Prosthetic joint infections (PJI) are frequent complications of arthroplasties. Their treatment is made complex by the rapid formation of bacterial biofilms, limiting the effectiveness of antibiotic therapy.
KEYWORDS: Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, biofilms, dispersin B, endonuclease, cellulase, prosthetic joint infection, antibiotics, coagulase-negative staphylococci, enzymes, prosthesis infections
ABSTRACT
Prosthetic joint infections (PJI) are frequent complications of arthroplasties. Their treatment is made complex by the rapid formation of bacterial biofilms, limiting the effectiveness of antibiotic therapy. In this study, we explore the effect of a tri-enzymatic cocktail (TEC) consisting of an endo-1,4-β-d-glucanase, a β-1,6-hexosaminidase, and an RNA/DNA nonspecific endonuclease combined with antibiotics of different classes against biofilms of Staphylococcus aureus, Staphylococcus epidermidis, and Escherichia coli grown on Ti-6Al-4V substrates. Biofilms were grown in Trypticase soy broth (TSB) with 10 g/liter glucose and 20 g/liter NaCl (TGN). Mature biofilms were assigned to a control group or treated with the TEC for 30 min and then either analyzed or reincubated for 24 h in TGN or TGN with antibiotics. The cytotoxicity of the TEC was assayed against MG-63 osteoblasts, primary murine fibroblasts, and J-774 macrophages using the lactate dehydrogenase (LDH) release test. The TEC dispersed 80.3 to 95.2% of the biofilms’ biomass after 30 min. The reincubation of the treated biofilms with antibiotics resulted in a synergistic reduction of the total culturable bacterial count (CFU) compared to that of biofilms treated with antibiotics alone in the three tested species (additional reduction from 2 to more than 3 log10 CFU). No toxicity of the TEC was observed against the tested cell lines after 24 h of incubation. The combination of pretreatment with TEC followed by 24 h of incubation with antibiotics had a synergistic effect against biofilms of S. aureus, S. epidermidis, and E. coli. Further studies should assess the potential of the TEC as an adjuvant therapy in in vivo models of PJI.
INTRODUCTION
Prosthetic joint infections (PJI) are catastrophic complications of joint replacement surgeries. With a prevalence of 2.0% to 2.4% in patients following a total knee or total hip replacement (1), PJIs represent one of the leading causes of failure for arthroplasties (2).
The most prevalent species encountered in PJIs are Staphylococcus aureus, coagulase-negative staphylococci, streptococci, and Gram-negative bacilli (3). These species are known to adhere to the surfaces of implants and to form complex communities embedded in an extracellular matrix called biofilms. In these structures, bacteria become tolerant to antibiotics up to concentrations 1,000 times the MIC of planktonic cells (4–6).
Surgical debridement, implant retention, and antibiotic therapy (DAIR) is the preferred treatment strategy of acute (less than 3 weeks of symptoms) PJI (7), with better functional outcomes (8) and lower costs (9, 10) than alternative strategies. However, the observed failure rate is considerable (11, 12). These failures are partly explained by an insufficient biofilm removal during the debridement surgery, as was shown in vitro by Urish et al. (13) and Poilvache et al. (14).
As a consequence, the need for new adjuvant treatments to facilitate the dispersal of biofilms has been recognized in the literature (15–19). Among mechanical and pharmaceutical approaches, the use of hydrolytic enzymes targeting specific components of the biofilm matrix has been suggested (19). The dispersal potential of single enzymes targeting individual matrix components, such as extracellular DNA (eDNA), exopolysaccharides, or extracellular proteins, has been demonstrated by previous authors in in vitro models (20–23). Moreover, the synergy of the combination of these enzymes with antibiotics (24, 25) or antiseptics (26, 27) against biofilms has been reported in previous studies.
However, these previous studies did not address the variety in matrix composition among bacterial species, as they focused on the effect of single enzymes on selected species. For this reason, a broad-spectrum tri-enzymatic cocktail (hereafter referred to as TEC for tri-enzymatic cocktail), composed of an endo-1,4-β-d-glucanase, a β-1,6-hexosaminidase, and a recombinant RNA/DNA nonspecific endonuclease (see Materials and Methods), was tested, targeting the matrix of Escherichia coli, Enterococcus faecalis, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, and Staphylococcus epidermidis biofilms (patent numbers WO2017/211737 A1 [28] and BE2020/5726 [29]).
In this study, we investigated the dispersal effect of this tri-enzyme cocktail against Staphylococcus aureus, Staphylococcus epidermidis, and Escherichia coli monomicrobial biofilms grown in vitro on titanium alloy surfaces similar to prosthetic implants and examined its synergy in combination with a broad selection of antibiotics belonging to different classes recommended for PJI treatment (7) at concentrations relevant to human use.
RESULTS
Antibiotic susceptibility of planktonic bacteria and of biofilms.
The MICs and minimal biofilm eradication concentrations (MBECs) are shown in Table 1. The strains were susceptible to all tested antibiotics according to the CLSI breakpoints, except for the ATCC 33591 methicillin-resistant S. aureus (MRSA) strain, which was resistant to doxycycline. For this reason, doxycycline was not used against ATCC 33591 biofilms for the next experiments. The MICs in cation-adjusted Mueller-Hinton broth (MHB-ca) and Trypticase soy broth (TSB) with 10 g/liter glucose and 20 g/liter NaCl (TGN) were within two dilutions of each other. Most of the MBECs exceeded the MICs by a hundred- or thousand-fold factor, to the notable exception of daptomycin, linezolid, and moxifloxacin against the strain ATCC 33591 in MHB-ca. The MBECs could not be established in TGN for the strain ATCC 47076, as we observed inconsistent results across replicates. All MBECs were found to be higher than the concentrations chosen for the coupon assays (Table 2).
TABLE 1.
MICs and minimal biofilm eradication concentrationsa
| Antibiotic | Assay | MIC or MBEC value (mg/liter) |
|||||
|---|---|---|---|---|---|---|---|
| ATCC 33591 |
ATCC 35984 |
ATCC 47076b |
|||||
| MHB-ca | TGN | MHB-ca | TGN | MHB-ca | TGN | ||
| Daptomycin | MIC | 1 | 1 | 1 | 1 | ||
| MBEC | 4 | >1,024 | >1,024 | >1,024 | |||
| Doxycycline | MIC | 16 | 16 | 0.25 | 0.25 | ||
| MBEC | 128 | 512 | |||||
| Linezolid | MIC | 2 | 1 | 2 | 1 | ||
| MBEC | 8 | >1,024 | >1,024 | >1,024 | |||
| Moxifloxacin | MIC | 0.125 | 0.125 | 0.06 | 0.125 | ||
| MBEC | 2 | >1,024 | >1,024 | >1,024 | |||
| Rifampin | MIC | 0.004 | 0.004 | 0.004 | 0.004 | ||
| MBEC | 8 | 2 | 128 | 4 | |||
| Vancomycin | MIC | 1 | 8 | 2 | 8 | ||
| MBEC | >1,024 | >1,024 | >1,024 | >1,024 | |||
| Ceftazidime | MIC | 0.25 | 0.125 | ||||
| MBEC | >1,024 | ||||||
| Ciprofloxacin | MIC | 0.016 | 0.03 | ||||
| MBEC | 128 | ||||||
| Colistin | MIC | 0.5 | 0.25 | ||||
| MBEC | >1,024 | ||||||
| Meropenem | MIC | 0.03 | 0.06 | ||||
| MBEC | >256 | ||||||
Values in bold indicate an MIC superior to the CLSI breakpoint values (50).
The MBEC assay results for the ATCC 47076 strain were unreliable and were not included.
TABLE 2.
Antibiotics experimental concentrations and rationales
| Antibiotic | Concn (mg/liter) | Rationalea |
|---|---|---|
| Daptomycin | 30 | |
| Doxycycline | 1.5 | |
| Linezolid | 5 | Cminb |
| Moxifloxacin | 1.5 | |
| Rifampin | 2.5 | |
| Vancomycin | 2 | Target trough concn (20 mg/liter) |
| Ceftazidime | 3.8 | Cmin |
| Ciprofloxacin | 1.625 | |
| Colistin | 2.5 | |
| Meropenem | 0.25 | Cmin |
Pharmacokinetic and PK/PD information obtained from the EUCAST rationales for clinical breakpoint documents or from the literature (references in Table S1 in the supplemental material).
Cmin, minimum concentration of drug in serum.
Kinetics of TEC effect.
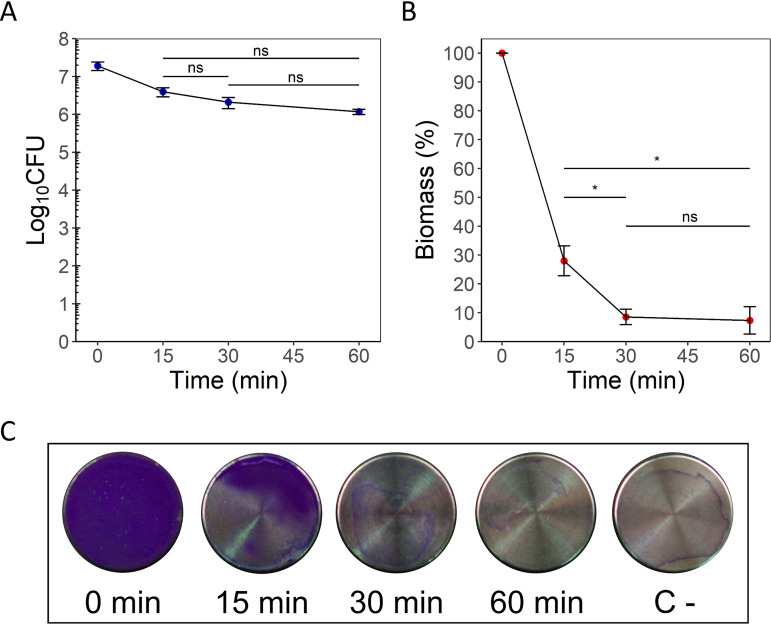
The effect of TEC on ATCC 33591 MRSA biofilms is illustrated in Fig. 1 for CFU counts (Fig. 1A) and biomass (Fig. 1B and C). The CFU counts decreased modestly after 15 min of incubation (−0.68 log10 CFU, P = 0.0365 from time zero), and no statistically significant differences were observed between the subsequent incubation times. A sharp decrease in the total biomass was observed after a 15-min incubation (−72.0% from time zero, P = 3.3e−06) and appeared to be maximal after 30 min (−95.82% from time zero, P = 7.0e−07; −19.45% from 15 min, P = 0.014). These effects were corroborated by macroscopic observation of the crystal violet-stained samples (Fig. 1C). Based on these results, an incubation time of 30 min with TEC was adopted for the next experiments.
FIG 1.
Kinetics of biofilm dispersal by TEC against MRSA S. aureus ATCC 33591 biofilms. (A) Log10 CFU counts per coupon. (B) Total biomass assay (expressed as the percentage of time zero control). (C) Pictures of biofilm samples exposed to TEC for the indicated times (C-, negative control [coupon incubated in sterile culture medium]) stained with crystal violet. The values indicate the means of triplicate experiments (± standard error). Statistical analysis, repeated measure ANOVA, followed by paired t tests with Holm correction. *, P < 0.05.
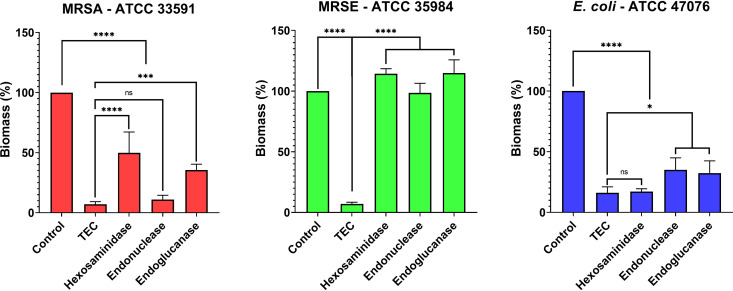
Effect of individual enzymes compared to the TEC.
In a preliminary experiment, the effect of the individual enzymes composing the TEC was tested against biofilms of each of the three species in comparison with the cocktail (Fig. 2). The TEC was found to be at least equivalent to the most effective single enzyme against all three species. While we did not observe any significant biofilm dispersal when treating methicillin-resistant S. epidermidis (MRSE) ATCC 35984 biofilms with the individual enzymes, exposure to the TEC resulted in a 93% reduction of the biomass (P < 0.0001). The RNA/DNA endonuclease was found to be equivalent to the TEC against MRSA ATCC 33591, and the β-1,6-hexosaminidase was equivalent to the TEC against E. coli ATCC 47076.
FIG 2.
Effect of a 30-min treatment with the individual enzymes on the biomass of ATCC 33591, ATCC 35984, and ATCC 47076 biofilms and comparison to the TEC. Hexosaminidase, β-N-acetylhexosaminidase; endonuclease, DNA/RNA endonuclease; endoglucanase, endo-1,4-β-d-glucanase. Data expressed as percentage of the control. The values indicate the means of triplicate experiments (± standard error). Statistical analysis, two-way ANOVA followed by Holm-Sidàk post hoc test. NS, no significant difference; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Effect of TEC and of antibiotics alone or in combination against biofilms.
Biofilms exposed to TEC for 30 min were washed and reincubated in medium supplemented or not with antibiotics at clinically relevant concentrations (Table 2).
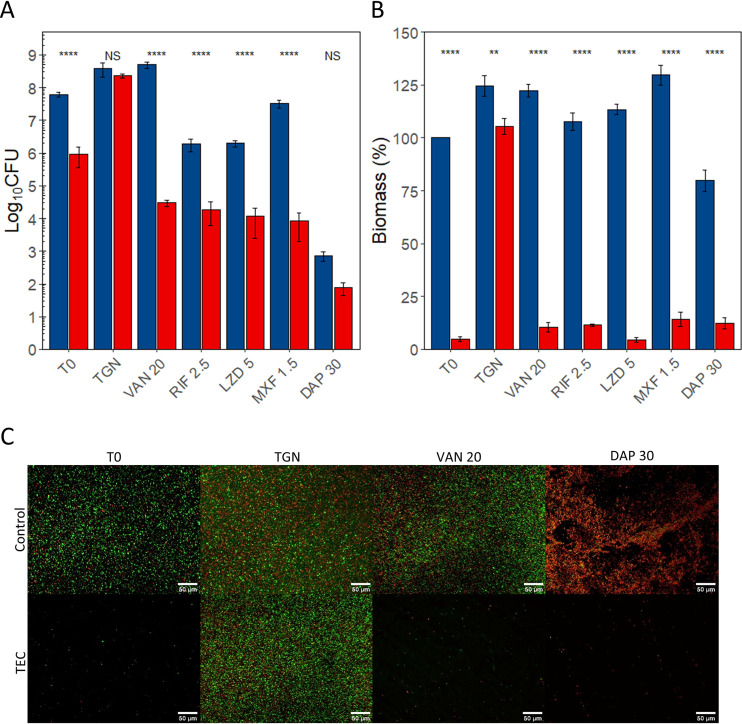
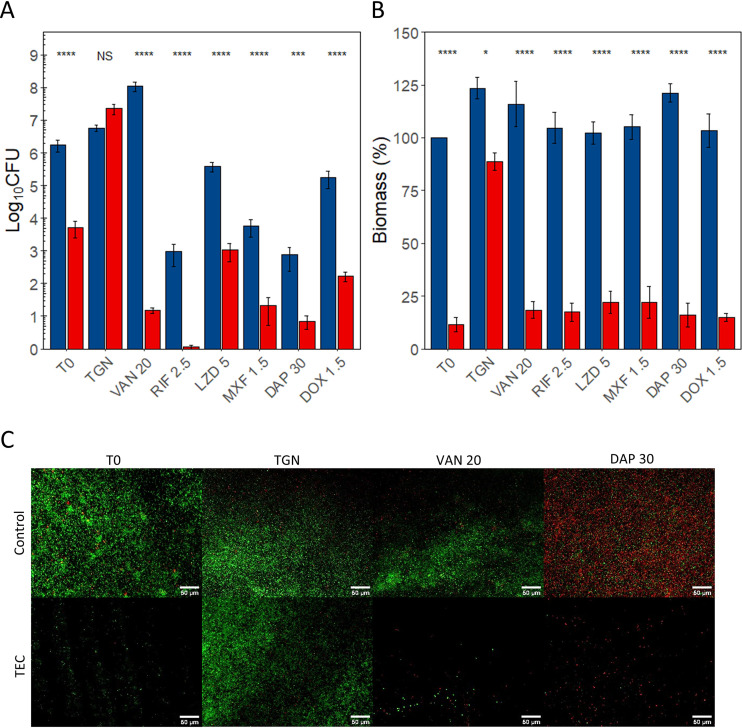
The results are illustrated in Fig. 3 (MRSA ATCC 33591), Fig. 4 (MRSE ATCC 35984), and Fig. 5 (E. coli ATCC 47076) for CFU counts (Fig. 3A, Fig. 4A, and Fig. 5A), biomass (Fig. 3B, Fig. 4B, and Fig. 5B), and fluorescence microscopy (Fig. 3C, Fig. 4C, and Fig. 5C). The details of the comparisons are shown in Tables S2 to S7 in the supplemental material.
FIG 3.
Effect of the combination of the TEC with antibiotics on MRSA ATCC 33591 biofilms. (A) CFU counts. (B) Biomass. (C) Fluorescence microscopy. (A and B) Blue, controls; red, samples treated for 30 min with TEC. T0, no reincubation; TGN, 24 h reincubation in TGN; others, 24 h reincubation in TGN with antibiotics at the indicated concentrations (see Table 1). CFU counts expressed in log10 CFU per coupon, and biomass expressed in percentage of T0 controls. The values indicate the means of triplicate experiments (± standard error). Statistical analysis, two-way ANOVA followed by Tukey HSD comparing control samples to samples exposed to the enzymatic cocktail. NS, no significant difference; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Green, living cells (SYTO 9); red, dead cells (propidium iodide). ×20 magnification. Scale bars represent 50 μm. VAN, vancomycin; RIF, rifampin; LZD, linezolid; MXF, moxifloxacin; DAP, daptomycin.
FIG 4.
Effect of the combination of the TEC with antibiotics on MRSE ATCC biofilms. (A) CFU counts. (B) Biomass. (C) Fluorescence microscopy. (A and B) Blue, controls; red, samples treated for 30 min with TEC. T0, no reincubation; TGN, 24 h reincubation in TGN; others, 24 h reincubation in TGN with antibiotics at the indicated concentrations (see Table 1). CFU counts expressed in log10 CFU per coupon, and biomass expressed in percentage of T0 controls. The values indicate the means of triplicate experiments (± standard error). Statistical analysis, two-way ANOVA followed by Tukey HSD comparing control samples to samples exposed to the enzymatic cocktail. NS, no significant difference; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Green, living cells (SYTO 9); red, dead cells (propidium iodide). ×20 magnification. Scale bars represent 50 μm. VAN, vancomycin; RIF, rifampin; LZD, linezolid; MXF, moxifloxacin; DAP, daptomycin; DOX, doxycycline.
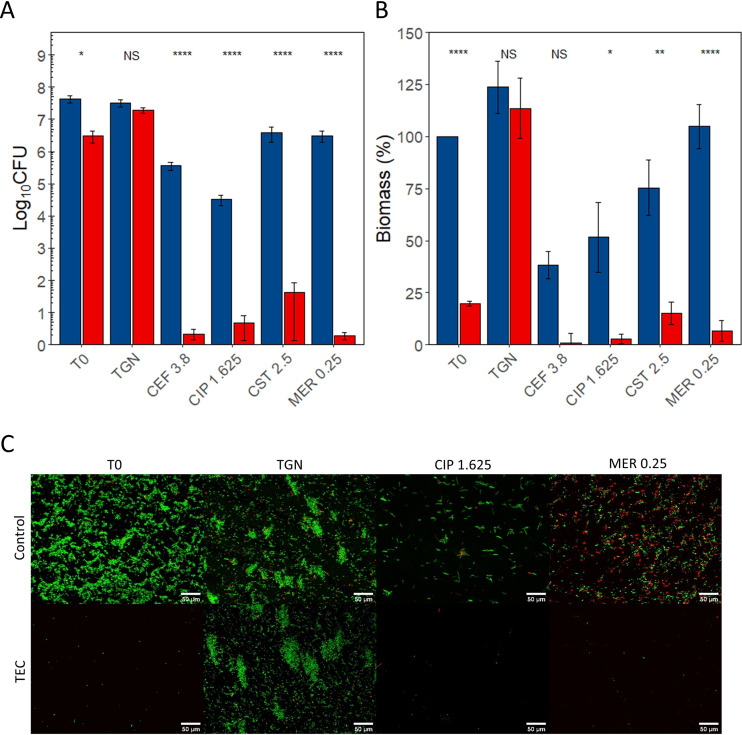
FIG 5.
Effect of the combination of the TEC with antibiotics on E. coli ATCC 47076 biofilms. (A) CFU counts. (B) Biomass. (C) Fluorescence microscopy. (A and B) Blue, controls; red, samples treated for 30 min with TEC. T0, no reincubation; TGN, 24 h reincubation in TGN; others, 24 h reincubation in TGN with antibiotics at the indicated concentrations (see Table 1). CFU counts expressed in log10 CFU per coupon, biomass expressed in percentage of T0 controls. The values indicate the means of triplicate experiments (± standard error). Statistical analysis, two-way ANOVA followed by Tukey HSD comparing control samples to samples exposed to the enzymatic cocktail. NS, no significant difference; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Green, living cells (SYTO 9); red, dead cells (propidium iodide). ×20 magnification. Scale bars represent 50 μm. CEF, ceftazidime; CIP, ciprofloxacin; CST, colistin; MER, meropenem.
A 30-min exposure of the coupons to TEC significantly reduced the CFU counts and biomass of all strains. The CFU counts were reduced by 2.04 log10 in strain ATCC 33591, 2.54 log10 in strain ATCC 35984, and 1.24 log10 in strain ATCC 47076. Biomass was reduced by 95.2% in strain ATCC 33591, 88.6% in strain ATCC 35984, and 80.3% in strain ATCC 47076.
A 24-h reincubation of control samples was not associated with significant changes in CFU counts. Their biomass remained stable in strains ATCC 35984 and ATCC 47076 but increased by 24.4% in strain ATCC 33591.
Similarly, the CFU counts of the samples treated with the enzyme cocktail and then reincubated for 24 h in TGN were similar to those of nontreated samples for all three strains. The biomass was however significantly lower than that for control group samples for ATCC 33591 (−19.1%) and ATCC 35984 (−34.7%) biofilms.
The effect of a 24-h exposure to antibiotics on control biofilms was variable versus CFU counts and generally weak or nonexistent versus the biomass when compared to initial controls (statistical analyses shown in Tables S2 to S7 in the supplemental material).
Against staphylococci, vancomycin (20 mg/liter) caused a 1.84 log10 CFU increase in the CFU counts of MRSE ATCC 35984 biofilms but no changes MRSA ATCC 33591. Conversely, the biomass of MRSA samples increased by 22.3% but did not change significantly in MRSE samples. Rifampin (2.5 mg/liter) caused a significant reduction of the CFU counts in both strains (ATCC 33591, −1.59 log10 CFU; ATCC 35984, −3.39 log10 CFU) but did not affect the biomass of either strain. Linezolid (5 mg/liter) had a limited effect on the CFU counts of strain ATCC 33591 (−1.50 log10) and did not affect the CFU counts of strain ATCC 35984 nor the biomass of both strains. Moxifloxacin (1.5 mg/liter) impacted both strains differently despite similar MICs. In ATCC 33591 samples, the CFU counts did not change, and the biomass increased by 29.7%, while in ATCC 35984, a 2.52 log10 CFU decrease was observed and the biomass remained stable. Daptomycin (30 mg/liter) had a strong effect on the CFU counts of both strains (ATCC 33591, −4.99 log10; ATCC 35984, −3.50 log10) as well as a moderate effect on the biomass of strain 33591 (−20.2%), while the biomass of strain ATCC 35984 remained unchanged. Doxycycline (1.5 mg/liter) was used against MRSE ATCC 35984 only as MRSA ATCC 33591 was resistant. No significant effect was observed for either CFU counts or biomass.
Against E. coli ATCC 47076 biofilms, ceftazidime (3.8 mg/liter) and ciprofloxacin (1.625 mg/liter) had a significant effect on CFU counts (ceftazidime, −2.06 log10; ciprofloxacin, −3.21 log10) and biomass (ceftazidime, −61.7%; ciprofloxacin, −48.33%) while colistin (2.5 mg/liter) and meropenem (0.25 mg/liter) did not.
Turning now to the results for biofilms of the three strains successively exposed to TEC and antibiotics at set concentrations, we observed significant reductions of CFU counts and biomass compared to those of samples treated only with antibiotics, except for the CFU counts of MRSA ATCC 33591 samples treated with daptomycin (which were already markedly decreased by the antibiotic alone) and the biomass of E. coli ATCC 47076 treated with ceftazidime (for which the difference between samples preexposed or not to the TEC cocktail did not reach significance). The difference in CFU counts between the samples preexposed to the enzymes and the control group samples was always higher than 2 log10 and exceeded 3 log10 in MRSA ATCC 33591 samples treated with vancomycin (−4.22 log10, P < 0.0001) or moxifloxacin (−3.87 log10, P < 0.0001), in MRSE ATCC 35984 samples treated with vancomycin (−6.84 log10, P < 0.0001), and in all E. coli ATCC 470476 samples exposed to any of the 4 tested antibiotics (ceftazidime, −5.23 log10, P < 0.0001); ciprofloxacin, −3.98 log10, P < 0.0001; colistin, −5.57 log10, P < 0.0001; meropenem, −6.11 log10, P < 0.0001).
The difference in biomass between samples treated with antibiotics alone or with the combination of TEC and antibiotics exceeded 80% in most conditions, except for strain ATCC 33591 samples treated with daptomycin and strain ATCC 47076 samples treated with ciprofloxacin and colistin.
The two-way analysis of variance (ANOVA) (Tables S8 to S13 in the supplemental material) showed significant interaction parameters in all cases when considering the exposure to the enzyme cocktail and the reincubation of the samples as factors. This suggests that the tri-enzyme cocktail and the antibiotics have a synergistic effect on both CFU counts and biomass of all three strains.
The analysis of biofilms using fluorescence microscopy after LIVE/DEAD staining (Fig. 3C, Fig. 4C, and Fig. 5C) illustrates the observed CFU counts and biomass assays. The treatment of samples with the TEC dispersed an important proportion of the bacterial cells, which were enough to regrow a biofilm after 24 h of reincubation in TGN. This recovery appeared to be inhibited by the reincubation with antibiotics. The aspect of control samples exposed during 24 h to vancomycin, ciprofloxacin, and meropenem appeared unchanged from the samples incubated in TGN, keeping a majority of live cells (green), while the samples incubated with daptomycin exhibited a high proportion of dead cells (red).
Cytotoxicity tests.
The cytotoxicity of the tri-enzyme cocktail for different types of eukaryotic cells was then evaluated by measuring the release of the cytosolic enzyme lactate dehydrogenase (LDH) in the culture medium after 1-h and 24-h exposures to TEC at two concentrations (concentration used against biofilms and a 1:1 dilution). At 1 h, no significant toxicity was detected (values of <5% in all cases). At 24 h, the release of the cytosolic enzyme lactate dehydrogenase (LDH) reached a mean percentage (95% confidence interval) of 3.0% (−1.1–7.0) for primary murine fibroblasts, −1.6% (−4.7–1.4) for MG-63 human osteosarcoma cells, and −3.2% (−17.0–10.7) for J774 murine macrophages.
DISCUSSION
This study found that a tri-enzymatic cocktail containing two glycoside hydrolases (an endo-1,4-β-d-glucanase and a β-1,6-hexosaminidase) and a nonspecific RNA/DNA endonuclease had a significant dispersal effect on mature biofilms of S. aureus, S. epidermidis, and E. coli reference strains grown on titanium alloy and that the successive treatment of these biofilms with this enzyme cocktail and antibiotics at concentrations relevant to the clinical setting was synergistic, resulting in significantly lower residual CFU counts than those with either individual treatment.
While the potential of glycoside hydrolases and DNase I as biofilm dispersal agents has been previously described in the literature (19, 22), the originality of our work resides in the association of enzymes targeting biofilm matrix components found in biofilms of varied species (30), which is hypothesized to help sidestep the limited spectrum of individual enzymes caused by the significant variations in matrix contents observed among bacterial species and strains (31, 32). Indeed, the results of individual enzyme assays performed in this study underline the added value of combining enzymes as was done in the TEC. The β-1,6-hexosaminidase used in this study is a dispersin B homolog targeting the poly-β-1,6-N-acetylglucosamine (PNAG) component of the biofilm matrix (33, 34). PNAG is an important constituent of the matrix of multiple species’ biofilms and is required for the formation of biofilm by S. epidermidis (32, 35, 36) and S. aureus strains expressing the icaABCD operon (32, 37) and of E. coli strains expressing the pgaABCD locus (31, 38). Dispersin B was shown to disperse the biofilms of PNAG-producing strains of multiple species, including those included in our study (20, 39, 40). Cellulase is as endo-1,4-β-d-glucanase targeting cellulose, which is an important constituent of the biofilm matrix of Enterobacter spp. (41), including E. coli (42). It has been shown to disperse E. coli (27) and Pseudomonas aeruginosa (21) biofilms in vitro. The recombinant nonspecific endonuclease targets both DNA and RNA phosphodiester bonds (43). DNA is a common component of the biofilm matrix of most bacterial species, including S. aureus, S. epidermidis, and E. coli (44). While the biofilm dispersal properties of this enzyme had not been previously investigated, it has been favorably compared to the widely studied DNase I (22) for the hydrolysis of DNA in sputum (45).
The dispersal of biofilms by enzymes similar to the ones constituting TEC was shown in previous publications to reduce the tolerance of bacterial biofilms to antibiotics (21, 25, 46) and antiseptics (27). However, limitations related to the use of single enzymes have been reported. Izano et al. (39) observed that DNase I did not disperse biofilms of S. epidermidis contrarily to S. aureus and that dispersin B had an inverse effect by dispersing S. epidermidis biofilms and leaving S. aureus biofilms intact. Accordingly, they found that the dispersal of these biofilms by the appropriate enzyme (DNase I for S. aureus of dispersin B for S. epidermidis) improved the effect of the detergent cetylpyridinium chloride on CFU counts (39).
Our results illustrate the broad spectrum of the tri-enzymatic cocktail, which stems from the combination of enzymes with different activity spectrums (30). Furthermore, these results support the hypothesis of a synergy of enzymatic dispersal and a broad selection of antibiotics at concentrations clinically relevant to the treatment of PJIs against biofilms. As could be expected from the observed MBEC values, the antibiotic concentrations used for the coupon assays had a minimal effect on control biofilms, but their combination with an enzymatic pretreatment resulted in significant reductions of culturable cell numbers as well as total biomass in nearly all combinations of bacterial species and antibiotics.
The cytotoxicity of TEC to different cell lines relevant to the joint environment (macrophages, fibroblasts, osteoblasts) was found to be negligible after a 24-h exposure. An explanation to this observation could be that the targets of these enzymes are either not produced by mammalian cells (cellulose) or irrelevant to their proliferation (eDNA and PNAG).
These results should however be interpreted with caution, as our study presents some limitations. We limited ourselves to the study of biofilms of reference strains. While this facilitates the replication of our results by other teams in comparison to clinical strains, the loss of diversity in gene expression could mask the effect of the variability in matrix composition observed in some species (47). The antibiotic concentrations considered in our study did not take into account the serum protein binding. Among the drugs used in this study, doxycycline and daptomycin show a high serum protein binding, which could lead to an overestimation of their effects in our experimental setting. However, the albumin concentration in synovial fluid is three to four times inferior to the serum concentration (48). This difference in albumin concentrations could lead to a higher free fraction of drugs in the synovial fluid than in serum. For this reason, the total serum concentration appeared to be a reasonable estimate that limited the risk of underestimating the effects of antibiotics. The in vitro nature of our experiments limits extrapolation to the clinical setting, as it does not represent the diversity of the situations encountered in a clinical setting nor does the use of set concentrations of antibiotics reproduce the complex pharmacokinetics observed in the bone and joint environment (49).
However, these observations support our hypothesis that a treatment approach combining local administration, such as an intraarticular injection, of a broad-spectrum enzymatic cocktail and antibiotics could prove to be beneficial in a PJI setting where the variety of pathological bacterial species encountered is important (3). Further studies in more refined models, such as animal PJI models, should therefore be undertaken to confirm our in vitro results and determine an adequate strategy around this approach in complement to DAIR.
MATERIALS AND METHODS
Bacterial strains, antibiotics, and enzymes.
(i) Strains. Methicillin-resistant Staphylococcus aureus ATCC 33591, methicillin-resistant Staphylococcus epidermidis ATCC 35984, and Escherichia coli ATCC 47076 were used as representatives of the bacterial species usually encountered in PJI.
(ii) Antibiotics. The following antibiotics were used as laboratory standards: ciprofloxacin HCl (potency, 93.9%) and moxifloxacin HCl (potency, 90.9%) from Bayer AG (Leverkusen, Germany); colistin (potency, 79.6%) and doxycycline (potency, 90.2%) from Sigma-Aldrich (Saint-Louis, MO, USA); and linezolid (potency, 99.2%) from RibX Pharmaceuticals (Morristown, NJ, USA). The following antibiotics were used as powders for injections in humans: ceftazidime (potency, 72.5%; Panpharma France, Luitré, France), daptomycin (potency, 100%; MSD, Puteaux, France), meropenem (potency, 92%; Hospira Benelux BVBA, Brussels, Belgium), rifampin (potency, 100%; Aventis, Schiltigheim, France), and vancomycin (potency, 97.5%; Mylan, Hoeilaart, Belgium). The antibiotic concentrations were set on the basis of the pharmacokinetic/pharmacodynamic (PK/PD) characteristics of the antibiotics studied and their pharmacokinetic parameters in humans described in the EUCAST rationales for clinical breakpoints or in the literature (Table 2).
(iii) Tri-enzymatic cocktail. The tri-enzymatic cocktail (TEC; OrthenzyQure) used in this study was designed and provided by OneLife SA (Louvain-la-Neuve, Belgium). It consists of a sterile Tris-buffered (pH = 7) aqueous solution containing 400 U/ml of aspecific DNA/RNA endonuclease from Serratia marcescens (c-LEcta GmbH, Leipzig, Germany), 50 U/ml of endo-1,4-β-d-glucanase from Aspergillus niger (Sigma), and 0.06 U/ml of β-N-acetylhexosaminidase from Actinobacillus pleuropneumoniae (GIGA, Liège, Belgium) (composition described in patent publication numbers WO2017/211737 [28] and BE2020/5726 [29]). The TEC was stored at 4°C and was heated at 37°C for 30 min before use. The TEC is available upon request at OneLife SA for research purposes (under R&D reference OL-RD1000/OrthenzyQure).
In order to assay the biofilm dispersal effect of the individual enzymes, OneLife provided solutions of the individual enzymes at concentrations identical to those used in the TEC.
Susceptibility testing.
MICs were determined by microdilution following the CLSI protocol (50) in MHB-ca and TGN (TSB + 2% NaCl + 1% glucose), i.e., the medium used for growing biofilms. MICs of daptomycin were tested in MHB-ca and TGN containing 50 mg/liter of Ca2+.
MBECs were assayed by growing biofilms in Calgary biofilm devices (CBD; Innovotech, Edmonton, Canada) in MHB-ca and TGN. Bacterial suspensions were prepared in phosphate-buffered saline (PBS), starting from overnight cultures on Trypticase soy agar (TSA) from frozen stocks. The suspensions were adjusted to a turbidity of 0.5 McFarland and diluted 1:10 in medium. A total of 150 μl was added per well of the CBDs. The CBDs were incubated for 24 h at 37°C under a continuous rotation of 50 rpm to allow biofilm formation. Antibiotic dilutions in the two media—with 50 mg/liter of Ca2+ when assaying daptomycin—were prepared in new 96-well plates at concentrations ranging from 1,024 mg/liter to 0.5 mg/liter (256 mg/liter to 0.125 mg/liter for meropenem). The lids of the CBDs were rinsed twice in PBS before being transferred to the plates containing the antibiotics. The plates were then incubated for 24 h at 37°C with a continuous rotation of 50 rpm. At the end of the antibiotic challenge, the lids were rinsed twice in PBS before being transferred to new plates containing plain medium. The plates were then sonicated for 10 min (Branson 5510 ultrasonic bath; Emerson Electric, Saint-Louis, MO, USA). New sterile lids were placed, and the plates were incubated for 24 h at 37°C without agitation. At the end of the incubation period, the plates were examined for bacterial growth, and optical density measures at 650 nm (OD650) were performed using a SpectraMax M3 spectrophotometer (Molecular Devices, San Jose, CA, USA).
Biofilm culture.
Bacterial suspensions were prepared in PBS, starting from overnight cultures on TSA from frozen stocks. The suspensions were adjusted to an OD620 of 0.5 (∼8.5 log10 CFU/ml) using a CECIL 2021 spectrophotometer (Cecil Instruments Ltd, Milton, United-Kingdom). The suspension was then diluted 1:100 in TGN. The biofilms were grown on titanium alloy (Ti-6Al-4V) coupons (BioSurface, Bozeman, MN, USA) in 12-well plates using 3 ml of the final suspension per coupon. The plates were incubated for 24 h (ATCC 33591 and ATCC 35984) or 48 h (ATCC 47076) at 37°C, with a constant orbital shaking of 50 rpm. In the case of strain ATCC 47076 biofilms, the medium was changed after 24 h. These culture times were selected, as they allowed the biofilms to reach steady state over the study period and were determined in previous growth kinetic experiments for S. aureus (14). The culture duration period for S. epidermidis was derived from these results. Growth kinetic experiments were performed in this study for E. coli biofilms and showed that a steady state was reached after 48 h of culture. Samples were analyzed for CFU counts and biomass every 24 h for the growth kinetic experiments.
Biofilm treatments.
(i) Individual enzyme assays. The biofilm samples were rinsed twice with sterile PBS. They were then transferred to 24-well plates filled with either 1 ml of TEC or 1 ml of one of the individual enzymes at 37°C. The plates were then incubated for 30 min at 37°C. At the end of the incubation period, the biofilms were rinsed twice in sterile PBS and assayed for total biomass (see below).
(ii) Combination of enzymatic cocktail and antibiotics. The biofilm samples were separated into a control group and an enzyme cocktail group. The coupons of both groups were rinsed twice in sterile PBS before being submitted to the treatments. The tri-enzyme cocktail group samples were transferred to a 24-well plate, in which the wells were previously filled with 1 ml of enzyme cocktail at 37°C. The plates were then incubated at 37°C with no agitation for either 15, 30, and 60 min (when studying the kinetics of activity) or 30 min (other experiments). After incubation, the samples were rinsed twice in sterile PBS before either immediate analysis (see below for analysis technique descriptions) or reincubation. Control group samples were directly rinsed in sterile PBS. All samples were then reincubated for 24 h at 37°C under a constant orbital shaking of 50 rpm in TGN or TGN containing antibiotics at set concentrations (TGN + 50 mg/liter of Ca2+ for samples exposed to daptomycin) (see Table 2) in 12-well plates containing 3 ml of medium per well.
Biofilms analysis.
The coupons were rinsed twice in sterile PBS and then analyzed using the following techniques.
(i) CFU counts. Coupons were placed in 15-ml conical tubes (Greiner Bio-One International GmbH, Kremsmünster, Austria) filled with 2 ml of sterile PBS. In order to detach the biofilms and suspend the bacteria, the tubes were then vortexed for 30 s at maximum intensity (Vortex-Genie 2; Scientific Industries, Inc., Bohemia, NY, USA), sonicated for 5 min (Branson 5510 Ultrasonic bath, Emerson Electric, Saint-Louis, MO, USA), and vortexed again for 30 s. The suspension was then sampled and serially diluted before plating on TSA. The TSA plates were then incubated for 24 to 48 h at 37°C until the colonies reached an adequate size. CFU counts were then obtained using an automated method (image acquisition using Gel Doc XR+ and image processing using Quantity One (Bio-Rad, Hercules, CA, USA).
(ii) Biomass assay. Coupons were dried overnight at 60°C. They were then stained with 1 ml of 1% crystal violet solution for 10 min (Sigma-Aldrich Corp.). The coupons were then rinsed twice using deionized water to remove the excess dye. Afterward, they were placed for 1 h in 24-well plates filled with 1 ml of 66% acetic acid solution (Merck KGaA, Darmstadt, Germany) to resolubilize the dye. The coupons were removed from the wells, and the absorbance of the solution was read at 570 nm using a SpectraMax M3 spectrophotometer (Molecular Devices, San Jose, CA, USA).
(iii) Fluorescence microscopy. Coupons were rinsed twice in sterile deionized water to avoid interference from phosphate ions. They were then stained using the Filmtracer LIVE/DEAD biofilm viability kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s instructions. Coupons were then rinsed again twice in sterile deionized water before imaging using an Axio Imager Z1 microscope fitted with an ApoTome 1 attachment (Zeiss, Oberkochen, Germany) at a ×20 magnification. The images were postprocessed using FIJI (51, 52), using the maximum intensity projection technique, and by adjusting the brightness of the channels.
Cytotoxicity assays.
(i) Cell culture. J774 murine macrophages (Sandoz Forschung Laboratories, Vienna, Austria) (53), MG-63 human osteoblasts (ATCC CRL-1427), and primary murine fibroblasts (54) were used for the cytotoxicity assays. The cells were grown in RPMI 1640 or Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (all products from Thermo Fisher Scientific Inc., Waltham, MA, USA) at 37°C in a 5% CO2 atmosphere. The cells were used for the assays between passages 2 and 5.
The cells were cultured initially in 250-cm³ cell culture flasks before being transferred to 96-well plates at a concentration of 104 cells per well. The plates were incubated for 24 h at 37°C with 5% CO2 to allow cellular adhesion to the wells before testing.
(ii) Assay.Cytotoxicity was assessed by measuring the release of the cytoplasmic enzyme lactate dehydrogenase (LDH) in the culture medium, using the cytotoxicity detection kit (Sigma-Aldrich). The wells were rinsed with sterile PBS before adding 100 μl of medium with 1% decomplemented fetal bovine serum. A total of 100 μl of the following test substances were then added to the wells (final concentrations): TEC (concentration used against biofilms and half of this concentration), Triton X-100 solution (1%, positive control), medium with 1% decomplemented serum (negative control). Three empty wells per experiment were filled with 200 μl of medium with 1% decomplemented serum to measure the background signal.
The plates were then incubated for 1 h or 24 h at 37°C in a 5% CO2 atmosphere. A total of 100 μl of the supernatant of each well was aspirated and transferred to a new 96-well plate and added by 100 μl of the extemporaneously prepared LDH assay solution as per the manufacturer’s instructions. The plates were then incubated for 30 min at room temperature in the dark. The light absorbance was then measured at 490 nm and 650 nm using a SpectraMax M3 spectrophotometer (Molecular Devices, San Jose, CA, USA).
Data transformation and statistical analysis.
CFU counts were normalized using a base 10 logarithmic transformation before analysis. Absorbance values of the biomass assays were transformed into percentages of the T0 (no reincubation) controls after subtracting the average value of negative controls. The absorbance values of the LDH assays were transformed into percentages using the following equation:
The statistical analysis was then performed using R (version 3.6.1) and the package car (version 3.0.6) or in GraphPad 8.4.3 (GraphPad Software, San Diego, CA). The TEC kinetics experiment data were analyzed using repeated measures ANOVA followed by paired t tests with Holm correction. The data relating to the combined effect of TEC and antibiotics were analyzed using two-way ANOVA after assessing the normality of residues with QQ plots and the equality of variances using the Levene test. The Tukey honestly significant difference (HSD) test was used to perform pairwise comparisons. The synergy of treatments was defined as a statistically significant interaction parameter of the two-way ANOVAs (55). The threshold for alpha errors is 0.05. The data reported in the text are the means with 95% confidence interval.
Data availability.
The data for the experiments reported in this publication are available upon request to the corresponding author.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alix Mangin for his technical help and OneLife SA for providing the tri-enzymatic cocktail for academic research purposes and its R&D team for their intellectual contribution to the project.
H.P. designed and performed the experiments and data analysis. The other authors provided helpful suggestions regarding experiment design or performance. All authors contributed to the writing of the manuscript.
The experiments presented in this manuscript were funded by the Région Wallonne through a Win2Wal grant (Orthenzy; grant number 1810122) and the F.R.S.-FNRS (Fonds de la Recherche Scientifique – FNRS) (grants T.0189.16 and J.0162.19). H.P. is the beneficiary of a FRIA (Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture) grant of the FRS-FNRS.
F.V.B. is a research director of the FRS-FNRS.
We declare that we have no affiliations or involvement with any entity with financial interests related to the content of this manuscript.
REFERENCES
- 1.Kurtz SM, Lau E, Watson H, Schmier JK, Parvizi J. 2012. Economic burden of periprosthetic joint infection in the United States. J Arthroplasty 27:61–65. doi: 10.1016/j.arth.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Bozic KJ, Kamath AF, Ong K, Lau E, Kurtz S, Chan V, Vail TP, Rubash H, Berry DJ. 2015. Comparative epidemiology of revision arthroplasty: failed THA poses greater clinical and economic burdens than failed TKA. Clin Orthop Relat Res 473:2131–2138. doi: 10.1007/s11999-014-4078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal VK, Bakhshi H, Ecker NU, Parvizi J, Gehrke T, Kendoff D. 2014. Organism profile in periprosthetic joint infection: pathogens differ at two arthroplasty infection referral centers in Europe and in the United States. J Knee Surg 27:399–406. doi: 10.1055/s-0033-1364102. [DOI] [PubMed] [Google Scholar]
- 4.Arciola CR, Campoccia D, Montanaro L. 2018. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol 16:397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 5.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3:MB-0010-2014. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osmon DR, Berbari EF, Berendt AR, Lew D, Zimmerli W, Steckelberg JM, Rao N, Hanssen A, Wilson WR, Infectious Diseases Society of America. 2013. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 56:e1–e25. doi: 10.1093/cid/cis803. [DOI] [PubMed] [Google Scholar]
- 8.Grammatopoulos G, Bolduc ME, Atkins BL, Kendrick BJL, McLardy-Smith P, Murray DW, Gundle R, Taylor AH. 2017. Functional outcome of debridement, antibiotics and implant retention in periprosthetic joint infection involving the hip: a case-control study. Bone Joint J 99-B:614–622. doi: 10.1302/0301-620X.99B5.BJJ-2016-0562.R2. [DOI] [PubMed] [Google Scholar]
- 9.Peel TN, Dowsey MM, Buising KL, Liew D, Choong PFM. 2013. Cost analysis of debridement and retention for management of prosthetic joint infection. Clin Microbiol Infect 19:181–186. doi: 10.1111/j.1469-0691.2011.03758.x. [DOI] [PubMed] [Google Scholar]
- 10.Klouche S, Sariali E, Mamoudy P. 2010. Total hip arthroplasty revision due to infection: a cost analysis approach. Orthop Traumatol Surg Res 96:124–132. doi: 10.1016/j.otsr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Urish KL, Bullock AG, Kreger AM, Shah NB, Jeong K, Rothenberger SD, Infected Implant Consortium. 2018. A multicenter study of irrigation and debridement in total knee arthroplasty periprosthetic joint infection: treatment failure is high. J Arthroplasty 33:1154–1159. doi: 10.1016/j.arth.2017.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunutsor SK, Beswick AD, Whitehouse MR, Wylde V, Blom AW. 2018. Debridement, antibiotics and implant retention for periprosthetic joint infections: a systematic review and meta-analysis of treatment outcomes. J Infect 77:479–488. doi: 10.1016/j.jinf.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Urish KL, DeMuth PW, Craft DW, Haider H, Davis CM, III. 2014. Pulse lavage is inadequate at removal of biofilm from the surface of total knee arthroplasty materials. J Arthroplasty 29:1128–1132. doi: 10.1016/j.arth.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Poilvache H, Ruiz-Sorribas A, Sakoulas G, Rodriguez-Villalobos H, Cornu O, Van Bambeke F. 2020. Synergistic effects of pulsed lavage and antimicrobial therapy against Staphylococcus aureus biofilms in an in-vitro model. Front Med (Lausanne) 7:527. doi: 10.3389/fmed.2020.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriarty TF, Kuehl R, Coenye T, Metsemakers WJ, Morgenstern M, Schwarz EM, Riool M, Zaat SAJ, Khana N, Kates SL, Richards RG. 2016. Orthopaedic device-related infection: current and future interventions for improved prevention and treatment. EFORT Open Rev 1:89–99. doi: 10.1302/2058-5241.1.000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoodley P, Ehrlich GD, Sedghizadeh PP, Hall-Stoodley L, Baratz ME, Altman DT, Sotereanos NG, Costerton JW, Demeo P. 2011. Orthopaedic biofilm infections. Curr Orthop Pract 22:558–563. doi: 10.1097/BCO.0b013e318230efcf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arciola CR, Campoccia D, Speziale P, Montanaro L, Costerton JW. 2012. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33:5967–5982. doi: 10.1016/j.biomaterials.2012.05.031. [DOI] [PubMed] [Google Scholar]
- 18.Romano CL, Toscano M, Romano D, Drago L. 2013. Antibiofilm agents and implant-related infections in orthopaedics: where are we? J Chemother 25:67–80. doi: 10.1179/1973947812Y.0000000045. [DOI] [PubMed] [Google Scholar]
- 19.Vuotto C, Donelli G. 2019. Novel treatment strategies for biofilm-based infections. Drugs 79:1635–1655. doi: 10.1007/s40265-019-01184-z. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleming D, Chahin L, Rumbaugh K. 2017. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother 61:e01998-16. doi: 10.1128/AAC.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming D, Rumbaugh KP. 2017. Approaches to dispersing medical biofilms. Microorganisms 5:15. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan S, Zapotoczna M, Stevens NT, Humphreys H, O'Gara JP, O'Neill E. 2017. Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections. J Hosp Infect 96:177–182. doi: 10.1016/j.jhin.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan JB, LoVetri K, Cardona ST, Madhyastha S, Sadovskaya I, Jabbouri S, Izano EA. 2012. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J Antibiot (Tokyo) 65:73–77. doi: 10.1038/ja.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darouiche RO, Mansouri MD, Gawande PV, Madhyastha S. 2009. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother 64:88–93. doi: 10.1093/jac/dkp158. [DOI] [PubMed] [Google Scholar]
- 27.Lim ES, Koo OK, Kim MJ, Kim JS. 2019. Bio-enzymes for inhibition and elimination of Escherichia coli O157:H7 biofilm and their synergistic effect with sodium hypochlorite. Sci Rep 9:9920. doi: 10.1038/s41598-019-46363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siala MW, Vanzieleghem MT. 14 December 2017. Composition comprising at least one enzyme and at least one microbicidal molecule for the prevention or treatment of post-implant infections. International patent WO2017/211737A1.
- 29.Fontaine L, Lardinois A, Chalhoub H. 16 October 2020. Composition para-pharmaceutique ou pharmaceutique administrable à un être vivant, de préférence un être humain, comprenant au moins une enzyme pour le traitement et/ou la prévention d'infections bactériennes impliquant la formation de biofilm. Patent BE2020/5726.
- 30.Siala W, Hoche A, Van Bambeke F, Vanzieleghem T. 2018. Activity of combinations of an enzymatic cocktail (CDD) with antibiotics against biofilms of clinical isolates of ESKAPE pathogens, abstr. 28th European Congress of Clinical Microbiology and Infectious Diseases, Madrid, Spain, 21 to 24 April 2018.
- 31.Horvat M, Pannuri A, Romeo T, Dogsa I, Stopar D. 2019. Viscoelastic response of Escherichia coli biofilms to genetically altered expression of extracellular matrix components. Soft Matter 15:5042–5051. doi: 10.1039/c9sm00297a. [DOI] [PubMed] [Google Scholar]
- 32.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JKM, Ragunath C, Kaplan JB, Mack D. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol 185:4693–4698. doi: 10.1128/jb.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JKM, Ramasubbu N. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol 186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriology 178:175–183. doi: 10.1128/JB.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post V, Harris LG, Morgenstern M, Mageiros L, Hitchings MD, Meric G, Pascoe B, Sheppard SK, Richards RG, Moriarty TF. 2017. Comparative genomics study of Staphylococcus epidermidis isolates from orthopedic-device-related infections correlated with patient outcome. J Clin Microbiol 55:3089–3103. doi: 10.1128/JCM.00881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gowrishankar S, Kamaladevi A, Balamurugan K, Pandian SK. 2016. In vitro and in vivo biofilm characterization of methicillin-resistant Staphylococcus aureus from patients associated with pharyngitis infection. Biomed Res Int 2016:1289157. doi: 10.1155/2016/1289157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Preston JF, III, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186:2724–2734. doi: 10.1128/jb.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, III, Romeo T. 2005. Depolymerization of beta-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol 187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zogaj X, Bokranz W, Nimtz M, Römling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun 71:4151–4158. doi: 10.1128/iai.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 43.Nestle M, Roberts WK. 1969. An extracellular nuclease from Serratia marcescens. I. Purification and some properties of the enzyme. J Biol Chem 244:5213–5218. doi: 10.1016/S0021-9258(18)63648-8. [DOI] [PubMed] [Google Scholar]
- 44.Jakubovics NS, Shields RC, Rajarajan N, Burgess JG. 2013. Life after death: the critical role of extracellular DNA in microbial biofilms. Lett Appl Microbiol 57:467–475. doi: 10.1111/lam.12134. [DOI] [PubMed] [Google Scholar]
- 45.Vafina G, Zainutdinova E, Bulatov E, Filimonova MN. 2018. Endonuclease from Gram-negative bacteria Serratia marcescens is as effective as Pulmozyme in the hydrolysis of DNA in sputum. Front Pharmacol 9:114. doi: 10.3389/fphar.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waryah CB, Wells K, Ulluwishewa D, Chen-Tan N, Gogoi-Tiwari J, Ravensdale J, Costantino P, Gökçen A, Vilcinskas A, Wiesner J, Mukkur T. 2017. In vitro antimicrobial efficacy of tobramycin against Staphylococcus aureus biofilms in combination with or without DNase I and/or dispersin B: a preliminary investigation. Microb Drug Resist 23:384–390. doi: 10.1089/mdr.2016.0100. [DOI] [PubMed] [Google Scholar]
- 47.O'Gara JP. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 48.Levick JR. 1981. Permeability of rheumatoid and normal human synovium to specific plasma proteins. Arthritis Rheum 24:1550–1560. doi: 10.1002/art.1780241215. [DOI] [PubMed] [Google Scholar]
- 49.Landersdorfer CB, Bulitta JB, Kinzig M, Holzgrabe U, Sorgel F. 2009. Penetration of antibacterials into bone: pharmacokinetic, pharmacodynamic and bioanalytical considerations. Clin Pharmacokinet 48:89–124. doi: 10.2165/00003088-200948020-00002. [DOI] [PubMed] [Google Scholar]
- 50.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 51.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyderman R, Pike MC, Fischer DG, Koren HS. 1977. Biologic and biochemical activities of continuous macrophage cell lines P388D1 and J774.1. J Immunol 119:2060–2066. [PubMed] [Google Scholar]
- 54.Tulkens P, Beaufay H, Trouet A. 1974. Analytical fractionation of homogenates from cultured rat embryo fibroblasts. J Cell Biol 63:383–401. doi: 10.1083/jcb.63.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slinker BK. 1998. The statistics of synergism. J Mol Cell Cardiol 30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for the experiments reported in this publication are available upon request to the corresponding author.