Abstract
Inflammatory bowel diseases (IBD) develop via convergence of environmental, microbial, immunological, and genetic factors. Alterations in the gut microbiota have been associated with development and progression of IBD, but it is not clear which populations of microbes are involved or how they might contribute to IBD. We review the genetic and environmental factors affecting the gut microbiota, the roles of gut microbes and their bioproducts in the development and clinical course of IBD, and strategies by which microbiome-based therapies can be used to prevent, manage, and eventually cure IBD. We discuss research findings that help bridge the gap between the basic sciences and clinical application.
Keywords: intestinal microbiome, host-microbe interactions, colitis, mucosal inflammation, dysbiosis, pathobiont, immune regulation
Inflammatory Bowel Diseases – Emerging global diseases having a microbial basis
A microbial basis for Inflammatory Bowel Diseases (IBD) has been suspected as far back as the 19th century when IBD were first described by Samuel Wilks in 1859.1,2 These heterogenous complex immune disorders of the gastrointestinal (GI) tract present as two major clinical phenotypes: ulcerative colitis (UC) and Crohn’s disease (CD). UC primarily involves confluent inflammation of the colonic mucosa, whereas CD is often transmural, patchy, and can involve any part of the GI tract from mouth to anus.3 IBD likely arise from a convergence of host genetic, microbial, and environmental factors, each necessary, but not sufficient by themselves to cause disease. Carrying a genetic risk variant, for example, does not necessarily forebode the development of disease. Furthermore, given that IBD have only risen to their modern incidence and prevalence in industrializing societies within the last century, this rise is not likely to have arisen from genetic drift or natural selection.4 Rather, rapid changes in societal norms, lifestyle, diet, and environment brought about by human actions are likely causing or contributing to the rising tide of these diseases, particularly among individuals who are genetically susceptible. The membership and function of the human gut microbiome are particularly sensitive to these dietary and environmental shifts, exacerbating immune imbalance and promoting the development of IBD in genetically prone individuals. While there is wide consensus that IBD have a microbial basis, many gaps in knowledge remain, particularly regarding the mechanistic intricacies that link environment, genetics, and gut microbes to the development of disease. Here, we present a selective review and commentary on the microbial basis of IBD to develop a conceptual framework for the risk factors, epidemiology, and etiopathogenesis of inflammatory bowel diseases. This framework will provide a better understanding into the causative factors to help advance the field and lay a path for the development of microbiome-based interventions to prevent, manage, and eventually cure IBD.
What evidence supports a microbial basis for human IBD?
Experimental IBD models typically involve either genetic or chemical induction of disease. In almost all of these models, under germ-free conditions, disease either does not develop at all or is significantly attenuated, suggesting that microbes are essential for the development of intestinal inflammation in IBD.5 Moreover, the gut microbiota of patients with active disease exhibit alterations to bacterial diversity, composition, and/or abundance compared to healthy individuals.6 As there are many clinical phenotypes of IBD, a single microbial community profile that accounts for all disease types is highly unlikely. That said, there are microbial patterns shared among IBD patients such as reduced microbial diversity, decreased relative abundance of Firmicutes, and an increase in Proteobacteria.7–11 In a longitudinal study of UC patients with ileal pouch anal anastomosis (IPAA), for example, sequential sampling of the ileal pouch mucosa show that most patients develop immune activation and inflammation early on before the appearance of endoscopic or histologic changes.12,13 In addition, numerous studies point towards disruptions of the gut microbiome arising from environmental and/or host factors which create mismatches in host-microbe interactions to promote the development of IBD.14,15
It should also be noted that, to date, most studies of the gut dysbiosis associated with various forms of IBD have been focused on bacteria, without much consideration of the potential role of other members of the gut microbiome such as fungi, bacteriophage, and Archaea. Some studies have suggested that alterations in the virome and mycobiome have a role in the pathogenesis of human IBD.16–18 Bacteriophages (phages), for example, are prokaryotic viruses that only infect bacteria and archaea, and are the most prevalent viruses in the human gastrointestinal tract. Though gut phages demonstrate low intrapersonal and high interpersonal variation, there appear to be shared phage populations among individuals globally, raising the possibility that the “healthy” gut phageome is significantly diminished in IBD patients.17,19,20 Moreover, IBD patients harbor an increased abundance of phages belonging to the Caudovirales order, but the clinical significance of this is unclear.21 Less is known about Archaea, which include methanogens, in human IBD, other than that they appear to be less abundant during active disease.22 On the other hand, both experimental and clinical studies have implicated fungi in the pathogenesis of IBD. IBD patients display increased abundance of Candida albicans but a decreased abundance of Saccharomyces cerevisae.23 A commensal skin yeast, Malessezia restricta, was found in the colonic mucosa of CD patients and exacerbated colitis in mice by stimulating an anti-fungal signaling molecule, CARD9.24 Greater fungal diversity was found in CD patients compared to controls, suggesting the mycobiota could be involved in the pathogenesis of IBD.25 In addition, anti-Saccharomyces cerevisiae antibody (ASCA) is the most robust biomarker of CD.26–30 ASCA targets mannan, the polysaccharide component of the fungal cell wall. ASCA generation and targeting is thought to indicate fungal invasion of the intestinal epithelium. Similarly, mice have increased susceptibility to chemically-induced colitis when they lack Dectin-1, an innate pattern-recognition receptor for β-1,3-glucan, another fungal cell wall component.31 The development of colitis in these mice is believed to be due to the increased Candida invasion of epithelial cells. Furthermore, polymorphisms in Dectin-1 and CARD9 have been linked to severe forms of ulcerative colitis and Candidiasis.32–35 Despite the increasing connections between fungi and phage with IBD, much remains to be learned about their contributions to IBD pathogenesis.
What defines gut dysbiosis in IBD?
It is estimated that there are trillions of microbial organisms in the human gut that have a symbiotic relationship with their hosts, fulfilling essential functions in healthy individuals such as nutrition, host defense, and immune development.36–38 In most healthy individuals, 99% of the intestinal microbiota is composed of Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, with Firmicutes and Bacteroidetes together accounting for about 90% of the microbiota.39–42 Firmicutes and Bacteroidetes in conjunction with oligosaccharide-fermenting bacteria (e.g. Bifidobacterium) are able to produce short-chain fatty acids (SCFAs) by fermenting dietary plant fibers that are naturally indigestible by humans.43,44 SCFAs, specifically butyrate and propionate, are energy sources for the colonic epithelium and have been shown to play key roles in regulating intestinal immune homeostasis.45
Although there are two major dominant phyla, there are thousands of different bacterial species and an even greater number of strains in each individual. By taxonomic criteria, the membership and relative abundance of the gut microbiota varies considerably from individual to individual. However, their core functions, e.g. those important for microbial fitness and adaptation to the various niches along the GI tract, are more similar.38,39 Some of these functions, such as conversion of primary to secondary bile acids and generation of SCFAs, are important for immune regulation and metabolic homeostasis.44,46–49 Loss or reduction of these functions can potentially impact host-microbe interactions essential for intestinal mucosal and immune homeostasis. Thus, despite the large inter-individual differences in taxonomic representation, functional criteria may be more useful for distinguishing relative states of health and disease in IBD.37,39,50
Gut dysbiosis occurs when the diversity, composition, and/or functions of the intestinal microbiome are disrupted, negatively impacting the individual, for instance through the loss of intestinal homeostasis and inappropriate immune activation.50–52 IBD patients display reductions in biodiversity (mostly Firmicutes), decreased stability, and an expansion of Proteobacteria such as Enterobacteriaceae, Bilophila, and certain members of Bacteroidetes.7,10,11,53 Loss of biodiversity may lead to a loss or reduction of key functions necessary for maintaining intestinal barrier integrity and regulating the host immune system, potentially resulting in inflammation and increased immune responses.54,55 There is also an increase in mucolytic bacteria and pathogenic bacteria, leading to the degradation of the mucosal barrier to allow for increased penetration of pathogens into the intestinal tissues.56–59
While gut dysbiosis is associated with IBD, it has been difficult to determine if these changes are a cause or consequence of these diseases. It is also possible these alterations, while not the main drivers of IBD development, contribute to the evolution and progression of these diseases by amplifying and sustaining the immune/inflammatory process. (Figure 1) By doing so, they promote tissue injury and impaired wound healing, disease complications (e.g. fibrosis and strictures, fistula formation, abscess, extraintestinal disease manifestations), and the decline or even extinction of key commensal microbiota that would be needed to restore immune and intestinal homeostasis.
Figure 1. Factors influencing dysbiotic and chronic inflammatory state in IBD.
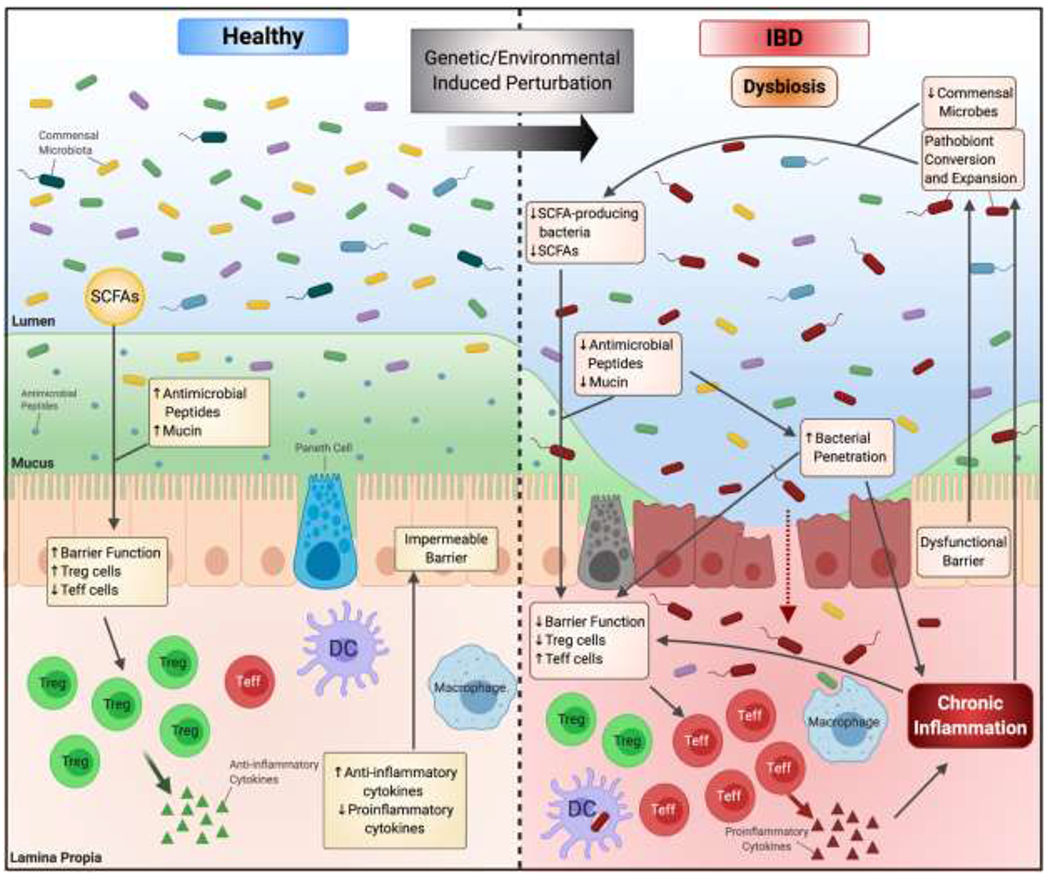
In healthy individuals, commensal bacteria provide functions that benefit the host. (left) Certain bacteria produce short-chain fatty acids (SCFAs) such as butyrate to regulate intestinal homeostasis. Antimicrobial peptides produced by Paneth cells and the intact mucus layer prevent bacterial invasion of the epithelial layer. SCFAs are an energy source for the epithelial cells and help maintain a functional barrier. SCFAs also provide anti-inflammatory properties by facilitating differentiation of regulatory T-cells (Treg) and effector T-cells (Teff). However, in IBD patients, perturbations due to genetic and/or environmental factors lead to dysbiosis of the gut microbiota. (right) IBD patients display a reduction of commensal bacteria, including SCFA-producing bacteria. This leads to decrease in SCFAs and their beneficial effects including the ability to modulate Treg and Teff differentiation. The imbalance of Treg and Teff cells leads to increased production of proinflammatory cytokines. Due to the perturbations, the mucosal layer is damaged, leading to a dysfunctional barrier. This allows for bacterial penetration, triggering an inflammatory cascade against the invading bacteria. Furthermore, pathobionts, commensal bacteria that can display pathogenic properties given the right opportunity, emerge and expand in the inflammatory conditions. This continues to promote inflammatory conditions, allowing for the adapted pathobionts to survive and further limiting commensal growth. The loss of commensals, emergence of pathobionts, and the imbalance of immune regulation leads to a chronic inflammatory state.
What determines the development of IBD dysbiosis?
Genetic Susceptibility and Immune Regulation:
Genome-wide association studies (GWAS) led to the discovery of genes and gene loci associated with IBD susceptibility.60–62 Many of these genes encode for important components for sensing and adapting to changes in the gut microbiome. For example, two of the major IBD susceptibility genes include NOD2 and ATG16L1.63,64 These genes are involved in the autophagy pathway, a major component in pathogen sensing and clearance to ensure only the correct microbes remain in the host. Many of them play roles in other defense pathways, thus mutations in these genes could affect broadly crucial host defenses and immune mechanisms. For example, bacterial sensor NOD2 is a receptor for muramyl dipeptide, a constituent of both Gram-positive and -negative bacteria, and plays a role in many cell types in addition to its primary role in autophagy. NOD2 mutations lead to impaired epithelial clearance of invasive bacteria, dysfunction of Paneth cells, and decreased antimicrobial peptide production.65,66
Aside from impaired microbial sensing ability and bacterial clearance, IBD gene variants may also alter intestinal immune homeostasis by disrupting the intestinal epithelial barrier. The colonic mucosal barrier is composed of a firmly attached and mostly sterile inner layer and an outer layer with varying mucus thickness.67 A healthy mucus layer will permit some microbial attachment to the mucus, but does not permit the microbes from reaching the intestinal epithelium. As NOD2 mutations lead to defects in components necessary for an intact mucus layer, the disrupted barrier combined with decreased antimicrobial peptide production allows for the expansion of pathogenic bacteria, but decreases in overall diversity and reduced abundance of beneficial bacteria such as the butyrate-producing genus Roseburia.68–70 Notably, NOD2 expression is also reduced in genetically wild-type germ-free mice and is restored through supplementation of commensal bacteria, highlighting the complex microbe-by-genotype interactions that characterize IBD.71
Mutations in these IBD-associated genes may lead to defects in immune responses including T-cell differentiation (IL10, IL21), TH17 cell maintenance (IL23R, JAK2, PTPN2), and NF-κB activation (TNF-signaling genes).72–74 Defects in immune inhibitory pathways impair immune cells from properly responding, leading to aberrant immune responses such as release of pro-inflammatory factors and dampened anti-inflammatory responses. The dysregulation of inflammatory mediators influences the chronic inflammatory state characteristic of IBD pathogenesis.
Polymorphisms in these IBD susceptibility genes alter the gut microbiome and cause imbalances in critical host-microbe interactions which increase the risk of IBD development. These genes could act as genetic biomarkers to identify individuals at higher risk of developing IBD to potentially treat the disease before onset of symptoms. Despite the success of identifying many significantly associated susceptibility genes through GWAS, these genes only account for <25% of predicted heritability.61 While GWAS is proficient at identifying common risk alleles, less well represented and rare risk alleles are less likely to be detected. Thus, further genetic analyses will be needed to comprehensively discover these uncommon variants to better utilize susceptibility genes as predictive biomarkers.
Diet:
Of the many changes that have arisen through Westernization, shifts in diet have been particularly significant and are associated with risk for IBD. Increased variety, availability, affordability, processing, and formulation of foods made possible through improved socioeconomic standards have resulted in increased consumption of animal-based, high calorie, high fat, and processed sugar diets low in dietary fiber.75,76 Unsurprisingly, changes in diet have led to decreased diversity and impaired microbial function, and often override other determinants of gut microbiota composition including genetics and mucus deterioration.77–80 Mice fed a typical Western diet exhibited changes in the colonic mucus characteristic of IBD pathogenesis, which were associated with shifts in intestinal microbial communities.54,80 Microbial diversity in composition and function are negatively impacted by diets with reduced dietary fiber, which may not be recoverable even with resumption of fiber in the diet.78
Some commensal microbes are able to naturally ferment indigestible dietary fiber to form SCFAs. SCFAs including acetate, propionate, and butyrate are energy sources for colonic epithelial cells and help maintain intestinal homeostasis.81,82 SCFAs maintain intestinal homeostasis by facilitating production and differentiation of colonic regulatory T cells (Treg).83,84 For example, Faecalibacterium prausnitzii was found to have anti-inflammatory properties through its ability to produce butyrate, allowing for Treg and Th17 regulation.85,86 IBD patients display a reduction in SCFA-producing bacteria like F. prausnitzii and butyrate production, and recovery of SCFA-producing bacteria or the SCFA itself led to amelioration of inflammatory environment.87,88
Bile acids:
Bile acids are emulsifiers needed for fat absorption and digestion and come in many forms. They also have profound effects in shaping the gut microbiome.89 Their relative proportion in secreted bile is highly influenced by dietary composition and their conversion from primary to secondary acids by gut bacteria is important to make them bioactive ligands for downstream receptors such as anti-inflammatory signaling receptor FXR and TGR5.90–93 The ratio of conjugated to deconjugated bile acids also plays a role in health and disease; a shift towards excess conjugated bile acids may promote inflammatory conditions and can lead to blooms of potentially harmful bacteria. UC and CD patients display bile acid malabsorption and secondary bile acid deficiency which can lead to persistent diarrhea.92,94,95
A study using genetically susceptible IL-10-deficient mice fed a saturated fat diet identified a diet-induced bloom of Bilophila wadsworthia, a bile-tolerant commensal microbe, which increased the incidence of spontaneous colitis in these mice.53 Another study examined human subjects on either a plant-based or animal-based diet and compared their fecal microbial communities.78 They found an increase in bile-tolerant organisms and decreases in plant polysaccharide metabolizing Firmicutes. Furthermore, reductions in the Ruminococcaceae family in UC pouch patients led to reductions in secondary bile acids.92 These studies show that alterations to the microbial community structure due to diet and nutrients can be associated with intestinal inflammation and higher incidence of colitis.53,92
Barrier integrity and impact of mucosal inflammation:
As gut microbes evolved to adapt to the changing gut environment, the host immune system likely co-evolved to allow selective engraftment by microbes that benefit the host. The symbiotic relationship between the gut microbes and host is in particularly delicate balance at the intestinal mucosa, a primary interface between host and microbe where proper separation is essential for intestinal homeostasis.67 A healthy mucus layer will permit some microbial attachment to the mucus, but deters most microbes from reaching the surface of the intestinal epithelium. Additionally, the secretion of antimicrobial peptides and IgA helps to prevent microbial invasion of the mucus layer.96,97 In IBD, many of these factors break down. The intestinal epithelium becomes more permeable due to defects or downregulation of processes that regulate tight junctions, mucus produced by goblet cells, transepithelial transport, nutrient digestion and absorption, and mucosal restitution.54,98–101 The defective intestinal barrier allows for increased penetration of bacteria and closer contact with the intestinal epithelial surface and beyond.
These changes to the mucosal barrier lead in turn to shifts in mucosal bacterial communities. Mucolytic, or mucin-degrading, bacteria are able to use mucins as an energy source and release sugars from the glycosylated mucins for other bacteria to potentially use.102 Akkermansia muciniphila, a mucolytic commensal, is generally abundant in healthy microbiota but reduced in the guts of IBD patients. While the mucolytic commensal populations diminish, the overall mucosal bacteria population increases due to increased infiltration of the damaged barrier by invading commensal bacteria.103 Moreover, one study showed that compositional changes in the mucosal bacterial community occurred prior to changes in the fecal microbial community and the onset of symptoms.104 These studies show the inappropriate presence of microbes at the mucosal surface could be a potential cause of increased inflammation.
While it is not clear whether the mis-localization of commensals into the mucosal layer is causative or adaptive to the depleting mucus layer, it is known that there are certain bacteria that directly exacerbate epithelial barrier dysfunction. Adherent-invasive Escherichia coli (AIEC) have a heightened ability to adhere to gastrointestinal epithelial cells and are present at increased abundance in IBD patients.58 Contrastingly from the invading commensals, AIEC have the capacity to further disrupt intestinal barrier integrity by producing an α-hemolysin and stimulating the release of pro-inflammatory cytokines.58,105,106 AIEC isolated from CD patients have demonstrated the capacity to replicate in macrophages, further evading killing by immune cells while initiating an inflammatory cascade.107
As the epithelial barrier is the first line of defense against invading microbes, the disruption of the barrier is highly detrimental and a key factor in IBD pathogenesis. With the loss of the protective mucus layer, a downstream cascade of events leads to an inflammatory reaction to the invading bacteria. It is not well-understood if the initial barrier dysfunction arises as a reaction to the invasive and adherent mucosal bacteria or if an alternative factor is responsible, and the microbes are simply adapting to the changing environment. Nevertheless, an intact, microbially-impermeable mucosal barrier is necessary to maintain intestinal homeostasis and the perturbation of this barrier increases the risk of colitis.
Developmental factors:
Community assembly in early life is complex and impacted by many factors such as genetics, environment, and interspecies interactions. These early life exposures and developmental programs are essential for developing a stable, yet resilient microbial community that promotes good health.108–110 As a consequence, other non-resident microbes encountered later in life may be rejected by fitness filters set by both gut microbiota and host immune system early in life. Conversely, perturbations of the gut microbiome that compromise gut microbial diversity, fitness, and function have been shown to become more susceptible to invasion and colonization.111 In addition, studies have shown long-lasting effects of dysbiosis of the early-life microbiome result in the inability of the gut immune system to develop properly.108,109 The development of immune tolerance to both “self” and to acquired gut microbes takes place during a window of time early in life, after which the imprinting process is set.112 Supporting this notion is the finding that a vertically transferred gut dysbiosis acquired by pups from antibiotic-treated, IL-10 gene-deficient dams are at a significantly greater risk for developing spontaneous and DSS-induced colitis.15 Furthermore, the consequences of dysbiosis could not be fixed by late life exposure to commensal microbes. Besides antibiotics, other factors such as formula feeding, Caesarian-sections, and diet can also disrupt the early-life microbiome and infant immune development.113–120 Studies show that by 1-2 years of age, the infant microbiota begins to resemble a profile characteristic of the adult microbiota.121–124 Genetically predisposed individuals may therefore have a brief developmental window of time to correct their microbiota before the gut microbiome and host immunity are fully developed. Interventions to engraft key microbes that were missing in early life are less likely to be effective past this developmental window.125–127
Are there disease-promoting gut microbiota that trigger the onset of IBD?
There has been no consensus or definitive evidence that IBD are caused by classical pathogens. Pathogens are generally not indigenous to the gut microbiota and typically possess virulence mechanisms that take advantage of weaknesses in host physiology. Most are infectious and typically affect a broader range of individuals than just patients with IBD. Thus, pathogens in the traditional sense are not likely to be the main cause of IBD. More likely, particularly based on recent reports, the culprits may actually be part of the commensal microbiota involving populations that can display pathogenic properties given the right opportunity, context, and circumstance. These microbes have been called “pathobionts”, although the term has shortcomings in accurately reflecting the pleomorphic states of microbes that can be both beneficial and harmful.128 Pathobionts are difficult to detect by current measures of microbial membership, genome sequence, or even function, especially if performed outside of physiological context. Underscoring the latter point, it is hypothesized that colonization with these microbes in a healthy, non-IBD susceptible host will not cause disease. B. wadsworthia, for example, is a bile-tolerant, sulfite-reducing Proteobacteria that blooms and causes colitis in IL-10-gene deficient mice fed a high saturated fat diet.53 It has also been reported to be associated with human IBD.78 Yet, it is found in the commensal microbiota of healthy, non-IBD subjects, and when engrafted into the gut microbiota of wild type, non-IBD-prone mice, fails to cause colitis.
However, in IBD-prone subjects, the inflammatory gut environment may impose a fitness cost on some commensal microbes, and at the same time, provide a selective advantage for other microbes that can survive in a proinflammatory environment. These microbes could contribute to and sustain the inflammatory process to impair the recovery of other commensal microbes by taking advantage of the inflamed environment. For example, oxygen levels and free oxygen radicals increase in the inflamed gut environment.129,130 Oxygen-tolerant microbes and microbes able to utilize oxygen radicals are selected for survival. E. coli is able to use nitric oxide as an electron acceptor, giving AIEC a competitive advantage against gut community members during states of inflammation.131 Overall in dysbiotic conditions, functions for survival in a hostile inflammatory environment, such as oxidative stress tolerance, metabolite uptake, and carbohydrate metabolism, are upregulated for higher fitness.50,132
Bacteroides fragilis has been implicated as a potential pathobiont of IBD as it is found in more than 60% of the biofilm mass of IBD patients despite being a low-abundance member of the commensal microbiota.133 Furthermore, an ileal pouchitis study of UC patients showed B. fragilis bloomed in many subjects before and during the development of pouchitis.12 Despite the harsh inflammatory conditions, B. fragilis is able to bloom and persist, due to its ability to adapt and respond to this environment making it a prime pathobiont candidate. One potential factor for its survival could be the wide range of capsular polysaccharide coats at its disposal, although each individual capsular polysaccharide function has not yet been fully studied.134,135
Currently, it is still unclear whether changes in the commensal microbiome population in IBD are a cause or effect of the inflammatory environment. Even if they are not causative, do these microbes actively contribute to and sustain the heightened immune and inflammatory state? Equally unclear is whether the emergence of the pathobionts has a larger impact on IBD pathogenesis than the loss of commensal microbes and their homeostatic functions.
Does the reduction of key commensal gut microbes cause or contribute to human IBD?
Commensal microbes play a large role in maintaining proper immune and intestinal homeostasis with specific host-microbe interactions. These microbes provide key functions in digestion and nutrition, pathogen limitation, immune regulation, development, and mucosal properties including barrier function, generation of antimicrobial peptides, mucus, and repair. Inflammatory conditions that suppress these beneficial populations and effects can potentially trigger or contribute to IBD.
Microbial metabolic activity is crucial in combating intestinal inflammation, and reduction of key microbes involved in these activities allows for this chronic inflammatory state. As mentioned above, SCFAs produced by bacteria such as F. prausnitzii have anti-inflammatory properties that protect the intestinal epithelium.55 In patients with active IBD, the reduction in F. prauznitzii and the associated reduction of this metabolic function critically impacts intestinal homeostasis.136 Functional reduction may in fact be more relevant than the taxonomic identity of reduced bacterial populations during IBD: suppression of other butyrate-producing bacteria such as Roseburia hominis or Eubacterium rectale similarly leads to reduced SCFA levels, impaired immune regulation, and poor colonic mucosal health.11,137 Microbial tryptophan metabolism provides another example illustrating the critical interactions between microbial metabolic products and host health. Some commensals are able to produce indole compounds from tryptophan catabolism to activate the host aryl hydrocarbon receptor (AHR). AHR modulates differentiation and activity of T-cells, and AHR expression is dampened in IBD patients.138 CARD9 deficient mice displayed intestinal inflammation due to the inability of the microbiota to produce AHR agonists from tryptophan, and this inflammation was attenuated after adding tryptophan-metabolizing Lactobacillus or AHR agonists.139 Furthermore, Clostridium-colonized mice expressed high levels of indoleamine 2,3-dioxygenase (IDO) which catabolizes tryptophan to kynurenine which also activates the AHR.140,141 IDO expression increases during gut colonization, suggesting microbiota directly and indirectly modulate AHR agonists to affect immune regulation for a positive host-microbe relationship.142
The metabolic products of microbes in the gut also contribute to a complex web of inter-microbial interactions. The ecological concept of a foundation or keystone species in determining and sustaining stability and resilience of the gut microbiome applies to both health and disease. While both terms have been interchangeably used, a foundation species is often the most abundant and is able to physically modify the environment to produce and maintain habitats that support other organisms. A keystone species has a disproportionately large effect on its environment relative to its abundance. These species play a critical role in maintaining the structure of an ecological community by directly or indirectly affecting the abundance of many other organisms in the ecosystem. Loss of either the keystone or foundation species has strong destabilizing effects, leading to reduction in biodiversity and decreased resistance to invasion.143 Currently, there are few studies regarding potential keystone or foundation species in IBD, but if these roles do exist in the context of the gut microbiota, the clinical and translational value in understanding their ecology cannot be overstated.
Bacteroidetes is an abundant phylum in healthy individuals and is known to decrease in IBD patients.50,52 Some members of the Bacteroides genus have been shown to exhibit beneficial anti-inflammatory functions.144–146 Moreover, many Bacteroides are able to digest complex polysaccharides and release simple carbohydrate products for utilization by other bacteria.147,148 Previous studies show Bacteroides to be highly connected in ecological networks of the gut microbiota, and removal of these species harms the network.143,149 Bacteroides are therefore hypothesized as potential foundation species that act to maintain the gut microbial community. A recent study from our group has shown a previously uncharacterized murine strain of Bacteroides, Bacteroides sp. CL1-UC, to be an integral component to reduce risk of spontaneous colitis of IL-10 gene-deficient mice when engrafted into a dysbiotic gut in early life.125 Engraftment of this single strain early in life restored, to a significant extent, the development of the gut microbiome and host immune tolerance. Losing or failing to acquire key foundation species, especially early in life, could have a lasting impact on the gut microbiome, particularly in complex immune disorders like IBD. Once the community has been established, priority effects makes it difficult for even other keystone species to engraft into a stable, yet dysbiotic microbial community.150
Understanding microbial metabolic functions as well as the individual contribution of each microbe to the community will help clarify the roles necessary to maintain a healthy microbiome. Currently, it is difficult to determine whether the gain of function (pathobiont bloom) or the loss of function (losing commensals) has a larger impact on IBD pathogenesis. (Figure 1) However, both of these concepts show the complexity in predicting IBD development and progression. As both the emergence of pathobionts and the loss of commensals have significant impact on creating and maintain an inflammatory environment, researchers must better understand the intricate host-microbe and microbe-microbe interactions.
What present and future microbiome-based interventions are there for IBD?
Current therapeutic approaches for IBD involve surgical and medical modalities that each have their limitations. The development of microbial interventions for IBD is being intensely investigated, but the promise of this approach remains unrealized. Given that the gut microbiomes among individual are very different, it is unlikely that a single formulation simply based on taxonomical criteria is likely to work in everyone. A more logical approach is one based on functional criteria to restore health to both gut microbiota and host. For example, butyrate-producing bacteria have been shown to be beneficial and anti-inflammatory, but would it matter if F. prausnitzii is chosen over R. hominis? As mentioned above, some microbes may play a larger role in community assembly (i.e. keystone and foundation species), thus these microbes may be needed to properly restore the microbial balance.
Fecal microbial transplantation (FMT) and probiotics are currently the main microbial therapies for IBD. Both aim to re-introduce beneficial microbes into the dysbiotic gut of the patient to restore intestinal balance, but one involves transplanting fecal matter from a healthy donor and the other involves introducing a defined live microbial consortium.151,152 FMT appears potentially effective in UC patients, but multiple doses and preclearance of indigenous gut microbiota with antibiotics may be needed to achieve a sustainable effect. The long-term effects of these treatments are unknown.153–158 A recent technical review on probiotics in UC or CD patients showed little or no evidence of efficacy.159 The effectiveness and sustainability of these treatments may be related to the ability of FMT and probiotics to engraft in the recipient gut, which is extremely difficult because of the stability and resilience of pre-existing microbiota.160 In addition, with uncontrolled, active disease, most commensal microbes would find the environment too hostile to be fit.52 Prebiotics are dietary fiber supplementations that stimulate growth of specific, putatively beneficial bacteria already present in the gut.161 Prebiotics do not require live bacterial transfer and are easier to administer, but the efficacy depends on the fiber choice and the growth of gut microbes that can metabolize it. While promising, long-term effects on humans and gut microbiota remain unknown. Synbiotics involve supplementing a probiotic with a nutritional source (prebiotic) to provide a competitive advantage for the probiotic strain to increase their colonization chances.162 However, without engraftment, probiotics and synbiotics would likely have to be give frequently in large doses, making compliance and adherence by patients more difficult. Live bacteria probiotics are harder to control and do not guarantee production of beneficial compounds. Furthermore, once engrafted, the probiotic strain may no longer be of use, but may continue to fill a niche that could otherwise be occupied by a different beneficial strain. Postbiotics bypass the need for live bacteria and use bioactive microbe-derived small molecules.163 Thus, they have the advantage of controlled dosage without the concerns about engraftment. However, the development of postbiotics is still in its infancy and further research is needed to understand their mechanisms of action and long-term health effects. Currently, probiotics, synbiotics, and postbiotics in IBD models have limited data and require more stringently controlled trials.136,159,161–165 Postbiotics have yet to pass clinical trials, but experimental models demonstrate potential therapeutic potential.136
Genetically engineered microbes using synthetic biology represent a future therapeutic approach for IBD. Modified bacteria could act as biosensors or delivery vehicles to monitor and/or react to molecules being produced in the gut and potentially deliver therapeutics at disease sites.166–169 However, as live microbes, biosafety issues with containment and toxicity have to be considered. Bacteria kill-switches have been added into some synthetic bacteria, but overall their effects on humans still remain unknown.170
What needs to be done to better understand the role of gut microbes in IBD?
Despite the fact that there is a microbial basis in IBD, research in this area has been hampered by technical, clinical, and conceptual challenges. It remains unclear and controversial, for example, whether IBD-associated dysbiosis causes these diseases or is merely a consequence of them.171–173 Most studies to date have also been cross-sectional in design, relying on samples acquired from a single time point without associated clinical metadata such as the state (active vs quiescent) and type of IBD (CD vs UC), regional involvement (small vs large intestine), diet, medications, etc. Without this information, these data become difficult to interpret. By incorporating a time-series of region-specific specimen collection from both host and microbiota, much more can be gained from clinical investigations that seek to identify pathobionts, functional markers, and host pathways and targets that may have a role in disease pathogenesis. In addition, patients serve as their own controls, obviating the problems of genetic, environmental, and microbiome variation among individuals. As an example, clinically useful host and microbial biomarkers could be identified that predict responsiveness to treatment before initiation of a therapeutic intervention, thereby ensuring better clinical outcomes. To identify potential gut microbial communities or specific populations that cause or trigger IBD or disease relapses, samples and clinical metadata should be obtained before and after the onset of disease. In most instances, we simply lack the ability to know who will develop inflammatory bowel diseases and when. However, there are opportunities where very high-risk subjects, such as first-degree relatives of IBD patients, can be studied before the onset of disease.174 For example, studies have shown that nearly half of UC patients who undergo colectomy and IPAA will develop an inflammatory condition of the ileal pouch, called pouchitis, within 2-3 years from surgery.12,175 This work revealed the emergence of pathobionts before and during the development of pouchitis, one example being B. fragilis, which is otherwise a minor constituent of the healthy gut microbiome. A second finding was the identification of an anomalous host gene response to microbial stimuli that was only seen in the pouch and not the pre-pouch mucosa, i.e. the segment of ileum above the pouch that is usually disease-free. Interestingly, the same transcriptomic response was observed in a subset of CD patients, suggesting certain IBD patients display specific responses to the gut microbiota, rendering them at higher risk, especially if microbial factors are encountered that can trigger the onset of disease.176 Going forward, attention to study design is extremely important to gain leads that may provide insight into causality and consequence in IBD.
Adding to the challenge of human studies is that most studies have been performed on fecal microbiota, which arguably only represent the luminal microbial population of the distal colon, and not microbiota of other areas where active disease may be (e.g. cecum, upper GI tract). Substantiation is needed to support claims that fecal analysis can be used to identify biomarkers of region-specific IBD, but this may be technically limiting with the need for colonic lavage (which skews the gut microbial populations) and the inability to reach certain regions of the bowel endoscopically. It is also well known that there are large inter-individual differences in membership and metagenomics of gut microbiota particularly at the subphylum level which makes it difficult to identify meaningful associations even with large diverse subject populations. While much has been gained through cultivation-independent next generation sequencing approaches, they have their limits as well. Analyses of 16S ribosomal DNA sequences only provide information about what microbial populations are most represented in a sample. With the clustering platforms currently used for data analysis, operational taxonomic units (OTUs), or clusters of similar sequence variants of the 16S rDNA marker gene sequence defined by having 97% sequence homology, are limited to identifying broad shifts and states in microbial membership that can only be resolved to the genus level. This is inadequate, as microbial populations belonging to the same species can vary widely in genomic content and function. Moreover, 16S rDNA profiles provide no functional information. Attempts to extrapolate functional information based on genomic sequence information of reference strains are likely to be misleading, as genomes and functional properties among similar species and strains in different contexts can vary considerably.177,178 Metagenomic data (microbial DNA) acquired through shotgun sequencing provides much more information, but even this data is limited because the function of most microbial genes remains unclear. As a consequence, functional profiles drawn from short read sequences are based on annotation of only 10-20% of the total sequences which skew interpretation to most highly represented genes that are shared across the microbiome, many mediating basic functions such as metabolism, propagation, and fitness. What is becoming increasingly apparent is the growing need to understand the microbial basis of IBD at a microbial strain, genomic, and functional level and in the proper context and circumstance. Metatranscriptomic (mRNA expression) and metabolomic profiles, particularly when analyzed along with metagenomic and whole genome sequences of particular strains of interest can be extremely useful in providing a high level of resolution by combining compositional and functional profiles. However, having associated metadata from the patient side such as gene/RNA/protein expression, histology, and single cell genomics would provide a much better picture of functional impact that the microbiota may be having at that particular point of time and stage of disease. To determine if observed correlations are actually related, experimental models involving gnotobiotic animal technologies have been frequently used to evaluate the functional impact of human gut microbiota and individual microbial strains. Human gut microbiota, for instance, readily colonize the guts of germ-free mice, but whether the observed relationships between human gut microbes and their murine host are representative of those in the human gut remains unclear.173 Arguably, human IBD is very unique and individual, and it is unlikely that any of the existing animal models of experimental IBD will faithfully recapitulate the milieu, host-microbe relationships, and etiopathophysiological underpinnings of human IBD. In this regard and because functional testing in human subjects is difficult for both technical and ethical reasons, patient-derived intestinal organoids may provide a useful platform to test specific hypotheses between matched sets of the patient microbiota.179 Microfluidic systems using the “organ-on-a-chip” model may also be a potential method to mimic IBD conditions using live patient-derived cells.180 Ultimately, a look back to the patient using newer technologies that are emerging like single cell genomics, spatial transcriptomes, super-resolution microscopy, etc. can provide further validation for the in vitro and experimental findings. This type of iterative, multi-prong approach will become increasingly necessary to reconstruct events that will lead to a better understanding of the microbial basis of IBD.
Knowledge gained through proper study design, the emergence of novel and enabling technologies, advances in bioinformatics analytics, and more representative experimental models will be transformative and have immediate clinical application. We will be able to identify individuals at high risk for the development of IBD, in whom early interventions such as correction of gut dysbiosis can be implemented to prevent disease. The identification of specific microbial populations, strains, and genomic element translates into highly specific therapeutics that can target those microbes specifically without collateral damage to the rest of the gut microbiome. The discovery of novel, effective microbiome-based biotherapeutics will become part of our therapeutic armamentarium. Similarly, mitigation strategies can be developed to protect host pathways that are targeted by pathobionts. Finally, panels of host and microbiome biomarkers can be developed that will guide physicians when determining best therapies for effective disease management, better clinical outcomes, and even cure.
Acknowledgements:
We thank Ashley Sidebottom and Megan Kennedy for their suggestions, review, and edits of this manuscript.
Grant Support:
The authors’ research is supported by the following grants from NIDDK (RC2DK122394, R01DK47722, R01DK113788, NIH T32 DK07074, and the Center for Interdisciplinary Study of Inflammatory Intestinal Diseases (P30 DK42086). Additional support has been provided by the Gastrointestinal Research Foundation (GIRF) of Chicago, the David and Ellen Horing Research Fund, and the Helmsley Charitable Trust.
Abbreviations:
- AHR
aryl hydrocarbon receptor
- AIEC
adherent-invasive Escherichia coli
- ASCA
anti-Saccharomyces cerevisiae antibody
- FMT
fecal microbial transplantation
- GI
gastrointestinal
- GWAS
genome-wide association studies
- IDO
indoleamine 2,3-dioxygenase
- IPAA
ileal pouch anal anastomosis
- OTU
operational taxonomic unit
- SCFAs
short-chain fatty acids
Footnotes
Disclosures: Eugene B. Chang is the co-founder and Chief Medical Officer for AVnovum Therapeutics.
References
- 1.Kirsner JB. The historical basis of the idiopathic inflammatory bowel diseases. Inflamm Bowel Dis 1995;1:2–26. [Google Scholar]
- 2.Hawkins HP. An address on the natural history of ulcerative colitis and its bearing on treatment. Br Med J 1909;1:765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn S, Eisenstein S. Inflammatory Bowel Disease Presentation and Diagnosis. Surg Clin North Am 2019. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan GG, Ng SC. Understanding and Preventing the Global Increase of Inflammatory Bowel Disease. Gastroenterology 2017;152:313–321.e2. [DOI] [PubMed] [Google Scholar]
- 5.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: Antibiotics, probiotics, and prebiotics. Gastroenterology 2004;126:1620–1633. [DOI] [PubMed] [Google Scholar]
- 6.Lucas López R, Grande Burgos MJ, Gálvez A, et al. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. Apmis 2017;125:3–10. [DOI] [PubMed] [Google Scholar]
- 7.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano A, Umeno J, Okamoto Y, et al. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J Gastroenterol Hepatol 2018;33:1590–1597. [DOI] [PubMed] [Google Scholar]
- 9.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004;53:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishino K, Nishida A, Inoue R, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol 2018;53:95–106. [DOI] [PubMed] [Google Scholar]
- 11.Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–1283. [DOI] [PubMed] [Google Scholar]
- 12.Vineis JH, Ringus DL, Morrison HG, et al. Patient-specific Bacteroides genome variants in pouchitis. MBio 2016;7:e01713–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Dalal S, Antonopoulos D, et al. Early Transcriptomic Changes in the Ileal Pouch Provide Insight into the Molecular Pathogenesis of Pouchitis and Ulcerative Colitis. Inflamm Bowel Dis 2017;23:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011;145:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyoshi J, Bobe AM, Miyoshi S, et al. Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep 2017;20:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepage P, Colombet J, Marteau P, et al. Dysbiosis in inflammatory bowel disease: A role for bacteriophages? Gut 2008;57:424–425. [DOI] [PubMed] [Google Scholar]
- 17.Duerkop BA, Kleiner M, Paez-Espino D, et al. Murine colitis reveals a disease-associated bacteriophage community. Nat Microbiol 2018;3:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes A, Wu M, McNulty NP, et al. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci U S A 2013;110:20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manrique P, Bolduc B, Walk ST, et al. Healthy human gut phageome. Proc Natl Acad Sci U S A 2016;113:10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuo T, Lu XJ, Zhang Y, et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol 2008;8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limon JJ, Tang J, Li D, et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019;25:377–388.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol 2008;43:831–841. [DOI] [PubMed] [Google Scholar]
- 26.Barnes RMR, Allan S, Taylor-Robinson CH, et al. Serum antibodies reactive with Saccharomyces cerevisiae in inflammatory bowel disease: Is IgA antibody a marker for crohn’s disease? Int Arch Allergy Immunol 1990;92:9–15. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie H, Main J, Pennington CR, et al. Antibody to selected strains of Sacharomyces cerevisiae (baker’s and brewer’s yeast) and Candida albicans in Crohn’s disease. Gut 1990;31:536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giaffer MH, Clark A, Holdsworth CD. Antibodies to Saccharomyces cerevisiae in patients with Crohn’s disease and their possible pathogenic importance. Gut 1992;33:1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinton JF, Sendid B, Reumaux D, et al. Anti-Saccharomyces cerevisiae manna antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: Prevalence and diagnostic role. Gut 1998;42:788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard ML, Lamas B, Liguori G, et al. Gut fungal microbiota: The Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis 2015;21:656–665. [DOI] [PubMed] [Google Scholar]
- 31.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science 2012;336:1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia XM, Tang B, Zhu L Le, et al. CARD9 mediates Dectin-1-induced ERK activation by linking Ras-GRF1 to H-Ras for antifungal immunity. J Exp Med 2014;211:2307–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014;14:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanternier F, Mahdaviani SA, Barbati E, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol 2015;135:1558–1568.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckburg PB, Bik EM, Bernstein CN, et al. Microbiology: Diversity of the human intestinal microbial flora. Science 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science 2008;320:1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andoh A Physiological role of gut microbiota for maintaining human health. Digestion 2016;93:176–181. [DOI] [PubMed] [Google Scholar]
- 42.Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol 2015;50:495–507. [DOI] [PubMed] [Google Scholar]
- 43.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: A new clinical frontier. Gut 2016;65:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc 2003;62:67–72. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad MS, Krishnan S, Ramakrishna BS, et al. Butyrate and glucose metabolism by colonocytes in experimental colitis in mice. Gut 2000;46:493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- 47.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science 2013;341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyce SA, Gahan CGM. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annu Rev Food Sci Technol 2016;7:313–333. [DOI] [PubMed] [Google Scholar]
- 49.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006;47:241–259. [DOI] [PubMed] [Google Scholar]
- 50.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79–R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carding S, Verbeke K, Vipond DT, et al. Dysbiosis of the gut microbiot a in disease. Microb Ecol Heal Dis 2015;26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank DN, Amand AL St., Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alipour M, Zaidi D, Valcheva R, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohn’s Colitis 2016;10:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002;122:44–54. [DOI] [PubMed] [Google Scholar]
- 57.Chassaing B, Darfeuillemichaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011;140:1720–1728.e3. [DOI] [PubMed] [Google Scholar]
- 58.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004;127:412–421. [DOI] [PubMed] [Google Scholar]
- 59.Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013;502:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franke A, McGovern DPB, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–606. [DOI] [PubMed] [Google Scholar]
- 64.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005;307:731–734. [DOI] [PubMed] [Google Scholar]
- 66.Hisamatsu T, Suzuki M, Reinecker HC, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 2003;124:993–1000. [DOI] [PubMed] [Google Scholar]
- 67.Johansson MEV, Holmén Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 2011;108:4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 2014;6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mondot S, Barreau F, Nabhani Z Al, et al. Altered gut microbiota composition in immune-impaired Nod2−/− mice. Gut 2012;61:634–635. [DOI] [PubMed] [Google Scholar]
- 70.Imhann F, Vich Vila A, Bonder MJ, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A 2009;106:15813–15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009;361:2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Momozawa Y, Mni M, Nakamura K, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 2011;43:43–47. [DOI] [PubMed] [Google Scholar]
- 74.Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet 2005;14:3499–3506. [DOI] [PubMed] [Google Scholar]
- 75.Dolan KT, Chang EB. Diet, gut microbes, and the pathogenesis of inflammatory bowel diseases. Mol Nutr Food Res 2017;61:1600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rizzello F, Spisni E, Giovanardi E, et al. Implications of the westernized diet in the onset and progression of IBD. Nutrients 2019;11:1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carmody RN, Gerber GK, Luevano JM, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015;17:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ussar S, Griffin NW, Bezy O, et al. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab 2015;22:516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schroeder BO, Birchenough GMH, Ståhlman M, et al. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 2018;23:27–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mariadason JM, Barkla DH, Gibson PR. Effect of short-chain fatty acids on paracellular permeability in Caco- 2 intestinal epithelium model. Am J Physiol - Gastrointest Liver Physiol 1997;272:G705–12. [DOI] [PubMed] [Google Scholar]
- 82.Wong JMW, Souza R De, Kendall CWC, et al. Colonic health: Fermentation and short chain fatty acids. In: Journal of Clinical Gastroenterology. Vol 40.; 2006:235–243. [DOI] [PubMed] [Google Scholar]
- 83.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013;500:232–236. [DOI] [PubMed] [Google Scholar]
- 84.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science 2013;341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang M, Zhou Q, Dorfman RG, et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol 2016;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou L, Zhang M, Wang Y, et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm Bowel Dis 2018;24:1926–1940. [DOI] [PubMed] [Google Scholar]
- 87.Martin R, Chain F, Lu J, et al. Tu1988 Impact of the Commensal Bacterium Faecalibacterium prausnitzii in a Non Active Inflammation Murine Model. Gastroenterology 2013;144:S-897–S-898. [Google Scholar]
- 88.Varela E, Manichanh C, Gallart M, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther 2013;38:151–161. [DOI] [PubMed] [Google Scholar]
- 89.Best N van, Rolle-Kampczyk U, Schaap FG, et al. Bile acids drive the newborn’s gut microbiota maturation. Nat Commun 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gadaleta RM, Erpecum KJ Van, Oldenburg B, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011;60:463–472. [DOI] [PubMed] [Google Scholar]
- 91.Islam KBMS, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011;141:1773–1781. [DOI] [PubMed] [Google Scholar]
- 92.Sinha SR, Haileselassie Y, Nguyen LP, et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 2020;27:659–670.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wahlström A, Sayin SI, Marschall HU, et al. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab 2016;24:41–50. [DOI] [PubMed] [Google Scholar]
- 94.Fitzpatrick LR, Jenabzadeh P. IBD and Bile Acid Absorption: Focus on Pre-clinical and Clinical Observations. Front Physiol 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tiratterra E, Franco P, Porru E, et al. Role of bile acids in inflammatory bowel disease. Ann Gastroenterol 2018;31:266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kinnebrew MA, Pamer EG. Innate immune signaling in defense against intestinal microbes. Immunol Rev 2012;245:113–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tollin M, Bergman P, Svenberg T, et al. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides 2003;24:523–530. [DOI] [PubMed] [Google Scholar]
- 98.Schmitz H, Barmeyer C, Fromm M, et al. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 1999;116:301–309. [DOI] [PubMed] [Google Scholar]
- 99.Kucharzik T, Walsh SV., Chen J, et al. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am J Pathol 2001;159:2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soderholm JD, Peterson KH, Olaison G, et al. Epithelial permeability to proteins in the noninflamed ileum of Crohn’s disease? Gastroenterology 1999;117:65–72. [DOI] [PubMed] [Google Scholar]
- 101.Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract 2008;62:762–769. [DOI] [PubMed] [Google Scholar]
- 102.Belzer C, Chia LW, Aalvink S, et al. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. MBio 2017;8:e00770–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 2010;105:2420–2428. [DOI] [PubMed] [Google Scholar]
- 104.Glymenaki M, Singh G, Brass A, et al. Compositional Changes in the Gut Mucus Microbiota Precede the Onset of Colitis-Induced Inflammation. Inflamm Bowel Dis 2017;23:912–922. [DOI] [PubMed] [Google Scholar]
- 105.Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004;127:80–93. [DOI] [PubMed] [Google Scholar]
- 106.Subramanian S, Rhodes JM, Hart CA, et al. Characterization of epithelial IL-8 response to inflammatory bowel disease mucosal E. coli and its inhibition by mesalamine. Inflamm Bowel Dis 2008;14:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Subramanian S, Roberts CL, Hart CA, et al. Replication of colonic crohn’s disease mucosal Escherichia coli isolates within macrophages and their susceptibility to antibiotics. Antimicrob Agents Chemother 2008;52:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Milani C, Duranti S, Bottacini F, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev 2017;81:e00036–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhuang L, Chen H, Zhang S, et al. Intestinal Microbiota in Early Life and Its Implications on Childhood Health. Genomics, Proteomics Bioinforma 2019;17:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015;17:690–703. [DOI] [PubMed] [Google Scholar]
- 111.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nabhani Z Al, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol 2020;13:183–189. [DOI] [PubMed] [Google Scholar]
- 113.Salminen S, Gibson GR, McCartney AL, et al. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 2004;53:1388–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. Cmaj 2013;185:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Azad MB, Konya T, Persaud RR, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG An Int J Obstet Gynaecol 2016;123:983–993. [DOI] [PubMed] [Google Scholar]
- 116.O’Sullivan A, Farver M, Smilowitz JT. The Influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights 2015;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rigas A, Rigas B, Glassman M, et al. Breast-feeding and maternal smoking in the etiology of Crohn’s disease and ulcerative colitis in childhood. Ann Epidemiol 1993;3:387–392. [DOI] [PubMed] [Google Scholar]
- 118.Baron S, Turck D, Leplat C, et al. Environmental risk factors in paediatric inflammatory bowel diseases: A population based case control study. Gut 2005;54:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bager P, Simonsen J, Nielsen NM, et al. Cesarean section and offspring’s risk of inflammatory bowel disease: A national cohort study. Inflamm Bowel Dis 2012;18:857–862. [DOI] [PubMed] [Google Scholar]
- 120.Li Y, Tian Y, Zhu W, et al. Cesarean delivery and risk of inflammatory bowel disease: A systematic review and meta-analysis. Scand J Gastroenterol 2014;49:834–844. [DOI] [PubMed] [Google Scholar]
- 121.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota Ruan Y, ed. PLoS Biol 2007;5:1556–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016;8:343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Robertson RC, Manges AR, Finlay BB, et al. The Human Microbiome and Child Growth–First 1000 Days and Beyond. Trends Microbiol 2019;27:131–147. [DOI] [PubMed] [Google Scholar]
- 125.Miyoshi J, Miyoshi S, Delmont TO, et al. Early-life microbial intervention reduces colitis risk promoted by antibiotic-induced gut dysbiosis. bioRxiv 2020:2020.03.11.987412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Laforest-Lapointe I, Arrieta MC. Patterns of early-life gut microbial colonization during human immune development: An ecological perspective. Front Immunol 2017;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012;336:489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jochum L, Stecher B. Label or Concept – What Is a Pathobiont? Trends Microbiol 2020;S0966-842X:30104–9. [DOI] [PubMed] [Google Scholar]
- 129.Zhu H, Li YR. Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: Updated experimental and clinical evidence. Exp Biol Med 2012;237:474–480. [DOI] [PubMed] [Google Scholar]
- 130.Rigottier-Gois L Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J 2013;7:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013;339:708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 2013;14:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 2005;43:3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krinos CM, Coyne MJ, Weinacht KG, et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 2001;414:555–558. [DOI] [PubMed] [Google Scholar]
- 135.Cui HL, Lee SM, VanLare JM, et al. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci U S A 2008;105:3951–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Russo E, Giudici F, Fiorindi C, et al. Immunomodulating Activity and Therapeutic Effects of Short Chain Fatty Acids and Tryptophan Post-biotics in Inflammatory Bowel Disease. Front Immunol 2019;10:2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vermeiren J, Abbeele P van den, Laukens D, et al. Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol Ecol 2012;79:685–696. [DOI] [PubMed] [Google Scholar]
- 138.Monteleone I, Rizzo A, Sarra M, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011;141:237–48. [DOI] [PubMed] [Google Scholar]
- 139.Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011;331:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seok SH, Ma ZX, Feltenberger JB, et al. Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J Biol Chem 2018;293:1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aidy S El, Derrien M, Aardema R, et al. Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef Microbes 2014;5:67–77. [DOI] [PubMed] [Google Scholar]
- 143.Trosvik P, Muinck EJ de. Ecology of bacteria in the human gastrointestinal tract--identification of keystone and foundation taxa. Microbiome 2015;3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ramakrishna C, Kujawski M, Chu H, et al. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat Commun 2019;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zocco MA, Ainora ME, Gasbarrini G, et al. Bacteroides thetaiotaomicron in the gut: Molecular aspects of their interaction. Dig Liver Dis 2007;39:707–712. [DOI] [PubMed] [Google Scholar]
- 146.Delday M, Mulder I, Logan ET, et al. Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of Crohn’s disease. Inflamm Bowel Dis 2019;25:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Macfarlane GT, Englyst HN. Starch utilization by the human large intestinal microflora. J Appl Bacteriol 1986;60:195–201. [DOI] [PubMed] [Google Scholar]
- 148.Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe 2008;4:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One 2014;9:e10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol 2018;15:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grehan MJ, Borody TJ, Leis SM, et al. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J Clin Gastroenterol 2010;44:551–561. [DOI] [PubMed] [Google Scholar]
- 152.Hill C, Guarner F, Reid G, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014;11:506–514. [DOI] [PubMed] [Google Scholar]
- 153.Paramsothy S, Paramsothy R, Rubin DT, et al. Faecal microbiota transplantation for inflammatory bowel disease: A systematic review and meta-analysis. J Crohn’s Colitis 2017;11:1180–1199. [DOI] [PubMed] [Google Scholar]
- 154.Rossen NG, Fuentes S, Spek MJ Van Der, et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015;149:110–118.e4. [DOI] [PubMed] [Google Scholar]
- 155.Paramsothy S, Nielsen S, Kamm MA, et al. Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis. Gastroenterology 2019;156:1440–1454.e2. [DOI] [PubMed] [Google Scholar]
- 156.Moayyedi P, Surette MG, Kim PT, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015;149:102–109.e6. [DOI] [PubMed] [Google Scholar]
- 157.Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 2017;389:1218–1228. [DOI] [PubMed] [Google Scholar]
- 158.Shi Y, Dong Y, Huang W, et al. Fecal microbiota transplantation for ulcerative colitis: A systematic review and meta-analysis. PLoS One 2016;11:e0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Preidis GA, Weizman AV., Kashyap PC, et al. AGA Technical Review on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology 2020;S0016-5085:34732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee STM, Kahn SA, Delmont TO, et al. Tracking microbial colonization in fecal microbiota transplantation experiments via genome-resolved metagenomics. Microbiome 2017;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017;8:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: A randomised controlled pilot trial. Gut 2005;54:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsilingiri K, Rescigno M. Postbiotics: What else? Benef Microbes 2013;4:101–107. [DOI] [PubMed] [Google Scholar]
- 164.Jonkers D, Stockbrügger R. Probiotics and inflammatory bowel disease. J R Soc Med 2003;96:167–171. [PMC free article] [PubMed] [Google Scholar]
- 165.Steed H, MacFarlane GT, Blackett KL, et al. Clinical trial: The microbiological and immunological effects of synbiotic consumption - A randomized double-blind placebo-controlled study in active Crohn’s disease. Aliment Pharmacol Ther 2010;32:872–883. [DOI] [PubMed] [Google Scholar]
- 166.McKay R, Ghodasra M, Schardt J, et al. A platform of genetically engineered bacteria as vehicles for localized delivery of therapeutics: Toward applications for Crohn’s disease. Bioeng Transl Med 2018;3:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meyer AJ, Segall-Shapiro TH, Glassey E, et al. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat Chem Biol 2019;15:196–204. [DOI] [PubMed] [Google Scholar]
- 168.Benbouziane B, Ribelles P, Aubry C, et al. Development of a Stress-Inducible Controlled Expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J Biotechnol 2013;168:120–129. [DOI] [PubMed] [Google Scholar]
- 169.Steidler L, Neirynck S, Huyghebaert N, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 2003;21:785–789. [DOI] [PubMed] [Google Scholar]
- 170.van de Poel I, Robaey Z. Safe-by-Design: from Safety to Responsibility. Nanoethics 2017;11:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ni J, Wu GD, Albenberg L, et al. Gut microbiota and IBD: Causation or correlation? Nat Rev Gastroenterol Hepatol 2017;14:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Khan I, Ullah N, Zha L, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Walter J, Armet AM, Finlay BB, et al. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell 2020. [DOI] [PubMed] [Google Scholar]
- 174.Santos MPC, Gomes C, Torres J. Familial and ethnic risk in inflammatory bowel disease. Ann Gastroenterol 2018;31:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stocchi L, Pemberton JH. Pouch and pouchitis. Gastroenterol Clin North Am 2001;30:223–241. [DOI] [PubMed] [Google Scholar]
- 176.Weiser M, Simon JM, Kochar B, et al. Molecular classification of Crohn’s disease reveals two clinically relevant subtypes. Gut 2018;67:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Meyer F, Trimble WL, Chang EB, et al. Functional predictions from inference and observation in sequence-based inflammatory bowel disease research. Genome Biol 2012;13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Angus HCK, Butt AG, Schultz M, et al. Intestinal Organoids as a Tool for Inflammatory Bowel Disease Research. Front Med 2020;6:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014;32:760–772. [DOI] [PubMed] [Google Scholar]


