic lipid | Plasmids encoding genes with or without Cre recombinase were transfected into Salmonella as a platform for oral DNA vaccination against Newcastle disease virus in poultry. Plasmid containing Cre recombinase allowed for the in vivo generation of minicircle encoding either eGFP or HN. Chickens orally inoculated with Salmonella transfected with Cre/eGFP-containing plasmid contained significantly more eGFP in liver than plasmid without Cre. Chickens that received Cre/HN inoculation were protected against challenge with NDV significantly more than chickens inoculated with Salmonella containing HN plasmid alone. | [35] |
| Minicircle Adenovirus |
Unknown NA |
Bcl-2 | Cationic lipid | Percentage of NSCs overexpressing Bcl-2 was comparable when using adenovirus or minicircle but minicircle-treated cells lost expression faster. NSCs treated with adenovirus or minicircle overexpressing Bcl-2 were partially rescued from transplant-associated insults. | [38] |
| Minicircle Plasmid Minicircle Plasmid |
3,088 7,100 4,618 8,581 |
GFP GFP/Sox9 |
Cationic lipid | Percent GFP+ was increased ~10-fold in canine, equine, and rat MSCs following transfection with GFP minicircle over an equal mass of GFP plasmid. Sox9 was successfully expressed in canine MSCs after transfection with Sox9 minicircle in vitro. | [40] |
| Minicircle Plasmid |
1,715 3,531 |
VEGF | Electroporation/microporation | Transfection with either plasmid or minicircle did not change expansion potential, differentiation capacity, or immunophenotype of MSCs, but transfection with minicircle led to 2.5-fold more VEGF transcripts, greater VEGF production, and improved angiogenic potential of MSCs in vitro. | [41] |
| Minicircle Plasmid Lentivirus |
4,129 8,133 NA |
hPAX7/GFP | Cationic lipid | Repeated transfection with hPAX7 minicircle generated myogenic progenitors that could terminally differentiate, but their transplantation resulted in limited engraftment. Formation of hPAX7+ myogenic progenitors using lentivirus remains the more efficient platform for generation of myogenic progenitors. | [42] |
| Minicircle Plasmid Minicircle Plasmid |
3,400* 6,100* 2,300* 4,700* |
Venus fluorescent protein SB100X transposase |
Nucleofection | CD34+ HSPC electroporated with minicircle encoding Sleeping Beauty components resulted in increased cell viability, enhanced transient gene delivery, and higher rates of stable gene integration over equimolar amounts of plasmid expressing these components. | [43] |
Italicized values are estimated lengths provided by the authors of those studies.
This study focused on DNA vector length.
This study sought to use plasmids harboring Cre recombinase and the gene of interest for the in vivo production of minicircles (carrying the gene of interest without Cre recombinase) by using Cre expression to recombine the originally transfected plasmid, effectively amounting to the delivery of both plasmid and minicircle to the Salmonella cells receiving plasmid encoding Cre recombinase.
AAV: Adeno-associated virus; ALCL: Anaplastic large cell lymphoma; ALK: Anaplastic lymphoma kinase; Bcl-2: B-cell lymphoma 2; CAR: Chimeric antigen receptor; Cas9: Clustered regularly interspaced short palindromic repeats-associated protein 9; CD19: Cluster of differentiation 19; CD34: Cluster of differentiation 34; CFU: Colony forming unit; Cre: Causes recombination; eGFP: Enhanced green fluorescent protein; gen.: Generation; GFP: Green fluorescent protein; hESCs: Human embryonic stem cells; His: Histidine-tagged; HN: Hemagglutinin neuraminidase; hPAX7: Human paired box 7; HSPC: Hematopoietic stem and progenitor cells; IFN: Interferon; LHNPs: Liposome-templated hydrogel nanoparticles; MSCs: Mesenchymal stem cells; NA: Not applicable; NDV: Newcastle disease virus; NK: Natural killer; NSCs: Neural stem cells; PLK1: Polo-like kinase 1; Ref.: Reference; SB100X: Sleeping Beauty 100X transposase; shRNA: Short hairpin RNA; siRNA: Small interfering RNA; Sox: Sex-determining region Y-box transcription factor 9; TK/NTR: Thymidine kinase/nitroreductase; VEGF: Vascular endothelial growth factor.
There are several types of non-viral minimized DNA vectors in pre-clinical use (reviewed in [1]). Here, we will highlight recent advances for minicircles [1,7,8] and minivectors [1,9,10]. Several different methods exist for the production of these vectors [7,8,11], but common to most is the use of bacteria to propagate plasmids. Bacteria are induced to express enzymes that catalyze recombination of these parental plasmids. This reaction excises the bacterial propagation sequences into a separate molecule (the ‘miniplasmid’) that can be removed either by endonuclease-mediated degradation in the bacteria [12] or by size-exclusion chromatography [11,13]. Complete removal of unrecombined parent plasmid, miniplasmid, immunogenic endotoxin, and bacterial genomic DNA is laborious and time-consuming, yet essential. Recently, a production method was developed that relies upon a multiplex PCR protocol for minicircle formation [6]. This method circumvents the use of bacteria, eliminating the need for removal of bacterial contaminants and, thus, can be completed in hours versus days. The product vectors, dubbed ‘bacteria-free minicircles,’ could be a useful tool for gene therapy, but production scale-up may still be an issue [6].
In common, minicircles and minivectors are double-stranded, circular, supercoiled DNA vectors encoding therapeutic sequences. One key difference between the two is that minivectors employ a more rigorous purification method that takes advantage of the small size of the minivectors generated, allowing for complete removal of the larger miniplasmid contaminant. Additional advantages include increased negative supercoiling and the ability to generate vectors as small as a few hundred base pairs [9,10].
The reduced size of minimized DNA vectors allows for the delivery of many more therapeutic molecules per given unit of mass. Therefore, much less mass of DNA is required to deliver an equivalent number of molecules. Minimized DNA vectors may thus be advantageous for delivering higher doses of a potential therapy without evidence of the cytotoxic effects that prohibit the use of higher doses of plasmids. Less mass of vector also means less delivery vehicle and thus reduction of another potential source of toxicity. The decreased toxicity and decreased immunogenicity of minimized DNA vectors, and especially of minivectors, may help mitigate some of the adverse effects observed in gene therapy clinical trials, such as in the recent X-linked myotubular myopathy clinical trials that used adeno-associated virus (AAV) [14–16].
Exciting pre-clinical work with non-viral minimized DNA vectors has continued since our last review in 2017 [1], bringing the field closer to realizing the hope of wide-spread clinical success. In this brief update, we summarize these new developments, concentrating on two key applications where progress has been most impressive—cancer therapy and stem cell therapy. We also present a new idea stemming from an improved understanding of DNA structure. With support from computational simulation data to illustrate the feasibility of the approach, we demonstrate that it may be possible to manipulate the shape of DNA vectors for selective tissue or cell targeting, and/or increased cellular uptake.
USING MINIMIZED DNA VECTORS FOR CANCER THERAPY
To date, the field that has probably benefitted most from minimized DNA vector technology is that of cancer therapy, particularly in the development of chimeric antigen receptor T cells (reviewed in [17]). Chimeric antigen receptors (CARs), so named because they artificially fuse antigen-binding domains to specific cell-activating domains [18], have brought the gene therapy field some of its first clinical and commercial achievements (e.g., Kymriah®, Yescarta®). Although CAR T cell therapy has been successful, particularly for hematological malignancies [19], improvements are still needed. The therapy can be immunogenic and the protocol for developing and delivering the T cells is expensive, complicated, and takes several weeks. Non-viral minimized DNA vectors could replace the viral vectors used to engineer autologous (or allogeneic) CAR T cells [20], resulting in cheaper, faster, and safer production. Indeed, minicircles encoding a CD44-CAR have been electroporated into T cells to engineer them against hepatocellular carcinoma. The resultant CD44-CAR T cells resembled normal T cells in cytokine profile and phenotype, specifically lysed CD44+ cell lines and not CD44- cell lines, and suppressed tumor growth in vivo compared to controls [21]. This result was important as it demonstrated the efficacy of minicircle-generated CAR T cells against a solid tumor, which is more challenging to treat than the diffuse lymphomas treated previously [19].
Similar breakthroughs of minicircle-generated CAR T cells have also been reported for prostate cancer [22] and colorectal cancer [23]. Cheng et al. (2019) successfully generated anti-CD19 CAR T cells via electroporation with minicircles generated using the bacteria-free production method described above [6]. The resultant CAR T cells decreased tumor burden in mice with at least the same efficacy as lentiviral-generated CAR T cells carrying the same anti-CD19 CAR genes [6]. Furthermore, Batchu et al. (2019) engineered CAR natural killer (NK) cells capable of killing pancreatic cancer cells in vitro using a combination of minicircles encoding a mesothelin CAR and Sleeping Beauty transposition [3]. CAR T cell therapy requires the ex vivo modification of autologous T cells from each individual. In contrast NK cells, because their cytolytic activity is antigen-independent, can be taken from healthy donors and engineered in advance of therapy. This process creates an off-the-shelf product that saves both time and money. Of all the minicircle-based applications currently in development, use of the non-viral Sleeping Beauty transposon system for the safe and reasonably effective generation of CAR T cells is probably the closest to achieving clinical efficacy [20,24].
Various other minicircle-based strategies have emerged for breast cancer [4,25], brain cancer [5], ovarian cancer [26], nasopharyngeal carcinoma [27] and other applications (Table 2). Kanada et al. (2019) developed a method that uses microvesicles to deliver minicircles encoding prodrug converting enzymes [4]. The expressed enzymes convert co-delivered prodrugs into cytotoxic agents that kill tumor cells. Minicircles were also combined with calcium phosphate nanoneedles for ovarian cancer [26] and others used liposome-templated hydrogel nanoparticles to deliver both Cas9 protein and minicircles encoding guide RNA intravenously to tumor cells in the brain [5]. When polo-like kinase 1 was targeted for inhibition in brain cancer cells, tumor burden was decreased and survival of mice increased [5]. Finally, in Wu et al., (2017) nasopharyngeal carcinoma cells were targeted by way of a commonly expressed Epstein–Barr virus antigen (EBNA1) that selectively triggers the expression of a microRNA that inhibits nasopharyngeal carcinoma cell growth and metastasis [27].
TABLE 2.
Studies using minicircles for the development of novel therapies.
| Sequence encoded | Vector length | Transfection method | Outcomes | Ref. |
|---|---|---|---|---|
| 3rd generation anti-CD44 CAR |
NR | Electroporation | Minicircle-generated anti-CD44-CAR T cells expressed CAR molecules with strong hepatocellular carcinoma tumor suppression activity in vitro and overcame tumor microenvironment barriers in mice. | [21] |
| 3rd generation anti-PSCA CAR |
4,575 | Electroporation | Unlike normal T cells, minicircle-generated PSCA CAR T cells had high cytokine secretion, strong antitumor effects, infiltrated tumor tissue, and persisted up to 28 days in mice. | [22] |
| 3rd generation NKG2D CAR |
NR | Electroporation | Minicircle-generated NKG2D CAR T cells demonstrated efficient and specific cytotoxic activity against human colorectal cancer in vitro and in vivo. | [23] |
| TIPE2 | NR | Hydrodynamic tail vein injection | Minicircle-mediated TIPE2 expression inhibited breast cancer cell proliferation and promoted in vivo anti-tumor immune responses by boosting CD8+ T cell and NK cell function. | [25] |
| Anti-EpCAM/CD3 | NR | Calcium phosphate nanoneedle-mediated cell perforation | Minicircle-mediated expression of an anti-EpCAM/CD3 bispecific antibody showed significant anti-cancer effects in vivo and increased survival of a xenograft mouse model of human ascites ovarian cancer by simultaneously conjugating immune cells and cancer cells. | [26] |
| miR-31 5p | NR | Cationic lipid | Minicircle transfection resulted in miRNA expression levels comparable to that of a lentiviral vector system used to generate cell lines stably expressing miR-31; This study validated WDR5 inhibition as a novel therapeutic option for nasopharyngeal carcinoma. | [27] |
| KLF4 | NR | IV injection | Minicircle-mediated KLF4 overexpression validated the role of KLF4 in the development and pathogenesis of inflammatory arthritis because it led to severe autoimmune arthritis in mice. KLF4 inhibition regulates the apoptosis of FLS and their expression of matrix metalloproteinases and proinflammatory cytokines. | [30] |
| Anti-alpha-synuclein shRNA | NR | RVG exosomes | Delivery of an anti-alpha-synuclein shRNA minicircle provided stable and prolonged gene downregulation and decreased aggregation of alpha-synuclein in the brain of a mouse model of Parkinson’s disease, improving clinical symptoms. | [31] |
| CBS | 2,336 | Hydrodynamic tail vein injection | Delivery of naked minicircle encoding CBS partially corrected metabolic and phenotypic defects in a mouse model of CBS deficiency. | [32] |
| sTNFR2-Fc | 3,000 | Electroporation | Minicircle-transfected MSCs produced the biologic TNFα inhibitor etanercept in vitro and had anti-inflammatory effects when injected into a collagen-induced rheumatoid arthritis mouse model. | [33] |
| IFNα IFNλ.3 |
1,656 1,677 |
Cationic lipid | Minicircles encoding liver-specific cytokine, IFNλ3, exhibited strong anti-HBV activity in transfected HBV-infected hepatocytes in vitro and suppressed viral antigen expression and viral DNA replication. | [34] |
| Bcl-2/GFP | NR | Electroporation/magnetofection | Minicircles encoding Bcl-2 attached to magnetic nanoparticles for in vivo transfection stimulated bone regeneration through the transient expression of Bcl-2, which prevented apoptosis of cell implants and promoted cell survival. | [36] |
| Sox9/Sox6/shANGPTL4 | NR | Cationic polymer | PEI minicircle particles encoding Sox9, Sox6, and shRNA against ANGPTL4 promoted chondrogenesis in vitro and suppressed osteoarthritis in mice. | [37] |
| GFP | 1,552 | Magnetofection | Neural stem cells engineered with minicircles in conjunction with magnetic nanoparticles were successfully grown and propagated on a novel neurosurgical-grade biomaterial scaffold with no adverse effects on key regenerative parameters. | [39] |
| BMP2/RFP TGFβ3/RFP |
7,300* 7,500* |
Cationic lipid | MSC-like, human iPSC-derived outgrowth cells transfected with two minicircles encoding TGFβ3 and BMP2, respectively, differentiated into the chondrogenic lineage and rescued osteochondral defects in rat models. | [58] |
| Sox2/Oct4/NanogLin28/GFP | NR | Cationic lipid | Transfection of Oct4, Sox2, Lin28, and Nanog-encoding minicircles to reprogram B16F10 murine melanoma cells resulted in incomplete reprogramming of cancer cells that did not form teratomas (an indicator of complete reprogramming). These cells, however, still displayed the characteristics of cancer stem cells and formed smaller, less aggressive tumors than the parental cell line. | [59] |
Italicized values are estimated lengths provided by the authors of those studies.
Bcl-2: B-cell lymphoma 2; BMP2: Bone morphogenic protein 2; CAR: Chimeric antigen receptor; CBS: Cystathionine β synthase; CD3: Cluster of differentiation 3; CD44: Cluster of differentiation 44; CD8: Cluster of differentiation 8; EpCAM: Epithelial cell adhesion molecule; FLS: Fibroblast-like synoviocytes; GFP: Green fluorescent protein; HBV: Hepatitis B virus; IFN: Interferon; IV: Intravenous; IPSCs: Induced pluripotent stem cells; KLF4: Kruppel-like factor 4; Lin28: Abnormal cell lineage 28; miRNA or miR: microRNA; MSCs: Mesenchymal stem cells; NK: Natural killer; NKG2D: Natural killer group 2 member D; NR: Not reported; Oct4: Octamer binding transcription factor 4; PEI: Polyethyleneimine; PSCA: Prostate stem cell antigen; Ref.: Reference; RFP: Red fluorescent protein; RVG: Rabies virus glycoprotein peptide; shRNA: Short hairpin RNA; shANGPLT4: Short hairpin angiopoietin-like protein 4; Sox: Sex-determining region Y-box transcription factor; sTNFR2-Fc: Soluble tumor necrosis factor receptor 2; TGFβ3: Transforming growth factor beta 3; TIPE2: Tumor necrosis factor alpha induced protein 8 family 2; TNF: Tumor necrosis factor; WDR5: WD repeat domain 5.
Minimized DNA vectors have had a broad range of applicability throughout the cancer field and their use has also helped make headway in other disease areas, such as retinal disorders [28,29], rheumatoid arthritis [30], Parkinson’s disease [31], and inborn errors of metabolism [32]. They have even been used for the endogenous production of biologics [33]. Their impact has also been felt in the areas of, among others, anti-viral treatments [34], vaccination [35], and regenerative medicine [36,37].
USING MINIMIZED DNA VECTORS FOR STEM CELL THERAPY & STEM CELL REPROGRAMMING
Regenerative medicine uses autologous (or allogeneic) stem cells for the repair or replacement of damaged or diseased tissue. A major limitation to this approach has been associated with the use of integrating viruses, such as retroviruses or lentiviruses, to deliver the appropriate enabling therapeutic genes to stem cells. The potential for insertional mutagenesis is high, which could lead to disastrous downstream consequences. Minimized DNA vectors have been tested as a replacement for viral vectors to mitigate these safety issues. Several varieties of stem cells have been successfully manipulated using minicircles, including neural stem cells [38,39], mesenchymal stem cells [40,41], skeletal myogenic progenitors [42], and hematopoietic stem cells [43]. Most frequently this work has been done in mouse and human cells, but canine and equine cells have also been used [40].
Minicircles have further been used to enable stem cell reprogramming, which refers to the process of reverting mature, differentiated cells into pluripotent stem cells capable of expanding indefinitely and differentiating into all other cell types in the body (called induced pluripotent stem cells or iPSCs). iPSCs are classically produced using somatic cells transduced with integrating viruses that carry genes for the cellular reprogramming factors needed to induce reversal of the developmental state [44]. As with the other types of stem cells described above, however, the use of integrating viruses renders iPSCs produced in this manner inappropriate for clinical translation. Indeed, chimeric mice generated from iPSCs produced with virus and then injected into blastocysts formed tumors [45].
The persistent safety issues surrounding integrating viruses have spurred research into alternative approaches (reviewed in [46]). In addition to minimized DNA vectors, other non-viral [46,47] methods for stem cell reprogramming include plasmids, mRNA [48,49], microRNA [50,51], and transposon systems, such as Sleeping Beauty (the components of which can be encoded on either plasmids or minimized vectors) for the safer genomic integration of DNA sequences. The use of non-integrating viruses such as Sendai virus [52], adenovirus [53], AAV [54], and measles virus [55], has also been explored. Fortunately, insertional mutagenesis is not required for the production of iPSCs [53], and thus it is feasible to use non-integrating vectors. Even with most non-integrating viruses, however, there is still a small chance for genomic integration and even gene expression from integrated vector DNA beyond the point at which reprogramming has taken place [56]. Other difficulties with viruses include immunogenicity, limits on the size of the therapeutic insert, and variable tropism, which makes it so that some viral systems will not work well with some cell types. Other approaches to stem cell reprogramming forgo the use of genetic material entirely (thoroughly reviewed in [57]). These methods, however, are still very technically challenging and often result in a low yield of usable cells.
Non-viral minimized DNA gene therapy approaches provide a valuable option for stem cell reprogramming as they are safer and less complex to use. For example, minicircles expressing bone morphogenetic protein 2 and transforming growth factor beta 3 were used in a strategy for cartilage treatment and regeneration. Mesenchymal stem cell-like, human iPSC-derived outgrowth cells transfected with minicircles successfully differentiated into cells of the chondrogenic lineage. Chondrogenic pellets derived from these cells also corrected defects in a rat osteochondral defect model [58]. In an interesting development for cancer treatment, minicircles were used to reprogram murine melanoma cells. The reprogramed cancer cells were less malignant than non-reprogrammed cancer cells, as evidenced by a smaller proportion of cells in S-phase and by the formation of smaller tumors in mice [59].
It is important to keep pushing stem cell/iPSC research forward because these cells are critically needed for drug screening, organ and tissue generation, and disease modeling. Enabling the study of the patient-specific basis of disease also further advances personalized medicine. While not without challenges [42], minimized DNA vectors should continue to advance this field.
THE DIFFICULTY OF TRANSLATING NON-VIRAL MINIMIZED DNA VECTORS TO THE CLINIC
Despite the encouraging successes described above, significant hurdles have slowed the advancement of minimized DNA vectors into the clinic. One hurdle has been the achievement of high quality yet cost-effective scale-up of the vectors. Fortunately, gains are being made in improving vector yields and in minimizing contaminants [60–62], which will ultimately lower the cost of production (briefly reviewed in [63]).
Viral vectors are generally more efficient than non-viral vectors at delivering a genetic payload. Perhaps reflecting this difference, nearly two-thirds of gene therapy clinical trials are based on viral rather than non-viral methods [24]. Transient gene expression from non-viral vectors is another hurdle. For example plasmids, which thus far have been the most commonly employed non-viral vector in clinical trials [1], are prone to silencing [64–66] and have generally failed to afford long-enough lasting benefit in patients [67,68]. Minimized DNA vectors are much less susceptible to transgene silencing than plasmids and are capable of producing long-lasting gene expression [2–6]. Substituting plasmids with minimized DNA vectors should provide the benefit of stable and pro-longed gene expression.
Physical or chemical means are required to carry non-viral DNA vectors into cells [1,69–74]. Once inside the cell, vectors must also enter the nucleus to express the encoded therapeutic cargo. Nuclear trafficking of DNA, however, is a complex and not yet fully understood process [75]. In the cell cytoplasm, DNA associates with proteins to facilitate migration toward the microtubule organizing center and the nuclear envelope [76]. If the DNA vector delivered is large, organelles and translation machinery in the cytoplasm prevent free diffusion inside the cells [64,77] and across the nuclear membrane pore channel [78]. DNA ≥ 2,000 bp is unable to diffuse into the perinuclear space [79]. In addition, the inner diameter of the nuclear pore complex is ~40–42.5 nm [75,80–82]. Hence, small and compact DNA particles are more likely than plasmids to successfully traverse the cell, avoid degradation, and diffuse through the nuclear pore. Minivectors, for example, are typically shorter than 2,000 bp in length and can be as small as ~40 nm in diameter [83], facilitating passage through cell and nuclear membranes.
DESIGN OF NANOPARTICLES FOR IMPROVED CELLULAR UPTAKE
The field of nanotechnology takes advantage of the benefits provided by nanometer-sized particles [84], and the advances made in this field could potentially be used to inform the design of the next generation of minimized DNA vectors. Nanoparticle size is important not only for cellular internalization but also for retention [85,86], as persistence can have major implications for therapeutic delivery and gene expression. Cancer-targeting nanoparticles less than 100 nm in diameter freely diffuse through tumor pores and accumulate within tumors [86,87]. Based on thermodynamic modeling studies of ligand-coated nanoparticles, the optimal particle size for cellular uptake should be between 25–30 nm [88]. Maximum in vitro uptake of polystyrene and gold particles in cultured HeLa cells was achieved when particles were between 25–42 nm [89] and 50 nm [90], respectively. 50 nm was also the most effective size for uptake of silver nanoparticles by red blood cells [91]. In 3-D cultures, fluorescently labeled carboxylic acid-modified nanoparticle beads ≥ 100 nm were restricted from cellular uptake, whereas particles ≤ 40 nm were not [92]. In vivo, drug-silica nanoconjugate particles of 50 nm display maximum tissue retention and deep tumor penetration [93]. Gold nanoparticles of 15 and 50 nm are even able to effectively cross the blood–brain barrier [94] and accumulate faster in tumors than particles ≥ 60 nm. For larger tumor volumes, however, the larger nanoparticles accumulated better [95]. Whereas smaller particles are generally more effective, particles that are too small are not. Inert nanoparticles with diameter < 10 nm are quickly eliminated by the kidneys [96,97]. RNA nanoparticles of < 5 nm are also promptly cleared after injection in mice [98]. Even variations as small as 2 nm may change biodistribution [97].
Nanoparticle shape also influences cellular and nuclear uptake [99,100]. Filamentous particles are more effective at cellular uptake than spherical systems [86]. Nanoparticles with sharp edges escape endosomes faster, avoid exocytosis, and persist longer inside cells than those with rounded edges [101]. Similarly, gold nanotriangles are more readily taken up by cells than nanorods or nanostars [100]. Structural differences of polyethyleneimine/DNA nanoparticles dictate their cellular uptake mechanism (macropinocytosis-mediated versus clathrin-mediated endocytosis), thus resulting in different transfection and gene expression levels [77]. In vivo, particle shape affects venous circulation, biodistribution, cellular uptake [102], and influences tumor penetration [103]. Uptake of nanoparticles by cells is dependent not only on size and shape, but also surface area, flexibility, and charge [97,104,105]. Altering these parameters targets nanoparticles to different tumors, tissues, and cell types [96].
Nanotechnology has the potential to address chronic diseases through controlled, site-specific delivery of precise medicine [84,106–109], as well as through the development of multimodality agents with both imaging and therapeutic capabilities [85,108,110,111]. Nanoparticles have great potential for the treatment of cancer and other diseases [85,96]. Obstacles still remain, however. The materials that make up some nanoparticles contain heavy metals, which may be toxic [91,109,110,112], and systemic delivery of nanoparticles is difficult [106].
DESIGN OF DNA MINIVECTOR NANOPARTICLES
Since the concept was first proposed in the 1980s, significant progress has been made in constructing nanostructures made of DNA [113,114]. DNA is remarkably stable [115–117] and the strict rules for pairing between bases allow for the rational design of increasingly complex DNA nanostructures [118,119]. Current DNA nanotechnology applications include construction of structural lattices, scaffolds, molecular machines, bio-sensors, and targeted drug delivery systems [113,120–122]. The properties of DNA make it suitable for the construction of a nearly limitless choice of nanostructures that can be further controlled and modified by a variety of DNA-acting enzymes [120,123,124].
Although assembling DNA into complex nanostructures holds promise for clinical applications (reviewed in [120]), the process of making these structures is far from straight-forward. First, a large number of DNA fragment components are typically required to build these structures, which increases the likelihood of incorrect assembly. Second, annealing products correctly requires very long folding times, limiting throughput. Third, products need to be purified, but protocols for purification have not been fully optimized. Fourth, the procedures required for annealing and purification are difficult to scale up, resulting in low yield. Finally, even though composed of DNA, none of these structures are themselves the ‘active’ therapeutic component, but rather serve as the carrier for, or foundation of, delivery or construction of other nanoparticles [125–128]. Indeed, many of the breakthroughs in nanotechnology for gene therapy are based upon the design of synthetic nanoparticles as delivery vehicles for nucleic acid payloads into the cell and none have yet focused on modification of the therapeutic-encoding DNA vector itself.
With diameters of around 40 nm [83], supercoiled minivectors can be made small enough such that their diameter is within a nanoparticle size range [10,83]. Furthermore, when complexed with delivery vehicle, for example, poly-L-lysine-polyethylene glycol, minivectors are highly homogenous, monodisperse, and adopt a needle-shaped conformation; comparatively, plasmids are not nanoparticle-sized and adopt highly heterogeneous shapes (Figure 1). These parameters are all important for cellular and nuclear entry.
FIGURE 1. Transmission electron micrographs of three DNA vector sizes.
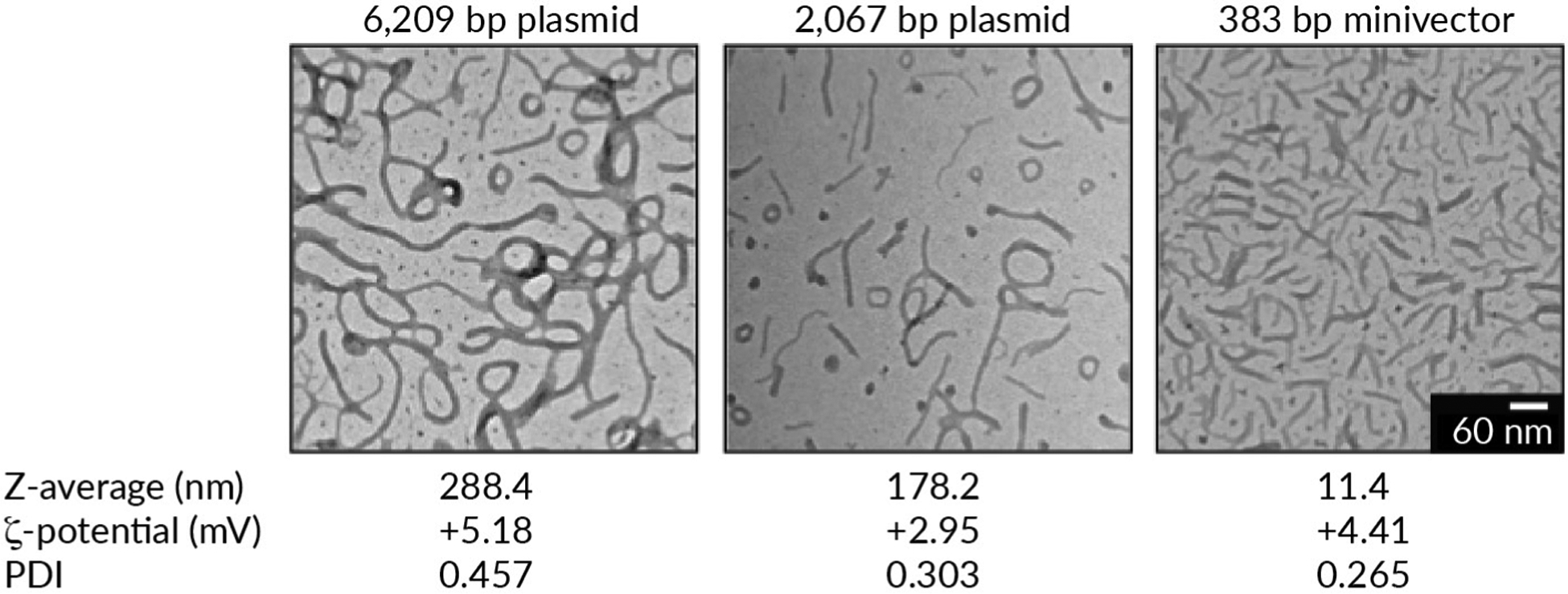
Poly-L-lysine-polyethylene glycol and DNA were complexed at a nitrogen:phosphate ratio of 2:1. Z-average (a measure of particle size), ζ-potentials (a measure of the degree of electrostatic repulsion between adjacent particles), and polydispersity index (PDI, a measure of the amount of variability in the particle size distribution) values were determined using dynamic light scattering using a Malvern Zetasizer Nano (data courtesy of Dr Jin Wang, Dr Fude Feng, and Dr. Daniel J Catanese, Jr.).
Could DNA minivectors be both the nanoparticle and the genetic payload? By using DNA supercoiling and adding ‘bend site’ sequences, it seems possible. Certain DNA sequences are much more flexible than others [129,130]. Additionally, because single-stranded DNA is more flexible than double-stranded DNA [131], disruptions to base pairing can generate hyperflexible hinges to facilitate bending [132–135]. The propensity for base pair disruption in supercoiled DNA is also sequence-dependent [136]. Based on these principles, certain sequences are more likely to bend, either because they are intrinsically more flexible, or because of disruptions to base pairing [Fogg et al., 2020, submitted]. The placement of bend sites could influence the 3-D structure of supercoiled DNA molecules. Therefore, by modifying the DNA sequence, we hypothesize that it should be possible to manipulate minivector DNA shape with supercoiling.
We demonstrated the feasibility of this approach by simulating, using established computational models [137–140], the effect of engineering three bend sites in a supercoiled 336 bp minivector and predicted that this should cause the DNA to adopt three-lobed shapes [141]. Because the high compaction of rod-shaped minivectors may offer improved cellular uptake, we reasoned that introducing a bend site diametrically (180°) opposite another bend site in the 336 bp minivector could result in a strong mechanical correlation between the two sequences. If correct, the two sites should then facilitate bending at the two apices of the rod to stabilize the rod-shaped conformation (Figure 2A).
FIGURE 2. Generating custom minimized DNA vector shapes.
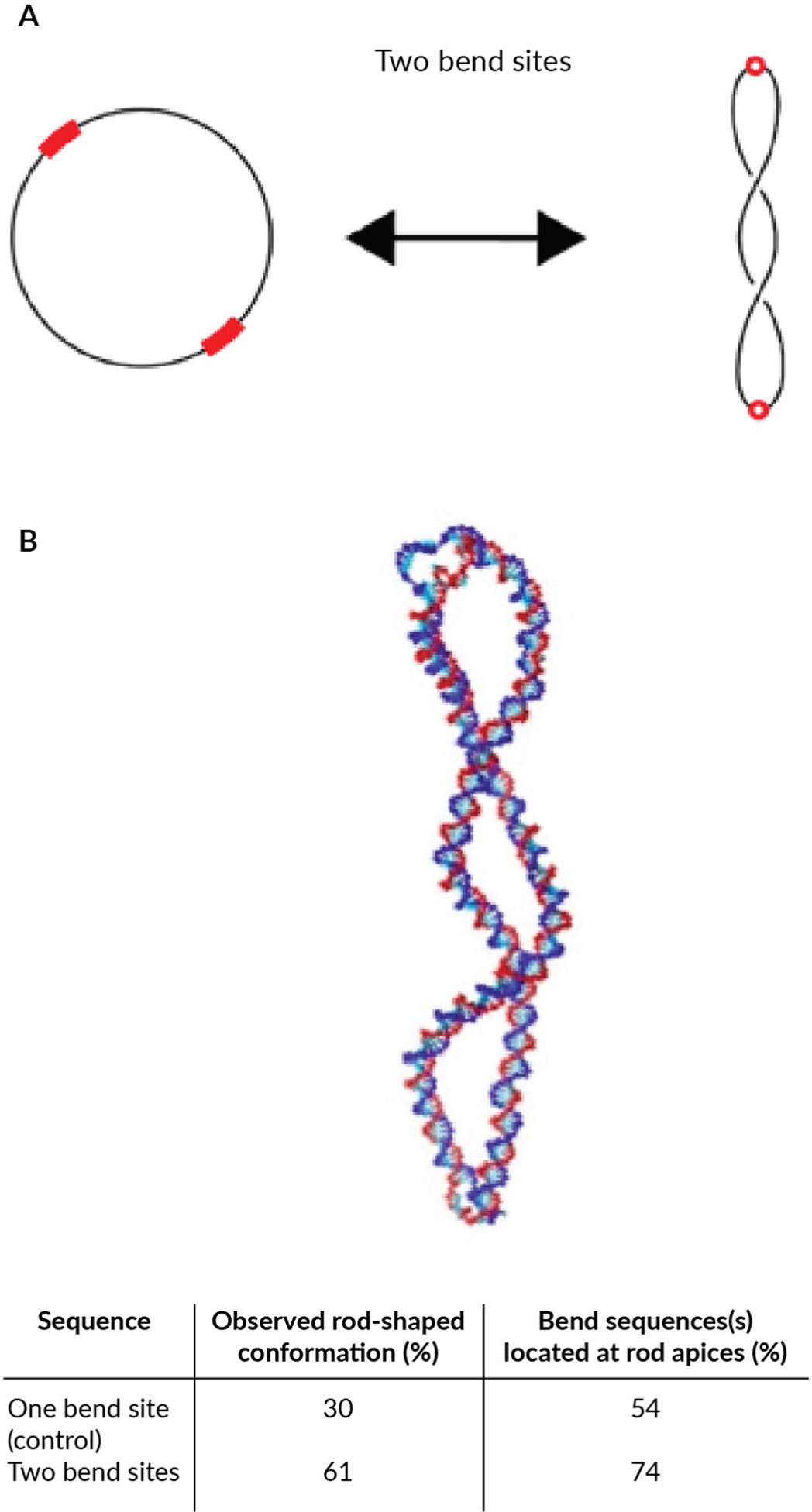
(A) Schematic representation of the predicted effect of adding bend sites. Bend sites (red) are flexible, which should localize them to superhelical apices with supercoiling. (B) Representative image from the coarse-grained simulations showing the rod-shaped conformation, (the conformation observed most frequently with two bend sites), and summary of how frequently rod-shaped conformations were observed during the simulations, and of these rod-shaped conformations, what percent had the bend site(s) localized to the apices.
Using coarse-grained molecular dynamics simulations with oxDNA [142], we found that the unmodified (no added bend site) negatively supercoiled minivector sequence formed a rod-shaped conformation 30% of the time simulated across 10 independent simulations (Figure 2B). This prediction is in good agreement with the fraction of rod-shaped conformations observed directly in this minivector [83]. When the modified minivector sequence containing one bend site opposite the other bend site was simulated, the fraction of rod-shaped conformations observed increased to 61%. We observed the predicted bend sites localizing to the apices of these rod-shaped conformations as well as base pair disruption accompanying the bend sites (Figure 2B). In the simulations, once the rod-shaped conformation formed with bend sites at the apices, it was typically stable for the remainder of the simulation. Minivectors with the unmodified sequence (with a single bend site) fluctuated among multiple different conformations. Simulations, therefore, predict that we can use circularity, DNA supercoiling, and sequence to enrich for certain nanoparticle shapes.
Simulations suggest that it may be possible to design at least two different novel DNA shapes (rod-shaped and three-lobed conformations). These two shapes have potential for targeted therapy. Lung tissue selectively accumulates star-shaped over spherical gold nanoparticles [143]. Rod-shaped are more amenable to cellular transfection in clinically relevant breast cancer cell lines compared to spherical polystyrene nanoparticles [144]; the authors of this study [144] speculated that the increased surface area of rods allows for more contact with the cell membrane. Nanoparticles with higher aspect ratios (i.e., much longer than they are wide, as in the rod-shape) also seem to be more effective at avoiding clearance through phagocytosis—an important pharmacokinetic characteristic [145–147]. Specific shapes of non-viral minimized DNA vectors could thus exhibit tissue specificity and improved cellular uptake, with implications for targeted therapies.
CONCLUSIONS
The recent pre-clinical results summarized here showcase the benefits of using minimized DNA vectors for therapeutic purposes. There is still plenty of room for improvement in vector design and in advancing non-viral minimized DNA vectors to the clinic. Difficulties remain in production scale-up and in getting DNA vectors into cells efficiently. One avenue for improvement takes advantage of two important features of nanotechnology: particle size and shape. The smallest minimized DNA vectors (minivectors) fall within the range of ideal sizes for cellular uptake. Strategically placed bend sites in supercoiled minivectors may enable specific nanoparticle conformations that could one day prove beneficial for gene therapy and targeted nanomedicine.
Acknowledgements:
We thank Dr. Jin Wang (Baylor College of Medicine, Houston, TX, USA), Dr. Fude Feng (previously Baylor College of Medicine; currently Nanjing University, Nanjing, China), and Dr. Daniel J. Catanese, Jr. (previously Baylor College of Medicine; currently Rice University, Houston, TX, USA) for generating the transmission electron micrographs and dynamic light scattering data shown in Figure 1.
Funding declaration:
The authors acknowledge funding (to Dr Lynn Zechiedrich) from National Institutes of Health Grant R01 GM115501 and from Baylor College of Medicine.
Footnotes
Disclosure and potential conflicts of interest: Drs Lirio M Arévalo-Soliz, Jonathan M Fogg, and Lynn Zechiedrich are co-inventors on issued and pending patents covering the minivector technology in this paper. Cinnamon L Hardee, Nathan R Corman and Cameron Noorbakhsh declare no conflict of interest.
Contributor Information
Lirio M Arévalo-Soliz, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA; Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA; Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Cinnamon L Hardee, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA; Interdepartmental Program in Integrative Molecular and Biomedical Sciences, Baylor College of Medicine, Houston, TX 77030, USA.
Jonathan M Fogg, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA; Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA; Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Nathan R Corman, Rural Medical Education Program, University of Illinois College of Medicine, Rockford, IL 61107, USA.
Cameron Noorbakhsh, Weiss School of Natural Sciences, Rice University, Houston, TX 77005, USA.
Lynn Zechiedrich, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX 77030, USA; Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA; Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, TX 77030, USA; Interdepartmental Program in Integrative Molecular and Biomedical Sciences, Baylor College of Medicine, Houston, TX 77030, USA.
REFERENCES
- 1.Hardee C, Arévalo-Soliz L, Hornstein B, Zechiedrich L. Advances in non-viral DNA vectors for gene therapy. Genes 2017; 8(2): 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levacic AK, Morys S, Kempter S, Lächelt U, Wagner E. Minicircle versus plasmid DNA delivery by receptor-targeted polyplexes. Hum. Gene Ther 2017; 28(10): 862–74. [DOI] [PubMed] [Google Scholar]
- 3.Batchu RB, Gruzdyn OV, Tavva PS et al. Engraftment of mesothelin chimeric antigen receptor using a hybrid Sleeping Beauty/minicircle vector into NK-92MI cells for treatment of pancreatic cancer. Surgery 2019; 166(4): 503–8. [DOI] [PubMed] [Google Scholar]
- 4.Kanada M, Kim BD, Hardy JW et al. Microvesicle-mediated delivery of minicircle DNA results in effective gene-directed enzyme prodrug cancer therapy. Mol. Cancer Ther 2019; 18(12): 2331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, Liu F, Chen Y et al. Targeted delivery of CRISPR/Cas9-mediated cancer gene therapy via liposome-templated hydrogel nanoparticles. Adv. Funct, Mater 2017; 27(46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C, Tang N, Li J et al. Bacteria-free minicircle DNA system to generate integration-free CAR-T cells. J. Med. Genet 2019; 56(1): 10–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspar V, de Melo-Diogo D, Costa E et al. Minicircle DNA vectors for gene therapy: advances and applications. Expert Opin. Biol. Ther 2015; 15(3): 353–79. [DOI] [PubMed] [Google Scholar]
- 8.Minicircle Schleef M. and Miniplasmid DNA Vectors: The future of nonviral and viral gene transfer. Weinheim, Germany: John Wiley & Sons; 2013. [Google Scholar]
- 9.Zhao N, Fogg JM, Zechiedrich L, Zu Y. Transfection of shRNA-encoding Minivector DNA of a few hundred base pairs to regulate gene expression in lymphoma cells. Gene Ther. 2011; 18(3): 220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catanese DJ, Fogg JM, Schrock DE, Gilbert BE, Zechiedrich L. Supercoiled minivector DNA resists shear forces associated with gene therapy delivery. Gene Ther. 2012; 19(1): 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogg JM, Kolmakova N, Rees I et al. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens. Matter 2006; 18(14): S145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kay MA, He C-Y, Chen Z-Y. A simple and rapid minicircle DNA vector manufacturing system. Nat. Biotechnol 2010; 28(12): 1287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida AM, Eusébio D, Queiroz JA, Sousa F, Sousa A. The use of size-exclusion chromatography in the isolation of supercoiled minicircle DNA from Escherichia coli lysate. J. Chromatogr. A 2020; 1609: 460444. [DOI] [PubMed] [Google Scholar]
- 14.Audentes Therapeutics News. Audentes Therapeutics provides update on the ASPIRO clinical trial evaluating AT132 in patients with X-linked myotubular myopathy: https://www.audentestx.com/press_release/audentes-therapeutics-provides-update-on-the-aspiro-clinical-trial-evaluating-at132-in-patients-with-x-linked-myotubular-myopathy
- 15.Paulk N Gene Therapy: It’s Time to talk about high-dose AAV. GEN - Genetic Engineering and Biotechnology News. 2020: https://www.genengnews.com/commentary/gene-therapy-its-time-to-talk-about-high-dose-aav [Google Scholar]
- 16.High-dose AAV gene therapy deaths. Nat. Biotechnol 2020; 38(8): 910–910. [DOI] [PubMed] [Google Scholar]
- 17.Hong M, Clubb JD, Chen YY. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell. 2020; 14: 473–88. [DOI] [PubMed] [Google Scholar]
- 18.Kuwana Y, Asakura Y, Utsunomiya N et al. Expression of chimeric receptor composed of immunoglobulin-derived V resions and T-cell receptor-derived C regions. Biochem. Biophys. Res. Commun 1987; 149(3): 960–8. [DOI] [PubMed] [Google Scholar]
- 19.Cortés-Hernández A, Alvarez-Salazar EK, Soldevila G (2021) Chimeric antigen receptor (CAR) T cell therapy for cancer. Challenges and opportunities: An overview. In: Robles-Flores M. (eds) Cancer Cell Signaling. Methods in Molecular Biology 2021; 2174. Humana, NY, USA: 10.1007/978-1-0716-0759-6_14. [DOI] [PubMed] [Google Scholar]
- 20.Hudecek M, Gogishvili T, Monjezi R et al. Minicircle-based engineering of chimeric antigen receptor (CAR) T cells. Recent Results Cancer Res. 2016; 209: 37–50. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Ye X, Ju Y et al. Minicircle DNA-mediated CAR T cells targeting CD44 suppressed hepatocellular carcinoma both in vitro and in vivo. Onco Targets Ther. 2020; 13: 3703–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J, Gao F, Geng S et al. Minicircle DNA-engineered CAR T cells suppressed tumor growth in mice. Mol. Cancer Ther 2020; 19(1): 178–86. [DOI] [PubMed] [Google Scholar]
- 23.Deng X, Gao F, Li N et al. Antitumor activity of NKG2D CAR-T cells against human colorectal cancer cells in vitro and in vivo. Am. J. Cancer Res 2019; 9(5): 945–58. [PMC free article] [PubMed] [Google Scholar]
- 24.Hudecek M, Izsvák Z, Johnen S, Renner M, Thumann G, Ivics Z. Going non-viral: the Sleeping Beauty transposon system breaks on through to the clinical side. Crit. Rev. Biochem. Mol. Biol 2017; 52(4): 355–80. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Liu L, Cao S, Zhu Y, Mei Q. Gene delivery of TIPE2 inhibits breast cancer development and metastasis via CD8 + T and NK cell-mediated antitumor responses. Mol. Immunol 2017; 85: 230–7. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Chen G, Pang X et al. Calcium phosphate nanoneedle based gene delivery system for cancer genetic immunotherapy. Biomaterials 2020; 250: 120072. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Tan X, Lin J et al. Minicircle-oriP-miR-31 as a novel EBNA1-specific miRNA therapy approach for nasopharyngeal carcinoma. Hum. Gene Ther 2017; 28(5): 415–27. [DOI] [PubMed] [Google Scholar]
- 28.Gallego I, Villate-Beitia I, Martínez-Navarrete G et al. Non-viral vectors based on cationic niosomes and minicircle DNA technology enhance gene delivery efficiency for biomedical applications in retinal disorders. Nanomedicine 2019; 17: 308–18. [DOI] [PubMed] [Google Scholar]
- 29.Barnea-Cramer AO, Singh M, Fischer D et al. Repair of retinal degeneration following ex vivo minicircle DNA gene therapy and transplantation of corrected photoreceptor progenitors. Mol. Ther 2020; 28(3): 830–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi S, Lee K, Jung H et al. Kruppel-like factor 4 positively regulates autoimmune arthritis in mouse models and rheumatoid arthritis in patients via modulating cell survival and inflammation factors of fibroblast-like synoviocyte. Front. Immunol 2018; 9: 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izco M, Blesa J, Schleef M et al. Systemic exosomal delivery of shRNA minicircles prevents Parkinsonian pathology. Mol. Ther 2019; 27(12): 2111–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H-O, Gallego-Villar L, Grisch-Chan HM, Häberle J, Thöny B, Kruger WD. Treatment of cystathionine β-synthase deficiency in mice using a minicircle-based naked DNA vector. Hum. Gene Ther 2019; 30(9): 1093–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park N, Rim YA, Jung H et al. Etanercept-synthesising mesenchymal stem cells efficiently ameliorate collagen-induced arthritis. Sci. Rep 2017; 7: 39593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo X, Chen D, Cai Q et al. Minicircle DNA vector expressing interferon-lambda-3 inhibits hepatitis B virus replication and expression in hepatocyte-derived cell line. BMC Mol. Cell Biol 2020; 21: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Gao X, Xu K et al. A Novel Cre recombinase-mediated in vivo minicircle DNA (CRIM) vaccine provides partial protection against Newcastle disease virus. Appl Environ Microbiol. 2019; 85(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brett E, Zielins ER, Luan A et al. Magnetic nanoparticle-based upregulation of B-cell lymphoma 2 enhances bone regeneration. Stem Cells Transl Med. 2017; 6(1): 151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong S-Y, Kang M-L, Park J-W, Im G-I. Dual functional nanoparticles containing SOX duo and ANGPT4 shRNA for osteoarthritis treatment. J. Biomed. Mater. Res. B Appl. Biomater 2020; 108(1): 234–42. [DOI] [PubMed] [Google Scholar]
- 38.Mooney R, Majid AA, Mota D et al. Bcl-2 overexpression improves survival and efficacy of neural stem cell-mediated enzyme prodrug therapy. Stem Cells Int. 2018; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finch L, Harris S, Solomou G et al. Safe nanoengineering and incorporation of transplant populations in a neurosurgical grade biomaterial, DuraGen PlusTM, for protected cell therapy applications. J. Control Release 2020; 321: 553–63. [DOI] [PubMed] [Google Scholar]
- 40.Tidd N, Michelsen J, Hilbert B, Quinn JC. Minicircle mediated gene delivery to canine and equine mesenchymal stem cells. Int. J. Mol. Sci 2017; 18(4): 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serra J, Alves CPA, Brito L et al. Engineering of human mesenchymal stem/stromal cells with vascular endothelial growth factor–encoding minicircles for angiogenic ex vivo gene therapy. Hum. Gene Ther 2018; 30(3) :316–29. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Oliveira VKP, Yamamoto A, Perlingeiro RCR. Generation of skeletal myogenic progenitors from human pluripotent stem cells using non-viral delivery of minicircle DNA. Stem Cell Res. 2017; 23: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holstein M, Mesa-Nuñez C, Miskey C et al. Efficient non-viral gene delivery into human hematopoietic stem cells by minicircle sleeping beauty transposon vectors. Mol. Ther 2018; 26(4): 1137–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126(4): 663–76. [DOI] [PubMed] [Google Scholar]
- 45.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 2007; 448(7151): 313–7. [DOI] [PubMed] [Google Scholar]
- 46.Haridhasapavalan KK, Borgohain MP, Dey C et al. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 2019; 686: 146–59. [DOI] [PubMed] [Google Scholar]
- 47.Hamann A, Nguyen A, Pannier AK. Nucleic acid delivery to mesenchymal stem cells: a review of nonviral methods and applications. J. Biol. Eng 2019; 13(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badieyan ZS, Evans T. Concise Review: Application of chemically modified mRNA in cell fate conversion and tissue engineering. Stem Cells Transl. Med 2019; 8(8): 833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L, Lin C. mRNA-based genetic reprogramming. Mol. Ther 2019; 27(4): 729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MA, He L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays 2012;34(8):670–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Zhuang L, Lin C-P. Roles of MicroRNAs in establishing and modulating stem cell potential. Int. J. Mol. Sci 2019; 20(15): 3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn Acad. Ser. B Phys. Biol. Sci 2009; 85(8): 348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science 2008; 322(5903): 945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weltner J, Anisimov A, Alitalo K, Oton-koski T, Trokovic R. Induced pluripotent stem cell clones reprogrammed via recombinant adeno-associated virus-mediated transduction contain integrated vector sequences. J. Virol 2012; 86(8): 4463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Driscoll CB, Tonne JM, El Khatib M, Cattaneo R, Ikeda Y, Devaux P. Nuclear reprogramming with a non-integrating human RNA virus. Stem Cell Res. Ther 2015; 6(1): 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harui A, Suzuki S, Kochanek S, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol 1999; 73(7): 6141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Higuchi A, Ling Q-D, Kumar SS et al. Generation of pluripotent stem cells without the use of genetic material. Lab. Invest 2015; 95(1): 26–42. [DOI] [PubMed] [Google Scholar]
- 58.Rim YA, Nam Y, Park N et al. Chondrogenic differentiation from induced pluripotent stem cells using non-viral minicircle vectors. Cells 2020; 9(3): 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Câmara DAD, Porcacchia AS, Costa AS, Azevedo RA, Kerkis I. Murine melanoma cells incomplete reprogramming using non-viral vector. Cell Prolif. 2017; 50(4): e12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ata-abadi NS, Rezaei N, Dormiani K, Nasr-Esfahani MH. Production of minicircle DNA vectors using site-specific recombinases. In: Eroshenko N (Ed.). Site-Specific Recombinases. New York, NY: Springer New York. 2017; 325–39. [DOI] [PubMed] [Google Scholar]
- 61.Alves CPA, Šimčíková M, Brito L, Monteiro GA, Prazeres DMF. Production and purification of supercoiled minicircles by a combination of in vitro endonuclease nicking and hydrophobic interaction chromatography. Hum. Gene Ther. Methods 2018; 29(4): 157–68. [DOI] [PubMed] [Google Scholar]
- 62.Almeida AM, Queiroz JA, Sousa F, Sousa A. Minicircle DNA purification: Performance of chromatographic monoliths bearing lysine and cadaverine ligands. J Chrom B. 2019; 1118–1119: 7–16. [DOI] [PubMed] [Google Scholar]
- 63.Almeida AM, Queiroz JA, Sousa F, Sousa Â. Minicircle DNA: The future for DNA-based vectors? Trends Biotechnol. 2020; 38(10): 1047–51. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen J, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle? Acc. Chem. Res 2012; 45(7): 1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen ZY, He CY, Meuse L, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004; 11(10): 856–64. [DOI] [PubMed] [Google Scholar]
- 66.Chen Z-Y, Riu E, He C-Y, Xu H, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008; 16(3): 548–56. [DOI] [PubMed] [Google Scholar]
- 67.Alton EWFW, Boyd AC, Porteous DJ et al. A Phase I/IIa safety and efficacy study of nebulized liposome-mediated gene therapy for cystic fibrosis supports a multidose trial. Am. J. Respir. Crit. Care Med 2015;192(11):1389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLachlan G, Baker A, Tennant P et al. Optimizing aerosol gene delivery and expression in the ovine lung. Mol. Ther 2007; 15(2): 348–54. [DOI] [PubMed] [Google Scholar]
- 69.Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr. Gene Ther 2002; 2(2): 183–94. [DOI] [PubMed] [Google Scholar]
- 70.Lam AP, Dean DA. Progress and prospects: nuclear import of nonviral vectors. Gene Ther. 2010; 17(4): 439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lechardeur D, Verkman A, Lukacs G. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Deliv Rev. 2005; 57(5): 755–67. [DOI] [PubMed] [Google Scholar]
- 72.Miller AM, Dean DA. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv. Drug Deliv. Rev 2009; 61(7–8): 603–13. [DOI] [PubMed] [Google Scholar]
- 73.Reis LG dos, Svolos M, Hartwig B, Windhab N, Young PM, Traini D. Inhaled gene delivery: a formulation and delivery approach. Expert Opin. Drug Deliv 2017; 14(3): 319–30. [DOI] [PubMed] [Google Scholar]
- 74.van Haasteren J, Li J, Scheideler OJ, Murthy N, Schaffer DV. The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat. Biotechnol 2020; 38(7): 845–55. [DOI] [PubMed] [Google Scholar]
- 75.Burns LT, Wente SR. Trafficking to uncharted territory of the nuclear envelope. Curr. Opin. Cell Biol 2012; 24(3): 341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai H, Lester GMS, Petishnok LC, Dean DA. Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep 2017; 37(6): BSR20160616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang W, Kang X, Yuan B et al. Nano-structural effects on gene transfection: large, botryoid-shaped nanoparticles enhance DNA delivery via macropinocytosis and effective dissociation. Theranostics 2019; 9(6): 1580–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu G, Li D, Pasumarthy MK, Kowalczyk TH et al. Nanoparticles of compacted DNA transfect postmitotic cells. J. Biol. Chem 2003; 278(35): 32578–86. [DOI] [PubMed] [Google Scholar]
- 79.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem 2000; 275(3): 1625–9. [DOI] [PubMed] [Google Scholar]
- 80.Knockenhauer KE, Schwartz TU. The nuclear pore complex as a flexible and dynamic gate. Cell 2016; 164(6): 1162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kabachinski G, Schwartz TU. The nuclear pore complex – structure and function at a glance. J. Cell Sci 2015; 128(3): 423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin DH, Hoelz A. The structure of the nuclear pore complex (an update). Annu. Rev. Biochem 2019; 88: 725–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Irobalieva RN, Fogg JM, Catanese DJ et al. Structural diversity of supercoiled DNA. Nat. Commun 2015; 6: 8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soares S, Sousa J, Pais A, Vitorino C. Nanomedicine: principles, properties, and regulatory issues. Front. Chem 2018; 6: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ficai D, Ficai A, Andronescu E. Advances in cancer treatment: Role of nanoparticles. In: Soloneski Sonia and Larramendy Marcelo L. (Eds). Nanomaterials – Toxicity and risk assessment. 2015. July 15. IntechOpen, DOI: 10.5772/60665. [DOI] [Google Scholar]
- 86.Wang J, Byrne JD, Napier ME, DeSimone JM. More effective nanomedicines through particle design. Small 2011; 7(14): 1919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.National Cancer Institute. Benefits of nanotechnology for cancer. 2017: https://www.cancer.gov/nano/cancer-nanotechnology/benefits
- 88.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv. Mater 2009; 21(4): 419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lai SK, Hida K, Man ST et al. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials 2007; 28(18): 2876–84. [DOI] [PubMed] [Google Scholar]
- 90.Chithrani BD, Ghazani AA, Chan WCW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006; 6(4): 662–8. [DOI] [PubMed] [Google Scholar]
- 91.Chen LQ, Fang L, Ling J, Ding CZ, Kang B, Huang CZ. Nanotoxicity of silver nanoparticles to red blood cells: Size dependent adsorption, uptake, and hemolytic activity. Chem Res Toxicol. 2015; 28(3): 501–9. [DOI] [PubMed] [Google Scholar]
- 92.Ng CP, Pun SH. A Perfusable 3D cell–matrix tissue culture chamber for in situ evaluation of nanoparticle vehicle penetration and transport. Biotechnol. Bioeng 2008; 99(6): 1490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang L, Yang X, Yin Q et al. Investigating the optimal size of anticancer nanomedicine. Proc. Natl Acad.Sci. USA 2014; 111(43): 15344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sonavane G, Tomoda K, Makino K. Biodistribution of colloidal gold nanoparticles after intravenous administration: effect of particle size. Colloids Surf B Biointerfaces 2008; 66(2): 274–80. [DOI] [PubMed] [Google Scholar]
- 95.Sykes EA, Dai Q, Sarsons CD et al. Tailoring nanoparticle designs to target cancer based on tumor pathophysiology. Proc. Natl Acad.Sci. USA 2016; 113(9): E1142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoshyar N, Gray S, Han H, Bao G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine (Lond). 2016; 11(6): 673–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crist R, McNeil S. Nanotechnology for treating cancer: Pitfalls and bridges on the path to nanomedicines. National Cancer Institute. 2015:: https://www.cancer.gov/research/key-initiatives/ras/ras-central/blog/2015/nanomedicines [Google Scholar]
- 98.Jasinski DL, Li H, Guo P. The effect of size and shape of RNA nanoparticles on biodistribution. Mol. Ther 2018; 26(3): 784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salatin S, Dizaj SM, Khosroushahi AY. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol Int. 2015; 39(8): 881–90. [DOI] [PubMed] [Google Scholar]
- 100.Xie X, Liao J, Shao X, Li Q, Lin Y. The effect of shape on cellular uptake of gold nanoparticles in the forms of stars, rods, and triangles. Sci. Rep 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chu Z, Zhang S, Zhang B et al. Unambiguous observation of shape effects on cellular fate of nanoparticles. Sci. Rep 2014; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu X, Vo C, Taylor M, Smith BR. Non-spherical micro- and nanoparticles in nanomedicine. Mater Horiz. 2019; 6(6): 1094–121. [Google Scholar]
- 103.Zhang Y-R, Lin R, Li H-J, He W, Du J-Z, Wang J. Strategies to improve tumor penetration of nanomedicines through nanoparticle design. WIREs Nanomed. Nanobiotechnol 2019; 11(1): e1519. [DOI] [PubMed] [Google Scholar]
- 104.Dasgupta S, Auth T, Gompper G. Shape and orientation matter for the cellular uptake of nonspherical particles. Nano Lett. 2014. February 12;14(2):687–93. [DOI] [PubMed] [Google Scholar]
- 105.Agarwal R, Singh V, Jurney P, Shi L, Sreenivasan SV, Roy K. Mammalian cells preferentially internalize hydrogel nanodiscs over nanorods and use shape-specific uptake mechanisms. Proc. Natl Acad.Sci. USA 2013; 110(43): 17247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Anis HA. Gene therapy in the era of nanotechnology/a review of current data. J. Cancer Prev. Curr. Res 2019; 10(1): 2. [Google Scholar]
- 107.Riley MK, Vermerris W. Recent advances in nanomaterials for gene delivery—A review. Nanomaterials (Basel). 2017; 7(5): 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roy I, Stachowiak MK, Bergey EJ. Non-viral gene transfection nanoparticles: Function and applications in brain. Nanomedicine 2008; 4(2): 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patra JK, Das G, Fraceto LF et al. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnology 2018; 16(1): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond). 2008; 3(5): 703–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vines JB, Yoon J-H, Ryu N-E, Lim D-J, Park H. Gold nanoparticles for photothermal cancer therapy. Front Chem. 2019; 7: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lombardo D, Kiselev MA, Caccamo MT. Smart nanoparticles for drug delivery application: Development of versatile nanocarrier platforms in biotechnology and nanomedicine. Vol. 2019, J. Nanomater 2019; e3702518. [Google Scholar]
- 113.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol 1982; 99(2): 237–47. [DOI] [PubMed] [Google Scholar]
- 114.Fan C, Li Q. Advances in DNA nanotechnology. Small 2019; 15(26): 1902586. [DOI] [PubMed] [Google Scholar]
- 115.Dabney J, Knapp M, Glocke I et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad.Sci. USA 2013; 110(39): 15758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meyer M, Fu Q, Aximu-Petri A et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 2014; 505(7483): 403–6. [DOI] [PubMed] [Google Scholar]
- 117.Orlando L, Ginolhac A, Zhang G et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 2013; 499(7456): 74–8. [DOI] [PubMed] [Google Scholar]
- 118.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009; 37(15): 5001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Benson E, Lolaico M, Tarasov Y, Gådin A, Högberg B. Evolutionary refinement of DNA nanostructures using coarse-grained molecular dynamics simulations. ACS Nano 2019; 13(11): 12591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seeman NC, Sleiman HF. DNA nanotechnology. Nature Rev. Mater 2017; 3(1): 1–23. [Google Scholar]
- 121.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature 2006; 440(7082): 297–302. [DOI] [PubMed] [Google Scholar]
- 122.Dong Y, Mao Y. DNA Origami as scaffolds for self-assembly of lipids and proteins. ChemBioChem. 2019; 20(19): 2422–31. [DOI] [PubMed] [Google Scholar]
- 123.Ong HS, Rahim MS, Firdaus-Raih M, Ramlan EI. DNA tetrominoes: the construction of DNA nanostructures using self-organised heterogeneous deoxyribonucleic acids shapes. PLoS ONE 2015; 10(8): e0134520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu Y, Niemeyer CM. From DNA nanotechnology to material systems engineering. Adv. Mater 2019; 31(26): 1806294. [DOI] [PubMed] [Google Scholar]
- 125.Chandrasekaran AR, Levchenko O. DNA nanocages. Chem. Mater 2016; 28(16): 5569–81. [Google Scholar]
- 126.Vindigni G, Raniolo S, Ottaviani A et al. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano 2016; 10(6): 5971–9. [DOI] [PubMed] [Google Scholar]
- 127.Liu J, Song L, Liu S et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 2018; 18(6): 3328–34. [DOI] [PubMed] [Google Scholar]
- 128.Xie N, Liu S, Fang H et al. Three-dimensional molecular transfer from DNA nanocages to inner gold nanoparticle surfaces. ACS Nano 2019; 13(4): 4174–82. [DOI] [PubMed] [Google Scholar]
- 129.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol Cell. 2004;14(3): 355–62. [DOI] [PubMed] [Google Scholar]
- 130.Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theory Comput 2006; 2(3): 685–95. [DOI] [PubMed] [Google Scholar]
- 131.Smith SB, Cui Y, Bustamante C. Over-stretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996; 271(5250): 795–9. [DOI] [PubMed] [Google Scholar]
- 132.Forties RA, Bundschuh R, Poirier MG. The flexibility of locally melted DNA. Nucleic Acids Res. 2009; 37(14): 4580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tomonaga T, Michelotti GA, Libutti D, Uy A, Sauer B, Levens D. Unrestraining genetic processes with a protein-DNA hinge. Mol. Cell 1998; 1(5): 759–64. [DOI] [PubMed] [Google Scholar]
- 134.Yan J, Marko JF. Localized single-stranded bubble mechanism for cyclization of short double helix DNA. Phys. Rev. Lett 2004; 93(10): 108108. [DOI] [PubMed] [Google Scholar]
- 135.Yan J, Kawamura R, Marko JF. Statistics of loop formation along double helix DNAs. Phys. Rev. E Stat. Nonlin. Soft Matter Phys 2005;71(6 Pt 1): 061905. [DOI] [PubMed] [Google Scholar]
- 136.Bi C, Benham CJ. WebSIDD: Server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinformatics 2004; 20(9): 1477–9. [DOI] [PubMed] [Google Scholar]
- 137.Pettersen EF, Goddard TD, Huang CC et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 2004; 25(13): 1605–12. [DOI] [PubMed] [Google Scholar]
- 138.Ouldridge TE, Louis AA, Doye JPK. Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J. Chem. Phys 2011; 134(8): 085101. [DOI] [PubMed] [Google Scholar]
- 139.Šulc P, Romano F, Ouldridge TE, Rovigatti L, Doye JPK, Louis AA. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys 2012; 137(13): 135101. [DOI] [PubMed] [Google Scholar]
- 140.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996; 14(1): 33–8. [DOI] [PubMed] [Google Scholar]
- 141.Wang Q, Irobalieva RN, Chiu W et al. Influence of DNA sequence on the structure of minicircles under torsional stress. Nucleic Acids Res. 2017; 45(13): 7633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Snodin BEK, Randisi F, Mosayebi M et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys 2015; 142(23): 234901. [DOI] [PubMed] [Google Scholar]
- 143.Talamini L, Violatto MB, Cai Q et al. Influence of size and shape on the anatomical distribution of endotoxin-free gold nanoparticles. ACS Nano. 2017; 11(6): 5519–29. [DOI] [PubMed] [Google Scholar]
- 144.Barua S, Yoo J-W, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl Acad.Sci. USA 2013; 110(9): 3270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sharma G, Valenta DT, Altman Y et al. Polymer particle shape independently influences binding and internalization by macrophages. J. Control Rel 2010; 147(3): 408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Champion JA, Katare YK, Mitragotri S. Particle shape: A new design parameter for micro- and nanoscale drug delivery carriers. J. Control Rel 2007; 121(1–2): 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Champion JA, Mitragotri S. Role of target geometry in phagocytosis. Proc. Natl Acad. Sci. USA 2006; 103(13): 4930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]


