Abstract
In Southeast Asia, surveillance at live bird markets (LBMs) has been identified as crucial for detecting avian influenza viruses (AIV) and reducing the risk of human infections. However, the design of effective surveillance systems in LBMs remains complex given the rapid turn-over of poultry. We developed a deterministic transmission model to provide guidance for optimizing AIV surveillance efforts. The model was calibrated to fit one of the largest LBMs in northern Vietnam at high risk of low pathogenic H7N9 virus introduction from China to identify the surveillance strategy that optimizes H7N9 detection. Results show that (i) using a portable diagnostic device would slightly reduce the number of infected birds leaving the LBM before the first detection, as compared to a laboratory-based diagnostic strategy, (ii) H7N9 detection could become more timely by sampling birds staying overnight, just before new susceptible birds are introduced at the beginning of a working day, and (iii) banning birds staying overnight would represent an effective intervention to reduce the risk of H7N9 spread but would decrease the likelihood of virus detection if introduced. These strategies should receive high priority in Vietnam and other Asian countries at risk of H7N9 introduction.
Keywords: transmission model, low pathogenic H7N9 avian influenza virus, surveillance strategies, live-bird markets
1. Introduction
New avian influenza A virus (AIV) strains continue to pose serious clinical and economic challenges to global public and animal health, and leading to substantial economic losses. Since 2013, a novel avian-origin H7N9 virus has emerged in eastern China, causing severe respiratory disease and fatalities in humans [1,2]. China experienced five epidemic waves from 2013 to 2017, during which the number of reported cases has increased significantly, reaching over 1600 human infections [3,4]. During the first four waves, the H7N9 AIV circulating in Chinese poultry were low-pathogenic (LP) with asymptomatic infection in poultry [4,5]. During the fifth wave in 2017, a highly pathogenic (HP) H7N9 AIV variant emerged [6–8]. Such rapid increase in the number of H7N9 human infections and the emergence of H7N9 as HP AIV variant have raised fear of a pandemic threat [9].
In Asia, live bird markets (LBMs) are known as high-risk places for the transmission, evolution and maintenance of AIV [10,11]. In China, most human H7N9 cases have been associated with previous exposure to poultry at LBMs [12–14]. In Chinese LBMs, H7N9 virus has been extensively detected in chickens [15,16], which are the primary source of H7N9 infection in humans [17,18]. Frequent interactions among different poultry species and humans at LBMs provide an ideal interface for transmission of AIV and emergence of new variants by mixing AIV from different sources [12,16,19].
Systematic surveillance at LBMs remains essential for detecting novel AIV and reducing the risk of human infections [16,20,21]. Optimizing AIV surveillance strategies in LBMs has mainly focused so far on identifying the most sensitive sample materials [11,22] and on increasing diagnostic assay sensitivity [23]. However, lack of knowledge remains on how the sampling design (i.e. sampling time and sample size) could be optimized to maximize the probability to detect AIV, given the rapid turn-over of poultry populations in LBMs. Moreover, the cost of surveillance, which is a key parameter for policy decision and surveillance design, should be taken into consideration when assessing different surveillance strategies, especially in low- and middle-income settings.
Given the sharp increase in the number of H7N9 outbreaks in China in 2013–2017 and regular cross-border trade of birds originating from China [24], the rapid detection of emerging AIV was crucial to minimize public health and economic impact in Vietnam. The current surveillance programme for H7N9 in Vietnam involved biweekly random sampling of chickens in LBMs with value chain linkages to China. All samples were transported to an official diagnostic laboratory where they were consecutively screened for M, H7 and N9 genes using RT-PCR. As an alternative to this surveillance strategy, a portable PCR device has been introduced recently to improve H7N9 detection and response capacities in Vietnam [25,26]. This device can be directly deployed in LBMs and allows virus detection within 7 h after sampling, while the laboratory-based surveillance protocol takes on average 72 h due to logistic reasons.
In the light of the introduction of this innovative diagnostic tool, the general objective of this study was to compare the weekly costs and effectiveness of different surveillance strategies at LBMs, with the view to identify the strategy that optimizes H7N9 detection. To do this, a deterministic transmission model of AIV including environmental shedding and bird trading was developed and calibrated to fit one of the largest LBMs in northern Vietnam at high risk of LP H7N9 introduction from China.
2. Material and methods
2.1. Structure of the within-LBM H7N9 transmission model
2.1.1. Dynamics of bird trading in the LBM
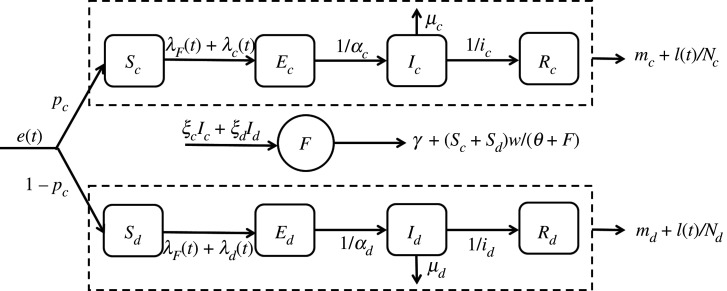
It was assumed that chickens and ducks were introduced into the LBM at an entry rate e(t) that depended on the time of the day, and moved out for slaughter and trade at a time-varying exit rate l(t) (figure 1; electronic supplementary material, appendix). The model formulation also allowed a proportion of the bird populations to stay overnight in the LBM and therefore to contribute potentially to the maintenance and amplification of the virus. Among the birds leaving the market, it was considered that a given proportion was traded to farms and other LBMs. These birds were assumed to be those representing the highest risk for secondary spread outside the studied LBM.
Figure 1.
Flux diagram of the full dynamic transmission model. Parameters involved in the formulation of transition rates between compartments are: e(t) being the entry rate, pc (resp. 1 − pc) being the proportion of chickens (resp. ducks), λF being the force of infection due to environmental contamination, λc (resp. λd) being the force of infection due to contacts with infectious chickens (resp. ducks), αc (resp. αd) being the average duration of the latent period in chickens (resp. ducks), µc (resp. µd) being the mortality rate due to H7N9 infection in chickens (resp. ducks), ic (resp. id) being the average duration of the infectious period in chickens (resp. ducks), mc (resp. md) being the natural mortality rate for chickens (resp. ducks), l(t) being the exit rate, Nc (resp. Nd) being the total number of chickens (resp. ducks), ξc (resp. ξd) being the excretion rate of infectious doses by chickens (resp. ducks), Ic (resp. Id) being the number of infectious chickens (resp. ducks), γ being the virus inactivation rate, Sc (resp. Sd) being the number of susceptible chickens (resp. ducks), w being the contact rate with one infectious dose in the environment, θ being the number of infectious doses that are necessary to infect a bird and F being the number of infectious doses present in the environment.
2.1.2. Dynamics of H7N9 transmission in the LBM
Once the virus was introduced into an LBM, chickens and ducks were assumed to pass through four infection stages: susceptible (Sc and Sd), infected but not yet infectious (Ec and Ed), infectious (Ic and Id) and recovered (Rc and Rd). All birds that were brought into the LBM were assumed to be susceptible. Susceptible birds (S) could become infected (E) during an average latent period duration (αd and αc) after which they were infectious (I) during an average infectious period duration (id and ic). Subsequently, they could die due to H7N9 infection (with a rate µc and µd) or recovered (R) from the infection. Note that all birds (whatever the compartment S, E, I or R) could leave the LBM for slaughter and trade at time-varying exit rates l(t) or could die from natural causes other than H7N9 infection (with a rate mc and md). Viral persistence of H7N9 in the environment has been recently reported [27] and several studies have demonstrated the importance of environmental transmission as a driver of AIV outbreaks [28–30]. Therefore, it was assumed that infectious ducks and chickens could contaminate the environment (F) by excreting infectious doses at an excretion rate ξd and ξc, respectively, with ξd < ξc, since ducks were reported excreting less H7N9 virus than chickens [24]. The amount of infectious doses in the environment was assumed to decrease at an inactivation rate γ.
The force of infection, determining the rate at which susceptible birds moved to the infected compartment (E), was defined as the sum of the forces of infection due to environmental contamination (indirect transmission) and due to contacts with infectious birds (direct transmission) [31]. Firstly, the force of infection due to environmental contamination (λF) was adapted from [32,33] using Hill-type function:
with w being the contact rate with one infectious dose in the environment, θ being the number of infectious doses that are necessary to infect a bird and F being the number of infectious doses present in the environment. Note that an infection via the contaminated environment could occur in the absence of infectious birds. Secondly, the force of infection due to contacts with infectious chickens (λc) and ducks (λd) was given by
with βj being the infection rate for chickens (j = c) and ducks (j = d).
The full dynamic transmission model is defined by the flux diagram in figure 1 (see electronic supplementary material, appendix, for the full mathematical specification of the model).
2.2. Calibration of the within-LBM H7N9 transmission model
2.2.1. Parameters for the bird trading process
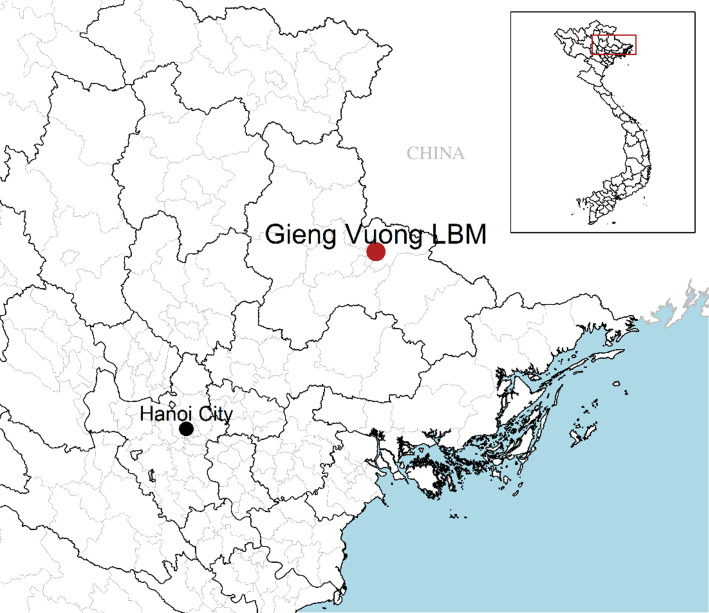
The parameters related to the population dynamics of birds were derived from field observations conducted at Giếng Vuông LBM, one of the largest LBMs in Lang Son province in northern Vietnam (figure 2). This LBM was identified by Vietnamese veterinary services as at high risk of H7N9 introduction due to potential cross-border trade of birds from China [24], and thus represented a relevant candidate for this study. A questionnaire survey was developed to collect information from LBM managers and traders on population dynamics of birds. Interviews were conducted in Vietnamese language by a native Vietnamese speaker proficient in English seconded by the first author and were facilitated by representatives from the District Veterinary Station (DVS) and the Sub-Department of Animal Health (SDAH). The LBM manager and staff were interviewed together, with answers agreed upon consensus while eight poultry traders were randomly selected and individually interviewed. Direct observations were made during the visits and used to cross-check interviewees' answers. Due to the nature of the study and the low risk posed to the participants, formal approval from an ethics committee was not a requirement at the time of the study. The questionnaire is available upon request to the corresponding author.
Figure 2.
Geographical location of Giếng Vuông LBM, Lang Son province, northern Vietnam.
2.2.2. Parameters for the H7N9 transmission process
Most of the parameters related to H7N9 transmission were derived from the published literature. Values and references are summarized in table 1 and details can be found in electronic supplementary material, appendix. Based on the transmission model, the basic reproduction number (R0), defined as the average number of secondary infections caused by a single infectious individual in a fully susceptible population [31], was formulated as a function of all model parameters using a next-generation matrix approach [40]. Assuming that the R0 had been estimated around 4.1 for H7N9 bird-to-bird transmission in LBMs [34], the lower bound value for w was thus estimated at 10−4 (electronic supplementary material, appendix). The sensitivity analysis was performed for parameters for which limited information was available from the published literature (i.e. Q, ξ and w) and presented in detail in electronic supplementary material, appendix.
Table 1.
Parameter values related to H7N9 transmission model.
| parameter | description | value (unit) | subtype | reference |
|---|---|---|---|---|
| dt | step-time | hour | ||
| mc | natural mortality rate for chickens attributable to other causes than H7N9 infection (per hour) | 10−4 | — | — |
| md | natural mortality rate for ducks attributable to other causes than H7N9 infection (per hour) | 10−4 | — | — |
| βc | infection rate for chickens (per hour) | 0.02 | H7N9 | [34] |
| βd | infection rate for ducks (per hour) | βc . K | H7N9 | [24,27] |
| K | ratio of infection rate for ducks versus chickens | 0.8 | H7N9 | [24,27] |
| αc | average duration of the latent period for chickens (hours) | 14.9 | H5N1 | [35] |
| αd | average duration of the latent period for ducks (hours) | 14.9 | H5N1 | [35] |
| µc | mortality rate for chickens due to infection (per hour) | 10−4 | H7N9 | [27,36,37] |
| µd | mortality rate for ducks due to infection (per hour) | 10−4 | H7N9 | [27] |
| ic | average duration of the infectious period for chickens (hours) | 192 | H7N9 | [24] |
| id | average duration of the infectious period for ducks (hours) | 120 | H7N9 | [24] |
| γ | inactivation rate (per hour) | 0.01 | AIV | [38,39] |
| ξc | number of infectious doses excreted by chickens (per hour) | 1 | — | —a |
| ξd | number of infectious doses excreted by ducks (per hour) | ξc . Q | — | —a |
| Q | ratio of excretion rate for ducks versus chickens | 0.8 | H7N9 | [24,27]a |
| w | contact rate with one infectious dose (per hour) | 10−4 | — | —a |
| θ | number of infectious doses to infect chickens and ducks | 1 | — | — |
aSee electronic supplementary material, appendix, for sensitivity analysis regarding these parameter assumptions.
2.3. Simulation of the within-LBM H7N9 transmission model
The model formulation was embedded within a deterministic framework. It assumed homogeneous mixing, i.e. birds uniformly and randomly contact each other, be they ducks or chickens. The model was initialized by introducing one infectious chicken at the moment of the day with the highest entry rate in the LBM and used to simulate the number of birds in each infection stage. The model was implemented in the R programming language [41].
2.4. Cost-effectiveness of surveillance strategies
2.4.1. Definition of the different surveillance strategies
The following surveillance strategies were incorporated into the model to assess their cost-effectiveness.
-
(1)
The laboratory-based surveillance strategy (strategy 1): this baseline strategy corresponds to the current surveillance programme for H7N9 in LBMs considered at high risk of H7N9 introduction in Vietnam. It involves the random sampling of 40 oropharyngeal swabs on chickens only, as they have been shown to be more susceptible to H7N9 infection than ducks [24]. Chickens are usually sampled twice a week at times between 8.00 and 10.00. All samples are then transported to an official diagnostic laboratory where they are all screened for the M gene using RT-PCR; those positive for the M gene are subsequently tested for the H7 gene using RT-PCR. As planned by the procedure, those positive for the H7 gene should be tested for the N9 gene. However, according to the World Animal Health Information System (WAHIS), H7 viruses have not yet been detected in Vietnam so testing for N9 was not included in the calculation of surveillance costs. On average, the delay between sampling and diagnostic test result communication is around 72 h.
-
(2)
The optimized laboratory-based surveillance strategy (strategy 2): this first alternative strategy is similar to strategy 1 except that samples are collected at the time of the day that maximizes the probability of sampling at least one infectious bird, i.e. at the time of the highest within-LBM prevalence of infection, as predicted by the model.
-
(3)
The portable PCR surveillance strategy (strategy 3): this second alternative strategy is similar to strategy 1 except that all samples are directly analysed on site for the H7 gene using the portable PCR device [25,26], assuming an average delay of 7 h between sampling and diagnostic test result communication.
-
(4)
The optimized portable PCR surveillance strategy (strategy 4): this third alternative strategy is similar to strategy 3 except that samples are collected at the time of the day that maximizes the probability of sampling at least one infectious bird.
These four different surveillance strategies were coupled with three different sampling frequencies: once a week, twice a week and every day. The surveillance strategies were also assessed assuming that the LBM policy could change and forbid the presence of birds staying overnight, as this intervention strategy has been shown to be an effective control option for AIV [21,42].
2.4.2. Comparison of the surveillance strategies
For a given combination of surveillance strategy, sampling frequency and overnight staying policy, the effectiveness was described by estimating the most likely day of detection post-introduction and the number of infected or infectious birds traded to farms or other LBMs at the most likely day of detection. The most likely day of detection was defined as the day when the cumulative probability of detection of at least one infectious bird became greater than 50% (electronic supplementary material, appendix). The weekly costs of H7N9 surveillance were also estimated for each strategy, with cost parameters provided by the SDAH (electronic supplementary material, appendix and table S1).
3. Results
3.1. Parameters for the LBM bird trading model
Table 2 presents parameter values related to the population dynamics of birds at Giếng Vuông LBM. This was a large LBM, with a high average daily number of birds (n = 7000). The predominant traded bird species reported was chicken (80%), with 25% of total number of birds staying overnight. The highest percentage of birds entering and leaving the LBM was reported to be at times between 2.00 and 6.00 and between 6.00 and 8.00, respectively.
Table 2.
Parameter values related to population dynamics of birds at Giếng Vuông LBM, Lang Son province, northern Vietnam.
| parameter | value |
|---|---|
| average daily number of birds entering the LBM | 7000 |
| percentage of birds present in the LBM | chickens: 80% ducks: 20% |
| percentage of birds staying overnight | 25% |
| percentage of birds per type of destination | to trading places, including farms and other LBMs: 40% to slaughter places, including slaughter houses, restaurants and end-consumers: 60% |
| percentage of birds entering the LBM per time slot | 2.00–3.00: 20% 3.00–4.00: 20% 4.00–5.00: 20% 5.00–6.00: 20% 6.00–7.00: 10% 7.00–8.00: 10% |
| percentage of birds leaving the LBM per time slot | 4.00–5.00: 5% 5.00–6.00: 5% 6.00–7.00: 20% 7.00–8.00: 20% 8.00–9.00: 15% 9.00–10.00: 15% 10.00–11.00: 10% 11.00–12.00: 10% |
3.2. Within-LBM H7N9 transmission model
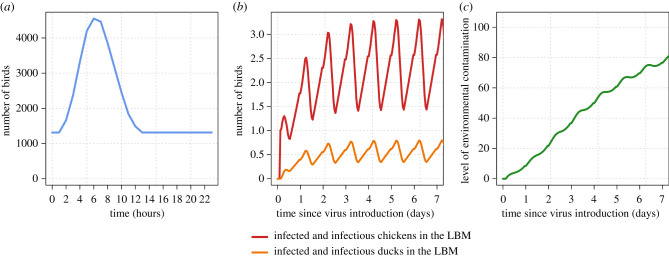
Assuming an overnight stay of birds, the daily dynamic of infection was expected to reach a regular pattern around two to three days after virus introduction (figure 3). At night, when the overall population within the LBM is closed (no entry, no exit), the virus spread between the overnight-staying birds, resulting in the decrease in the number of susceptible birds and the increase in the number of infected or infectious birds. Given the model parameters, the virus was able to persist in the environment in the long term, likely due to the presence of birds staying overnight in the LBM allowing virus amplification. The level of environmental contamination was expected to reach a plateau after several days. When implementing a ban on the presence of birds staying overnight, the number of infected birds and the environmental contamination level remained very limited, resulting in the fade-out of the epidemic within the first few days (electronic supplementary material, appendix and figure S1). However, note that this intervention likely reduced the chance of virus detection before it disappears (see below).
Figure 3.
Population dynamics of birds (a), estimated number of infected and infectious birds (b) and level of environmental contamination (c) at Giếng Vuông LBM, Lang Son province, Vietnam.
3.3. Cost-effectiveness of surveillance strategies
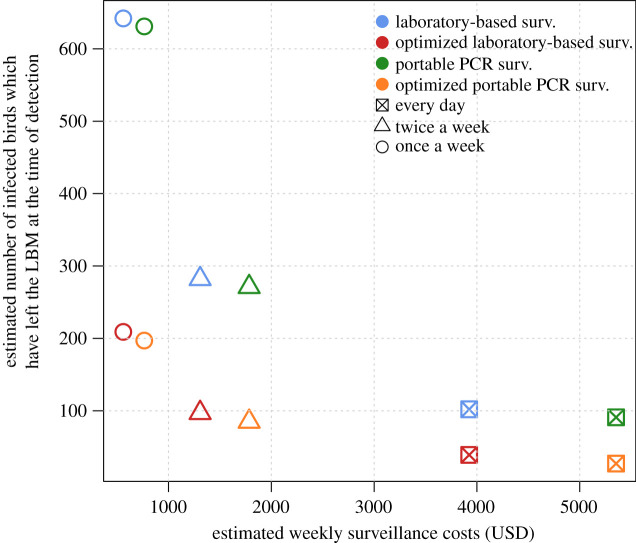
Based on model simulations, the time of the day when the prevalence was at its maximum, i.e. when the probability of sampling at least one infectious bird in the studied LBM was at its peak, was estimated at 1.00. This corresponds to the moment just before new susceptible birds are introduced into the LBM at the beginning of a working day. In figure 4, the laboratory-based surveillance strategy involving the sampling of chickens twice a week (blue triangle) corresponds to the baseline strategy for the surveillance of H7N9 in LBMs in Vietnam. Using its equivalent optimized strategy (red triangle) decreased the number of infected birds by approximately 60% for the same weekly surveillance cost (1308 USD) (electronic supplementary material, appendix and table S2). Using the optimized laboratory-based surveillance strategy once a week (red circle) decreased both the number of birds and the costs by approximately 30% and 60%, respectively. Similar results were obtained when comparing the portable PCR surveillance strategy (green) and its equivalent optimized strategy (orange). Also, when compared with the optimized laboratory-based surveillance strategy (red), using the optimized portable PCR surveillance strategy (orange) decreased the number of infected birds by approximately 10–30% (depending on the sampling frequency) but increased the weekly surveillance costs by approximately 30%.
Figure 4.
Estimated weekly costs (in USD) and estimated number of infected birds which have left the LBM at the day of detection for the different surveillance strategies at Giếng Vuông LBM, Lang Son province, Vietnam.
The sensitivity analysis (electronic supplementary material, appendix and figures S2, S3 and S4) showed that changes in the three parameter values for which no information was available in the literature (w, Q and ξ) did not impact the time of the day that maximized the probability of sampling at least one infectious bird (i.e. 1.00) and the effectiveness of the surveillance strategies.
4. Discussion
Model outputs suggested that H7N9 transmission could be sustained for weeks following the introduction of a single infectious bird through virus amplification and transmission within the LBM among birds staying overnight, which is consistent with results from a previous study on H5N1 [42]. The LBM system thus acted as a reservoir of infection for newly introduced susceptible birds. The model predicted that, using the current laboratory-based surveillance strategy for H7N9 in LBMs, several hundreds of infected birds would have already been sold to farms or other LBMs by the time the virus is expected to be detected in the LBM. A portable PCR method to detect H7N9 virus at the LBM with similar sensitivity (98%) and specificity (100%) to laboratory-based PCR assays was recently introduced with the aim of reducing the time delay between sampling and diagnostic test result communication [26]. Model outputs showed that using this alternative surveillance strategy twice a week would lead to similar surveillance costs and to only a slightly improved effectiveness as compared to the current laboratory-based surveillance strategy.
Model outputs also indicated that both laboratory-based and portable PCR surveillance strategies could be further optimized by sampling the birds staying at night just before new susceptible birds are introduced at the beginning of an opening LBM day. However, this would involve sampling and testing birds very early in the morning, which should be discussed with local communities and stakeholders as it may not be feasible in certain settings. Another limitation to consider is that scaling up surveillance systems involving a portable PCR device could be challenging due to constraints on the maximum number of samples that can be tested at once, while economies of scale can be achieved under laboratory-based strategies.
Birds infected with H7N9 generally show mild clinical signs but they can excrete the virus for 5 to 8 days [24]. Traders reported keeping birds for a few days within the LBM until being sold, housing them in cages overnight. These birds staying for longer time at LBMs are more likely to get infected and play an important role in the maintenance and amplification of H7N9 within the LBM. Model outputs showed that banning birds overnight at LBM resulted in the fade-out of the epidemic within the first few days but reduced the chance of virus detection before fade-out. Importantly, this intervention strategy could represent a less disruptive, more sustainable and effective measure than LBM closure, especially since the latter approach requires the virus to be detected what can take weeks (figure 4). Furthermore, LBM closure has been associated with substantial costs for the poultry industry and the emergence of illegal trade in some LBMs or neighbourhoods, making disease monitoring and control more difficult [43–45].
Given the diversity of poultry species present in LBMs, one should be cautious when attempting to generalize these results as variation in H7N9 transmission may be caused by different levels of susceptibility to H7N9 infection. Indeed, susceptibility to H7N9 has been reported to vary between species, with chickens being generally considered to be more susceptible than ducks [27,30]. Moreover, spent hens have been identified as one of the main poultry categories imported from China to Vietnam for consumption [46]. Limited information was available on the different species present within the LBMs, preventing more information to be integrated into the model. The model formulation assumed homogeneous mixing, meaning that birds could uniformly and randomly contact each other in the LBM, which was considered as an acceptable assumption given the high number of bird stalls, the limited physical separations and distances between stalls, the frequent handling of birds by sellers and buyers in LBMs. Another limitation of the model was the limited information regarding H7N9 virus survival in the environment. Environmental contamination by H7N9 in LBMs has been documented with positive samples retrieved in faeces, cages and floor, but also in de-feathering machines and chopping tools [10]. However, little is known on the virus inactivation rate across time for various environmental factors, such as temperature and humidity. While no information was available on the number of excreted infectious doses and on the contact rate with one infectious dose, the sensitivity analysis showed that the assumptions related to these parameters did not impact the conclusions of this study. Finally, note that model outputs showed that the virus was able to persist in the environment in the long term, suggesting that the sensitivity of PCR surveillance strategies based on environmental sampling needs to be further investigated.
The Giếng Vuông LBM is one of the 17 LBMs in northern Vietnam that had been identified as at high risk of becoming H7N9 infected due to potential cross-border trade of birds from China. This is a unique LBM with its proper structure and functioning. Thus, its bird population dynamics differ from those of other LBMs, resulting in different optimized times of sampling. Moreover, its risk of becoming H7N9 infected has significantly decreased following the implementation of a vaccination campaign for H7N9 in China in 2017 [47]. Thus, to allow stakeholders to apply this analytical framework to other LBM systems and AIV, a Web application was developed using the shiny package [48] and made publicly available with a graphical user interface in html format at: https://envt-inra.shinyapps.io/optimia/. The Web application allowed model simulation outputs to be adapted to other LBMs identified by Vietnamese veterinary services as at high risk of H7N9 introduction but which were not described in this paper for clarity reasons. Such Web application applied in LBMs could help in reducing zoonotic and pandemic risks posed by similar emerging AIV.
Acknowledgements
The authors are very grateful to all the LBM managers, staff and traders who responded to the questionnaire. They also gratefully acknowledge representatives from District Veterinary Station (DVS) and Sub-Department of Animal Health (SDAH) for facilitating the interviews and providing detailed information on the surveillance and sampling strategies of LP H7N9 in Vietnam. They are also very grateful to the Epidec group (ENVT-INRAE, France) for providing advice on the Web application and to Guillaume Fournié (RVC, UK) and Benjamin Roche (IRD, France) for providing useful comments on this research study.
Data accessibility
A Web application was developed using the shiny package and made publicly available with a graphical user interface in html format at: https://envt-inra.shinyapps.io/optimia/. The Web application allowed to provide model simulation outputs adapted to other LBMs identified by Vietnamese veterinary services as at high risk of H7N9 introduction but which were not described in this paper for clarity reasons. The data are provided in the electronic supplementary material.
Authors' contributions
C.G., D.T. and T.V. designed and coordinated the study. T.V. and C.G. carried out the analysis. C.S. and R.D.-D. participated in the analysis. C.G. and T.N.T.T. collected field data. C.G. wrote the manuscript. T.V., D.T., T.C., C.S., R.D.-D., M.P., D.R., T.N.T.T., K.I., L.P.T. and P.P. critically revised the manuscript. C.G. and T.C. developed the Web-based application. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
The authors declare no financial and non-financial competing interests.
Funding
The research was funded by the United States Agency for International Development (USAID) and the FEDER/Région Occitanie Recherche et Sociétés 2018—AI-TRACK.
References
- 1.Gao R, et al. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888-1897. ( 10.1056/NEJMoa1304459) [DOI] [PubMed] [Google Scholar]
- 2.Kageyama T, et al. 2013. Genetic analysis of novel avian A (H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 18, 20453. [PMC free article] [PubMed] [Google Scholar]
- 3.Lam TT-Y, et al. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502, 241-244. ( 10.1038/nature12515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S, Gu M, Liu D, Cui J, Gao GF, Zhou J, Liu X. 2017. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 25, 713-728. ( 10.1016/j.tim.2017.06.008) [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Xiao H, Wu Y, Liu D, Qi X, Shi Y, Gao GF. 2014. H7N9: a low pathogenic avian influenza A virus infecting humans. Curr. Opin. Virol. 5, 91-97. ( 10.1016/j.coviro.2014.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi W, et al. 2018. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J. Virol. 15, 92. ( 10.1186/s12985-018-0993-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L, et al. 2017. Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus, China, 2017. Emerg. Infect. Dis. 23, 1355-1359. ( 10.3201/eid2308.170640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang M, et al. 2017. Epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus in Guangdong, 2016 to 2017. Euro Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 22, 30568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pu Z, Xiang D, Li X, Luo T, Shen X, Murphy RW, Liao M, Shen Y. 2018. Potential pandemic of H7N9 avian influenza A virus in human. Front. Cell Infect. Microbiol. 8, 414. ( 10.3389/fcimb.2018.00414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang M, He J, Song T, Rutherford S, Wu J, Lin J, Huang G, Tan X, Zhong H. 2015. Environmental sampling for avian influenza A(H7N9) in live-poultry markets in Guangdong, China. PLoS ONE 10, e0126335. ( 10.1371/journal.pone.0126335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Indriani R, et al. 2010. Environmental sampling for avian influenza virus A (H5N1) in live-bird markets, Indonesia. Emerg. Infect. Dis. 16, 1889-1895. ( 10.3201/eid1612.100402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao C, Cui L, Zhou M, Hong L, Gao GF, Wang H. 2013. Live-animal markets and influenza A (H7N9) virus infection. N. Engl. J. Med. 368, 2337-2339. ( 10.1056/NEJMc1306100) [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet Lond. Engl. 381, 1916-1925. ( 10.1016/S0140-6736(13)60903-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, et al. 2014. Relationship between domestic and wild birds in live poultry market and a novel human H7N9 virus in China. J. Infect. Dis. 209, 34-37. ( 10.1093/infdis/jit478) [DOI] [PubMed] [Google Scholar]
- 15.Wu J, et al. 2015. Seasonality of avian influenza A(H7N9) activity and risk of human A(H7N9) infections from live poultry markets. J. Infect. 71, 690-693. ( 10.1016/j.jinf.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, et al. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet Lond. Engl. 383, 541-548. ( 10.1016/S0140-6736(13)61904-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husain M. 2014. Avian influenza A (H7N9) virus infection in humans: epidemiology, evolution, and pathogenesis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 28, 304-312. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, et al. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341, 410-414. ( 10.1126/science.1240532) [DOI] [PubMed] [Google Scholar]
- 19.Gilbert M, et al. 2014. Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia. Nat. Commun. 5, 4116. ( 10.1038/ncomms5116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Tang X, Yang Z, Wang M, Zheng B. 2014. Enhanced disinfection and regular closure of wet markets reduced the risk of avian influenza A virus transmission. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 58, 1037-1038. ( 10.1093/cid/cit951) [DOI] [PubMed] [Google Scholar]
- 21.Leung YHC, Lau EHY, Zhang LJ, Guan Y, Cowling BJ, Peiris JSM. 2012. Avian influenza and ban on overnight poultry storage in live poultry markets, Hong Kong. Emerg. Infect. Dis. 18, 1339-1341. ( 10.3201/eid1808.111879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vergne T, Meyer A, Long PT, Elkholly DA, Inui K, Padungtod P, Newman SH, Fournié G, Pfeiffer DU. 2019. Optimising the detectability of H5N1 and H5N6 highly pathogenic avian influenza viruses in Vietnamese live-bird markets. Sci. Rep. 9, 1031. ( 10.1038/s41598-018-37616-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan L, et al. 2016. Detection of influenza A virus from live-bird market poultry swab samples in China by a pan-IAV, one-step reverse-transcription FRET-PCR. Sci. Rep. 22, 30015. ( 10.1038/srep30015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, Suarez DL. 2014. Role of poultry in the spread of novel H7N9 influenza virus in China. J. Virol. 88, 5381-5390. ( 10.1128/JVI.03689-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schar D, Padungtod P, Tung N, O'Leary M, Kalpravidh W, Claes F. 2019. New frontiers in applied veterinary point-of-capture diagnostics: toward early detection and control of zoonotic influenza. Influenza Other Respir. Viruses 13, 618-621. ( 10.1111/irv.12648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inui K, et al. 2019. A field-deployable insulated isothermal RT-PCR assay for identification of influenza A (H7N9) shows good performance in the laboratory. Influenza Other Respir. Viruses 13, 610-617. ( 10.1111/irv.12646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidaña B, et al. 2018. Transmission and immunopathology of the avian influenza virus A/Anhui/1/2013 (H7N9) human isolate in three commonly commercialized avian species. Zoonoses Public Health 65, 312-321. ( 10.1111/zph.12393) [DOI] [PubMed] [Google Scholar]
- 28.Roche B, Lebarbenchon C, Gauthier-Clerc M, Chang C-M, Thomas F, Renaud F, van der Werf S, Guegan JF. 2009. Water-borne transmission drives avian influenza dynamics in wild birds: the case of the 2005–2006 epidemics in the Camargue area. Infect. Genet. Evol. 9, 800-805. ( 10.1016/j.meegid.2009.04.009) [DOI] [PubMed] [Google Scholar]
- 29.Mata MA, Greenwood P, Tyson R. 2019. The relative contribution of direct and environmental transmission routes in stochastic Avian flu epidemic recurrence: an approximate analysis. Bull. Math. Biol. 81, 4484-4517. ( 10.1007/s11538-018-0414-6) [DOI] [PubMed] [Google Scholar]
- 30.Wang R-H, Jin Z, Liu Q-X, Koppel Jvd, Alonso D. 2012. A simple stochastic model with environmental transmission explains multi-year periodicity in outbreaks of avian flu. PLoS ONE 7, e28873. ( 10.1371/journal.pone.0028873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson RM, May RM. 1979. Population biology of infectious diseases: part I. Nature 280, 361-367. ( 10.1038/280361a0) [DOI] [PubMed] [Google Scholar]
- 32.Codeço CT. 2001. Endemic and epidemic dynamics of cholera: the role of the aquatic reservoir. BMC Infect. Dis. 1, 1. ( 10.1186/1471-2334-1-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paroissin C, Cazelles B, de Lara M, Delmas J-F, Guégan J-F. 2005. Modeling environmental impacts of plankton reservoirs on cholera population dynamics. ESAIM: Proc. 14, 156-173. ( 10.1051/proc:2005013) [DOI] [Google Scholar]
- 34.Hsieh Y-H, Wu J, Fang J, Yang Y, Lou J. 2014. Quantification of bird-to-bird and bird-to-human infections during 2013 novel H7N9 avian influenza outbreak in China. PLoS ONE 9, e111834. ( 10.1371/journal.pone.0111834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouma A, Claassen I, Natih K, Klinkenberg D, Donnelly CA, Koch G, Van Boven M. 2009. Estimation of transmission parameters of H5N1 avian influenza virus in chickens. PLoS Pathog. 5, e1000281. ( 10.1371/journal.ppat.1000281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ku KB, Park EH, Yum J, Kim HM, Kang YM, Kim JC, Kim JA, Kim HS, Seo SH. et al. 2014. Transmissibility of novel H7N9 and H9N2 avian influenza viruses between chickens and ferrets. Virology 450–451, 316-323. [DOI] [PubMed] [Google Scholar]
- 37.Spackman E, Pantin-Jackwood M, Swayne DE, Suarez DL, Kapczynski DR. 2015. Impact of route of exposure and challenge dose on the pathogenesis of H7N9 low pathogenicity avian influenza virus in chickens. Virology 477, 72-81. ( 10.1016/j.virol.2015.01.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown JD, Swayne DE, Cooper RJ, Burns RE, Stallknecht DE. 2007. Persistence of H5 and H7 avian influenza viruses in water. Avian Dis. 51(1 Suppl.), 285-289. ( 10.1637/7636-042806R.1) [DOI] [PubMed] [Google Scholar]
- 39.Handel A, Brown J, Stallknecht D, Rohani P. 2013. A multi-scale analysis of influenza A virus fitness trade-offs due to temperature-dependent virus persistence. PLoS Comput. Biol. 9, e1002989. ( 10.1371/journal.pcbi.1002989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Driessche P, Watmough J. 2002. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29-48. ( 10.1016/S0025-5564(02)00108-6) [DOI] [PubMed] [Google Scholar]
- 41.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 42.Fournié G, Guitian F, Mangtani P, Ghani A. 2011. Impact of the implementation of rest days in live bird markets on the dynamics of H5N1 highly pathogenic avian influenza. J. R. Soc. Interface 8, 1079-1089. ( 10.1098/rsif.2010.0510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, et al. 2016. Effect of live poultry market interventions on influenza A(H7N9) virus, Guangdong, China. Emerg. Infect. Dis. 22, 2104-2112. ( 10.3201/eid2212.160450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peiris JSM, Cowling BJ, Wu JT, Feng L, Guan Y, Yu H, Leung GM. 2016. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. Lancet Infect. Dis. 16, 252-258. ( 10.1016/S1473-3099(15)00502-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fournié G, Pfeiffer DU. 2014. Can closure of live poultry markets halt the spread of H7N9? Lancet Lond Engl. 383, 496-497. ( 10.1016/S0140-6736(13)62109-1) [DOI] [PubMed] [Google Scholar]
- 46.Desvaux S, Nguyen C, Vu D, Henriquez C, Ky V, Roger F, Fenwick S, Goutard F. 2016. Risk of introduction in northern Vietnam of HPAI viruses from China: description, patterns and drivers of illegal poultry trade. Transbound. Emerg. Dis. 63, 389-397. ( 10.1111/tbed.12279) [DOI] [PubMed] [Google Scholar]
- 47.Zeng X, Tian G, Shi J, Deng G, Li C, Chen H. 2018. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci. China Life Sci. 61, 1465-1473. ( 10.1007/s11427-018-9420-1) [DOI] [PubMed] [Google Scholar]
- 48.Potter G, Wong J, Alcaraz I, Chi P. 2016. Web application teaching tools for statistics using R and shiny. Technol. Innov. Stat. Educ. 9, 1-33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A Web application was developed using the shiny package and made publicly available with a graphical user interface in html format at: https://envt-inra.shinyapps.io/optimia/. The Web application allowed to provide model simulation outputs adapted to other LBMs identified by Vietnamese veterinary services as at high risk of H7N9 introduction but which were not described in this paper for clarity reasons. The data are provided in the electronic supplementary material.