Abstract
Small regulatory RNAs are essential and ubiquitous riboregulators that are the key mediators of RNA interference (RNAi). They include microRNAs (miRNAs) and short‐interfering RNAs (siRNAs), classes of ∼22 nucleotide RNAs. miRNAs and siRNAs bind to Argonaute proteins and form effector complexes that regulate gene expression; in animals, this regulation occurs primarily at the post‐transcriptional level. In this review, we will discuss our current understanding of how miRNA and siRNAs are generated and how they function to silence gene expression, focusing on animal and, in particular, mammalian miRNAs.
Keywords: microRNA, miRNA, Argonaute, Ago, Dicer, RNA/intereference, RNAi, miRNP, RISC, siRNA, mRNA transcription
An overview of how animal microRNAs (miRNAs) and short‐interfering RNAs (siRNAs) are generated and function that concisely captures the essence of the miRNA and RNAi field (24) is shown in Figure 1. Details are discussed in the remainder of this review.
Figure 1.

Biogenesis and function of microRNAs (miRNAs) and short‐interfering RNAs (siRNAs). As is true for almost all RNAs, miRNAs and siRNAs are derived from larger precursor RNAs. The precursor for miRNAs and siRNAs is double‐stranded (ds) RNA. In the case of miRNAs, the immediate precursor RNA is termed pre‐miRNA, adopts a hairpin structure and has a 5′‐phosphate and a 2‐nucleotide, 3′ overhang. In the case of siRNAs, the precursor is long dsRNA. Both are processed by the Dicer nuclease into duplexes termed miRNA or siRNA duplexes. The mature miRNA or siRNA is then released from the miRNA or siRNA duplex and binds to an Ago protein, to form a core effector complex that is commonly known as miRNP or RISC. miRNAs or siRNAs then act as specificity determinants to deposit the Ago proteins that they are bound to (RISCs, RNA‐induced silencing complex; miRNPs) onto their RNA targets, which are typically mRNAs. The deposited miRNP/RISC silences gene expression by suppressing the translation of the targeted mRNA and/or by destabilizing the targeted mRNA. Ago, Proteins that bind directly to miRNAs and siRNAs and are the core protein components of mitNPs/RISCs; Dicer, dsRNA endonuclease; miRNP, RiboNucleoProtein containing miRNA and an Ago protein.
miRNA GENES: ORGANIZATION, TRANSCRIPTION AND NUCLEAR PROCESSING
The official miRNA database (miRBase) currently lists ∼540 human miRNAs (31). Most of the miRNA genes are transcribed by RNA Polymerase II (13, 49), although a cluster of human miRNAs interspersed within Alu elements in Chromosome 19 have recently been shown to utilize RNA Polymerase III for their transcription (13). The primary miRNA transcripts are termed pri‐miRNAs (47). Many miRNA genes tend to be in close proximity to other miRNAs, and are transcribed as polycistronic transcripts (47). According to the genomic region that a miRNA resides in, miRNAs can be grouped into several categories: intronic miRNAs in protein‐coding genes; exonic miRNAs in non‐coding genes; intronic miRNAs in non‐coding genes (84). Many mammalian miRNAs lie within introns of protein‐coding genes and have the same transcription pattern as that of the protein‐coding genes in which they reside (84). When located in imprinted regions, some miRNAs (eg, mir‐127 and mir‐136) have also been found to exhibit parental specific expression pattern as that of imprinted genes. Very interestingly, the prototype of imprinted genes—H19 non‐coding RNA––has recently been shown to give rise to miR‐675 in both humans and mice (12).
A typical monocistronic pri‐miRNA is composed of a double‐stranded stem of ∼33 base pairs, a terminal loop and two flanking unstructured single‐stranded segments (Figure 2). After transcription, a pri‐miRNA is first cropped by the microprocessor into a ∼70 nucleotide (nt) hairpin‐like precursor miRNA (pre‐miRNA; Figure 2; (16, 29, 35, 45, 48. The core components of the microprocessor are the RNase III enzyme Drosha (48) and a double‐stranded (ds) RNA binding protein termed DGCR8/Pasha (16, 29, 35, 45. Drosha liberates the double‐stranded stem from the remainder of the pri‐miRNA by cleaving proximal and distal of the stem, and thus generates a pre‐miRNA that has a 5′ monophosphate and a 3′2‐nt hydroxyl overhang (Figure 2) (48). Very interestingly, DGCR8 has recently been shown to be a heme‐binding protein (22). Binding to heme dimerizes DGCR8, substantially enhances the affinity of DGCR8 to pri‐miRNAs and the cleavage of pri‐miRNA by microprocessor (22). Cleavage of a pri‐miRNA by microprocessor begins with DGCR8 recognizing the ssRNA‐dsRNA junction typical of a pri‐miRNA (36). Then, Drosha is brought close to its substrate through interaction with DGCR8 and cleaves the stem of a pri‐miRNA ∼11 nt away from the two single‐stranded segments (36). Although microprocessor is already sufficient for conversion of a pri‐miRNA into a pre‐miRNAs in vitro, cleavage of pri‐miRNA in vivo does not depend on Drosha and DGCR8 only, but also on other accessory proteins, such as the RNA binding protein hnRNP A1 (32) and the p68 and p72 RNA helicases (26). Gene‐targeting experiments have demonstrated the importance of p68 and p72 for the biogenesis of a subset of mouse miRNAs (26). In p68 or p72 knock‐out embryos (knock‐out of p68 in mice causes embryonic lethality while knock‐out of p72 causes neonatal death), the expression of a subset of miRNAs is severely compromised (26). In another recent study, hnRNP A1 has been shown to be specifically required for the processing of pri‐miR‐18a in a context‐dependent manner (32).
Figure 2.

Overview of microRNA (miRNA) processing.
In addition to the classical biogenesis pathway that depends on microprocessor, a subclass of pre‐miRNAs, pre‐miRNA/introns (mirtrons), have recently been shown to depend on the RNA splicing machinery for their biogenesis in Drosophila, Caenorhabditis elegans and mammals (6, 68, 85). Mirtrons are derived from certain debranched introns that fold into hairpin structures with 5′ monophosphates and 3′ 2‐nt hydroxyl overhangs, which mimic the structural hallmarks of pre‐miRNAs and enter the miRNA‐processing pathway (68, 85). The discovery of mirtrons suggests that any RNA, with a size comparable to a pre‐miRNA and all the structural features of a pre‐miRNA, can be utilized by the miRNA processing machinery, and potentially give rise to a functional miRNA.
After a pre‐miRNA is released from a pri‐miRNA in the nucleus, it is exported by RanGTP and Exportin‐5 (Exp‐5) to the cytoplasm (Figure 2) (9, 57, 100). After a pre‐miRNA is exported to the cytoplasm, RanGTP is hydrolyzed by RanGAP to RanGDP, and the pre‐miRNA is released from Exp‐5. Exp‐5 is also important for stabilizing pre‐miRNAs in the nucleus. When Exp‐5 is knocked down by siRNAs, the levels of pre‐miRNAs are reduced not only in the cytoplasm, but also in the nucleus, suggesting that binding of pre‐miRNAs to Exp‐5 protects them from degradation (100).
CYTOPLASMIC PROCESSING OF PRE‐miRNAs AND ASSEMBLY OF MIRNAS INTO EFFECTOR COMPLEXES
In the cytoplasm, another RNAse III endonuclease termed Dicer is responsible for dicing pre‐miRNAs into short RNA duplexes termed miRNA duplexes (2, 3) (7). The RNA strand of the miRNA duplex that is complementary to the mature miRNA is depicted with a star symbol (miRNA*; Figure 3). Dicer can also process long dsRNA into small RNA duplexes (7), and these small RNAs (33) are termed siRNA duplexes (19). In addition to two RNase III signature domains, mammalian Dicer has a N‐terminal ATPase/helicase domain, a DUF 283 domain, a PAZ domain and a C‐terminal dsRNA binding domain (dsRBD) (76, 102, 103). Biochemical experiments have revealed that both PAZ domain and dsRBD are essential for the interaction of Dicer with pre‐miRNAs and long dsRNAs (103). PAZ domain functions in recognizing the 2‐nt 3′ overhang signature generated by Drosha, while dsRBD is critical for binding long dsRNAs (103). After capturing pre‐miRNAs or long dsRNAs with its PAZ domain and dsRBD, Dicer dimerizes its two RNase III domains intra‐molecularly to form a single processing center, and cuts the stem of pre‐miRNAs or long dsRNAs ∼22 nt away from their termini at positions separated by 2 nts, which generates 3′ 2‐nt termini (103) (2, 3). In the case of long dsRNA, subsequent to the first cleavage, Dicer can use its PAZ domain to place it again at the termini of the dsRNA and dice processively from the termini (76, 102). Although the functions of the C‐terminal domains of Dicer are clear now, the roles of the N‐terminal ATPase/helicase domain and DUF 283 domain of Dicer still remain elusive.
Figure 3.
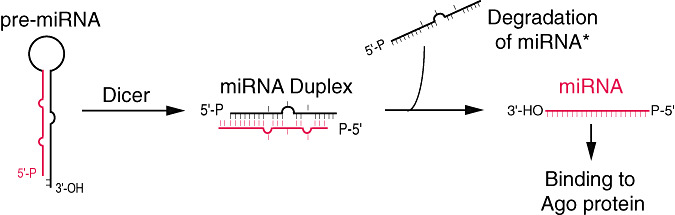
Precursor miRNA (Pre‐miRNA) processing: an RNA view.
After Dicer processing, the miRNA duplex is unwound and the mature miRNA binds to an Argonaute (Ago) protein in a process that is referred to as miRNA loading or assembly, while the miRNA* is degraded (2, 3).
The Argonaute family is a diverse family of proteins, each containing characteristic domains termed PAZ and PIWI. The Argonaute family can be phylogenetically divided into the Ago and Piwi protein families based on similarities to Arabidopsis AGO1 and Drosophila Piwi proteins, respectively (14). miRNAs bind Ago proteins whereas Piwi proteins bind a newly discovered class of small RNAs known as piwi‐interacting RNAs (piRNAs), which are almost exclusively expressed in the germline [reviewed in (3, 40)].
The miRNA/Ago ribonucleoprotein that is formed represents the core component of the effector complexes that mediate miRNA function and is known as miRNP (65). siRNA duplex unwinding also leads to loading of one strand to an Ago protein, and the resultant complex is referred to as RNA‐induced silencing complex (RISC) (34). The siRNA strand that is bound to Ago proteins is termed the guide strand, whereas the opposite strand is referred to as the passenger strand (Figure 4) [reviewed in Tomari and Zamore (91)]. miRNPs and RISCs are functionally equivalent.
Figure 4.

RNA‐induced silencing complex (RISC) assembly in Drosophila melanogaster.
A primary determinant of which the two strands of an miRNA duplex or an siRNA duplex will be loaded on Ago proteins is the inherent thermodynamic asymmetry of the miRNA or siRNA duplex. The RNA strand whose 5′ end is less stably bound to the opposite strand will be loaded to Ago proteins and forms the mature miRNA or siRNA (Figure 4) (91, 92).
The factors and mechanisms that sense the thermodynamic asymmetry of siRNA duplexes have been elucidated in Drosophila melanogaster. Flies contain two Dicer proteins (Dcr‐1 and Dcr‐2) with distinct functions. Dcr‐1 is responsible for miRNA production and is an essential protein that is required for fly development (50), whereas Dcr‐2 is required for siRNA production from long dsRNA, but flies lacking Dcr‐2 develop normally (50). After siRNA duplexes are generated by Dcr‐2, they interact with the RISC loading complex (RLC) that is comprised of Dcr‐2 and its dsRNA binding partner R2D2 (Figure 4) (54, 91, 92). The Dcr‐2/R2D2 heterodimer examines the base‐pairing extent of siRNA duplexes and of miRNA/miRNA* duplexes (the latter generated by Dcr‐1), and sorts them into different D. melanogaster (dm) Ago proteins. miRNA/miRNA* duplexes that typically have mismatches and thus have limited double‐stranded areas, have low affinity to R2D2, and are preferentially loaded into dmAgo1‐RISC (25, 93). In contrast, with extensive complementarity between its two strands, siRNAs have high affinity to R2D2 and are loaded into dmAgo2 (Figure 4) (25, 93). Once a siRNA is bound by the Dcr‐2/R2D2 heterodimer, R2D2 senses the thermodynamic differences in the base‐pairing stabilities of the 5′ ends of siRNA duplexes, and binds to the end with the greatest double‐stranded character (Figure 4) (92). During loading of miRNA/miRNA* or siRNA duplexes into Ago proteins, the miRNA or the guide strand of a siRNA is dissociated from the miRNA* or the passenger strand differentially. The passenger strand of a siRNA is cleaved by dmAgo2, and is released from the guide strand (63, 78). On the contrary, many miRNA/miRNA* duplexes have mismatches in the central region (positions 9–11) of the duplexes, and thus dmAgo2 cannot release the miRNA* strand by cleavage. Instead, many miRNA/miRNA* duplexes have mismatches in the seed region (positions 2–8 of the mature miRNA), and the binding affinity in the seed region of a miRNA/miRNA* duplex is much weaker than that of an siRNA duplex. Accordingly, a miRNA* strand is separated from a miRNA strand by a bypass mechanism, although the detail of the bypass mechanism is still unclear at this point.
Humans and other mammals contain a single Dicer gene and miRNP, and RISC assembly has many similarities but also important differences to RISC assembly in flies. In humans, miRNP assembly is accomplished by a protein complex termed the miRNA RISC Loading Complex (miRLC) (Figure 5) (59). The miRLC is a multiprotein complex whose core components are Ago and Dicer proteins (30, 59). The miRLC is devoid of miRNAs and processes miRNAs from pre‐miRNAs, and loads mature miRNAs to Ago proteins (59). The miRLC is then disassembled and the core miRNP (miRNA‐Ago ribonucleoprotein) is generated (Figure 5) (59). However, the details of miRNP assembly in humans are unknown.
Figure 5.

miRNP assembly in humans.
AGO PROTEINS, THE MAJOR MEDIATORS OF miRNA AND siRNA FUNCTION
The major mediators of miRNA and siRNA function are Ago proteins, which are ∼95 kDa proteins that contain N‐terminal and Mid‐domains in addition to the PAZ and PIWI domains. Ago proteins are highly conserved from metazoans to fission yeast. Most organisms possess multiple paralogs, the exception being the fission yeast, Schizosaccharomyces pombe, which has a single representative (14). Humans and mice contain four Ago proteins (Ago1–4).
Structural and biochemical analyses have shown that the ∼130‐amino‐acid PAZ domain contains an oligonucleotide‐binding fold that allows the protein to bind the single‐stranded 2‐nt 3′ terminal overhangs characteristic of small RNAs processed by Dicer (52). Furthermore, crystallographic studies of archaeal Ago family members have demonstrated that the PIWI domain of these proteins adopts a three‐dimensional fold similar to that of the RNase H family of endonucleases (70, 89, 101). RNase H proteins are characterized by a structural motif consisting of three β sheets surrounded by α helices (98), and they cleave RNA in RNA–DNA duplexes. Interestingly, only certain Ago proteins, including the Ago2 protein in humans, possess the triad of residues required for catalytic activity. The archaeal crystal structures, together with mutational analysis, have revealed that two aspartate residues (D557 and D669) and one histidine residue (H807) are required for the catalytic function of the human Ago2 protein (53, 83). Interestingly, only mammalian Ago2 is catalytically active, whereas the other Ago proteins are not, even though some of them contain the catalytic triad.
Co‐crystals of archaeal Ago proteins complexed with siRNAs have demonstrated that PIWI domains contain a conserved structural element that serves to bind and anchor the 5′ phosphate of small RNAs (58, 71). Additionally, recent analyses of the Ago Mid‐domain have revealed the presence of sequence motifs similar to the cap‐binding domain of the eukaryotic initiation factor eIF4E (42); the significance of this domain is discussed below.
FUNCTIONS OF miRNPs AND RISCs
miRNAs base‐pair with miRNA recognition elements (MREs) found in their mRNA targets (typically in the 3′ untranslated region—3′UTR) and deposit their bound Ago proteins onto mRNA targets. The result is translational repression of the targeted mRNA, often followed by mRNA destibilization or endonucleolytic cleavage of the targeted mRNA. The exact molecular function is dependent upon how extensive the complementarity of the miRNA or siRNA is with its mRNA target and which Ago protein is deposited on the mRNA target.
If an miRNA or siRNA bound to Ago2 pairs with extensive complementarity with a cognate mRNA target, then the mRNA is cleaved at a position across from nucleotides 10 and 11 of the miRNA (or siRNA), while the miRNA remains intact (Figure 6) (19, 38, 53). This cleavage event produces 5′‐phosphate and 3′‐hydroxyl terminal products, characteristic of other RNase H‐like enzymes (61, 87). The target mRNA is subsequently degraded via routine cellular pathways (Figure 6). Target mRNA cleavage by miRNAs is the major mechanism of regulation by plant miRNAs (18, 55). In animals, however, there are very few examples of miRNAs that regulate their mRNA targets by cleavage (99); rather, the predominant silencing mode of animal miRNAs is to repress the translation of their mRNA targets and/or to destabilize them without endonucleolytic cleavage (1, 23).
Figure 6.

Target RNA cleavage by Ago2‐containing miRNP.
Early studies in the worm C. elegans demonstrated that the small non‐coding RNAs lin‐4 and let‐7 (now known to be miRNAs) represented imperfect, complementary matches to sequences within the 3′UTRs of their respective target mRNAs (46, 80). These interaction between the miRNA and target mRNA furthermore resulted in decreased target protein levels (96) without affecting the stability of the mRNA (69). This profile of a significant reduction in protein level without a proportionate reduction in target mRNA levels became a hallmark of miRNA function.
We now know that the vast majority of animal miRNAs base‐pair with imperfect complementarity with their mRNA targets. Experimental and bioinformatics approaches have shown that the most important determinant of target RNA recognition by a miRNA is perfect or near‐perfect complementarity between the proximal (5′) region of the miRNA and the mRNA, also known as the “seed” region or the “nucleus” (Figure 7A) (10, 17, 41, 44, 51, 77). Base‐pairing between the 3′ portion of the miRNA and the mRNA target is not always essential for repression, but strong base‐pairing within this region can partially compensate for weaker seed matches or enhance repression (11, 41). Additionally, multiple MREs for the same, or different, miRNAs within the same 3′UTR can function cooperatively to enhance repression (17, 43). Spacing of the seed sites within the 3′UTR may play a significant role in the cooperative action of miRNAs (86). Finally, sequences adjacent to MREs (94) and the secondary structure of the 3′UTR of the target mRNA affect binding of miRNAs; miRNPs cannot efficiently “unwind” structured RNA areas, and thus miRNAs cannot bind to sites that are embedded in such structured areas (2, 56). Applying such miRNA binding rules to computational algorithms predicts that a large fraction (∼1/3) of the human mRNA transcriptome may be targeted by miRNAs [reviewed in Bartel and Chen (5)]. However, additional binding configurations between miRNAs and their targets may be functional, thus raising the fraction of mRNAs that may potentially be regulated by miRNAs (64, 82).
Figure 7.

A. Principles of miRNA binding to target RNA. Binding of the miRNA to the target mRNA always occurs in the presence of an Ago protein; the protein was omitted to highlight the nucleotides that base‐pair. Base‐pairing between nucleotides 2 and 8 of an miRNA (an area known as “seed” or “proximal” or nucleus”) and its cognate mRNA target (MRE, miRNA recognition elements) is essential for binding of most miRNAs to their targets. Base‐pairing of other nucleotides of a miRNA and its target also occurs and may be required when complementarity in the proximal area is not perfect, or to enhance binding or function. B. Mechanisms of repression of targeted mRNA by miRNPs.
Regulation of translation efficiency may occur during initiation, elongation or termination [for review see Gebauer and Hentze (27)]. In general, most translation repressors target translation initiation, the rate‐limiting step (81). The initiation of translation is an extensively regulated, multi‐step process that begins with the binding of eukaryotic initiation factor 4E (eIF4E) with the 5′ terminal 7‐methyl guanosine cap (m7G) that is present in all mRNAs (81). The initiation factor eIF4G interacts with eIF4E and helps to recruit the small ribosomal subunit via the 40S ribosome binding factor eIF3. Additional interaction of eIF4G with the poly(A)‐binding protein directs the formation of a circularized mRNA that enhances translation efficiency and allows regulatory sequences within the 3′UTR to affect translation initiation (81). The 40S ribosome then scans the mRNA and associates with the 60S ribosome at the AUG codon to begin protein synthesis (81). The formation of translation competent mRNP complexes can be monitored by sucrose‐gradient centrifugation. mRNAs actively undergoing translation associate with multiple ribosomes, forming dense particles known as polyribosomes or polysomes.
Initial studies suggested that miRNA‐mediated translational repression occurred at a step following the initiation of translation. This was based on the observation that, in C. elegans, lin‐14 mRNA, a target of the lin‐4 miRNA, could be found associated with mRNAs in polysomes, according to sucrose gradient sedimentation analysis (69, 88). miRNAs also associate with polysomes in both C. elegans and mammalian cells (39, 66, 69). More recent studies have lent additional support to the model of miRNA‐mediated repression occurring at a post‐initiation step (60, 67, 73) (Figure 7B). miRNA‐mediated translational repression results in decreased levels of the targeted protein. Thus, the model of miRNA function occurring at a post‐initiation step would require the release of and subsequent destruction of the resulting polypeptide. To date, no study has identified the accumulation of a nascent polypeptide corresponding to mRNAs repressed by miRNAs (67).
Recently, an alternative explanation for the sedimentation of some miRNAs as heavy particles in Drosophila has been proposed with the discovery of “pseudo‐polysomes”. These are heavy RNP particles that contain miRNAs and cognate mRNA targets, but they are inert and do not affect translation of the targeted mRNA (90). Whether repressed messages can be found in “pseudo‐polysomes” in mammals or nematodes has not yet been addressed. Finally, it should be noted that it is unknown whether all mRNAs bound by miRNPs are undergoing active repression at all times. Thus, it is possible that miRNPs will be found with a variety of mRNP complexes, both repressed and non‐repressed.
A second model proposes that miRNA‐mediated translational repression occurs at the initiation step (Figure 7B). Using human cells and reporter constructs targeted by either endogenous or exogenous miRNAs, two groups initially discovered that the m7G cap of an mRNA is required for efficient translational repression (37, 75). miRNA‐repressed messages sedimented in light fractions in polysome analyses, indicating that their translation was inhibited at the level of initiation (8, 75). Ago proteins are necessary for miRNA‐mediated translational repression. This was effectively shown in a study that enabled Ago proteins, expressed as fusion proteins to the phage λN‐peptide, to be artificially tethered to a reporter construct bearing several binding sites for the λN‐peptide in its 3′UTR (74). This system, which is presumably miRNA‐independent, results in repression of the reporter construct translation without significantly affecting the reporter mRNA level (74). Interestingly, Ago proteins have recently been shown to bear sequence similarities to the cap‐binding domain of eIF4E (42). Specifically, the two conserved tryptophan residues (W56 and W102) within eIF4E that are crucial for binding of the m7G cap are conserved within Ago proteins as phenylalanine residues (F470 and F505 in human Ago2) (42). Furthermore, using the Ago tethering system described above demonstrated that the ability for Ago2 protein to repress translation is dependent upon these residues (42). This study provides a molecular model to explain the cap‐dependent inhibition of translation initiation by Ago2 protein by proposing that binding of Ago2 to the m7G cap of an mRNA target precludes binding of eIF4E, thus inhibiting initiation of translation (42). Recent studies using in vitro translation systems to study the function of miRNAs provide strong evidence that miRNAs repress the initiation of translation in an m7G cap‐dependent manner (62, 90, 95). In particular, one study shows that, in vitro, translation initiation is the first step that is inhibited by a miRNA, and that there is a competition for access to the m7G cap of mRNAs by miRNPs, which repress initiation, and the eIF4E/G complex that promotes initiation. (62). Interestingly, miRNA‐dependent mRNA deadenylation is also recapitulated in vitro (95).
But other steps in the mechanism of miRNA‐mediated repression may still await discovery. Another recent study focusing on the translational inhibitory protein eIF6 has found, via biochemical and genetic studies, that this protein might also play an important role in miRNA‐mediated translational repression, perhaps at a step downstream of Ago function (15).
The fate of mRNAs targeted by miRNAs is becoming clearer, although no less complex. For example, although early studies in C. elegans proposed that the miRNAs let‐7 and lin‐4 repressed target mRNAs without affecting their stability, it has since been demonstrated that significant degradation of these mRNAs occurs (4). For many mRNAs, this accelerated degradation appears to be due secondary to miRNA‐mediated stimulation of target deadenylation (28, 97) (Figure 7B). However, degradation of targeted messages is not a universal finding; additionally, studies of the endogenous cationic amino acid transporter 1 (CAT‐1) mRNA, a target of the liver‐specific miR‐122, have shown that the repression of some miRNA targets can be reversed under certain conditions (8).
What causes some miRNA targets to be degraded and others preserved is currently unknown, although Ago proteins have been shown to interact with proteins involved in RNA degradation, including the GW182 protein and proteins that are involved in decapping mRNAs (79). Indeed, an emerging topic of study is the relationship between the microRNA pathway and cytoplasmic foci known as Processing bodies (P‐bodies) or GW‐bodies. P‐bodies (for reviews see Eulalio et al, Parker and Sheth (21, 72) are subcellular structures thought to assist in the translational repression and storage or degradation of mRNAs. They are characterized by possessing an abundance of enzymes involved in deadenylation, decapping and degradation of mRNAs. P‐bodies are notably devoid of both ribosomes and translation initiation factors except eIF4E and its transporter, eIF4E‐T. P‐bodies contain miRNPs and repressed mRNAsm, and they likely form as a consequence of mRNA repression, including miRNA‐dependent repression (20, 21).
The ultimate fate of a miRNA‐targeted mRNA likely depends on many factors in addition to Ago proteins, such as proteins that interact with miRNPs. It also depends on competition between the miRNA‐dependent silencing pathways and other cellular pathways. The best example to date comes from studies of the aforementioned miR‐122. Under normal conditions, the translation of CAT‐1 mRNA is inhibited, and the CAT‐1 mRNA is targeted to P‐bodies as a result of repression by miR‐122 (8). However, various stress conditions have been shown to abrogate this repression, resulting in loss of CAT‐1 mRNA from P‐bodies and the return of these messages to polysomes (8).
Finally, it should be noted that, to date, virtually nothing is known about the regulation of mammalian miRNAs or Ago proteins. Thus, ultimately, a model where miRNAs function through a variety of mechanisms, including the inhibition of both translation initiation and post‐initiation is plausible. An important challenge for the future is to understand how miRNPs are regulated and how they intersect with other cellular pathways that regulate mRNA translation, stability, localization and metabolism.
ACKNOWLEDGMENTS
We apologize to those whose work could not be cited because of space limitations. Supported by NIH Grants to KF (F30NS054396) and ZM (GM0720777, NS053839, P30‐HD026979), and by the Philadelphia Foundation to ZM.
REFERENCES
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. [DOI] [PubMed] [Google Scholar]
- 2. Ameres SL, Martinez J, Schroeder R (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130:101–112. [DOI] [PubMed] [Google Scholar]
- 3. Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi‐piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. [DOI] [PubMed] [Google Scholar]
- 4. Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R et al (2005) Regulation by let‐7 and lin‐4 miRNAs results in target mRNA degradation. Cell 122:553–563. [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5:396–400. [DOI] [PubMed] [Google Scholar]
- 6. Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC (2007) Mammalian mirtron genes. Mol Cell 28:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366. [DOI] [PubMed] [Google Scholar]
- 8. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA‐mediated translational repression in human cells subjected to stress. Cell 125:1111–1124. [DOI] [PubMed] [Google Scholar]
- 9. Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP‐dependent dsRNA‐binding protein that mediates nuclear export of pre‐miRNAs. RNA 10:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36. [DOI] [PubMed] [Google Scholar]
- 11. Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA‐target recognition. PLoS Biol 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cai X, Cullen BR (2007) The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13:313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10:1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16:2733–2742. [DOI] [PubMed] [Google Scholar]
- 15. Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA et al (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447:823–828. [DOI] [PubMed] [Google Scholar]
- 16. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432:231–235. [DOI] [PubMed] [Google Scholar]
- 17. Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7:512–520. [DOI] [PubMed] [Google Scholar]
- 19. Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21‐ and 22‐nucleotide RNAs. Genes Dev 15:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eulalio A, Behm‐Ansmant I, Schweizer D, Izaurralde E (2007) P‐body formation is a consequence, not the cause, of RNA‐mediated gene silencing. Mol Cell Biol 27:3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eulalio A, Behm‐Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post‐transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22. [DOI] [PubMed] [Google Scholar]
- 22. Faller M, Matsunaga M, Yin S, Loo JA, Guo F (2007) Heme is involved in microRNA processing. Nat Struct Mol Biol 14:23–29. [DOI] [PubMed] [Google Scholar]
- 23. Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS (2005) Post‐transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 15:331–341. [DOI] [PubMed] [Google Scholar]
- 24. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans. Nature 391:806–811. [DOI] [PubMed] [Google Scholar]
- 25. Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD (2007) Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer‐1. Cell 130:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K et al (2007) DEAD‐box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9:604–611. [DOI] [PubMed] [Google Scholar]
- 27. Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K et al (2006) Zebrafish MiR‐430 promotes deadenylation and clearance of maternal mRNAs. Science 312:75–79. [DOI] [PubMed] [Google Scholar]
- 29. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N et al (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240. [DOI] [PubMed] [Google Scholar]
- 30. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640. [DOI] [PubMed] [Google Scholar]
- 31. Griffiths‐Jones S, Saini HK, Dongen SV, Enright AJ (2007) miRBase: tools for microRNA genomics. Nucleic Acids Res, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guil S, Caceres JF (2007) The multifunctional RNA‐binding protein hnRNP A1 is required for processing of miR‐18a. Nat Struct Mol Biol 14:591–596. [DOI] [PubMed] [Google Scholar]
- 33. Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952. [DOI] [PubMed] [Google Scholar]
- 34. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146–1150. [DOI] [PubMed] [Google Scholar]
- 35. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha‐DGCR8 complex in primary microRNA processing. Genes Dev 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK et al (2006) Molecular basis for the recognition of primary microRNAs by the Drosha‐DGCR8 complex. Cell 125:887–901. [DOI] [PubMed] [Google Scholar]
- 37. Humphreys DT, Westman BJ, Martin DI, Preiss T (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102:16961–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hutvagner G, Zamore PD (2002) A microRNA in a multiple‐turnover RNAi enzyme complex. Science 297:2056–2060. [DOI] [PubMed] [Google Scholar]
- 39. Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM et al (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA 101:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim VN (2006) Small RNAs just got bigger: Piwi‐interacting RNAs (piRNAs) in mammalian testes. Genes Dev 20:1993–1997. [DOI] [PubMed] [Google Scholar]
- 41. Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z et al (2004) A combined computational‐experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kiriakidou M, Tan GS, Lamprinaki S, De Planell‐Saguer M, Nelson PT, Mourelatos Z (2007) An mRNA m7G cap binding‐like motif within human Ago2 represses translation. Cell 129:1141–1151. [DOI] [PubMed] [Google Scholar]
- 43. Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500. [DOI] [PubMed] [Google Scholar]
- 44. Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post‐transcriptional regulation. Nat Genet 30:363–364. [DOI] [PubMed] [Google Scholar]
- 45. Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 46. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 75:843–854. [DOI] [PubMed] [Google Scholar]
- 47. Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21:4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. [DOI] [PubMed] [Google Scholar]
- 49. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH et al (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ et al (2004) Distinct roles for Drosophila Dicer‐1 and Dicer‐2 in the siRNA/miRNA silencing pathways. Cell 117:69–81. [DOI] [PubMed] [Google Scholar]
- 51. Lewis BP, Shih IH, Jones‐Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798. [DOI] [PubMed] [Google Scholar]
- 52. Lingel A, Simon B, Izaurralde E, Sattler M (2004) Nucleic acid 3′‐end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11:576–577. [DOI] [PubMed] [Google Scholar]
- 53. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ et al (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 54. Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP et al (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925. [DOI] [PubMed] [Google Scholar]
- 55. Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow‐like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056. [DOI] [PubMed] [Google Scholar]
- 56. Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14:287–294. [DOI] [PubMed] [Google Scholar]
- 57. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303:95–98. [DOI] [PubMed] [Google Scholar]
- 58. Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′‐end‐specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maniataki E, Mourelatos Z (2005) A human, ATP‐independent, RISC assembly machine fueled by pre‐miRNA. Genes Dev 19:2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maroney PA, Yu Y, Fisher J, Nilsen TW (2006) Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol 13:1102–1107. [DOI] [PubMed] [Google Scholar]
- 61. Martinez J, Tuschl T (2004) RISC is a 5′ phosphomonoester‐producing RNA endonuclease. Genes Dev 18:975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T et al (2007) MicroRNA inhibition of translation initiation in vitro by targeting the cap‐binding complex eIF4F. Science 317:1764–1767. [DOI] [PubMed] [Google Scholar]
- 63. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (2005) Passenger‐strand cleavage facilitates assembly of siRNA into Ago2‐containing RNAi enzyme complexes. Cell 123:607–620. [DOI] [PubMed] [Google Scholar]
- 64. Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM et al (2006) A pattern‐based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217. [DOI] [PubMed] [Google Scholar]
- 65. Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L et al (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nelson PT, Hatzigeorgiou AG, Mourelatos Z (2004) miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10:387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nottrott S, Simard MJ, Richter JD (2006) Human let‐7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13:1108–1114. [DOI] [PubMed] [Google Scholar]
- 68. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA‐class regulatory RNAs in Drosophila. Cell 130:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Olsen PH, Ambros V (1999) The lin‐4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN‐14 protein synthesis after the initiation of translation. Dev Biol 216:671–680. [DOI] [PubMed] [Google Scholar]
- 70. Parker JS, Roe SM, Barford D (2004) Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J 23:4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Parker JS, Roe SM, Barford D (2005) Structural insights into mRNA recognition from a PIWI domain‐siRNA guide complex. Nature 434:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25:635–646. [DOI] [PubMed] [Google Scholar]
- 73. Petersen CP, Bordeleau ME, Pelletier J, Sharp PA (2006) Short RNAs repress translation after initiation in mammalian cells. Mol Cell 21:533–542. [DOI] [PubMed] [Google Scholar]
- 74. Pillai RS, Artus CG, Filipowicz W (2004) Tethering of human Ago proteins to mRNA mimics the miRNA‐mediated repression of protein synthesis. RNA 10:1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E et al (2005) Inhibition of translational initiation by Let‐7 MicroRNA in human cells. Science 309:1573–1576. [DOI] [PubMed] [Google Scholar]
- 76. Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21:5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rajewsky N, Socci ND (2004) Computational identification of microRNA targets. Dev Biol 267:529–535. [DOI] [PubMed] [Google Scholar]
- 78. Rand TA, Petersen S, Du F, Wang X (2005) Argonaute2 cleaves the anti‐guide strand of siRNA during RISC activation. Cell 123:621–629. [DOI] [PubMed] [Google Scholar]
- 79. Rehwinkel J, Behm‐Ansmant I, Gatfield D, Izaurralde E (2005) A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA‐mediated gene silencing. RNA 11:1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE et al (2000) The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906. [DOI] [PubMed] [Google Scholar]
- 81. Richter JD, Sonenberg N (2005) Regulation of cap‐dependent translation by eIF4E inhibitory proteins. Nature 433:477–480. [DOI] [PubMed] [Google Scholar]
- 82. Rigoutsos I, Huynh T, Miranda K, Tsirigos A, McHardy A, Platt D (2006) Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci USA 103:6605–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ et al (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12:340–349. [DOI] [PubMed] [Google Scholar]
- 84. Rodriguez A, Griffiths‐Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Saetrom P, Heale BS, Snove O, Jr , Aagaard, L , Alluin J, Rossi JJ (2007) Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res 35:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schwarz DS, Tomari Y, Zamore PD (2004) The RNA‐induced silencing complex is a Mg2+‐dependent endonuclease. Curr Biol 14:787–791. [DOI] [PubMed] [Google Scholar]
- 88. Seggerson K, Tang L, Moss EG (2002) Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin‐28 after translation initiation. Dev Biol 243:215–225. [DOI] [PubMed] [Google Scholar]
- 89. Song JJ, Smith SK, Hannon GJ, Joshua‐Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 90. Thermann R, Hentze MW (2007) Drosophila miR2 induces pseudo‐polysomes and inhibits translation initiation. Nature 447:875–878. [DOI] [PubMed] [Google Scholar]
- 91. Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19:517–529. [DOI] [PubMed] [Google Scholar]
- 92. Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004) A protein sensor for siRNA asymmetry. Science 306:1377–1380. [DOI] [PubMed] [Google Scholar]
- 93. Tomari Y, Du T, Zamore PD (2007) Sorting of Drosophila small silencing RNAs. Cell 130:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004) The C. elegans microRNA let‐7 binds to imperfect let‐7 complementary sites from the lin‐41 3′UTR. Genes Dev 18:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wakiyama M, Takimoto K, Ohara O, Yokoyama S (2007) Let‐7 microRNA‐mediated mRNA deadenylation and translational repression in a mammalian cell‐free system. Genes Dev 21:1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin‐14 by lin‐4 mediates temporal pattern formation in C. elegans . Cell 75:855–862. [DOI] [PubMed] [Google Scholar]
- 97. Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103:4034–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang W, Steitz TA (1995) Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3:131–134. [DOI] [PubMed] [Google Scholar]
- 99. Yekta S, Shih IH, Bartel DP (2004) MicroRNA‐directed cleavage of HOXB8 mRNA. Science 304:594–596. [DOI] [PubMed] [Google Scholar]
- 100. Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G et al (2005) Crystal structure of A. aeolicus argonaute, a site‐specific DNA‐guided endoribonuclease, provides insights into RISC‐mediated mRNA cleavage. Mol Cell 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21:5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68. [DOI] [PubMed] [Google Scholar]


