Abstract
Acute inflammatory exacerbations (AIE) represent precipitous deteriorations of a number of chronic lung conditions, including pulmonary fibrosis (PF), chronic obstructive pulmonary disease and asthma. AIEs are marked by diffuse and persistent polycellular alveolitis that profoundly accelerate lung function decline and mortality. In particular, excess monocyte mobilization during AIE and their persistence in the lung have been linked to poor disease outcome. The etiology of AIEs remains quite uncertain, but environmental exposure and genetic predisposition/mutations have been identified as two contributing factors. Guided by clinical evidence, we have developed a mutant model of pulmonary fibrosis leveraging the PF-linked missense isoleucine to threonine substitution at position 73 [I73T] in the alveolar type-2 cell-restricted Surfactant Protein-C [SP-C] gene [SFTPC]. With this toolbox at hand, the present work investigates the role of peripheral monocytes during the initiation and progression of AIE-PF. Genetic ablation of CCR2+ monocytes (SP-CI73TCCR2KO) resulted in improved lung histology, mouse survival, and reduced inflammation compared to SP-CI73TCCR2WT cohorts. FACS analysis of CD11b+CD64-Ly6Chi monocytes isolated 3 d and 14 d after SP-CI73T induced injury reveals dynamic transcriptional changes associated with “Innate Immunity’ and ‘Extracellular Matrix Organization’ signaling. While immunohistochemical and in situ hybridization analysis revealed comparable levels of tgfb1 mRNA expression localized primarily in parenchymal cells found nearby foci of injury we found reduced effector cell activation (C1q, iNOS, Arg1) in SP-CI73TCCR2KO lungs as well as partial colocalization of tgfb1 mRNA expression in Arg1+ cells. These results provide a detailed picture of the role of resident macrophages and recruited monocytes in the context of AIE-PF driven by alveolar epithelial dysfunction.
Keywords: alveolar type-2 cell, Sftpc I73T surfactant protein-C I73T mutant, idiopathic pulmonary fibrosis, chemokine receptor-2, monocyte-derived alveolar macrophages, acute exacerbation of PF
Introduction
Pulmonary fibrosis (PF) is a devastating degenerating disease characterized by failure to properly resolve inflammation, heterogeneous disruption of alveolar and bronchiolar architecture, and irreversible scarring (1–3). Despite the intrinsic ability of the lung parenchyma to withstand repeated bouts of injury, persistent and widespread stress induced by endogenous (functional mutations) and/or exogenous (infection, toxicant exposure) sources promotes aberrant epithelial-immune and epithelial-mesenchymal communication. In this context, inflammation has been widely studied as an essential aspect of fibrogenic scarring and PF progression, with so called “acute inflammatory exacerbations” (AIE) strongly linked to lung function decline and mortality (4–6). Both clinical and experimental evidence indicate that mononuclear myeloid cell function reflects disease outcome, whereby disproportionate mobilization of peripheral monocytes, as well as their persistence in the lung as monocyte-derived alveolar macrophages, are associated with poor outcome (7–10).
Our understanding of the phenotype and function of resident and monocyte-derived macrophages and infiltrating monocytes in the context of chronic injury and fibrosis has been revolutionized in the past decade (8, 10–16). As a result, a growing research area is now dedicated to comprehend the interplay between ontogeny and polarization from birth to adulthood, in healthy and disease state (16–19). Leveraging chemical-induced fibrosis (i.e., bleomycin, asbestos) and innovative lineage tracing systems defined peripheral monocytes as a dynamic mixture of populations that has the capacity to differentiate into alveolar macrophage-like cells, yet remains incompetent in their ability to terminate/resolve inflammation, a function central to “true” resident alveolar macrophages (9, 14). With this in mind, it is essential to comprehend monocyte biology to fully appreciate their role in injury resolution and tissue remodeling occurring during AIE-PF (9, 20).
The etiological, temporal, and spatial complexity of PF has thus far represented an almost insurmountable roadblock to overcome. Epidemiological observation of familial cohorts of PF supports the notion that parenchymal mutations contribute to PF pathogenesis and progression, and provides a workable platform to develop translationally relevant models of PF (2, 21–23). In particular, mutations associated with key functional genes (i.e., telomere function, or pulmonary surfactants) are shown to robustly generate a fibrotic phenotype (3, 24). The most common mutation in the alveolar epithelial-restricted gene encoding for surfactant protein C (SP-C), the missense substitution g.1286T>C leading to isoleucine to threonine substitution at position 73 in the SFTPC proprotein (“SP-CI73T”), has been linked to extensive tissue remodeling driven by aberrant macroautophagy and mitophagy function (25, 26). We have previously demonstrated that allelic insertion of the mutant SP-CI73T generates a viable strain producing hypomorphic levels of mutant SP-C (≈20% of SP-CWT expression). This is associated with relatively moderate inflammation and lung remodeling (23). Thanks to its inducible nature, SP-C mutant levels can be tripled, thereby overwhelming epithelial cell capacity to cope with stress and systemic response. Previous evidence indicates that SP-CI73T epithelial cells coordinate peripheral myeloid cell recruitment and activation, with their pro-inflammatory/pro-fibrotic activation is responsible for the propagation of the injury and fibrogenesis. In an effort to complement this dataset, we further provided initial proof of concept evidence that peripheral cell recruitment alters the trajectory of fibrotic disease induced by SP-CI73T injury (27). Several clinical and experimental lines (SP-CI73T, bleomycin, radiation, asbestosis) of evidence draws strong correlation between excess monocyte mobilization and the outcome of inflammatory driven fibrosis, with specific chemokine axes responsible for the recruitment of distinct subsets (CCR2/CCL2, CCR4/CCL17, CX3CR1/CX3CL1) (28–32).
Building on these notions, the present work progressively narrows the cellular target responsible for lung remodeling during acute inflammatory exacerbations of PF from blood monocytes described in previous literature from our group and others (27, 33), to the Ly6C+ subset, and then the CCR2 subset. RNA-sequencing analysis of Ly6Chi cells identified an early and persistent pro-inflammatory and pro-fibrotic phenotype during SP-CI73T induced injury, while targeted depletion of the CCR2 subset (SP-CI73TCCR2KO) demonstrates that these cells are primarily involved in coordinating the degree of tissue damage downstream of the injury cue. Furthermore, use of in situ hybridization techniques provide evidence for spatial localization of activated immune cells with respect to fibrotic foci (tgfb1). Together, these data deliver an important piece of the puzzle, highlighting the contribution of specific monocyte/macrophages subsets in the initiation and progression of PF exacerbation.
Materials and Methods
Reagents
Tamoxifen (non-pharmaceutical grade) was purchased from Sigma-Aldrich (St Louis, MO). Giemsa cytological stain was purchased from Sigma-Aldrich. Antibodies used for in situ hybridization, immunohistochemical and flow cytometric analysis were: tgfb1 (Cat # 407751, Advanced Cell Diagnostics, ACD); RFP (Cat # ab62341; 1:1000, Abcam, Cambridge, MA); Arg1 (Cat # ab91279; 1:1500, Abcam); iNOS (Cat # ab15323; 1:200, Abcam); CD64 (Cat # bs-3511R; 1:250, Bioss Antibodies, Woburn, MA); CCR2 (Cat # EPR20844; 1:400, Abcam); phospho-SMAD2/3 (Cat # PA5-37636; 1.500, Invitrogen CX3CR1 (Cat # bs-1728R; Bioss Antibodies); CD64 (Cat # bs-3511R; Bioss Antibodies); CD125/IL-5RA (Cat # bs-2601R, Bioss Antibodies); C1q (Cat # A0136; 1:500, Dako/Agilent Technologies, Santa Clara, CA); CD16/32 (clone 93; eBiosciences, San Diego, CA), CD11b (clone # M1/70; eFluo450, eBiosciences); Fixable Viability dye (Cat # 65-0865-14; eFluo780, eBiosciences); SigF (clone S17007L; PE-CF594, BD Biosciences, San Jose, CA); CD45 (clone 30-F11; PerCP5.5, Biolegend, San Diego, CA); CD11c (clone # N418; BV705, Biolegend); Ly6G (clone # 1A8; AF700, Biolegend); Ly6C (clone HK1.4, BV510, Biolegend); CD64 (clone X54-5/7.1; PE/Cy7, Biolegend); CD43 (clone # S11; PE, Biolegend); CD3 (clone # 17A2; BUV395, Biolegend). All other reagents were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA), or Sigma-Aldrich.
Murine Model of SP-CI73T Induced Lung Injury
Tamoxifen inducible SP-CI73T mice were generated as previously reported (9). Briefly, the SP-CI73T founder line (expressing a Neomycin cassette) was crossed with a mouse line expressing an estrogen receptor (ER)-2 controlled Flp-O recombinase strain knocked into the Rosa26 locus (Jackson Laboratory, Bar Harbor, ME) to generate the inducible-SP-CI73TFlp line. adult homozygote SP-CI73TFlp mice received tamoxifen (175 mg/kg in corn oil, oral gavage) at 8-12 weeks of age. Both male and female animals were used for the studies. Control groups mice are represented as pooled data from tamoxifen treated SP-CI73T not expressing Flp-O recombinase or oil (vehicle) treated Flp-O expressing SP-CI73T mice. To knock out monocyte subsets, these mice were then crossed to homozygosity with CCR2KO (Stock No: 004999, Jackson Laboratories) and CCR2RFP (Stock No: 017586, Jackson Laboratories) lines. All mice were housed under pathogen free conditions in AALAC approved barrier facilities at the Perelman School of Medicine (University of Pennsylvania), and Skaggs College of Pharmacy, University of Utah. All experiments were approved by the Institutional Animal Care and Use Committee at the University of Utah and Pennsylvania.
Lung Histology, Histochemistry, and In Situ Hybridization
Whole lungs were fixed by tracheal instillation of 10% neutral buffer formalin at a constant pressure (25 cm H2O). Following paraffin embedding, 6 µm sections were cut and stained with Hematoxylin & Eosin (H&E) by the Associated Regional and University Pathologists Inc., at the University of Utah. Immunostaining of deparaffinized tissue sections was performed as previously described (34). Briefly, after antigen retrieval using citrate buffer (10.2 mM sodium citrate, pH 6.0, for 20 minutes) and quenching of endogenous peroxidase with 3% hydrogen peroxide in methanol (30 minutes), non-specific binding was blocked with 10% goat or rabbit serum according to primary antibody origin. Appropriate serum/IgG controls each diluted in blocking buffer were applied for overnight incubation at 4°C in a humidified chamber. Following incubation with biotinylated secondary antisera (Vectastain Elite ABC kit, Vector Labs, Burlingame, CA) for 30 minutes (room temperature), staining was visualized using a Peroxidase Substrate Kit DAB (Vector Labs) and counterstained with Harris Modified Hematoxylin (Thermo Fisher Scientific, Inc.). In other studies, in situ hybridization was performed prior to immunohistochemical staining. Briefly, paraffin embedded sections were deparaffinized in xylene and 100% EtOH. This was followed by peroxidase quenching in H2O2 (10’, away from light), antigen retrieval (RNAscope® Target Retrieval Reagent, ACD), and protease IV treatment (RNAscope® Protease IV Reagent, ACD). Tgfb1 probe was then incubated for 2 h in hybridization oven (40°C), a step followed by a series of signal amplification steps and chromogenic development as indicated by manufacturer protocol. Slides were then washed and immunohistochemistry blocking step resumed as described above.
Bronchoalveolar Lavage Fluid (BALF) Analysis
BALF was collected from mice using five sequential lavages of 1 ml sterile saline and processed for analysis as previously described (9). Briefly, cell pellets obtained by centrifuging BALF samples at 400 × g for 6 minutes were re-suspended in 1 ml of PBS, and total cell counts determined using a NucleoCounter (New Brunswick Scientific, Edison, NJ). Differential cell counts were determined manually from BALF cytospins stained with modified Giemsa for 20 minutes to identify macrophages, lymphocytes, eosinophils and neutrophils.
Multiplex Cytokine Analysis
First-return aliquots of cell-free BALF were analyzed for CCL2 levels using a Luminex platform (Millipore Sigma, Burlington, MA) by the Human Immunology Core at Perelman School of Medicine.
Flow Cytometry and Cell Sorting for Identification of Immune Populations
Following BALF collection, lungs were cleared of blood by cardiac perfusion with saline solution, removed from the chest cavity, minced, and transferred into a 50 ml conical tube and incubated (37°C, 30’) in DMEM + 5% FBS + 2 mg/ml Collagenase D (Cat #11088866001, Roche, Indianapolis, IN). Digested lungs were passed through 70-μm nylon mesh to obtain a single-cell suspension, counted and mixed with ACK Lysis Buffer (Thermo Fisher Scientific) to remove any remaining red blood cells. BALF and tissue cell pellet (1X106 cells) were resuspended in 100µl staining buffer (PBS+0.1% sodium azide) and incubated with anti-mouse CD16/32 antibody (Fc block, eBiosciences, San Diego, CA) for 10 min at 4°C to block nonspecific binding. This was followed by 30-minute incubation with fluorescently-tagged antibodies or appropriate isotype controls (0.25–1.5 µg/106 cells) for 30 minutes (4°C). Cells were then spun and resuspended in staining buffer for viability staining (30 minutes at 4°C). Cells were fixed in 2% paraformaldehyde and analyzed with an LSR Fortessa (BD Biosciences, San Jose, CA) or FACS ARIA (BD Biosciences) for cell sorting experiments. Inflammatory monocytes (SigF+CD11c-CD11b+Ly6C+) were identified following forward and side scatter selection of singlet CD45+ viable cells. To ensure cell sorting of a purified population of monocytes pregating/exclusion of resident alveolar macrophages (SigF+ CD11b-CD11c+), eosinophils (SigFintCD11b+CD11c-), neutrophils (Ly6G+) and lymphocytes (CD3+), based on gating strategy modified from our group and others (27, 35, 36). Gating strategy is shown in Supplementary Figure 1 . All analysis was performed using FlowJo software (FlowJo, LLC, Ashland, Oregon).
RNA Sequencing Preparation and Analysis
Total RNA was extracted from fresh frozen cell pellets using Qiagen RNeasy Plus Universal mini kit following manufacturer’s instructions (Qiagen, Hilden, Germany). Extracted RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina following manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were first enriched with Oligo(dT) beads. Enriched mRNAs were fragmented for 15 minutes at 94°C. First strand and second strand cDNAs were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3' - ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. The sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were pooled and clustered on 1 lane of a flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq4000 instrument according to manufacturer’s instructions. The samples were sequenced using a 2x150bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification. Analysis of RNA counts was performed using R (3.6.3) (37). Differential gene expression analysis was conducted using the hciR package (38). Fast Gene Set Enrichment Analysis (fgsea) was used for gene set enrichment analysis with the Reactome database (39). Data were deposited in NCBI’s Gene Expression Omnibus (40) and are accessible through GEO Series accession number GSE166300 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166300).
Statistics
All data are presented with dot-plots and group mean ± SEM unless otherwise indicated. Statistical analyses were performed with Prism GraphPad 9.0 (GraphPad Software, San Diego, CA). Student’s t-test were used for paired data; for analyses involving multiple groups, one-way or two-way analysis of variance (ANOVA) was performed with post hoc testing as indicated. Survival analyses were performed using Log Rank (Mantel-Cox) test. In all cases statistical significance was considered at p ≤ 0.05.
Results
SP-CI73T Induced Injury Is Linked to Monocyte Activation
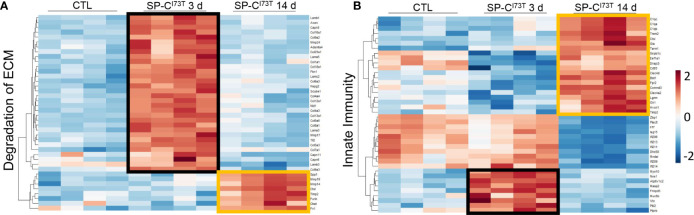
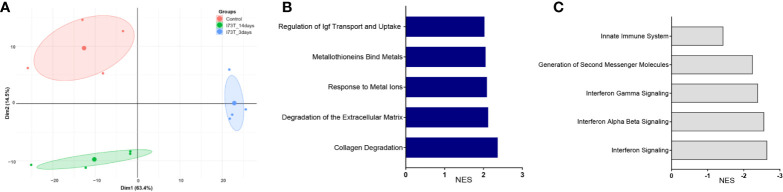
We have previously shown that lung injury generated by induction of mutant SP-CI73T expression is accompanied by dynamic changes in SigF+CD11bint resident alveolar macrophage and CD11b+Ly6Chi infiltrating monocyte mobilization and activation (27). We therefore used comparable gating strategy to sort CD11b+Ly6Chi inflammatory monocytes from collagenase digested tissue 3 d and 14 d post induction, times coordinated with initiation and peak of inflammatory exacerbations. Principle component analysis (PCA) revealed transcriptional variance (Dim1: 63,4%; Dim2: 14,5%) between the control group, consisting of oil treated SP-CI73T mice, an acute inflammatory state (3 d post tamoxifen induced SP-C mutant induction) and late inflammation/early remodeling (14 d) ( Figure 1A ). Pathway analysis of CD11b+Ly6Chi monocyte isolated 3 d post injury indicated transcriptional changes (positive and negative normalized enrichment score, NES) related to survival and extracellular matrix homeostasis ( Supplementary Figure 2 ). By 14 d, signaling pathways associated with extracellular matrix remodeling displayed sustained positive NES scores ( Figure 1B ), while those associated with inflammatory activation (i.e., innate immunity, interferon signaling) showed a negative NES ( Figure 1C ). Further examination of significantly altered genes belonging to ‘Degradation of extracellular matrix’ gene set revealed metalloproteinases (mmp17, mmp24) and collagen genes (col6a1, col1a1, col8a1, col4a3, col2a1) to be significantly increased at 3 d ( Figure 2A ). By comparison, analysis of Ly6Chi monocyte transcripts at 14 d highlighted downregulation of all the above-mentioned transcripts, paired with increases in a distinct subset of matrix/matrix degradation, including spp1, mmp19, mmp14, timp2, and fn1 ( Figure 2A , black boxes). Analysis of gene sets linked to ‘Innate immunity’ outlined expression signatures exclusive to 3 d (nos1, frmpd3; masp2) and 14 d (c1qa/c, cd93, clec4a2; and downregulated expression of plac8, irf7, zbp1) post injury ( Figure 2B , black and orange boxes). Similar examination of ‘Interferon signaling’ also identified genes distinctively expressed acutely (trim2/6/17, irf6) or at 14 d (socs3, and marked reductions in stat1, trim34a, irf7) ( Supplementary Figure 3 , black boxes).
Figure 1.
Extracellular matrix and innate immunity expression analysis of flow cytometry sorted CD11b+Ly6Chi monocytes following SP-CI73T induced injury. Differentially expressed genes (fold change > 1.5 and false discovery rate [q-value] < 0.05) were analyzed using Reactome database to highlight enriched signaling pathways. N = 4 was utilized in each study condition. (A) Heat-map showing expression of significantly regulated genes involved in ‘Degradation of Extracellular Matrix (ECM)’. (B) Heat-map showing expression of significantly regulated genes involved in ‘Innate Immunity’. Note black boxes represent genes uniquely expressed 3 d after SP-CI73T injury; orange boxes represent genes uniquely expressed 14 d after SP-CI73T injury.
Figure 2.
RNA sequencing analysis of flow cytometry sorted cells shows temporal dynamics in CD11b+Ly6Chi monocytes function following SP-CI73T induced injury. RNA-sequencing of cell sorted CD11b+Ly6Chi monocytes from control (CTL, oil treated SP-CI73T mice) or SP-CI73T mice 3 d and 14 d after injury. Total RNA was prepared and analyzed by RNA-seq. N = 4 was utilized in each study condition. (A), Two-dimension principle component analysis (PCA) of gene expression variance among monocytes accumulating in the lung in control (CTL) or SP-CI73T mice 3 d and 14 d after injury. (B) Positively enriched pathways at 3 d post injury (normalized enrichment score, NES); (C) Negatively enriched pathways at 14 d post injury.
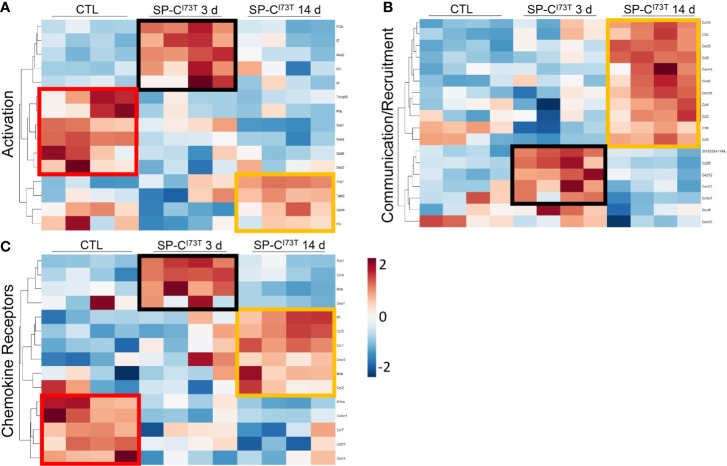
As a complement to our unbiased pathway analysis, a gene list extrapolated from literature in pulmonary fibrosis examined dynamic changes in monocyte activation, recruitment, and surface receptor expression after SP-CI73T induced injury (8, 9, 13, 14, 41) ( Figures 3A–C ). We found that Ly6Chi monocytes isolated from control lung digests expressed high levels of transcription factors involved in the inflammatory response (Stat1, Stat4 and Stat5), displayed a unique chemokine receptor repertoire (cx3cr1, ccr2, ccr7, cxcr4, cxcr5, il10ra), and negligible levels of chemokine/cytokine ligands compared to cells isolated after SP-CI73T induced injury ( Figures 3A–C , red boxes). By comparison, Ly6Chi monocytes accumulating in the lung 3 d post injury expressed nos2, il12b and il7 ( Figure 3A , black boxes) and a battery of chemokine/cytokine ligands (ccl28, cxcl12, cxcl17, cx3cl1) ( Figure 3B , black boxes), and receptors (folr1, ccr4 and il5ra) ( Figure 3C , black boxes). This was further changed by 14 d, which was coordinated with high levels of anti-inflammatory genes (arg1, tgm2, apoe) recruitment factors (ccl2/3/4/6/8/9/12, cxcl2/14/16), as well as a unique receptor repertoire characterized by il7r, ccr1, ccr5, il4ra ( Figures 3A–C , orange boxes).
Figure 3.
Targeted gene expression analysis of flow cytometry sorted CD11b+Ly6Chi monocytes following SP-CI73T induced injury. (A-C) Differential gene expression of factors associated with (A) activation, (B) cell communication/recruitment, or (C) chemokine receptor. Heat maps show significant genes. Gene selection was based on previous literature (9, 42, 43). Criteria for significance were set at fold change > 1.5 and false discovery rate (q-value) < 0.05. N = 4 was utilized in each study condition. Note red boxes represent genes uniquely expressed in control populations; black boxes represent genes uniquely expressed 3 d after SP-CI73T injury; orange boxes represent genes uniquely expressed 14 d after SP-CI73T injury.
CCR2 Monocyte Depletion Reduced Injury and Inflammatory Induced by SP-CI73T Expression
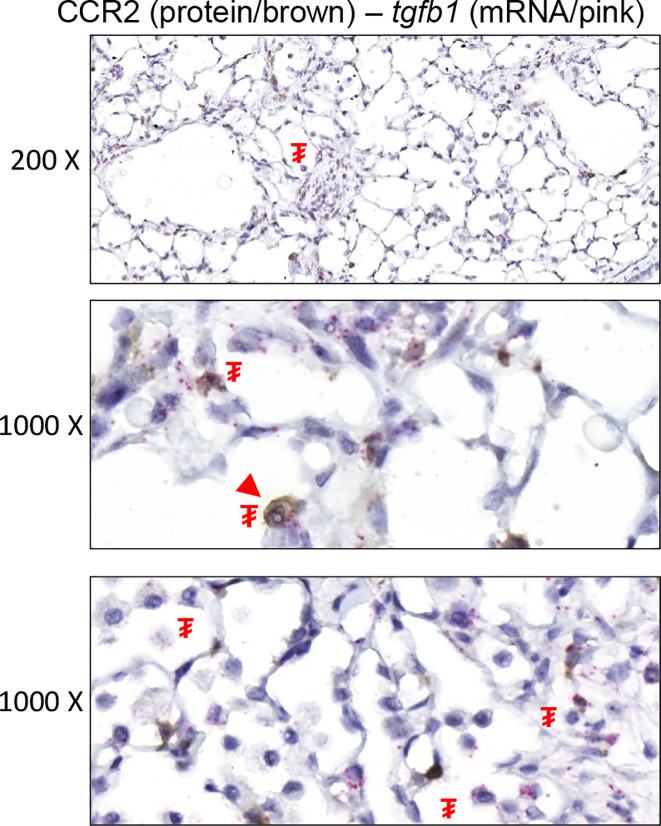
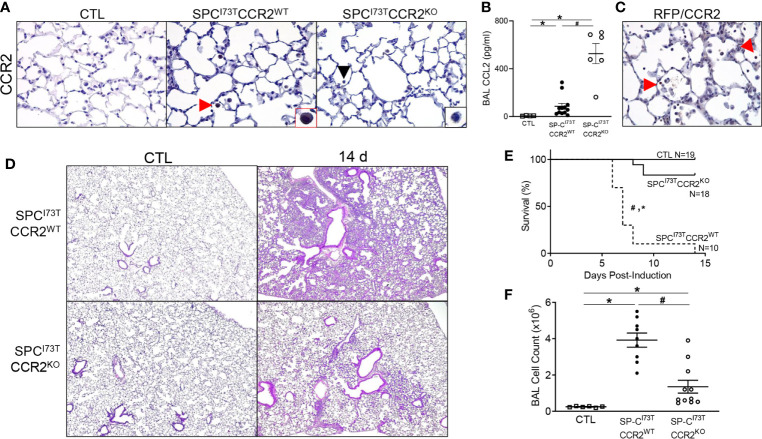
Previous work conducted by our laboratory preliminarily linked early influx of peripheral myeloid cells to lung disease outcome following mutant SP-C induction (27). Rather than investigate the fibrogenic potential of the broader monocyte population, represented by Ly6C expression, we opted to limit our search to a subset of monocytes expressing CCR2, a marker linked with lung injury and fibrosis (33, 44). RNA in situ hybridization analysis indicated that CCR2+ cells do not express the master regulator of fibrosis, tgfb1; rather, we noted it to be predominant parenchymal (epithelial and mesenchymal), while CCR2+ cells accumulated in the proximity of these tgfb1-rich foci of injury ( Figure 4 ). To test whether ablation of the CCR2+ pool results in disease modifying effects on SP-CI73T induced injury, we opted to utilize a knock out model directly targeting CCR2 monocytes (SP-CI73TCCR2KO). We confirmed that CCR2+ cells accumulate in the lung following SP-CI73T induced injury, and that this response is not observed in SP-CI73TCCR2KO mice (black arrowhead, Figure 5A ). Notably, despite CCR2 receptor depletion, levels of MCP1/CCL2 were significantly increased compared to CCR2WT counterparts, whereas CCL17 and CX3CL1, chemokines also involved in monocyte recruitment, were not altered ( Figure 5B and not shown). Histochemical analysis for RFP in lung sections isolated from mice expressing red fluorescent protein in lieu of CCR2 (SP-CI73TCCR2RFP) 14 d post injury also revealed accumulation of RFP/CCR2 positive cells, thus indicating that monocytes expressing CCR2 may be recruited through mechanisms independent of CCR2 ( Figure 5C ). Next, we performed histopathological analysis of SP-CI73TCCR2WT and SP-CI73TCCR2KO lungs 14 d post injury. Unsurprisingly, in control conditions (SP-C wild type and oil treated SP-CI73T mutant lines) CCR2 monocyte ablation was not associated with any architectural alterations ( Figure 5D , left panels). As previously described, tamoxifen induction of SP- CI73TCCR2WT mice was associated with extensive perivascular and alveolar inflammatory cell infiltration and alveolar architecture disruption at 14 d ( Figure 5D , right panels) (23, 27). By comparison, both inflammatory cell clustering within the alveolar space and early fibrotic remodeling were lessened in CCR2 depleted mice. Consistent with these observations, CCR2 monocyte ablation was also linked to reduction in animal mortality (100% SP-CI73TCCR2WT vs. 16.7% SP-CI73TCCR2KO, at 2 weeks) and bronchoalveolar lavage (BAL) cell count ( Figures 5E, F ).
Figure 4.
Tgfb1 expression in CCR2+ cells following SP-CI73T induced injury. In situ hybridization (tgfb1/pink) combined with immunohistochemistry (CCR2/brown) analysis of SP-CI73TCCR2WT lung 14 d after injury. ₮ indicates area expressing tgfb1; arrowhead indicates CCR2 staining cells. Representative 200x (top) and 1000x (bottom) images from N = 3 separate animals are shown.
Figure 5.
Effects of genetic CCR2 monocyte ablation on lung injury, survival and inflammation following SP-CI73T mutant induced injury. (A) Immunohistochemical analysis of control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO lung sections were immunostained with antibody to CCR2. Red arrowheads indicate myeloid cells staining for CCR2. Insets show magnified cells; box color matches that of respective arrowhead. Black arrowheads indicate negative CCR2 staining in mononuclear cells in SP-CI73TCCR2KO cohorts. Images shown are representative of 3-5 animals per group. Magnification: 400x. (B) CCL2 ELISA of BAL fluid from control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO mice 14 d following tamoxifen administration. Data are represented as mean ± SEM (N=4-13). (C) Histochemical analysis of SP-CI73TCCR2RFP lung sections 14 d post injury immunostained with antibody to RFP/CCR2. Arrowheads indicate cells expressing the receptor (D) Hematoxylin & Eosin stained sections of control (CTL, tamoxifen treated SP-CWT, CCR2KO, or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO lungs 14 d post injury. Magnification: 400x. (E) Kaplan–Meier survival analysis from control (tamoxifen treated SP-CWT or oil treated SP-CI73T mice) SP-CI73TCCR2WT and SP-CI73TCCR2KO. Mice found dead or displaying body weight loss equating >25% of stating weigh for 2 consecutive days. *p<0.05 compared to control mice; #p<0.05 compared to SP-CI73TCCR2WT mice by Log-Sum (Mantel-Cox) Rank test. (F) BAL fluid cell counts from control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO (N =6-11) 14 d following SP-CI73T induced lung injury. Data are represented as mean ± SEM. *p<0.05 compared to control mice; #p<0.05 compared to SP-CI73TCCR2WT mice by One-Way ANOVA, using Tukey post-hoc test.
CCR2 Monocyte Depletion Alters Inflammatory Cell Recruitment and Activation
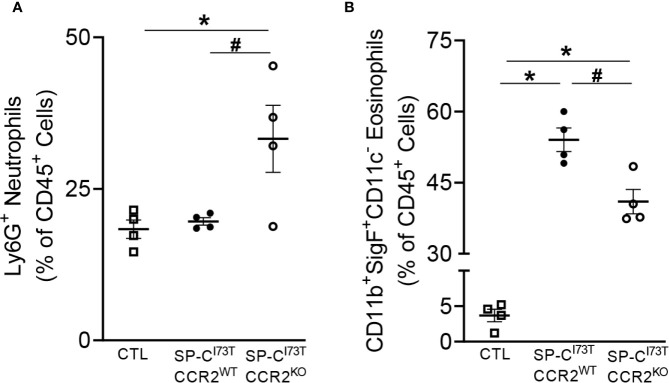
Flow cytometric analysis of lung tissue digest and manual cytospin counts of BAL cells revealed dynamic changes in neutrophil and eosinophil accumulation following CCR2 monocyte depletion. Flow cytometric analysis of tissue digests revealed no changes in the relative abundance of alveolar macrophages (SigFhiCD11bloCD11c+) and lymphocytes (CD3+) in SP-CI73TCCR2KO mice. Comparatively, accumulation of Ly6G+ neutrophils almost doubled in SP-CI73TCCR2KO mice (18.35 ± 1.5% in CTL; 19.63 ± 0.61% in SP-CI73TCCR2WT; 33.25 ± 5.6% in SP-CI73TCCR2KO), while overall eosinophilia was reduced at 14 d post injury ( Figures 6A, B ).
Figure 6.
CCR2 monocyte depletion results in altered polymorphonucleated cell influx following SP-CI73T induced injury. Changes in relative abundance of tissue (A) Ly6G+ neutrophils and (B) SigF+CD11c-CD11b+ eosinophils isolated by enzymatic digested (collagenase D) from control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO 14 d following injury. Data are represented as mean ± SEM (N = 4). All analysis was considered significant *p<0.05 compared to control mice; #p<0.05 compared to SP-CI73TCCR2WT mice by One-Way ANOVA, using Tukey post- hoc test.
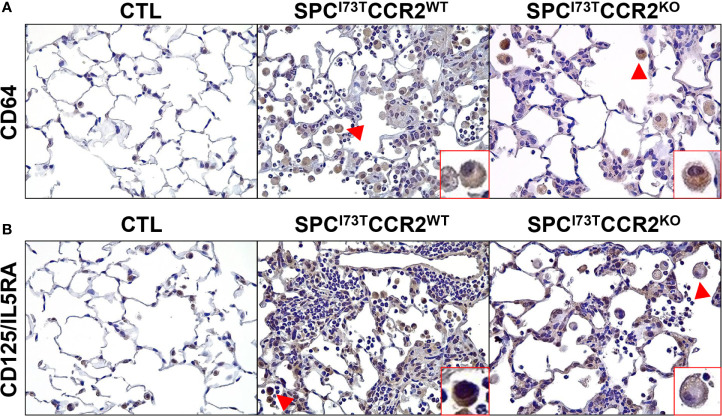
Immunohistochemical analysis, alone or in combination with in situ hybridization, was used to examine changes in myeloid cell maturation (CD64, Figure 7A ), surface receptor repertoire (IL5R and CX3CR1 Figure 7B and Supplementary Figure 4 ), and lung inflammatory state (iNOS, Arg1, tgfb1, Figures 8 , 9 ) following depletion of CCR2 monocytes in SP-CI73T mice. Ablation of CCR2+ cells dampened the number of CD64+ mature macrophages accumulating within foci of injury (arrowheads, Figure 7A and Supplementary Figure 5A ). Similarly, SP-CI73TCCR2KO mice significantly reduced the abundance of macrophages expressing IL-5RA receptor (CD125), a response likely to be guided by increases in IL-5 expression and secretion by epithelial cells during SP-CI73T induced injury ( Figure 7B and Supplementary Figure 5B ) (27). Differential expression of chemokine receptors allows us to define functionally distinct monocyte/macrophage populations. In this context, CCR2 and CX3CR1 have been used to discriminate the mobilization of Ly6Chi monocytes from the bone marrow and their accumulation at the site of injury, respectively (45). Ablation of CCR2+ monocyte did not affect accumulation of CX3CR1+ cells at 3 d post injury; by 14 d, we noted increases in the number of CX3CR1 expressing cells but not the relative intensity in individual cell expression ( Supplementary Figure 4 , 5C ). There was no difference in monocytes/macrophages maturation and activation between CCR2 wild type and knock outs in non-remodeled regions of the lung (data not shown).
Figure 7.
Effects of CCR2 monocyte ablation on monocyte/macrophage maturation and recruitment following SP-CI73T induced injury. Histochemical analysis of control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO lung sections 14 d post injury immunostained with antibody to (A) CD64 and (B) CD125/IL5RA. Binding was visualized using a Vectastain kit. Arrowheads indicate cells expressing the receptor. Insets show magnified cells; box color matches that of respective arrowhead. Original magnification, 400x; inset magnification, 750x. Representative sections from 3 mouse/group are shown.
Figure 8.
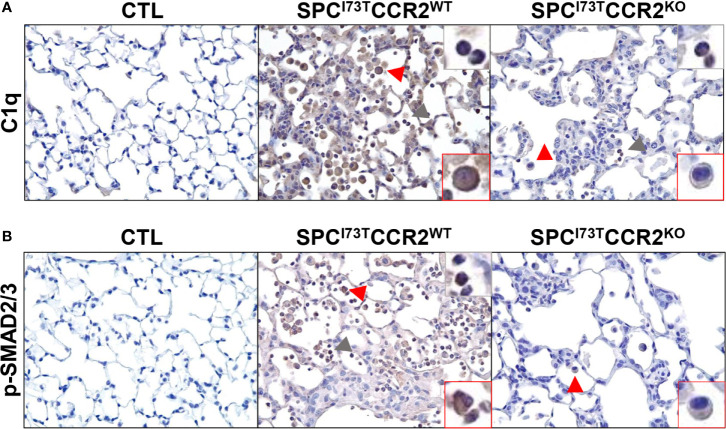
Effects of CCR2 monocyte ablation on inflammatory and fibrotic pathways following SP-CI73T induced injury. Histochemical analysis of control (CTL, tamoxifen treated SP-CWT or oil treated SP-CI73T mice), SP-CI73TCCR2WT and SP-CI73TCCR2KO lung sections 14 d post injury immunostained with antibody to (A) C1q and (B) p-SMAD2/3. Binding was visualized using a Vectastain kit. Red arrowheads indicate macrophage expression; grey arrowheads indicate non-macrophage expression. Insets show magnified cells; box color matches that of arrowhead. Original magnification, 400x; inset magnification, 750x. Representative sections from 3 mice/group are shown.
Figure 9.
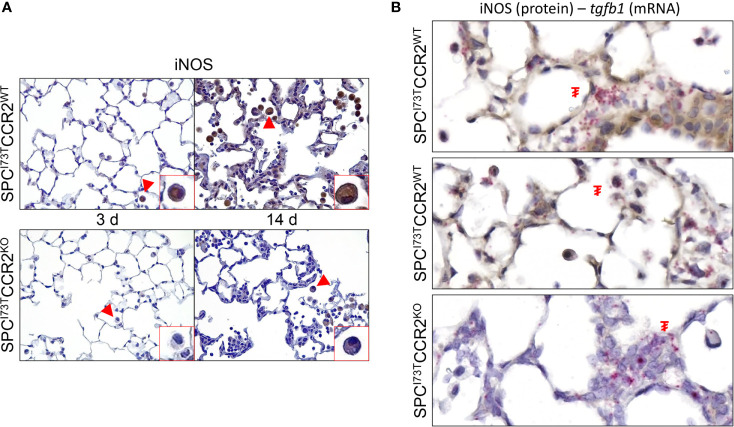
Effects of CCR2 monocyte ablation on pro-inflammatory activation following SP-CI73T induced injury. Histochemical analysis alone or in combination with in situ hybridization of SP-CI73TCCR2WT and SP-CI73TCCR2KO lung following SP-CI73T induced injury. Sections were immunostained with antibody to (A) iNOS. Binding was visualized using a Vectastain kit. Insets show magnified cells; box color matches that of arrowhead. Original magnification, 400x; inset magnification, 750x. (B) IHC + in situ hybridization (iNOS+ Tgfb1). mRNA visualization is shown in pink. Protein expression was visualized using a DAB Vectastain kit (brown). Arrowheads indicate cells expressing the receptor. ₮ indicate mRNA expressing cells. Original magnification, 1000x. Representative region is shown (N = 3).
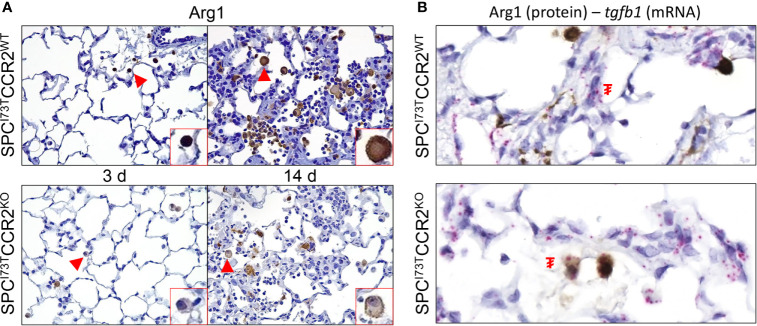
We then shifted our histochemical analysis towards macrophage activation. Guided by our RNA-sequencing results, we found increases in expression of the inflammation-linked complement component 1q (C1q) in all immune subsets and parenchymal cells ( Figure 8A ). By comparison, phosphorylation of SMAD2/3, a protein downstream of the TGFβ1 signaling pathway, was restricted to inflammatory cells ( Figure 8B ). Expression of these markers in CCR2 depleted cohorts revealed almost complete ablation of these responses, with sparse polymorphonucleated cell expression of C1q ( Figure 8A , grey arrowhead). Consistent with these results, we analyzed expression of the canonical pro-inflammatory activation marker, iNOS. While its expression was negligible at baseline ( Supplementary Figure 6A ), iNOS expression was progressively amplified in SP-CI73TCCR2WT lungs at 3 d and 14 d post injury ( Figure 9A , top panels). Notably, we found no iNOS expression at 3 d in SP-CI73TCCR2KO lungs; 14 d post injury, parenchymal and inflammatory cell expression was visible but significantly reduced ( Figure 9A ). Analysis of iNOS protein expression with in situ hybridization demonstrated accumulation of iNOS+ cells in proximity of foci of injury enriched in tgfb1 mRNA expression ( Figure 9B ). As described in Figure 9A , no iNOS+ cells were visible in SP-CI73TCCR2KO, but Tgfb1 expression was not affected by CCR2 monocyte ablation. We also found higher Arg1 expression (both number of expressing cells and relative intensity) in SP-CI73TCCR2WT mice, both at 3 d and 14 d after SP-CI73T induced lung injury ( Supplementary Figure 6B and Figure 10A ). Analysis of fibrotic foci, identified by higher intracellular tgfb1 density, revealed no colocalization with ARG1positive cells in SP-CI73TCCR2WT mice, but we observed a number of double positive cells in the CCR2 knock out cohorts ( Figure 10B ).
Figure 10.
Effects of CCR2 monocyte ablation on anti-inflammatory activation following SP-CI73T induced injury. Histochemical analysis alone or in combination with in situ hybridization of SP-CI73TCCR2WT and SP-CI73TCCR2KO lung following SP-CI73T induced injury. Sections were immunostained with antibody to (A) Arg1 or (B) Arg1+Tgfb1. mRNA visualization is shown in pink. Protein expression was visualized using a DAB Vectastain kit (brown). Arrowheads indicate cells expressing the receptor. ₮ indicate mRNA expressing cells. Original magnification, 1000x. Representative region is shown (N = 4).
Discussion
Pulmonary fibrosis represents the end-result of an aberrantly resolved inflammatory state. Early signals of stress are initially coordinated by the lung parenchyma, whereby both resident and peripheral immune subsets are central in the subsequent remodeling. These so-called “acute inflammatory exacerbations” are centrally responsible for propagating the injury and triggering rapid histological and functional decline through scarring and alveolar remodeling (honeycombing), events that ultimately accelerate patient death (4). Although we broadly comprehend these responses, there are substantial knowledge gaps related to the specific mechanisms by which exacerbations are initiated, the factors regulating individual thresholds of disease, and the nuances of studying a spatially heterogeneous injury. As a result of these obstacles, the first line of therapy against PF relies on broad-spectrum agents (corticosteroids, cytokine modulators, anti-fibrotics) that only target the symptoms (46–48). To refine our approach to disease interventions during disease defining processes such as exacerbations of PF, it is pivotal that we dynamically identify and functionally characterize inflammatory cell populations. In these studies, we provide transcriptional profiling of Ly6C+ peripheral myeloid populations during initiation and progression of inflammatory exacerbations triggered by excess mutant SP-CI73T. Furthermore, we provide evidence that a small subpopulation of bone marrow monocytes identified by their CCR2 expression, (CCR2+) monocytes represent a valuable candidate responsible for shifting disease trajectory.
The vast majority of experimental models of PF rely on exogenous stressors (bleomycin, radiation, asbestos, silica) to generate a robust fibrogenic response. Parenchymal mutations of key functional regulators (surfactant function, proteostasis, telomere and mitochondrial maintenance) have been abundantly mapped in PF patients (33, 49–52). Mutations of the SFTPC gene have been linked to varying degrees of fibrotic disease in adults and pediatric patients (2, 3, 53). In particular, the isoleucine to threonine substitution at position 73 in the SFTPC proprotein represents the most common (25, 26). Previous experimental evidence compellingly showed that mistrafficking of the SP-CI73T mutant leads to epithelial macroautophagy block, polycellular alveolitis, and parenchymal injury consistent of an acute exacerbation (8). Regardless of the model driving fibrogenic response, monocytes/macrophages have been shown to be linked to all phases of the injury process (8, 9, 13, 20, 27). Consistent with this evidence, our RNA sequencing analysis provides comprehensive assessment of the time-related changes in Ly6C+ inflammatory monocytes (also pregated as CD11c-CD64-CD11b+) phenotype during initiation and progression of SP-CI73T induced injury. Pathway analysis confirms that peripheral monocytes participate in the initiation of the inflammatory response 3 d post induction (‘cytokine signaling’, ‘innate immunity’), followed by negative enrichment scores for ‘interferon signaling’ and ‘innate immunity’ at 14 d, an observation that supports a time related phenotypic switch in these populations over time. By comparison, signaling pathways related to tissue remodeling (‘collagen formation’, ‘collagen degradation’, and ‘ECM reorganization’) are progressively increased. In support of this notion, gene expression analysis for ‘degradation of ECM’ signaling displays a shift away from signatures canonically seen in fibrosis (col1a2, col2a1, mmp17 and mmp24) (54), while favoring fn, spp1, timp2, mmp14 and mmp19 expression. Our data also provides a degree of overlap with other literature describing a pro-remodeling role of monocytes and monocytes-derived alveolar macrophages in fibrogenesis induced by bleomycin (9, 13). By comparison, RNA in situ hybridization analysis (tgfb1) combined with CCR2 (protein) staining did not fully corroborate these sequencing results. This observation could due to a limitation in our analysis being restricted to evaluation of a small fraction of monocytes (those expressing CCR2). We rather found tgfb1 expression primarily in the epithelium/mesenchyme, with a number of CCR2- mononuclear myeloid cells expressing tgfb1. Consistent with the notion that tfgb1 is involved in fibrosis, we noted significant expression localized within areas of remodeled tissue (55).
While highly informative, our study is not without limitations. For instance, there is clear antibody bias associated with flow cytometric sorting of myeloid populations which could have excluded subgroups of macrophages and monocytes that would not fit the criteria of expression (i.e., Ly6Cint/lo expressing cells). To reduce deviation from the literature, we followed previously published protocols for tissue dissociation and phenotypic characterization (9, 35). Furthermore, the methodology utilized for this work (whole body CCR2KO strain) cannot differentiate between “true” resident alveolar macrophages and monocyte populations that acquire such phenotype through maturation. As we move forward, our goal is to provide an unbiased approach that accounts for, or at least attempts to, transcriptional trajectories corresponding to biological processes like monocyte maturation (i.e., single cell sequencing with pseudotime) (56, 57).
A number of reports describe the effects of pharmacological depletion of phagocytic cells (clodronate liposomes) in lung injury and fibrosis (9, 27). Though cleared from the body in a matter of hours (58), we previously presented pathophysiological benefits of intravenous liposome administration lasting up to 14 d post SP-CI73T injury. This was juxtaposed to the protective role of resident alveolar macrophages receiving intratracheal clodronate liposome during the initiation of SP-CI73T induced injury (27). Guided by those results, our RNA sequencing analysis of sorted SigF-CD64-CD11b+Ly6C+ monocytes confirms extensive pro-inflammatory and pro-fibrotic activation state. Building on these notions, we opted to focus even further our analysis, by targeting (ablate) a relatively well-established subset of bone marrow-derived inflammatory monocytes (59, 60), identified by their expression of the Monocyte Chemoattractant Protein-1 receptor, CCR2+, and examining its effects in SP-CI73T mice. While this monocyte subset accounts for a small proportion of the monocytic milieu, they are known to participate to the initiation of the inflammatory response and thus promote the exacerbation of injury to a fibrotic phenotype (33, 44). The MCP-1/CCR2 axis has been previously shown to be involved in monocyte egression from bone marrow into the peripheral blood and the site of injury (60, 61). Notably, the aberrantly elevated MCP-1 levels found in the BAL of SP-CI73TCCR2KO cohorts, indicate that in the absence of adequate monocyte response the pulmonary system compensates by exceeding its normal MCP-1 output. Histochemical analysis of homozygous SP-CI73TCCR2RFP mice, in which functional CCR2 was replaced by red fluorescent protein (CCR2RFP), indicates that alternative mechanisms of inflammatory monocyte recruitment may be in place. Our findings exclude two established monocyte recruitment pathways (CCL17 and CX3CL1), thus leaving non-canonical chemokine axes still in play (CXCL12-CXCR4, CCL20-CCR6 and CCL5-CCR5) (62). Furthermore, immunohistochemical analysis reveals accumulation of mature (CD64), activated (Arg1, iNOS, C1q, p-SMAD2/3) macrophages accumulating from the periphery (CX3CR1, CD125/IL5RA). Notably, this aberrant monocyte/macrophage accumulation was solely noted within foci of injury of SP-CI73TCCR2WT mice, while their numbers were comparable to those of SP-CI73TCCR2KO cohorts in non-inflamed areas. This distinct response is likely connected to ablation of highly destructive monocytes in the initial inflammatory response (3 d post SP-CI73T injury), which dampens subsequent bouts of peripheral immune cell recruitment downstream of the injury (14 d). This notion is supported by our findings of reduced size of remodeled foci and inflammatory cell congestion in the alveolar compartment (in particular eosinophils), improved survival, and lower BAL cell counts in SP-CI73TCCR2KO mice. Notable was the surge in Ly6G+ neutrophils in SP-CI73TCCR2KO mice 14 d post injury, an effects possibly linked to the increase in MCP-1 noted in these cohorts (63), and previously observed in other models of lung injury leveraging CCR2 monocyte depletion (64, 65). Together, these data indicate a clear shift in SP-CI73T induced inflammatory responses following CCR2 ablation, which may be mediated by non-canonical inflammatory paths that will require additional analysis (66).
Examination of injury heterogeneity is vital in the context of PF. To this end, we first confirmed our RNA-sequencing results indicating increases in fibrotic pathways including TGFβ1 via histochemical analysis of its downstream effector SMAD2/3 and then studied the spatial correlation of activated (Arg1, iNOS) inflammatory cells with respect to fibrotic foci using RNA in situ hybridization. Our data shows reduced numbers of iNOS+ and Arg1+ cells in the lungs of SP-CI73CCR2KO mice, a notion consistent with reduced inflammatory burden in this cohort. While the limited colocalization between activation molecules (Arg1 and iNOS) and tgfb1 mRNA indicate that neither of those two subsets is involved in direct fibrogenic signaling, we noted increases in the numbers of positive inflammatory cells in areas enriched with tgfb1. This observation is indicative of increased recruitment and perhaps communication between parenchymal and inflammatory cells during early fibrotic remodeling, a notion that we aim to test moving forward.
To conclude, we have provided evidence supporting a pro-injury and pro-fibrotic role of peripheral monocytes (both Ly6C+ and CCR2+) in the initiation and progression of acute inflammatory exacerbations of PF induced by epithelial stress. Using knock out strains and histological techniques we identify activated monocytes/macrophages as disease modifying cells accumulating in proximity to fibrotic foci. In addition, RNA in situ hybridization and sequencing analysis demonstrated their potential to promote extracellular matrix remodeling, while highlighting they are not directly involved in fibrogenic factor production (tgfb1). Taken together, this work advances our understanding of epithelial-immune cell crosstalk in PF.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/geo/, GSE166300.
Ethics Statement
The animal study was reviewed and approved by IACUC. All mice were housed under pathogen free conditions in AALAC approved facility.
Author Contributions
AV designed and analyzed all experiments; AV, YT and BA performed all sample collection; SC performed bulk RNA seq analysis; JK, SM, and MFB provided input on study design and interpretation of the results. All authors contributed to the article and approved the submitted version.
Funding
Research reported in this publication was supported by NIEHS R01ES032553 (AV), VA Merit Review 1I01BX001176 (MB), and NIH R01 HL145408 (MB). JK was supported by NIH K08HL150226, Francis Family Foundation Parker B. Francis Fellowship, and PFF Scholar of the Pulmonary Fibrosis Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MFB is the Albert M. Rose Established Investigator of the Pulmonary Fibrosis Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.665818/full#supplementary-material
Abbreviations
AIE, acute inflammatory exacerbations; AT2, alveolar type-2 cell; SftpcI73T surfactant protein-C I73T mutant; ILD, interstitial lung disease; IPF/PF, idiopathic pulmonary fibrosis; BALF, bronchoalveolar lavage fluid; FACS, fluorescence-activated cell sorting; CCR2, chemokine receptor-2; MoAMs, monocyte-derived alveolar macrophages.
References
- 1. Cameron HS, Somaschini M, Carrera P, Hamvas A, Whitsett JA, Wert SE, et al. A Common Mutation in the Surfactant Protein C Gene Associated With Lung Disease. J Pediatr (2005) 146(3):370–5. 10.1016/j.jpeds.2004.10.028 [DOI] [PubMed] [Google Scholar]
- 2. Abou Taam R, Jaubert F, Emond S, Le Bourgeois M, Epaud R, Karila C, et al. Familial Interstitial Disease With I73T Mutation: A Mid- and Long-Term Study. Pediatr Pulmonol (2009) 44(2):167–75. 10.1002/ppul.20970 [DOI] [PubMed] [Google Scholar]
- 3. Crossno PF, Polosukhin VV, Blackwell TS, Johnson JE, Markin C, Moore PE, et al. Identification of Early Interstitial Lung Disease in an Individual With Genetic Variations in ABCA3 and SFTPC. Chest (2010) 137(4):969–73. 10.1378/chest.09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hambly N, Cox G, Kolb M. Acute Exacerbations of Idiopathic Pulmonary Fibrosis: Tough to Define; Tougher to Manage. Eur Respir J (2017) 49(5):1700811. 10.1183/13993003.00811-2017 [DOI] [PubMed] [Google Scholar]
- 5. Leuschner G, Behr J. Acute Exacerbation in Interstitial Lung Disease. Front Med (2017) 4:176–6. 10.3389/fmed.2017.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Juarez MM, Chan AL, Norris AG, Morrissey BM, Albertson TE. Acute Exacerbation of Idiopathic Pulmonary Fibrosis-a Review of Current and Novel Pharmacotherapies. J Thorac Dis (2015) 7(3):499–519. 10.3978/j.issn.2072-1439.2015.01.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scott MKD, Quinn K, Li Q, Carroll R, Warsinske H, Vallania F, et al. Increased Monocyte Count as a Cellular Biomarker for Poor Outcomes in Fibrotic Diseases: A Retrospective, Multicentre Cohort Study. Lancet Respir Med (2019) 7(6):497–508. 10.1016/S2213-2600(18)30508-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights Into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med (2019) 199(12):1517–36. 10.1164/rccm.201712-2410OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-Derived Alveolar Macrophages Drive Lung Fibrosis and Persist in the Lung Over the Life Span. J Exp Med (2017) 214(8):2387–404. 10.1084/jem.20162152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-Cell RNA-seq Reveals Ectopic and Aberrant Lung-Resident Cell Populations in Idiopathic Pulmonary Fibrosis. Sci Adv (2020) 6(28):eaba1983. 10.1126/sciadv.aba1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith LC, Venosa A, Gow AJ, Laskin JD, Laskin DL. Transcriptional Profiling of Lung Macrophages During Pulmonary Injury Induced by Nitrogen Mustard. Ann N Y Acad Sci (2020) 1480(1):146–54. 10.1111/nyas.14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ardain A, Marakalala MJ, Leslie A. Tissue-Resident Innate Immunity in the Lung. Immunology (2020) 159(3):245–56. 10.1111/imm.13143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-Based Analysis of Lung Single-Cell Sequencing Reveals a Transitional Profibrotic Macrophage. Nat Immunol (2019) 20(2):163–72. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joshi N, Watanabe S, Verma R, Jablonski RP, Chen C-I, Cheresh P, et al. Single-Cell RNA-seq Reveals Spatially Restricted Multicellular Fibrotic Niches During Lung Fibrosis. bioRxiv (2019) p:569855. 10.1101/569855 [DOI] [Google Scholar]
- 15. Liegeois M, Legrand C, Desmet CJ, Marichal T, Bureau F. The Interstitial Macrophage: A Long-Neglected Piece in the Puzzle of Lung Immunity. Cell Immunol (2018) 330:91–6. 10.1016/j.cellimm.2018.02.001 [DOI] [PubMed] [Google Scholar]
- 16. Schyns J, Bureau F, Marichal T. Lung Interstitial Macrophages: Past, Present, and Future. J Immunol Res (2018) 2018:5160794–5160794. 10.1155/2018/5160794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evren E, Ringqvist E, Willinger T. Origin and Ontogeny of Lung Macrophages: From Mice to Humans. Immunology (2020) 160(2):126–38. 10.1111/imm.13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New Insights Into the Multidimensional Concept of Macrophage Ontogeny, Activation and Function. Nat Immunol (2016) 17(1):34–40. 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 19. Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, et al. Alveolar Macrophages Develop From Fetal Monocytes That Differentiate Into Long-Lived Cells in the First Week of Life Via GM-CSF. J Exp Med (2013) 210(10):1977. 10.1084/jem.20131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joshi N, Watanabe S, Verma R, Jablonski RP, Chen C-I, Cheresh P, et al. A Spatially Restricted Fibrotic Niche in Pulmonary Fibrosis is Sustained by M-CSF/M-CSFR Signalling in Monocyte-Derived Alveolar Macrophages. Eur Respir J (2020) 55(1):1900646. 10.1183/13993003.00646-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, et al. Adult-Onset Pulmonary Fibrosis Caused by Mutations in Telomerase. Proc Natl Acad Sci U.S.A. (2007) 104(18):7552–7. 10.1073/pnas.0701009104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomas AQ, Lane K, Phillips J, 3rd3rd, Prince M, Markin C, Speer M, et al. Heterozygosity for a Surfactant Protein C Gene Mutation Associated With Usual Interstitial Pneumonitis and Cellular Nonspecific Interstitial Pneumonitis in One Kindred. Am J Respir Crit Care Med (2002) 165(9):1322–8. 10.1164/rccm.200112-123OC [DOI] [PubMed] [Google Scholar]
- 23. Nureki SI, Tomer Y, Venosa A, Katzen J, Russo SJ, Jamil S, et al. Expression of Mutant Sftpc in Murine Alveolar Epithelia Drives Spontaneous Lung Fibrosis. J Clin Invest (2018) 128(9):4008–24. 10.1172/JCI99287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Povedano JM, Martinez P, Flores JM, Mulero F, Blasco MA. Mice With Pulmonary Fibrosis Driven by Telomere Dysfunction. Cell Rep (2015) 12(2):286–99. 10.1016/j.celrep.2015.06.028 [DOI] [PubMed] [Google Scholar]
- 25. Brasch F, Griese M, Tredano M, Johnen G, Ochs M, Rieger C, et al. Interstitial Lung Disease in a Baby With a De Novo Mutation in the SFTPC Gene. Eur Respir J (2004) 24(1):30–9. 10.1183/09031936.04.00000104 [DOI] [PubMed] [Google Scholar]
- 26. Hawkins A, Guttentag SH, Deterding R, Funkhouser WK, Goralski JL, Chatterjee S, et al. A non-BRICHOS SFTPC Mutant (SP-C(I73T)) Linked to Interstitial Lung Disease Promotes a Late Block in Macroautophagy Disrupting Cellular Proteostasis and Mitophagy. Am J Physiol Lung Cell Mol Physiol (2015) 308(1):L33–47. 10.1152/ajplung.00217.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Venosa A, Katzen J, Tomer Y, Kopp M, Jamil S, Russo SJ, et al. Epithelial Expression of an Interstitial Lung Disease–Associated Mutation in Surfactant Protein-C Modulates Recruitment and Activation of Key Myeloid Cell Populations in Mice. J Immunol (2019) 202(9):2760–71. 10.4049/jimmunol.1900039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shinoda H, Tasaka S, Fujishima S, Yamasawa W, Miyamoto K, Nakano Y, et al. Elevated CC Chemokine Level in Bronchoalveolar Lavage Fluid is Predictive of a Poor Outcome of Idiopathic Pulmonary Fibrosis. Respiration (2009) 78(3):285–92. 10.1159/000207617 [DOI] [PubMed] [Google Scholar]
- 29. Capelli A, Di Stefano A, Gnemmi I, Donner CF. CCR5 Expression and CC Chemokine Levels in Idiopathic Pulmonary Fibrosis. Eur Respir J (2005) 25(4):701–7. 10.1183/09031936.05.00082604 [DOI] [PubMed] [Google Scholar]
- 30. Yogo Y, Fujishima S, Inoue T, Saito F, Shiomi T, Yamaguchi K, et al. Macrophage Derived Chemokine (CCL22), Thymus and Activation-Regulated Chemokine (CCL17), and CCR4 in Idiopathic Pulmonary Fibrosis. Respir Res (2009) 10:80. 10.1186/1465-9921-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belperio JA, Dy M, Murray L, Burdick MD, Xue YY, Strieter RM, et al. The Role of the Th2 Cc Chemokine Ligand CCL17 in Pulmonary Fibrosis. J Immunol (2004) 173(7):4692. 10.4049/jimmunol.173.7.4692 [DOI] [PubMed] [Google Scholar]
- 32. Inoue T, Fujishima S, Ikeda E, Yoshie O, Tsukamoto N, Aiso S, et al. CCL22 and CCL17 in Rat Radiation Pneumonitis and in Human Idiopathic Pulmonary Fibrosis. Eur Respir J (2004) 24(1):49. 10.1183/09031936.04.00110203 [DOI] [PubMed] [Google Scholar]
- 33. Young LR, Gulleman PM, Short CW, Tanjore H, Sherrill T, Qi A, et al. Epithelial-Macrophage Interactions Determine Pulmonary Fibrosis Susceptibility in Hermansky-Pudlak Syndrome. JCI Insight (2016) 1(17):e88947. 10.1172/jci.insight.88947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Venosa A, Malaviya R, Gow AJ, Hall L, Laskin JD, Laskin DL. Protective Role of Spleen-Derived Macrophages in Lung Inflammation, Injury, and Fibrosis Induced by Nitrogen Mustard. Am J Physiol Lung Cell Mol Physiol (2015) 309(12):L1487–98. 10.1152/ajplung.00276.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow Cytometric Analysis of Macrophages and Dendritic Cell Subsets in the Mouse Lung. Am J Respir Cell Mol Biol (2013) 49(4):503–10. 10.1165/rcmb.2013-0086MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nureki SI, Tomer Y, Venosa A, Katzen J, Russo SJ, Jamil S, et al. Expression of Mutant Sftpc in Murine Alveolar Epithelia Drives Spontaneous Lung Fibrosis. J Clin Invest (2018) 128(9):4008–24. 10.1172/JCI99287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. R Core Team R. R: A Language and Environment for Statistical Computing. Vienna, Austria: R foundation for statistical computing; (2013). [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-seq Data With Deseq2. Genome Biol (2014) 15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korotkevich G, Sukhov V, Sergushichev A. Fast Gene Set Enrichment Analysis. bioRxiv (2021) 060012. 10.1101/060012 [DOI] [Google Scholar]
- 40. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res (2002) 30(1):207–10. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez FO, Sica A, Mantovani A, Locati M. Macrophage Activation and Polarization. Front Biosci (2008) 13:453–61. 10.2741/2692 [DOI] [PubMed] [Google Scholar]
- 42. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol (2004) 25(12):677–86. 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 43. Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity (2016) 44(3):450–62. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Francis M, Groves AM, Sun R, Cervelli JA, Choi H, Laskin JD, et al. Editor’s Highlight: CCR2 Regulates Inflammatory Cell Accumulation in the Lung and Tissue Injury Following Ozone Exposure. Toxicol Sci (2017) 155(2):474–84. 10.1093/toxsci/kfw226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boivin N, Menasria R, Gosselin D, Rivest S, Boivin G. Impact of Deficiency in CCR2 and CX3CR1 Receptors on Monocytes Trafficking in Herpes Simplex Virus Encephalitis. J Gen Virol (2012) 93(Pt 6):1294–304. 10.1099/vir.0.041046-0 [DOI] [PubMed] [Google Scholar]
- 46. Behr J, Richeldi L. Recommendations on Treatment for IPF. Respir Res (2013) 14(Suppl 1):S6–6. 10.1186/1465-9921-14-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richeldi L, Davies H, Spagnolo P, Luppi F. Corticosteroids for Idiopathic Pulmonary Fibrosis. Cochrane Database Syst Rev (2003) 3:CD002880. 10.1002/14651858.CD002880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Papiris SA, Manali ED, Kolilekas L, Triantafillidou C, Tsangaris I, Kagouridis K. Steroids in Idiopathic Pulmonary Fibrosis Acute Exacerbation: Defenders or Killers? Am J Respir Crit Care Med (2012) 185(5):587–8. 10.1164/ajrccm.185.5.587 [DOI] [PubMed] [Google Scholar]
- 49. Stevens PA, Pettenazzo A, Brasch F, Mulugeta S, Baritussio A, Ochs M, et al. Nonspecific Interstitial Pneumonia, Alveolar Proteinosis, and Abnormal Proprotein Trafficking Resulting From a Spontaneous Mutation in the Surfactant Protein C Gene. Pediatr Res (2005) 57(1):89–98. 10.1203/01.PDR.0000147567.02473.5A [DOI] [PubMed] [Google Scholar]
- 50. Selman M, King TE, Pardo A. Idiopathic Pulmonary Fibrosis: Prevailing and Evolving Hypotheses About its Pathogenesis and Implications for Therapy. Ann Intern Med (2001) 134(2):136–51. 10.7326/0003-4819-134-2-200101160-00015 [DOI] [PubMed] [Google Scholar]
- 51. Diaz de Leon A, Cronkhite JT, Katzenstein A-LA, Godwin JD, Raghu G, Glazer CS, et al. Telomere Lengths, Pulmonary Fibrosis and Telomerase (TERT) Mutations. PloS One (2010) 5(5):e10680. 10.1371/journal.pone.0010680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bilgili H, Białas AJ, Górski P, Piotrowski WJ. Telomere Abnormalities in the Pathobiology of Idiopathic Pulmonary Fibrosis. J Clin Med (2019) 8(8):1232. 10.3390/jcm8081232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beers MF, Hawkins A, Maguire JA, Kotorashvili A, Zhao M, Newitt JL, et al. A Nonaggregating Surfactant Protein C Mutant is Misdirected to Early Endosomes and Disrupts Phospholipid Recycling. Traffic (Copenhagen Denmark) (2011) 12(9):1196–210. 10.1111/j.1600-0854.2011.01223.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cutroneo KR, White SL, Phan SH, Ehrlich HP. Therapies for Bleomycin Induced Lung Fibrosis Through Regulation of TGF-beta1 Induced Collagen Gene Expression. J Cell Physiol (2007) 211(3):585–9. 10.1002/jcp.20972 [DOI] [PubMed] [Google Scholar]
- 55. Yue X, Shan B, Lasky JA. Tgf-β: Titan of Lung Fibrogenesis. Curr Enzyme Inhibition (2010) 6(2):10.2174/10067. 10.2174/10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion Pseudotime Robustly Reconstructs Lineage Branching. Nat Methods (2016) 13(10):845–8. 10.1038/nmeth.3971 [DOI] [PubMed] [Google Scholar]
- 57. Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, et al. Alveolar Regeneration Through a Krt8+ Transitional Stem Cell State That Persists in Human Lung Fibrosis. Nat Commun (2020) 11(1):3559. 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Rooijen N, Kors N, vd Ende M, Dijkstra CD. Depletion and Repopulation of Macrophages in Spleen and Liver of Rat After Intravenous Treatment With Liposome-Encapsulated Dichloromethylene Diphosphonate. Cell Tissue Res (1990) 260(2):215–22. 10.1007/BF00318625 [DOI] [PubMed] [Google Scholar]
- 59. Hammond MD, Taylor RA, Mullen MT, Ai Y, Aguila HL, Mack M, et al. Ccr2+ Ly6C(hi) Inflammatory Monocyte Recruitment Exacerbates Acute Disability Following Intracerebral Hemorrhage. J Neurosci Off J Soc Neurosci (2014) 34(11):3901–9. 10.1523/JNEUROSCI.4070-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of Splenic Reservoir Monocytes and Their Deployment to Inflammatory Sites. Science (2009) 325(5940):612–6. 10.1126/science.1175202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gelman AE, Okazaki M, Sugimoto S, Li W, Kornfeld CG, Lai J, et al. CCR2 Regulates Monocyte Recruitment as Well as CD4 T1 Allorecognition After Lung Transplantation. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surgeons (2010) 10(5):1189–99. 10.1111/j.1600-6143.2010.03101.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Argyle D, Kitamura T. Targeting Macrophage-Recruiting Chemokines as a Novel Therapeutic Strategy to Prevent the Progression of Solid Tumors. Front Immunol (2018) 9:2629–9. 10.3389/fimmu.2018.02629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fujimura N, Xu B, Dalman J, Deng H, Aoyama K, Dalman RL. CCR2 Inhibition Sequesters Multiple Subsets of Leukocytes in the Bone Marrow. Sci Rep (2015) 5(1):11664. 10.1038/srep11664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gaupp S, Pitt D, Kuziel WA, Cannella B, Raine CS. Experimental Autoimmune Encephalomyelitis (EAE) in CCR2(-/-) Mice: Susceptibility in Multiple Strains. Am J Pathol (2003) 162(1):139–50. 10.1016/S0002-9440(10)63805-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chu HX, Arumugam TV, Gelderblom M, Magnus T, Drummond GR, Sobey CG. Role of CCR2 in Inflammatory Conditions of the Central Nervous System. J Cereb Blood Flow Metab Off J Int Soc Cereb Blood Flow Metab (2014) 34(9):1425–9. 10.1038/jcbfm.2014.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang J, Agarwal M, Ling S, Teitz-Tennenbaum S, Zemans RL, Osterholzer JJ, et al. Diverse Injury Pathways Induce Alveolar Epithelial Cell CCL2/12, Which Promotes Lung Fibrosis. Am J Respir Cell Mol Biol (2020) 62(5):622–32. 10.1165/rcmb.2019-0297OC [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/geo/, GSE166300.