Abstract
Background
Early-life exposures to antibiotics may increase the risk of developing childhood asthma. However, little is known about the mechanisms linking antibiotic exposures to asthma. We hypothesized that changes in the nasal airway microbiota serve as a causal mediator in the antibiotics–asthma link.
Methods
In a population-based birth-cohort study in Finland, we identified longitudinal nasal microbiota profiles during age 2–24 months using 16S rRNA gene sequencing and an unsupervised machine learning approach. We performed a causal mediation analysis to estimate the natural direct effect of systemic antibiotic treatments during age 0–11 months on risks of developing physician-diagnosed asthma by age 7 years and the natural indirect (causal mediation) effect through longitudinal changes in nasal microbiota.
Results
In our birth cohort of 697 children, 8.0% later developed asthma. Exposure to ≥2 antibiotic treatments during age 0–11 months was associated with a 4.0% increase in the absolute risk of developing asthma (absolute increase, 95% CI, .9–7.2%; P = .006). The unsupervised clustering approach identified 6 longitudinal nasal microbiota profiles. Infants with a larger number of antibiotic treatments had a higher risk of having a profile with early Moraxella sparsity (per each antibiotic treatment, adjusted RRR, 1.38; 95% CI, 1.15–1.66; P < .001). This effect of antibiotics on asthma was partly mediated by longitudinal changes in the nasal microbiota (natural indirect effect, P = .008), accounting for 16% of the total effect.
Conclusions
Early exposures to antibiotics were associated with increased risk of asthma; the effect was mediated, in part, by longitudinal changes in the nasal airway microbiota.
Keywords: airway microbiota, antibiotics, asthma, causal mediation, children
In this population-based birth-cohort study, we found that antibiotic treatments during infancy were associated with increased risk of developing childhood asthma. Through causal mediation analysis, the antibiotics–asthma link was mediated, in part, by longitudinal changes in the nasal microbiota.
(See the Editorial Commentary by Rosas-Salazar and Hartert on pages 1555–6.)
Asthma affects approximately 9% of children in the United States [1] and Western Europe [2, 3]. Concurrently, the rate of inappropriate antibiotic use in children remains high [4, 5], despite discouragement by national and international guidelines [6, 7]. Several epidemiological studies have reported a dose-dependent association between antibiotic exposures during the first years of life and subsequent asthma development [8–11], with broad-spectrum antibiotics associated with a higher risk of asthma [8]. Yet, despite the clinical and research relevance, little is known about the factors mediating the antibiotics–asthma link.
The literature has indicated that the microbiota plays major roles in immune development during early childhood and the development of various disease conditions, including atopy and asthma [3, 12–14]. In addition to the gut microbiota, the airway microbiota also modulates immune responses in the airway niche [12] and contributes to the risk and severity of acute respiratory infections (ARIs) [15, 16], wheezing [17, 18], and asthma [19–21] in childhood. Antibiotic exposures cause temporary alterations in the microbiota, thereby making major effects during the early life—a critical period for the microbiota–host interplay that shapes long-term physiological functions [12]. Accordingly, it has been hypothesized that the antibiotics-related microbiota changes may mediate the effects of antibiotics on the development of childhood asthma [18]. However, no study has investigated the potential mediator role of airway microbiota in the link of interest. Recent advent of causal mediation analysis methods now enables us to disentangle the mediation effects in such complex interrelations [22].
To address this knowledge gap, by applying a causal mediation approach to the data of a population-based birth-cohort study, we tested the hypothesis that the exposure to systemic antibiotics during infancy contributes—through longitudinal changes in the nasal airway microbiota—to the development of childhood asthma.
METHODS
Design, Setting, and Participants
In a population-based birth-cohort study—the Steps to the Healthy Development and Well-being of Children (STEPS) Study—families of Finnish children born in the Hospital District of Southwest Finland between January 2008 and April 2010 were enrolled during pregnancy or soon after birth. Details of the study design, testing, and analysis are described in the Supplementary Methods. Briefly, as part of the STEPS Study, children were enrolled into a subcohort with an intensive follow-up for ARIs from birth to age 24 months and followed for development of asthma until age 7.5 years [15, 23]. No selection criteria other than language (Finnish- or Swedish-speaking family) were applied to recruiting the families in the STEPS Study. Patient demographics; family history; pre-, peri-, and postnatal history; and environmental information were collected from the National Birth Registry, by structured questionnaires, and with a diary. The Ministry of Social Affairs and Health and the Ethics Committee of the Hospital District of Southwest Finland approved the study. Parents of participating children gave their written, informed consent.
Exposure
The exposure of interest was systemic antibiotic use during infancy (age 0–11 months). Antibiotic treatments were classified in therapeutic classes (narrow- vs broad-spectrum) as previously described [4]. Narrow-spectrum penicillins (amoxicillin, phenoxymethylpenicillin, benzylpenicillin, and ampicillin), first-generation cephalosporins, sulfonamides, and nitrofurantoin were considered narrow-spectrum antibiotics. All other antibiotics—including combination of β-lactam and β-lactamase inhibitors (eg, amoxicillin-clavulanate) and macrolides—were considered broad-spectrum [4]. Data on antibiotic use were captured through multiple sources. Parents were instructed to record all respiratory and other symptoms as well as physician visits with antibiotic treatments into a daily diary during infancy. Families were also instructed to visit the study clinic during ARIs, and children were examined by a study physician using a structured form. Data on emergency department visits, outpatient visits, and hospitalizations during infancy with antibiotic treatments were retrieved from medical and prescription records of the Hospital District of Southwest Finland
Mediator
The mediator of interest was longitudinal changes in the nasal airway microbiota during age 2–24 months. Briefly, using a standardized protocol [15], nasal swab specimens were collected by study personnel at scheduled visits at age 2, 13, and 24 months at healthy state.. The 16S ribosomal RNA (rRNA) gene-sequencing methods were adapted from those developed for the National Institutes of Health Human Microbiome Project [24]. The 16S rDNA V4 region was amplified by polymerase chain reaction and sequenced on the MiSeq platform (Illumina, San Diego, CA) using the 2 × 250-bp paired-end protocol. Sequencing reads were merged using USEARCH v7.0.1090. 16S rRNA gene sequences were clustered into operational taxonomic units (OTUs) at a similarity cutoff value of 97% using UPARSE [25]. OTUs were determined by mapping centroids to the SILVA database.
To identify profiles of longitudinal patterns in the nasal microbiota during age 2–24 months, we applied a longitudinal k-means clustering approach [26] based on correlation distance [27] to the individual longitudinal trajectories based on relative abundances of the 100 most common genera, which accounted for 99% of overall abundance. The number of profiles was chosen based on Calinski-Harabasz methods and clinical plausibility [26]. The derived longitudinal profiles were used as the mediator in the causal mediation analysis.
Outcome
The outcome of interest was physician-diagnosed asthma, defined as a diagnosis of asthma in the medical records from age 6.5 to 7.5 years (asthma at age 7 years) with or without a prescription of inhaled corticosteroids for asthma at the same age. Physician diagnosis of asthma was retrieved from the medical records (Supplementary Table 1) and asthma medication use from nationwide electronic prescription records.
Statistical Analysis
First, a directed acyclic graph (DAG) (Figure 1) was constructed to represent our proposed model linking the exposure (antibiotic treatments) to the outcome (asthma development) with the mediator (longitudinal microbiota profiles) and potential confounders (sex, parental history of asthma, household siblings, breastfeeding during age 0–2 months, and frequency of ARIs during age 0–11 months). The model was constructed based on clinical plausibility and a priori knowledge [8, 10–12, 18, 28]. Next, to examine the association between frequency of antibiotic treatments and derived longitudinal microbiota profiles, multinomial regression models adjusting for confounders were constructed.
Figure 1.
Causal DAG of the proposed mediation model. Abbreviations: ARI, acute respiratory infection; DAG, directed acyclic graph.
To examine the direct and indirect effects in a counterfactual framework, causal mediation analysis was performed [22, 29, 30]. This method enables us to examine the extent to which the effect of exposure on the outcome is direct (direct effect) and to what extent it is mediated by the mediator (indirect effect). Specifically, the natural direct effect represents how much asthma risk would change if the patient were set to be exposed versus to be unexposed, but for each individual the longitudinal microbiota pattern was kept at the level it would have taken in the absence of exposure [22, 29, 30]. The natural indirect effect represents how much asthma risk would change if the patient were set to be exposed but the longitudinal microbiota pattern was changed from the level it would take if unexposed to the level it would take if exposed [22, 29, 30]. In the causal mediation analysis, the number of antibiotic treatments during age 0–11 months was dichotomized based on the distribution of data: 0–1 antibiotic treatment and 2 or more antibiotic treatments (the highest quartile). Additionally, to improve the interpretability of inferences, the longitudinal microbiota profiles were further consolidated into the profile with the highest Moraxella abundance (low-risk profile [with regard to asthma risk; Supplementary Table 2]) versus other profiles (high-risk profile). Stratification by Moraxella genus was chosen based on its dominance of the nasal microbiota and the literature reporting the relations of Moraxella with ARIs, wheezing, and asthma [15–19]. To account for confounding, we used inverse probability weighting for marginal structural models, which creates a pseudo-population where the exposed and unexposed are exchangeable as the exposure is independent from the measured confounders [31, 32].
In sensitivity analyses, the analysis was repeated with any use of broad-spectrum antibiotics during age 0–11 months as the exposure, use of a different cutoff for antibiotic exposure (0–2 vs ≥3), restriction of antibiotic treatments to age 0–2 months, and use of a different mediator categorization (the lowest Moraxella abundance vs other profiles), separately. Data were analyzed using R version 3.6.1.
RESULTS
Study Sample
Of 923 children in the STEPS respiratory cohort, 886 (96%) children had data on antibiotic treatments during age 0–11 months (Supplementary Figure 1). A total of 2261 nasal samples were collected at age 2, 13, and 24 months; and 2172 (96%) met the quality-control requirements and had sufficient sequence depth for 16S rRNA gene sequencing (rarefaction cutoff, 2023 reads per sample). Children with data on antibiotic exposures and longitudinal microbiota data were included in the current analysis (analytical cohort, n = 697 children with 1923 nasal samples). Medical records and electronic prescription data were available for 99% of the analytic cohort and 56 (8.0%) developed physician-diagnosed asthma by age 7 years. Inhaled corticosteroid prescription was documented in 93% of these children with asthma. Children in the analytical cohort and those in the nonanalytical cohort were similar in most baseline characteristics and outcome (Supplementary Table 3).
Antibiotic Use During Age 0–11 Months
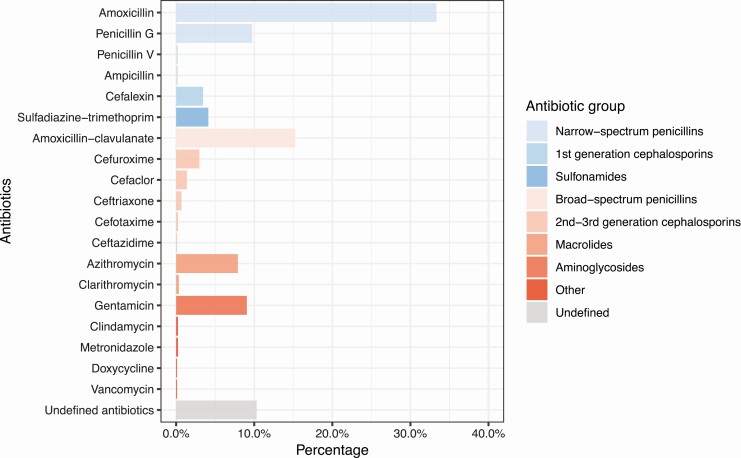
Overall, while 48% of children did not receive any systemic antibiotics, 52% received antibiotics during age 0–11 months (median, 1 per child; interquartile range [IQR], 0–2). Of the used antibiotics (Figure 2), 39% were classified as broad-spectrum antibiotics. During age 0–11 months, 32% of children used at least 1 course of broad-spectrum antibiotics. The most commonly used antibiotic was amoxicillin (33%), followed by amoxicillin-clavulanate (15%), penicillin G (10%), gentamicin (9%), and azithromycin (8%). Compared with children with 0–1 antibiotic treatment, those with 2 or more antibiotic treatments were more likely to have household siblings and ARIs (both P < .001) (Table 1).
Figure 2.
Exposure to antibiotic treatments during age 0–11 months in 697 children enrolled in the STEPS cohort. Narrow-spectrum antibiotics are presented in blue and broad-spectrum antibiotics in red. Abbreviation: STEPS, Steps to the Healthy Development and Well-being of Children.
Table 1.
Baseline Characteristics of 697 Children in the STEPS Cohort by Number of Antibiotic Treatments During Age 0–11 Months
| 0–1 Antibiotic Treatment (n = 501; 72%) | ≥2 Antibiotic Treatments (n = 196; 28%) | |
|---|---|---|
| Male sex | 255 (51) | 114 (58) |
| Maternal history of asthma | 36 (7) | 16 (8) |
| Parental history of asthma | 56 (11) | 30 (15) |
| Maternal smoking during pregnancy | 23 (5) | 9 (5) |
| Birth by cesarean delivery | 72 (14) | 18 (9) |
| Prematurity (<37 weeks) | 24 (5) | 6 (3) |
| Low birth weight (<2500 g) | 17 (3) | 4 (2) |
| Small for gestational age | 11 (2) | 3 (2) |
| Household sibling | 183 (37) | 119 (61) |
| Breastfed during age 0–2 monthsa | 391 (78) | 164 (84) |
| Parental smokingb | 59 (12) | 29 (15) |
| Eczema by age 13 months | 69 (14) | 39 (20) |
| Day care at age 13 months | 111 (22) | 43 (22) |
| Number of ARIs during age 0–11 months, median (IQR) | 4 (2–6) | 7 (5–10) |
Data are no. (%) of children unless otherwise indicated.
Abbreviations: ARI, acute respiratory infection; IQR, interquartile range; STEPS, Steps to the Healthy Development and Well-being of Children.
aData on breastfeeding available for 630 (90%) children.
bData on parental smoking available for 557 (80%) children.
Longitudinal Nasal Microbiota Profiles
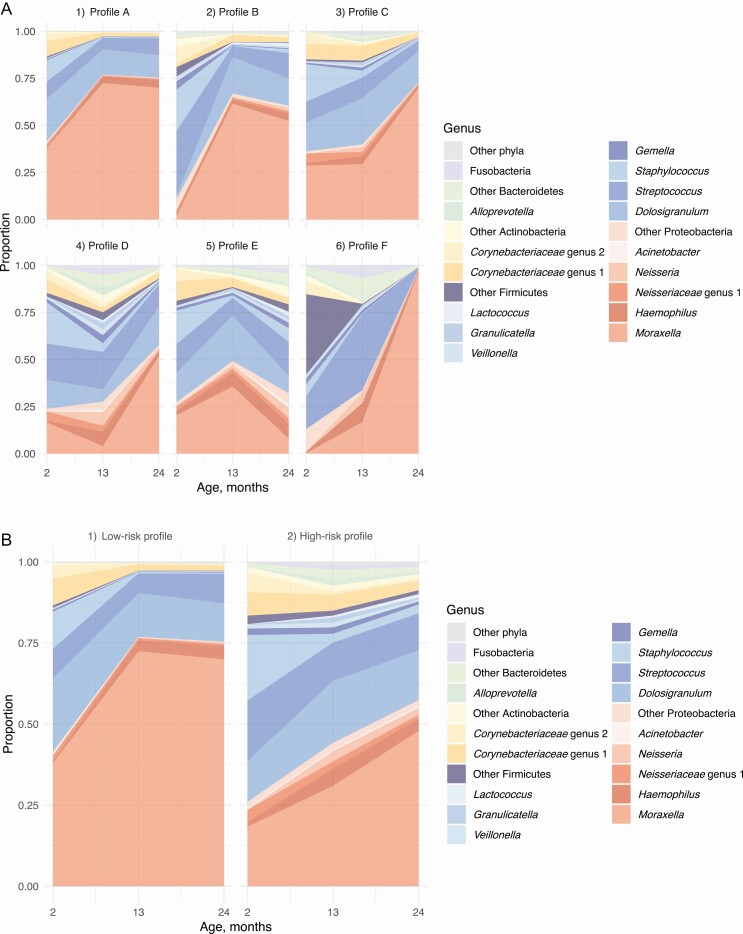
Nasal microbiota testing at age 2, 13, and 24 months detected 30 phyla and 1386 genera. The microbiota was dominated by Firmicutes at age 2 months (53%), followed by Proteobacteria at age 13 (58%) and 24 (64%) months. Clustering of longitudinal nasal microbiota data during age 2–24 months identified 6 longitudinal profiles (Figure 3A). Children with a larger number of antibiotic treatments had higher risks of having profile D with early Moraxella sparsity than profile A with persistent Moraxella dominance (reference) (per each antibiotic treatment, adjusted relative rate ratio [RRR], 1.38; 95% confidence interval [CI], 1.15–1.66; P < .001) (Table 2 and Supplementary Table 4). Similarly, broad-spectrum antibiotic treatments were associated with significantly higher risks of having profile D (per each antibiotic treatment, adjusted RRR, 1.74; 95% CI, 1.31–2.30; P < .001) (Table 2 and Supplementary Table 5). When using dichotomized nasal microbiota profiles (Figure 3B and Supplementary Table 6), both any antibiotic and broad-spectrum antibiotic treatments were associated with a higher probability of having a high-risk profile (both P < .01) (Supplementary Table 7).
Figure 3.
Longitudinal nasal microbiota profiles during age 2–24 months in 697 children enrolled in the STEPS cohort. A, Six longitudinal nasal microbiota profiles were identified using the k-means clustering method: (1) profile A (reference) with persistent Moraxella dominance with high Dolosigranulum as well as low Streptococcus and Staphylococcus abundances, n = 279 (40%); (2) profile B with Streptococcus-to-Moraxella transition, n = 84 (12%); (3) profile C with early Dolosigranulum/Corynebacteriaceae dominance, n = 139 (20%); (4) profile D with early Moraxella sparsity with its subsequent increase as well as persistently high Streptococcus abundance, n = 100 (14%); (5) profile E with mixed longitudinal patterns, n = 92 (13%); (6) profile F, n = 3 (0.4%). Relative abundances of the 15 most abundant genera are shown with the other genera categorized into the 5 most abundant phylum groups. Color codes of genera are based on taxonomic annotation at the phylum level: red, Proteobacteria; blue, Firmicutes; yellow, Actinobacteria; and green, Bacteroidetes. B, For the mediation analysis, longitudinal nasal microbiota profiles were dichotomized to (1) low-risk profile with persistent Moraxella dominance, early high Dolosigranulum as well as low Streptococcus and Staphylococcus abundances (reference, profile A), n = 279 (40%) and (2) high-risk profile with early Moraxella sparsity, early low Dolosigranulum as well as high Streptococcus and Staphylococcus abundances (profiles B–F), n = 418 (60%). Abbreviation: STEPS, Steps to the Healthy Development and Well-being of Children.
Table 2.
Association of Antibiotic Treatments During Age 0–11 Months With Longitudinal Nasal Microbiota Profiles During Age 2–24 Months
| RRR (95% CI), per Each Antibiotic Treatment | ||
|---|---|---|
| Longitudinal Microbiota Profiles During Age 2–24 Months (Dependent Variable) | Antibiotic Treatments During Age 0–11 Months (Exposure) | Broad-spectrum Antibiotic Treatments During Age 0–11 Monthsa (Exposure) |
| Profile A with persistent Moraxella dominance (n = 279, 40%) | Reference | Reference |
| Profile B with Streptococcus-to-Moraxella transition (n = 84, 12%) | 1.16 (.92–1.45) | 1.16 (.80–1.67) |
| Profile C with early Dolosigranulum/Corynebacteriaceae dominance (n = 139, 20%) | 1.20 (1.01–1.43) | 1.16 (.87–1.55) |
| Profile D with early Moraxella sparsity with its subsequent increase (n = 100, 14%) | 1.38 (1.15–1.66) | 1.74 (1.31–2.30) |
| Profile E with mixed longitudinal patterns (n = 92, 13%) | 1.20 (.98–1.48) | 1.30 (.94–1.79) |
Longitudinal clustering of nasal microbiota during age 2–24 months identified 6 distinct profiles. Of these, the profile F included only 3 children and was excluded from this analysis. To examine the association between frequency of antibiotic treatments and derived longitudinal microbiota profiles, multinomial logistic regression models adjusting for potential confounders (sex, parental history of asthma, household siblings, breastfeeding during age 0–2 months, and acute respiratory infections during age 0–11 months) were constructed. Profile A with persistent Moraxella dominance (low-risk profile) was used as the reference. Results of unadjusted analysis are shown in Supplementary Tables 4 and 5.
Abbreviations: CI, confidence interval; RRR, relative rate ratio.
aNarrow-spectrum antibiotics were defined as narrow-spectrum penicillins (amoxicillin, phenoxymethylpenicillin, benzylpenicillin, and ampicillin), first-generation cephalosporins, and sulfonamides. All other antibiotics were defined as broad-spectrum antibiotics, including broad-spectrum penicillins (eg, amoxicillin-clavulanate), second- and third-generation cephalosporins, macrolides, and aminoglycosides.
Causal Mediation Analysis
Of the children who used 0–1 antibiotic treatment during age 0–11 months, 6.8% developed asthma. In contrast, of those who underwent 2 or more antibiotic treatments, 11.2% developed asthma (Supplementary Table 8). The prevalence of asthma by longitudinal microbiota profile is shown in Supplementary Table 2. In the mediation analysis, children with 2 or more antibiotic treatments had a 4.0% increase in the absolute risk of developing asthma (total effect; 95% CI, .9–7.2%; P = .006), with a direct effect of a 3.3% increase (95% CI, .4–6.4%; P = .03) (Table 3). The effect of antibiotics on asthma risk was mediated, in part, by the longitudinal changes in the nasal microbiota (low vs high-risk nasal microbiota profiles; P = .008), accounting for 16.2% (95% CI, 3.1–65.0%) of total effect. Likewise, a broad-spectrum antibiotic treatment during age 0–11 months was associated with a 3.6% increase in the absolute risk of developing asthma (95% CI, .6–6.7%; P = .02), with a significant indirect effect (P = .01), accounting for 13.9% (95% CI, 1.7–64.9%) of total effect (Table 3). In the sensitivity analyses using a different cutoff for antibiotic treatments (0–2 vs ≥3), restricting antibiotic use to age 0–2 months (with a limited statistical power), and using different mediator categorization, the results did not materially change (Supplementary Tables 9–11).
Table 3.
Effect of Antibiotic Treatments During Age 0–11 Months on Risk of Developing Asthma by Age 7 Years Mediated by Longitudinal Patterns of Nasal Microbiota
| Absolute Risk Difference (95% CI), % | ||
|---|---|---|
| Antibiotic Treatments (≥2) | Broad-spectrum Antibiotic Treatment (≥1) | |
| Total effect | 4.0 (.9–7.2) | 3.6 (.6–6.7) |
| Natural direct effect | 3.3 (.4–6.4) | 3.1 (.1–6.1) |
| Natural indirect effect | .7 (.1–1.4)a | .5 (.1–1.1)b |
n = 623. Causal mediation analysis estimating the total and direct effects of antibiotic exposure during age 0–11 months on the risk of developing asthma by age 7 years as well as indirect effect by longitudinal changes in nasal microbiota during age 2–24 months (low-risk profile with persistent Moraxella dominance vs high-risk profile with early Moraxella sparsity). Inverse probability weighting with marginal structural models was used in the mediation analysis to account for potential confounders (ie, sex, parental history of asthma, household siblings, breastfeeding during age 0–2 months, and acute respiratory infections during age 0–11 months).
Abbreviation: CI, confidence interval.
aProportion of indirect effect by antibiotic treatments was 16.2% (95% CI, 3.1–65.0%).
bProportion of indirect effect by broad-spectrum antibiotic treatments was 13.9% (95% CI, 1.7–64.9%).
DISCUSSION
In this population-based birth-cohort study with longitudinal characterization of nasal airway microbiota, we found that antibiotic treatments during age 0–11 months had dose-dependent effects on nasal airway microbiota patterns in the first 2 years of life, with broad-spectrum antibiotics having larger effects. Specifically, antibiotic treatments were associated with higher risks of having a nasal microbiota profile with early Moraxella sparsity. Furthermore, the causal mediation analysis demonstrated that antibiotic treatments were associated with increased risks of childhood asthma and that the link was mediated, in part, by the longitudinal changes in nasal microbiota. To the best of our knowledge, this is the first study that has demonstrated the role of airway microbiota during early childhood in mediating the antibiotics–asthma link.
In agreement with our findings, epidemiological research reported relations of early-life antibiotic use with higher risks of asthma development [8–11]. For example, recent analysis of claims-based data reported dose-dependent associations between antibiotic prescription fills during infancy and higher risk of asthma [8]. However, the causal inferences on the antibiotics–asthma link in previous registry-based studies are potentially limited through confounding by indication, particularly by ARIs [33]. Our findings are buttressed by the intensive ascertainment of antibiotic treatments with their indications in a population-based study and the rigorous adjustment of confounders including ARIs. Additionally, the observed relationships between antibiotic treatments and altered composition of airway microbiota are also consistent with previous studies. For example, studies reported antibiotics-related reductions in Moraxella [34], Dolosigranulum [35], and Corynebacterium [18, 35] as well as increases in Streptococcus and Haemophilus [18] in airway microbiota. For example, a single-center trial of 39 infants with respiratory syncytial virus bronchiolitis reported that 2-week treatment with azithromycin depleted Moraxella at the end of treatment [34]. While studies collectively suggested aberrant (and, at least, transient) effects of antibiotic exposures on the airway microbiota, the literature has remained uncertain about the long-term effects of antibiotic exposures on the longitudinal airway microbiota patterns and their relations with chronic airway morbidities, such as childhood asthma. This large birth-cohort study of healthy children builds on previous studies, and extends them by demonstrating, for the first time, the mediation role of longitudinal nasal microbiota changes in the antibiotics–asthma link.
Underlying mechanisms of the antibiotics–nasal microbiota–asthma relationship warrant further clarification. It is possible that antibiotic exposures altered not only the airway microbiota composition but also its function, thereby contributing to asthma risk. For example, in gut microbiota research, dysbiosis was found to be related to enriched proinflammatory microbe-derived metabolites—eg, 12,13-diHome—and asthma risk [13]. Intra-abdominal treatments of 12,13-diHome in mice reduced regulatory T cells and induced pulmonary inflammation [36]. In the airway microbiota research, the exact mechanisms linking the airway microbiota—including Moraxella—to childhood airway diseases (eg, ARI, incident asthma) remain uncertain [17–19, 21, 34, 37]. Similar to our finding, studies showed relations of Moraxella sparsity with higher morbidity [16, 38]. For example, a multicenter prospective study of 1016 US infants with ARI showed that Moraxella sparsity was associated with higher severity, which was also validated in an independent cohort [38]. Additionally, in a case-control study of 461 children aged less than 5 years in the Netherlands, sparsity of several different Moraxella spp was associated with lower respiratory infection [16]. By contrast, other studies reported that dominance/presence by Moraxella (and other bacteria, such as Streptococcus) was associated with higher risks of respiratory morbidities (eg, recurrent wheeze [17], incident asthma [19], and asthma exacerbation [21, 37]). For example, in post hoc analysis of a clinical trial of 254 school-age children with asthma, Zhou et al [37] reported that transition to Moraxella cluster during loss of asthma control was associated with higher risks of exacerbations. These apparent discrepancies between studies may be attributable to the difference in study design (eg, birth cohort vs trials), population (healthy infants vs school-age asthmatics), setting (sampling at healthy state vs at ARI), testing (eg, 16S rRNA gene sequencing vs culture), and analysis (eg, longitudinal vs cross-sectional analysis). Regardless, studies showed that altered upper airway microbiota is associated with unique airway metabolome profiles [39] and that airway microbes elicit a unique host response—eg, M. catarrhalis with increased T-helper (TH) 1 cytokine profile [40] and Haemophilusinfluenzae with mixed TH1/TH2/TH17 inflammatory response [41]. Our observations should advance research into mechanisms that underlie the antibiotics–microbiota–asthma link.
Interestingly, our data also demonstrated that most of the total effect—the effect of antibiotic exposures on asthma development—was mediated by pathways other than the longitudinal changes in nasal microbiota. There are several potential mechanisms for this observation. First, it is possible that the effect was mediated by the changes in gut microbiota. Indeed, antibiotics are known to have long-term aberrant effects on the gut microbiota [42], which not only has a large microbial mass but plays major roles in immune development [12]. The unique composition of gut microbiota has also been linked to risks of developing childhood asthma [3, 14]. Second, antibiotics may have contributed to asthma risk through their immunomodulatory effects. Third, it is also possible that our dichotomization of longitudinal nasal microbiota patterns might have resulted in loss of information in compensation for the greater interpretability of high-dimensional microbiota data. Notwithstanding the complexity, the identification of longitudinal nasal microbiota patterns as a mediator in the antibiotics–asthma link is an important finding.
Our study has several potential limitations. First, we analyzed upper airway microbiota while asthma involves lower airways. Yet, nasal sampling is a noninvasive procedure in young infants and previous studies have reported that the upper airway microbiota reliably represents the lower airway microbiota [16, 43]. Second, some nasal samples were excluded from analysis due to insufficient sequence depth. However, the analytical and nonanalytical cohort did not materially differ, arguing against substantial selection bias. Third, there was a partial temporal overlap between antibiotic exposures and baseline microbiota data. Yet, there was no overlap with the subsequent (age 13-month and 24-month) data that comprise the longitudinal patterns. Additionally, the sensitivity analysis restricting antibiotic treatment to age 0–2 months demonstrated consistent inferences. Fourth, as with any observational study, causal inferences may be confounded by unmeasured factors (eg, gut microbiota, child’s genetics), while we applied robust adjustment for confounding (eg, adjustment for parental history of asthma accounts for, at least partially, confounding by the child’s genetics). Our data should also provide insights into future studies that may improve confounding control. Finally, we must be cautious in generalizing the inferences to other populations (eg, young American children) until they are externally validated. Yet, the observed relationships in this population-based cohort study are clinically and biologically plausible.
CONCLUSIONS
In this population-based birth-cohort study, we found that exposure to antibiotic treatments during age 0–11 months had dose-dependent relations with longitudinal nasal airway microbiota patterns during the first 2 years of life, with broad-spectrum antibiotics having a larger impact. We also found that antibiotic exposures were associated with increased risks of developing asthma, which was mediated, in part, by longitudinal changes in nasal microbiota. For clinicians, these findings lend additional support to the current guidelines that discourage unnecessary use of antibiotics, particularly in young children. Furthermore, our observations should not only facilitate further investigations into the complex interplay between antibiotic exposures, microbiota, and host response but they also offer new avenues for the prevention of childhood asthma.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank all the families who participated in this study, the midwives for their help in recruiting the families, and the whole STEPS Study team for assistance with data collection; Hanna Lagström, PhD, for her many contributions to the STEPS Study; study nurses Niina Lukkarla, Petra Rajala, and Mira Katajamäki for their assistance in the study clinic; Tamara Teros-Jaakkola, MD, for her assistance with the data collection; and Anne Kaljonen, MS, for her assistance with data handling. The authors thank the Finnish Functional Genomics Center, supported by University of Turku, Åbo Akademi University, and Biocenter Finland. The authors also thank Joseph F. Petrosino, PhD, and Nadim A. Ajami, PhD, at Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine (Houston, TX) for 16S rRNA gene-sequencing analysis and Orianne Dumas, PhD, French Institute of Health and Medical Research (France) for her help with the statistical analysis.
Financial support. This work was supported by the University of Turku; the Abo Akademi University; the Turku University Hospital; the Academy of Finland (grants numbers 123571, 140251, 277535, and 324926); the Emil Aaltonen Foundation; the Finnish Medical Foundation; the Päivikki and Sakari Sohlberg Foundation; the Foundation for Pediatric Research; the Allergy Research Foundation; the Paulo Foundation; Research Funds from Specified Government Transfers, Hospital District of Southwest Finland; the Tampere Tuberculosis Foundation; the Foundation of the Finnish Anti-Tuberculosis Association; the Orion Research Foundation; and the Juho Vainio Foundation.
Potential conflicts of interest. K. H. reports grants from the National Institutes of Health, Novartis, and Teva, outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Moorman JE, Akinbami LJ, Bailey CM, et al. National surveillance of asthma: United States, 2001–2010. Vital Health Stat 2012; 3:1–58. [PubMed] [Google Scholar]
- 2. Lai CK, Beasley R, Crane J, Foliaki S, Shah J, Weiland S; International Study of Asthma and Allergies in Childhood Phase Three Study Group . Global variation in the prevalence and severity of asthma symptoms: phase three of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2009; 64:476–83. [DOI] [PubMed] [Google Scholar]
- 3. Stokholm J, Blaser MJ, Thorsen J, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018; 9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poole NM, Shapiro DJ, Fleming-Dutra KE, Hicks LA, Hersh AL, Kronman MP. Antibiotic prescribing for children in United States emergency departments: 2009–2014. Pediatrics 2019; 143:e20181056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van de Maat J, van de Voort E, Mintegi S, et al. Antibiotic prescription for febrile children in European emergency departments: a cross-sectional, observational study. Lancet Infect Dis 2019; 19:382–91. [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention. Antibiotic use in the United States, 2018 update: progress and opportunities. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention, 2019. [Google Scholar]
- 7. European Commission. EU g uidelines for the prudent use of antimicrobials in human health (2017/C212/01). Available at: https://ec.europa.eu/health/amr/sites/amr/files/amr_guidelines_prudent_use_en.pdf. Accessed 20 November 2019.
- 8. Donovan BM, Abreo A, Ding T, et al. Dose, timing, and type of infant antibiotic use and the risk of childhood asthma. Clin Infect Dis 2019. doi:10.1093/cid/ciz448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ahmadizar F, Vijverberg SJH, Arets HGM, et al. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatr Allergy Immunol 2017; 28:430–7. [DOI] [PubMed] [Google Scholar]
- 10. Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy 2015; 45:137–45. [DOI] [PubMed] [Google Scholar]
- 11. Marra F, Marra CA, Richardson K, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics 2009; 123:1003–10. [DOI] [PubMed] [Google Scholar]
- 12. Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol 2019; 20:1279–90. [DOI] [PubMed] [Google Scholar]
- 13. Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015; 7: 307ra152. [DOI] [PubMed] [Google Scholar]
- 15. Toivonen L, Hasegawa K, Waris M, et al. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 2019; 74:592–9. [DOI] [PubMed] [Google Scholar]
- 16. Man WH, van Houten MA, Mérelle ME, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 2019; 7:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teo SM, Tang HHF, Mok D, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 2018; 24:341–52.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007; 357:1487–95. [DOI] [PubMed] [Google Scholar]
- 20. Kim BS, Lee E, Lee MJ, et al. Different functional genes of upper airway microbiome associated with natural course of childhood asthma. Allergy 2018; 73:644–52. [DOI] [PubMed] [Google Scholar]
- 21. McCauley K, Durack J, Valladares R, et al. ; National Institute of Allergy and Infectious Diseases–sponsored Inner-City Asthma Consortium . Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol 2019; 144:1187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernán M, Robins J.. Causal inference: what if. Boca Raton, FL: Chapman & Hall/CRC, 2020. [Google Scholar]
- 23. Toivonen L, Forsstrom V, Waris M, Peltola V. Acute respiratory infections in early childhood and risk of asthma at 7 years of age. J Allergy Clin Immunol 2019; 143:407–10.e6. [DOI] [PubMed] [Google Scholar]
- 24. Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012; 486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013; 10:996–8. [DOI] [PubMed] [Google Scholar]
- 26. Genolini C, Alacoque X, Sentenac M, Arnaud C. kml and kml3d: R packages to cluster longitudinal data. J Stat Softw 2015; 65:1–34. doi:10.18637/jss.v065.i04 [Google Scholar]
- 27. James G, Witten D, Hastie T, Tibshirani R.. An introduction to statistical learning with applications in R. New York: Springer, 2017. [Google Scholar]
- 28. Abreo A, Gebretsadik T, Stone CA, Hartert TV. The impact of modifiable risk factor reduction on childhood asthma development. Clin Transl Med 2018; 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robins JM, Greenland S. Identifiability and exchangeability for direct and indirect effects. Epidemiology 1992; 3:143–55. [DOI] [PubMed] [Google Scholar]
- 30. Pearl J. Direct and indirect effects. In: Proceedings of the 17th Conference on Uncertainty in Artificial Intelligence. San Francisco, CA: Morgan Kaufmann Publishers Inc, 2001:411–20. [Google Scholar]
- 31. Robins JM, Hernán MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000; 11:550–60. [DOI] [PubMed] [Google Scholar]
- 32. Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology 2000; 11:561–70. [DOI] [PubMed] [Google Scholar]
- 33. Örtqvist AK, Lundholm C, Kieler H, et al. Antibiotics in fetal and early life and subsequent childhood asthma: nationwide population based study with sibling analysis. BMJ 2014; 349:g6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou Y, Bacharier LB, Isaacson-Schmid M, et al. Azithromycin therapy during respiratory syncytial virus bronchiolitis: upper airway microbiome alterations and subsequent recurrent wheeze. J Allergy Clin Immunol 2016; 138:1215–9.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med 2017; 196:1582–90. [DOI] [PubMed] [Google Scholar]
- 36. Levan SR, Stamnes KA, Lin DL, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol 2019; 4:1851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y, Jackson D, Bacharier LB, et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat Commun 2019; 10:5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hasegawa K, Mansbach JM, Ajami NJ, et al. ; MARC-35 Investigators . Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016; 48:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stewart CJ, Mansbach JM, Wong MC, et al. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. a multiomic analysis. Am J Respir Crit Care Med 2017; 196:882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amedei A, Della Bella C, Niccolai E, et al. Moraxella catarrhalis-specific Th1 cells in BAL fluids of chronic obstructive pulmonary disease patients. Int J Immunopathol Pharmacol 2009; 22:979–90. [DOI] [PubMed] [Google Scholar]
- 41. Følsgaard NV, Schjørring S, Chawes BL, et al. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med 2013; 187:589–95. [DOI] [PubMed] [Google Scholar]
- 42. Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016; 7:10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marsh RL, Kaestli M, Chang AB, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016; 4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.