Suppression of the recA SOS response gene and reactive oxygen species (ROS) overproduction have been shown, separately, to enhance fluoroquinolone activity and lethality. Their putative synergistic impact as a strategy to potentiate the efficacy of bactericidal antimicrobial agents such as fluoroquinolones is unknown.
KEYWORDS: resistance reversion, quinolones, SOS response, recA, oxidative stress, detoxification systems
ABSTRACT
Suppression of the recA SOS response gene and reactive oxygen species (ROS) overproduction have been shown, separately, to enhance fluoroquinolone activity and lethality. Their putative synergistic impact as a strategy to potentiate the efficacy of bactericidal antimicrobial agents such as fluoroquinolones is unknown. We generated Escherichia coli mutants that exhibited a suppressed ΔrecA gene in combination with inactivated ROS detoxification system genes (ΔsodA, ΔsodB, ΔkatG, ΔkatE, and ΔahpC) or inactivated oxidative stress regulator genes (ΔoxyR and ΔrpoS) to evaluate the interplay of both DNA repair and detoxification systems in drug response. Synergistic sensitization effects, ranging from 7.5- to 30-fold relative to the wild type, were observed with ciprofloxacin in double knockouts of recA and inactivated detoxification system genes. Compared to recA knockout, inactivation of an additional detoxification system gene reduced MIC values up to 8-fold. In growth curves, no growth was evident in mutants doubly deficient for recA gene and oxidative detoxification systems at subinhibitory concentrations of ciprofloxacin, in contrast to the recA-deficient strain. There was a marked reduction of viable bacteria in a short period of time when the recA gene and other detoxification system genes (katG, sodA, or ahpC) were inactivated (using absolute ciprofloxacin concentrations). At 4 h, a bactericidal effect of ciprofloxacin was observed for ΔkatG ΔrecA and ΔahpC ΔrecA double mutants compared to the single ΔrecA mutant (Δ3.4 log10 CFU/ml). Synergistic quinolone sensitization, by targeting the recA gene and oxidative detoxification stress systems, reinforces the role of both DNA repair systems and ROS in antibiotic-induced bacterial cell death, opening up a new pathway for antimicrobial sensitization.
INTRODUCTION
Widespread and indiscriminate use of antibiotics in human and veterinary medicine and agriculture has become a global public health problem (1, 2), and the need to take measures to avoid returning to the preantibiotic era is now urgent (3). Efforts to overcome the problem of resistance have focused mainly on modifying existing antibiotics by circumventing molecular mechanisms conferring resistance (4). Various strategies are being studied to combat this emergence, including searching for new antibiotics, use of adjuvants that increase sensitivity, genome editing technologies, and phage therapy (4–9). While such efforts might be efficacious against resistant strains, new resistance mechanisms often arise in the process of adaptation to new antimicrobial agents (10, 11). Hence, it becomes necessary to discover how to reverse resistance or to sensitize bacteria to old antibiotics and block their evolution (9).
Enterobacterales bacteria such as Escherichia coli are among the most common causes of community and nosocomial infections. Fluoroquinolones are used for empirical and directed therapy in infections caused by E. coli (12). Fluoroquinolones are broad-spectrum antimicrobials whose mechanism of action is related to the inhibition of two enzymes essential for bacterial viability, DNA gyrase and topoisomerase type IV, which are involved in DNA synthesis. Target mutations in the topoisomerases cause resistance to these antimicrobial agents (13). Plasmid-mediated quinolone resistance mechanisms have also been described (14).
Microorganisms have the ability to evolve rapidly to ensure survival, which has led to high levels of antibiotic resistance. The SOS response is a coordinated cellular response to genotoxic damage, and it can contribute to this evolution (low-fidelity polymerases). Some antibiotics induce SOS by a variety of molecular mechanisms. Specifically, fluoroquinolones activate the SOS response caused by DNA damage to arrest replication forks (15, 16). A number of studies have recently demonstrated the role of RecA protein activation, which is associated with the SOS response in E. coli, in the emergence of mutations in gyrA and parC that lead to resistance (15, 17–19). LexA protein represses the SOS regulon, which consists of more than 50 genes, many associated with different repair mechanisms (16, 20, 21). When DNA damage occurs, single-strand DNA activates RecA, which forms nucleofilaments around it and induces autoproteolysis of LexA and activation of the SOS response. Sequential induction of genes could facilitate the transition from high-fidelity to lower-fidelity DNA repair mechanisms (16, 21). Recent studies have demonstrated that suppression of the SOS response enhances the bactericidal activity of antimicrobials such as quinolones in bacteria with specific resistant mechanisms (17, 22).
It is also known that bactericidal agents are involved in oxidative stress since, under aerobic conditions, they produce reactive oxygen species (ROS). Downstream of their target-specific interactions, bactericidal antibiotics induce complex redox alterations that contribute to cellular damage and death, thus supporting their involvement in antibiotic lethality (23–25). In vitro studies have shown a protective effect against ROS, produced by antibiotic, of some antioxidant molecules, such as ascorbic acid and N-acetylcysteine (25, 26). Bacteria also have multiple oxidative detoxification systems, such as superoxide dismutase (SOD) and catalase, to combat oxidative stress (27, 28). Specifically, E. coli has three types of SOD (SodA, SodB, and SodC), two types of catalase (KatG and KatE), which remove O2– and H2O2, respectively, and an alkyl hydroperoxide reductase (made of two subunits, AhpF and AhpC), which reduces NADH while removing H2O2. Transcriptional regulation is controlled by oxidative stress regulators such as OxyR or general stress response regulators such as RpoS. During oxidative stress, OxyR increases the expression of katG and ahpC genes, while RpoS regulates the expression of katE and sodC (29).
Hence, suppression of the SOS response and overproduction of ROS have been shown, separately, to enhance the activity and lethality of fluoroquinolones. Nevertheless, the use of their putative synergistic effect as a strategy to reinforce the efficacy of fluoroquinolones remains an open question. To address this point, we generated a wide collection of E. coli mutants that exhibited a suppressed SOS response (ΔrecA) in combination with inactivated ROS detoxification system genes (ΔsodA, ΔsodB, ΔkatG, ΔkatE, ΔsodA, and ΔahpC) or inactivated oxidative stress regulatory genes (ΔoxyR and ΔrpoS) to evaluate the interplay of both DNA repair/recombination and detoxification systems in the antimicrobial drug response. Our comprehensive analysis opens up a new strategy for bacterial sensitization by double-targeting SOS response and oxidative stress.
RESULTS
Quinolone sensitization by targeting the recA SOS response gene and oxidative stress response.
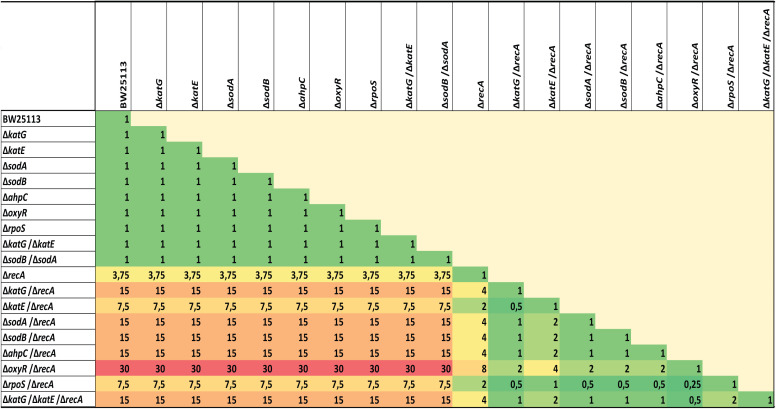
We generated E. coli mutants that exhibited the deleted recA gene in combination with suppressed ROS detoxification system genes (ΔsodA, ΔsodB, ΔkatG, ΔkatE, and ΔahpC) or genes regulating oxidative stress (ΔoxyR and ΔrpoS) to evaluate the interplay of both DNA repair and detoxification systems in antimicrobial drug response (Table 1; see Table S1 in the supplemental material).
TABLE 1.
Heat map of the fold decrease in ciprofloxacin MICs of different recA and detoxification system inactivationsa
MIC values represent the means of at least three independent determinations performed on separate days. The raw MIC values of all strains are shown in Table S1 in the supplemental material.
We confirmed first that the recA knockout produces the expected perturbation of the SOS response in both the wild-type BW25113 and the various detoxification system knockouts. Figure S1 represents the results obtained for each isogenic group used in this study. No fluorescence was observed when recA was inactivated, confirming the perturbation of the SOS response (together with DNA repair and recombination). In terms of MIC values, as expected, the ciprofloxacin MICs were 3.75-fold lower for the ΔrecA mutant compared to wild-type BW25113 (Table 1, Table S1). No reductions in ciprofloxacin MIC were observed when any of the detoxification system genes were inactivated, including single (ΔkatG, ΔkatE, ΔsodA, ΔsodB, ΔahpC, ΔoxyR, and ΔrpoS) and double knockouts (ΔkatG ΔkatE and ΔsodB ΔsodA) compared to wild-type BW25113.
Notably, in general, a synergistic sensitization effect was observed in double knockouts of recA (with SOS inactivation deficient for DNA repair and recombination) and the different ROS detoxification system genes that were inactivated (see Table S1). Fractional inhibitory concentration (FIC) values were below 0.5 for double and triple gene deletion mutants, 0.16 (ΔoxyR ΔrecA) and 0.32 (ΔkatG ΔrecA, ΔsodA ΔrecA, ΔsodB ΔrecA, ΔahpC ΔrecA, and ΔkatG ΔkatE ΔrecA). Only ΔkatE ΔrecA and ΔrpoS ΔrecA show an additive effect, as their FIC value was 0.63; the value was located in the range of 0.5 to ≤1. The process of sensitization was similarly efficient across all double combinations, including the effector detoxification genes (sodA, sodB, katG, katE, sodA, and ahpC) and genes regulating oxidative stress (oxyR and rpoS), which ranged from 7.5-fold to 30-fold compared to wild-type BW25113 (Table 1). The highest sensitization was found for the ΔrecA ΔoxyR double knockout, with a ciprofloxacin MIC value of 0.0005 mg/liter (Table 1, Table S1). For most of the combinations, sensitization increased about 15-fold compared to wild-type BW25113. Compared to values for the recA knockout, inactivation of an additional detoxification system gene reduced MIC values between 2-fold and 8-fold. Here, we show that, in terms of MIC, recA gene suppression combined with inactivation of multiple detoxification system genes sensitizes to ciprofloxacin in a synergistic way.
As expected, the effect of synergistic sensitization to ciprofloxacin following recA gene suppression in combination with inactivation of detoxification system genes was not evident under anaerobic conditions (Table S1).
In addition, we also tried to evaluate the impact of this strategy on an evolved E. coli mutant strain harboring acquired quinolone resistance mechanisms (BW15 strain in Table S1). This mutant harbored changes affecting the gyrA, gyrB, parE, and marR genes, including a D87G substitution in the gyrA gene. The modifications produced a ciprofloxacin-resistant phenotype with a MIC value of 2 mg/liter according to CLSI and EUCAST breakpoints (30, 31). As expected, the recA-deficient BW15 derivative involved a change of category to susceptible-intermediate or areas of technical uncertainty (ATU) according to CLSI and EUCAST breakpoints (MIC of 0.5 mg/liter). Unexpectedly, no further sensitization was apparent when the recA gene and detoxification system genes (katG or sodA) were inactivated in this resistant mutant (Table S1).
Besides synergistic sensitization found for ciprofloxacin, in terms of MICs, was not always reproduced for other fluoroquinolones in the multiple genetic combinations assayed (Table S1). Based on these data, the rest of the study focused on the synergistic effects of sensitization to ciprofloxacin as a result of recA gene suppression in combination with inactivation of detoxification systems.
Monitoring bacterial in vitro growth in the presence of quinolones.
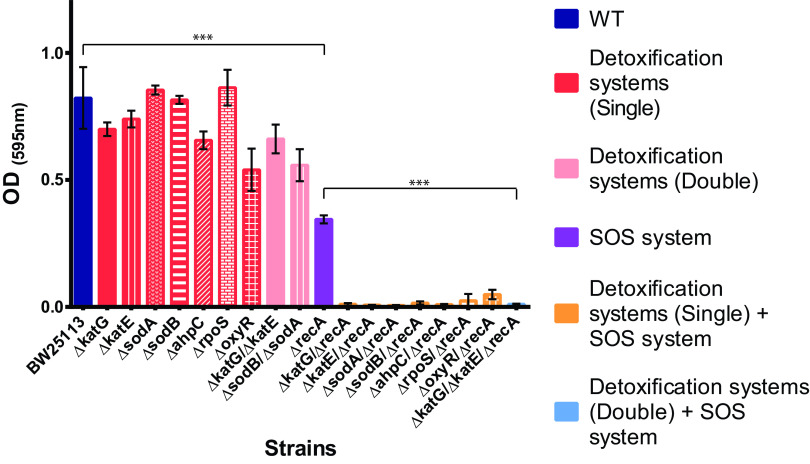
In vitro growth curves were analyzed at subinhibitory concentrations of ciprofloxacin (0.001 mg/liter, 1/15× MIC based on the susceptibility of the BW25113 strain with the unmodified recA gene or ROS detoxification system genes) (Fig. 1). At this concentration, growth (after 20 h) was observed for the wild type and mutants with deficient detoxification systems (optical density [OD] values ranged from 0.54 to 0.86). As expected, reduced growth was observed for the recA-deficient strain (OD value, 0.35) (P < 0.001 compared to wild-type BW25113; OD value, 0.82). Interestingly, no growth was found for mutants doubly deficient in the recA gene and oxidative detoxification systems (OD of less than 0.05), including effector detoxification genes (sodA, sodB, katG, katE, sodA, and ahpC) and regulatory genes of oxidative stress (oxyR and rpoS) (P < 0.001 compared to recA-deficient BW25113) (Fig. 1).
FIG 1.
The impact of recA gene inactivation and detoxification system suppression on cell growth in the presence of sublethal quinolone concentrations. OD values for E. coli BW25113 and all genetic combinations with ciprofloxacin concentrations of 0.001 mg/liter at 20 h. Data are represented as the mean of at least four independent measurements. The OD of all constructions was similar in the nonantibiotic control (data not shown). Standard deviations are indicated by error bars. Significant P values are noted (***, P < 0.001).
Figure S2 shows full monitoring of in vitro growth curves at subinhibitory concentrations (0.0005 to 0.002 mg/liter) of ciprofloxacin for each isogenic group. Under these conditions, no growth or a very marked delay in growth was observed in mutants doubly deficient in the recA gene and oxidative detoxification systems compared to recA-deficient BW25113. For example, for effector detoxification genes such as the katE group (Fig. S2B), at concentrations of 0.001 mg/liter of ciprofloxacin, only the BW25113 strain and ΔkatE grew regularly at both 8 h and 16 h (this behavior was stable after 24 h). Significantly delayed growth was observed for ΔrecA, and no growth was observed for the ΔkatE ΔrecA combination. A similar pattern was obtained for the rest of the effector and regulator detoxification genes (Fig. S2).
These data support the efficacy of combining the inactivation of the recA gene with detoxification system genes in order to prevent bacterial growth in the presence of sublethal concentrations of ciprofloxacin over shorter and longer periods of time.
The recA gene and oxidative stress response suppression enhances ciprofloxacin bactericidal activity.
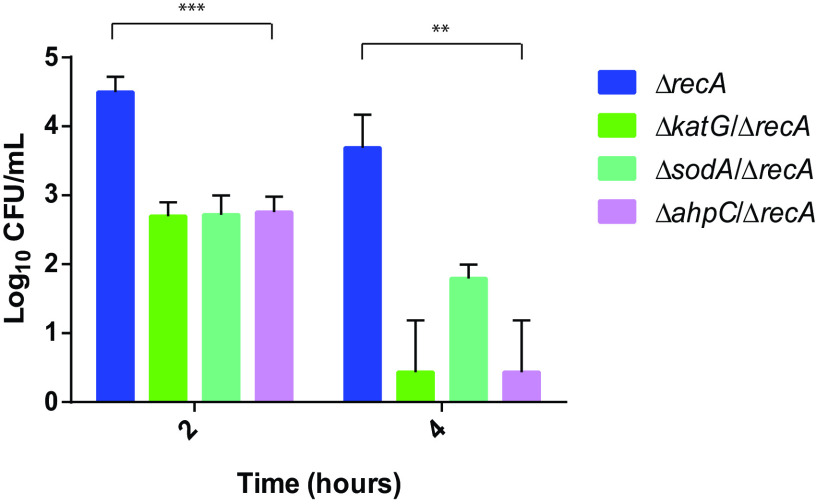
To show the synergistic impact of suppressing the recA gene and the oxidative stress response in terms of bacterial viability, time-kill curves were obtained for each isogenic group (recA, katG, sodA, and ahpC) (Fig. 2 and Fig. S3). At 1× MIC relative to the ΔrecA MIC value of ciprofloxacin (0.004 mg/liter), a marked reduction in viable bacteria was observed in a short period of time (2 and 4 h) when the recA gene and other ROS detoxification system genes (katG, sodA, or ahpC) were inactivated (Fig. 2). At 2 h, a bacteriostatic effect (drop of <3 log10 CFU/ml) was observed for the three detoxification system genes (although a marked difference was observed compared to the single ΔrecA mutant, up to Δ1.8 log10 CFU/ml for the ΔkatG ΔrecA genes) (P < 0.001). At 4 h, however, a bactericidal effect (drop of >3 log10 CFU/ml) was observed for the double mutants ΔkatG ΔrecA and ΔahpC ΔrecA compared to the single ΔrecA mutant (Δ3.4 log10 CFU/ml for the ΔkatG ΔrecA and ΔahpC ΔrecA genes) (P < 0.01) (Fig. 2).
FIG 2.
Combination of recA gene inactivation and detoxification system suppression inactivation enhances synergistically bactericidal activity of quinolones. Viable bacterial counts of E. coli ΔrecA, E. coli ΔkatG ΔrecA, E. coli ΔsodA ΔrecA, and E. coli ΔahpC ΔrecA in time-kill assays at ciprofloxacin (CIP) concentrations of 1× MIC ΔrecA (0.004 mg/liter) at 2 and 4 h. Data are represented as the means of at least three independent measurements. E. coli BW25113 was not included because the antibiotic concentration used was under its MIC, so its behavior was similar to the nonantibiotic control. Viable bacterial counts of all constructions were similar in the nonantibiotic control (data not shown). Error bars represent standard deviations. Significant P values are noted (**, P < 0.01; ***, P < 0.001).
At 1× MIC relative to the wild-type MIC of ciprofloxacin, 0.015 mg/liter (Fig. S3), a marked reduction in viable bacteria was also observed after a short and long period of time (2 to 8 h) when the recA gene together with other detoxification system genes (sodA or ahpC) was inactivated, compared to the single recA mutant. Nevertheless, this concentration appeared to be too high to demonstrate differences in the case of the katG gene group (BW25113, ΔkatG, ΔrecA, and ΔkatG ΔrecA). Under these conditions, a drop of 1.79 log10 CFU/ml was observed at 4 h for the ΔahpC ΔrecA double mutant compared to the single ΔrecA mutant (Fig. S3). Interestingly, after 8 h, no viable bacteria were detected for the single ΔrecA mutant and the ΔkatG ΔrecA, ΔsodA ΔrecA, and ΔahpC ΔrecA double mutants compared to wild-type E. coli BW25113 and single mutants for detoxification system genes.
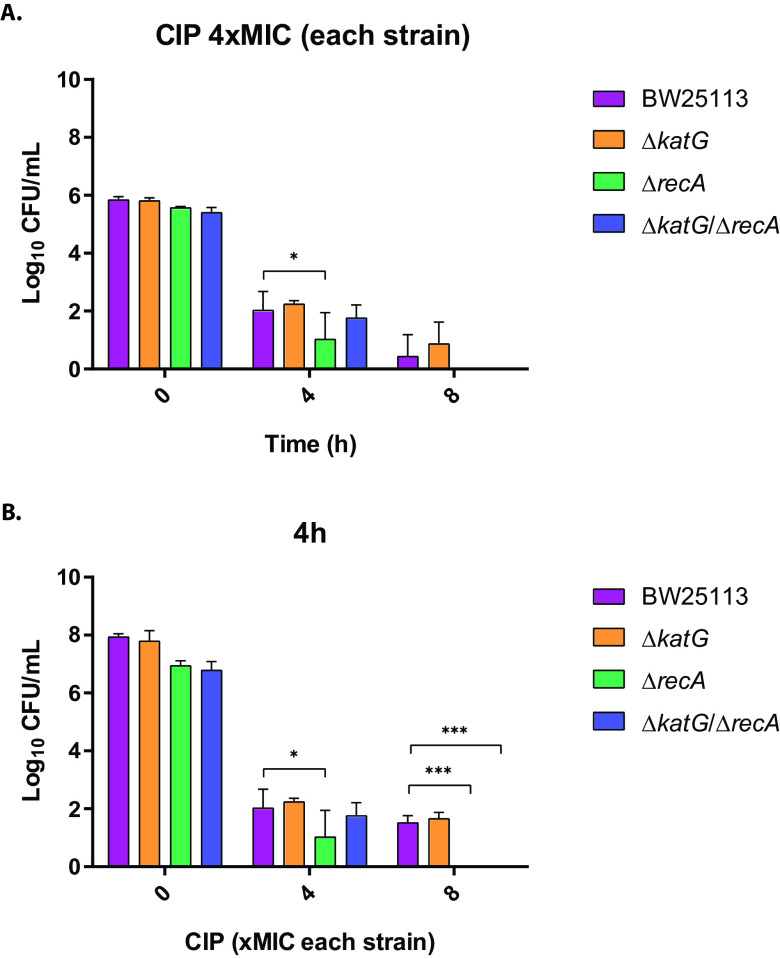
From another point of view, we also determined bacterial viability, specifically for the isogenic group KatG, at normalized drug concentrations according to the MICs. At a ciprofloxacin concentration of 4× MIC relative to each strain (0.06 mg/liter for BW25113 and ΔkatG; 0.015 mg/liter for ΔrecA and 0.004 mg/liter for ΔkatG ΔrecA) at 0 h, 4 h, and 8 h, no significative differences were observed between strains, with the exception of ΔrecA relative to control strain BW25113 (P < 0.05) (Fig. 3A). Also, at 8× MIC relative to each strain (0.125 mg/liter for BW25113 and ΔkatG, 0.03 mg/liter for ΔrecA, and 0.008 mg/liter for ΔkatG ΔrecA) at 4h, significant differences were observed between ΔrecA and ΔkatG ΔrecA relative to control strain BW25113 (P < 0.001) (Fig. 3B), but not between them. These data support that observed differences in lethality could be due to initial differences observed in terms of MIC values.
FIG 3.
(A and B) Viable bacterial counts of E. coli BW25113 with recA gene and katG detoxification system inactivation alone or in combinations in time-kill assays at (A) ciprofloxacin (CIP) concentrations of 4× MIC of each strain (normalized MICs, 0.06 mg/liter for BW25113 and ΔkatG, 0.015 mg/liter for ΔrecA, and 0.004 mg/liter for ΔkatG ΔrecA) at 0 h, 4 h, and 8 h and (B) ciprofloxacin concentrations of 4× MIC and 8× MIC of each strain (normalized MICs, 0.125 mg/liter for BW25113 and ΔkatG, 0.03 mg/liter for ΔrecA, and 0.008 mg/liter for ΔkatG ΔrecA) and the nonantibiotic control at 4 h. Data are represented as the means of at least three independent measurements. Standard deviations are shown. CIP, ciprofloxacin. Significant P values are noted (*, P < 0.05; ***, P < 0.001).
Additionally, we determined how antioxidants, such as l-ascorbic acid, affects drug lethality when detoxification systems are intact or suppressed in a recA-deficient background (Fig. S4). We found that pretreatment with l-Ascorbic acid provided at least 1 log10 of protection from cell death induced by ciprofloxacin (1× MIC of ΔrecA) at 4 h posttreatment in a recA-deficient background. The protection was higher when, in addition to a recA-deficient background, other detoxification system genes (katG or ahpC) were inactivated (almost 2 log10 of protection). Minor differences were observed for the ΔsodA ΔrecA double mutant compared to the ΔrecA mutant. Notably, l-ascorbic acid slightly increased the MIC against ciprofloxacin (2- to 4-fold, data not shown), supporting again that differences in lethality could be due in part to differences observed in MIC values.
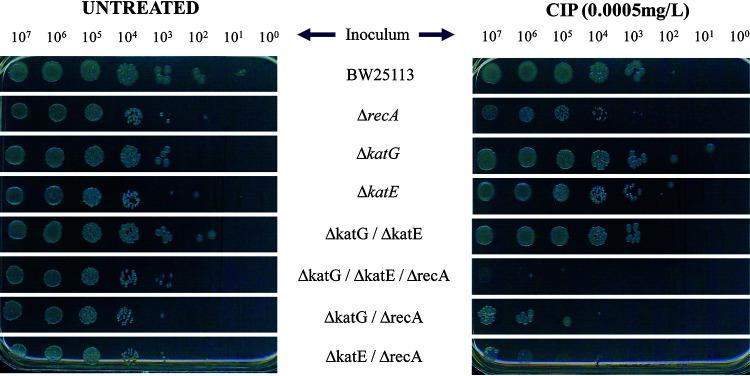
Bacterial survival of E. coli mutants after treatment with ciprofloxacin under aerobic conditions was also evaluated using spot test assays with multiple inocula (Fig. 4). Under these conditions, the ΔrecA mutant was more sensitive than wild-type BW25113 to the presence of ciprofloxacin. Furthermore, double and triple mutants with a combination of the recA gene and detoxification system genes (ΔkatG ΔrecA, ΔkatE ΔrecA, and ΔkatG ΔkatE ΔrecA) were more sensitive than the single ΔrecA mutant. Higher susceptibility was not observed for mutants with deletions only in the detoxification system genes (Fig. 4).
FIG 4.
Survival of E. coli mutants by spot test assays treated with ciprofloxacin under aerobic conditions. Overnight liquid cultures were serially diluted and plated on LB agar with or without ciprofloxacin at concentrations of 0.0005 mg/liter. The results shown are representative of at least three independent experiments.
Under these conditions, the recA gene and oxidative stress response suppression lead to a higher bactericidal effect in E. coli compared to recA gene inactivation alone.
Increased ROS production in the recA gene and oxidative stress response-suppressed backgrounds.
At the test concentration (0.004 mg/liter of ciprofloxacin, 1× MIC ΔrecA), only minor, nonsignificant differences in ROS production were observed for single detoxification system gene mutants compared to the wild-type BW25113 (Fig. S5). The tested concentration was too low to show obvious differences in ROS production. However, as expected, at this concentration of ciprofloxacin, strong significant differences (P < 0.001) were observed in the ΔrecA mutant versus wild-type BW25113. In general, a trend toward increased ROS production was observed with the combination of recA gene and detoxification system suppression in double and triple mutants compared to the single ΔrecA mutant. This comparison was significant in the case of ΔkatG ΔrecA (P < 0.05), ΔrpoS ΔrecA (P < 0.001), and ΔkatG ΔkatE ΔrecA (P < 0.05) mutants (Fig. S5). This association between increased ROS production and inactivation of the recA gene-detoxification systems following treatment with a bactericidal antibiotic support that ROS may actively contribute to the bactericidal activity of fluoroquinolones.
DISCUSSION
Bacterial stress responses, such as the SOS response, play an important role in adaptation and acquired bacterial resistance to antibiotics. The key regulators of the SOS response (LexA and RecA) have been proposed as an attractive target for increasing bacterial sensitivity to antibiotics and combating the emergence of resistance. Systematically altering the bacterial SOS activity, mainly through recA gene inactivation, has been revealed as a possible therapeutic strategy for potentiating bactericidal antibiotics such as quinolones against susceptible and resistant E. coli. Beyond this, recA inactivation modifies DNA repair and recombination events (17, 22, 32). Several compounds have also been shown to inhibit the ATPase activity of RecA in vitro (26, 33, 34). Phthalocyanine tetrasulfonates were recently characterized as an in vivo RecA inhibitor (35). ROS, on the other hand, are formed in cells exposed to three major classes of bactericidal antibiotics: quinolones, β-lactams, and aminoglycosides. This observation led to the theory that downstream formation of ROS is a common part of the killing mechanism of bactericidal antimicrobials (36). According to this line of thought, a secondary effect of exposure to these antibiotics is ROS formation through a common chain of events, which is an equally important and possibly more lethal part of the killing mechanism than the primary effect (25, 37–39). Hence, given this new way of thinking about the response of bacterial populations to lethal stress, efforts to target oxidative stress pathways as adjuvant antimicrobial therapy could increase lethality (40). In light of this literature, and even though much clinical investigation remains to be delivered on ROS therapy, in vitro work on infection models and early clinical evaluations looks promising (41, 42).
In this study, we used a wide collection of mutants to evaluate the impact of the recA gene (affecting the SOS response and DNA repair) and multiple ROS detoxification systems on the activity of quinolones such as ciprofloxacin. We showed that the combination of recA inactivation in combination with detoxification system gene inactivation produced a synergistic sensitization effect in terms of FIC index and susceptibility to ciprofloxacin. Furthermore, combined suppression of the recA gene and oxidative stress response leads to a higher bactericidal effect in E. coli than recA gene inactivation alone. Our results represent an important advance in the design and improvement of possible strategies to generate adjuvants to treatment with bactericidal antimicrobials.
The process of sensitization, using this double targeting strategy, was equally efficient across all genes, including effector detoxification genes and those regulating oxidative stress, and ranged from 7.5-fold to 30-fold relative to wild-type BW25113 (Table 1). Although these differences were moderate, they could play a significant role in therapeutic failure, bearing in mind the concentration-dependent character of these antimicrobials, whose predictors of efficacy in vivo are Cmax/MIC and AUC/MIC (43, 44).
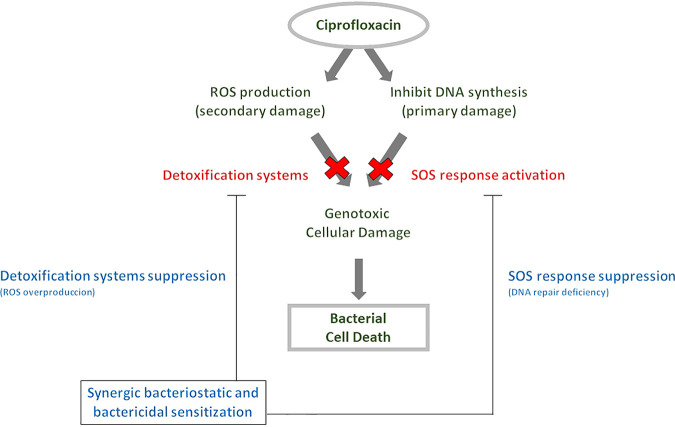
In terms of kinetic assays, both growth curves and killing curves supported the synergistic effect of sensitization to ciprofloxacin through recA gene suppression in combination with inactivation of detoxification system genes (Fig. 1 and 2). Rapid quinolone-mediated killing was increased in a recA-deficient E. coli background after detoxification system inactivation (for katG, sodA, and ahpC) when absolute concentrations were used (not normalized to MICs). A bactericidal effect (drop of >3 log10 CFU/ml) was observed for the ΔkatG ΔrecA and ΔahpC ΔrecA double mutants relative to the single ΔrecA mutant (Fig. 2). However, these differences were not evident when high killing concentrations normalized to MIC values were used (Fig. 3), indicating that mechanistic differences are not attributable to the lethality process. In addition, these data support a synergistic sensitization effect for ciprofloxacin, confirming that ROS are relevant factors in the quinolone-mediated lethality mode (25, 40, 45). More importantly, synergistic lethality was enhanced in the absence of DNA repair systems like those under the control of the SOS response (Fig. 5). Hence, even highly lethal fluoroquinolones can be improved by finding ways to increase lethality through the SOS response and ROS accumulation. ROS-stimulating and SOS response inhibitor adjuvants may enhance the lethality of quinolones and possibly other antimicrobials. Accordingly, some antioxidants, such as l-ascorbic acid, provide protection against antibiotic lethality (25).
FIG 5.
Scheme describing synergistic quinolone sensitization by targeting the SOS response and oxidative stress.
It is important to mention that a wide range of ciprofloxacin concentrations were initially used in this study, some of them being excessively lethal (Fig. S6 and S7). The results of the conditions that showed the greatest differences were selected to elaborate this study.
In summary, we highlight synergistic quinolone sensitization by targeting the recA gene and oxidative detoxification stress systems. These data reinforce the role of DNA repair systems and ROS in antibiotic-induced bacterial cell death and how both pathways can act synergistically in the quinolone sensitization process. Our detailed analysis opens up a new strategy for sensitizing drug resistance by targeting the recA gene and oxidative stress.
MATERIALS AND METHODS
Strains, growth conditions, and antimicrobial agents.
Wild-type E. coli BW25113 was used as the starting strain for all constructions (Table S1). E. coli BW25113 (wild-type) single-gene deletion mutants (ΔrecA, ΔkatG, ΔkatE, ΔsodA, ΔsodB, ΔahpC, ΔoxyR, and ΔrpoS) were selected from the KEIO collection (46). Double-gene deletion mutants (ΔkatG ΔrecA, ΔkatE ΔrecA, ΔsodA ΔrecA, ΔsodB ΔrecA, ΔahpC ΔrecA, ΔoxyR ΔrecA, ΔrpoS ΔrecA, ΔkatG ΔkatE, and ΔsodB ΔsodA) and the triple-gene deletion mutant (ΔkatG ΔkatE ΔrecA) were generated by P1vir phage transduction and by excision of the kanamycin resistance cassette with the plasmid pCP20 expressing the Flp recombinase (47, 48). Although some triple knockouts such as ΔsodA ΔsodB ΔrecA were planned at the start of the study, they proved not to be viable after several attempts.
Liquid or solid Luria broth (LB) medium, Mueller-Hinton broth (MHB), and M9 minimal medium were used. Strains were grown at 37°C. Ciprofloxacin was used in all assays (Sigma-Aldrich, Madrid, Spain). Kanamycin (Sigma-Aldrich) at 30 mg/liter was used for plasmid maintenance. Expression of ciprofloxacin-induced GFP into the kanamycin-resistant pMSrecA-gfp vector was used to detect recA transcription (see Table S2) (17, 49).
MICs.
MICs were determined in triplicate for each bacterial strain under conditions of aerobiosis and anaerobiosis, using two techniques, the gradient strip technique and broth microdilution following CLSI reference methods (30).
FIC index analysis.
The FIC index was calculated for a double gene deletion mutant with the equation FIC = MIC (double gene deletion mutant)/MIC (recA gene deletion mutant) + MIC (double gene deletion mutant)/MIC (ROS detoxification gene deletion mutant). In the case of the triple gene deletion mutant, the FIC index was calculated with the equation FIC = MIC (triple gene deletion mutant)/MIC (recA gene deletion mutant) + MIC (triple gene deletion mutant)/MIC (double gene deletion mutant ΔkatG ΔkatE). The combination of two systems was considered to be synergistic when the FIC value was ≤0.5, additive when it was 0.5 to ≤1, indifferent when it was 1 to 4, and antagonistic when it was >4, following the criteria of (50).
Bacterial growth rates.
Transparent 96-well flat-bottom plates containing 200 μl of sublethal concentrations (1/8× MIC to 1/32× MIC of wild-type) of ciprofloxacin in MHB were prepared as previously described using an Infinite 200 PRO plate reader (Tecan, Madrid, Spain) (17). At least four biological replicates were measured for each condition in at least two independent assays.
Time-kill curve assays.
To show the synergistic effect of suppression of RecA protein and oxidative detoxification systems on bacterial viability when challenged by ciprofloxacin, time-kill assays were performed with each isogenic group, based on RecA inactivation, specific detoxification system suppression, or combinations. LB was used with ciprofloxacin concentrations of 1× MIC of the wild type and 1× MIC of ΔrecA (ciprofloxacin concentrations were relative to MICs for strains harboring the unmodified recA gene or ΔrecA mutant, respectively). Additionally, bacterial viability, specifically for the isogenic group katG, at normalized ciprofloxacin concentrations according to the MICs (4× MIC and 8× MIC relative to the MICs of each strain, normalized MICs) were also evaluated. Growth in drug-free broth was evaluated in parallel as a control. Cultures were incubated at 37°C with shaking at 180 rpm. An initial inoculum of 106 CFU/ml was used in all experiments, and viable cells were determined at 0, 2, 4, 6, 8, and 24 h by colony counting.
Spot test.
Fresh overnight cultures in LB were diluted to achieve an optical density at 625 nm (OD625nm) of 0.1 (ca. 108 CFU/ml) and then serially diluted in saline solution (1%). A 5-μl volume of each dilution was spotted on LB agar plates with 1/8× MIC (0.0005 mg/liter) of ciprofloxacin (relative to the ΔrecA mutant) under aerobic conditions. After 20 h of incubation at 37°C, spots were checked for growth and compared with spots on the control LB agar plates.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism 6 software. Student’s t test was used for statistical evaluation when two groups were compared. Differences were considered significant when P values were <0.05.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Plan Nacional de I+D+i 2013‐2016 and the Instituto de Salud Carlos III (projects PI14/00940 and PI17/01501), Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI; RD16/0016/0001 and REIPI RD16/0016/0009) cofinanced by the European Development Regional Fund “A Way to Achieve Europe,” operative program Intelligent Growth 2014‐2020. Sara Diaz-Diaz is supported by a PFIS grant from the Instituto de Salud Carlos III (FI18/00086).
We have no transparency declarations to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Aarestrup FM. 2005. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin Pharmacol Toxicol 96:271–281. 10.1111/j.1742-7843.2005.pto960401.x. [DOI] [PubMed] [Google Scholar]
- 2.Witte W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996–997. 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum PC. 2012. 2012 and beyond: potential for the start of a second pre-antibiotic era? J Antimicrob Chemother 67:2062–2068. 10.1093/jac/dks213. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093. 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito R, Tomich AD, McElheny CL, Mettus RT, Sluis-Cremer N, Doi Y. 2017. Inhibition of fosfomycin resistance protein FosA by Phosphonoformate (foscarnet) in multidrug-resistant Gram-negative pathogens. Antimicrob Agents Chemother 61:e01424-17. 10.1128/AAC.01424-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin DM, Koskella B, Lin HC. 2017. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8:162–173. 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bikard D, Barrangou R. 2017. Using CRISPR-Cas systems as antimicrobials. Curr Opin Microbiol 37:155–160. 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Baym M, Stone LK, Kishony R. 2016. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351:aad3292. 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Méhi O, Bogos B, Csörgő B, Pál F, Nyerges A, Papp B, Pál C. 2014. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol Biol Evol 31:2793–2804. 10.1093/molbev/msu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer AC, Kishony R. 2013. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet 14:243–248. 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vila J, Sáez-López E, Johnson JR, Römling U, Dobrindt U, Cantón R, Giske CG, Naas T, Carattoli A, Martínez-Medina M, Bosch J, Retamar P, Rodríguez-Baño J, Baquero F, Soto SM. 2016. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev 40:437–463. 10.1093/femsre/fuw005. [DOI] [PubMed] [Google Scholar]
- 13.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Martínez JM, Machuca J, Cano ME, Calvo J, Martínez-Martínez L, Pascual A. 2016. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat 29:13–29. 10.1016/j.drup.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 16.Blázquez J, Rodríguez-Beltrán J, Matic I. 2018. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu Rev Microbiol 72:209–230. 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 17.Recacha E, Machuca J, Díaz de Alba P, Ramos-Güelfo M, Docobo-Pérez F, Rodriguez-Beltrán J, Blázquez J, Pascual A, Rodríguez-Martínez JM. 2017. Quinolone resistance reversion by targeting the SOS response. mBio 8:e00971-17. 10.1128/mBio.00971-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blázquez J, Couce A, Rodríguez-Beltrán J, Rodríguez-Rojas A. 2012. Antimicrobials as promoters of genetic variation. Curr Opin Microbiol 15:561–569. 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Thi TD, Lopez E, Rodriguez-Rojas A, Rodriguez-Beltran J, Couce A, Guelfo JR, Castaneda-Garcia A, Blazquez J. 2011. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother 66:531–538. 10.1093/jac/dkq496. [DOI] [PubMed] [Google Scholar]
- 20.Fernández de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. 2002. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol 35:1560–1572. 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 21.Simmons LA, Foti JJ, Cohen SE, Walker GC. 2008. The SOS regulatory network. EcoSal Plus 2008. 10.1128/ecosalplus.5.4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mo CY, Manning SA, Roggiani M, Culyba MJ, Samuels AN, Sniegowski PD, Goulian M, Kohli RM. 2016. Systematically altering bacterial SOS activity under stress reveals therapeutic strategies for potentiating antibiotics. mSphere 1:e00163-16. 10.1128/mSphere.00163-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 24.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100—E2109. 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Rosado AI, Valencia EY, Rodríguez-Rojas A, Costas C, Galhardo RS, Rodríguez-Beltrán J, Blázquez J. 2019. N-acetylcysteine blocks SOS induction and mutagenesis produced by fluoroquinolones in Escherichia coli. J Antimicrob Chemother 74:2188–2196. 10.1093/jac/dkz210. [DOI] [PubMed] [Google Scholar]
- 27.Van Acker H, Coenye T. 2017. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol 25:456–466. 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Ezraty B, Gennaris A, Barras F, Collet J-F. 2017. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15:385–396. 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 29.Chiang SM, Schellhorn HE. 2012. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys 525:161–169. 10.1016/j.abb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. M100-S29.CLSI, Wayne, PA. [Google Scholar]
- 31.EUCAST. 2020. Clinical breakpoints and epidemiological cut-off values. http://www.eucast.org/clinical_breakpoints/.
- 32.Recacha E, Machuca J, Díaz-Díaz S, García-Duque A, Ramos-Guelfo M, Docobo-Pérez F, Blázquez J, Pascual A, Rodríguez-Martínez JM. 2018. Suppression of the SOS response modifies spatiotemporal evolution, post-antibiotic effect, bacterial fitness and biofilm formation in quinolone-resistant Escherichia coli. J Antimicrob Chemother 74:66–73. 10.1093/jac/dky407. [DOI] [PubMed] [Google Scholar]
- 33.Bellio P, Brisdelli F, Perilli M, Sabatini A, Bottoni C, Segatore B, Setacci D, Amicosante G, Celenza G. 2014. Curcumin inhibits the SOS response induced by levofloxacin in Escherichia coli. Phytomedicine 21:430–434. 10.1016/j.phymed.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Mo CY, Culyba MJ, Selwood T, Kubiak JM, Hostetler ZM, Jurewicz AJ, Keller PM, Pope AJ, Quinn A, Schneck J, Widdowson KL, Kohli RM. 2018. Inhibitors of LexA autoproteolysis and the bacterial SOS response discovered by an academic–industry partnership. ACS Infect Dis 4:349–359. 10.1021/acsinfecdis.7b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam MK, Alhhazmi A, DeCoteau JF, Luo Y, Geyer CR. 2016. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem Biol 23:381–391. 10.1016/j.chembiol.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giroux X, Su W-L, Bredeche M-F, Matic I. 2017. Maladaptive DNA repair is the ultimate contributor to the death of trimethoprim-treated cells under aerobic and anaerobic conditions. Proc Natl Acad Sci U S A 114:11512–11517. 10.1073/pnas.1706236114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi N, Gruber CC, Yang JH, Liu X, Braff D, Yashaswini CN, Bhubhanil S, Furuta Y, Andreescu S, Collins JJ, Walker GC. 2017. Lethality of MalE-LacZ hybrid protein shares mechanistic attributes with oxidative component of antibiotic lethality. Proc Natl Acad Sci U S A 114:9164–9169. 10.1073/pnas.1707466114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ter Kuile BH, Hoeksema M. 2018. Antibiotic killing through incomplete DNA repair. Trends Microbiol 26:2–4. 10.1016/j.tim.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Hong Y, Zeng J, Wang X, Drlica K, Zhao X. 2019. Post-stress bacterial cell death mediated by reactive oxygen species. Proc Natl Acad Sci U S A 116:10064–10071. 10.1073/pnas.1901730116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memar MY, Ghotaslou R, Samiei M, Adibkia K. 2018. Antimicrobial use of reactive oxygen therapy: current insights. Infect Drug Resist 11:567–576. 10.2147/IDR.S142397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dryden M. 2018. Reactive oxygen species: a novel antimicrobial. Int J Antimicrob Agents 51:299–303. 10.1016/j.ijantimicag.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 43.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 26:1–10. 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 44.Wright DH, Brown GH, Peterson ML, Rotschafer JC. 2000. Application of fluoroquinolone pharmacodynamics. J Antimicrob Chemother 46:669–683. 10.1093/jac/46.5.669. [DOI] [PubMed] [Google Scholar]
- 45.Hong Y, Li Q, Gao Q, Xie J, Huang H, Drlica K, Zhao X. 2020. Reactive oxygen species play a dominant role in all pathways of rapid quinolone-mediated killing. J Antimicrob Chemother 75:576–585. 10.1093/jac/dkz485. [DOI] [PubMed] [Google Scholar]
- 46.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006–0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomason L, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3:623–638. 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 50.Mackay ML, Milne K, Gould IM. 2000. Comparison of methods for assessing synergic antibiotic interactions. Int J Antimicrob Agents 15:125–129. 10.1016/S0924-8579(00)00149-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.