Abstract
In Pompe disease, the deficiency of the lysosomal enzyme acid alpha-glucosidase (GAA) causes skeletal and cardiac muscle weakness, respiratory failure, and premature death. While enzyme replacement therapy using recombinant human GAA (rhGAA) can significantly improve patient outcomes, detailed disease mechanisms and incomplete therapeutic effects require further studies. Here we report a three-dimensional primary human skeletal muscle (“myobundle”) model of infantile-onset Pompe disease (IOPD) that recapitulates hallmark pathological features including reduced GAA enzyme activity, elevated glycogen content and lysosome abundance, and increased sensitivity of muscle contractile function to metabolic stress. In vitro treatment of IOPD myobundles with rhGAA or adeno-associated virus (AAV)-mediated hGAA expression yields increased GAA activity and robust glycogen clearance, but no improvements in stress-induced functional deficits. We also apply RNA sequencing analysis to the quadriceps of untreated and AAV-treated GAA−/− mice and wild-type controls to establish a Pompe disease-specific transcriptional signature and reveal novel disease pathways. The mouse-derived signature is enriched in the transcriptomic profile of IOPD vs. healthy myobundles and partially reversed by in vitro rhGAA treatment, further confirming the utility of the human myobundle model for studies of Pompe disease and therapy.
Subject terms: Musculoskeletal models, Metabolic disorders
Wang et al. describe the development of a 3D in vitro model of human skeletal muscle that recapitulates the pathophysiology of Pompe disease. They further define a Pompe disease-specific transcriptional signature, confirm its presence in the Pompe myobundles, and detect a partial reversal of the signature upon in vitro treatment of myobundles with rhGAA.
Introduction
Pompe disease (glycogen storage disease type II, or acid maltase deficiency; OMIM 232300) is an autosomal-recessive disorder of metabolism caused by mutations in the lysosomal hydrolase, acid alpha-glucosidase gene (GAA). GAA normally breaks down lysosomal glycogen, and its dysfunction results in glycogen buildup in striated and smooth muscles, cardiac complications, progressive muscle atrophy, and respiratory failure1. The disease is categorized into two major types based on age of onset: infantile-onset Pompe disease (IOPD) and late-onset Pompe disease (LOPD) correlated with the level of residual enzyme activity2,3. Patients with the classic form of IOPD have little to no enzyme activity and develop hypertrophic cardiomyopathy and a progressive respiratory failure, leading to death within 1 year of age in absence of effective treatment. Patients with LOPD have greater GAA activity, but still have significant skeletal and smooth muscle involvement, difficulty walking, and progressively reduced respiratory function. Skeletal muscle weakness is common in both IOPD and LOPD4,5, driven by physical disruption of myofibrillar architecture related to enlarged and ruptured lysosomes, accumulated autophagosomes, and cellular debris6,7. However, the molecular mechanisms underlying the human disease are incompletely understood.
The current standard of care treatment for Pompe disease, enzyme replace therapy (ERT), consisting of frequent intravenous infusions of recombinant human GAA (rhGAA), reduces muscle glycogen and can improve muscle function and patient quality of life8–11. However, ERT has several limitations, including high cost12, attenuated efficacy due to neutralizing antibodies13, inefficient muscle targeting, and variable patient response14. Therefore, both improved understanding of the mechanisms driving the disease and new therapies with enhanced long-term efficacy are needed to improve patient outcomes.
Current in vivo studies of Pompe disease mainly utilize GAA−/− mice15 that display a disease phenotype in between that of IOPD and LOPD, exhibiting glycogen accumulation but having delayed-onset of skeletal muscle weakness15,16 and only mild cardiac symptoms16. With almost 600 reported mutations in Pompe patients17 and the added complexity of modifier genes that can significantly alter disease severity even for the same mutation18, patient-specific human cell culture systems are necessary to model the wide spectrum of Pompe disease phenotypes. While two-dimensional (2D) cultures of patient-derived human myotubes have been utilized for this purpose19,20, they are not amenable to long-term culture, lack maturity relative to three-dimensional (3D) culture systems, and do not allow measurements of contractile function (twitch and tetanic contractions)21. Previously, we have developed a tissue-engineered 3D model of functional human skeletal muscle termed myobundles, using primary myoblasts22,23 or pluripotent stem cells24. These myobundles exhibit several features of native skeletal muscle (positive force-frequency relationship, length-tension relationship, mature dystrophin localization)22,23, adapt structurally and metabolically to electrical stimuli22, and demonstrate clinically relevant responses to tested drugs23. However, studies of Pompe disease in biomimetic functional 3D skeletal muscle culture systems have not yet been reported.
Here we sought to develop the first in vitro 3D model of human skeletal muscle with Pompe disease. We generated functional myobundles made from primary muscle cells derived from multiple patients with IOPD and healthy donors. Under standard culture conditions, IOPD myobundles exhibited GAA deficiency, elevated glycogen, and lysosomal enlargement, but displayed functional characteristics similar to those of healthy myobundles. Under stress conditions, including lysosomal perturbation, inhibition of glycogen phosphorylase, and glucose starvation, Pompe myobundles showed reduced tetanic force production, decreased fatigue resistance, and impaired glycogen mobilization, respectively. A 7-day treatment of IOPD myobundles with rhGAA or with adeno-associated virus (AAV)-mediated GAA expression significantly reduced glycogen accumulation, but did not rescue stress-induced functional deficits.
In addition, gene set enrichment analysis (GSEA) of RNA sequencing (RNA-seq) results indicated that IOPD myobundles exhibited transcriptomic changes characteristic of Pompe disease, including downregulation of gene sets involved in skeletal muscle contraction, increased endoplasmic reticulum stress, and reduced utilization of specific metabolic pathways. We also defined a muscle transcriptional signature of Pompe disease by comparing GAA−/− and wild-type mice, confirmed the presence of this signature in IO Pompe myobundles, and detected a partial reversal of the signature upon in vitro treatment of myobundles with rhGAA.
Results
Structural and functional characterization of IO Pompe myobundles
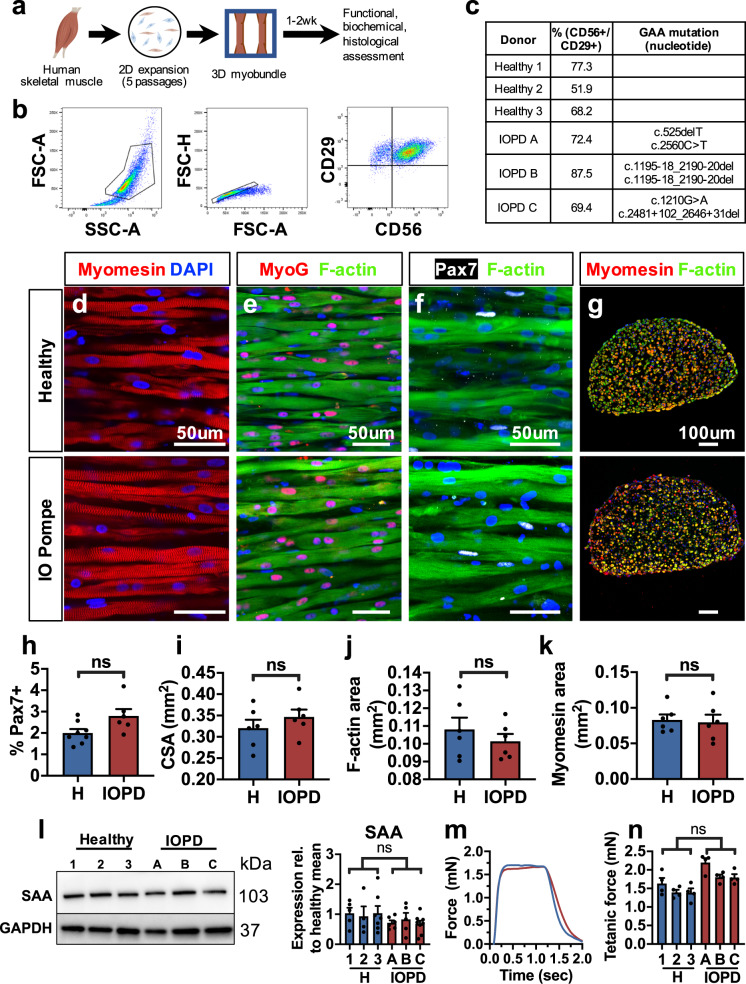
To generate a 3D in vitro model of Pompe disease muscle, we have applied our method to generate functional human myobundles from primary muscle cells (Fig. 1a)23. Cells were isolated following enzymatic digestion and outgrowth culture from muscle biopsies of multiple healthy and IOPD donors and expanded for five passages. The myogenic cell fraction of expanded cells was quantified using flow cytometry for the myoblast surface antigens CD56+CD29+ (Fig. 1b, c)25,26 and found to be similar between healthy and Pompe disease cells (65.8 ± 7.4% and 76.4 ± 5.6% myoblasts, respectively). Passaged cells were embedded into a fibrin-based gel and differentiated for 2 weeks to form 3D tissue-engineered myobundles22,23. Similar to healthy myobundles, IOPD myobundles consisted of aligned, cross-striated, multinucleated myotubes that expressed the transcription factor myogenin, indicating terminal differentiation, while harboring a pool of Pax7+ satellite-like cells (Fig. 1d–g). The percentage of Pax7+ cells was similar between healthy and IOPD myobundles (Fig. 1h), consistent with previous histological assessment of human muscle biopises27. Morphological analysis revealed no difference in total, F-actin+, or myomesin+ cross-sectional areas (CSA) between healthy and IOPD myobundles (Fig. 1i–k), and, along with no difference in sarcomeric α-actinin (SAA) protein expression (Fig. 1l), revealed that IOPD satellite cells retained normal capacity for in vitro expansion, myofiber formation, and muscle differentiation.
Fig. 1. Comparison of IOPD vs. healthy myobundle structure and function.
a Schematic of experimental flow from primary muscle cell isolation to myobundle fabrication and assessment. b Representative flow cytometry analysis of a muscle cell population expanded for five passages from IOPD patient biopsy, highlighting CD56+/CD29+ cell fraction. c Percentage of CD56+/CD29+ cells in expanded muscle cells and corresponding GAA mutations shown for each donor. d, e Representative longitudinal sections of healthy and IOPD myobundles differentiated in 3D culture for 2 weeks and stained for myomesin (d), myogenin and F-actin (e), and Pax7 and F-actin (f); nuclei are labeled with DAPI (blue). g Representative myobundle cross-sections stained for myomesin, F-actin, and DAPI. h–k, Quantification of immunostainings for (h) percentage of Pax7+ nuclei in total myobundle nuclei (n = 6–8), i total cross-sectional area (CSA) of myobundles (n = 6), j F-actin-positive CSA (n = 6), and k myomesin-positive CSA (n = 6). l Representative western blots and quantified protein expression (normalized to GAPDH) of sarcomeric α-actinin (SAA) in myobundles from three healthy (1, 2, 3) and three IOPD (A, B, C) donors (n = 4–8 myobundles per donor). m, n Representative 20 Hz tetanic force traces and n quantified force amplitudes in myobundles from healthy and IOPD donors (n = 4 myobundles per donor). Data: mean ± SEM. ns not significant.
We then assessed the force-generating capacity of myobundles from three healthy and three IOPD donors. Twitch responses of healthy and IOPD myobundles showed similar shapes and no significant differences in amplitude, time to peak twitch, or half relaxation time (Supplementary Fig. 1a–d). To compare the maximum force-generating capacities, tissues were stimulated at 20 Hz for 1 s to induce tetanus and, similar to twitch response, tetanic forces produced by healthy and IOPD myobundles were found to be comparable (Fig. 1m, n). Myobundles were also tested under a fatigue protocol in which tissues were held in tetanus for 30 s and their decline in generated contractile force was measured (Supplementary Fig. 1e)22. Greater resistance to fatigue is associated with reduced glycolysis and increased oxidative metabolism28. Healthy and IOPD myobundles displayed similar fatigue resistance based on two metrics: (1) drop in tension during stimulation and (2) impulse (area under the curve) normalized by peak force to cumulatively assess the force decline during fatigue testing (Supplementary Fig. 1f, g). Together, these results demonstrated that under normal culture conditions, 2-week old IOPD myobundles exhibited similar satellite cell abundance, muscle structure, and contractile properties to those of age-matched healthy myobundles.
Reduced GAA activity and elevated glycogen in IOPD myobundles
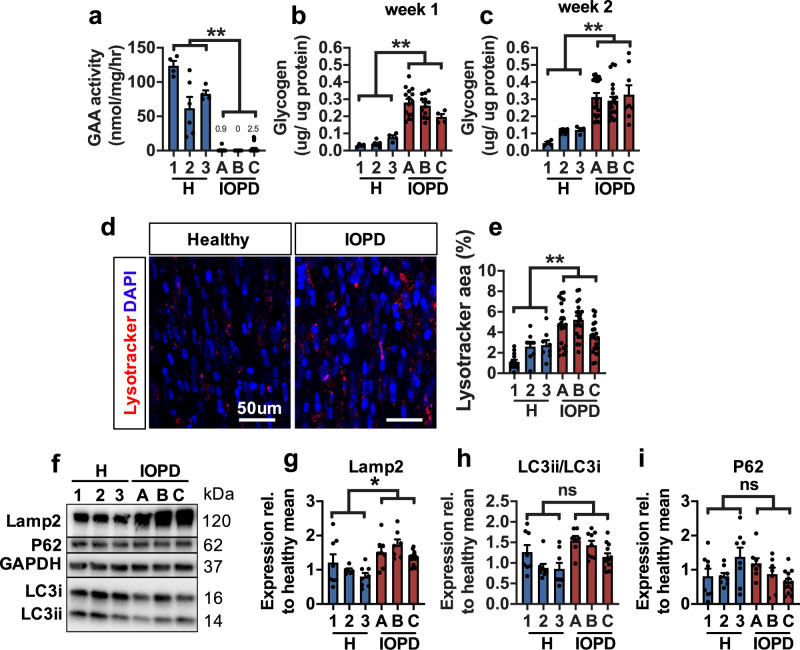
In Pompe disease, impaired lysosomal GAA activity results in the buildup of glycogen in skeletal muscles, which prompted us to examine GAA activity and glycogen levels in healthy and IO Pompe myobundles (shown normalized by myobundle protein content). GAA activity in healthy myobundles was comparable to previously reported values from healthy human muscle biopsies29 and to other in vitro skeletal muscle models19,20 (Fig. 2a). Compared to these values, IOPD myobundles exhibited severely reduced GAA activity (Fig. 2a), consistent with human studies29. In addition, IOPD myobundles exhibited 4.9- and 3.1-fold greater glycogen content than healthy myobundles at week 1 and 2 of 3D differentiation, respectively (Fig. 2b, c). These results were unchanged when normalization was performed by cellular (approximated by GAPDH) instead of total myobundle protein (Supplementary Fig. 2). We also fabricated functional 3D human myobundles using myoblasts from two patients with late-onset Pompe disease (LOPD). The LOPD myobundles reduced GAA activity, yet not as low as IOPD myobundles (Supplementary Fig. 3a) and, unlike IOPD myobundles, did not exhibit elevated glycogen content (Supplementary Fig. 3b, c). This lack of phenotype in LOPD myobundles may be due to the longer disease progression timeline in LOPD, and is consistent with the observation that ~20% of patients with LOPD do not develop elevated glycogen30,31.
Fig. 2. Biochemical and morphological characteristics of IOPD myobundles.
a GAA activity in 3 healthy (1, 2, 3) and 3 IOPD (A, B, C) myobundles (n = 4–21 myobundles per donor). b, c Glycogen content after b 1 week (n = 4–12 myobundles per donor) and c 2 weeks (n = 4–21 myobundles per donor) of 3D differentiation. d, e Representative images of healthy and IOPD myobundles stained live with lysotracker and DAPI at 2 weeks of 3D differentiation (d) and corresponding quantification of percentage of myobundle area positive for lysotracker (e) (n = 8–21 myobundles per donor). f–i Representative western blots (f) and quantified protein expression of g Lamp2, h LC3-II/LC3-I ratio, and i p62/SQSTM1 (normalized to GAPDH) (n = 5–13 myobundles per donor). Data: mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant.
Characterization of disease markers in IOPD myobundles
Glycogen buildup associated with IOPD causes lysosome swelling and dysfunction, resulting in impaired autophagy, autophagosome accumulation, and aggregation of proteins originally targeted for degradation32,33. To track lysosome abundance in myobundles, we stained 2-week-differentiated live myobundles with lysotracker, a pH-sensitive dye (Fig. 2d). IOPD myobundles showed 2.2-fold larger total area occupied by lysosomes compared to healthy myobundles (Fig. 2e) as well as augmented expression of the disease biomarker lysosome-associated membrane protein 2 (Lamp2, Fig. 2f, g), overall indicating increased lysosome abundance. Additionally, we assessed levels of microtubule-associated protein 2 light chain 3 (LC3-II), an autophagosome-specific protein that is converted from cytosolic LC3-I during autophagosome membrane formation and plays a key role in selecting cargo for degradation34. Impaired autophagy in Pompe disease results in an increased LC3-II/LC-I ratio and expression of p62/SQSTM135, a multifunctional stress-inducible scaffold protein. After 2 weeks of differentiation, IOPD myobundles did not exhibit increased LC3-II/LC3-I ratio (Fig. 2f, h) or p62 expression (Fig. 2f, i) relative to healthy myobundles, consistent with findings in cultured myotubes derived from GAA−/− mice36.
Differential sensitivity of IOPD myobundles to lysosomal and metabolic stress
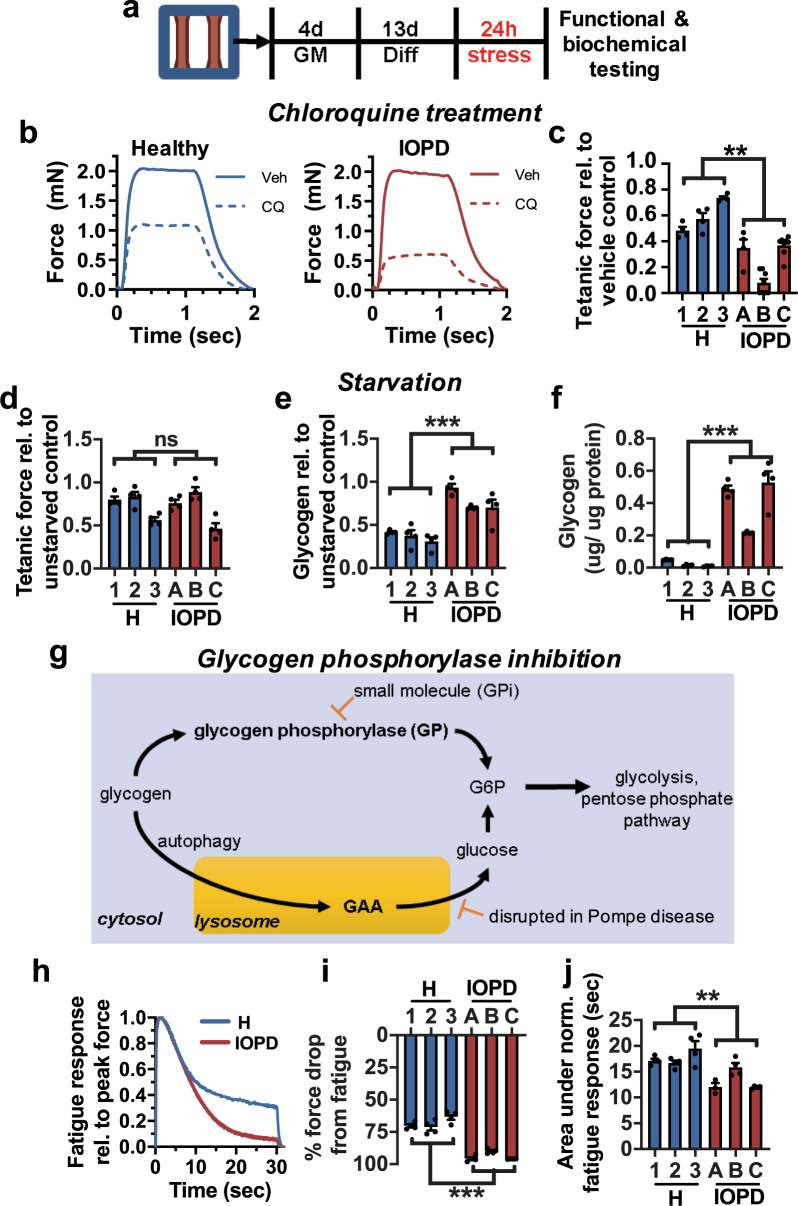
Considering that under standard conditions IOPD myobundles showed no contractile deficit, we tested if they would reveal altered functional responses to stress in the form of chloroquine treatment, glucose starvation, or glycogen phosphorylase inhibition applied for 24 h at day 13 of differentiation (Fig. 3a). Chloroquine inhibits autophagosome-lysosome fusion and disrupts lysosomal membrane integrity37,38. When applied at 5 µM, it reduced force generation in the IOPD group to a greater extent than in the healthy group (Fig. 3b, c), reflecting the increased functional sensitivity of IOPD myobundles to lysosomal stress39. Glucose starvation for 24 h was applied to metabolically stress the myobundles, testing their ability to access stored glycogen. Although IOPD and healthy myobundles showed similar resulting tetanic forces (Fig. 3d), IOPD myobundles exhibited significantly impaired glycogen breakdown and mobilization compared to healthy myobundles as indicated by greater total remaining glycogen (Fig. 3e) and reduced glycogen depletion (Fig. 3f). Furthermore, in healthy native skeletal muscle, the cytoplasm rather than the lysosome is the primary site of glycogen storage40,41; thus, we hypothesized that the predominant reliance on the cytoplasmic glycogen degradation in myobundles may mask a pathological phenotype that depends on deficits in lysosomal glycogen degradation. We thus blocked cytoplasmic glycogenolysis for 24 h in healthy and IOPD myobundles by inhibiting the rate-limiting enzyme glycogen phosphorylase with the small molecule inhibitor (GPi) CP-91149 (Fig. 3g)42,43. While without the GPi, healthy and IOPD myobundles showed similar fatigue test responses (Supplementary Fig. 1e–g). After 10 μM GPi treatment, IOPD myobundles showed significantly reduced fatigue resistance compared to healthy myobundles (Fig. 3h–j), thus revealing another stress-induced functional deficit brought about by the underlying differences in overall glycogen metabolism.
Fig. 3. Response of myobundles to metabolic stress.
a Schematic of experimental design: myobundles were cultured in growth media (GM) for 4 days and then differentiated for 13 days prior to 24 h of stress. b Representative force traces following 24 h of treatment with vehicle (Veh) or 5 µM chloroquine (CQ) in 3 healthy (1, 2, 3) and 3 IOPD (A, B, C). c Tetanic force after 24 h of treatment with 5 µM chloroquine shown relative to vehicle control (n = 4–8 myobundles per donor). d Tetanic force in myobundles after 24 h of glucose starvation relative to unstarved control (n = 4 myobundles per donor). e Glycogen content remaining after 24 h of glucose starvation (n = 4 myobundles per donor). f Fraction of glycogen remaining after 24 h of glucose starvation relative to unstarved control (n = 4 myobundles per donor). g Schematic of glycogen metabolism in skeletal muscle through lysosomal GAA and rate-limiting cytosolic glycogen phosphorylase (GP) pathways. h Representative force traces during fatigue test (20 Hz stimulation for 30 s) normalized to value of peak force, shown after 24 h of treatment with the GP inhibitor CP-91149 (10 µM). i, j Percent force decline at the end of fatigue stimulation (i, n = 4 myobundles per donor), and area under normalized force curve during fatigue stimulation (j, n = 4 myobundles per donor) after 24 h of treatment with the GP inhibitor. Data: mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant.
Recombinant GAA treatment of IOPD myobundles
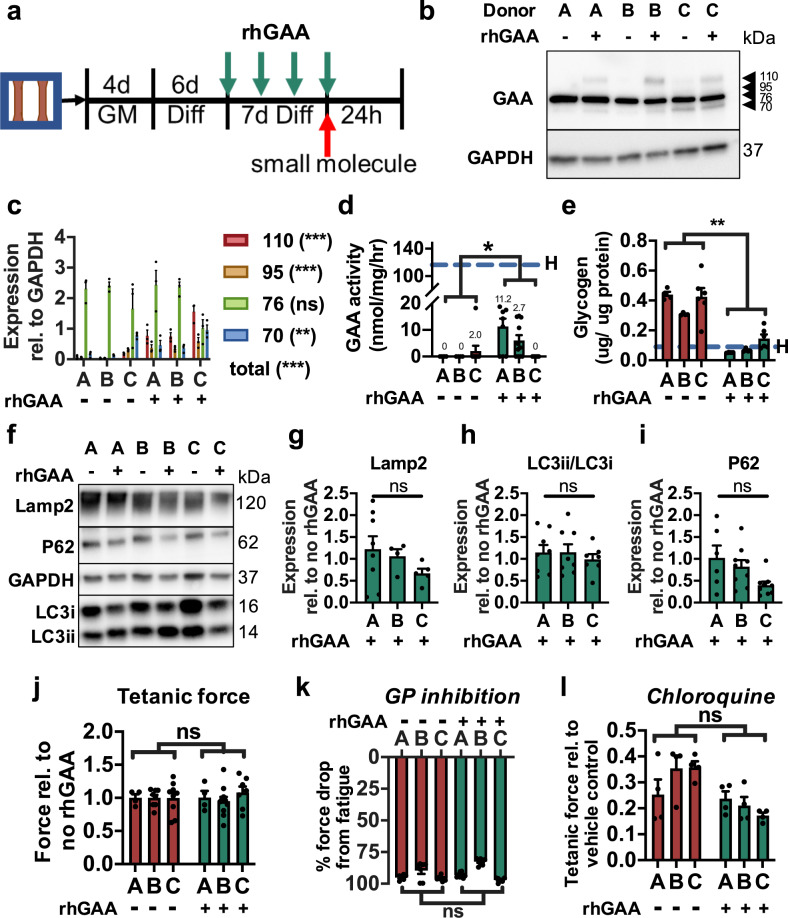
The current standard of care for Pompe disease is ERT, wherein rhGAA enzyme is delivered biweekly to patients to reduce glycogen accumulation44,45. Successful delivery relies on endocytosis of the 110-kDa rhGAA enyzme and its transport to the lysosomes, during which several proteolytic processing steps cleave rhGAA to its mature and most catalytically active 76-kDa and 70-kDa isoforms46,47. To study the response of IOPD myobundles to ERT, we provided rhGAA at 2 µg/mL in fresh culture media every two days between differentiation days 7–14 (Fig. 4a). In examining the distribution of GAA isoforms by western blot, we found that rhGAA treatment resulted in successful uptake of the enzyme as indicated by greater levels of the 110-, 95-, and 70-kDa GAA isoforms but no significant increase in the 76-kDa isoform (Fig. 4b, c). This lack of increase in the 76-kDa band may be due to the already high expression of endogenous (dysfunctional) 76-kDa GAA in untreated myobundles, with a possibility that a fraction of the band was still contributed by rhGAA-derived (functional) 76-kDa isoform. Notably, rhGAA treatment resulted in a disproportionate increase in the immature 110-kDa isoform (11.7-fold versus no treatment) relative to the increase in the mature 70-kDa isoform (1.7-fold), possibly due to impaired trafficking of the enzyme observed in more advanced stages of the disease48,49. In comparison, rhGAA treatment of healthy myobundles resulted in neither disproportionate nor individual increases in any of the GAA isoforms (Supplementary Fig. 4a, b), suggesting that unknown feedback mechanisms may regulate homeostatic levels of GAA protein. Importantly, rhGAA treatment of IOPD myobundles significantly increased GAA enzyme activity, albeit below healthy levels (Fig. 4d), and reduced glycogen content by 77%, near levels measured in healthy myobundles (Fig. 4e). This 1-week rhGAA treatment was insufficient to significantly reduce Lamp2 expression (Fig. 4f, g), the LC3-II/LC3-I ratio (Fig. 4f, h), or p62 expression (Fig. 4f, i). rhGAA treatment also did not improve tetanic force generation (Fig. 4j) under standard culture conditions, attenuate GPi-induced decline in fatigue (Fig. 4k), or reduce chloroquine-induced loss of tetanic force (Fig. 4l).
Fig. 4. Response of IOPD myobundles to rhGAA treatment.
a Schematics depicting protocol for rhGAA treatment of IO myobundles. b, c, Representative western blot of GAA isoforms in rhGAA treated (+) and untreated (−) myobundles from 3 IOPD patients (A, B, C) (b) and quantification of isoform expression relative to GAPDH (c, n = 3 myobundles per group). d, e GAA activity (d, n = 4–11 myobundles per group) and glycogen content (e, n = 4–6 myobundles per group) in IO myobundles after 1 week of rhGAA or no treatment. Dashed H-line denotes mean value across all healthy donors. f–i Representative western blot (f) and quantification of g Lamp2 expression, h LC3-II/LC3-I expression ratio, and i p62 expression (n = 4–9 myobundles per group) in IO myobundles after 1 week of rhGAA or no treatment. j Tetanic force of IO myobundles after 1 week of rhGAA treatment shown relative to no treatment (n = 4–11 myobundles per group). k Percent force decline at the end of fatigue stimulation after 24 h of GPi exposure following 1-week rhGAA treatment or no treatment (n = 6–8 myobundles per group). l Tetanic force after 24 h of chloroquine exposure following 1-week rhGAA treatment or no treatment, shown relative to vehicle control (n = 4 myobundles per group). Data: mean ± SEM. *p < 0.05; *p < 0.01; ns, not significant between rhGAA-treated and untreated myobundles.
Effect of AAV-mediated GAA gene therapy on IOPD myobundles
Gene therapy is a promising single-treatment approach to restore robust GAA expression to diseased tissue50. Overexpression of GAA via intramuscular injection of recombinant (r) AAV vectors increases tissue GAA activity and reduces muscle glycogen in GAA−/− mice51. To explore the effect of AAV gene therapy in our in vitro model, healthy and IOPD myobundles were transduced during myobundle fabrication with rAAV9 vectors encoding either hGAA51 or eGFP (control) driven by the muscle-specific regulatory cassette MHCK752. After 2 weeks of 3D differentiation, successful myobundle transduction was confirmed by observing strong eGFP fluorescence in immunostained muscle fibers (Supplementary Fig. 5a). The rAAV9-MHCK7hGAA transduced IOPD myobundles showed both robust increase in GAA activity (Supplementary Fig. 5b) and a ~75% decrease in glycogen content (Supplementary Fig. 5c), reaching levels measured in healthy myobundles. Nevertheless, consistent with the rhGAA supplementation, vector treatment did not improve tetanic force production under standard conditions (Supplementary Fig. 5d) or reduce muscle fatigue and force loss in GPi-induced (Supplementary Fig. 5e) and chloroquine-induced (Supplementary Fig. 5f) stress conditions, respectively.
Transcriptomic analysis of GAA−/− (KO) mice and identification of a Pompe disease signature
To better understand the molecular signature underlying the Pompe disease phenotype and response to therapy, RNA sequencing (RNA-Seq) analysis was performed on the quadriceps of 6-month-old wild-type (WT) and GAA−/− (KO) mice. As expected, KO mice showed lower GAA activity and greater glycogen accumulation than WT mice (Supplementary Fig. 6a, b). Transcriptomic analysis revealed 2244 genes that were significantly differentially expressed (|log2(fold-change)| ≥ 1, padj < 0.05) between WT and KO mice. Gene set enrichment analysis (GSEA) using the gene ontology (GO) annotation to investigate the significant functional categories describing coordinated gene expression revealed both expected and novel descriptors of Pompe disease (q < 10−3), including reduced protein translation (GO:0006414, GO:0006415), impaired mitochondrial function and oxidative metabolism (GO: 0006119), reduced ATP biosynthesis (GO:0006754), and increased cAMP signaling (GO:0046058) (Supplementary Fig. 6c), some of which are consistent with previous studies in GAA−/− mice4,39,53,54. From the RNA-Seq data set, we also generated a Pompe disease signature that consisted of the top 50% of altered genes with |log2FC| ≥ 2 and padj < 0.05, of which 243 were downregulated and 39 were upregulated in GAA−/− vs. WT mice (Supplementary Data 1a, b). Using the Reactome database (https://reactome.org/), constituent genes in each set were confirmed to span multiple categories consistent with Pompe disease phenotype, including muscle contraction, autophagy, amino acid and lipid metabolism, extracellular matrix organization, and protein metabolism4,5,32,55,56.
Effects of AAV-GAA gene therapy on GAA−/− mouse transcriptomics and Pompe disease signature
To understand the transcriptomic consequences of hGAA gene therapy in GAA KO mice, we applied liver depot gene therapy with an rAAV8 vector to induce liver-restricted expression and continuous secretion of hGAA into the blood and produce immune tolerance57,58, leading to increased muscle GAA activity and reduced glycogen content (Supplementary Fig. 6a, b). GSEA of AAV-treated and untreated KO mouse muscles showed that vector treatment caused significant (FDR < 0.15) improvement in oxidative phosphorylation (GO:0006119, q = 1.3 × 10−2; GO:0022900, q = 1.6 × 10−2), skeletal muscle contraction (GO:0003009, q = 1.5 × 10−2; GO:0036379, q = 1.5 × 10−2), and cholinergic muscle stimulation (GO:0007271, q < 1 × 10−3; GO:0042165, q = 2.4 × 10−2), and significantly (FDR < 0.15) reduced Golgi vesicle-mediated transport (GO:0006891, q = 5.3 × 10−3; GO:0006896, q = 2.4 × 10−2) and ubiquitin-related catabolism (GO:0071947, q = 6.5 × 10−2; GO:0031461, q = 2.5 × 10−2) (Supplementary Fig. 6d). These results suggested AAV therapy-induced reversal of multiple pathways affected by Pompe disease5,32,53,59. Specifically, of the 2244 genes that were significantly differentially expressed between KO and WT mice, 1006 (44.8%) were significantly reversed due to AAV treatment (Supplementary Fig. 6e). Transcriptome-wide analysis of the two comparisons (1: KO versus WT and 2: AAV-treated KO versus untreated KO) showed a negative correlation (Spearman’s correlation coefficient, −0.76; p < 2.2 × 10−16) (Supplementary Fig. 6f, g), further suggesting a reversal in the pathological transcriptional phenotype in GAA−/− mice following AAV treatment. Changes in six of the affected genes were further validated using qPCR (Supplementary Fig. 6h). GSEA of AAV-treated versus untreated KO mice also showed significant reversal in the disease signature (Supplementary Fig. 6i, j) described above. Specifically, the gene set encompassing the genes upregulated in untreated KO mice were downregulated in AAV-treated KO mice (q < 1 × 10−3) and vice versa (q < 1 × 10−3) (Supplementary Fig. 6i, j).
Transcriptomic comparison of healthy, IOPD, and rhGAA-treated IOPD myobundles
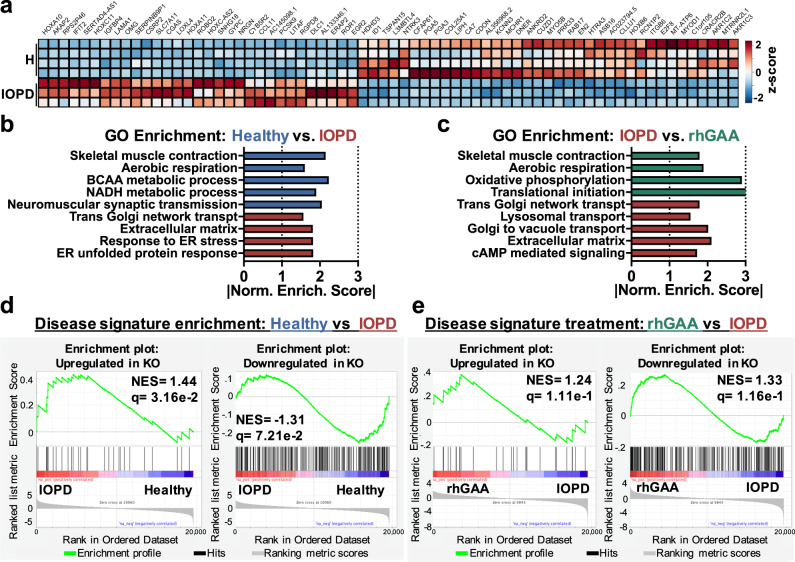
RNA sequencing analysis of myobundles was performed to further understand differences among healthy, IOPD, and rhGAA-treated IOPD myobundles and relate these findings to those obtained from GAA−/− mice. When comparing healthy and IOPD myobundles, 68 genes exhibited significantly different gene expression (|log2FC| ≥ 1, padj < .05) (Fig. 5a). Five of these genes were selected for validation using qPCR, and showed the expected relationship between healthy and IOPD groups (Supplementary Fig. 7). Compared to healthy myobundles, IOPD myobundles exhibited increased expression of genes associated with extracellular matrix (GO:0031012, q = 4.5 × 10−2) and reduced expression of genes associated with skeletal muscle contraction (GO:0003009, q = 4.4 × 10−3), including those related to sarcomeric A-band (GO:0031672, q = 4.2 × × 10−2), I-band (GO:0031674, q = 3.8 × 10−3), and actin-mediated contraction (GO:0070252, q = 5.4 × 10−3) (Fig. 5b). In addition to association with contractile dysfunction, IOPD myobundles were associated with endoplasmic reticulum stress (GO:0034976, q = 4.3 × 10−2) and unfolded protein response (GO:1900101, q = 4.3 × 10−2), characteristic of misfolded protein accumulation (Fig. 5b)60. Interestingly, neuromuscular synaptic transmission (GO:0007274, q = 7.3 × 10−3) was also reduced in IOPD myobundles (Fig. 5b) and the constituent genes involved in the core enrichment of significant neuromuscular gene sets were confirmed to be highly represented in human skeletal muscle according to the Genotype-Tissue Expression (GTEx) database. This finding is consistent with the involvement of neuromuscular deficit in Pompe disease patients and mice61,62. Moreover, several metabolic pathways were also downregulated in IOPD myobundles, including aerobic (GO:0009060, q = 1.2 × 10−1), NADH (GO:0006734, q = 2.6 × 10−2), branched-chain amino acid (GO:0009081, q = 7.5 × 10−4), and pyruvate (GO:0006090, q = 4.9 × 10−2) metabolism. These findings are consistent with previous associations of IOPD with dysregulated mTOR signaling and reduced protein synthesis4,5,63, dysfunctional mitochondria and oxidative metabolism39,53, and muscle weakness. When collecting the GO terms identified in the healthy versus IOPD GSEA, examining individually significant genes (padj < 0.05) within these terms, and tracking their response to rhGAA treatment, significant reversal at the individual gene level was not observed (Supplementary Fig. 8). However, at the gene set level, comparing IOPD versus rhGAA-treated IOPD revealed that rhGAA treatment reversed the extracellular matrix term (GO:0031012, q = 6.5 × 10−4) while also being significantly associated with increased expression of genes involved in protein translation (GO:0006413, q < 1 × 10−3; GO:0006414, q < 1 × 10−3) and protein synthesis (GO:0002181, q < 1 × 10−3), endoplasmic reticulum function (GO:0072599, q < 1 × 10−3), oxidative metabolism (GO:0006119, q < 1 × 10−3) and aerobic respiration (GO:0009060, q = 6.8 × 10−3), and skeletal muscle contraction (GO:0003009, q = 1.7 × 10−2) (Fig. 5c).
Fig. 5. Transcriptomic changes in myobundles as a result of IOPD and rhGAA treatment.
a, RNA-seq heatmap of genes with significantly different (padj < .05, |log2FC| ≥ 1) expression in IOPD vs. healthy myobundles. b GSEA on RNA-seq data to identify GO terms enriched in healthy (blue) or IOPD (red) myobundles plotted against normalized enrichment score (NES) (FDR < 0.15). c GSEA on RNA-seq data to identify GO terms enriched in rhGAA-treated (green) or untreated (red) IOPD myobundles (FDR < 0.15). d Enrichment plots of myobundles (IOPD vs. healthy) using disease signature gene sets consisting of the top 50% of the most significantly altered genes (padj < 0.05, |log2FC|≥2 in KO vs. WT RNA-seq), 39 of which were upregulated (left) and 243 downregulated (right) in KO vs. WT mice. e Enrichment plots of Pompe disease reversal in rhGAA-treated vs. untreated IOPD myobundles.
Disease signature enrichment in IOPD myobundles and attenuation with rhGAA therapy
To further determine whether myobundles recapitulated the disease phenotype resulting from complete loss of GAA in skeletal muscle, IOPD and healthy myobundles were tested for enrichment of the disease-specific transcriptional signature identified in mice (Supplementary Data 1). Interestingly, both up- and downregulated gene sets were significantly represented in IO Pompe myobundles (q = 3.16 × 10−2, q = 7.21 × 10−2) (Fig. 5d). Furthermore, we tested the ability of rhGAA treatment to reverse the mouse transcriptomic signature of disease in IOPD myobundles and found significantly reversed expression of the gene set downregulated in GAA−/− mice (q = 1.16 × 10−1) but not upregulated in GAA−/− mice (Fig. 5e). Consistent with the clinical studies8,44, these results suggest that rhGAA therapy yields only a partial rescue of IOPD phenotype in human skeletal muscle, signifying the need for improved treatment modalities.
Discussion
In this study, we described the first in vitro three-dimensional human skeletal muscle model of infantile-onset Pompe disease, determined an IOPD disease signature in GAA−/− vs. WT mice, and applied the developed IOPD myobundle model to study in vitro responses of human skeletal muscle to recombinant protein and AAV-based hGAA treatments. The IOPD myobundles exhibited low GAA activity and elevated glycogen content, consistent with previous studies in 2D cell culture19,20,64, GAA−/− mice in vivo45,65, and clinical data9,44. Compared to 2D culture systems, 3D tissue-engineered skeletal muscle permits extended culture time, formation of more mature myofibers, and assessment of muscle function21. The ability to measure contractile force, in particular, allowed us to identify metabolic stress conditions that revealed distinct functional responses (tetanic force production, fatigue resistance) between healthy and IOPD myobundles. The reproducibility and robustness of these results were ensured using muscle cells from multiple patients and healthy donors. We further validated this human in vitro model by performing transcriptomic analysis which showed that IOPD myobundles exhibited reduced expression of genes involved in skeletal muscle contraction and increased expression of genes involved in endoplasmic reticulum stress, consistent with the transcriptomic changes found in GAA−/− vs. wild-type mice. In vitro rhGAA therapy in myobundles partially reversed the transcriptomic disease signature identified in mice without correcting stress-revealed functional deficit. Together, these results establish a new in vitro human skeletal muscle model to improve understanding and treatment of Pompe disease.
In IOPD, GAA deficiency results in glycogen buildup in muscle fibers, followed by lysosome enlargement and rupture44. In our system, after 2 weeks of 3D differentiation, GAA-deficient IOPD myobundles showed 3-fold greater glycogen content relative to healthy myobundles (reaching levels measured in Pompe muscle biopsies66), along with significantly increased lysosomal area and Lamp2 expression, indicative of lysosome enlargement (Fig. 2d–g). A secondary consequence of lysosome dysfunction is impaired autophagy, which leads to autophagosome buildup, accumulation of ubiquitinated proteins6, and disruption of muscle architecture67. Based on assessment of the LC3-II/LC3-I ratio, autophagosome accumulation was not observed in IOPD myobundles, consistent with findings in cultured IOPD skeletal muscle cells64 and cardiomyocytes59. While the predominance of autophagosome buildup was found in isolated muscle fibers from LO rather than IOPD patients68, a separate study showed increased LC3-II and p62 expression in IOPD vs. healthy muscle69, suggestive of impaired autophagy and protein aggregation. Our GSEA analysis revealed that IOPD myobundles exhibited pathological upregulation of gene sets involved in endoplasmic reticulum stress, previously shown to induce autophagy in LOPD patient fibroblasts60. Moreover, 24 h treatment with chloroquine (which blocks autophagosome-lysosome fusion and further disrupts lysosome function37) revealed a greater vulnerability to functional loss in IOPD than healthy myobundles (Fig. 3a–c). Furthermore, given that IOPD patients require a median time of 4 months to develop muscle weakness70, these results suggest that IOPD myobundles may require a longer culture time and application of various metabolic challenges4,71,72 to accelerate the acquisition of an advanced pathological phenotype with significant autophagic buildup.
Given that extensive autophagosomes can directly impede myofiber contractions73,74, the lack of autophagic buildup in IOPD myobundles likely explains the lack of contractile deficit observed under baseline culture conditions (Fig. 1n), which could be further attributed to the immaturity of myobundles relative to native muscle, evident from their expression levels of myosin heavy chain isoforms and calcium-handling genes24. Nevertheless, under baseline conditions IOPD myobundles exhibited downregulation of gene sets involved in skeletal muscle contraction and sarcoplasmic reticulum function1,39 (Fig. 5b), similar to a microarray study of muscle biopsies from IO Pompe and healthy donors75. Interestingly, examining the core enrichment for the muscle contraction gene set (GO:0006941) in healthy versus IOPD myobundles revealed differentially expressed genes related to muscle excitability (SCN5A, SCN3B, KCNE2), neuronal excitation (CHRNA1, CHRNB1, CHRND), and Na+/K+ ATPase (ATP1A1, ATP1A2). These findings suggest that replicating at least some aspects of advanced Pompe disease in vitro may require functional innervation76, as achieved in co-cultures of engineered muscle and motoneurons77.
Current enzyme replacement therapy (ERT) for Pompe disease consists of intravenous delivery of rhGAA (Myozyme®/Lumizyme®; alglucosidase alfa), which increases muscle GAA activity, reduces glycogen accumulation, and improves muscle function and patient survival9,44,78. Consistent with clinical results and previous in vitro studies64,79, 7-day rhGAA treatment yielded successful enzyme uptake (Fig. 4b, c) and strong reduction of glycogen content in IO Pompe myobundles (Fig. 4e), but unlike long-term clinical ERT80, it only modestly increased GAA activity (Fig. 4d), primarily via improved abundance of the mature 70-kDa GAA isoform (Fig. 4b, c). Interestingly, rhGAA treatment of healthy myobundles did not increase expression of any GAA isoforms (Supplementary Fig. 4a, b), which indicates the potential existence of a negative feedback regulating GAA protein level. The incomplete therapeutic effects induced by modestly increased GAA activity were also evident from the lack in reversal of lysosomal accumulation or increased vulnerability to GP inhibition and chloroquine (Fig. 4k, l). While rhGAA treatment was unable to significantly correct the expression of individual genes identified within the enriched GO terms of IOPD versus healthy myobundles (Supplementary Fig. 8), gene set analysis showed rhGAA treatment significantly upregulated gene sets known to be downregulated in disease (related to oxidative metabolism39,53, muscle contraction, and protein synthesis4 pathways) (Fig. 5c). Studies in murine GAA KO myotubes in vitro and GAA−/− mice in vivo suggested that lysosome size reduction occurs in a dose-dependent manner and requires high rhGAA concentrations81, while several weeks of ERT may be needed to fully rescue dysregulated glycogen metabolism82. Accordingly, re-normalization of GAA activity, lysosome abundance, and sensitivity to GP inhibition and chloroquine in IOPD myobundles may require increased duration and dose of rhGAA treatment or development of novel means to improve rhGAA affinity for83 or expression of45 mannose-6-phosphate receptor, which mediates muscle uptake of extracellular GAA.
Interestingly, rAAV9-MHCK7hGAA transduction of IOPD myobundles produced levels of GAA activity comparable to those of healthy myobundles (Supplementary Fig. 5b), but despite glycogen clearance (Supplementary Fig. 5c), it did not improve contractile response to GP inhibition or chloroquine (Supplementary Fig. 5e, f). While complete functional recovery of muscle may be achieved with longer AAV vector treatment, it may also require the effective rescue of GAA activity in non-muscle cells which in myobundles were not transduced using the MHCK7 promoter. For example, fibroblasts in Pompe disease are also affected by GAA deficiency, showing glycogen accumulation, increased sensitivity to endoplasmic reticulum stress-induced autophagy, and impaired extracellular uptake of rhGAA60,84 and may contribute to muscle pathology via paracrine or juxtacrine effects. Alternatively, although unlikely, the three IOPD donors used in this study may not represent the typical functional response to rhGAA or rAAV9 vector therapy, given the range of clinical outcomes observed in response to ERT8,44.
Our RNA-Seq analyses of GAA−/− and WT mouse quadriceps provided further mechanistic insights in Pompe disease, while allowing us to define a disease-specific transcriptomic signature for additional validation of the human model. Consistent with previous studies39,53, GAA−/− quadriceps showed significant upregulation of L-type calcium channel subunits (e.g., Cacng7 (Supplementary Fig. 6h), Cacna1c, and Cacna2d4) and downregulation of gene sets involved in oxidative metabolism and mitochondrial function (Supplementary Fig. 6c). Furthermore, GAA−/− muscle exhibited downregulation of protein synthesis gene sets (Supplementary Fig. 6c), consistent with the muscle atrophy and protein loss observed in GAA−/− mice and Pompe disease patients where post-translational modifications may serve to further regulate cellular protein metabolism85. Changes in other less-investigated pathways suggested additional mechanisms underlying Pompe disease, including negative regulation of Notch (involved in GAA expression86), increased cAMP (involved in glycogen metabolism87), and increased PKC signaling. A Pompe disease signature composed of the top significantly altered genes (upregulated and downregulated; Supplementary Data 1) was largely reversed by liver-specific AAV-hGAA treatment (Supplementary Fig. 6e–j), which increased enrichment of oxidative phosphorylation, synaptic transmission, and muscle contraction gene sets (Supplementary Fig. 6d). Importantly, human IOPD myobundles also exhibited significant enrichment for altered gene sets present in the mouse disease signature (Fig. 4d) and consistent with histological and functional results, showed a partial reversal of the signature gene sets after 7-day rhGAA treatment (Fig. 4e). Along with studies in clenbuterol-treated LOPD patients88, our results confirmed the utility of RNA-Seq analysis to understand treatment effects in Pompe disease.
In summary, we introduce an in vitro 3D human skeletal muscle model of infantile-onset Pompe disease that displays: (1) morphological and biochemical features observed in patients’ biopsies, (2) contractile deficits revealed under metabolic stress, (3) transcriptomic signature consistent with that of a mouse model, and (4) partial phenotype rescue with in vitro recombinant protein or AAV-mediated treatments. We anticipate that with further development, human IOPD myobundles will hold the potential to serve as a novel pre-clinical platform for mechanistic studies and therapeutic testing in Pompe disease.
Methods
Human myoblast isolation and expansion
Ten human skeletal muscle samples were obtained from standard needle biopsies or surgical discards using protocols approved by the Duke University Institutional Review Board. Of the healthy muscle donors 1–5 (12/F, 16/M, 15/M, F/18, F/14 (age/sex), respectively), the largest biopsies and cell yields were obtained for donors 1–3 and were consequently used in all performed assays. Muscle samples from healthy donors 4 and 5 were only used in select assays (GAA activity, glycogen content, RNA sequencing). The IOPD samples (A, B, C) came respectively from 1 male (6 months old, CRIM-negative), and 2 female (7 years old, CRIM-negative; 13 years old, CRIM-positive) patients. The LOPD samples (a, b) came from 1 male (46 years old; c.-32-13T>G, c.2238G>A) and 1 female (30 years old; c.-32-13T>G, c.2481+102_2646+31) patient. Human skeletal muscle cells were isolated as described previously22,23. Briefly, muscle samples were washed in PBS, minced, and enzymatically digested in 0.05% trypsin for 30 min. After FBS neutralization of the digestion buffer, muscle pieces were centrifuged, resuspended in growth media (GM; low-glucose DMEM, 10% FBS, dexamethasone, EGF, and 1× antibiotic-antimycotic), and pre-plated for 2 h to reduce fibroblast contamination. Unattached muscle fragments were then gathered and replated on Matrigel (BD Biosciences)-coated flasks for outgrowth and cell expansion (passage 0) in GM. Cells were frozen at p2, p3, and p4 in 90% FBS and 10% DMSO and stored in liquid nitrogen. After the fifth passage, cells were incorporated into a fibrin-based gel for myobundle construction.
Fabrication and culture of tissue-engineered human myobundles
Myobundles were formed within polydimethylsiloxane (PDMS) molds containing two semi-cylindrical wells (7-mm long, 2-mm diameter), cast from 3D-machined Teflon masters23. PDMS molds were coated with 0.2% (w/v) pluronic F-127 (Invitrogen) for 1 h at room temperature to prevent hydrogel adhesion. Laser-cut Cerex frames positioned around the two wells were used to anchor the ends of the myobundles and facilitate handling. A cell solution (7.5 × 105 cells in 17.2 μL media per bundle + 2 μL of 50 U/mL thrombin in 0.1% BSA in PBS) and a gelling solution (3 μL media + 10 μL Matrigel + 10 μL of 15 mg/mL fibrinogen in PBS) were prepared in separate vials on ice for up to eight myobundles per vial. Gelling solution was added to the cell solution, mixed thoroughly, and injected into the PDMS wells to polymerize at 37 °C for 30 min. Resulting myobundles were cultured dynamically on a rocker (0.5 Hz) in GM supplemented with 1.5 mg/mL 6-aminocaproic acid (ACA, Sigma) for 4 days. The media was then switched to differentiation media (DM) consisting of a custom low-glucose, low-amino acid media89 to mimic physiological amino acid concentrations, with 1× N2 serum supplement (Thermo Fisher), and 2 mg/mL ACA. The media was exchanged every other day.
Metabolic stress induction in myobundles
Both healthy and IOPD myobundles were exposed to 24 h small molecule treatment between differentiation days 13 and 14. Glycogen phosphorylase was inhibited using 10 µM CP-91,149 (Cayman) and lysosomal stress was induced using 5 µM chloroquine (Sigma, C6628). For 24 h glucose starvation, myobundles were cultured in low-amino acid media with N-2 supplement (as described above) without glucose. All biochemical and functional measurements were performed at the end of the 24 h stress period.
Recombinant GAA treatment of myobundles
Recombinant human acid alpha glucosidase (rhGAA, alglucosidase alfa, Myozyme®/Lumizyme®) was obtained from Sanofi Genzyme and added at 2 µg/mL between differentiation weeks 1 and 2 during every-other-day media change. Untreated control received the application of media only.
AAV vector preparation and treatment of myobundles
AAV vectors were prepared as described previously and purified by centrifugation with a cesium chloride gradient65. Briefly, HEK293 cells were transfected with an AAV vector plasmid, the packaging plasmid90, and pAdHelper (Stratagene, La Jolla, CA). After 48 h, cells were harvested and freeze-thawed three times. AAV vectors were isolated by sucrose cushion pelleting, followed by two cesium chloride gradient centrifugation steps. AAV stocks were dialyzed against three changes of phosphate-buffered saline with 5% sorbitol added to the third dialysis, and aliquots were stored at −80 °C until use. The viral vectors were handled under the guideline of Biohazard Safety Level 2 which is published by the NIH. For myobundle experiments, IOPD myoblasts were mixed with either rAAV9-MHCK7eGFP (control) or rAAV9-MHCK7hGAA at 2.5 × 103 vector genomes/cell during myobundle fabrication.
Isometric force measurements in myobundles
Contractile properties of myobundles were measured using a custom, temperature-controlled force measurement setup described previously22,23. Briefly, single myobundles were anchored on one end to an immobilized PDMS block and on the opposite end to a suspended PDMS float connected to a force transducer. Tissues were stimulated using 40 V/cm, 5 ms electrical pulses via parallel platinum electrodes to generate twitch (single pulse) and tetanic (20 Hz for 1 s) contractions. A computer-controlled motorized linear actuator (Thor labs) connected to the PDMS float was used to stretch myobundles to a length (1.12 × culture length) that yielded the maximum generated force. Recorded force traces were analyzed for peak twitch or tetanus amplitude, time to peak twitch, and half-relaxation time using a custom MATLAB program22,23.
GAA activity assay
Individual myobundles were resuspended in 180 µL of water plus protease inhibitor (Sigma) and were lysed using the Bullet Blender (Next Advance). Protein concentration was measured using a BCA protein assay. To measure GAA activity, 15 µL of sample, 2.14 µL of 70 mM 4-MU-a-D (Sigma M9766), and 82.86 µL of citrate-phosphate buffer (pH = 3.7) were incubated in a 96-well plate at 37 °C for 1 h. Stop buffer (200 µL of 0.5 M EDTA buffer pH = 11.8) was then added to halt the reaction, and fluorescence at 450 nm was measured using a plate reader. 4MU (Sigma M1381)-containing wells with concentrations from 100 nM to 20 µM 4MU were run in parallel to generate a standard curve for calculating product generation during the reaction. 100 µL of each standard was added prior to incubation, and 200 µL stop buffer was added to each well after incubation, diluting the concentration of the standard 1:3. As a baseline for zero activity, a 15 µL water sample was incubated with 2.14 µL of 70 mM 4MU-a-D and 82.86 µL of citrate-phosphate buffer.
Glycogen content assay
Samples were isolated as described for the GAA activity assay, diluted 1:10 in sodium acetate buffer (0.1 M, pH 5.5), and heated at 100 °C for 3 min to inactivate endogenous amylases. Each sample was distributed into two wells with 50 µL sample in each. One µL of 120 U/mL amyloglucosidase (Sigma Aldrich, cat. 10113) was added to one well for 45 min. Both wells then received a solution containing 1 µL of 100 U/mL glucose oxidase (Sigma Aldrich, cat. G6125), 1 µL of 10 U/mL horseradish peroxidase (Sigma Aldrich cat. 516531), 0.5 µL of 10 mM ADHP (Setareh Biotech, cat. 6773), and 47.5 µL of PBS for 30 min. Glycogen standards (Cayman Chemical, cat. 700481) were generated and treated in parallel to produce standard curves for each assay. Absorbance at 560 nm or fluorescence at 540/590 nm for low-concentration samples was measured as a readout.
Immunohistochemistry
Cell monolayers were fixed in 4% paraformaldehyde in PBS for 10 min and myobundles were fixed in 2% paraformaldehyde in PBS overnight at 4 °C91. Following fixation, samples were washed in PBS followed by blocking in 5% chick serum with 0.3% Triton-X 100. Primary and secondary antibodies were applied overnight at 4 °C. Images were acquired using the Zeiss 880 inverted confocal microscope. For live imaging of lysosomes, lysotracker was incubated at 1:10,000 lysotracker (Thermo Fisher) for 15 min and washed in PBS before imaging. For cross-sectional images, fixed tissues were cut in half, frozen in OCT blocks using liquid nitrogen, and cut at 10 µm increments. Primary and secondary antibody information can be found in Supplementary Table 1.
Western blotting
Western blotting was performed as previously described22,23. Briefly, protein was isolated in RIPA lysis and extraction buffer with protease inhibitor (Sigma) and phosphatase inhibitor cocktail (Roche), lysed using a Bullet Blender (Next Advance), and protein concentration was determined using a BCA assay (Thermo Fisher). Western blots were performed using Bio-Rad Mini-PROTEAN gels and transferred using a Pierce 2000 powerblot semi-dry transfer system (Thermo Fisher). Primary antibodies were applied overnight at 4 °C before application of HRP-conjugated anti-mouse (1:20,000) and HRP-conjugated anti-rabbit antibody (1:5000; SCBT). Chemiluminescence was performed using Clarity Western ECL substrate (Bio-Rad). Images were acquired using a Bio-Rad Chemidoc and analyzed using Image Studio. Original blots are included in Supplementary Fig. 9. Samples were first normalized to GAPDH expression. In healthy versus IOPD comparisons, samples were normalized a second time such that the mean of healthy means was 1. In rhGAA-treated versus untreated IOPD comparisons, the rhGAA group was normalized to its respective IOPD donor control.
Flow cytometry
Muscle cells expanded for five passages were fixed in 4% paraformaldehyde for 10 min and stained with CD56 PE-Cy7 (1:100, BD Biosciences) and CD29 APC (1:100, BD Biosciences) in 3% FBS for 30 min on ice followed by three PBS wash steps. Samples were run on a BD FACSCanto II and analyzed in FlowJo. Cells were gated based on size and granularity to exclude debris, followed doublet removal, before recording CD56 and CD29 intensities. Additional antibody information can be found in Supplementary Table 1.
GAA-KO mice
All animal procedures were performed under the guidelines of the Duke University Institutional Animal Care and Use Committee. Two-month-old GAA-KO male mice with a C57BL/6 background were administrated the liver-specific hGAA expression vector AAV2/8-LSPhGAApA57 (2 × 1012 vg/kg) via tail vein injection. Control treatment group consisted of mice injected with PBS. After 4 months, the mice were euthanized and the quadriceps were harvested, along with the quadriceps of age-matched wild-type C57BL/6 mice. The collected quadriceps were split for RNA extraction and for assaying GAA activity and glycogen content58,65.
RNA extraction and quantitative RT-PCR
RNA was isolated from mouse tissue and human myobundles using the Bullet Blender (Next Advance) and the RNeasy Fibrous Tissue Mini Kit (Qiagen), following by RNA reverse-transcription using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR for muscle-related genes was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Primer information can be found in Supplementary Table 2.
RNA sequencing and gene set enrichment analysis (GSEA)
Two RNA-seq datasets (mouse and human) were generated from isolated mRNA. The samples were processed by the Duke Center for Genomic and Computational Biology in the same manner unless otherwise specified. RNA-seq data were processed using the fastp toolkit92 (human) or TrimGalore toolkit93 (mouse) to trim low-quality bases and Illumina sequencing adapters from the 3′ end of the reads. Only reads that were 20 nt or longer after trimming were kept for further analysis. Reads were mapped to the human (version GRCh38v93) or mouse (version GRCm38v73) genome and transcriptome94 using the STAR RNA-seq alignment tool95. Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the featureCounts tool96 (human) or HTSeq tool97 (mouse). Only genes that had at least 10 reads in any given library were used in subsequent analysis. Normalization and differential expression were carried out using the DESeq298 Bioconductor99 package within the R statistical programming environment. For the human Pompe myobundle rhGAA-treated versus untreated analysis, we included personID as a cofactor in the model. The false discovery rate was calculated using the Benjamini-Hochberg procedure to control the false-positive rate during multiple hypothesis testing. Gene set enrichment analysis100 was performed to identify gene ontology terms and pathways associated with altered gene expression for each of the comparisons performed with FDR cutoff 0.15. The Pompe disease signature was generated using the top 50% of differentially expressed genes with |log2FC| ≥ 2 and padj < 0.05 in the mouse WT versus GAA−/− comparison, and these genes were then divided based on up- or downregulation (Supplementary Data 1a, b). To test the enrichment of this signature in the human myobundle system, mouse genes were first mapped to human orthologs using the biomaRt101 package in R, which was followed by GSEA100. The human orthologs (HGNC symbols) are listed alongside the mouse disease signature in Supplementary Data 1a, b.
Statistics and reproducibility
Data are expressed as mean ± SEM. Unless noted otherwise, statistical comparisons were made using the mixed-effects model Yij = αi + β + εij, where Y is the response variable for the jth measurement of individual i, αi is the random effect for individual i, β is the fixed effect (i.e., IOPD vs. healthy or treated vs. untreated IOPD), and random error (εij). Significance for the fixed effects was calculated using a Wald test for p < 0.05 using the lme4 package in R.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank the Duke Center for Genomic and Computational Biology and Dr. Jianhong Ou for their contributions to RNA-seq processing and data analysis. rhGAA (Lumizyme®) was supplied by Sanofi Genzyme. This work was supported by NIH Grants AR055226, AR065873, and R01AR065873 from the National Institute of Arthritis and Musculoskeletal and Skin Disease, and grant UH3TR002142 from the NIH Common Fund for the Microphysiological Systems Initiative. The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Author contributions
J.W. and N.B. designed experiments and wrote the manuscript. J.W., C.J.Z., A.K., S.T., A.L.C., and L.M. optimized culture conditions and performed in vitro myobundle experiments. C.J.Z. and S.T. performed glycogen content and GAA activity, immunofluorescence staining, and confocal imaging on myobundles. S.H. performed mouse experiments, including mouse tissue glycogen content and GAA activity analysis. RNA-seq analysis was performed by the Duke Center for Genomic and Computational Biology. P.S.K., D.D.K., and N.B. supervised the efforts, including the manuscript preparation.
Code availability
Data supporting the results of the main figures are available at 10.6084/m9.figshare.14173013.v1102.The sequencing data generated for this study have been deposited in Gene Expression Omnibus under the SuperSeries GSE159064 and SubSeries GSE159062 (human myobundle model) and GSE159063 (GAA KO mouse model). Data referenced in the “Discussion” of this study are available on Gene Expression Omnibus GSE57980 and GSE38680. Any remaining information can be obtained from the corresponding author upon reasonable request.
Competing interests
P.S.K. has received research/grant support from Sanofi Genzyme, Valerion Therapeutics, and Amicus Therapeutics. P.S.K. has received consulting fees and honoraria from Sanofi Genzyme, Amicus Therapeutics, Vertex Pharmaceuticals, and Asklepios Biopharmaceutical, Inc. (AskBio). P.S.K. is a member of the Pompe and Gaucher Disease Registry Advisory Board for Sanofi Genzyme, Amicus Therapeutics, and Baebies. D.D.K. has received research/grant support from Sanofi Genzyme, Sangamo Therapeutics, Pharming, Viking Therapeutics, and Amicus Therapeutics. D.D.K. has received consulting fees and honoraria from Sanofi Genzyme, Amicus Therapeutics, Sangamo Therapeutics, Vertex Pharmaceuticals and Asklepios Biopharmaceutical, Inc. (AskBio). D.D.K. and P.S.K. have equity in AskBio, which is developing gene therapies for Pompe disease. D.D.K., P.S.K., and Duke University might benefit financially if the experimental treatments discussed here prove effective and are successful commercially. All other authors have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02059-4.
References
- 1.Ausems MG, et al. Frequency of glycogen storage disease type II in The Netherlands: implications for diagnosis and genetic counselling. Eur. J. Hum. Genet. 1999;7:713–716. doi: 10.1038/sj.ejhg.5200367. [DOI] [PubMed] [Google Scholar]
- 2.Mehler M, DiMauro S. Residual acid maltase activity in late-onset acid maltase deficiency. Neurology. 1977;27:178–184. doi: 10.1212/WNL.27.2.178. [DOI] [PubMed] [Google Scholar]
- 3.Kroos M, Hoogeveen-Westerveld M, van der Ploeg A, Reuser AJ. The genotype-phenotype correlation in Pompe disease. Am. J. Med. Genet. Part C., Semin. Med. Genet. 2012;160C:59–68. doi: 10.1002/ajmg.c.31318. [DOI] [PubMed] [Google Scholar]
- 4.Lim JA, et al. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol. Med. 2017;9:353–370. doi: 10.15252/emmm.201606547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shemesh A, et al. Suppression of mTORC1 activation in acid-alpha-glucosidase-deficient cells and mice is ameliorated by leucine supplementation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R1251–R1259. doi: 10.1152/ajpregu.00212.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raben N, Wong A, Ralston E, Myerowitz R. Autophagy and mitochondria in Pompe disease: nothing is so new as what has long been forgotten. Am. J. Med Genet C. Semin Med. Genet. 2012;160C:13–21. doi: 10.1002/ajmg.c.31317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nascimbeni AC, Fanin M, Tasca E, Angelini C, Sandri M. Impaired autophagy affects acid alpha-glucosidase processing and enzyme replacement therapy efficacy in late-onset glycogen storage disease type II. Neuropath. Appl. Neuro. 2015;41:672–675. doi: 10.1111/nan.12214. [DOI] [PubMed] [Google Scholar]
- 8.Kishnani PS, et al. Recombinant human acid [alpha]-glucosidase: major clinical benefits in infantile-onset Pompe disease. Neurology. 2007;68:99–109. doi: 10.1212/01.wnl.0000251268.41188.04. [DOI] [PubMed] [Google Scholar]
- 9.Kishnani PS, et al. Early treatment with alglucosidase alpha prolongs long-term survival of infants with Pompe disease. Pediatr. Res. 2009;66:329–335. doi: 10.1203/PDR.0b013e3181b24e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, N. et al. Clinical and molecular disease spectrum and outcomes in patients with infantile-onset Pompe disease. J Pediatr.216, 44–50.e5. (2019). [DOI] [PubMed]
- 11.Gungor D, et al. Survival and associated factors in 268 adults with Pompe disease prior to treatment with enzyme replacement therapy. Orphanet J. Rare Dis. 2011;6:34. doi: 10.1186/1750-1172-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien YH, Hwu WL. A review of treatment of Pompe disease in infants. Biologics. 2007;1:195–201. [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries JM, et al. Pompe disease in adulthood: effects of antibody formation on enzyme replacement therapy. Genet Med. 2017;19:90–97. doi: 10.1038/gim.2016.70. [DOI] [PubMed] [Google Scholar]
- 14.van der Ploeg AT, et al. A randomized study of alglucosidase alfa in late-onset Pompe’s disease. N. Engl. J. Med. 2010;362:1396–1406. doi: 10.1056/NEJMoa0909859. [DOI] [PubMed] [Google Scholar]
- 15.Raben N, et al. Targeted disruption of the acid alpha-glucosidase gene in mice causes an illness with critical features of both infantile and adult human glycogen storage disease type II. J. Biol. Chem. 1998;273:19086–19092. doi: 10.1074/jbc.273.30.19086. [DOI] [PubMed] [Google Scholar]
- 16.Raben N, Nagaraju K, Lee E, Plotz P. Modulation of disease severity in mice with targeted disruption of the acid alpha-glucosidase gene. Neuromuscul. Disord. 2000;10:283–291. doi: 10.1016/S0960-8966(99)00117-0. [DOI] [PubMed] [Google Scholar]
- 17.Peruzzo P, Pavan E, Dardis A. Molecular genetics of Pompe disease: a comprehensive overview. Ann. Transl. Med. 2019;7:278. doi: 10.21037/atm.2019.04.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Filippi P, et al. Genotype-phenotype correlation in Pompe disease, a step forward. Orphanet. J. Rare Dis. 2014;9:102. doi: 10.1186/s13023-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato Y, et al. TFEB overexpression promotes glycogen clearance of Pompe disease iPSC-derived skeletal muscle. Mol. Ther. Methods Clin. Dev. 2016;3:16054. doi: 10.1038/mtm.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Wal E, et al. Large-scale expansion of human iPSC-derived skeletal muscle cells for disease modeling and cell-based therapeutic strategies. Stem Cell Rep. 2018;10:1975–1990. doi: 10.1016/j.stemcr.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandenburgh HH, Karlisch P, Farr L. Maintenance of highly contractile tissue-cultured avian skeletal myotubes in collagen gel. Vitr. Cell Dev. Biol. 1988;24:166–174. doi: 10.1007/BF02623542. [DOI] [PubMed] [Google Scholar]
- 22.Khodabukus A, et al. Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials. 2019;198:259–269. doi: 10.1016/j.biomaterials.2018.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. eLife. 2015;4:e04885. doi: 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao L, Qian Y, Khodabukus A, Ribar T, Bursac N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 2018;9:126. doi: 10.1038/s41467-017-02636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illa I, Leon-Monzon M, Dalakas MC. Regenerating and denervated human muscle fibers and satellite cells express neural cell adhesion molecule recognized by monoclonal antibodies to natural killer cells. Ann. Neurol. 1992;31:46–52. doi: 10.1002/ana.410310109. [DOI] [PubMed] [Google Scholar]
- 26.Boldrin L, Morgan JE. Human satellite cells: identification on human muscle fibres. PLoS Curr. 2012;3:RRN1294. doi: 10.1371/currents.RRN1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaaf GJ, et al. Lack of robust satellite cell activation and muscle regeneration during the progression of Pompe disease. Acta Neuropathol. Commun. 2015;3:65. doi: 10.1186/s40478-015-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degens H, Veerkamp JH. Changes in oxidative capacity and fatigue resistance in skeletal muscle. Int. J. Biochem. 1994;26:871–878. doi: 10.1016/0020-711X(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 29.Bergsma AJ, et al. Identification and characterization of aberrant GAA pre-mRNA splicing in pompe disease using a generic approach. Hum. Mutat. 2015;36:57–68. doi: 10.1002/humu.22705. [DOI] [PubMed] [Google Scholar]
- 30.Winkel LP, et al. The natural course of non-classic Pompe’s disease; a review of 225 published cases. J. Neurol. 2005;252:875–884. doi: 10.1007/s00415-005-0922-9. [DOI] [PubMed] [Google Scholar]
- 31.Barohn RJ, McVey AL, DiMauro S. Adult acid maltase deficiency. Muscle Nerve. 1993;16:672–676. doi: 10.1002/mus.880160614. [DOI] [PubMed] [Google Scholar]
- 32.Raben N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenney DL, Benarroch EE. The autophagy-lysosomal pathway: general concepts and clinical implications. Neurology. 2015;85:634–645. doi: 10.1212/WNL.0000000000001860. [DOI] [PubMed] [Google Scholar]
- 34.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkoy G, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 36.Spampanato C, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stauber WT, Hedge AM, Trout JJ, Schottelius BA. Inhibition of lysosomal function in red and white skeletal muscles by chloroquine. Exp. Neurol. 1981;71:295–306. doi: 10.1016/0014-4886(81)90090-X. [DOI] [PubMed] [Google Scholar]
- 38.Mauthe M, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim JA, Li L, Kakhlon O, Myerowitz R, Raben N. Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy. 2015;11:385–402. doi: 10.1080/15548627.2015.1009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geddes R, Stratton GC. The influence of lysosomes on glycogen metabolism. Biochem. J. 1977;163:193–200. doi: 10.1042/bj1630193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lerin C, et al. Regulation of glycogen metabolism in cultured human muscles by the glycogen phosphorylase inhibitor CP-91149. Biochem. J. 2004;378:1073–1077. doi: 10.1042/bj20030971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halse R, Fryer LG, McCormack JG, Carling D, Yeaman SJ. Regulation of glycogen synthase by glucose and glycogen: a possible role for AMP-activated protein kinase. Diabetes. 2003;52:9–15. doi: 10.2337/diabetes.52.1.9. [DOI] [PubMed] [Google Scholar]
- 44.Thurberg BL, et al. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for Pompe disease. Lab. Invest. 2006;86:1208–1220. doi: 10.1038/labinvest.3700484. [DOI] [PubMed] [Google Scholar]
- 45.Koeberl DD, et al. Enhanced efficacy of enzyme replacement therapy in Pompe disease through mannose-6-phosphate receptor expression in skeletal muscle. Mol. Genet. Metab. 2011;103:107–112. doi: 10.1016/j.ymgme.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wisselaar HA, Kroos MA, Hermans MM, van Beeumen J, Reuser AJ. Structural and functional changes of lysosomal acid alpha-glucosidase during intracellular transport and maturation. J. Biol. Chem. 1993;268:2223–2231. doi: 10.1016/S0021-9258(18)53985-5. [DOI] [PubMed] [Google Scholar]
- 47.Moreland RJ, et al. Lysosomal acid alpha-glucosidase consists of four different peptides processed from a single chain precursor. J. Biol. Chem. 2005;280:6780–6791. doi: 10.1074/jbc.M404008200. [DOI] [PubMed] [Google Scholar]
- 48.Fukuda T, et al. Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann. Neurol. 2006;59:700–708. doi: 10.1002/ana.20807. [DOI] [PubMed] [Google Scholar]
- 49.Chien YH, Hwu WL, Lee NC. Pompe disease: early diagnosis and early treatment make a difference. Pediatr. Neonatol. 2013;54:219–227. doi: 10.1016/j.pedneo.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Smith BK, et al. Phase I/II trial of adeno-associated virus-mediated alpha-glucosidase gene therapy to the diaphragm for chronic respiratory failure in Pompe disease: initial safety and ventilatory outcomes. Hum. Gene Ther. 2013;24:630–640. doi: 10.1089/hum.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun B, et al. Correction of multiple striated muscles in murine Pompe disease through adeno-associated virus-mediated gene therapy. Mol. Ther. 2008;16:1366–1371. doi: 10.1038/mt.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salva MZ, et al. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 2007;15:320–329. doi: 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- 53.Selak MA, et al. Mitochondrial activity in Pompe’s disease. Pediatr. Neurol. 2000;23:54–57. doi: 10.1016/S0887-8994(00)00145-4. [DOI] [PubMed] [Google Scholar]
- 54.Sato Y, et al. Metabolomic profiling of pompe disease-induced pluripotent stem cell-derived cardiomyocytes reveals that oxidative stress is associated with cardiac and skeletal muscle pathology. Stem Cells Transl. Med. 2017;6:31–39. doi: 10.5966/sctm.2015-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Gijt JP, van Capelle CI, Oosterhuis JW, van der Ploeg AT, van der Wal KG. Gingival overgrowth in Pompe disease: a case report. J. Oral. Maxillofac. Surg. 2011;69:2186–2190. doi: 10.1016/j.joms.2011.03.070. [DOI] [PubMed] [Google Scholar]
- 56.Verity MA. Infantile Pompe’s disease, lipid storage, and partial carnitine deficiency. Muscle Nerve. 1991;14:435–440. doi: 10.1002/mus.880140509. [DOI] [PubMed] [Google Scholar]
- 57.Franco LM, et al. Evasion of immune responses to introduced human acid alpha-glucosidase by liver-restricted expression in glycogen storage disease type II. Mol. Ther. 2005;12:876–884. doi: 10.1016/j.ymthe.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 58.Han SO, et al. Low-dose liver-targeted gene therapy for Pompe disease enhances therapeutic efficacy of ERT via immune tolerance induction. Mol. Ther. Methods Clin. Dev. 2017;4:126–136. doi: 10.1016/j.omtm.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raval KK, et al. Pompe disease results in a Golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J. Biol. Chem. 2015;290:3121–3136. doi: 10.1074/jbc.M114.628628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimada Y, et al. Endoplasmic reticulum stress induces autophagy through activation of p38 MAPK in fibroblasts from Pompe disease patients carrying c.546G>T mutation. Mol. Genet Metab. 2011;104:566–573. doi: 10.1016/j.ymgme.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Corti M, et al. Altered activation of the tibialis anterior in individuals with Pompe disease: Implications for motor unit dysfunction. Muscle Nerve. 2015;51:877–883. doi: 10.1002/mus.24444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sidman RL, et al. Temporal neuropathologic and behavioral phenotype of 6neo/6neo Pompe disease mice. J. Neuropathol. Exp. Neurol. 2008;67:803–818. doi: 10.1097/NEN.0b013e3181815994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mobarhan S, Pintozzi RL, Damle P, Friedman H. Treatment of acid maltase deficiency with a diet high in branched-chain amino acids. JPEN J. Parenter. Enter. Nutr. 1990;14:210–212. doi: 10.1177/0148607190014002210. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida T, et al. A skeletal muscle model of infantile-onset pompe disease with patient-specific iPS cells. Sci. Rep. 2017;7:13473. doi: 10.1038/s41598-017-14063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han SO, Li S, Koeberl DD. Salmeterol enhances the cardiac response to gene therapy in Pompe disease. Mol. Genet. Metab. 2016;118:35–40. doi: 10.1016/j.ymgme.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkel LP, et al. Enzyme replacement therapy in late-onset Pompe’s disease: a three-year follow-up. Ann. Neurol. 2004;55:495–502. doi: 10.1002/ana.20019. [DOI] [PubMed] [Google Scholar]
- 67.Hesselink RP, Schaart G, Wagenmakers AJ, Drost MR, van der Vusse GJ. Age-related morphological changes in skeletal muscle cells of acid alpha-glucosidase knockout mice. Muscle Nerve. 2006;33:505–513. doi: 10.1002/mus.20482. [DOI] [PubMed] [Google Scholar]
- 68.Raben N, et al. Differences in the predominance of lysosomal and autophagic pathologies between infants and adults with Pompe disease: implications for therapy. Mol. Genet. Metab. 2010;101:324–331. doi: 10.1016/j.ymgme.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nascimbeni AC, Fanin M, Masiero E, Angelini C, Sandri M. Impaired autophagy contributes to muscle atrophy in glycogen storage disease type II patients. Autophagy. 2012;8:1697–1700. doi: 10.4161/auto.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leslie, N. & Bailey, L. in GeneReviews((R)), (eds. M.P. Adam et al.) (Seattle, WA, 1993).
- 71.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeBosch BJ, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 2016;9:ra21. doi: 10.1126/scisignal.aac5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu S, et al. Impaired organization and function of myofilaments in single muscle fibers from a mouse model of Pompe disease. J. Appl Physiol. 2010;108:1383–1388. doi: 10.1152/japplphysiol.01253.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drost MR, Hesselink RP, Oomens CW, van der Vusse GJ. Effects of non-contractile inclusions on mechanical performance of skeletal muscle. J. Biomech. 2005;38:1035–1043. doi: 10.1016/j.jbiomech.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 75.Palermo AT, et al. Transcriptional response to GAA deficiency (Pompe disease) in infantile-onset patients. Mol. Genet Metab. 2012;106:287–300. doi: 10.1016/j.ymgme.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 76.DeRuisseau LR, et al. Neural deficits contribute to respiratory insufficiency in Pompe disease. Proc. Natl Acad. Sci. USA. 2009;106:9419–9424. doi: 10.1073/pnas.0902534106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osaki T, Uzel SGM, Kamm RD. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 2018;4:eaat5847. doi: 10.1126/sciadv.aat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amalfitano A, et al. Recombinant human acid alpha-glucosidase enzyme therapy for infantile glycogen storage disease type II: results of a phase I/II clinical trial. Genet. Med. 2001;3:132–138. [PubMed] [Google Scholar]
- 79.Huang HP, et al. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum. Mol. Genet. 2011;20:4851–4864. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- 80.Klinge L, et al. Safety and efficacy of recombinant acid alpha-glucosidase (rhGAA) in patients with classical infantile Pompe disease: results of a phase II clinical trial. Neuromuscul. Disord. 2005;15:24–31. doi: 10.1016/j.nmd.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 81.Takikita S, Myerowitz R, Zaal K, Raben N, Plotz PH. Murine muscle cell models for Pompe disease and their use in studying therapeutic approaches. Mol. Genet. Metab. 2009;96:208–217. doi: 10.1016/j.ymgme.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor KM, et al. Dysregulation of multiple facets of glycogen metabolism in a murine model of Pompe disease. PLoS ONE. 2013;8:e56181. doi: 10.1371/journal.pone.0056181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu, S. et al. Improved efficacy of a next-generation ERT in murine Pompe disease. JCI Insight4, e125358 (2019). [DOI] [PMC free article] [PubMed]
- 84.Cardone M, et al. Abnormal mannose-6-phosphate receptor trafficking impairs recombinant alpha-glucosidase uptake in Pompe disease fibroblasts. Pathogenetics. 2008;1:6. doi: 10.1186/1755-8417-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim JA, Sun B, Puertollano R, Raben N. Therapeutic benefit of autophagy modulation in Pompe disease. Mol. Ther. 2018;26:1783–1796. doi: 10.1016/j.ymthe.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan B, Raben N, Plotz P. The human acid alpha-glucosidase gene is a novel target of the Notch-1/Hes-1 signaling pathway. J. Biol. Chem. 2002;277:29760–29764. doi: 10.1074/jbc.M204721200. [DOI] [PubMed] [Google Scholar]
- 87.Berdeaux R, Stewart R. cAMP signaling in skeletal muscle adaptation: hypertrophy, metabolism, and regeneration. Am. J. Physiol. Endocrinol. Metab. 2012;303:E1–E17. doi: 10.1152/ajpendo.00555.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koeberl DD, et al. Correction of biochemical abnormalities and improved muscle function in a phase I/II clinical trial of clenbuterol in Pompe disease. Mol. Ther. 2018;26:2304–2314. doi: 10.1016/j.ymthe.2018.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kondash, M.E., Ananthakumar, A., Khodabukus, A., Bursac, N. & Truskey, G.A. Glucose uptake and insulin response in tissue-engineered human skeletal muscle. Tissue Eng. Regen. Med.17, 801–813 (2020). [DOI] [PMC free article] [PubMed]
- 90.Gao GP, et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Juhas M, Bursac N. Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells. Biomaterials. 2014;35:9438–9446. doi: 10.1016/j.biomaterials.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krueger, F. Trim galore. A wrapper tool. Cutadapt FastQC consistently apply Qual. adapter trimming FastQ files516, 517 (2015).
- 94.Kersey PJ, et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 97.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huber W, Carey V, Davis S, Hansen KD, Morgan M. The Bioconductor channel in F1000Research. F1000Res. 2015;4:217. doi: 10.12688/f1000research.6758.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 102.Wang, J. & Bursac, N. dryad_main_figure_raw_pompe_data.xlsx. figshare. 10.6084/m9.figshare.14173013.v1 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data supporting the results of the main figures are available at 10.6084/m9.figshare.14173013.v1102.The sequencing data generated for this study have been deposited in Gene Expression Omnibus under the SuperSeries GSE159064 and SubSeries GSE159062 (human myobundle model) and GSE159063 (GAA KO mouse model). Data referenced in the “Discussion” of this study are available on Gene Expression Omnibus GSE57980 and GSE38680. Any remaining information can be obtained from the corresponding author upon reasonable request.