Abstract
HLA polymorphism is one of the genetic factors that may be associated with variations in susceptibility to COVID-19 infection. In this study, the frequency of HLA alleles among Saudi patients infected with COVID-19 was examined. The association with infection susceptibility and mortality was evaluated. This study included 135 Saudi COVID-19-infected patients (106 recovered and 29 died) who were admitted to hospitals because of their symptoms, and 135 healthy controls. HLA class I (A, B, C) and class II (DRB1, DQB1) genotyping was performed using the molecular method (PCR-rSSO). In this study, there was a significant increase in the frequency of HLA-A*01, B*56 and C*01 among infected patients compared to the control group (12.1% vs. 5.2%, p = 0.004, 3.7% vs. 0%, p = 0.006, 4.4% vs. 1.5%, p = 0.042, respectively). Moreover, there was a significant increase in the frequency of HLA-A*03 and C*06 among fatal patients compared to infected patients (13.8% vs. 5.7%, p = 0.036, 32.8% vs. 17.5%, p = 0.011, respectively). In terms of HLA class II, HLA-DRB1*04 was significantly higher in the control group compared to infected patients (27.4% vs. 16.3%, p = 0.002), while HLA-DRB1*08 was significantly higher in the infected group compared to the control (4.8% vs. 0.7%, p = 0.004). After statistical correction of the p value, A*01, B*56, DRB1*04 and DRB1*08 remained statistically significant (pc = 0.04, pc = 0.03, pc = 0.014 and pc = 0.028). This initial data suggested that individual HLA genotypes might play a role in determining susceptibility to COVID-19 infection and infection outcome. However, examining a larger sample size from different populations is required to determine a powerful association for clinical application.
Keywords: HLA polymorphism, COVID-19, Saudi, Susceptibility, Mortality
1. Introduction
A novel β-coronavirus strain which was identified in Wuhan, China in December 2019 caused more than 30 million infections and in excess of one million deaths by October 2020 (https://www.worldometers.info/coronavirus/). The new virus, called Severe Acute Respiratory Syndrome coronavirus-2 (SARS-COV2), mainly infects the respiratory system, causing flu-like symptoms and acute respiratory distress in mild cases. In severe cases, multi-organ dysfunction and death can occur (Guo et al., 2020). The virus showed different geographical distributions, with significantly more cases and deaths reported in some areas compared to others. Additionally, there were variations in the severity of the infection between different individuals, with some patients experiencing mild or even no symptoms, while others needed intensive care support (Guo et al., 2020). The most affected areas were America with more than 16 million cases, followed by Asia with more than 10 million cases and Europe with more than 6 million cases reported by October 1, 2020. In this report, India was the most affected country in Asia with more than 6 million cases reported, while the lowest number of cases was in Saudi Arabia (<500,000 cases) (https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-ca-s). Although different factors such as age, comorbidity, and adaptive immunity might play a role in viral infections, genetic factors have a fundamental role in determining individuals’ susceptibility to any infectious agents (Jordan et al., 2020).
Human Leukocyte Antigen (HLA) molecules have been extensively studied as a genetic factor that might affect virus progression or resistance (Lin et al., 2003; Blackwell et al., 2009). Because of the role of HLA molecules in antigen presentation, host immune response will vary according to the selected peptide presented by individual HLA molecules. The interaction between the HLA/peptide complex and T-lymphocytes can induce an efficient immune response to eradicate the virus, while other interactions might induce weak T-lymphocyte activation leading to infection progression (Blackwell et al., 2009). Therefore, HLA polymorphism between individuals or populations can have a role in the response to certain viruses. In other words, viruses which might be fatal to certain populations may be less severe in others depending on their genetic variance and the type of HLA molecules. Many research groups have studied the association between HLA genotypes and respiratory viral infections, however no definite association has been identified between a specific HLA type and susceptibility to viral infection (Wang et al., 2011; Keicho et al., 2009), (Røder et al., 2008). This could be attributed to the small sample size of these studies or to polymorphism of HLA alleles, where more than one antigen might be involved in infection susceptibility for certain populations. In this study, the frequency of HLA class I (A, B, C) and HLA class II (DR, DQ) alleles between Saudi COVID-19-positive patients was examined. The positive group was divided into infected but recovered patients and fatal patients, and these were compared to a control group. To our knowledge, this is the first study from an Arab country to consider the association between HLA genotypes and COVID-19 infection susceptibility and mortality.
2. Materials and methods
2.1. Research subjects
This study included 135 COVID-19-positive patients. All patients were Saudi and were admitted to MOH government hospitals due to their symptoms. Patients were diagnosed by a polymerase chain reaction test for viral RNA from a nasopharyngeal swab. Of those, 106 patients were classified as having mild/moderate symptoms and suffering from fever, cough, headache and mild dyspnea. Patients were admitted to the hospitals for at least 5 days before recovery and discharge. Twenty-nine patients were classified as severe/critical cases and were admitted in the ICU; these patients died due to low oxygen saturation and acute respiratory distress syndrome. The study also included 135 control subjects who were healthcare workers from the same region as the patients. The control group was defined as individuals with no history of symptoms for the past 3 months, i.e. either asymptomatic or negative. The study was performed after obtaining appropriate ethical approval from the Ministry of Health, Saudi Arabia, research ethical committee, IRP registration number with KACST, KSA: H-02-J-002. Blood samples were collected from participants after they signed a consent form.
2.2. Genomic DNA extraction
Blood samples were collected using EDTA tubes for DNA extraction. DNA extraction was performed using an EZ1 Advanced XL instrument (Qiagen, USA) located at King Fahd Medical Research Center, King Abdul-Aziz University, Jeddah. Samples with a DNA concentration of more than 25 ng/μl and DNA purity of more than 1.8 for a 260/280 ratio were used for the HLA genotyping. DNA was stored at −80 °C until HLA genotyping.
2.3. HLA genotyping
Both HLA class I and HLA class II genotyping was performed at a low resolution typing using the reverse SSO-PCR method (One Lambda, USA). In brief, the method is based on the amplification of an HLA-specific locus using a biotinylated primer. An HLA-specific allele was then determined using oligonucleotide probes attached to multiplex polystyrene beads as a detection method. The beads were run on Luminex FLEXMAP 3D (Luminex Corporation, USA), at King Fahd Medical Research Center, Jeddah. HLA alleles assignment was performed using HLA fusion software 4.3 (One Lambda, USA).
2.4. Statistical analysis
The control and infected groups were tested for Hardy-Weinberg equilibrium using Guo and Thomson method implemented in Pypop (www.pypop.org) (Solberg et al., 2008). Absolute count and percentage were applied to describe allele frequency. Arithmetic mean and standard deviation were used to describe continuous variables (e.g. age). The chi-squared test was applied to test for the association between two categorical variables. Fisher's exact test was performed in case of having a frequency of less than 5 in one or more of the cells. One-way analysis of variance and post-hoc Tukey's test were applied to compare the mean of and between the three compared groups. Statistical significance was determined at p < 0.05. Statistical analyses were performed using the Statistical Package for Social Sciences, version 25 software. The Bonferroni correction was applied to the p value and calculated by multiplying by the number of alleles with a frequency >5%. Relative risks with corresponding 95% confidence intervals (95% CI) were calculated according to Woolf's formula (Woolf, 1955). Relative risks <1 indicated protection while >1 indicated an increased risk.
3. Results
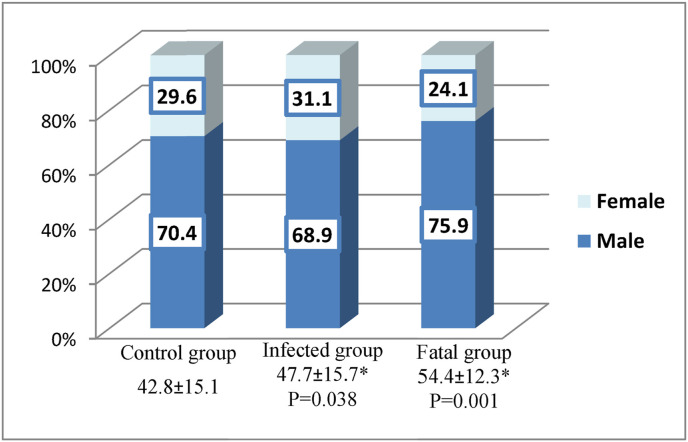
The study included 135 Saudi COVID-19-positive patients divided into two subgroups: 106 patients with COVID-19 infection who were admitted at Jeddah hospitals because of their symptoms and who recovered, and 29 infected patients who died (fatal). Those were compared to 135 healthy volunteers as a control group. The results of HLA-A, B, C, DRB1 and DQB1 genotyping for both groups were tested for Hardy-Weinberg equilibrium, Appendix A, Appendix A The control group was fit for the equilibrium while there was a significant deviation from the Hardy-Weinberg equilibrium at the HLA-B and HLA-C loci among the infected patients. Haplotypes of the control and infected groups were described in Appendix A, Appendix A with a comparison between the most common haplotypes in Appendix A, Appendix A. There were no differences among the three groups with respect to gender distribution, as shown in Fig. 1 . However, COVID-19 infection showed a male preference as 70% of the infected group was male. The ages of subjects in the control group were significantly lower than the infected patients group (p = 0.038) and the fatal group (p = 0.001), with no statistical significance between the infected and fatal groups.
Fig. 1.
Age and gender distribution of the study participants. Age was represented in the X-axis as mean ± standard deviation. Statistical analysis was performed between groups using one-way ANOVA followed by a post-hoc Tukey's test. *Statistically significant compared to the control group.
3.1. HLA allele frequency
3.1.1. HLA-A allele frequency between groups
HLA-A genotypes were compared among the three groups (control, infected and fatal). The HLA-A frequency in each group is shown in Table 1 . The most commonly reported HLA-A alleles among the control group were A*02 (26.3%), A*24 (10.4%), and A*26 (10%), while among the infected group they were A*02 (22.9%), A*01 (12.3%) and A*68 (8.5%). Among the fatal group, the most common HLA-A alleles were A*02 (24.1%), A*03 (13.8%), and A*68 (12.1%). The frequency of HLA-A*01 was significantly higher in the infected group compared to the control group (12.3% vs. 5.2%, p = 0.004). The increase in the frequency of this allele remained statistically significant after correction of the p value (pc = 0.032). However, the frequency of HLA-A*01 in the infected group was not statistically different from the fatal group (p = 0.968). The HLA-A*24 frequency was significantly higher in the control and infected groups compared to the fatal group (10.4% vs. 1.7%, p = 0.021; 8.1% vs. 1.7%, p = 0.029, respectively). Additionally, HLA-A*26 was significantly higher in the control group compared to the infected group (p = 0.021). On the other hand, the frequency of HLA-A*03 was higher among the fatal group than the infected group (13.8% vs. 5.7%, p = 0.036). The frequency of this allele was not significant between the control and infected groups or between the control and fatal groups (p = 0.871, p = 0.143, respectively). After Bonferroni correction, the differences in these alleles between groups were non-significant.
Table 1.
Frequency of HLA-A genotype between control, COVID-19 infected and fatal groups. Data was analyzed using the chi-squared test or Fisher's exact test, in case of having a frequency of less than 5 in one or more of the cells at p < 0.05. ⱶChi-squared test, *Fisher's exact test, NA: Not applicable. Significant p values are in bold.
| Control N = 270 N (%) |
COVID-19 infected N = 270 N (%) |
Fatal N = 58 N (%) |
Control vs. COVID-19 infected P value |
Control vs. fatal P value |
COVID-19 Infected vs. fatal P value |
|
|---|---|---|---|---|---|---|
| A*01 | 14 (5.2) | 33 (12.3) | 7 (12.1) | 0.004ⱶ | 0.052ⱶ | 0.968ⱶ |
| A*02 | 71 (26.3) | 62 (22.9) | 14 (24.1) | 0.552ⱶ | 0.734ⱶ | 0.990ⱶ |
| A*03 | 21 (7.8) | 20 (7.4) | 8 (13.8) | 0.871ⱶ | 0.143ⱶ | 0.036ⱶ |
| A*11 | 10 (3.7) | 7 (2.6) | 1 (1.7) | 0.460ⱶ | 0.390* | 0.535* |
| A*23 | 16 (5.9) | 15 (5.6) | 5 (8.6) | 0.853ⱶ | 0.447ⱶ | 0.199ⱶ |
| A*24 | 29 (10.7) | 22 (8.1) | 1 (1.7) | 0.373ⱶ | 0.021* | 0.029* |
| A*26 | 26 (9.6) | 13 (4.8) | 2 (3.4) | 0.021ⱶ | 0.082* | 0.444ⱶ |
| A*29 | 5 (1.9) | 3 (1.1) | 1 (1.7) | 0.476* | 0.713* | 0.517ⱶ |
| A*30 | 15 (5.6) | 21 (7.8) | 2 (3.4) | 0.301ⱶ | 0.384* | 0.130ⱶ |
| A*31 | 9 (3.3) | 13 (4.8) | 2 (3.4) | 0.384ⱶ | 0.609* | 0.444ⱶ |
| A*32 | 11 (4.1) | 16 (5.9) | 3 (5.2) | 0.676ⱶ | 0.464* | 0.556ⱶ |
| A*33 | 15 (5.6) | 15 (5.6) | 2 (3.4) | NA | 0.394* | 0.338ⱶ |
| A*34 | 2 (0.7) | 2 (0.7) | 1 (1.7) | NA | 0.443* | 0.384ⱶ |
| A*66 | 1 (0.4) | 0 (0.0) | 0 (0.0) | 0.563* | 0.823* | NA |
| A*68 | 22 (8.1) | 23 (8.5) | 7 (12.1) | 0.876* | 0.340ⱶ | 0.274ⱶ |
| A*69 | 0 (0.0) | 1 (0.37) | 1 (1.7) | 0.5638 | 0.177* | 0.215* |
| A*74 | 3 (1.1) | 4 (1.5) | 1 (1.7) | 0.704* | 0.543* | 0.622* |
3.1.2. HLA-B allele frequency between groups
Table 2 shows that the most common HLA-B alleles in the control and infected groups were B*51 (19.3% and 17.4%, respectively), B*50 (13% and 13.7%, respectively) and B*35 (11.9% and 10%, respectively), whereas among the fatal group, the most common HLA-B alleles were B*50 (20.7%), B*51 (15.5%), B*15 and B*38 (6.9%). HLA-B*07 was significantly higher in the control group than in the infected group (6.7% vs. 3%, p = 0.044); this became non-significant after p value correction (pc = 0.22). On the other hand, B*56 and B*57 were significantly higher in the infected group compared to the control group (3.7% vs. 0.0%, p = 0.006 and 2.8% versus 0.4%, p = 0.019), respectively. B*38 and B*47 were observed significantly among the fatal group compared to the control group (6.9% vs. 1.5%, p = 0.036 and 3.4% vs. 0.0%, p = 0.031), respectively. B*47 was observed only among the fatal group and was not present in the other groups (3.4% versus 0.0%, p = 0.046). However, only HLA-B*56 remained statistically significant after p value correction (pc = 0.03).
Table 2.
Frequency of HLA-B genotype between control, COVID-19 infected and fatal groups. Data was analyzed using the chi-squared test or Fisher's exact test, in case of having a frequency of less than 5 in one or more of the cells at p < 0.05. ⱶChi-squared test, *Fisher's exact test, NA: Not applicable. Significant p values are in bold.
| Control N = 270 N (%) |
COVID-19 infected N = 270 N (%) |
Fatal N = 58 N (%) |
Control vs. COVID-19 infected P value |
Control vs. fatal P value |
COVID-19 Infected vs. fatal P value |
|
|---|---|---|---|---|---|---|
| B*07 | 18 (6.7) | 8 (3.0) | 3 (5.2) | 0.044ⱶ | 0.473* | 0.234* |
| B*08 | 16 (5.9) | 19 (7.0) | 1 (1.7) | 0.600ⱶ | 0.163* | 0.056* |
| B*13 | 8 (3.0) | 7 (2.6) | 1 (1.7) | 0.793ⱶ | 0.507* | 0.535* |
| B*14 | 6 (2.2) | 3 (1.4) | 0 (0.0) | 0.313* | 0.308* | 0.483* |
| B*15 | 9 (3.3) | 16 (5.9) | 4 (6.9) | 0.152ⱶ | 0.181* | 0.463* |
| B*18 | 7 (2.6) | 5 (1.5) | 1 (1.7) | 0.361* | 0.572* | 0.622* |
| B*27 | 5 (1.9) | 3 (1.4) | 0 (0.0) | 0.476* | 0.375* | 0.483* |
| B*35 | 32 (11.9) | 27 (10.0) | 3 (5.2) | 0.490ⱶ | 0.098* | 0.125* |
| B*37 | 3 (1.1) | 8 (3.0) | 2 (3.4) | 0.128* | 0.215* | 0.542* |
| B*38 | 4 (1.5) | 9 (3.3) | 4 (6.9) | 0.160* | 0.036* | 0.103* |
| B*39 | 8 (3.0) | 7 (2.6) | 2 (3.4) | 0.793ⱶ | 0.553* | 0.465* |
| B*40 | 10 (3.7) | 5 (1.9) | 3 (5.2) | 0.190ⱶ | 0.412* | 0.068* |
| B*41 | 8 (3.0) | 4 (1.5) | 2 (3.4) | 0.243* | 0.553* | 0.203* |
| B*42 | 3 (1.1) | 3 (1.1) | 1 (1.7) | NA | 0.543* | 0.517* |
| B*44 | 3 (1.1) | 5 (2.4) | 0 (0.0) | 0.476* | 0.557* | 0.295* |
| B*45 | 0 (0.0) | 2 (0.7) | 1 (1.7) | 0.558* | 0.177* | 0.384* |
| B*46 | 1 (0.4) | 1 (0.37) | 1 (1.7) | NA | 0.323* | 0.215* |
| B*47 | 0 (0.0) | 2 (0.8) | 2 (3.4) | 0.558* | 0.031* | 0.046* |
| B*48 | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0.563* | NA | 0.785* |
| B*49 | 6 (2.2) | 7 (2.6) | 1 (1.7) | 0.779ⱶ | 0.641* | 0.535* |
| B*50 | 35 (13.0) | 37 (13.7) | 12 (20.7) | 0.800ⱶ | 0.128ⱶ | 0.067ⱶ |
| B*51 | 52 (19.3) | 47 (17.4) | 9 (15.5) | 0.578ⱶ | 0.506ⱶ | 0.668ⱶ |
| B*52 | 8 (3.0) | 4 (1.5) | 1 (1.7) | 0.243* | 0.507* | 0.622* |
| B*53 | 12 (4.4) | 9 (3.3) | 0 (0.0) | 0.504ⱶ | 0.093* | 0.109* |
| B*55 | 2 (0.7) | 1 (0.4) | 0 (0.0) | 0.563* | 0.677* | 0.785* |
| B*56 | 0 (0.0) | 10 (3.7) | 0 (0.0) | 0.006* | NA | 0.085* |
| B*57 | 1 (0.4) | 8 (2.8) | 2 (3.4) | 0.019* | 0.082* | 0.542* |
| B*58 | 11 (4.1) | 9 (3.0) | 1 (0.0) | 0.824ⱶ | 0.341* | 0.328* |
| B*73 | 0 (0.0) | 1 (0.4) | 0 (0.0) | 0.563* | NA | 0.785* |
| B*81 | 1 (0.4) | 0 (0.0) | 1 (0.0) | NA | 0.323* | 0.215* |
| B*82 | 1 (0.4) | 1 (0.4) | 0 (0.0) | NA | 0.823* | 0.785* |
3.1.3. HLA-C allele frequency between groups
From Table 3 , the most common HLA-C alleles in the control and infected groups were C*07 (19.6% and 23.7%, respectively), C*06 (19.3% and 20.7%, respectively) and C*04 (15.9% and 13.7%, respectively), whereas among the fatal group, the most common alleles were C*06 (32.8%), C*07 (27.6%) and C*15 (10.3%). C*01 was significantly reported in the infected group compared to the control group (4.4% vs. 1.5%, p = 0.042), with no statistical difference from the fatal group. On the other hand, C*16 was significantly higher in the control group than in the infected group (8.1% vs. 3.3%, p = 0.016) but with no statistical difference between the infected and fatal groups. The frequency of the HLA-C*04 allele was significantly lower in the fatal group compared to both the control group and the infected group (5.2% vs. 15.9%, p = 0.020, 5.2% vs. 13.7%, p = 0.021, respectively). The frequency of the HLA-C*06 allele was significantly higher in the fatal group compared to the control group (32.8% vs. 19.3%, p = 0.024) and to the infected group (32.8% vs. 17.5%, p = 0.011). There were no statistical differences between groups after p value correction.
Table 3.
Frequency of HLA-C genotype between control, COVID-19 infected and fatal groups. Data was analyzed using the chi-squared test or Fisher's exact test in case of having a frequency of less than 5 in one or more of the cells, at p < 0.05. ⱶChi-squared test, *Fisher's exact test, NA: Not applicable. Significant p values are in bold.
| Control N = 270 N (%) |
COVID-19 infected N = 270 N (%) |
Fatal N = 58 N (%) |
Control vs. COVID-19 infected P value |
Control vs. fatal P value |
COVID-19 Infected vs. fatal P value |
|
|---|---|---|---|---|---|---|
| C*01 | 4 (1.5) | 12 (4.4) | 1 (1.7) | 0.042* | 0.625* | 0.229* |
| C*02 | 4 (1.5) | 5 (1.9) | 0 (0.0) | 0.737* | 0.457* | 0.295* |
| C*03 | 13 (4.8) | 13 (4.8) | 3 (5.2) | NA | 0.561* | 0.556* |
| C*04 | 43 (15.9) | 37 (13.7) | 3 (5.2) | 0.586ⱶ | 0.020* | 0.021* |
| C*05 | 4 (1.5) | 4 (1.5) | 1 (1.7) | NA | 0.625* | 0.622* |
| C*06 | 52 (19.3) | 56 (20.7) | 19 (32.8) | 0.667ⱶ | 0.024ⱶ | 0.011ⱶ |
| C*07 | 53 (19.6) | 64 (23.7) | 16 (27.6) | 0.251ⱶ | 0.177ⱶ | 0.433ⱶ |
| C*08 | 7 (2.6) | 6 (2.2) | 0 (0.0) | 0.779ⱶ | 0.253* | 0.231* |
| C*12 | 14 (5.2) | 14 (5.1) | 3 (5.2) | NA | 0.648* | 0.648* |
| C*14 | 6 (2.2) | 10 (3.7) | 0 (0.0) | 0.310ⱶ | 0.308* | 0.085* |
| C*15 | 36 (13.3) | 33 (12.2) | 6 (10.3) | 0.699ⱶ | 0.537ⱶ | 0.622ⱶ |
| C*16 | 22 (8.1) | 9 (3.3) | 3 (5.2) | 0.016ⱶ | 0.322* | 0.300* |
| C*17 | 11 (4.1) | 6 (2.2) | 3 (5.2) | 0.218ⱶ | 0.464* | 0.116* |
| C*18 | 1 (0.4) | 1 (0.4) | 0 (0.0) | NA | 0.823* | 0.785* |
3.1.4. HLA class II allele frequency between groups
The most common HLA-DRB1 alleles in the control and infected groups were DRB1*04 (27.4% and 16.3%, respectively), DRB1*03 (15.9% and 15.1%, respectively) and DRB1*07 (11.1% and 15.6%, respectively). The most common HLA-DRB1 alleles reported in the fatal group were DRB1*07 (19%), DRB1*03 (13.8%) and DRB1*04, DRB1*11 & DRB1*13 (12.1%). DRB1*04 was significantly higher in the control group compared to both the infected group (27.4% vs. 16.3%, p = 0.002, pc = 0.014) and the fatal group (27.4% vs. 12.1%, p = 0.014, pc = 0.098) with no statistical significance between the infected and fatal groups. DRB1*08 was significantly higher in the infected group compared to the control group (5.2% vs. 0.7%, p = 0.004, pc = 0.028) with p value of 0.189 when compared to the fatal group. Table 4 shows the frequency of HLA-DRB1 in each group.
Table 4.
Frequency of HLA-DRB1 genotype between control, COVID-19 infected and fatal groups. Data was analyzed using the chi-squared test or Fisher's exact test, in case of having a frequency of less than 5 in one or more of the cells at p < 0.05. ⱶChi-squared test, *Fisher's exact test, NA: Not applicable. Significant p values are in bold.
| Control N = 270 N (%) |
COVID-19 infected N = 270 N (%) |
Fatal N = 58 N (%) |
Control vs. COVID-19 infected P value |
Control vs. fatal P value |
COVID-19 Infected vs. fatal P value |
|
|---|---|---|---|---|---|---|
| DRB1*01 | 9 (3.3) | 9 (3.3) | 3 (5.2) | NA | 0.359* | 0.300* |
| DRB1*03 | 43 (15.9) | 41 (15.1) | 8 (13.8) | 0.812ⱶ | 0.684ⱶ | 0.739ⱶ |
| DRB1*04 | 74 (27.4) | 44 (16.3) | 7 (12.1) | 0.002ⱶ | 0.014ⱶ | 0.325ⱶ |
| DRB1*07 | 30 (11.1) | 42 (15.6) | 11 (19.0) | 0.129ⱶ | 0.101ⱶ | 0.419ⱶ |
| DRB1*08 | 2 (0.7) | 13 (4.8) | 1 (1.7) | 0.004* | 0.433* | 0.189* |
| DRB1*09 | 0 (0.0) | 4 (1.5) | 1 (1.7) | 0.176* | 0.177* | 0.622* |
| DRB1*10 | 11 (4.1) | 7 (2.6) | 1 (1.7) | 0.338ⱶ | 0.341* | 0.535* |
| DRB1*11 | 26 (9.6) | 23 (8.5) | 7 (12.1) | 0.653ⱶ | 0.575ⱶ | 0.274ⱶ |
| DRB1*12 | 2 (0.7) | 4 (1.5) | 1 (1.7) | 0.412* | 0.443* | 0.622* |
| DRB1*13 | 22 (8.1) | 35 (13.0) | 7 (12.1) | 0.687ⱶ | 0.340ⱶ | 0.819ⱶ |
| DRB1*14 | 1 (0.4) | 2 (0.7) | 0 (0.0) | 0.563* | 0.823* | 0.616* |
| DRB1*15 | 25 (9.3) | 24 (8.9) | 6 (10.3) | 0.881ⱶ | 0.798ⱶ | 0.660ⱶ |
| DRB1*16 | 25 (9.3) | 22 (8.1) | 5 (8.6) | 0.647ⱶ. | 0.878ⱶ | 0.882ⱶ |
Regarding HLA-DQB1 frequency, the most common alleles in the control group were DQB1*02 (31.5%), DQB1*03 (30.4%) and DQB1*05 (18.9%), whereas among the infected group, the most frequently reported alleles were DQB1*02 (36.2%), DQB1*03 (32.2%) and DQB1*06 (25.5%). In the fatal group, the most common alleles were DQB1*02 (29.3%), DQB1*03 (22.4%), DQB1*05 (20.7%) and DQB1*06 (20.7%). There were no statistical differences in the frequency of HLA-DQB1 between groups. Table 5 shows the frequency of HLA-DQB1 between different groups.
Table 5.
Frequency of HLA-DQB1 genotype between control, COVID-19 infected and fatal groups. Data was analyzed using the chi-squared test or Fisher's exact test in case of having a frequency of less than 5 in one or more of the cells at p < 0.05. ⱶChi-squared test, *Fisher's exact test, NA: Not applicable.
| Control N = 270 N (%) |
COVID-19 infected N = 270 N (%) |
Fatal N = 58 N (%) |
Control vs. COVID-19 infected | Control vs. fatal | COVID-19 Infected vs. fatal | |
|---|---|---|---|---|---|---|
| DQB1*02 | 85 (31.5) | 81 (30) | 17 (29.3) | 0.709ⱶ | 0.746ⱶ | 0.952ⱶ |
| DQB1*03 | 82 (30.4) | 74 (27.4) | 13 (22.4) | 0.448ⱶ | 0.225ⱶ | 0.336ⱶ |
| DQB1*04 | 8 (3.0) | 9 (3.3) | 4 (6.9) | 0.805ⱶ | 0.144* | 0.103* |
| DQB1*05 | 51 (18.9) | 47 (17.4) | 12 (20.7) | 0.655ⱶ | 0.752ⱶ | 0.513ⱶ |
| DQB1*06 | 44 (16.3) | 57 (21.1) | 12 (20.7) | 0.151ⱶ | 0.420ⱶ | 0.929ⱶ |
3.1.5. Association between HLA genotype and relative risk of COVID-19 infection
The risks associated with the identified alleles and increased incidence of infection were determined, see Table 6 . The relative risk was calculated using the most common HLA in the control group as a reference. Compared to A*02, participants with A*01 were at significant risk of being infected (relative risk “RR” = 1.47, 95% CI: 1.14–1.90, p = 0.011). Considering B*51 as a reference category, patients with B*56 were at almost double the risk of being infected (RR = 2.11, 95% CI: 1.71–2.59, p = 0.001). As opposed to C*07, patients with C*16 had a lower significant risk of infection (RR = 0.53, 95% CI: 0.30–0.94, p = 0.015). Compared to DRB1*04, patients with DRB1*08 were at higher risk of becoming infected with SARS-COV2 (RR = 2.32, 95% CI: 1.71–3.16, p < 0.001).
Table 6.
HLA genotype predictors of infection. Common HLA in the control group was used as a reference genotype to assess the relative risk at p < 0.05.
| Control | COVID-19 infected | RR (95% CI) | p-value | |
|---|---|---|---|---|
| A*02a A*01 A*26 |
17 (26.3) 14 (5.2) 27 (10.0) |
65 (24.1) 33 (12.2) 13 (4.8) |
1.0 1.47 (1.14–1.90) 0.68 (0.42–1.10) |
– 0.011 0.104 |
| B*51a B*07 B*56 B*57 |
52 (19.3) 18 (6.7) 0 (0.0) 1 (0.4) |
47 (17.4) 8 (3.0) 10 (3.7) 8 (3.0) |
1.0 0.65 (0.35–1.20) 2.11 (1.71–2.59) 1.87 (1.37–2.55) |
– 0.182 0.001 0.032 |
| C*07a C*01 C*16 |
53 (19.6) 4 (1.5) 22 (8.1) |
64 (23.7) 12 (4.4) 9 (3.3) |
1.0 1.37 (0.99–1.90) 0.53 (0.30–0.94) |
– 0.178 0.015 |
| DRB1*04a DRB1*08 |
74 (27.4) 2 (0.7) |
44 (16.3) 13 (4.8) |
1.0 2.32 (1.71–3.16) |
– <0.001 |
RR: Relative risk.
CI: Confidence interval.
Reference category.
4. Discussion
The COVID-19 pandemic is still causing a significant number of cases worldwide with a stable rate of mortality not exceeding 7% (Lu et al., 2020). The infection is creating pressure on an economic level due to business disruptions and the burden on healthcare institutions. The virus has spread to most countries at different rates of infection, with some countries reporting thousands of cases daily while others are reporting no more than one thousands cases in total (https://covid19.who.int/). Differences in the rate of infection between different geographical regions cannot be fully understood. HLA polymorphism, with more than 20,000 alleles identified for classical HLA class I and 7000 alleles identified for HLA class II, is one of the genetic factors that may affect individuals’ susceptibility or resistance to COVID-19 infection (https://www.ebi.ac.uk/ipd/imgt/hla/stats.html). Previous studies have shown an association between HLA alleles and susceptibility to various viral infections such as hepatitis, HIV, and respiratory infections (Liu et al., 2003; Hill, 1998; Neumann-Haefelin et al., 2006; Kim et al., 2011). There is conflicting data on the association between HLA and respiratory viral infections. Some studies have shown a positive correlation between classical HLA loci and previous respiratory infections such as Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS-COV). An early study performed on Taiwanese patients infected with SARS-COV1 showed that the HLA-B*46:01 genotype was common among infected patients (Lin et al., 2003). On the other hand, another study showed that patients with HLA-C*15 and HLA-DRB1*03 genotypes were more resistant to SARS infection (Wang et al., 2011). Furthermore, a study from China again showed no HLA contribution to the incidence of SARS infection (Xiong et al., 2008).
In this study, the genotype frequency of HLA, both class I and class II, was identified in a cohort of Saudi patients infected with COVID-19. The positive patients were subdivided into two groups according to the infection outcome: recovered and fatal. They were compared to a control group of healthcare workers with no evidence of infection. The aim of the study was to identify alleles responsible for infection susceptibility and alleles responsible for infection-related mortality. This kind of study might have implications for identifying individuals at higher risk of becoming infected, especially healthcare workers. The strength of our study is the inclusion of a reasonable sample size in addition to samples from fatal patients. Our findings showed a variation in HLA frequency among the three groups in both class I and class II alleles. A significant increase in the frequency of HLA-A*01, B*56 and C*01 between the control and infected groups was observed, while the frequency of HLA-A*03 genotype was higher among fatal patients than in the infected group. The differences in HLA-A*01 and B*56 frequencies between the two groups remain significant after p value correction. These results suggest that the HLA-A*01 and B*56 alleles might be candidates for increasing the incidence of COVID-19 infection. Both alleles were associated with a significant relative risk of infection, estimated as 1.47 and 2.11, respectively. The frequency of these antigens is less common in the Saudi population. Previous HLA data analysis of HLA allele frequencies from the Saudi bone marrow registry showed that the frequency of A*01 was around 7.1%, the frequency of HLA-A*03 was around 5.9% of the registered donors and the frequency of HLA-B*56 was low with a frequency reaching to 0.0007 (Jawdat et al., 2020). Similar allele frequencies of these two alleles were reported by another group analyzed the HLA allele frequencies of 45,457 stem cell registered donors (Alfraih et al., 2021). The most common specific alleles which were reported among Saudi population were A*01:01, A*01:03, HLA-A*03:01/02 and B*56:01 (Jawdat et al., 2020; Alfraih et al., 2021). In other Arab populations, the frequency of HLA-A*01 ranged from 15% (Tunisians) to 6% in some Arab populations (Emirates). On the other hand, the frequency of the HLA-A*03 allele was estimated to be 15% in the Iranian population and ranging to 6% in other Arab populations (Hajjej et al., 2018). These allele frequencies might be in parallel with our results, in which the low frequency of A*01 in the Saudi population might be associated with the low number of reported cases compared to the numbers reported worldwide. In contrast, HLA-A*01 is a common allele in some European countries with high COVID-19 infection rates, such as Italy with 10% allele frequency and Brazil with 15% frequency (Sacchi et al., 2019; Halagan et al., 2018). However, a definite HLA association with COVID-19 infection will not be drawn unless combined data from different areas is analyzed. There are limited studies with published data regarding the HLA genotype and COVID-19 infection. In addition, bioinformatics-based studies have been published which are focused on identifying HLA alleles that can bind specific viral peptides at different affinities (Nguyen et al., 2020; Lee and Koohy, 2020; Iturrieta-Zuazo et al., 2020). An early study from Wuhan, China reported the presence of the HLA-A*24 specific allele in four out of five patients infected with SARS-COV2 (Warren and Birol, 2020). Another study from China reported an increase in the frequency of HLA-C*07:19 and B*15:27 in a cohort of 82 COVID-19-infected patients compared to the control with no statistical difference reported at the level of HLA-A genotype between the two groups (Wang et al., 2020). However, these studies lack a sufficient number of cases to make a significant conclusion. This may cause variations in the results between studies. In our cohort, there was no significant difference in the frequency of HLA-A*24 between the control and infected groups. However, HLA-A*24 was significantly lower in the fatal group compared to the infected group (recovered) with insignificant p value after correction. Interestingly, a recent in-silico study reported the HLA-A*24 specific allele as an allele associated with efficient T-cell anti-viral response leading to virus eradication (Tomita et al., 2020). The involvement of this allele in COVID-19 recovery might be elucidated by examining more patients.
In this study, there was a significant difference in the frequency of HLA-C between groups. There was a significant decrease in the frequency of HLA-C*04 in the fatal group compared to the infected group, while a significant increase was seen in the frequency of HLA-C*06 in the fatal group compared to the infected group. Additionally, we reported a significant increase in the frequency of HLA-C*01 in the infected group compared to the control group. However, these results were non-significant after p value correction. A larger sample size in the future might elucidate this association clearly. A recent epidemiological study from Italy concluded that HLA-C*01 might be a genetic factor responsible for the variations in susceptibility to COVID-19 infection between different regions in Italy (Correale et al., 2020). HLA-C loci is well known as a ligand for killer immunoglobulin-like receptors (KIR) on natural killer cells (Biassoni et al., 1995). The interaction of a certain HLA-C/peptide complex with certain KIR is associated with inhibition of NK cell cytotoxic activity to target infected cells. In other viral infectious diseases, an association between HLA-C*04 and a KIR subset allele was associated with an increase in viral load and infection progression (Olvera et al., 2015). The frequency of HLA-C alleles in infected and fatal groups might be associated with a disturbance in the activity of the natural killer cells as described previously with other viral infections (Wauquier et al., 2019). However, this hypothesis will not be confirmed unless KIR genotyping is performed, which can be investigated in another study.
In this study, there was a significant increase in the frequency of HLA-DRB1*04 in the control group compared to the infected group with insignificant corrected p value. There was a significant increase in the frequency of HLA-DRB1*08 in the infected group compared to the control group, which remained significant after p value correction and was associated with a relative risk 2.32. The most common specific alleles reported in Saudi population were DRB1*04:01 (0.011) followed by DRB1*04:04 and DRB1*04:06 (0.0067). While the most common HLA-DRB1*08 alleles were DRB1*08:04 (0.013) followed by DRB1*08:03 (0.002) with low frequencies in both DRB1*08:02 and 08:06 (0.0002) (Jawdat et al., 2020). Although HLA class I is well known as a molecule that presents intracellular molecules, HLA class II can have a role in viral peptide presentation. A recent study showed that peptides from SARS-COV2 can bind HLA-DR molecules present on monocytes (Parker et al., 2020). Furthermore, differences in HLA class II frequency were also reported in a study from Italy with a cohort of 99 COVID-19-infected patients, in which an association between HLA‐DRB1*15:01, DQB1*06:02 and B*27:07 and the incidence of COVID-19 infection was suggested (Novelli et al., 2020). As mentioned earlier, a lack of consistency between studies might be attributed to sample size and population variations. In this study, we can show that certain HLA alleles, such as HLA-A*01, might be associated with increased incidence of infection by SARS-COV2, and certain alleles were observed only in the infected group, such as B*56, in addition to alleles identified mainly in the fatal group, such as HLA-A*03. However, these initial results need exploring with larger sample sizes from different regions and probably with higher resolution of typing.
5. Conclusions
In conclusion, individual HLA genotypes may be associated with susceptibility to COVID-19 infection and infection outcome. Examining a larger sample size and samples from different populations might identify a powerful association, which can be used as a risk factor in a screening protocol.
Funding
This study was kindly supported by King Abdul-Aziz City for Science and Technology (KACST) with a grant number 5-20-01-502-0009 via Fast track funding path for COVID-19.
Declaration of competing interest
“The authors declare no conflict of interest.”
Acknowledgments
The authors would like to thank Science and Technology Unit, Ministry Of Health, Kingdome of Saudi Arabia for their assistance. Many thanks to Dr. Howell Martin for his feedback.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.04.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alfraih F., et al. High-resolution HLA allele and haplotype frequencies of the Saudi Arabian population based on 45,457 individuals and corresponding stem cell donor matching probabilities. Hum. Immunol. 2021;82(2):97–102. doi: 10.1016/j.humimm.2020.12.006. [DOI] [PubMed] [Google Scholar]
- Biassoni R., et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J. Exp. Med. 1995;182(2):605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell J.M., Jamieson S.E., Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22(2):370–385. doi: 10.1128/CMR.00048-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correale P., et al. HLA-B*44 and C*01 prevalence correlates with Covid19 spreading across Italy. Int. J. Mol. Sci. 2020;21(15) doi: 10.3390/ijms21155205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Medical Research. 2020;7(1) doi: 10.1186/s40779-020-00240-0. 11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjej A., et al. The genetic heterogeneity of Arab populations as inferred from HLA genes. PloS One. 2018;13(3) doi: 10.1371/journal.pone.0192269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halagan M., et al. Immunogenetics; 2018. The Distribution of HLA Haplotypes in the Ethnic Groups that Make up the Brazilian Bone Marrow Volunteer Donor Registry (REDOME) p. 70. [DOI] [PubMed] [Google Scholar]
- Hill A.V. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- Iturrieta-Zuazo I., et al. Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: a pilot study in a cohort of Covid-19 Spanish patients. Clin. Immunol. 2020;219:108572. doi: 10.1016/j.clim.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawdat D., et al. HLA-A, -B, -C, -DRB1 -DQB1 and – DPB1 allele and haplotype frequencies of 28927 Saudi stem cell donors typed by sequencing. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.544768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R.E., Adab P., Cheng K.K. Covid-19: risk factors for severe disease and death. Bmj. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- Keicho N., et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009;70(7):527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.Y., et al. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology. 2011;140(2):686–696.e1. doi: 10.1053/j.gastro.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Research. 2020;9:145. doi: 10.12688/f1000research.22507.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4(1):9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., et al. Association of polymorphisms in human leukocyte antigen class I and transporter associated with antigen processing genes with resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 2003;187(9):1404–1410. doi: 10.1086/374394. [DOI] [PubMed] [Google Scholar]
- Lu L., et al. A comparison of mortality-related risk factors of COVID-19, SARS, and MERS: a systematic review and meta-analysis. J. Infect. 2020;81(4):e18–e25. doi: 10.1016/j.jinf.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C., et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43(3):563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- Nguyen A., et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94(13):e00510–e00520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A., et al. 2020. HLA Allele Frequencies and Susceptibility to COVID-19 in a Group of 99 Italian Patients. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera A., et al. The HLA-C*04: 01/KIR2DS4 gene combination and human leukocyte antigen alleles with high population frequency drive rate of HIV disease progression. Aids. 2015;29(5):507–517. doi: 10.1097/QAD.0000000000000574. [DOI] [PubMed] [Google Scholar]
- Parker R., et al. bioRxiv; 2020. Mapping the SARS-CoV-2 Spike Glycoprotein-Derived Peptidome Presented by HLA Class II on Dendritic Cells; p. 2020. 08.19.255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Røder G., et al. Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501. Acta crystallographica. Section F, Structural biology and crystallization communications. 2008;64(Pt 6):459–462. doi: 10.1107/S1744309108012396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchi N., et al. High-resolution analysis of the HLA-A, -B, -C and -DRB1 alleles and national and regional haplotype frequencies based on 120 926 volunteers from the Italian Bone Marrow Donor Registry. HLA. 2019;94(3):285–295. doi: 10.1111/tan.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O.D., et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 2008;69(7):443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y., et al. Association between HLA gene polymorphisms and mortality of COVID-19: an in silico analysis. Immunity, Inflammation and Disease. 2020;8(4):684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.F., et al. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24(5):421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- Wang W., et al. Distribution of HLA Allele Frequencies in 82 Chinese Individuals with Coronavirus Disease-2019 (COVID-19. 2020;vol. 96(2):194–196. doi: 10.1111/tan.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R.L., Birol I. HLA Predictions from the Bronchoalveolar Lavage Fluid Samples of Five Patients at the Early Stage of the Wuhan Seafood Market COVID-19 Outbreak. ArXiv. 2020 arXiv:2004.07108vol. 3. [Google Scholar]
- Wauquier N., et al. HLA-C-restricted viral epitopes are associated with an escape mechanism from KIR2DL2+ NK cells in Lassa virus infection. EBioMedicine. 2019;40:605–613. doi: 10.1016/j.ebiom.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf B. On estimating the relation between blood group and disease. Ann. Hum. Genet. 1955;19(4):251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Xiong Q., et al. Lack of association between HLA-A,-B and-DRB1 alleles and the development of SARS: a cohort of 95 SARS-recovered individuals in a population of Guangdong, Southern China. Int. J. Immunogenet. 2008;35:69–74. doi: 10.1111/j.1744-313X.2007.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.