Abstract
Phage therapy represents a possible treatment option to cure infections caused by multidrug-resistant bacteria, including methicillin and vancomycin-resistant Staphylococcus aureus, to which most antibiotics have become ineffective. In the present study, we report the isolation and complete characterization of a novel phage named JD219 exhibiting a broad host range able to infect 61 of 138 clinical strains of S. aureus tested, which included MRSA strains as well. The phage JD419 exhibits a unique morphology with an elongated capsid and a flexible tail. To evaluate the potential of JD419 to be used as a therapeutic phage, we tested the ability of the phage particles to remain infectious after treatment exceeding physiological pH or temperature. The activity was retained at pH values of 6.0–8.0 and below 50°C. As phages can contain virulence genes, JD419’s complete genome was sequenced. The 45509 bp genome is predicted to contain 65 ORFs, none of which show homology to any known virulence or antibiotic resistance genes. Genome analysis indicates that JD419 is a temperate phage, despite observing rapid replication and lysis of host strains. Following the recent advances in synthetic biology, JD419 can be modified by gene engineering to remove prophage-related genes, preventing potential lysogeny, in order to be deployed as a therapeutic phage.
Keywords: Staphylococcus aureus, bacteriophage, morphology, genome, host-range, phage therapy
Background
Antimicrobial resistance (AMR) presents a significant threat to humankind. A study estimates that in 2050 about 10 million people will die annually from untreatable bacterial infections if alternatives and novel antibiotics are not available (O’Neill, 2014). The rate of antibiotic-resistance in bacteria has been increasing over the decades due to the over-and misuse of antibiotics (Leptihn, 2019). So-called antimicrobial stewardship programs aim to optimize antibiotic use to decrease the emergence and spread of infections caused by multidrug-resistant organisms (Manohar et al., 2020b). Due to this and the lack of financial incentives for the pharmaceutical industry to develop new antibiotics, alternative treatment strategies are urgently needed. One such strategy is the deployment of bacterial viruses that infect and kill bacteria, also known as phage therapy (Manohar et al., 2020a). Phage therapy has been used successfully against many Gram-negative and -positive bacteria, including S. aureus but is not yet a standard therapeutic strategy (Abedon et al., 2011; Schooley et al., 2017; Maddocks et al., 2019). S. aureus is a particularly concerning pathogen as it exists as a harmless commensal. Yet, it can cause various types of infections, from skin abscesses to lethal bloodstream infections, often in hospitals. Phage therapy shows promising potential for treating methicillin- as well as vancomycin-resistant S. aureus (Moellering, 2012; Limbago et al., 2014) as phages can infect multidrug-resistant bacteria (Lobocka et al., 2012; Kazmierczak et al., 2014).
While members from other families have been reported, most Staphylococcus phages such as K, Twort, CH15, φSA012, etc. belong to the family of Myoviridae, which in some instances exhibit a broad-host-range that is considered beneficial for phage therapy (Cui et al., 2017b; Iwano et al., 2018). Podoviridae infecting S. aureus have also been reported (Culbertson et al., 2019) and have been used together with other phages in so-called phage cocktails (“Microgen cocktail”) for the treatment of wound and skin infections (McCallin et al., 2018; Kornienko et al., 2020). Staphylococcus phages belonging to the family of Siphoviridae have been found worldwide; however, many of them display a lysogenic life cycle while occasionally also carrying virulence genes and antibiotic-resistant genes, which are the undesired characteristics for therapeutic phages. Several reports have shown the potential use of Staphylococcus phages belonging to Siphoviridae alone or combined with chemical antibiotics to penetrate and remove biofilms to treat infections (Rahman et al., 2011; Ji et al., 2020).
With recent advancements in genome engineering, lysogenic phages represent a valuable therapeutic virus source and have been recently employed in clinical therapy (Dedrick et al., 2019). While many lytic phages have been characterized in the past, considerably fewer lysogenic phages have been described on the biochemical level due to their lifestyle complexity, with the exception of prophage sequence analyses in bacterial genomes (Goerke et al., 2009; Loh et al., 2020a) or the impact of prophages on their hosts (Ingmer et al., 2019). When properly characterized, lysogenic phages can be a treasure-trove for genetically modified therapeutic phages. In this work, we describe Staphylococcus phage JD419, a comparably “unstable lysogenic phage” as lysogeny seems not to be preferred, as the phage quickly undergoes replication and host cell lysis. The morphology of JD419 revealed that the phage belongs to the Siphoviridae. Whole-genome analysis of the Staphylococcus phage indicates that it does not contain any virulence factors nor antimicrobial resistance genes, which must be absent in therapeutic phages. Due to a remarkable killing ability and broad host range of JD419, this phage is an ideal target for genetic engineering to create a lytic phage by removing genes allowing integration and gene repression following an approach similar to Dedrick et al. (2019), as our study illustrates that Staphylococcus phage JD419 has the potential to be a high-value therapeutic phage for prophylaxis and therapeutic purposes.
Materials and Methods
Bacteria Isolates and Culture Conditions
A total of 135S. aureus isolates were obtained from the clinical samples, including blood, wound secretions, and sputum from patients with S. aureus infections in different hospitals in Shanghai, China. Strains were identified by the VITEK 2 Compact (Biomerieux); 76 strains were MSSA (methicillin-sensitive Staphylococcus aureus), 59 strains were MRSA (Methicillin-resistant Staphylococcus aureus), which were tested for resistance to cefoxitin; detailed information can be found in the Supplementary Material. Three standard strains, including N315, USA300, and Newman, which Han Yang kindly provided in Shanghai General Hospital, were also used. Isolates were grown in liquid LB (Luria-Bertani) medium at 37°C, on LB medium (1.5% agar), or in LB soft agar overlays (0.7% agar) (Sangon Biotech (Shanghai) Co., Ltd, China). The phage JD419 was initially found as plaques in the lawn of S. aureus clinical isolates MR-84 belonging to MLST 239 type, which was observed in the Molecular Microbiology laboratory in Shanghai Jiao Tong University School Medicine, Shanghai, China. Even without any chemicals such as Mitomycin C, clear plaques were observed.
The phage was purified three times from plaques using two-layered agar plates described previously (Cui et al., 2017a). High-titer phage stocks (2 × 109pfu/mL) were obtained through amplification in liquid LB medium containing 10 mM MgCl2 and 5 mM CaCl2 (Sangon Biotech (Shanghai) Co., Ltd.) (Cui et al., 2017a), for which S. aureus MR-84 was used as a host strain. 101 of 138 different strains of S. aureus were characterized using the MLST typing method, as previously described (Bao et al., 2013).
Staphylococcus Phage JD419 Amplification and Purification
First, phage JD419 was added to host cells of S. aureus strain MR-84 at a Multiplicity of Infection (MOI) of 0.01. After incubation at 37°C overnight, cell lysis -indicated by a decrease in the liquid culture’s turbidity- was observed. The lysate was then incubated with chloroform (final concentration was 2%) for 30 min under gentle shaking to kill the residual bacteria and release phage particles from bacterial cells. Enrichment and purification of phage particles were conducted as described previously (Cui et al., 2017a). Briefly, cell debris was removed by centrifugation at 6,500 rpm (Beckman, JA18.0) for 15 min, followed by the precipitation of phages in the supernatant using polyethylene glycol (PEG) 8000 (at 4°C overnight) and precipitated (final concentration 10% w/v) by centrifugation at 8,500 rpm, for 20 min (Beckman, JA18.0). The pellets were re-suspended in TM buffer [(Tris-Mg2+ Buffer) 10 mM Tris–HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 5 mM CaCl2)], vortexed, and PEG8000 was removed by adding the same volume of chloroform and vortexed, centrifuged at 4,000 g/min for 10 min. The supernatant contained high concentrations of phage to which CsCl was added (0.5 g/mL). The phages were then purified by centrifugation in a CsCl gradient (1.33, 1.45, 1.50, 1.70 g/cm3) in TM buffer in Ultra-Clear tubes (Beckman Coulter Inc., Fullerton, CA, United States). Finally, the band containing the enriched phages was taken out using a syringe, then dialyzed against TM buffer and stored at 4°C.
Electron Microscopy Imaging
After centrifugation in a CsCl gradient (described above), purified phage particles obtained by dialysis were collected by centrifugation at 33,000 × g for one hour (Beckman high-speed centrifuge and a JA-18.1 fixed-angle rotor) and washed twice in 0.1 × PBS (pH 7.4). The sample was deposited on carbon-coated copper grids and stained with 2% (w/v) potassium phosphotungstate (pH7.0). Phage photos were taken using a Hitachi H7500 transmission electron microscope (TEM) at 80 kV.
Host Range Determination
The host range of phage JD419 was analyzed by spotting serial dilutions of the phage on a two-layer soft agar lawn of S. aureus. Two microliters of a dilution series of phage lysate (≈108 PFU/ml) were gently pipetted on a Luria-Bertani (LB) medium plate overlay agar plate containing S. aureus (overlaid with S. aureus (OD600≈0.4) mixed in 0.7% top agar and cultured 30 min). Phage’s lytic ability was assessed based on the clarity of the inhibition zone as published previously (Cui et al., 2017a): clear, turbid, or faint. Lytic activity was confirmed by determining the phage’s ability to create single plaques after incubating serial dilutions of the phage with host bacteria using the soft-agar overlay technique. 138 strains of S. aureus were used to establish an estimate of the host range of phage JD419.
Determination of Adsorption Rate
Phage JD419 was added to an exponentially growing culture of S. aureus MR-84 at an MOI of 0.0005 and incubated at room temperature without shaking. An aliquot of 100 μL, containing a mixture of phage JD419 with S. aureus MR-84, was taken at different time points (1–30 min) 1, 3, 5, 7, 9, 12, 15, 18, 21, 24, 27, and 30 min respectively, and centrifuged immediately at 16,000 g for 30 s once collected. Afterward, the titers of phage JD419 in the supernatant were determined by counting the plaques on overlay agar plates. The adsorption rate of phage JD419 to the S. aureus MR-84 at the time of i was calculated by the formula of (N0-Ni)/N0∗100, with Ni representing the titer of phage in the supernatant after co-incubation for i minutes, and N0 is the titer of phage before co-incubation. The proportion of non-adsorbed phages to the number of phages used for infection and standard deviations were calculated.
One-Step Growth Curve and the Burst Size of Staphylococcus Phage JD419
Staphylococcus aureus cells at mid-log growth phase were harvested by centrifugation at 13,000g for 15 min and resuspended in fresh LB medium. Phage JD419 was added to the culture at an MOI of 0.01 and allowed to adsorb for 10 min at room temperature, then incubated at 37°C with gentle shaking. Aliquots were taken at different points (1–110 min) 1, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 100, and 110 min. After centrifugation, the phage titers were determined by counting plaques on overlay agar plates.
Phage JD419 was added to S. aureus MR-84 at an MOI of 0.01. The burst size was calculated using the formula (Nf-N0)/N0, where Nf represents the titer of phage JD419 at the beginning of the next growth circle, and N0 represents the titer of phage JD419 at the beginning of the former growth circle.
Stability of Staphylococcus Phage JD419
The pH stability of phage JD419 was assessed as follows: 100-fold dilutions of the initial phage solution containing 108 PFU/mL were added to TM buffer with pH values ranging from 2 to 12 incubated at 37°C for 1 h. Subsequently, the samples were diluted in TM buffer (pH 7.5), and the titer of each sample was determined. To address thermal stability, phage JD419 were incubated in TM buffer (pH7.5) at different temperatures (24°C, 37°C, 40°C, 50°C, 60°C, and 70°C) for 1 h, then the titer was determined.
Inhibition Growth Curve of Staphylococcus Phage JD419
Four milliliters of TSB (Oxiod; containing 5mM CaCl2 and 10mM MgSO4) in an L-shaped plastic test tube was inoculated (1%) with an overnight S. aureus (either N315 or MR-84) culture, Staphylococcus phage JD419 was added at an MOI of 100, 10, or 0 and incubated at 37°C without shaking. OD600 value at different time points (interval 30 min extending 8 h) was measured. Furthermore, an independent experiment involved the CFU of S. aureus in the group of MOI = 100 and 0 being counted by serial dilution of the mixture using the CFU forming assay in the TSA plate (TSA, Oxiod); they were measured by interval 2 h extending 12 h.
Genome Analysis
The DNA of Staphylococcus phage JD419 was extracted using an Aidlab kit (Aidlab Biotechnologies Co., Ltd, Beijing, China). The complete genome of Staphylococcus phage JD419 was obtained by 454 pyro-sequencing at the Chinese National Human Genome Center (Shanghai, China). Assembly of quality filtered reads was performed using the 454 Life Sciences Corporation platform. The sequence of JD419 was deposited under the nucleotide sequence accession number MT899504 (GenBank). tRNAs were detected using tRNAscan-SE software (Lowe and Eddy, 1997). To identify any virulence factors or antibiotic-resistant genes in the genome of Staphylococcus phage JD419, Blast searches were performed against the antibiotic resistance genes database (ARDB)1 (Liu and Pop, 2009) and the virulence factors database2. Genes with more than 70% coverage and a minimum identity of 30% were retained as results. The software PHACTS was used to determine whether Staphylococcus phage JD419 is a lytic or lysogenic (temperate) phage (McNair et al., 2012). Comparisons of complete genome sequences of other phages were performed with Mauve20150226 (Darling et al., 2010); Neighbor-joining trees were constructed using the major capsid proteins to analyze phylogeny JD419 by MEGA5. ORFs were predicted using several algorithms, including those of the software glimmer, RAST, and GeneMarkS (Delcher et al., 1999; Besemer and Borodovsky, 2005; Aziz et al., 2008). A two-tailed unpaired Student’s t-test was used for statistical analysis with GraphPad Prism Software and R. p-values of less than 0.05 were considered significant unless specifically indicated otherwise.
Results
Phage JD419 Has a Flexible Tail Attached to a Prolate Capsid
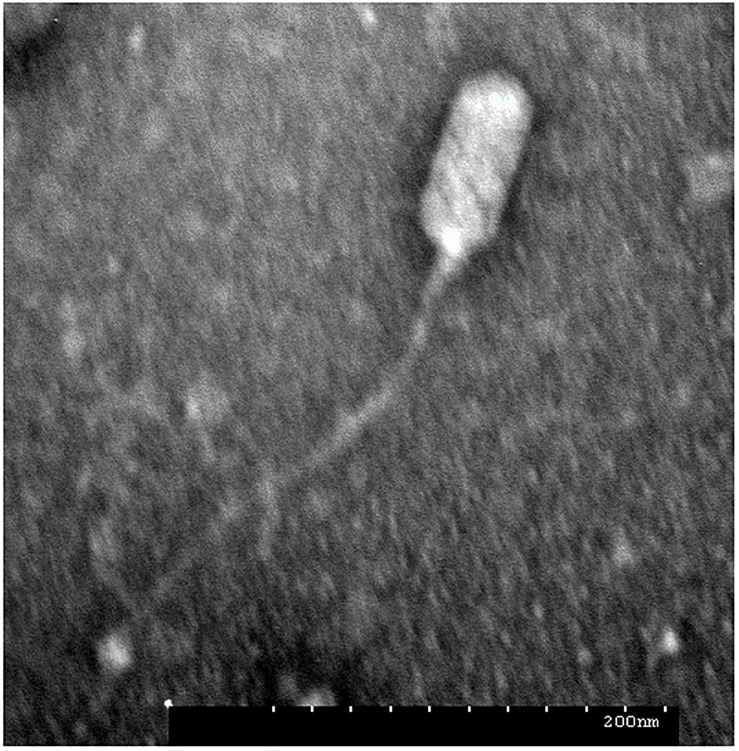
During routine culture of clinical S. aureus strains (Supplementary Table 1), plaques were observed in the lawn of isolate MR-84. Phages often occur together with their host, either as prophages or in natural prey-predator situations. The phage termed JD419 was isolated from the plaques and further purified three times. To address the morphology phage JD419 displays, we employed Transmission Electron Microscopy (TEM). Electron micrographs show that the tail is about 300 nm in length, while the head has a prolate geometry, elongated only in one direction. The length to width ratio is approximately 2, with a length of 100 ± 5 nm and a width of 50 ± 2 nm (Figure 1). A tail sheath between the head and tip of the tail is absent. The overall morphology indicates that phage JD419 is a member of the family Siphoviridae in the order Caudovirales, however, with an unusually elongated head. Table 1 lists phages with similar morphology described in the literature, which infect different bacteria or exhibit distant genetic relationships to JD419.
FIGURE 1.
Morphology of Staphylococcus phage JD419. The electron micrograph of phage JD419 shows that the tail is flexible and has a length of about 300 nm. The head of the phage is prolate, i.e., extended. The minor axis semi-diameter and major axis semi-diameter of its head are about 50 and 100 nm. Bar: 200 nm.
TABLE 1.
Sightings of phages with similar morphology to Staphylococcus phage JD419.
| Phage | Host | Head diameter (nm) | Tail length (nm) | References and comments |
| IME-EF1 | E. faecalis | Zhang et al., 2013 | ||
| P70 | L. monocytogenes | Schmuki et al., 2012 | ||
| 3A | S. aureus | 50 × 100 | 350 | http://www.phage.ulaval.ca |
| JD419 | S. aureus | This study | ||
| 4CbK | C. crescentus | 50 × 200 | 300 | Leonard et al., 1972 |
The Adsorption Rate of Phage JD419 to the Host Surface Is a Rapid Process
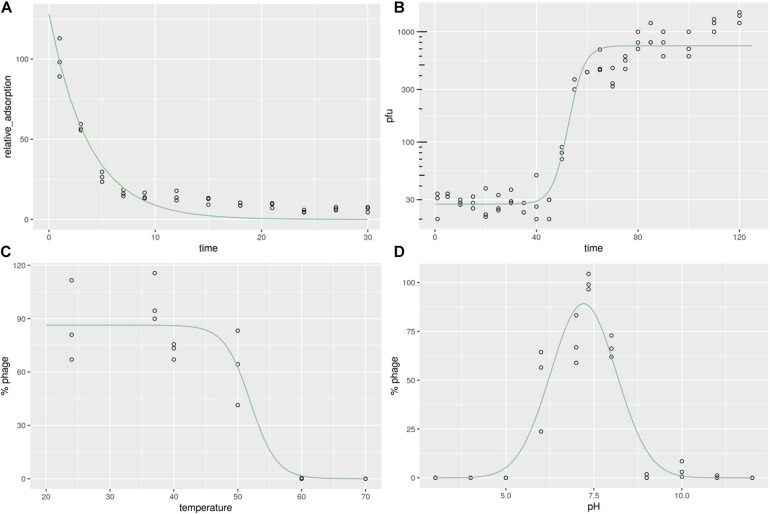
To calculate the phage adsorption rate to its bacterial host, we measured the time-dependent adsorption of phages to bacterial cells by simple co-incubation and subsequent counting of phage particles present in the supernatant. After incubation with its host strain MR-84 for 10 min, more than 80% of phages adhered to the bacterial surface, while after 20 min, more than 90% of phages remained attached to the host (Figure 2A). The observed adsorption rates could be fitted to an exponential decay reaction, with half of the phages attaching to the host cell within approximately 3 min, demonstrating the rapidity of this reaction, indicating a high affinity between phage and host.
FIGURE 2.
Characterization of Staphylococcus phage JD419. (A) Adsorption rate of Staphylococcus phage JD419 to its host S. aureus MR-84. The X-axis represents the incubation time of Staphylococcus phage JD419 and its host, and the Y-axis represents the percent of phage that its host had not absorbed (B) One-step growth curves of Staphylococcus phage JD419; X-axis represents co-culture time of Staphylococcus phage JD419 and MR-84, Y-axis represents the change in titer of phage JD419 in the mixture. (C) Thermal stability of phage JD419; X-axis represents different temperatures; Y-axis represents the relative titer of Staphylococcus phage JD419 to that the titer at 37°C;% stability = (N/N0) × 100, where N was the number of viable phages after one hour of incubation and N0 was the initial number of phages. (D) pH stability of Staphylococcus phage JD419; X-axis represents the different pH values; Y-axis shows the percentage stability relative to the PFU/mL at pH 7.45.
Phage Replication of JD419 Requires More Than the Typical Generation Time of the Host, Releasing About 30 Progeny Particles
When JD419 was used to infect a growing culture of the S. aureus host strain MR-84 with an MOI of 0.01, we observed phage particles released after a latent period of approximately 50 min. Complete lysis of all cells was observed at around 70 min. This data was fitted using a sigmoidal fit, showing that half of the phages have been fully released from the cells within 50 min, indicating that the host requires more than one generation time commonly observed for its host, S. aureus, when not infected by phages (Figure 2B). We also measured the number of phage particles that were released per cell. Here, we could determine a burst size of approximately 33 phages produced on average per cell.
Stability of Phage JD419 at Elevated Temperatures and Non-neutral pH Values
For a phage to be deployed for therapeutic use, the stability of the phage needs to be considered. Thermal stability assays show that JD419 remains an active infectious particle between 24 and 40°C (p > 0.05). By comparison, almost all phages were inactivated when incubated at 60°C for 1 h (p < 0.05) (Figure 2C). When data points were fitted with a sigmoidal function, the point at which half of the phages were inactivated lies at approximately 52°C (± 2.36).
The inactivation in basic or acidic solutions was measured within a pH of 3 to 12. Phage JD419 was stable at pH values between 6 and 8 (Figure 2D), with calculated optimum stability at pH 7.2 (± 0.073), using a Gaussian fitting function. Its stability makes JD419 suitable for most therapeutic applications except the oral route, as the stomach’s acidic environment would inactivate the phage.
Phage JD419 Exhibits a Broad Host Range
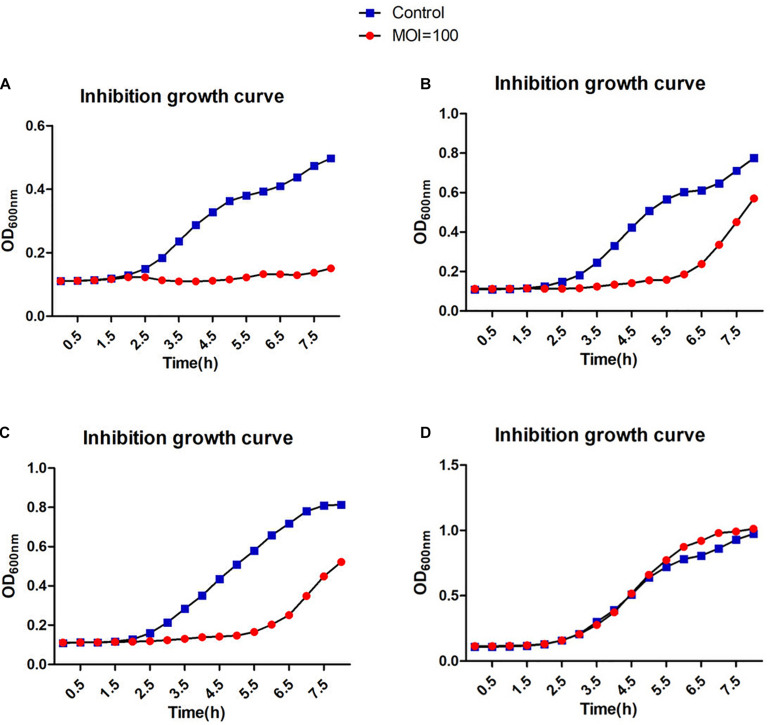
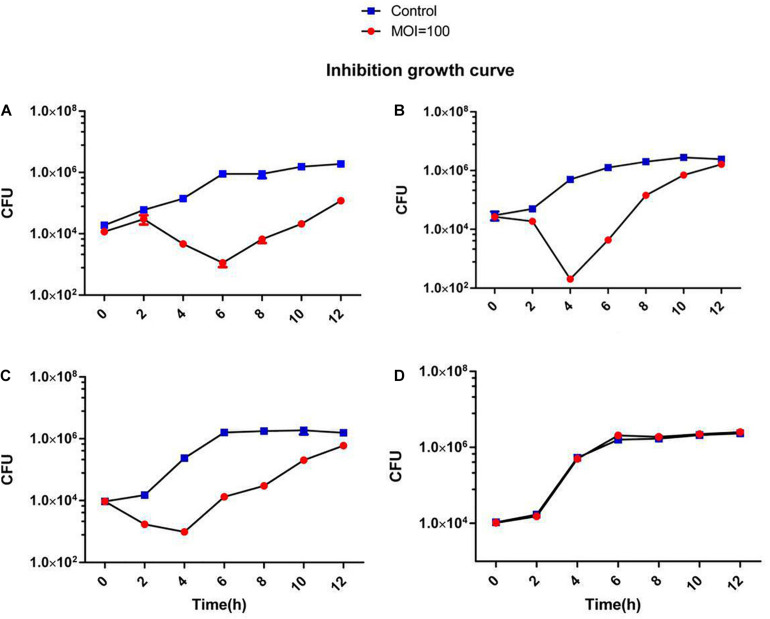
A broad host range would allow a therapeutic phage to be used on a large number of different strains. Therefore, it is advantageous to discover phages that can infect many clinical strains. We screened 138 different clinical S. aureus strains from two different hospitals in Shanghai, China. Many of the methicillin-resistant strains were classified according to their MLST types and included the prevalent ST239 and ST59 (Supplementary Figure 1). Phage JD419 can lyse 44.2% of tested strains (61/138). However, the extent of lysis was not identical for all strains. We categorized the phage host-range according to the visual appearance of single plaques (Supplementary Figure 1). While some of the strains exhibited plaque-like clearance zones when spotting phage lysate on lawns (albeit having a turbid appearance), some of these did not allow the formation of single plaques in an overlay assay, and these strains were therefore labeled as non-susceptible. The ability to infect and lysis efficiency showed no correlation with the MLST types (Supplementary Table 1). In addition to plaque formation, we also used four isolates of S. aureus to investigate the impact on growth in the presence of JD419. Here, we observed that at an MOI of 100 and 10 (Supplementary Figure 1), the growth of three phage-susceptible strains (N315, MR-84, MS-41) was inhibited, while a non-susceptible strain grew similar to the control in which phage was absent (Supplementary Figure 1 and Figures 3, 4).
FIGURE 3.
Inhibition assays of Staphylococcus phage JD419. The x-axis represents the co-culture time of phage JD419 and different strains of S. aureus, and the y-axis represents the change in OD600 in the mixture of Staphylococcus phage JD419 infecting different strains at the MOI = 100. Panel (A) represents strain N315; panel (B) represents strain MR-84; panel (C) represents strain MS-41; panel (D) represents strain MS-57.
FIGURE 4.
Inhibition assays of Staphylococcus phage JD419. The x-axis represents the co-culture time of phage JD419 and different strains of S.aureus, and the y-axis represents the change in CFU (5uL) in the mixture of Staphylococcus phage JD419 infecting different strains at the MOI = 100. Panel (A) represents strain N315; panel (B) represents strain MR-84; panel (C) represents strain MS-41; panel (D) represents strain MS-57.
Genome Analysis of Staphylococcus Phage JD419
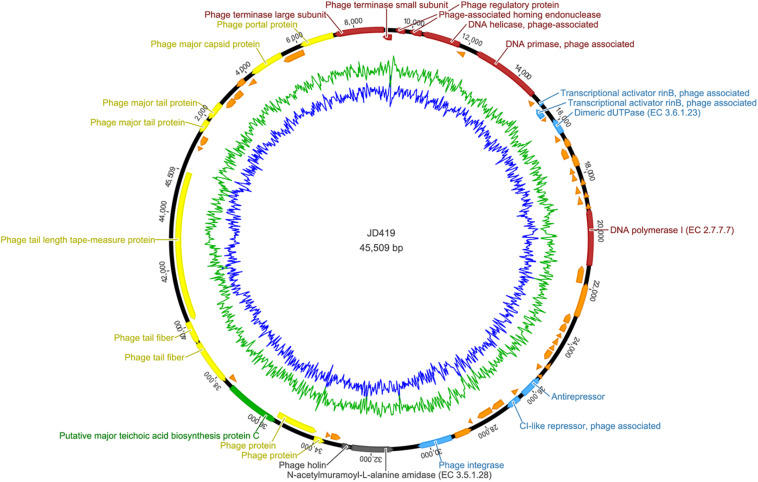
To assess the potential of phage JD419 to be used in phage therapy for treating S. aureus infections, we sequenced the complete genome of the bacterial virus. Our analysis showed that the phage has a circular genome with 45509 bp. Phage JD419 contains 65 putative open reading frames (ORFs). Most of the proposed ORFs have the same orientation, and several genes overlapped. Putative functions of the ORFs were assigned based on BLASTP analysis and domain searches. We categorized ORF encoded proteins into five modules: structure, DNA synthesis, lysis, regulation, and metabolism (Table 2 and Figure 5).
TABLE 2.
Predicted functional ORFs of Staphylococcus phage JD419.
| ORFs | Start | Stop | Strand | Function |
| JD419_ORF3 | 1889 | 1434 | − | Phage major tail protein |
| JD419_ORF4 | 2622 | 1981 | − | Phage major tail protein |
| JD419_ORF9 | 5305 | 4142 | − | Phage major capsid protein |
| JD419_ORF10 | 6090 | 5317 | − | Prophage Clp protease-like protein |
| JD419_ORF11 | 7312 | 6074 | − | Phage portal protein |
| JD419_ORF12 | 9008 | 7317 | − | Phage terminase large subunit |
| JD419_ORF13 | 9303 | 8998 | − | Phage terminase small subunit |
| JD419_ORF14 | 9748 | 9428 | − | Phage-associated homing endonuclease |
| JD419_ORF15 | 10342 | 9905 | − | Phage regulatory protein |
| JD419_ORF16 | 11713 | 10355 | − | DNA helicase, phage-associated |
| JD419_ORF18 | 14781 | 12334 | − | DNA primase, phage associated |
| JD419_ORF20 | 15253 | 15101 | − | Transcriptional activator rinB, phage associated |
| JD419_ORF21 | 15636 | 15250 | − | Transcriptional activator rinB, phage associated |
| JD419_ORF23 | 16412 | 15876 | − | Dimeric dUTPase |
| JD419_ORF35 | 21200 | 19239 | − | DNA polymerase I |
| JD419_ORF47 | 26300 | 25524 | − | Antirepressor |
| JD419_ORF49 | 26702 | 27016 | + | CI-like repressor, phage associated |
| JD419_ORF54 | 29205 | 30410 | + | Phage integrase |
| JD419_ORF55 | 32706 | 31252 | − | Phage lysin, N-acetylmuramoyl-L-alanine amidase |
| JD419_ORF56 | 33020 | 32718 | − | Phage holin |
| JD419_ORF61 | 37423 | 35513 | − | Putative major teichoic acid biosynthesis protein C |
| JD419_ORF63 | 39312 | 37729 | − | Phage tail fiber |
| JD419_ORF64 | 40145 | 39321 | − | Phage tail fiber |
| JD419_ORF65 | 45499 | 40145 | − | Phage tail length tape-measure protein |
FIGURE 5.
Complete genome sequence of Staphylococcus phage JD419. Genome map shows the predicted ORFs; arrows indicate the direction of transcription, proposed functional modules have been marked based on BLAST and domain search results.
Genes encoding for structural proteins, such as the major tail protein, the major capsid protein, a portal protein, tail fiber proteins, and a tail length tape-measure protein, were identified. ORFs involved in DNA replication including DNA helicase, DNA primase, DNA polymerase I, phage-associated homing endonuclease, phage terminase large subunit, phage terminase small subunits, phage integrase, a phage-associated homing endonuclease, and a gene with homology to a phage integrase were identified. Genes of holin and lysin (an N-acetylmuramoyl-L-alanine amidase) were also identified, both crucial for lysis. We found ORFs that might influence metabolic processes in the host. These genes coded, for example, a dimeric dUTPase, and a putative major teichoic acid biosynthesis protein C. We also identified ORFs involved in regulation, including the transcriptional activator rinB, CI-like repressor, and a prophage Clp protease-like protein, indicating JD419 is a temperate phage. According to the algorithm of the program PHACTS (on January 10, 2021), JD419 phage was confidently predicted as having a temperate lifestyle (Averaged Probability 0.694, Standard Deviation 0.043). While we could confirm that the genome of JD419 does not contain any known virulence factors, we were unable to identify putative roles or functions of a total of 40 of the 65 ORFs encoded proteins, as search algorithms solely reveal that the putative proteins had unknown functions. tRNAs, occasionally found in other large phages such as T4, have not been identified in the genome of JD419.
Discussion
The clinical use of suitable phages in treating infections might present a potential solution to the threat posed by antibiotic-resistant bacteria. S. aureus, which can exist as a harmless commensal bacterium, can cause severe and lethal infections that become increasingly difficult to treat, as many strains have acquired multidrug resistance. Bacteriophages can be used to treat infections caused by such strains, aiming to inactivate the pathogen if antibiotics are ineffective as a treatment. As in most cases, there is no sufficient time to isolate phages that can infect the particular strain that causes the infection; therefore, it is ideal to have phages that exhibit a broad host range at one’s disposal. In this work, we characterized the phage JD419, which can infect clinical strains of S. aureus. Its morphology is similar to other reported bacteriophages, such as IME-EF1, P70, and 3A (Leonard et al., 1972; Schmuki et al., 2012; Zhang et al., 2013). However, in general, phages with such elongated capsids and a flexible tail have rarely been described in the literature.
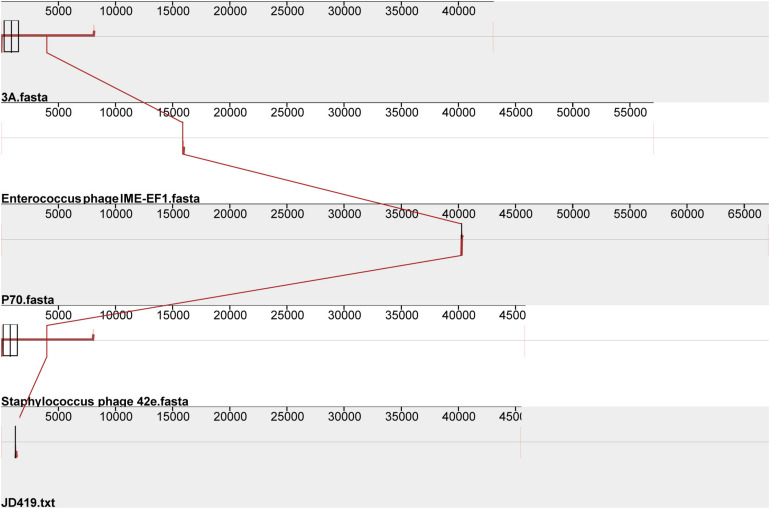
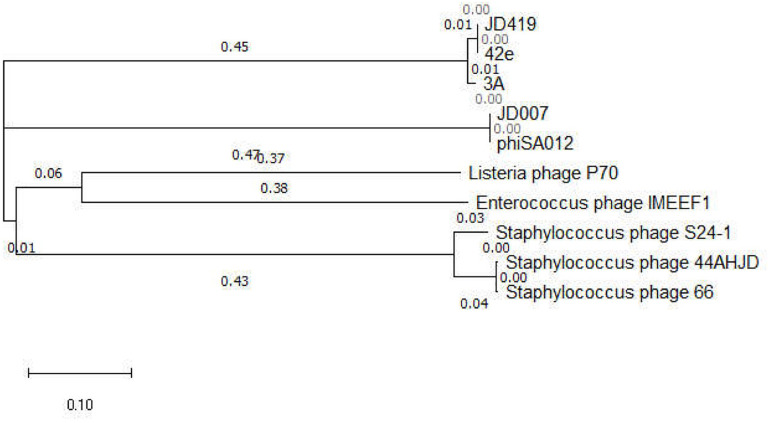
Interestingly, the three phages mentioned above with similar morphology infect Gram-positive bacteria and Gram-negative bacteria (Table 1). The phage IME-EF1 infects strains of Enterococcus feacalis (Zhang et al., 2013), phage P70 infects Listeria monocytogenes (Schmuki et al., 2012), phage 3A targets S. aureus (NC_007053.1), and phage 4CbK infects Caulobacter crescentus (Leonard et al., 1972). While it might be fair to assume that 3A is related to JD419, we find their identity to be at 95.98%, with a coverage of 82%; JD419 and 42e is 97.76%, however, with a coverage of 61%. When we compared the complete genome sequences with other morphologically similar phages, we did not find any close relatives among them (Figure 6). Interestingly, when analyzing distantly related phages of JD419, i.e., 42e, and 3A, the phages clustered in the same phylogenetic sub-branch when comparing their major capsid proteins only (Figure 7), which indicates that the phages are structurally related. It appears that the phages have acquired substantial differences in other genes required for infection and replication during evolution.
FIGURE 6.
Comparative genomic analysis of Staphylococcus phage JD419 with other phages with similar morphology. Staphylococcus phage 3A, Staphylococcus phage 42e, Enterococcus phage IME-EF1, and Listeria phage P70.
FIGURE 7.
Phylogenetic tree of bacteriophages with similar morphology to JD419 and other phages infecting S. aureus, based on the major capsid protein sequences. Phage JD007 and phiSA012 belong to the family of Myoviridae infecting S. aureus. Phage S24-1, 44AHJD, and 66 belong to the family of Podoviridae infecting S. aureus. Phage JD419, 42e, and 3A belong to the family of Siphoviridae infecting S. aureus.
Phages pose the risk of contributing to the virulence of the target bacterial strains, introducing antibiotic resistance; the deployment of phages carrying these undesired genes in phage therapy should thus be avoided (Cui et al., 2017b). Therefore, it is essential to analyze phage genomes before using them in the clinic (Cui et al., 2017b). In the 45 kb genome of JD419, we predicted 65 ORFs, none of which encode any known virulence genes that confer antibiotic resistance. However, the results revealed putative genes encoding proteins like the CI repressor, an anti-repressor, and integrase. Such proteins are present in temperate phages. Using the phage genome analysis program PHACTS, JD419 was confidently classified as a temperate phage. Further indication that JD419 can undergo lysogeny is the observation that lysis is incomplete in some strains, as observed in the inhibition zones formed in some instances were turbid. In the case of other clinical isolates, the plaques appear transparent (identical to strictly lytic phages). When studying JD419 and the strain MR-84, we observe that the phage induces replication and lysis while it appears not to enter a dormant prophage state. Before using JD419 in therapy, all prophage-related genes should be removed by genetic engineering to avoid potential lysogeny and enhance bactericidal activity. Such approaches have been reported for Mycobacterium (Dedrick et al., 2019) as well as for Salmonella (Shin et al., 2014), while there are reports that showed the potential and possible use of temperate phages in therapy (Chung et al., 2012; Monteiro et al., 2019). Once the issue with lysogeny has been solved, the microbiological and biochemical characterization of JD419 indicates its potential to be used for phage therapy: JD419 has a broad host range tested on mostly multidrug-resistant clinical S. aureus strains.
Phage JD419 can infect approximately 44% of tested isolates that belong to various MLST types. However, bactericidal activity toward different strains varied significantly. As shown in Figures 3, 4, JD419 could perturb some strains’ growth, yet lysis was not fast and efficient. A high MOI was chosen to ensure that more than one phage infects each bacterial cell simultaneously in the study. Growth inhibition of the host strain at a MOI = 10 was observed. As shown in Figure 3A and Supplementary Figure 1A, it could more persistently inhibit N315 at MOI = 100 than at MOI = 10. While this effect might be absent in a genetically modified JD419 where prophage genes have been removed, the phage should still be tested in vitro for its bactericidal activity toward a target S. aureus strain before using it for therapy (Cui et al., 2019). Establishing a phage library containing different kinds of phages covering most strains would overcome the general disadvantage of phages being less practical than antibiotics (Ding et al., 2018). To avoid the emergence of phage-resistant bacteria, a highly efficient phage or a so-called phage cocktail of efficient phages could be used in phage therapy. In addition to their ability to infect and kill a target strain, phages should display stability to withstand the preparation and application, i.e., in clinical solutions or formulations for subsequent patient administration. Phage JD419 exhibits a fair degree of stability toward pH and temperature, indicating that the phage might be a suitable candidate as a therapeutic phage. The lability toward acidic pH values indicates that the phage might need to be encapsulated if it was to be delivered through the oral route (Loh et al., 2020b); however, S. aureus infections are usually observed in environments that display a pH close to neutral, easily tolerated by JD419, such as in the epidermis or the blood.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT899504.
Author Contributions
ZC and XG conceived and designed the experiments. ZC and TF performed the experiments. TF, BL, SL, ML, MS, and ZC analyzed the data. ZC, KD, BL, TF, YZ, and MS contributed reagents, materials, and analysis tools. TF, BL, SL, and ZC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the outstanding medical youth program A of Shanghai General Hospital (No. 06N1702002) and the National Natural Science Foundation of China (Nos. 81872540 and 31500154).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.602902/full#supplementary-material
References
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. (2011). Phage treatment of human infections. Bacteriophage 1 66–85. 10.4161/bact.1.2.15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B., Chen Y. G., Zhang L., Na Xu Y. L., Wang X., Liu J., et al. (2013). Momordica charantia (Bitter Melon) reduces obesity-associated macrophage and mast cell infiltration as well as inflammatory cytokine expression in adipose tissues. PLoS One 8:e84075. 10.1371/journal.pone.0084075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Borodovsky M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33 W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I. Y., Sim N., Cho Y. H. (2012). Antibacterial efficacy of temperate phage-mediated inhibition of bacterial group motilities. Antimicrob. Agents Chemother. 56 5612–5617. 10.1128/aac.00504-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Feng T., Gu F., Li Q., Dong K., Zhang Y., et al. (2017a). Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai. China. Virol. J. 14:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Guo X., Dong K., Zhang Y., Li Q., Zhu Y., et al. (2017b). Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci. Rep. 7:41259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Guo X., Feng T., Li L. (2019). Exploring the whole standard operating procedure for phage therapy in clinical practice. J. Transl. Med. 17:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson E. K., Bari S. M. N., Dandu V. S., Kriznik J. M., Scopel S. E., Stanley S. P., et al. (2019). Draft Genome Sequences of Staphylococcus Podophages JBug18, Pike, Pontiff, and Pabna. Microbiol. Resour. Announc. 8: e00054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick R. M., Guerrero-Bustamante C. A., Garlena R. A., Russell D. A., Ford K., Harris K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25 730–733. 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27 4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Li Q., Guo M., Dong K., Zhang Y., Guo X., et al. (2018). Prevention of dermal abscess formation caused by Staphylococcus aureus using phage JD007 in nude mice. Front. Microbiol. 9:1553. 10.3389/fmicb.2018.01553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Pantucek R., Holtfreter S., Schulte B., Zink M., Grumann D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191 3462–3468. 10.1128/jb.01804-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmer H., Gerlach D., Wolz C. (2019). Temperate phages of Staphylococcus aureus. Microbiol. Spectr. 7. 10.1128/microbiolspec.GPP3-0058-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano H., Inoue Y., Takasago T., Kobayashi H., Furusawa T., Taniguchi K., et al. (2018). Bacteriophage PhiSA012 has a broad host range against Staphylococcus aureus and effective lytic capacity in a mouse mastitis model. Biology 7:8. 10.3390/biology7010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Liu Q., Wang R., Luo T., Guo X., Xu M., et al. (2020). Identification of a novel phage targeting methicillin-resistant Staphylococcus aureus In vitro and In vivo. Microb. Pathog. 149:104317. 10.1016/j.micpath.2020.104317 [DOI] [PubMed] [Google Scholar]
- Kazmierczak Z., Gorski A., Dabrowska K. (2014). Facing antibiotic resistance: Staphylococcus aureus phages as a medical tool. Viruses 6 2551–2570. 10.3390/v6072551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko M., Kuptsov N., Gorodnichev R., Bespiatykh D., Guliaev A., Letarova M., et al. (2020). Contribution of podoviridae and myoviridae bacteriophages to the effectiveness of anti-staphylococcal therapeutic cocktails. Sci. Rep. 10:18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard K. R., Kleinschmidt A. K., Agabian-Keshishian N., Shapiro L., Maizel J. V., Jr. (1972). Structural studies on the capsid of Caulobacter crescentus bacteriophage phiCbK. J. Mol. Biol. 71 201–216. 10.1016/0022-2836(72)90346-4 [DOI] [PubMed] [Google Scholar]
- Leptihn S. (2019). Welcome back to the pre-penicillin era. why we desperately need new strategies in the battle against bacterial pathogens. Infect. Microbes Dis. 1:33. 10.1097/im9.0000000000000009 [DOI] [Google Scholar]
- Limbago B. M., Kallen A. J., Zhu W., Eggers P., McDougal L. K., Albrecht V. S. (2014). Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J. Clin. Microbiol. 52 998–1002. 10.1128/jcm.02187-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Pop M. (2009). ARDB–antibiotic resistance genes database. Nucleic Acids Res. 37 D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobocka M., Hejnowicz M. S., Dabrowski K., Gozdek A., Kosakowski J., Witkowska M., et al. (2012). Genomics of staphylococcal twort-like phages–potential therapeutics of the post-antibiotic era. Adv. Virus Res. 83 143–216. 10.1016/b978-0-12-394438-2.00005-0 [DOI] [PubMed] [Google Scholar]
- Loh B., Chen J., Manohar P., Yu Y., Hua X., Leptihn S. (2020a). A biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front. Microbiol. 11:579802. 10.3389/fmicb.2020.579802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh B., Gondil V. S., Manohar P., Khan F. M., Yang H., Leptihn S. (2020b). Encapsulation and delivery of therapeutic phages. Appl. Environ. Microbiol. AEM.01979-20. 10.1128/AEM.01979-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S., Fabijan A. P., Ho J., Lin R. C. Y., Ben Zakour N. L., Dugan C., et al. (2019). Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 200 1179–1181. 10.1164/rccm.201904-0839le [DOI] [PubMed] [Google Scholar]
- Manohar P., Loh B., Athira S., Nachimuthu R., Hua X., Welburn S. C., et al. (2020a). Secondary bacterial infections during pulmonary viral disease: phage therapeutics as alternatives to antibiotics? Front. Microbiol. 11:1434. 10.3389/fmicb.2020.01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar P., Loh B., Nachimuthu R., Hua X., Welburn S. C., Leptihn S. (2020b). Secondary bacterial infections in patients with viral pneumonia. Front. Med. 7:420. 10.3389/fmed.2020.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallin S., Sarker S. A., Sultana S., Oechslin F., Brussow H. (2018). Metagenome analysis of Russian and Georgian pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 20 3278–3293. 10.1111/1462-2920.14310 [DOI] [PubMed] [Google Scholar]
- McNair K., Bailey B. A., Edwards R. A. (2012). PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28 614–618. 10.1093/bioinformatics/bts014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R. C., Jr. (2012). MRSA: the first half century. J. Antimicrob. Chemother. 67 4–11. 10.1093/jac/dkr437 [DOI] [PubMed] [Google Scholar]
- Monteiro R., Pires D. P., Costa A. R., Azeredo J. (2019). Phage therapy: going temperate? Trends Microbiol. 27 368–378. 10.1016/j.tim.2018.10.008 [DOI] [PubMed] [Google Scholar]
- O’Neill J. (2014). Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Review on Antimicrobial Resistance. [Google Scholar]
- Rahman M., Kim S., Kim S. M., Seol S. Y., Kim J. (2011). Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 27 1087–1093. 10.1080/08927014.2011.631169 [DOI] [PubMed] [Google Scholar]
- Schmuki M. M., Erne D., Loessner M. J., Klumpp J. (2012). Bacteriophage P70: unique morphology and unrelatedness to other listeria bacteriophages. J. Virol. 86 13099–13102. 10.1128/jvi.02350-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R. T., Biswas B., Gill J. J., Hernandez-Morales A., Lancaster J., Lessor L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61 e00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Lee J. H., Yoon H., Kang D. H., Ryu S. (2014). Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC. Appl. Environ. Microbiol. 80 374–384. 10.1128/aem.02279-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Mi Z., Yin X., Fan H., An X., Zhang Z., et al. (2013). Characterization of Enterococcus faecalis Phage IME-EF1 and its endolysin. PLoS One 8:e80435. 10.1371/journal.pone.0080435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT899504.