Abstract
An essential questions of gene regulation is how large number of enhancers and promoters organize into gene regulatory loops. Using transcription-factor binding enrichment as an indicator of enhancer strength, we identified a portion of H3K27ac peaks as potentially strong enhancers and found a universal pattern of promoter and enhancer distribution: At actively transcribed regions of length of ∼200–300 kb, the numbers of active promoters and enhancers are inversely related. Enhancer clusters are associated with isolated active promoters, regardless of the gene's cell-type specificity. As the number of nearby active promoters increases, the number of enhancers decreases. At regions where multiple active genes are closely located, there are few distant enhancers. With Hi-C analysis, we demonstrate that the interactions among the regulatory elements (active promoters and enhancers) occur predominantly in clusters and multiway among linearly close elements and the distance between adjacent elements shows a preference of ∼30 kb. We propose a simple rule of spatial organization of active promoters and enhancers: Gene transcriptions and regulations mainly occur at local active transcription hubs contributed dynamically by multiple elements from linearly close enhancers and/or active promoters. The hub model can be represented with a flower-shaped structure and implies an enhancer-like role of active promoters.
INTRODUCTION
How enhancers relay the regulatory information to their target promoters is an essential question in gene regulation. Millions of enhancers were predicted in the human and mouse genomes based on histone modification, transcription-factor (TF) binding, or bi-directional eRNA. The number of experimentally validated in vivo enhancer–promoter pairs is, however, at most, in the range of hundreds and, perhaps, only in the dozens.
Enhancers work within the context of chromatin domains. Interphase chromatin is organized into topologically associated domains (TADs) (1), with extensive chromatin interactions within each domain but few across neighboring domains. Promoter-enhancer interactions are usually formed through chromatin looping within a TAD (1–4), and the TAD boundaries impose regulatory constraints on enhancers, preventing ectopic gene activation (1,5–7). Interestingly, recent studies have found that disruption of chromatin topology on a large scale has only a modest effect on the transcriptome. Loss of the critical chromatin architecture protein cohesin eliminated all of the loop domains (in thousands), but only dozens of genes showed a >2 folder change in expression level (8). Diminishment of TAD structures in mouse liver cells by Nipbl knock-out resulted in only small transcriptome changes (9). A mutant drosophila genome with extensive rearranged chromatin architecture that disrupts many presumable long-range promoter-enhancer interactions also had a modest effect on the transcriptome (10). These results suggest that, although TAD structures are important for certain genes, regulation of the majority of genes occurs at sub-TAD or even finer genomic scales.
Adding to the complexity of promoter-enhancer interactions, disruption of many predicted enhancers has no effect on gene transcription. Recent research on a cellular genetic screen that tested more than 6,000 candidate enhancers (based on DNase, H3K27ac, p300 and GAGA1 binding) in human K562 cells detected only ∼500 possible target genes (11). A functional dissection of the predicted enhancer repertoire in human embryonic stem cells found that only a small fraction of regions marked by critical epigenetic marks or TF binding can function as enhancers (12).
How exactly tens of thousands of enhancers and a few thousand active promoters in a cell organize themselves into transcription-regulatory loops remains unclear. In the current study, we filter enhancers with the criteria of TF-binding enrichment and discover a pattern of active promoter and enhancer organization that would not be obvious if all H3K27ac peaks are counted as effective enhancers. We show that: At actively transcribed regions of length of ∼200–300 kb, the numbers of active promoters and enhancers are inversely related. Enhancer clusters tend to associate with single active promoters and clusters of active promoters associate with few distal enhancers. We propose a simple rule of spatial organization of regulatory elements: complicated interactions among active promoter and enhancers are mostly organized into clusters that can be represented with a flower-shaped transcription hub model. Our results provide insights for transcriptome robustness and, how the transcriptome is in large part decoupled from TAD structures.
MATERIALS AND METHODS
Analysis of ChIP-seq and RNA-seq data
We searched SRA and GEO for all ChIP-seq data of protein factors related to gene expression activation from mouse ESC and MEF cells. We aligned the ChIP-seq data to mm10 genome using bowtie2 (13) with default parameters. For peak calling, we used macs2 with a cutoff q-value of 0.01 (https://github.com/taoliu/MACS/). We used STAR (14) for RNA-seq mapping, with the following parameters: — outFilterMultimapNmax 20 -alignSJoverhangMin 8 -alignSJDBoverhangMin 1 -outFilterMismatchNoverReadLmax 0.04 -alignIntronMin 20 -alignIntronMax 1000000 -alignMatesGapMax 1000000.
Super enhancer (enhancer cluster) identification
Super enhancers were identified with ROSE (https://bitbucket.org/young_computation/rose/src/master/) on H3K27ac data of mouse ESC and MEF cells. ROSE was run with a stitching distance of 12500 bp and promoter exclusion zone of 4000 bp (TSS ± 2000 bp). This results in 1281 and 1337 SEs in ESC and MEF cells, respectively. We examined TF enrichment of these SEs. Some SEs overlap few or no TF or p300 ChIP-seq peaks, although their H3K27ac signals are picked by ROSE as SE. Therefore, we further filtered these SEs with TF enrichment. We identified all regulatory sites that overlap ≥n protein factor ChIP-seq peaks as H3K27ac_nPF (the protein factors are Nanog, Oct4, Sox2, Klf4, Esrrb, Tfcp2l1, Dppa2, E2f1, cMyc, p300, Med1, Brg1 in ESC and Cebpa, Cebpb, Fra1, Runx1, Klf4, cMyc, CBP, p300 and Brg1 in MEF cells). There are ∼22000 and ∼20700 H3K27ac_6PF in ESC and MEF, respectively. We selected SEs overlapping ≥2 distant H3k27ac_6PF. This filtering resulted in 893 and 913 SEs in ESC and MEF cells, respectively. Our 893 SEs in ESC overlap 85% of the 231 ESs from Whyte et al. (15), which proves the validity of our methods. Super enhancers were assigned to the expressed transcript (RPKM > 1) whose TSS is the nearest to the center of the SE (16).
Arithmetic methods for plots of the number of enhancers vs. promoters
For window selection
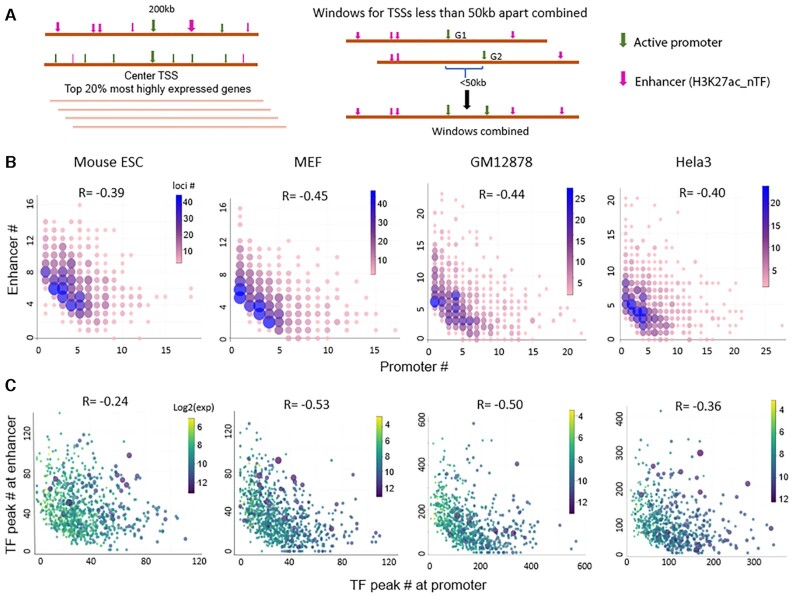
We selected genomic windows of 200 kb centered at TSSs of the top 20% most highly expressed genes. There are overlaps among some of these 200kb windows because some highly expressed genes are close or adjacent to each other. We combined windows with centers <50 kb apart, as shown in Figure 4A. For individual cases in which multiple window combinations result in a length of >300 kb, we manually examine the combinations and either separate one into two non-overlapping windows or ‘trim’ the ends so that the length of all windows is between 200 and 300 kb.
Figure 4.
The numbers of active promoters and distant enhancers are inversely related at a relatively fixed length of active transcribed regions. (A) Genomic windows of ∼200 kb centered at TSSs of the top 20% most highly expressed genes are selected, and windows with centers <50 kb apart are combined. (B) Plots of the number of active enhancers vs. active promoters. The color bars indicate the number of windows at each data point. (C) Plot of the number of total TF peaks at the active enhancers versus the number of total TF peaks at the active promoters. Each point represents one genomic window, and the color bars indicate total transcription level (log2(cpm)) at each window. R is the Spearman correlation coefficient.
For active promoter and enhancer counting
We count promoters of genes with expression level ≥1 RPKM as active promoters. For active genes with multiple TSSs, we observe that usually only one TSS is used in any given cell type, based on two pieces of evidence: One is the associated exon expression, and the other is the H3K27ac peak at the TSS. Therefore, in the case of one gene with multiple TSSs, we count only the promoter overlapping H3K27ac signal. For enhancer counting, we set a TF enrichment threshold so that only the H3K27ac peaks above the threshold, H3K27ac_nTF, will be counted as enhancers. We tried a different threshold, as n = 3, 4 or 5 for ESC and MEF cells (the number of H3K27ac_nTF is ∼25 000–40 000). With all three n values, there is an inverse relationship between the number of promoters and enhancers. In Figure 4, we choose the n value so that the total number of H3K27ac_nTF (including promoters) in each cell type is ∼20,000–25,000. The n values are 4, 4, 6 and 10 in mouse ESC, MEF cells, Hela3 cell line, and GM12878 cell line, respectively.
Hi-C data analysis
Raw observed Hi-C data in a 5kb resolution in GM12878 and K562 cell lines were retrieved from Rao's work (17). We followed the procedure described in Huang's work to filter for significant Hi-C interactions (18). First, we used the iterative correction and eigenvector decomposition (ICE) algorithm implemented in the Hi-Corrector package (19) to remove biases (20,21). Then, statistically significant interactions were identified by Fit-Hi-C (22) with the parameters -U = 2000000, -L = 5000 and using fixed FDRs of 1e–2, 1e–3 and 1e–5.
Using ENCODE TFBS ChIP-seq data (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform/), we selected strong regulatory elements based on TF enrichment: For the GM12878 cell line, we identified all H3K27ac peaks that overlap >10 TF peaks (H3K27ac_10TF, the same as used in the calculation in Figure 4), regardless of whether it is a promoter or enhancer, which results in ∼26,000 sites.
We separated annotated genes of the hg19 genome into gene groups based on a distance threshold of 100 kb. If the distance between two adjacent annotated genes (regardless of directionality) is <100 kb, then they belong to one gene group, otherwise, two groups, which results in 3290 gene groups. For each gene group with expression level >100 cpm, we located the center point and extended it into a 1 Mb long window, and overlapping ones are combined, which resulted in 823 non-overlapping regions. The above described H3K27ac_10TF sites within these 823 regions represent strong regulatory elements (REs), for which we tested our model of active transcription hub. For control sites selection, we screened each of the 823 regions from end to end and selected one 2kb-wide control site (not overlapping any TF ChIP-seq peaks) every 25 kb of ‘empty’ region (no REs), which resulted in a total of ∼22 000 REs (including ∼8600 promoters) and ∼6000 control sites.
We then examined the pairwise interactions among all of these sites with three different FDR cutoffs for Hi-C interactions: 1e–5, 1e–3 and 1e–2. There is great enrichment of interactions among REs compared to control sites at all three FDR cutoffs. Only cis (intra-chromosome) interactions were considered in this study, and interactions closer than 10 kb were excluded from analysis.
Definition of clusters from Hi-C data for the results in Figure 6E
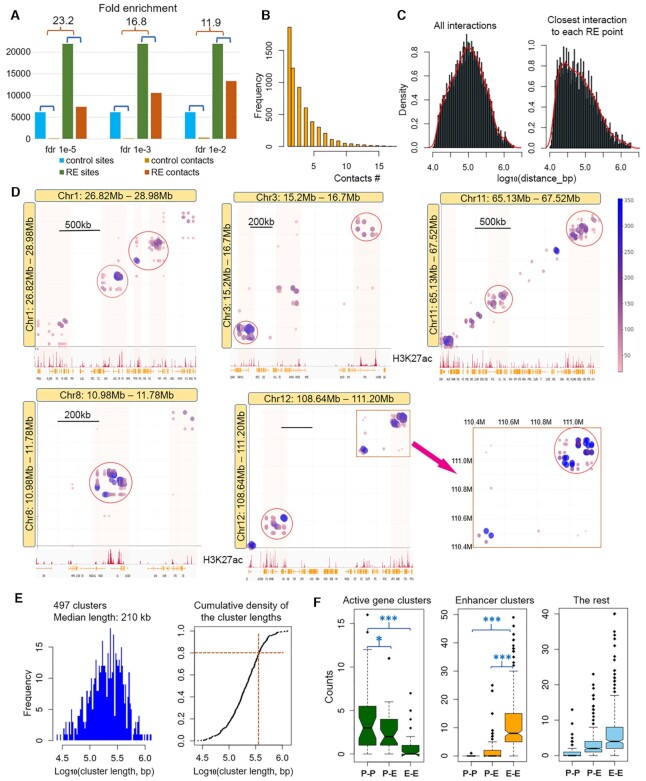
Figure 6.
Validation of the model with high resolution Hi-C data of GM12878 cell line. (A) Fold enrichment of interactions among strong regulatory elements (REs) compared to control sites, at different false discovery rate (FDR) values of filtering Hi-C data. FDR of 1e–3 is used for the of analysis. (B) Histogram of multiple contacts. (C) Density plots of distances between interacting pairs. (D) Visualization of clustered interactions among linearly close regulatory elements (active promoter and strong enhancers). Color bar shows the number of reads covering the interacting pairs. Some interacting clusters are circled in red. (E) Left: Distribution of linear genomic lengths of the 497 interacting clusters. Right: Cumulative density plot of the cluster lengths. Orange lines indicate 80% clusters have a length less than 350 kb. (F) Number of promoter–promoter (P–P), promoter–enhancer (P–E) and enhancer–enhancer (E–E) interacting pairs in three different groups of clusters: active gene clusters, enhancer clusters (SEs) and the rest.
An interacting cluster is a genomic region containing ≥4 interacting RE pairs and with on average ≥2 interacting pairs per 100 kb. Within a cluster, each interacting RE pair spans a region overlapping with at least one other RE pair or, share an RE site with at least one other RE pair (which means an RE site interacts with more than one other RE sites), so the cluster agrees with the flower-shape structure we proposed in Figure 5.
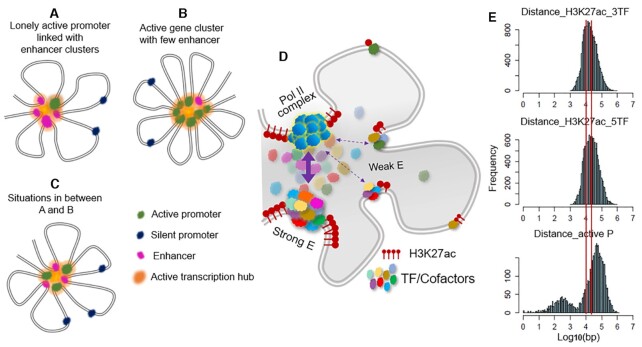
Figure 5.
A model of active transcription hub that unifies the roles of enhancer and active promoter. A–C: An active promoter and an enhancer can be considered similar types of regulatory elements, and gene transcriptions occur at active transcription hubs that consist of multiple linearly closely located elements brought into spatial proximity through DNA looping. (A) Isolated promoter with enhancer cluster. (B) Active multigene cluster with few enhancers. (C) Hybrid between A and B. (D) Active transcription hubs are dynamic with different enhancers and/or promoters contributing differently. (E) Histogram of distances between adjacent active promoters/enhancers.
RESULTS
We started with data from mouse embryonic stem cells (ESC) and mouse embryonic fibroblast (MEF) cells because these two cell types are usually prepared from early embryonic mouse tissues and are not transformed or exogenously immortalized cell lines that could differ greatly from their tissue of origin both genetically and phenotypically (23–25). We selected TF-ChIP data from public database based on the quantitative metrics of FRiP > 0.1 and NRF >0.8 on a minimum of 10M uniquely mapped reads for human and mouse genome (26). All of the datasets that we used for the presented results (RNA-seq, chromatin immunoprecipitation sequencing (ChIP-seq) of epigenetic marks, and proteins involved in transcription and regulation) are summarized in Supplementary Table S1. The high-quality data of RNA-seq and ChIP-seq of >20 TF/protein factors and histone marks enabled us to confidently identify putative enhancer elements with widely different levels of TF-binding enrichment (overlapping TF ChIP-seq peaks, not TFBS motif), reflecting enhancer strength, and to make an association between strong enhancers and linked genes.
25–30% of H3K27ac sites are highly enriched with TF binding
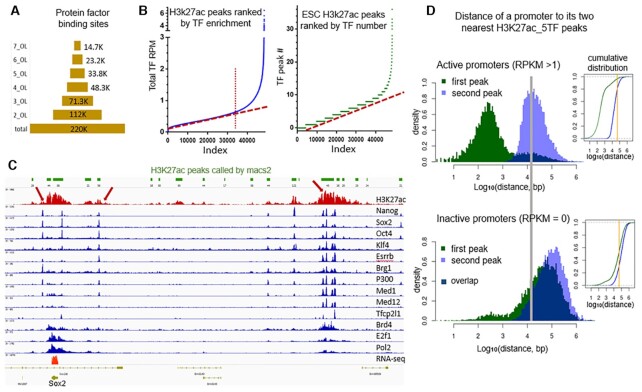
By aligning the ChIP-seq data of H3K4me3, H3K27ac, and all of the protein factors, we found that, although there are ∼40k to 100k peaks for each data set, only 15–30% of the peaks of each overlap multiple other factors (Supplementary Figure S1). Figure 1A shows the number of genomic sites bound by different numbers (1 to 7) of TFs in ESC. The genomic sites that are bound simultaneously by all major factors number in the range of ∼12k to 14k and are likely to be strong regulatory sites. This is in agreement with the concept that TFs seldom act alone and that a functional enhancer is often the result of the coordination among multiple TFs and other factors (27–29). Shown in Figure 1B are the H3K27ac peaks ranked by TF enrichment in mouse ESC. The total number of H3K27ac peaks is ∼48 000, yet <30% of H3K27ac peaks are highly enriched with TFs. In the example shown in Figure 1C, among ∼20 H3K27ac peaks, more than half of these peaks show no or little TF enrichment. All three H3K27ac regions highly enriched with TF binding (marked with red arrows) are experimentally validated enhancer sites (30,31), and the strongest one, which is ∼130 kb away from Sox2, has the greatest TF enrichment and is responsible for 90% of Sox2’s expression (31). Therefore, in the later quantitative analysis in this study, instead of assigning an enhancer to any called H3K27ac peak (not overlapping an active promoter), we mark the peaks with H3K27ac_nTFs. In order words, we assign how likely an H3K27ac peak region acts as effective enhancer by considering the number of TFs involved. By comparison, most studies in the field use all H3K27ac peaks, sometimes in combination with DHS or H3K4me1 peaks, as enhancers. We would like to mention that some previous studies such as Whyte et al. (15) and Hnisz et al. (16) showed plots of single signals like Med1 on enhancers with a similar shape to Figure 1B. However, their point is to use only the vertical part on the very right end of the curve to define super enhancers. Our message in the current study is clearly different.
Figure 1.
Enrichment of TFs at a small number of regulatory sites and active genes are associated with nearby enhancers. (A) Number of genomic sites that have different levels of TF enrichment (OL: overlapping). (B) H3K27ac peaks ranked by TF-peak enrichment; left: by total overlapping TF-signal intensity; right: by total number of overlapping TFs. (C) An example of enhancers associated with the only highly expressed gene nearby and TF-enrichment as reflecting enhancer strength. (D) Left: Histogram of the nearest (green) and the second nearest (purple) H3K27ac_5TF to the TSS of active genes (RPKM>1) and inactive genes (RPKM = 0) in mouse ESC. Dark blue area marks the overlapping part of the two peaks. Right: Cumulative distribution plot of the same data, green the nearest and blue the second nearest H3K27ac_5TF. The orange line marks a distance of 50 kb.
Active genes are associated with nearby H3K27ac peaks
Although enhancers can be a large distance away from their target promoters, we observe that active genes are associated with nearby H3K27ac peaks. For active genes with RPKM >1, the distance between their transcription start site (TSS) and the two nearest H3K27ac_5TF peaks (i.e. H3K27ac peak that overlaps at least five TF peaks) in mouse ESC is shown in Figure 1D (MEF cell results are similar). The first peak reflects the strong H3K27ac signal at promoters of active genes, and the second peak shows a preferred distance of ∼10–30 kb. Additionally, 80% of the second peaks are within 50 kb away from TSS (indicated with the orange line in the cumulative distribution plot). In this study, H3K27ac peaks overlapping promoter regions (the window of 2 kb upstream to 1 kb downstream of a TSS) are not considered as enhancers. For inactive genes (RPKM = 0), the distance between their promoters and the two nearest enhancer shows peaks at ∼60 kb and 200–250 kb. So far, most of the in vivo experimentally validated enhancer-gene pairs have their target genes as nearby highly expressed genes (30–36), although the nearest annotated gene might not. Again, as shown in the Figure 1C example, for all three strong enhancers, Sox2 is the only highly expressed gene nearby, although there are several other annotated genes in the region that are silent. An active gene with an enhancer nearby does not necessarily mean the enhancer works on that gene. Establishment of any enhancer-gene pair requires experimental validation.
Super enhancers (enhancer clusters) are mostly linked to isolated active promoters, regardless of the gene's cell-type specificity
Super enhancers (SE) are characterized with long stretches or clusters of H3K27ac peaks that are densely occupied by master regulators (15,16). The program ROSE was developed to identify SEs in any cell type based on H3K27ac ChIP-seq data (15). Originally, 231 SEs were identified in mouse ESC (15). The ROSE program was applied to our deep coverage ChIP-seq data of H3K27ac and resulted in 1,281 SEs. We further aligned the identified SEs on the ChIP-seq data of p300, Med1 and pluripotency-specific regulators Nanog, Oct4, Sox2, Klf4, Esrrb, Tfcp2l1. We then selected the SEs that contain at least two sites simultaneously bound by all of the major factors (see Methods for the details of SE selection). The additional filtering resulted in 893 super enhancers in mouse ESC.
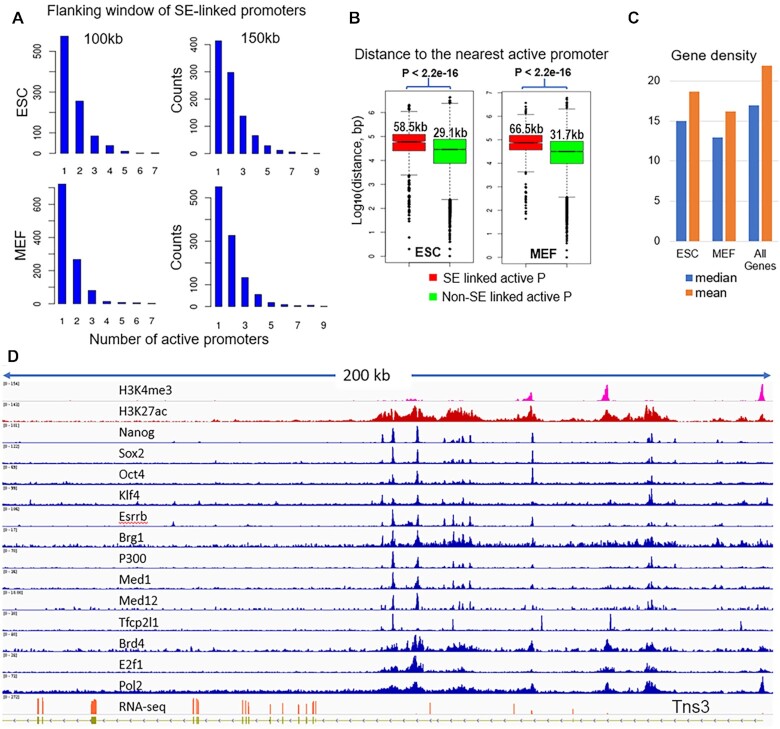
We found that a universal feature of the genes associated with SEs is that these genes usually are the only one or one of the two genes actively transcribed in the flanking region (∼100–150 kb). Figure 2A shows the distribution of active promoter counts within the 100 and 150 kb flanking windows of the SE-linked promoters. 86% and 74% of the 100kb and 150kb windows in ESC have only one or two active promoters, thus we call them isolated active promoters in this study. Frequently, there are other annotated genes in the region, but these are not expressed. We compared the distance to the nearest active promoter (RPKM > 1) from the SE-linked to that from non-SE-linked promoters and the former is significantly bigger (P < 2.2e–16, Wilcoxon rank sum test, Figure 2B). The median distance to the nearest active promoter from ESC SE-linked promoters is 58.5kb while that from non-SE-linked active promoters is 29.1 kb. We manually screened the highly expressed (RPKM > 10) genes possibly linked to SE(s) and the gene-SE pairs are listed in Supplementary Table S2. In addition to these well studied pluripotency master regulators (e.g. Nanog, Oct4, Sox2, Klf4, Esrrb), many non-ESC specific genes or housekeeping genes are surrounded by SEs, e.g. Tns3, Rcf5, Klf9, Ptma (Figure 2D and Supplementary Figure S2). The long stretch of H3K27ac and high TF enrichment of the SEs linked to non-ESC specific or housekeeping genes are comparable to those that surround the ESC master regulators.
Figure 2.
Enhancer clusters (or super enhancers) are mostly linked to isolated active promoters. (A) Distribution of the numbers of active promoters (RPKM > 1) within 100 and 150 kb flanking window of putative SE-linked promoters in ESC and MEF cells. When the x-axis value is 1, the SE-linked promoter is the only one in the region. (B) Distance to the nearest active promoters from SE linked promoters (red) and non-SE linked promoters (green) with the numbers as the median distance in each group. (C) Gene density in the flanking 1 Mb window of SE-linked genes in ESC, MEF and all annotated genes. (D) An example of enhancer clusters around a non-ES specific gene, Tns3, in mouse ESC.
SEs are generally referred to as defining the genes that determine cell lineage. When used on the deep coverage H3K27ac data here, the identified genes (by ROSE) extend to many housekeeping genes. Therefore, we will use the term enhancer cluster (EC), instead of super enhancer, for those identified by ROSE on the deep coverage data sets in this study. Similarly, our analysis of the data from MEF cells showed the same result. There are ∼900 ECs called with the ROSE program and further filtering of multiple TF binding. The universal feature of the active genes associated with ECs is that there is no or only one other active promoter within the flanking ∼100–150 kb window of its TSS (Figures 2A). The putative gene and EC pairs in MEF cells are also listed in Supplementary Table S2. Supplementary Figures S3 shows the examples of EC linked to non-MEF specific genes Wdr75, Tpt1, Rbpj and others.
We further examined the gene density around SE-linked genes to see whether those genes are in gene-poor regions defined at mega-base length scales, which usually contain less than five or six genes per Mb (37,38). The average gene number within the 1Mb flanking window is 18.7 and 16.2 for SE-linked genes in ESC and MEF whereas it is 21.9 for all mouse genes. The median gene number is 15 and 13 in ESC and MEF whereas it is 17 for all mouse genes (Figure 2C). Therefore, most of SE linked genes are not in gene-poor regions.
Active multigene clusters have few distant enhancers
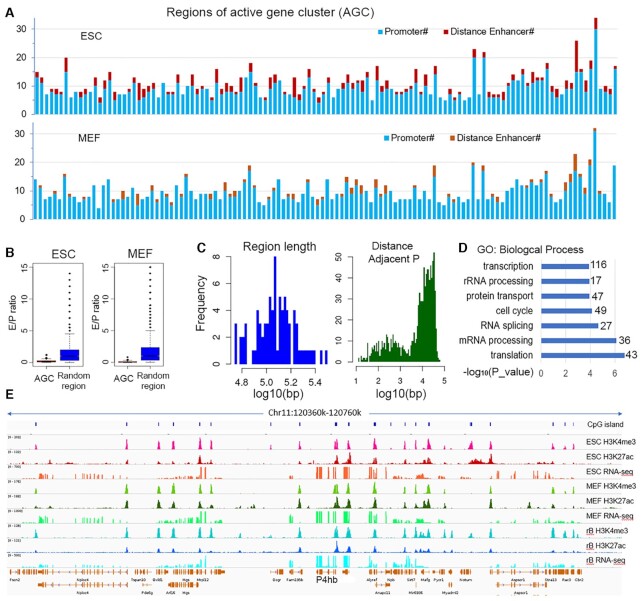
Interestingly, in contrast to the situation of single active gene(s) with enhancer clusters, we noticed that, at some genomic regions with densely packed active genes, there are usually few distant enhancers. We aligned the data from resting B cell with ESC and MEF cells together and noticed that most of these genes are active in all three cell types. To systematically examine the situation, we screened the whole genome for regions that have multiple active promoters with adjacent ones <40 kb apart. We identified 120 such active gene clusters (AGC) with ≥6 active promoters in each cluster. The 120 AGCs include ∼1050 genes that account for ∼10% of total active genes. Figure 3A shows the number of active promoters (RPKM > 1) and strong enhancers (H3K27ac_4TF) in these regions: Active promoters greatly outnumber the distant enhancers in both ESC and MEF cells. As a control, 10000 non-overlapping genomic regions of length ranging from 50 to 300 kb were randomly sampled and we selected the ∼3000 containing at least one active promoter (RPKM > 1). We examined the enhancer density (number of distant enhancers every 10 kb). In ESC the enhancer densities in AGC regions are overall higher than in the randomly selected regions while in MEF they are lower (Supplementary Figure S4A). However, the ratio of enhancers to active promoters in AGC regions are significantly lower compared to random regions in both cell types (P < 2.2e–16, Wilcoxon test) (Figure 3B). The length of the 120 AGC regions ranges from 60 to 320 kb, with a medium length of 120 kb, and the distance between adjacent active promoters is mainly 10–30 kb (Figure 3C). Remarkably, these genomic regions are enriched in housekeeping genes in the categories of basic cellular functions, such as transcription, mRNA processing, and protein transportation (Figure 3D). Figure 3E shows an example of active multigene clusters at chr11. The strong H3K27ac peaks in this type of region are almost always located around gene promoters (additional examples are shown in Supplementary Figure S4B).
Figure 3.
Few distant enhancers at active multigene clusters. (A) The number of active promoters (in blue) and distant enhancers (red for ESC and orange for MEF cells) at 120 AGC regions with ≥6 active promoters. Each bar represents one region. (B) Comparison of the ratio of distant enhancers to active promoters in AGC and random regions. (C) Distribution of length of the AGC regions (left) and distance between adjacent active promoters (right). (D) GO analysis of the genes. The numbers on the right are the gene counts in each GO category. (E) An example of active multigene clusters at chr11. From the top to bottom rows are CpG island, H3K4me3, H3K27ac and RNAseq signals of mouse ESC (top), MEF cell (middle), and resting B cells (bottom).
The number of active promoters and distant enhancers are inversely related
What we described above are two typical states—one (or two) active promoters linked with multiple enhancers compared to a state of multiple active promoters with few enhancers. This prompted us to examine the number of promoters and enhancers at actively transcribed regions, genome wide. We selected genomic windows of 200 kb centered at TSSs of the top 20% most highly expressed genes, and windows with centers <50 kb apart are combined, as shown in Figure 4A. The selection results in ∼1500 windows. We examined the expression levels of the genes whose promoters are located within these windows. These genes account for 70% and 76% of total mRNA expression of all the annotated genes in ESC and MEF, respectively. We then counted the number of active promoters and enhancers in these windows. We regard active promoters as those of genes with expression level >1RPKM. The enhancers are H3K27ac peaks that overlap at least four TF peaks, as H3K27ac_4TF, for mouse ESC and MEF cells (see Methods for the details of window and regulatory sites selection). Figure 4B shows plots of the number of enhancers versus the number of promoters in the windows that have ≥5 H3K27ac_4TF peaks. The color scheme represents the number of windows at each data point, as there can be multiple regions with the same number of promoters and enhancers. For example, there are 25 regions with one promoter, five enhancers and 47 regions with two promoters, and four enhancers in mouse ESC (Figure 4B, left panel). Approximately one quarter of regions have <5 H3K27ac_4TF peaks (including promoters), which are not included in the plots.
There is an overall inverse correlation between the number of promoters and enhancers in our selected active transcribed regions (Figure 4B): When there are only one or two promoters, often large number of enhancers are nearby, which is the case of isolated highly expressed genes associated with enhancer clusters. As the number of nearby active promoters increases, the number of enhancers decreases. When there are densely packed active promoters, e.g. >15 transcribed genes in a 200–300 kb region, there are few enhancers.
As stated earlier, we define the ‘strength’ of an enhancer with the number of overlapping TF peaks. Sites bound by more TFs presumably are ‘stronger’ than sites bound by less TFs, and we assume that this is similar for active promoters. For the same regions selected above, we calculated the number of TF peaks at each promoter and at each enhancer as a semi-quantification of the strength of a regulatory element. The sum of TF peaks of all enhancers is plotted against the sum of TF peaks of all promoters at each window (Figure 4C). The total ‘strength’ of promoters and enhancers at those windows also shows an inverse relationship.
We further analyzed the data from a few human cell lines for which many TF ChIP-seq data sets are available and observed similar trends for the numbers of promoters and enhancers (GM12787 and Hela3 in Figures 4B and C). These results were obtained by filtering the H3K27ac peaks with TF binding; only those with substantial TF enrichment are counted as enhancers. If we take all of the H3K27ac peaks as enhancers without considering the TF binding and perform the same calculation, the inverse relationship between the number of promoters and enhancers is not apparent (Supplementary Figure S5).
We considered if the same patterns observed in human and mouse genome also hold in lower organisms like Drosophila. Most studies about enhancers in Drosophila have been conducted at the whole organism level and investigate developmental processes. For our analysis, we choose the S2 cell line, the most used drosophila cell line. Since there are few TF or protein factor ChIP-seq data sets available in Drosophila cell lines, we used only H3K27ac signals as indicators of active enhancers. Drosophila has a compact genome. The gene lengths and intergenic distances of Drosophila is about one tenth of that of human or mouse (Supplementary Figure S6A). At window lengths of 50 or 100 kb, there is no obvious inverse relationship between the number of promoters and enhancers in Drosophila S2 cell line (Supplementary Figure S6B). At the majority of actively transcribed regions active promoters outnumber enhancers. The total number of enhancers in S2 cell is ∼5000 (based on H3K27ac peaks non-overlapping promoters) is less than that of active promoters, ∼9000 (RPKM > 1). The compactness of Drosophila genome makes a H3K27ac peak of several kilobases at a promoter region easily cover a large fraction of the whole gene and sometimes extend to the next gene (Supplementary Figure S6C). This explains, at least partially, why the number of distant enhancers in Drosophila is small, compared to tens of thousands in human/mouse genome. We postulate that large number of closely located active promoters greatly reduces the need for enhancers.
A model of active transcription hub that unifies the roles of active promoter and enhancer
Emerging evidence suggests that transcription occurs in phase-separated biomolecular condensates with dynamic features (39–44), which forms a local high concentration of protein components required for effective transcription. This was demonstrated at some super enhancer loci in mouse ESC (41). In addition, the transcriptive function of enhancers and the similarity between enhancers and promoters have been extensively investigated (45–49). Based on these two lines of study and our results here, we propose a simple model of transcription organization that unifies the role of promoters and enhancers: Gene transcriptions, especially high-level transcriptions, occur mainly at local active transcription hubs, in which a high concentration of required protein components come from multiple promoters and/or enhancers, which are linearly close to each other and are brought into three-dimensional (3D) proximity through looping. The model can be schematically represented with a flower-shaped structure (Figure 5).
Our results suggest that one promoter distant from other active promoters is rarely able to perform a task of high transcription by itself. It requires the ‘assistance’ of enhancers, characterized by multiple TF co-binding, and often long stretches of H3K27ac modification, to maintain an open chromatin structure. These sites are often identified as super enhancers or enhancer clusters (Figure 5A). As the number of nearby active promoters increases, the demand for the ‘assistance’ from enhancers decreases. In gene cluster regions where numerous genes are actively transcribed, active promoters are sufficient to create an active transcribing environment, largely precluding the need for enhancers (Figure 5B). A large fraction of transcribed regions can be considered hybrids between these two states (Figure 5C). Underlying this model is a concept not fully explored yet: an active promoter that is bound by transcription machine (e.g. Pol II complex with its co-factors, Mediator complex) may function as an enhancer to another active promoter nearby. Thus, a ‘lonely’ active promoter needs enhancer cluster, and multiple closely located active promoters obviate the need for enhancers, reflected by an overall inverse relationship between the number of promoters and enhancers.
We emphasize that active transcription hubs are dynamic, not a stable looping structure (although we draw the model thus due to the limits of 2D plotting). Interactions between active promoters and/or enhancers are essentially formed between protein components, and the formed biomolecular condensates or clusters range widely in average lifetime and size, with individual components phasing in and out dynamically (see live imaging (39,41,44,50,51)). Figure 5D shows some dynamic features of the model. For example, in the situation of one active promoter linked with an enhancer cluster, a strong enhancer featured by high-TF enrichment would interact with the promoter complex more strongly and more frequently than a weak enhancer, while all of the elements are dynamically part of the active hub.
We further examined the distance between adjacent regulatory elements (promoter and/or enhancers) in our model. For H3K27ac peaks above a certain threshold of TF enrichment (H3K27ac_3TF or H3K27ac_5TF, in mouse ESC and MEF cells), regardless of whether they are promoters or enhancers, the distribution of distance between adjacent elements exhibits a preference of ∼10–30 kb. The distance between adjacent active promoters (RPKM > 1) shows a peak at ∼50–60 kb (Figure 5E). It is possible that there is a plausible periodicity in chromatin looping, and when adjacent active promoters are too far apart, enhancers are more likely to form in between.
Validation of the model with Hi-C data
In order to validate the principle of our model that linearly close regulatory elements, active promoters, or strong enhancers form interacting clusters (the active transcription hubs), we searched for Hi-C data sets that meet two conditions: the first is that a high resolution enables confident detection of interactions between sites <40 kb apart, and the second is that multiple TF ChIP-seq data are available so that we can identify strong regulatory elements based on TF enrichment. A data set that meets the above two conditions is the Hi-C data of GM12878 cell line from the study by Rao et al. (17) and is used in the current study.
We used the ChIP-seq and RNA-seq data of the GM12878 cell line from the ENCODE study (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/encodeDCC/) and selected active transcribing regions (823 non-overlapping regions with expression level >100CPM, spanning a length of 0.6–13 Mb) and strong regulatory elements (REs) based on TF enrichment in these regions. Altogether, ∼22000 REs were selected, including ∼8600 promoters. We also selected ∼6000 control sites (no TF binding or H3K27ac) located between or next to the REs (see Methods for the details of active region and RE selection). We then examined all of the pairwise interactions among all of those sites. Figure 6A shows the number of interacting pairs among controls and REs versus the total number of control and RE sites. There is great enrichment of interactions among REs compared to control sites. At three different FDR thresholds of filtering Hi-C interactions, 1e–5, 1e–3 and 1e–2, the enrichment fold is 23.2, 16.8 and 11.9 (P-value = 5e–324, binomial test), respectively. Those interactions include ∼5% promoter–promoter, ∼21% promoter–enhancer and ∼74% enhancer–enhancer interactions (Supplementary Figure S7A). Because a change of the FDR threshold has marginal effects on our results, we used FDR 1e–3 for the remaining analysis, shown in Figure 6B–D.
We found that 68% of the RE sites with detected interactions interact with multiple other REs (Figure 6B). The distribution of distance between any pair of interacting sites shows a peak at ∼100 kb, and ∼90% is below 300 kb (Figure 6C, left panel). The distribution of distances between each RE site and its closest interacting partner shows a peak at ∼30 kb (Figure 6C, right panel), which is consistent with the earlier results calculated from data in mouse ESC and MEF cells, shown in Figure 5E. Figure 6D provides a visualization of interactions among the RE points at a few genomic regions. It demonstrates the clustering feature of interactions among active promoters and strong enhancers closely located. Some clustered interactions are circled with red in Figure 6D. These results strongly imply the presence of active transcription hubs and the underlying flower-shaped structure that we proposed in Figure 5. We further quantified the interacting clusters derived from the Hi-C data (see Materials and Methods part for definition of interacting clusters). From the ∼22000 selected RE sites we identified 497 clusters. The length of genomic regions each cluster spans ranges from 26 kb to 1.2 Mb with a median length of 210 kb. 80% cluster regions are less than 350 kb long (Figure 6E). We separated these regions into three groups: (i) active gene clusters (AGC) containing ≥6 active promoters with an average density of ≥1 promoter per 40 kb; (ii) enhancer clusters with one or two promoters; (iii) the rest. We examined the interacting pairs in the three groups. In AGC regions, there are significantly more promoter–promoter (P–P) interactions than the promoter–enhancer (P–E) and the enhancer–enhancer (E–E) interactions, while at enhancer cluster group there are mainly E–E and P–E interactions. In the third group, the situation is intermediate (Figure 6F). We did the same analysis in the K562 cell line and observed similar patterns. Due to the much lower read depth, however, the number of detected contacts from K562 is less than one-quarter of that from GM12878; nonetheless, there is a great enrichment of interactions in REs compared to control points (Supplementary Figures S7B and C).
Another line of supporting evidence for our model is the extensive promoter-promoter interactions revealed by promoter capture Hi-C studies (52–55). Supplementary Figure S8A provides an example of dense interactions among promoters in an active gene cluster where there are few distant enhancers (data from (53)). Most of the promoter-based interactions have a distance <200 kb (Supplementary Figure S8B). A possible explanation for the P–P interactions is that not only does an active promoter, bound by the basic transcription machine and multiple TFs, drive transcription, but also the proximity of multiple promoters is likely to generate a local active transcription hub with a high concentration of protein factors required for transcription. In this sense, active promoters in close spatial proximity might be considered enhancers for each other.
DISCUSSION
We demonstrated there is only a small percentage of putative enhancer sites co-bound by multiple TFs in spite of the widespread TF ChIP-seq peaks. Because a functional regulatory site often requires combinatorial actions of multiple protein factors (27–29), that may explain why the predicted regulatory sites based on one or two enhancer markers frequently show no enhancer function (11,12). The millions of candidate enhancers predicted with only H3K27ac or one or two additional markers could greatly outnumber those that are functional. Certainly, ChIP-seq data are now available for only ∼200 of ∼3000 of all human/mouse TFs. Our knowledge of the TF-binding landscape that relates to gene regulation will keep expanding as new data become available.
The concept of an active chromatin hub was suggested in 2003 (56) based on the study of the β-globin enhancers. The review article proposed that enhancer(s) and the target promoter are brought into physical proximity through chromatin looping (with silent genes looping out) as a basic mechanistic framework that underlies gene expression. This has evolved into a dynamic ‘hub and condensate’ model as studies in transcription condensates are gaining in evidence (39–42,57): Accumulation of Pol II, Mediator complex, transcription factors and other cofactors through liquid-liquid phase separation forms an active hub for gene transcription. The existing hub model, however, generally addresses the regulation of a single promoter with its enhancers, both promoter and enhancer in a traditional sense.
The active transcription hub model that we propose here incorporates several additional concepts: There is a unified role for active promoters and enhancers. Enhancers can drive transcriptions (45–49,58), whereas promoters also can function as enhancers (59,60). A study in Drosophila, using random insertion of reporter constructs, found that the activity of housekeeping gene promoters depends on the expression of their neighbors (61). This study suggests that active promoters in 3D proximity might function as enhancers for each other. There are about 8000–12 000 active genes (RPKM > 1) in a living cell and about 3000–7000 highly transcribed genes (RPKM > 10). In the research of Sabari et al. (41), ∼1000 puncta (fluorescent dots observed under the microscope) that contain Med1 or Brd4 were observed in mouse ESC. In Cho et al.’s (39) research, a few hundred Pol II and Mediator clusters with different sizes and lifetimes are observed, with the largest clusters as containing ∼200–400 molecules. High concentration of TFs, Pol II and cofactors at these condensates or clusters can be from enhancer clusters or multiple promoters (an active multigene cluster), whose transcriptions are highly coordinated, or a mix of promoters and enhancers. A recent study that uses multiway-4C detected two active genes simultaneously accommodated in the β-globin enhancer hub (62).
Another essential question of the transcription hub is where the promoters/enhancers that form a hub come from. Can they initiate from any site in a chromosome, as it is generally thought that an enhancer can act independent of distance? Using Hi-C data, we demonstrated that clustered and multiway (i.e. one element in contact with multiple other elements) interactions are formed among linearly close regulatory elements of active promoters or enhancers, mostly within 200–300 kb. This is consistent with experiments that showed the decoupling between TAD structure and the major part of transcriptome (8–10): The regulation of most genes happens at sub-TAD, or sub-sub-TAD scale. The multi-component of a transcription hub might partially underlie the robustness of transcriptome: Disturbance of an individual enhancer frequently has no effect on gene expression (63).
Complicated interactions among regulatory elements have been commonly observed in numerous 5C/Hi-C/promoter capture Hi-C experiments (1,4,54,64). To our knowledge, no previous Hi-C studies have reported or proposed how those interactions are further organized, besides that they usually occur within topologically associated domains (TADs). Our work proposes a model of how complicated interactions among promoters/enhancers are spatially organized into regulatory clusters. Our results, combined with those of experimental studies of transcription condensates, shed light on the essential question of E–P communication. TFs have the ability to form transcription activating phase-separated clusters with themselves or with Mediator complex through the TFs’ activation domain (AD) that contains low-complexity sequences (42,44). This could be a driving force to bring an active promoter and an enhancer (or two active promoters or two enhancers) together. The frequency at which two genomic loci encounter each other is inversely proportional to their linear distance (65). Therefore, linearly close active promoters and/or enhancers are much more likely to be brought into spatial proximity by their bound proteins than is the case with a distant element. The E–P communication might essentially be the ability of TFs and other transcription-related proteins to form clusters in a crowded nucleus. Non-expressed genes in the genomic regions of an active transcription hub might be due to repressive histone markers and repressive proteins bound at the promoters to prevent their participation in an active hub.
CONCLUSION
In this study, we identified a portion of histone H3K27ac peaks as potential strong enhancers, featured by combinatorial binding of multiple protein factors. We revealed an overall inverse relationship between the number of active promoters and strong enhancers in highly transcribed regions, which implies a general enhancer-like function of active promoters. We propose a general principle of spatial organizations of active promoters and enhancers: The basic units of gene transcriptions and regulations are local active transcription hubs consisting of multiple elements from linearly close enhancers or active promoters dynamically brought into 3D proximity through DNA looping. Our results provide explanations for the uncoupling of the transcriptome and TAD scale chromatin architecture and the transcriptome robustness that is resistant to disturbance of individual enhancers.
DATA AVAILABILITY
The sources and accession numbers of all the data sets, including ChIP-seq, RNAseq and Hi-C data, used in this study is summarized in Supplementary table S1.
Supplementary Material
Contributor Information
Iris Zhu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20892, USA.
Wei Song, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20892, USA.
Ivan Ovcharenko, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20892, USA.
David Landsman, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20892, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program at the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program at the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dixon J.R., Gorkin D.U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Plank J.L., Dean A.. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell. 2014; 55:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Symmons O., Pan L., Remeseiro S., Aktas T., Klein F., Huber W., Spitz F.. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell. 2016; 39:529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schoenfelder S., Fraser P.. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019; 20:437–455. [DOI] [PubMed] [Google Scholar]
- 5. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R.et al.. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franke M., Ibrahim D.M., Andrey G., Schwarzer W., Heinrich V., Schopflin R., Kraft K., Kempfer R., Jerkovic I., Chan W.L.et al.. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016; 538:265–269. [DOI] [PubMed] [Google Scholar]
- 8. Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D.et al.. Cohesin loss eliminates all loop domains. Cell. 2017; 171:305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N.A., Huber W., Haering C.H., Mirny L.et al.. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017; 551:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghavi-Helm Y., Jankowski A., Meiers S., Viales R.R., Korbel J.O., Furlong E.E.M.. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 2019; 51:1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gasperini M., Hill A.J., McFaline-Figueroa J.L., Martin B., Kim S., Zhang M.D., Jackson D., Leith A., Schreiber J., Noble W.S.et al.. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019; 176:1516. [DOI] [PubMed] [Google Scholar]
- 12. Barakat T.S., Halbritter F., Zhang M., Rendeiro A.F., Perenthaler E., Bock C., Chambers I.. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell. 2018; 23:276–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J., Li K., Cai W., Liu X., Zhang Y., Orkin S.H., Xu J., Yuan G.C.. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun. 2018; 9:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W., Gong K., Li Q., Alber F., Zhou X.J.. Hi-Corrector: a fast, scalable and memory-efficient package for normalizing large-scale Hi-C data. Bioinformatics. 2015; 31:960–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A.. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012; 9:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng C., Fu L.Y., Dong P.F., Deng Z.L., Li J.X., Wang X.T., Zhang H.Y.. The sequencing bias relaxed characteristics of Hi-C derived data and implications for chromatin 3D modeling. Nucleic Acids Res. 2013; 41:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ay F., Bailey T.L., Noble W.S.. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014; 24:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alge C.S., Hauck S.M., Priglinger S.G., Kampik A., Ueffing M.. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J. Proteome Res. 2006; 5:862–878. [DOI] [PubMed] [Google Scholar]
- 24. American Type Culture Collection Standards Development Organization Workgroup ASN. Cell line misidentification: the beginning of the end. Nat. Rev. Cancer. 2010; 10:441–448. [DOI] [PubMed] [Google Scholar]
- 25. Pan C., Kumar C., Bohl S., Klingmueller U., Mann M.. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics. 2009; 8:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P.et al.. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 28. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 175:598–599. [DOI] [PubMed] [Google Scholar]
- 29. MacArthur S., Li X.Y., Li J., Brown J.B., Chu H.C., Zeng L., Grondona B.P., Hechmer A., Simirenko L., Keranen S.V.et al.. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009; 10:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y., Rivera C.M., Ishii H., Jin F., Selvaraj S., Lee A.Y., Dixon J.R., Ren B.. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014; 9:e114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou H.Y., Katsman Y., Dhaliwal N.K., Davidson S., Macpherson N.N., Sakthidevi M., Collura F., Mitchell J.A.. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014; 28:2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blinka S., Reimer M.H. Jr, Pulakanti K., Rao S.. Super-enhancers at the nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep. 2016; 17:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shin H.Y., Willi M., HyunYoo K., Zeng X., Wang C., Metser G., Hennighausen L.. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016; 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moorthy S.D., Davidson S., Shchuka V.M., Singh G., Malek-Gilani N., Langroudi L., Martchenko A., So V., Macpherson N.N., Mitchell J.A.. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017; 27:246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hay D., Hughes J.R., Babbs C., Davies J.O.J., Graham B.J., Hanssen L., Kassouf M.T., Marieke Oudelaar A.M., Sharpe J.A., Suciu M.C.et al.. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016; 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wall L., deBoer E., Grosveld F.. The human beta-globin gene 3' enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 1988; 2:1089–1100. [DOI] [PubMed] [Google Scholar]
- 37. Dunham A., Matthews L.H., Burton J., Ashurst J.L., Howe K.L., Ashcroft K.J., Beare D.M., Burford D.C., Hunt S.E., Griffiths-Jones S.et al.. The DNA sequence and analysis of human chromosome 13. Nature. 2004; 428:522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K.et al.. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005; 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse I.I.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018; 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cisse I.I., Izeddin I., Causse S.Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., Darzacq X.. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013; 341:664–667. [DOI] [PubMed] [Google Scholar]
- 41. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018; 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henriques T., Scruggs B.S., Inouye M.O., Muse G.W., Williams L.H., Burkholder A.B., Lavender C.A., Fargo D.C., Adelman K.. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018; 32:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andersson R., Sandelin A., Danko C.G.. A unified architecture of transcriptional regulatory elements. Trends Genet. 2015; 31:426–433. [DOI] [PubMed] [Google Scholar]
- 47. Mikhaylichenko O., Bondarenko V., Harnett D., Schor I.E., Males M., Viales R.R., Furlong E.E.M.. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018; 32:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tippens N.D., Vihervaara A., Lis JT.. Enhancer transcription: what, where, when, and why. Genes Dev. 2018; 32:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis JT.. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014; 46:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen H., Levo M., Barinov L., Fujioka M., Jaynes J.B., Gregor T.. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 2018; 50:1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen J., Zhang Z., Li L., Chen B.C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E.et al.. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014; 156:1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J.et al.. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cairns J., Freire-Pritchett P., Wingett S.W., Varnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.M., Osborne C.et al.. CHiCAGO: robust detection of DNA looping interactions in capture Hi-C data. Genome Biol. 2016; 17:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A.et al.. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015; 47:598–606. [DOI] [PubMed] [Google Scholar]
- 55. Schoenfelder S., Furlan-Magaril M., Mifsud B., Tavares-Cadete F., Sugar R., Javierre B.M., Nagano T., Katsman Y., Sakthidevi M., Wingett S.W.et al.. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015; 25:582–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de Laat W., Grosveld F.. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003; 11:447–459. [DOI] [PubMed] [Google Scholar]
- 57. Furlong E.E.M., Levine M.. Developmental enhancers and chromosome topology. Science. 2018; 361:1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F.et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dao L.T.M., Galindo-Albarran A.O., Castro-Mondragon J.A., Andrieu-Soler C., Medina-Rivera A., Souaid C., Charbonnier G., Griffon A., Vanhille L., Stephen T.et al.. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 2017; 49:1073–1081. [DOI] [PubMed] [Google Scholar]
- 60. Medina-Rivera A., Santiago-Algarra D., Puthier D., Spicuglia S.. Widespread enhancer activity from core promoters. Trends Biochem. Sci. 2018; 43:452–468. [DOI] [PubMed] [Google Scholar]
- 61. Corrales M., Rosado A., Cortini R., van Arensbergen J., van Steensel B., Filion G.J.. Clustering of Drosophila housekeeping promoters facilitates their expression. Genome Res. 2017; 27:1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Allahyar A., Vermeulen C., Bouwman B.A.M., Krijger P.H.L., Verstegen M., Geeven G., van Kranenburg M., Pieterse M., Straver R., Haarhuis J.H.I.et al.. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 2018; 50:1151–1160. [DOI] [PubMed] [Google Scholar]
- 63. Osterwalder M., Barozzi I., Tissieres V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S.et al.. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018; 554:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sanyal A., Lajoie B.R., Jain G., Dekker J.. The long-range interaction landscape of gene promoters. Nature. 2012; 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Arensbergen J., van Steensel B., Bussemaker H.J.. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014; 24:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sources and accession numbers of all the data sets, including ChIP-seq, RNAseq and Hi-C data, used in this study is summarized in Supplementary table S1.