Abstract
Cilia sense and transduce extracellular signals via specific receptors. The intraflagellar transport (IFT) machinery mediates not only bidirectional protein trafficking within cilia but also the import/export of ciliary proteins across the ciliary gate. The IFT machinery is known to comprise two multisubunit complexes, namely, IFT-A and IFT-B; however, little is known about how the two complexes cooperate to mediate ciliary protein trafficking. We here show that IFT144–IFT122 from IFT-A and IFT88–IFT52 from IFT-B make major contributions to the interface between the two complexes. Exogenous expression of the IFT88(Δα) mutant, which has decreased binding to IFT-A, partially restores the ciliogenesis defect of IFT88-knockout (KO) cells. However, IFT88(Δα)-expressing IFT88-KO cells demonstrate a defect in IFT-A entry into cilia, aberrant accumulation of IFT-B proteins at the bulged ciliary tips, and impaired import of ciliary G protein–coupled receptors (GPCRs). Furthermore, overaccumulated IFT proteins at the bulged tips appeared to be released as extracellular vesicles. These phenotypes of IFT88(Δα)-expressing IFT88-KO cells resembled those of IFT144-KO cells. These observations together indicate that the IFT-A complex cooperates with the IFT-B complex to mediate the ciliary entry of GPCRs as well as retrograde trafficking of the IFT machinery from the ciliary tip.
INTRODUCTION
Primary cilia are microtubule-based projections from the surface of various eukaryotic cells that function as cellular antennae by sensing extracellular stimuli, such as fluid flow, and by receiving and transducing developmental signals, such as Hedgehog (Hh) signaling (Bangs and Anderson, 2017; Gigante and Caspary, 2020). To achieve these functions, specific proteins exist on the ciliary membrane, including G protein–coupled receptors (GPCRs) (Mukhopadhyay and Rohatgi, 2014; Anvarian et al., 2019; Nachury and Mick, 2019). The ciliary membrane and cilioplasm are distinguished from the plasma membrane and cytoplasm, respectively, by the presence of the ciliary gate, composed of transition fibers (TFs) and the transition zone (TZ), the latter of which acts as a diffusion/permeability barrier (Garcia-Gonzalo and Reiter, 2017; Nachury and Mick, 2019). Therefore, the biogenesis and functions of cilia rely on the controlled import and export of ciliary proteins across the TZ, as well as on their bidirectional trafficking along the axonemal microtubules. Owing to their crucial roles, defects in ciliary assembly and ciliary protein trafficking, as well as in TZ integrity, result in a wide range of hereditary diseases collectively referred to as the ciliopathies, including Bardet–Biedl syndrome (BBS), Joubert syndrome, and short-rib thoracic dysplasia (Braun and Hildebrandt, 2017; Reiter and Leroux, 2017).
Anterograde and retrograde trafficking of ciliary proteins along the axoneme and their import and export across the TZ are mediated by the intraflagellar transport (IFT) machinery, often referred to as IFT particles or trains (Kozminski et al., 1993, 1995; Rosenbaum and Witman, 2002; Taschner and Lorentzen, 2016; Nakayama and Katoh, 2020). The IFT machinery contains two multisubunit complexes, namely IFT-A and IFT-B. The IFT-B complex mediates anterograde trafficking from the base to the distal tip of cilia, powered by the kinesin-2 motor. On the other hand, the IFT-A complex mediates retrograde trafficking powered by the dynein-2 motor complex after the remodeling of IFT particles and motor exchange at the ciliary tip; in this context, it is noteworthy that intestinal cell kinase (ICK; recently renamed as CILK1 for ciliogenesis-associated kinase 1), which is a MAPK-like kinase that undergoes IFT-mediated anterograde trafficking, regulates retrograde trafficking by playing a crucial role in the turnaround event at the ciliary tip (Nakamura et al., 2020). In addition, we and others showed that the IFT-A complex and its adaptor protein TULP3 also regulate the import of membrane proteins, including GPCRs, into cilia (Mukhopadhyay et al., 2010; Badgandi et al., 2017; Hirano et al., 2017; Ye et al., 2018; Han et al., 2019). The role of IFT-A was further supported by a cryoelectron tomographic study of anterograde IFT particles in Chlamydomonas flagella, which showed that the IFT-A and IFT-B complexes contact each other extensively and associate with the flagellar membrane and axoneme, respectively (Jordan et al., 2018). In addition to these complexes, the BBSome composed of eight BBS proteins participates in the export of ciliary membrane proteins, including GPCRs, by connecting GPCRs to IFT particles (Liu and Lechtreck, 2018; Nozaki et al., 2018, 2019; Ye et al., 2018).
IFT particles originally purified from a Chlamydomonas flagellar fraction were found to be dissociated into IFT-A and IFT-B complexes (Piperno and Mead, 1997; Cole et al., 1998). A subsequent study confirmed that IFT-A proteins can be coimmunoprecipitated with IFT-B proteins from a Chlamydomonas flagellar fraction under certain conditions (Qin et al., 2004). We and others delineated the overall architectures of the IFT-A and IFT-B complexes (Taschner and Lorentzen, 2016; Nakayama and Katoh, 2018). The IFT-A complex can be divided into two subcomplexes (Mukhopadhyay et al., 2010; Behal et al., 2012; Hirano et al., 2017; Takahara et al., 2018). The noncore subcomplex comprises IFT139, IFT121, and IFT43, and the core subcomplex comprises IFT144, IFT140, and IFT122, and binds to TULP3, which mediates membrane association of the IFT-A complex and GPCR import into cilia (Mukhopadhyay et al., 2010; Badgandi et al., 2017). The IFT-B complex comprises 16 subunits and can be divided into the peripheral (IFT-B2) subcomplex composed of six subunits and the core (IFT-B1) subcomplex composed of 10 subunits; the two subcomplexes are connected by composite interactions involving IFT57–IFT38 (peripheral subunits) and IFT88–IFT52 (core subunits) (Boldt et al., 2016; Katoh et al., 2016; Taschner et al., 2016). We previously showed that the IFT88/IFT57/IFT52/IFT38 tetramer also serves as a docking site for heterotrimeric kinesin-II (Funabashi et al., 2018).
Despite the elucidation of the IFT-A and IFT-B architectures, little is known about how the two complexes are connected to function as IFT particles. We here found that IFT144–IFT122 from IFT-A and IFT88–IFT52 from IFT-B contribute to the IFT-A–IFT-B interface. More importantly, we showed that not only retrograde trafficking of the IFT machinery but also ciliary GPCR import depends on the firm interaction of the IFT-A complex with the IFT-B complex.
RESULTS
Subunits involved in the IFT-A– IFT-B interaction
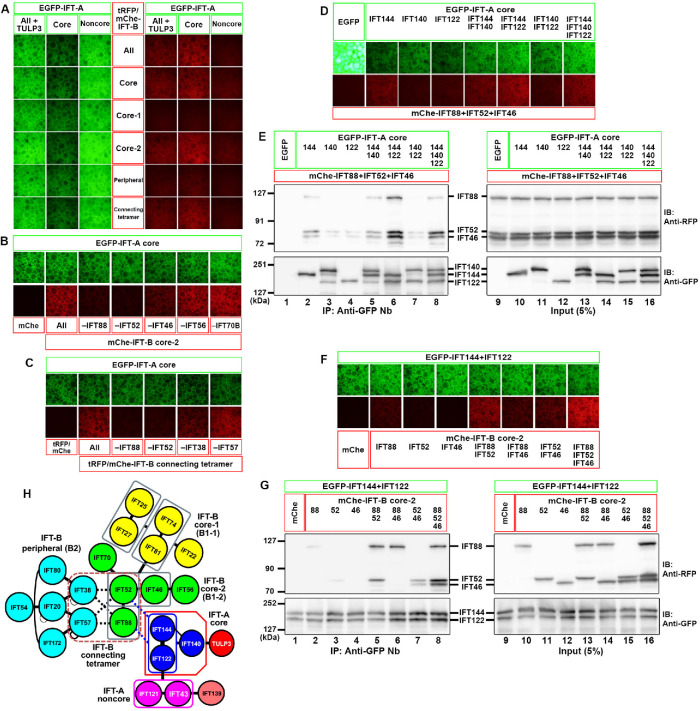
To identify the IFT-A–IFT-B interface, we utilized the visible immunoprecipitation (VIP) assay, which we developed as a versatile screening strategy that enables visual detection of not only binary but also one-to-many and many-to-many protein interactions (Katoh et al., 2015, 2016).
When lysates of cells coexpressing all six of the IFT-A subunits fused to enhanced green fluorescent protein (EGFP) and all 16 of the IFT-B subunits fused to mCherry (mChe) or TagRFP (tRFP) were processed for immunoprecipitation with glutathione S-transferase (GST)-tagged anti-GFP nanobodies (Nb) prebound to glutathione–Sepharose beads, red signals were detected on the precipitated beads (Figure 1A, top row), indicating coimmunoprecipitation of some of the IFT-B subunits with some of the IFT-A subunits. When the IFT-A subunits were grouped into those of the core (IFT144/IFT140/IFT122) and noncore (IFT139/IFT121/IFT43) subcomplexes, the core subunits fused to EGFP substantially coimmunoprecipitated the mChe/tRFP-fused IFT-B subunits (Figure 1A, top row). Among the IFT-B subunits, those of the core-2 subcomplex (IFT88/IFT70/IFT56/IFT52/IFT46) demonstrated a relatively strong interaction and those of the connecting tetramer (IFT88/IFT57/IFT52/IFT38) demonstrated a weak interaction with the IFT-A core subunits (Figure 1A, fourth and bottom rows).
FIGURE 1:
Determination of the interface between the IFT-A and IFT-B complexes. (A) Lysates of HEK293T cells coexpressing the indicated combinations of EGFP-fused IFT-A and tRFP/mChe-fused IFT-B subunits were subjected to the VIP assay. (B, C) The subtractive VIP assay was performed to identify IFT-B subunits that interact with IFT-A core subunits. Lysates of cells coexpressing EGFP-fused IFT-A core subunits and all but one (as indicated) subunits of the IFT-B core-2 subunits (B) or the connecting tetramer subunits (C) fused to tRFP/mChe were subjected to the VIP assay using GST-fused anti-GFP Nb. (D, E) Identification of the IFT-A subunits involved in the interaction with IFT88–IFT52–IFT46. Lysates of cells coexpressing mChe-fused IFT88+IFT52+IFT46 and EGFP-fused IFT-A subunits, as indicated, were subjected to the VIP assay (D) or immunoblotting analysis using anti-RFP and anti-GFP antibodies (E). (F, G) Identification of IFT-B subunits involved in the interaction with IFT144–IFT122. Lysates of cells coexpressing mChe-fused IFT88, IFT52, and/or IFT46, as indicated, and EGFP-IFT144+IFT122 were subjected to the VIP assay (F) or immunoblotting analysis (G). (H) Schematic representation of the mode of interaction between the IFT-A and IFT-B complexes, predicted from the data shown in Figure 1, A–G.
We then performed the subtractive VIP assay to determine which of the IFT-B core-2 subunits and connecting subunits contribute to its interaction with the IFT-A core subunits. Red signals were substantially decreased when IFT88 or IFT52 was omitted from the IFT-B core-2 subunits moderately decreased when IFT46 was omitted (Figure 1B), and slightly decreased when IFT88 or IFT52 was omitted from the connecting subunits (Figure 1C).
We then determined which of the IFT-A core subunits participate in its interaction with IFT-B by the additive VIP assay and immunoblotting analysis (Figure 1, D and E). Compared with any one of the IFT-A core subunits fused to EGFP (Figure 1E, lanes 2–4), strong signals were detected when both EGFP-IFT144 and EGFP-IFT122 were coexpressed together with mChe-fused IFT88+IFT52+IFT46 (Figure 1E, lane 6), indicating that the IFT144–IFT122 dimer of the IFT-A complex mainly participates in the IFT-A–IFT-B interaction.
We also determined which of the IFT-B subunits (IFT88, IFT52, and IFT46) participate in IFT144–IFT122 binding. As shown in Figure 1, F and G, no or very weak signals were detected when EGFP-fused IFT144 and IFT122 were coexpressed with either mChe-fused IFT88, IFT52, or IFT46 alone (Figure 1G, lanes 2–4). However, EGFP-IFT144+IFT122 firmly coimmunoprecipitated mChe-fused IFT88+IFT52 (Figure 1G, lane 5) and moderately coimmunoprecipitated IFT88+IFT46 and IFT52+IFT46 (Figure 1G, lanes 6 and 7). Coexpression of mChe-IFT46 with IFT88+IFT52 resulted in a marginal increase in its coprecipitation with EGFP-IFT144+IFT122 (Figure 1G, compare lane 8 with lane 5). These results indicate that the IFT88–IFT52 from IFT-B, and IFT144–IFT122 from IFT-A make major contributions to the IFT-A–IFT-B interface (Figure 1H) and that IFT46 may play an auxiliary role in the IFT-A–IFT-B interaction. However, these results do not necessarily exclude interactions involving other IFT-A and IFT-B subunits (see below), in view of a recent cryoelectron tomographic study of Chlamydomonas anterograde IFT trains, which suggests extensive contacts between the IFT-A and IFT-B complexes (Jordan et al., 2018).
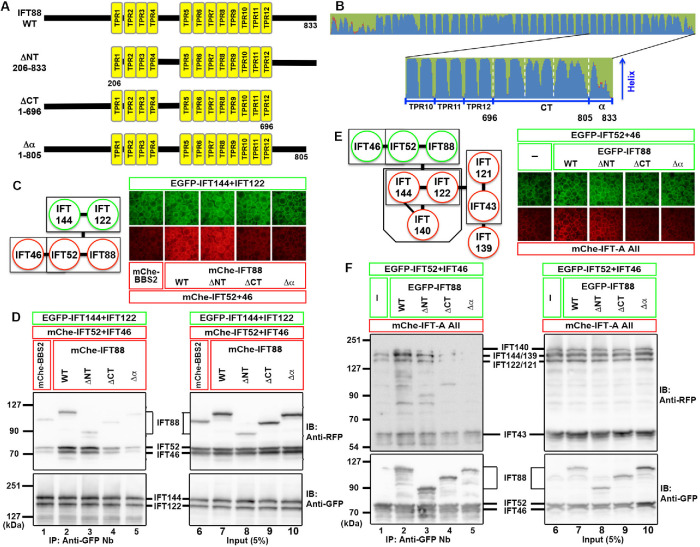
IFT88 requires its C-terminal α-helix region for its interaction with the IFT-A complex
We next aimed to identify mutants of IFT88 that are unable to interact with IFT-A subunits, in conjunction with IFT52 and IFT46. The IFT88 protein is predicted to contain 12 tetratricopeptide repeats (TPRs) in the middle region (Figure 2, A and B). When the region N-terminal to TPR1 was deleted, the resulting IFT88(ΔNT) construct, together with IFT52+IFT46, retained the ability to interact with IFT144–IFT122 (Figure 2, C and D, compare lanes 2 and 3). By contrast, binding of the IFT88(ΔCT) construct, in which the region C-terminal to TPR12 was deleted, to IFT144–IFT122 was decreased to a level comparable to the negative control (Figure 2D, compare lanes 1, 2, and 4). However, because IFT88(ΔCT) could not rescue the ciliogenesis defect of IFT88-knockout (KO) cells (see below), we made another construct, IFT88(Δα), in which the most C-terminal α-helix region comprising 28 amino acids was truncated (Figure 2, A and B). Similar to the IFT88(ΔCT) construct, IFT88(Δα), in conjunction with IFT52+IFT46, had a decreased ability to bind to IFT144–IFT122 (Figure 2, C and D, compare lanes 2 and 5).
FIGURE 2:
The C-terminal α-helix region of IFT88 is crucial for its interaction with the IFT-A complex. (A, B) Schematic representation of the IFT88 constructs. (A) Domain organization of IFT88(WT) and its deletion constructs. (B) Secondary structure of the C-terminal TPR and α-helix regions of the IFT88 protein, predicted by NetSurfP-1.0 (Petersen et al., 2009). (C, D) Lysates of cells coexpressing the mChe-fused IFT88 constructs, as indicated, plus IFT52 and IFT46, and EGFP-fused IFT144+IFT122 were subjected to the VIP assay (C) or immunoblotting analysis (D). (E, F) Lysates of cells coexpressing EGFP-fused IFT88 constructs, as indicated, plus IFT52 and IFT46, and all the IFT-A subunits (IFT144/IFT140/IFT139/IFT122/IFT121/IFT43) fused to mChe were subjected to the VIP assay (E) or immunoblotting analysis (F).
We then analyzed the interaction of these IFT88 constructs with the entire IFT-A complex (IFT144/IFT140/IFT139/IFT122/IFT121/IFT43) by the VIP assay and immunoblotting analysis (Figure 2, E and F). The results were essentially the same as those obtained using IFT144–IFT122. Namely, IFT88(ΔCT) and IFT88(Δα), combined with IFT52+IFT46, demonstrated decreased abilities to coimmunoprecipitate the IFT-A proteins, compared with IFT88(WT) and IFT88(ΔNT) (Figure 2F, compare lanes 4 and 5 with lanes 2 and 3); note that the bands for mChe-IFT144 and mChe-IFT139 and those for mChe-IFT122 and mChe-IFT121 were indistinguishable on the immunoblots, as described previously (Hirano et al., 2017; Takahara et al., 2019).
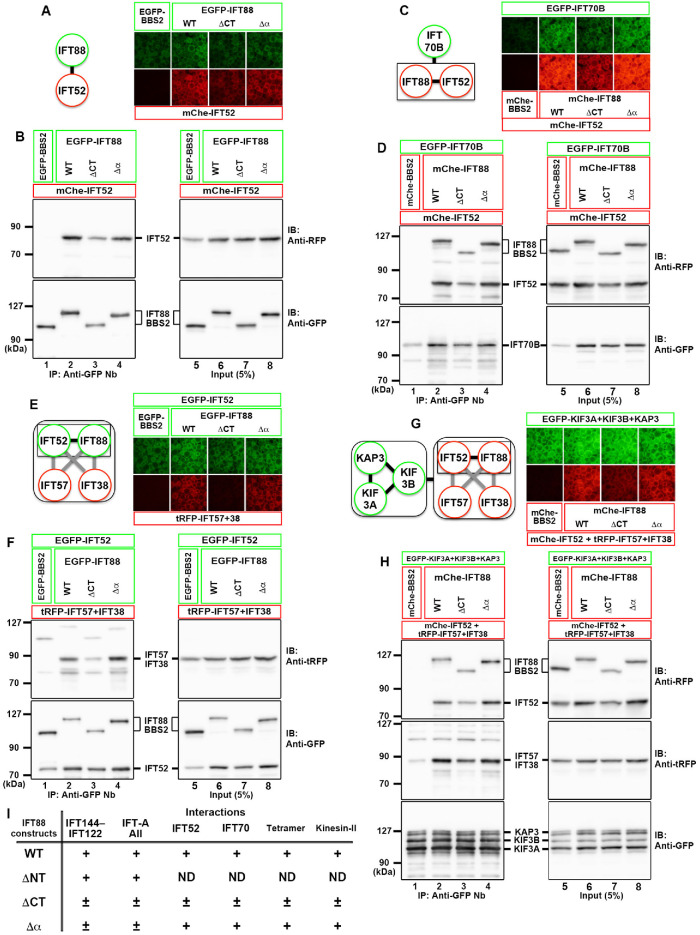
IFT88 acts as a hub subunit in the IFT-B complex, via interacting with several subunits. We therefore analyzed whether the IFT88(Δα) construct retained the ability to interact with other IFT-B subunits (see Figure 1H). 1) IFT88 forms a heterodimer with IFT52 (Katoh et al., 2016). As shown in Figure 3, A and B, IFT88(Δα) retained the ability to bind to IFT52 (Figure 3B, lane 4). On the other hand, IFT88(ΔCT) demonstrated a slightly decreased interaction with IFT52 (Figure 3B, compare lanes 2 and 3). 2) The IFT88–IFT52 dimer interacts with IFT70B (Takei et al., 2018). As shown in Figure 3, C and D, similar to IFT52, IFT88(Δα) retained the ability to bind to IFT70B (Figure 3D, lane 4), whereas IFT88(ΔCT) had a slightly reduced ability (Figure 3D, lane 3). 3) IFT88 and IFT52 from the core subcomplex and IFT57 and IFT38 from the peripheral subcomplex connect the two IFT-B subcomplexes (Katoh et al., 2016). As shown in Figure 3, E and F, IFT88(Δα) retained the ability to form the connecting tetramer, whereas IFT88(ΔCT) had a reduced ability (Figure 3F, lanes 2–4). 4) We previously showed that the connecting tetramer (IFT88/IFT57/IFT52/IFT38) interacts with heterotrimeric kinesin-II (KIF3A/KIF3B/KAP3) and that a slightly reduced interaction between them results in a severe ciliogenesis defect (Funabashi et al., 2018). As shown in Figure 3, G and H, the connecting tetramer containing IFT88(Δα) retained the ability to interact with heterotrimeric kinesin-II (Figure 3H, lane 4). By contrast, the IFT88(ΔCT)-containing tetramer had a slightly reduced ability to interact with kinesin-II (Figure 3H, compare lanes 2 and 3). Considering our recent observations (Funabashi et al., 2018), the slight reduction in the interaction of IFT88(ΔCT) with kinesin-II might be attributable to its inability to rescue the ciliogenesis defect of IFT88-KO cells (see Figure 4D), although we did not further characterize the IFT88(ΔCT) construct. Taking these results together, the IFT88(Δα) construct was compromised with regard to its interaction with IFT-A proteins among the analyzed interactions (Figure 3I).
FIGURE 3:
The C-terminal α-helix region of IFT88 does not play an important role in its interactions with other IFT-B subunits and kinesin-II. (A, B) Lysates of cells coexpressing EGFP-fused IFT88 constructs, as indicated, and mChe-IFT52 were subjected to the VIP assay (A) or immunoblotting analysis (B). (C, D) Lysates of cells coexpressing EGFP-IFT70B and mChe-fused IFT88 constructs, as indicated, plus IFT52 were subjected to the VIP assay (C) or immunoblotting analysis (D). (E, F) Lysates of cells coexpressing EGFP-fused IFT88 constructs, as indicated, plus IFT52 and tRFP-fused IFT57+IFT38 were subjected to the VIP assay (E) or immunoblotting analysis (F). (G, H) Lysates of cells coexpressing EGFP-fused kinesin-II subunits and the mChe-IFT88 constructs, as indicated, plus mChe/tRFP-fused IFT57+IFT52+IFT38 were subjected to the VIP assay (G) or immunoblotting analysis (H). Note that, in the immunoblotting analyses shown in B, D, F, and H, anti-RFP and anti-tRFP antibodies recognize mChe and tRFP, respectively. (I) Summary of the results of the interaction analyses shown in Figures 2 and 3. +, firm interaction; ±, weak interaction; ND, not determined.
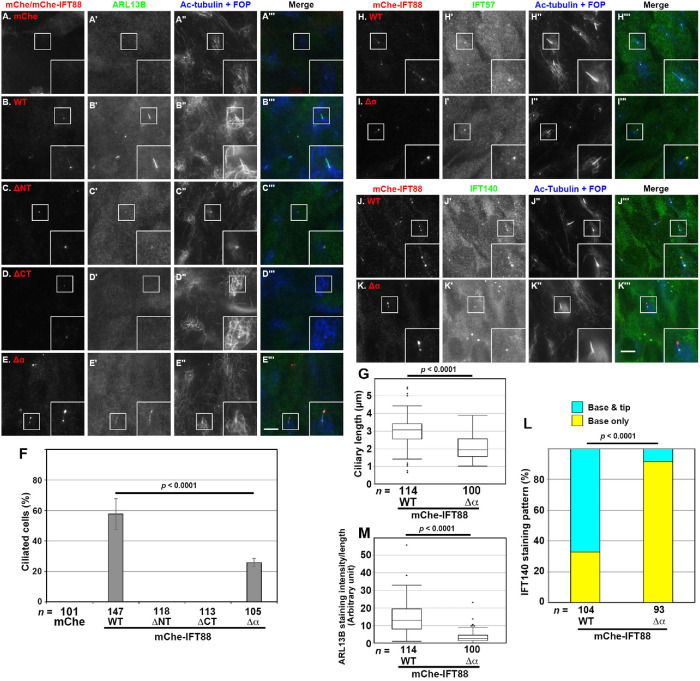
FIGURE 4:
Defects in ciliary localization of the IFT components and ARL13B in IFT88(Δα)-expressing IFT88-KO cells. (A–E) IFT88-KO cells stably expressing mChe as a negative control (A) or mChe-fused IFT88(WT) (B), IFT88(ΔNT) (C), IFT88(ΔCT) (D), or IFT88(Δα) (E) were serum-starved for 24 h to induce ciliogenesis, and immunostained for mChe (A–E), ARL13B (A′–E′), and Ac-tubulin+FOP (A′′–E′′). Scale bar, 5 µm. (F) Ciliated IFT88-KO cells expressing mChe or the mChe-fused IFT88 constructs shown in A–E were counted, and percentages of ciliated cells are expressed as bar graphs. Values are means ± SD of three independent experiments. In each set of experiments, 33 to 56 cells were analyzed, and the total numbers of cells analyzed (n) are shown. The p value was determined by the Student t test. (G, M) The length of cilia (G) and ciliary ARL13B staining intensity (M) in the IFT88-KO cells expressing mChe-IFT88(WT) or mChe-IFT88(Δα) were measured, and the ciliary length and the ciliary ARL13B staining intensity per length are represented as box-and-whisker plots. The boxes represent the 25th–75th percentiles (interquartile range [IQR]), and the median is indicated by a horizontal line. The whiskers show the minimum and maximum within 1.5 × IQR from the 25th and 75th percentiles, respectively. Outliers are indicated as dots. The total numbers of ciliated cells analyzed (n) are shown. P values were determined by the Student t test. (H–K) IFT88-KO cells stably expressing mChe-IFT88(WT) (H, J) or mChe-IFT88(Δα) (I, K) were immunostained for either IFT57 (H′, I′) or IFT140 (J′, K′), mChe (H–K), and Ac-tubulin and FOP (H′′–K′′). Scale bar, 5 µm. (L) Localization of IFT140 was classified as “ciliary base” or “ciliary base and tip,” and the cells in each category were counted. The percentages of these populations are represented as stacked bar graphs. Values are the means of three independent experiments. In each set of experiments, 30–37 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown. The p value was determined by the Pearson χ2 test.
IFT 88-KO cells expressing IFT88(Δα) demonstrate aberrant accumulation of the IFT-B complex at the ciliary tip and a defect in IFT-A entry into cilia
IFT88-KO RPE1 cells demonstrate the “cilia-lacking” phenotype (Katoh et al., 2017), indicating that integrity of the IFT-B complex is required for the trafficking of proteins essential for ciliogenesis, such as the αβ-tubulin dimer (Bhogaraju et al., 2014). We then investigated whether the IFT88 constructs analyzed above could rescue the ciliogenesis defect of IFT88-KO cells.
When mChe-IFT88(WT) was stably expressed in IFT88-KO cells by infection of a lentiviral vector, ciliogenesis depicted by staining for ARL13B and acetylated α-tubulin (Ac-tubulin) was restored (Figure 4, B′ and B′′). In these cells, mChe-IFT88(WT) was found mainly around the ciliary base and in smaller amounts at the tip (Figure 4B), as observed for endogenous IFT88 in control RPE1 cells (see Figure 5A). By total internal reflection fluorescence (TIRF) microscopy, mChe-IFT88(WT) – positive particles were observed to move along the cilium in both directions (Supplemental Video S1); the movement of IFT88(WT)-positive particles in IFT88-KO cells was essentially the same as that observed for IFT88(WT) expressed in control RPE1 cells (Supplemental Video S2). By contrast, the exogenous expression of mChe-fused IFT88(ΔNT) or IFT88(ΔCT) in IFT88-KO cells could not restore the ciliogenesis defect (Figure 4, C and D; also see Figure 4F). On the other hand, IFT88-KO cells expressing mChe-IFT88(Δα) were able to form cilia, as depicted by the staining for ARL13B and Ac-tubulin (Figure 4E). Thus, a firm interaction of the IFT-B complex with IFT-A is not necessarily required for ciliogenesis. However, the ciliogenesis efficiency of mChe-IFT88(Δα) –expressing cells was considerably lower (Figure 4F) and the ciliary length of these cells was shorter (Figure 4G) than those of mChe-IFT88(WT) –expressing cells.
FIGURE 5:
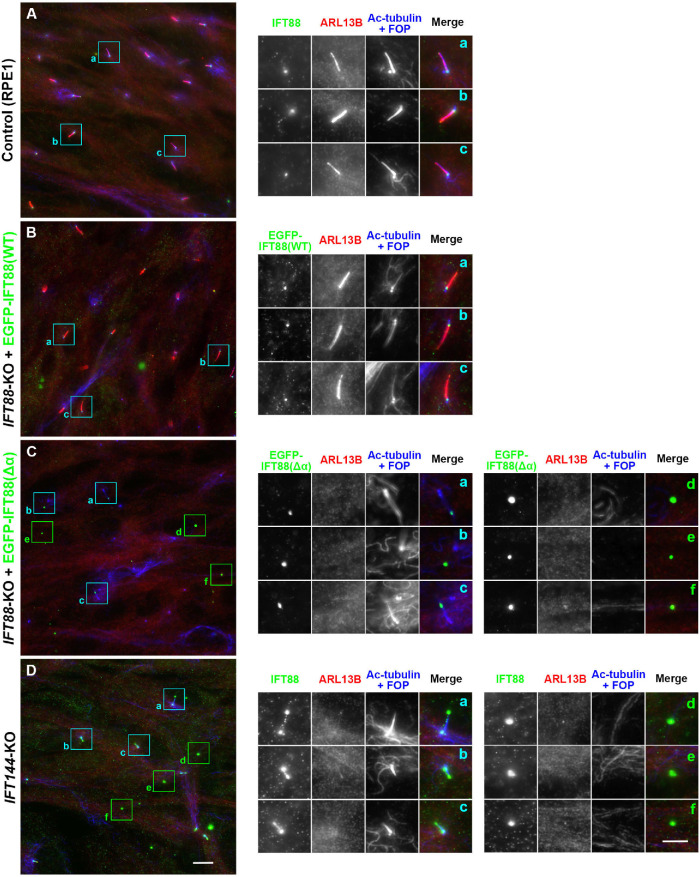
Presence of ECVs formed by IFT88(Δα)-expressing IFT88-KO cells and by IFT144-KO cells. Control RPE1 cells (A), IFT88-KO cells expressing EGFP-IFT88(WT) (B) or EGFP-IFT88(Δα) (C), or IFT144-KO cells (D) were serum-starved for 24 h to induce ciliogenesis and immunostained for IFT88, ARL13B, and Ac-tubulin+FOP. Scale bar, 10 µm. Images of the boxed regions enlarged 2.5 times are shown in a–f. Scale bar, 5 µm.
Movie S1.
TIRF microscopy of IFT88-KO cells expressing mChe-IFT88(WT)
Movie S2.
TIRF microscopy of control RPE1 cells expressing EGFP-IFT88(WT)
The most prominent difference between IFT88(WT)-expressing and IFT88(Δα)-expressing IFT88-KO cells was the localization of mChe-IFT88 itself. In contrast to mChe-IFT88(WT), mChe-IFT88(Δα) was markedly concentrated at the bulged distal tip (Figure 4E); this accumulation was not caused by the mChe-tag, because essentially the same accumulation was observed for EGFP-fused IFT88(Δα) expressed in IFT88-KO cells (see Figure 5C). Endogenous IFT57 (an IFT-B peripheral subunit) was also markedly concentrated at the distal tip (Figure 4I′) where mChe-IFT88(Δα) was also localized (Figure 4I), indicating accumulation of the entire IFT-B complex at the distal tip. IFT140 (an IFT-A subunit) was mainly found around the ciliary base, and a small proportion was found at the distal tip in IFT88(WT)-expressing IFT88-KO cells (Figure 4J′; also see Figure 4L), as observed in control RPE1 cells (e.g., see Hirano et al., 2017). By contrast, IFT140 signals were barely detectable at the tip in IFT88(Δα)-expressing IFT88-KO cells (Figure 4K′; also see Figure 4L). Overaccumulation of the IFT-B complex at the ciliary tip and impaired ciliary entry of the IFT-A complex observed in IFT88(Δα)-expressing IFT88-KO cells closely resembled our previous observations in IFT144-KO cells (Hirano et al., 2017). These observations suggest disengagement of the IFT-A complex from the anterograde IFT trains in IFT88(Δα)-expressing IFT88-KO cells.
Another difference between IFT88(WT)-expressing and IFT88(Δα)-expressing IFT88-KO cells was that the staining intensity for ciliary ARL13B was substantially lower in IFT88(Δα)-expressing cells (Figure 4M). Although we do not know the exact reason for this decreased ciliary ARL13B level, it may involve impaired entry of the IFT-A complex, as the ciliary ARL13B level is also decreased in IFT144-KO and TULP3-KO cells (Hirano et al., 2017; Han et al., 2019; Hwang et al., 2019; Legué and Liem, 2019).
TIRF microscopy showed that mChe-IFT88(Δα) signals at the bulged tip were rather static. Furthermore, mChe-IFT88(Δα) –positive signals moving along the cilium (Supplemental Video S3) appeared to be different from distinct particle-like signals of mChe-IFT88(WT) expressed in control RPE1 and IFT88-KO cells (Supplemental Videos S1 and S2) and rather resembled those of EGFP-IFT88(WT) expressed in IFT144-KO cells (Supplemental Video S4), in which the IFT-A complex cannot be assembled or loaded onto the anterograde trains due to the absence of the essential core subunit, IFT144 (Hirano et al., 2017). In view of the cryoelectron tomographic data of Chlamydomonas anterograde trains suggesting a stoichiometry of IFT-B:IFT-A of approximately 2:1 (Jordan et al., 2018), it is likely that the IFT88(Δα)-containing IFT machinery, owing to the lack of the IFT-A complex as in IFT144-KO cells, takes a configuration that is different from that of the normal IFT train (Supplemental Videos S1 and S2). These observations altogether indicate that, in IFT88(Δα)-expressing IFT88-KO cells, the IFT-B complex undergoes trafficking toward the distal tip, where it accumulates owing to impaired retrograde trafficking, because the IFT-A complex, even though it is intact, does not efficiently bind to the IFT-B complex to be loaded onto the anterograde train.
Movie S3.
TIRF microscopy of IFT88-KO cells expressing mChe-IFT88(Δα)
Movie S4.
TIRF microscopy of IFT144-KO cells expressing EGFP-IFT88(WT)
We have recently shown that in ICK-KO cells, the bulged ciliary tips containing aberrantly accumulated IFT components are torn off and released into the environment as extracellular vesicles (ECVs) (Nakamura et al., 2020). It is likely that proteins excessively accumulated at the ciliary tip, including components of the IFT machinery, caused by a defective turnaround event in the absence of ICK (Nakamura et al., 2020), are eliminated as ECVs to liberate cilia from the stress of their overaccumulation. We speculated that ECVs can also be released from the bulged tips of IFT88(Δα)-expressing IFT88-KO cells, as a block in retrograde trafficking caused the aberrant accumulation of IFT components.
In the case of ICK-KO cells, we noticed the presence of ECVs containing IFT components by observation of punctate structures that are positive for IFT88 but negative for the basal body marker FOP (recently renamed as CEP43). As RPE1 cells grow cilia not only on the dorsal surface but also on the ventral surface (Kukic et al., 2016), vesicles released from the ventral side can be trapped in the confined space between the cell surface and the coverslip. This was also the case for IFT88(Δα)-expressing IFT88-KO cells.
In the cases of control RPE1 cells (Figure 5A) and EGFP-IFT88(WT) –expressing IFT88-KO cells (Figure 5B), IFT88 signals were detected around the ciliary base, and ciliary ARL13B staining was clearly detected. In striking contrast, in addition to EGFP-IFT88(Δα) signals at the ciliary tips (Figure 5C, a–c), punctate structures representing ECVs with intense IFT88(Δα) signals but lacking FOP signals were often observed (Figure 5C, d–f). When the EGFP-IFT88(Δα) –expressing IFT88-KO cells were subjected to time-lapse recording, EGFP-IFT88(Δα) –positive vesicles were found to be released from the bulged tips (Supplemental Video S5).
Movie S5.
ECV formation from IFT88-KO cells expressing EGFP-IFT88(Δα)
As the phenotype of IFT88(Δα)-expressing IFT88-KO cells closely resembled that of IFT144-KO cells, which we reported previously (Hirano et al., 2017), we also analyzed IFT144-KO cells. As shown in Figure 5D, d–f, punctate structures positive for IFT88 but negative for FOP were observed. These observations together indicate that IFT-B proteins excessively accumulated in the bulged ciliary tips owing to disengagement of the IFT-A complex from the anterograde train are released as ECVs.
Lack of a firm IFT-A–IFT-B interaction impairs import of ciliary GPCRs
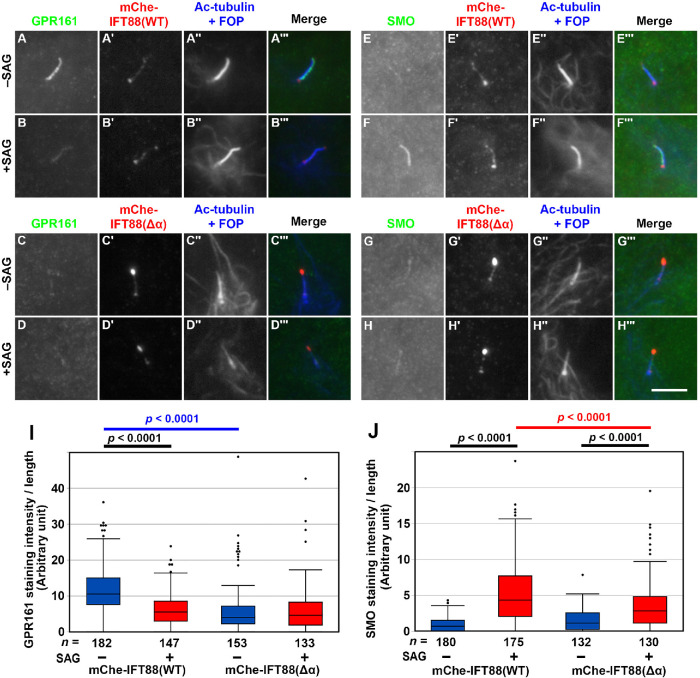
We then analyzed whether the lack of a firm IFT-A–IFT-B interaction affects the trafficking of GPR161 and Smoothened (SMO), which are class A and class F GPCRs, respectively, and are involved in negative and positive regulation of Hh signaling, respectively (Mukhopadhyay and Rohatgi, 2014; Anvarian et al., 2019; Nachury and Mick, 2019). GPR161 is localized on the ciliary membrane under basal conditions and exits cilia when Hh signaling is activated, whereas SMO is absent from cilia under basal conditions but enters cilia upon the activation of Hh signaling.
In IFT88(WT)-expressing IFT88-KO cells, GPR161 was found within cilia under basal conditions, whereas a significant proportion of GPR161 exited cilia upon treatment of cells with Smoothened Agonist (SAG) (Figure 6, A and B; also see Figure 6I). On the other hand, SMO was not found within cilia under basal conditions and entered cilia when cells were treated with SAG (Figure 6, E and F; also see Figure 6J). These results were essentially the same as those observed in control RPE1 cells (e.g., see Hirano et al., 2017; Tsurumi et al., 2019).
FIGURE 6:
Decreased entry of ciliary GPR161 and SMO in IFT88(Δα)-expressing IFT88-KO cells. (A–H) IFT88-KO cells stably expressing mChe-IFT88(WT) (A, B, E, F) or mChe-IFT88(Δα) (C, D, G, H) were serum-starved for 24 h and further cultured for 24 h in the absence (–SAG) or presence (+SAG) of 200 nM SAG. The cells were immunostained for either GPR161 (A–D) or SMO (E–H), mChe (A′–H′), and Ac-tubulin+FOP (A′′–H′′). Scale bar, 10 µm. (I, J) Fluorescence staining intensities of GPR161 (I) and SMO (J) within cilia were measured, and relative intensities per ciliary length are represented as box-and-whisker plots, as described in the legend to Figure 4, G and M. The p values were determined by the Student t test, to compare between cell lines and between cells with and without SAG treatment.
In striking contrast, GPR161 was present at a low level within cilia under not only SAG-stimulated but also basal conditions in IFT88(Δα)-expressing IFT88-KO cells (Figure 6, C and D; also see Figure 6I). On the other hand, entry of SMO into cilia upon SAG treatment was significantly decreased in IFT88(Δα)-expressing cells (Figure 6, G and H; also see Figure 6J). Thus, these results indicate that entry of GPR161 and SMO into cilia is impaired in the absence of a firm IFT-A–IFT-B interaction.
To address whether the defect in ciliary entry of GPCRs in IFT88(Δα)-expressing IFT88-KO cells is a more general event, we analyzed the localization of EGFP-fused SSTR3 expressed in IFT88(WT)-expressing and IFT88(Δα)-expressing IFT88-KO cells; in our previous study, we showed the lack of ciliary localization of SSTR3-EGFP in IFT144-KO cells (Hirano et al., 2017). As shown in Supplemental Figure S1, we could not detect the ciliary localization of SSTR3-EGFP in IFT88(Δα)-expressing IFT88-KO cells (Supplemental Figure S1B), unlike in IFT88(WT)-expressing IFT88-KO cells (Supplemental Figure S1A). Although the simultaneous expression of two distinct proteins (i.e., SSTR3-EGFP and either mChe-IFT88(WT) or mChe-IFT88(Δα)) by lentiviral vectors in IFT88-KO cells resulted in a substantial reduction in cell viability for an unknown reason, the lack of ciliary SSTR3-EGFP in IFT88(Δα)-expressing IFT88-KO cells was more robust than that in IFT88(WT)-expressing IFT88-KO cells (Supplemental Figure S1C).
DISCUSSION
In this study, we demonstrated two crucial roles of the interaction of the IFT-A complex with the IFT-B complex in ciliary protein trafficking. One role is associated with retrograde trafficking of the IFT-B complex. Namely without efficient loading of the IFT-A complex onto the anterograde IFT train, the IFT-B complex can undergo anterograde trafficking toward the distal tip, which is driven by heterotrimeric kinesin-II via its binding to the IFT-B connecting tetramer (Funabashi et al., 2018). However, the IFT-A-lacking train cannot undergo retrograde trafficking driven by dynein-2, which probably binds to the IFT-A complex. In this context, it is reasonable that IFT88(Δα)-expressing IFT88-KO cells phenocopy IFT144-KO cells, in which IFT-B complexes are accumulated at the distal tips (Hirano et al., 2017); that is, as IFT144 is an essential core subunit, the IFT-A complex cannot be assembled or loaded onto the anterograde train in its absence. It is also noteworthy that in this context, the overaccumulated IFT-B proteins in the bulged ciliary tips are released as ECVs, not only in IFT88(Δα)-expressing IFT88-KO cells but also in IFT144-KO cells. Thus, it is likely that the release of ECVs observed in the absence of ciliary entry of the IFT-A complex or in the absence of ICK (Nakamura et al., 2020) is a common mechanism to relieve cilia of the stress of excessive protein accumulation. For example, as mutations in the genes encoding proteins involved in retrograde trafficking, including subunits of the IFT-A and dynein-2 complexes and ICK, are known to cause skeletal ciliopathies, such as short-rib thoracic dysplasia and cranioectodermal dysplasia (Arts and Knoers, 2013 [updated 2018]; Schmidts, 2014; Paige Taylor et al., 2016; Zhang et al., 2018), ECV release is likely to occur at the cellular level in such cases. However, this type of ECV release might be different from what occurs under physiological conditions (Nagar et al., 2017; Phua et al., 2017; Hoang-Minh et al., 2018).
Second, the firm interaction of the IFT-A complex with the IFT-B complex is crucial for the import of ciliary GPCRs, including GPR161, SMO, and SSTR3. Therefore, it is also reasonable that IFT88(Δα)-expressing IFT88-KO cells demonstrate virtually the same phenotype as IFT144-KO cells (Hirano et al., 2017) and TULP3-knockdown cells (Mukhopadhyay et al., 2010; Badgandi et al., 2017), in which entry of GPCRs into cilia is impaired. TULP3 is likely to act as an adaptor that bridges membrane proteins and the IFT machinery by directly or indirectly binding to membrane proteins (Badgandi et al., 2017) and by directly binding to the IFT-A complex (Mukhopadhyay et al., 2010; Hirano et al., 2017). As TULP3 binds to the IFT-A core subcomplex IFT144/IFT140/IFT122 (Mukhopadhyay et al., 2010; Hirano et al., 2017), the IFT-B complex containing IFT88(Δα) cannot indirectly associate with TULP3. As the driving force for the import of GPCRs across the ciliary gate is provided by the kinesin-II motor, which directly binds to the IFT-B complex (Funabashi et al., 2018), disengagement of the IFT-A complex from the anterograde train, even though the IFT-A complex itself is intact, is likely to result in impaired GPCR import.
An issue to be addressed in the future is where the IFT machinery is assembled and where the kinesin-II motor and cargo proteins are loaded. In this context, it is interesting to note the superresolution imaging studies by us and others that indicated two distinct pools of IFT proteins at the ciliary base (Yang et al., 2015, 2018; Katoh et al., 2020); one at the TFs projecting from the basal body, where IFT88 appears to occupy the gaps between CEP164-positive blades with ninefold symmetry (Katoh et al., 2020), and the other in the TZ. We speculate that the TF pool represents the site for recruiting IFT proteins from the cell body, whereas the TZ pool may be a potential standby place of IFT particles, where IFT proteins from the TF pool and those from the retrograde trains can be mixed (Nakayama and Katoh, 2020).
Taking the results together, this study demonstrated that the IFT-A and IFT-B complexes play crucial roles in ciliary protein trafficking in a mutually dependent manner.
MATERIALS AND METHODS
Request a protocol through Bio-protocol (https://en.bio-protocol.org/e2687).
Plasmids, antibodies, reagents, and KO cell lines
Expression vectors for IFT proteins and their deletion constructs used in this study are listed in Supplemental Table S1; many of them were used in our previous studies (Katoh et al., 2015, 2016; Funabashi et al., 2017, 2018; Hirano et al., 2017; Takei et al., 2018; Nozaki et al., 2019). Antibodies used in this study are as follows: monoclonal mouse anti–Ac-α-tubulin (6-11B-1) from Sigma-Aldrich; polyclonal rabbit anti-IFT88 (13967-1-AP), anti-IFT57 (11083-1-AP), anti-IFT140 (17460-1-AP), anti-GPR161 (13398-1-AP), anti-ARL13B (17711-1-AP), and anti-mCherry (26765-1-AP) and monoclonal mouse anti-GFP (66002-1-Ig) from Proteintech; monoclonal mouse anti-ARL13B (N295B/66) from Abcam; monoclonal mouse anti-SMO (sc-166685) from Santa Cruz; monoclonal mouse anti-FOP (2B1) from Abnova; monoclonal mouse anti-RFP (3G5) from MBL Life Science; polyclonal rabbit anti-tRFP (AB233) from Evrogen; Alexa Fluor–conjugated secondary antibodies (A11034, A21131, A21127, A21137, and A21242) from Molecular Probes; and peroxidase-conjugated secondary antibodies (115-035-166 and 111-035-144) from Jackson ImmunoResearch. GST-tagged anti-GFP Nb prebound to glutathione–Sepharose 4B beads were prepared as described previously (Katoh et al., 2015, 2018). SAG was purchased from Enzo Life Sciences. The IFT88-KO cell line (#88-1-8) (Katoh et al., 2017) and the IFT144-KO cell line (#144-2-1) (Hirano et al., 2017) were established and characterized as described previously.
VIP assay and immunoblotting analysis
The VIP assay and subsequent immunoblotting analysis were performed as described previously (Katoh et al., 2015, 2016) with minor modifications (Nishijima et al., 2017); experimental details for the VIP assay were described elsewhere (Katoh et al., 2018). HEK293T cells were grown on a six-well plate, cotransfected with expression vectors for EGFP-fused and mChe/tRFP-fused proteins using Polyethylenimine Max (Polysciences), and cultured for 24 h in high-glucose DMEM (Nacalai Tesque) supplemented with 5% fetal bovine serum (FBS). The transfected cells were then lysed in 250 µl of HMDEKN cell-lysis buffer (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EDTA, 25 mM KCl, 0.05% NP-40) containing protease inhibitor cocktail (Nacalai Tesque). After 20 min on ice, the lysates were centrifuged at 16,100 × g for 15 min. The supernatants (200 µl) were transferred to a 0.2 ml eight-tube strip to which GST-tagged anti-GFP Nb bound to glutathione–Sepharose 4B beads (approximately 5 µl bed volume of the beads) was added and incubated for 1 h at 4°C with constant rotation of the tubes. After centrifugation of the tubes at 2,000 × g for 10 s, the precipitated beads were washed three times with lysis buffer (180 µl), transferred to a 96-well plate or a 96-well glass-bottom plate (AGC Techno Glass), and observed under a BZ-8000 all-in-one type microscope (Keyence) with a 20×/0.75 NA objective lens under fixed conditions (sensitivity ISO 400, exposure 1/30 or 1/10 s for green fluorescence; and sensitivity ISO 800, exposure 1/10, 1/5, or 1/2 s for red fluorescence), unless otherwise noted.
The beads conjugated with fluorescent proteins were then subjected to immunoblotting analysis. Proteins on the beads were separated by SDS–PAGE and electroblotted onto an Immobilon-P membrane (Merck Millipore). The membrane was blocked in 5% skimmed milk and incubated sequentially with primary antibody and peroxidase-conjugated secondary antibody. Protein bands were detected using the Chemi-Lumi One L kit (Nacalai Tesque).
Preparation of IFT88-KO cells stably expressing mChe-fused IFT88 constructs or SSTR3-EGFP
Lentiviral vectors for the expression of IFT88 constructs were prepared as described previously (Takahashi et al., 2012). In brief, pRRLsinPPT-mChe-IFT88 or its deletion construct was transfected into HEK293T cells together with packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE [ Thomas et al., 2009]; kind gifts from Peter McPherson, McGill University), and culture media were replaced 8 h after transfection. Culture media containing lentiviral particles were collected at 24, 36, and 48 h after transfection, passed through a 0.45-µm filter, and centrifuged at 32,000 × g at 4°C for 4 h using an R15A rotor and Himac CR22G centrifuge (Hitachi Koki). Precipitated lentiviral particles were resuspended in Opti-MEM (Invitrogen). IFT88-KO cells expressing the mChe-fused IFT88 construct were prepared by adding the lentiviral suspension to the culture medium, followed by a 24-h incubation. These cells were used for subsequent analyses. Expression of SSTR3-EGFP by infection of a lentiviral vector was performed as described previously (Hirano et al., 2017).
Immunofluorescence analysis, TIRF microscopy, and live cell imaging
Human telomerase reverse transcriptase–immortalized retinal pigment epithelial 1 (hTERT-RPE1) cells (CRL-4000; American Type Culture Collection) were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown on coverslips up to 100% confluence and starved for 24 h in Opti-MEM containing 0.2% bovine serum albumin.
Unless otherwise noted, immunofluorescence analysis was performed as described previously (Takahashi et al., 2012; Hirano et al., 2017; Nozaki et al., 2017). Cells on coverslips were fixed with 3% paraformaldehyde for 5 min at 37°C, permeabilized with 100% methanol for 5 min at −20°C, and washed three times with phosphate-buffered saline. The fixed/permeabilized cells were blocked with 10% FBS, incubated sequentially with primary and secondary antibodies diluted in Can Get Signal Immunostain Solution A (Toyobo) (for detection of IFT140 and SMO) or in 5% FBS (for detection of the other proteins), and observed using an Axiovert 200M microscope (Carl Zeiss) or an AxioObserver microscope (Carl Zeiss).
TIRF microscopy was performed as described previously (Takahashi et al., 2012; Kubo et al., 2015; Hamada et al., 2018). IFT88-KO cells expressing mChe-IFT88(WT) or mChe-IFT88(Δα) were serum-starved for 24 h on a glass-bottom culture dish (Greiner Bio-One), placed on a microscope stage prewarmed to 37°C, and observed using a TIRFM ECLIPSE Ti microscope (Nikon) at a video rate using NIS-Elements imaging software.
Live cell imaging to observe ECV release was performed as described previously (Takatsu et al., 2013; Nakamura et al., 2020). Briefly, IFT88-KO cells expressing EGFP-IFT88(Δα) were serum-starved for 24 h on a glass-bottom culture dish and observed under an Axio Observer microscope with an AxioCam 506 mono camera and a Plan-Apochromat 63×/1.4 NA oil immersion objective lens (Carl Zeiss). Z-stacks of seven images spaced 0.26 μm were acquired sequentially every 5 min. The region of interest was cropped and maximum-intensity projection images were generated and then converted into a movie file by Zen imaging software (Carl Zeiss).
Supplementary Material
Acknowledgments
We thank Peter McPherson for providing plasmids for the lentiviral production and Helena Akiko Popiel for critical reading of the manuscript. This work was supported in part by grants from the Japan Society for the Promotion of Science (grant numbers 19H00980 and 20H04904 to K.N. and 18H02403 to Y.K.); a grant of JRPs-LEAD with UKRI from the Japan Society for the Promotion of Science to K.N.; a grant from the Uehara Memorial Foundation to Y.K.; and the Kyoto University internal grant ISHIZUE to K. N.
Abbreviations used:
- Ac-tubulin
acetylated α-tubulin
- BBS
bardet–biedl syndrome
- ECV
extracellular vesicle
- EGFP
enhanced green fluorescent protein
- GPCR
G protein-coupled receptor
- GST
glutathione S-transferase
- hTERT-RPE1
human reverse transcriptase-immortalized retinal pigment epithelial 1
- ICK
intestinal cell kinase
- IFT
intraflagellar transport
- KO
knockout
- mChe
mCherry
- Nb
nanobody
- SAG
Smoothened Agonist
- SMO
Smoothened
- TF
transition fiber
- TIRF
total internal reflection fluorescence
- TPR
tetratricopeptide repeat
- tRFP
TagRFP
- TZ
transition zone
- VIP
visible immunoprecipitation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0556) on November 11, 2020.
REFERENCES
- Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST (2019). Cellular signaling by primary cilia in development, organ function and disease. Nat Rev Nephrol 15, 199–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts H, Knoers N (2013. [updated 2018]). Cranioectodermal dysplasia. In: GeneReviews [Internet], eds. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, Seattle: University of Washington, 24027799.. [PubMed] [Google Scholar]
- Badgandi HB, Hwang S, Shimada IS, Loriot E, Mukhopadhyay S (2017). Tubby family proteins are adaptors for ciliary trafficking of integral membrane proteins. J Cell Biol 216, 743–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs F, Anderson KV (2017). Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol 9, a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG (2012). Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem 287, 11689–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Weber K, Engel BD, Lechtreck K-F, Lorentzen E (2014). Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo binding sites? Bioessays 36, 463–467. [DOI] [PubMed] [Google Scholar]
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SEC, Horn N, Willer JR, Mans D, et al. (2016). An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun 7, 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Hildebrandt F (2017). Ciliopathies. Cold Spring Harb Perspect Biol 9, a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 144, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Michisaka S, Terada M, Sugawa M, Nakayama K (2017). Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol Biol Cell 28, 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Okazaki M, Sugawa M, Nakayama K (2018). Interaction of heterotrimeric kinesin-II with IFT-B-connecting tetramer is crucial for ciliogenesis. J Cell Biol 217, 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol 9, a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante ED, Caspary T (2020). Signaling in the primary cilum through the lens of the Hedgehog pathway. WIREs Dev Biol 9, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Tsurumi Y, Nozaki S, Katoh Y, Nakayama K (2018). Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol Biol Cell 29, 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Miyoshi K, Shikada S, Amano G, Wang Y, Yoshimura T, Katayama T (2019). TULP3 is required for localization of membrane-associated proteins ARL13B and INPP5E to primary cilia. Biochem Biophys Res Commun 509, 227–234. [DOI] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K (2017). Intraflagellar transport-A complex mediates ciliary entry as well as retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell 28, 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang-Minh LB, Dutra-Clarke M, Breunig JJ, Sarkisian MR (2018). Glioma cell proliferation is enhanced in the presence of tumor-derived cilia vesicles. Cilia 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S-H, Somatilaka BN, Badgandi H, Palicharla VR, Walker R, Shelton JM, Qian F, Mukhopadhyay S (2019). Tulp3 regulates renal cystogenesis by trafficking of cystoproteins to cilia. Curr Biol 29, 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, Pigino G (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20, 1250–1255. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Chiba S, Nakayama K (2020). Practical method for superresolution imaging of primary cilia and centrioles by expansion microscopy using an amplibody for fluorescence signal amplification. Mol Biol Cell 31, 2195– 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K (2017). Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol Biol Cell 28, 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nakamura K, Nakayama K (2018). Visible immunoprecipitation (VIP) assay: a simple and versatile method for visual detection of protein-protein interactions. Bio-protocol 8, e2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci 128, 2351–2362. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K (2016). Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem 291, 10962–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 131, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Kobayashi M, Nozaki S, Yagi C, Hatsuzawa K, Katoh Y, Shin H-W, Takahashi S, Nakayama K (2015). SNAP23/25 and VAMP2 mediate exocytic event of transferrin receptor-containing recycling vesicles. Biol Open 4, 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukic I, Rivera-Molina F, Toomre D (2016). The IN/OUT assay: a new tool to study ciliogenesis. Cilia 5, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué E, Liem KF Jr (2019). Tulp3 is a ciliary trafficking gene that regulates polycystic kidney disease. Curr Biol 29, 803–812. [DOI] [PubMed] [Google Scholar]
- Liu P, Lechtreck KF (2018). The Bardet-Biedl syndrome protein complex is an adaptor expanding the cargo of range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci USA 115, E934–E943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R (2014). G-protein-coupled receptors, Hedgehog signaling and primary cilia. Sem Cell Dev Biol 33, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24, 2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar AR, Goldstein JS, Herranz-Pérez V, Portran D, Ye F, Garcia-Verdugo J-M, Nachury MV (2017). An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Noguchi T, Takahara M, Omori Y, Furukawa T, Katoh Y, Nakayama K (2020). Anterograde trafficking of ciliary MAP kinase-like ICK/CILK1 by the intraflagellar transport machinery is required for intraciliary retrograde protein trafficking. J Biol Chem 295, 13363–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J Biochem 163, 155–164. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y (2020). Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit Rev Biochem Mol Biol 55, 179–196. [DOI] [PubMed] [Google Scholar]
- Nishijima Y, Hagiya Y, Kubo T, Takei R, Katoh Y, Nakayama K (2017). RABL2 interacts with the intraflagellar transport B complex and CEP19 and participates in ciliary assembly. Mol Biol Cell 28, 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Castro Araya RF, Katoh Y, Nakayama K (2019). Requirement of IFT-B–BBSome complex interaction in export of GPR161 from cilia. Biol Open 8, bio043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Kobayashi T, Nakayama K (2018). BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One 13, e0195005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K (2017). Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci 130, 563–576. [DOI] [PubMed] [Google Scholar]
- Paige Taylor S, Kunova Bosakova M, Varecha M, Balek L, Barta T, Trantirek L, Jelinkova I, Duran I, Vesela I, Forlenza KN, et al. (2016). An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum Mol Genet 25, 3998–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C (2009). A genetic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Setou M, Rohatgi R, Reiter JF, Ikegami K, Inoue T (2017). Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA 94, 4457–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol 164, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB (2002). Intraflagellar transport. Nat Rev Mol Cell Biol 3, 813–825. [DOI] [PubMed] [Google Scholar]
- Schmidts M (2014). Clinical genetics and pathobiology of ciliary chondrodysplasias. J Pediatr Genet 3, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M, Katoh Y, Nakamura K, Hirano T, Sugawa M, Tsurumi Y, Nakayama K (2018). Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum Mol Genet 27, 516–528. [DOI] [PubMed] [Google Scholar]
- Takahara M, Kunii M, Nakamura K, Harada A, Hirano T, Katoh Y, Nakayama K (2019). C11ORF74 interacts with the IFT-A complex and participates in ciliary BBSome localization. J Biochem 165, 257–267. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K (2012). Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci 125, 4049–4057. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Katoh Y, Ueda T, Waguri S, Murayama T, Takahashi S, Shin H-W, Nakayama K (2013). Mitosis-coupled, microtubule-dependent clustering of endosomal vesicles around centrosomes. Cell Struct Funct 38, 31–41. [DOI] [PubMed] [Google Scholar]
- Takei R, Katoh Y, Nakayama K (2018). Robust interaction of IFT70 with IFT52–IFT88 in the IFT-B complex is required for ciliogenesis. Biol Open 7, bio033241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E (2016). The intraflagellar transport machinery. Cold Spring Harb Perspect Biol 8, a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J 35, 773–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS (2009). Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem 284, 12410–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi Y, Hamada Y, Katoh Y, Nakayama K (2019). Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol Biol Cell 30, 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Chong WM, Wang W-J, Mazo G, Tanos B, Chen Z, Tran TMN, Chen Y-D, Weng RR, Huang C-E, et al. (2018). Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun 9, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C (2015). Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep 5, 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Nager AR, Nachury MV (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol 217, 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Paige Taylor S, Ennis HA, Forlenza KN, Duran I, Li B, Ortiz Sanchez JA, Nevarez L, Nickerson DA, Bamshad M, et al. (2018). Expanding the genetic architecture and phenotypic spectrum in the skeletal ciliopathy. Hum Mutat 39, 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.