Abstract
CD8+ T-cell infiltration and effector activity in tumors are correlated with better overall survival of patients, suggesting that the ability of T cells to enter and remain in contact with tumor cells supports tumor control. CD8+ T cells express the collagen-binding integrins CD49a and CD49b, but little is known about their function or how their expression is regulated in the tumor microenvironment (TME). Here, we found that tumor-infiltrating CD8+ T cells initially expressed CD49b, gained CD49a, and then lost CD49b over the course of tumor outgrowth. This differentiation sequence was driven by antigen-independent elements in the TME, although T-cell receptor (TCR) stimulation further increased CD49a expression. Expression of exhaustion markers and CD49a associated temporally, but not mechanistically. Intratumoral CD49a-expressing CD8+ T cells failed to upregulate TCR-dependent Nur77 expression, while CD69 was constitutively expressed, consistent with both a lack of productive antigen engagement and a tissue-resident memory-like phenotype. Imaging T cells in live tumor slices revealed that CD49a increased their motility, especially of those in close proximity to tumor cells, suggesting that it may interfere with T-cell recognition of tumor cells by distracting them from productive engagement, although we were not able to augment productive engagement by short-term CD49a blockade. CD49b also promoted relocalization of T cells at a greater distance from tumor cells. Thus, our results demonstrate that expression of these integrins impacts T-cell trafficking and localization in tumors via distinct mechanisms, and suggests a new way in which the TME, and likely collagen, could promote tumor-infiltrating CD8+ T-cell dysfunction.
Introduction
Survival of cancer patients is prolonged in those whose tumors are robustly infiltrated by T cells (1,2). T-cell representation in tumors depends on their ability to utilize homing mechanisms and to be retained and survive in the tumor microenvironment (TME)(3). Tumor infiltration by T cells depends on expression of homing receptor ligands VCAM-1, ICAM-1, E-selectin, and CXCR3-binding chemokines on tumor-associated vasculature (4–6). However, T-cell retention and localization in tumors is less well-studied and likely depends on antigen recognition, interaction with extracellular matrix (ECM) proteins, and expression of molecules that mediate egress into lymphatics (7–14). Specifically, interaction with the ECM could either retain T cells in tumors or sequester T cells in ECM-rich areas, preventing them from engaging with tumor cells (15,16). T-cell interactions with ECM proteins can be mediated by integrins such as α1β1 (CD49a) and α2β1 (CD49b), which predominantly bind to collagen type IV and type I, respectively (4,17–19). Blocking CD49a interaction with collagen in vivo decreases gut intraepithelial CD8+ T-cell number under homeostatic conditions and total T-cell numbers in mucosal tumors (20,21), suggesting that CD49a participates in T-cell retention in those tissues. Interestingly, CD49a signaling is associated with increased T-cell motility (22–24). Thus, interactions between collagens and integrins in tumors could determine T-cell number, localization, and ability to engage with target cells. Importantly, how CD49a and CD49b specifically impacts these processes remains to be elucidated.
CD49b is expressed on a fraction of effector T cells in the context of arthritis, influenza or LCMV infection, and in tumors, whereas CD49a expression is largely confined to effector cells localized to peripheral tissue sites (17,19,25,26) and after HSV infection (27). CD49a is also expressed on tissue-resident memory T cells (TRM) in lung, skin, and mucosal sites (27,28). Neither CD49a nor CD49b are expressed on naïve T cells or circulating memory T cells (17,29). T-cell receptor (TCR)-mediated activation is thus likely required for their expression, whereas CD49a may require an additional stimulus provided by the peripheral tissue microenvironment. We previously observed increased expression of CD49a and CD49b on human CD8+ T-cells after in vitro TCR stimulation, which was further enhanced by TGFβ, TNFα, and IL2 (29). Others found that expression of CD49a on T cells in mice depend on TGFβ, IL12, or Notch (27,30,31). However, it is unknown whether these are dominant mediators of CD49a and CD49b expression during an immune response in vivo, or in chronic inflammatory environments or tumors. Thus, it is important to further understand the regulation and expression dynamics of collagen-binding integrins CD49a and CD49b in vivo.
Here, we determined the dynamics of CD49a and CD49b integrin expression on CD8+ T cells during vaccine-induced and anti-tumor immune responses. We also tracked the differentiation of adoptively transferred integrin-expressing T cells in tumor-bearing hosts. We established separate roles of antigen-driven differentiation and the TME in controlling CD8+ T-cell expression of CD49a and CD49b. We also determined how integrin expression correlated with expression of exhaustion and TRM markers and T-cell functional capacity in tumors and in vitro. Our studies demonstrate an important role for these integrins in altering T-cell motility and localization in tumors that may suggest a new way in which the TME, and likely collagen, could promote tumor-infiltrating CD8+ T-cell dysfunction.
Methods
Patient samples
Metastatic melanoma surgical specimens were obtained from patients with informed written consent. Studies were approved by the University of Virginia Institutional Review Board (IRB10598), in accordance with the Declaration of Helsinki, the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS), the Belmont Report, and the U.S. Common Rule. Patients ranged between age 28–87 with stage IIB-IV melanoma, and the cohort included 12 males and 7 females. Melanoma samples were made into single-cell suspensions by mechanical separation and filtered through a 100 μm filter, within 4 hours of collection (median of approximately an hour). Single-cell suspensions were cryopreserved in 90% FCS and 10% DMSO using a controlled rate cell freezing container. Cells were cryopreserved in liquid nitrogen ranging from 1 up to 12 years. Samples were thawed in DNAse (100U/ml; Worthington Biochemical Corp.) containing media (RPMI 1640 with 5% heat-inactivated (HI) fetal calf serum (FCS)(Gibco) and counted on a Guava EasyCyte Plus benchtop flow cytometer (cell viability ranged between 33–78% with a median yield of 1.65E+07 viable cells). After thaw, cells were directly stained for flow cytometry.
Mice
C57BL/6 mice were from Charles River/NCI. Genetic strains, all on C57BL/6 background: CD2-dsRed (provided by Jordan Jacobelli, University of Colorado, Anschutz Medical Campus), Nur77-GFP reporter (C57BL/6-Tg(Nr4a1-EGFP/cre)820Khog/J)(32), OT-I transgenic (C57BL/6-Tg(TcraTcrb)1100Mjb/J), and Thy1.1 congenic mice (B6.PL-Thy1a/CyJ)(all from Jackson Laboratories) were bred and maintained in a pathogen-free facility at the University of Virginia. Six to 12-week-old OT-IxThy1.1 F1 and Nur77-GFP x (OT-IxThy1.1) F1 mice were the sources of OT-I cells used for adoptive transfer. All procedures were approved by the University of Virginia Animal Care and Use Committee in accordance with the NIH Guide for Care and Use of Laboratory Animals.
Cell lines
The poorly metastatic syngeneic C57BL/6-derived GFP+ breast tumor cell line Brpkp110 (33,34) was developed in 2014 and acquired by the Rutkowski lab in 2015. The Brpkp110 primary mammary tumor cell line was generated by culturing a mechanically dissociated B6 L-Stop-L-KRasG12Dp53flx/flxL-Stop-L-Myristoylated p110α−GFP/+ (35) primary breast tumor mass. Tumor cells were passaged 10 times before monoclonally deriving the Brpkp110 cell line. Clones were selected that retained epithelial morphology, including Brpkp110. Cells were authenticated by confirming positivity for estrogen receptor and progesterone receptor expression, and negativity for Her2 expression by immunohistochemistry and analysis by a pathologist. Tumors have been validated to exhibit cytokeratin 8, but not 5, positivity – resembling a Luminal A subtype. Tumor cells are kept at low passage number, and tested for mycoplasma frequently. BRPKp110 cells were cultured in RPMI-1640 (Corning) supplemented with 10% FBS, 0.05 mM β-mercaptoethanol (both from Sigma), 2 mM L-glutamine, and 10 mM sodium pyruvate (both from Gibco).
B16-F1 cells were obtained from the American Type Culture Collection (ATCC) sometime in 1999–2000 and transfected with cytoplasmic ovalbumin (OVA) using an expression vector provided by A. Lew (Walter & Eliza Hall Institute of Medical Research, Melbourne, Australia)(37). B16-F1 and OVA-transfected B16-F1 (B16-OVA) were cultured in RPMI-1640 (Corning) supplemented with 5% FBS (Sigma), 15 mM HEPES and 2 mM L-glutamine (both from Gibco). 10μg/ml Blasticidin (Gibco) was added to maintain OVA expression in B16-OVA. Cells were authenticated by visual confirmation of melanin pigment production in vitro and in vivo, and OVA expression confirmed by staining with the H-2Kb+OVA peptide specific antibody 25-D1.16. All cultured cells injected into mice were within 2–8 passages after thaw and mycoplasma free.
Tumor induction
BRPKp110, B16-F1, and B16-OVA cells (4×105) in 200 μL phosphate-buffered saline (PBS) were injected subcutaneously (SC) in the neck scruff of C57BL/6, Nur77-GFP reporter, or CD2-dsRed mice. Mice were monitored for weight loss, signs of distress and tumor size every 2–3 days. Where indicated, mice were injected intraperitoneally (IP) daily with 5 μg/mL FTY720 (Novartis) or saline control for the duration of tumor growth, or intravenously (IV) with 250 μg Brefeldin A (Sigma) 4–6 hours prior to harvest. At the time of harvest mice were euthanized and tumor, tumor-draining lymph node, non-tumor-draining lymph node, blood and/or spleen were collected and processed as outlined below.
Tumor cell lysates
BRPKp110 (day 28) and B16-OVA (day 14) tumors from C57BL/6 mice or cultured BRPKp110 cells were incubated in H2O for 20 minutes and sonicated (Sonic Dismembrator Model 500, Fisher Scientific, 27% amplitude) on ice for 20× 30 seconds. Lysates were centrifuged (10,000xg) for 20 minutes, filtered through a 0.2μm filter and adjusted to 1X PBS, 10% FBS, 0.05 mM β-mercaptoethanol (both from Sigma), 15 mM Hepes, 2 mM L-glutamine, 10 mM sodium pyruvate, 1X non-essential/essential amino-acids, and gentamicin (1 μg/mL) (all from Gibco). Protein concentrations were determined by BCA assay (Gibco). Lysates were used in in vitro T-cell activation assays below.
For cytokine analysis, BRPKp110 and B16-OVA tumors were harvested on day 14, incubated for 20 minutes on ice in 150 mM NaCl, 20 mM Tris, 1 mM EGTA, 1% Triton X-100 (all from Sigma), 1 mM EDTA (Fisher), Roche Complete Protease Inhibitor (1 tablet per 10ml buffer), and sonicated on ice 3x for 30 seconds (28% amplitude). Lysates were centrifuged (15,000xg) for 5 minutes. Protein concentration was determined by BCA assay, and samples of 20 μg were analyzed using a custom cytokine array (see Supplementary Table S2 for composition) and a MESO QuickPlex SQ 12 instrument (Meso Scale Discovery). Manufacturer-provided cytokine standards were used to determine cytokine concentrations in tumor lysates.
Analysis of tumor-infiltrating T cells
BRPKp110, B16-F1 and B16-OVA tumors from C57BL/6 or Nur77-GFP reporter mice were harvested in RPMI-1640 (Corning) supplemented with 2% FBS, 0.05 mM β-mercaptoethanol, 40 μg/mL DNase (all from Sigma), 15 mM HEPES, 2 mM L-glutamine, 10 mM sodium pyruvate, 1X essential and non-essential amino acids, gentamicin (1 μg/mL)(all from Gibco), and liberase™ (76 μg/mL)(Roche). Tumors were digested for 15 minutes at 37°C, manually homogenized, and filtered through 70 μm mesh (Miltenyi) to prepare single cell suspensions. The CD45+ fraction was enriched with CD45 MicroBeads mouse (Miltenyi) and analyzed by flow cytometry.
In vitro T-cell activation and culture
Single-cell suspensions of BRPKp110 tumors from C57BL/6 or Nur77-GFP reporter mice were prepared as described above. Single-cell suspensions of splenocytes from OT-IxThy1.1 or C57BL/6 mice were generated by manual homogenization, filtered through 70 μm mesh (Miltenyi), and treated with RBC lysis buffer (Sigma). CD8 T cells were magnetically enriched using CD8 (Ly-2) MicroBeads mouse (Miltenyi Biotec) using an AutoMACS instrument (tumor) or QuadroMACS magnets (both from Miltenyi) per the manufacturer’s protocol. Enriched CD8+ cells were cultured (1×106 cells/mL) in RPMI-1640 with 10% FBS, 15 mM Hepes, 2 mM L-glutamine, 10 mM sodium pyruvate, 1X essential and non-essential amino-acids, gentamicin (1 μg/mL), 0.05mM β-mercaptoethanol, human recombinant IL2 (120 IU/mL), and murine recombinant IL7 (10 ng/mL)(both from Peprotech). Tumor-derived cells were stimulated with CD3/CD28 T activator beads (10 μL/mL, Gibco) for 12 hours for assessment of Nur77-GFP or for 4–6 hours in presence of 10 μg/mL Brefeldin A for intracellular cytokine production, by flow cytometry. Splenocytes were stimulated with CD3/CD28 T activator beads for 48 hours and the beads were magnetically removed with a MagniSort Magnet (eBioscience). Cells were split to 1×106/mL with fresh medium every 2–3 days for a total of 7 days after initial harvest. Where indicated, 1×106/mL cells were subsequently cultured for 24 hours with tumor lysate (200 μg/mL) with or without anti-IL12 (10 ng/mL; BioXcell) or anti-TGFβ1–3 (10 μg/mL; BioXcell), or with recombinant mouse TNFα (20 ng/mLl), IL1β (10 ng/mL), IL33 (10 ng/mL), or human TGFβ1 (5 ng/mL)(all Peprotech). Cells were then evaluated for expression of CD49a and CD49b by flow cytometry.
Adoptive cell transfer
For analysis of responses to vaccination and tumor implantation, single-cell suspensions of splenocytes from naïve OT-IxThy1.1 or Nur77-GFPxOT-IxThy1.1 mice were prepared as described in the previous section, but were immediately injected IV (5×104) into Thy1.2+ C57BL/6 mice. The next day, recipient mice were either vaccinated IV with 500 μg ovalbumin (Sigma), 50 μg anti-CD40 (BioXcell), and 100 μg polyIC (InvivoGen), or were implanted SC with B16-OVA cells. Spleens were harvested at different time points, single cell suspensions prepared as described above, and cells were analyzed by flow cytometry. For analysis of CD49SP differentiation, spleens were harvested 5 days after the vaccination described above. Splenocytes were isolated and CD49a depleted and CD49b enriched using biotinylated CD49a- (Miltenyi) and CD49b-specific (HMα2, BioLegend) antibodies, anti-Biotin MicroBeads (Miltenyi) and QuadroMACS magnets (Miltenyi), per the manufacturer’s protocol. CD49a-depleted and CD49b-enriched Thy1.1+ cells (1×106) in PBS supplemented with 3,000U IL2 were injected IV into tumor-free or tumor-bearing (BRPKp110, B16-F1 or B16-OVA) mice. Spleen, blood and/or tumor were harvested as described above at 1, 4 or 7 days after adoptive transfer. Where indicated, mice were injected IP every day with 5 μL/mL FTY720 or saline control.
In vivo antibody blockade
BRPKp110 tumor cells (4×105) were injected SC into C57BL/6 mice. Mice were treated IP with anti-PD-1 (RMP1), anti-LAG-3 (C9B7W), and anti-TIM-3 (RMT2–23) or matching IgG isotype controls (HRPN and 2A3)(all from BioXCell, 250μg/mouse per antibody) 48 hours prior to tumor harvest at day 19. In other experiments, mice were treated IV with 200μg/mouse anti-CD49a (Ha31/8, BD Biosciences) 24 hours prior to tumor harvest at day 21.
Flow cytometry
Cells were Fc blocked with 1:1000 anti-CD16/CD32 (2.4G2, BioXcell) or human BD Fc Block (BD Biosciences) and stained with Live/Dead Fixable Aqua (Life Technologies) in PBS for 20 minutes at 4°C. Subsequently cells were stained with fluorescently labeled antibodies (Supplementary Table S1) in PBS supplemented with 2% FBS and 0.1% sodium azide for 30 minutes at 4°C. For extracellular stains only, cells were fixed in 2% paraformaldehyde (Thermo Scientific) for 10 minutes at 4°C. BD Cytofix/Cytoperm and BD Transcription Factor Staining kits were used for intracellular and intranuclear stains, according to manufacturer’s protocol. Cells were analyzed on Cytoflex (Beckman Coulter) or Attune (BD Biosciences) flow cytometers using FlowJo software. The gating strategy to define CD3+ CD8+ T cells is shown in Supplementary Fig. S1. In experiments that analyzed OT-I cells, CD3+ CD8+ T cells were subsequently gated on Thy1.1+ before assessing CD49a and CD49b expression. For direct ex vivo Nur77 assessment, Nur77 geometric mean fluorescent intensity (gMFI) on integrin-expressing populations was normalized to CD8+ T cells in non-draining lymph nodes (NDLN) and %Nur77 was determined after subtraction of values from CD8+ T cells from NDLN in the same mice.
Immunofluorescent microscopy
Tumors were placed in 4% paraformaldehyde for 1–2 hours, followed by 30% sucrose overnight. Sections were stained as described (38), with biotinylated collagen type I or collagen type IV antibodies (1:500, Southern Biotech) overnight at 4°C, followed by 1:1000 DyLight550-conjugated streptavidin (ThermoFisher), for 1 hour at room temperature. Images were obtained on a Zeiss AxioImager with Zen software.
Live tumor slice image acquisition and analysis
BRPKp110-GFP tumor cells (4×105) were implanted SC into CD2-dsRed mice. Tumors were harvested on day 21/22 and embedded in 2% low-melting point agarose (Lonza). Three 100–200 μm slices were cut at room temperature with a 51425 Tissue Slicer (Stoelting), placed in RPMI-1640 without phenol red (Gibco), and left untreated or treated with anti-CD49a (clone Ha31/8) or anti-CD49b (clone HMα2, BD Biosciences)(both 10 μg/mL) for 2 hours. Slices were mounted in a heating chamber with circulating oxygenated (bubbling 95% oxygen and 5% CO2) RPMI-1640 without phenol red at a flow rate of ~2 mL/min, and imaged using a Leica TCS SP8 two-photon microscope with a 25X water-immersion lens (0.9 Numerical Aperture) and a Coherent Chameleon laser at 880nm/25mW or below. Fluorescence was detected using a 565 nm dichroic mirror with 450/50 nm (for second harmonics generation (SHG) detection), 525/50 nm (for GFP detection) and 620/60 nm (for dsRed detection) emission filters. A 78–84 μm z-stack (3 μm step size) was collected every minute for 30 minutes, alternating between two fields every 30 seconds. Video image analysis was performed using Imaris (Bitplane) and MATLAB (Mathworks) software as described (39). T cells tracked for ≥10 minutes were used to obtain speed and interaction data. “Contact” interactions were defined as a minimum of 2 timepoints at <1 μm, whereas “sustained interactions” were defined as ≥10 minute at <1 μm.
Statistical analysis
Integrin-expressing subpopulations enumerated by flow cytometry were compared using a repeated-measures one-way ANOVA with Tukey’s multiple comparisons test. Time points and treatment groups were compared using a Welch’s corrected T-test. For live image analyses, all evaluated T-cell spots in individual slices from two tumors per treatment group were combined, and groups were compared using a one-way ANOVA with Tukey’s multiple comparisons test.
Results
CD49a+ and CD49b+ CD8+ T-cell subpopulations in B16 melanoma and BRPKp110 breast tumors
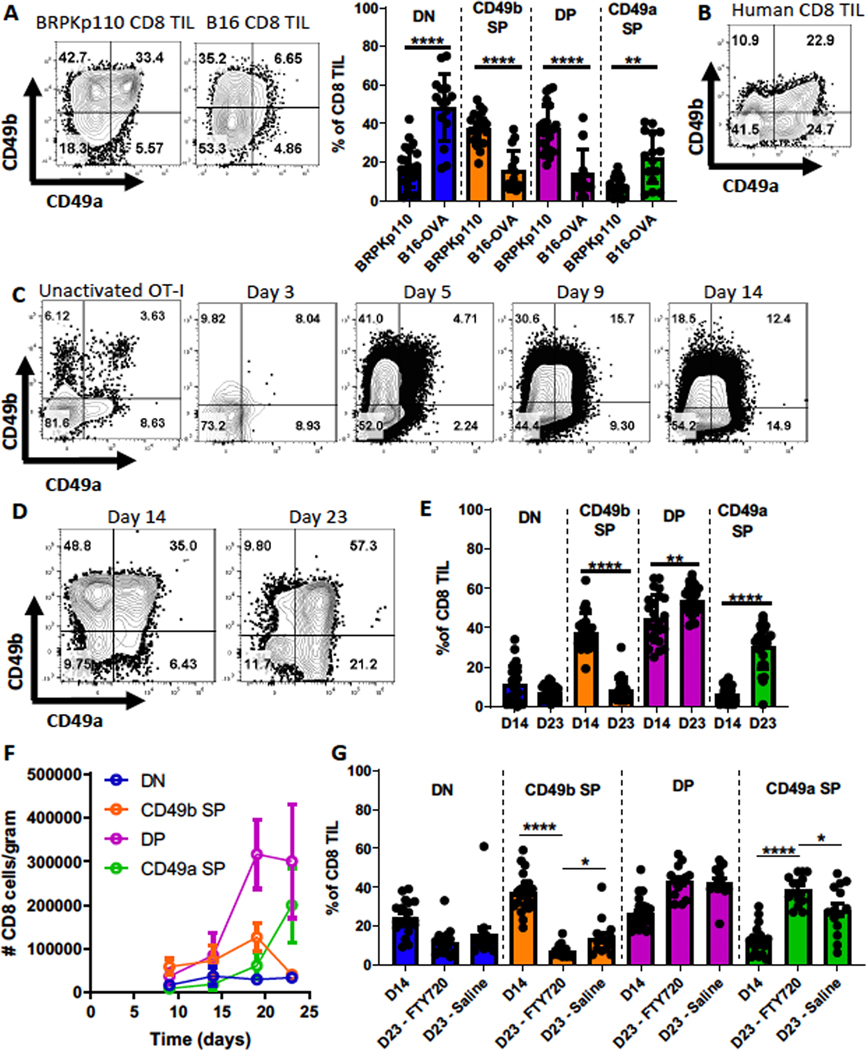
We previously found that CD49a and CD49b are expressed on CD8+ tumor-infiltrating lymphocytes (TILs) from human melanomas but are almost completely absent on CD8+ T cells from normal donor PBMCs or tumor-free lymph nodes from patients (29,40). Expression of these integrins identified subpopulations that varied in expression of perforin, CD127, PD-1, LAG-3 and TIM-3. To elucidate the development of these subpopulations, we utilized two murine tumor models: collagen-rich breast cancer BRPKp110 and collagen-poor melanoma B16. In both, we observed CD49a/CD49b double-negative (DN), CD49b single-positive (SP), CD49a/CD49b double-positive (DP), and CD49a SP populations (Fig. 1A). BRPKp110 contained proportionally more CD49b SP and DP cells than B16-OVA (Fig. 1A). Although the CD49b SP subpopulation was more abundant in both models than in human TILs (Fig. 1B), all subpopulations were present in both mouse and human TILs.
Figure 1. CD49b- and CD49a-expressing CD8+ T cells arise sequentially during immune responses.
(A) CD45+ cells from SC day 14 BRPKp110 breast carcinoma (n=21, 4 independent experiments) or B16-OVA melanoma (n=15, 3 independent experiments) were analyzed for CD49a and CD49b expression on CD3+CD8+ cells. Subsets were compared using repeated-measures (RM) one-way ANOVA and Tukey’s multiple comparisons tests. (B) Representative example of CD49a and CD49b expression on CD3+CD8+ cells from human metastatic melanoma lesions. CD8 T cells were analyzed in an earlier publication (29) and the data replotted here to enable a direct comparison with panel A. (C) Thy1.1+ OT-I splenocytes were transferred IV into C57BL/6 mice, which were subsequently immunized with OVA, polyIC, and anti-CD40. Representative CD49a and CD49b expression on Thy1.1+ CD3+ CD8+ splenocytes from OT-IxThy1.1 mice (unactivated OT-I, pre-transfer) or WT mice (post-transfer with OT-I and immunized), determined on the indicated days (n=3 mice/timepoint). (D-G) CD49a and CD49b expression on CD3+CD8+ cells from SC BRPKp110 tumors harvested on the indicated days was determined by flow cytometry. (E) n=23–24/group, 5 independent experiments. (F) n=6–8 mice/timepoint, 2 independent experiments. (G) Groups of BRPKp110-bearing mice received daily IP injections with either saline or FTY720, starting on day 14. n=14–20 mice/group, 3 independent experiments. (E-G) Groups were compared using Welch’s corrected T-test. Bar graphs and error bars indicate mean +/− standard deviation (SD). Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
CD49b- and CD49a-expressing CD8+ T cells appear sequentially
To understand the regulation of CD49a and CD49b expression during an immune response, we evaluated ovalbumin (OVA)-specific OT-I cells after immunization in tumor-free mice. Three days after activation, expression of CD49a and CD49b remained low and comparable to unactivated OT-I cells (Fig. 1C). By day 5, the fraction of OT-I cells expressing CD49b increased, whereas the fraction expressing CD49a did not increase until day 9 (Fig. 1C, Supplementary Fig. S2A). By day 14, an overall decline in cells expressing CD49b was observed (Fig. 1C). Next, we quantitated CD49a- and CD49b-expressing CD8+ TIL subpopulations during BRPKp110 tumor outgrowth. All subpopulations were evident 14 and 23 days after tumor implantation (Fig. 1D). However, the fraction of CD49b SP cells was higher on day 14, and the fractions of CD49a SP cells and DP cells were increased by day 23 (Fig. 1E–F). These transitions were accompanied by an early increase in the absolute number of CD49b SP and DP cells, followed by a decline in CD49b SP cells and an increase in the absolute number of CD49a SP cells (Fig. 1F). The absolute number of DN cells remained low throughout. Thus, these subpopulations varied over the course of an immune response.
Change in integrin expression occurs specifically in the TME
Our results suggested that during vaccine-induced and anti-tumor immune responses, either CD49b and CD49a are sequentially expressed on the same CD8+ TIL or independent subpopulations are sequentially generated. To test the latter hypothesis, we utilized FTY720 to block new T-cell infiltration from the periphery into tumors, starting 14 days after implantation. Despite blockade, the total CD8+ T-cell infiltrate was not changed at day 23 post-implantation (Supplementary Fig. S2B), but the proportion of CD49a SP cells still increased between day 14 and 23 (Fig. 1G). This increase was also significantly greater after FTY720 treatment compared to the saline control (Fig. 1G), potentially because entry of new CD49b SP cells was also blocked. These data suggest that CD49b SP cells differentiate into CD49a SP cells in the TME, with DP cells as a likely intermediate.
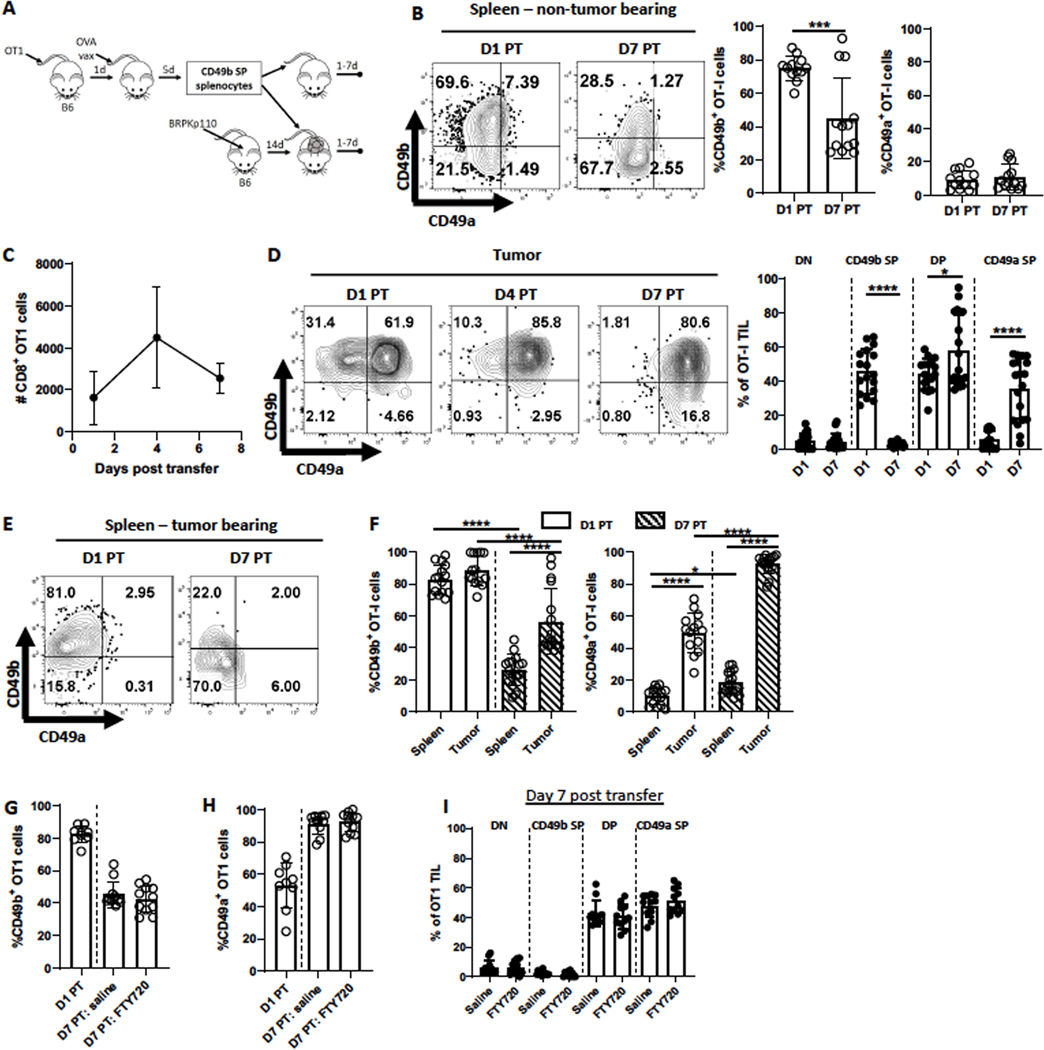
CD49a is upregulated on CD49b SP CD8+ T cells after entry into the TME
To determine whether the TME drives upregulation of CD49a on CD49b SP cells independent of cognate antigen, we transferred activated CD49b SP OT-I cells, purified from spleens of immunized mice (Fig. 1C), into either naïve, tumor-free, or established (day 14) OVA-negative BRPKp110 tumor-bearing mice (Fig. 2A). Seven days later, CD49b SP OT-I cells in non-tumor bearing mice had significantly diminished expression of CD49b but failed to upregulate CD49a (Fig. 2B). This contrasted with the upregulation of CD49a on these cells in the original immunized mice (Fig. 1C), suggesting it was upregulated by vaccination-associated environmental factors. CD49b SP OT-I cells in mice with BRPKp110 tumors trafficked to tumors consistently and numerously, despite the lack of OVA expression (Fig. 2C). CD49a was expressed on 50–60% of OT-I TILs within 24 hours and on virtually all cells after 7 days (Fig. 2D). At early times, most cells were DP, but a CD49a SP subpopulation was evident after 7 days, suggesting DP OT-I cells lose CD49b over time (Fig. 2D). In contrast, transferred CD49b SP OT-I cells isolated from the spleens of these tumor-bearing mice never upregulated CD49a and expressed significantly less CD49b (Fig. 2E–F), similar to their behavior in non-tumor bearing mice (Fig. 2B).
Figure 2. CD8+ T cells differentiate from CD49b SP to DP to CD49a SP in the TME.
(A) Experimental schematic. Details in Methods and Results. (B-I) Spleens from (B) tumor-free mice or (C-I) tumors or spleens from BRPKp110 tumor-bearing mice were harvested on the indicated days post transfer (PT) of CD49b SP Thy1.1+ OT-I effectors. (G-I) Mice received daily IP injections FTY720 or saline control starting at day 1 PT. n=9–11/group, 2 independent experiments. The number (C, n=6 mice/timepoint, 2 independent experiments) and CD49b and CD49a expression on accumulated Thy1.1+CD3+CD8+ cells (B, D, F; n=12–19 mice/timepoint, 4 independent experiments) was determined. Comparisons in (F) were done with an one-way ANOVA. Groups in all other panels were compared with a Welch’s corrected T-test. Bar graphs and error bars indicate mean +/− SD. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
These data did not exclude the possibility that OT-I cells upregulated CD49a elsewhere in the animal prior to infiltrating the tumor. Thus, we transferred activated CD49b SP OT-I cells into BRPKp110 tumor-bearing mice and treated them with FTY720 starting 1 day later. Despite reducing CD8+ T cells in blood (Supplementary Fig. S3A), FTY720 had no impact on OT-I TIL number (Supplementary Fig. 3B). FTY720 treatment also did not prevent the reduction in CD49b (Fig. 2G) or the induction of CD49a (Fig. 2H) expression on OT-I TILs, nor the proportions of SP and DP OT-I cells in tumors (Fig. 2I) or spleen (Supplementary Fig. S3C). Thus, CD49a was upregulated on CD49b SP cells after they enter tumors. Overall, our data indicate that activation-induced differentiation of CD8+ T cells was insufficient to induce CD49a and that it was dependent on antigen-independent environmental factors present in spleens of immunized mice and the TME.
Soluble factors in the TME, including IL12, upregulate CD49a expression on CD8+ T cells
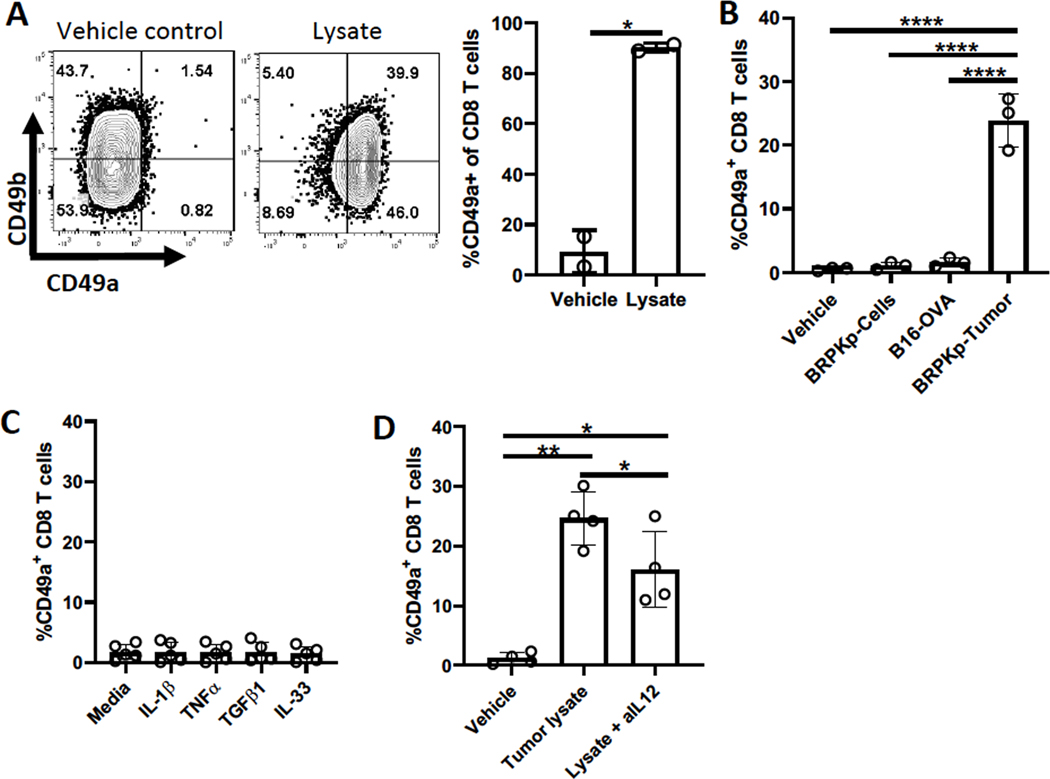
We hypothesized that factor(s) responsible for CD49a upregulation might be intrinsic to tumor cells. Thus, we cultured in vitro activated OT-I cells or bulk CD8+ T-cells with lysates from 28-day BRPKp110 tumors. The lysate upregulated CD49a on most cells within 24 hours, independent of CD49b status and without changing CD49b expression (Fig. 3A–B). However, activated CD8+ T cells cultured with lysate from 14-day B16-OVA tumors did not upregulate CD49a in 24 hours (Fig. 3B), consistent with the lower CD49a expression on CD8+ TILs from B16-OVA tumors (Fig. 1A). BRPKp110 tumor lysates contained high concentrations of several cytokines that were significantly (IL12/IL-23p40, IL33, TGFβ1, TGFβ2, and TGFβ3) or trending (TNFα and IL1β) higher than in B16-OVA lysates (Supplementary Table S2). No upregulation of CD49a was observed after the 24-hour culture of activated CD8+ T cells with recombinant IL33, TGFβ1, TNFα, or IL1β (Fig. 3C), although TGFβ1 diminished CD49b expression (Supplementary Fig. S4A). TGFβ1–3 blocking antibodies also did not alter CD49a upregulation by BRPKp110 tumor lysate in vitro (Supplementary Fig. S4A). However, IL12 blocking antibody significantly diminished induction of CD49a by BRPKp110 tumor lysate (Fig. 3D), suggesting that IL12, and potentially other unidentified factors, induced CD49a on CD8+ TILs in vivo.
Figure 3. Tumor-derived factor(s), including IL12, upregulate CD49a.
(A) Thy1.1+CD8+ OT-I cells were activated with CD3/CD28 beads for 48 hours in the presence of IL2 and IL7, cultured for 3 additional days with IL2 and IL7, and then for 24 hours with BRPKp110-derived tumor lysate. CD49a expression was determined by flow cytometry. (B-D) CD8+ splenocytes were activated and cultured as in (A), and then for 24 hours with (B) BRPKp110 culture lysate (BRPKp110-C), or lysates from B16-OVA or BRPKp110 tumors (BRPKp110-T); (C) the indicated cytokines; or (D) BRPKp110 tumor lysate with or without IL12 blocking antibody (aIL-12). Groups were compared with (A) a Welch’s corrected T-test or (B, D) one-way ANOVA. Bar graphs and error bars indicate mean +/− SD. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Antigen stimulation augments environmentally induced expression of CD49a
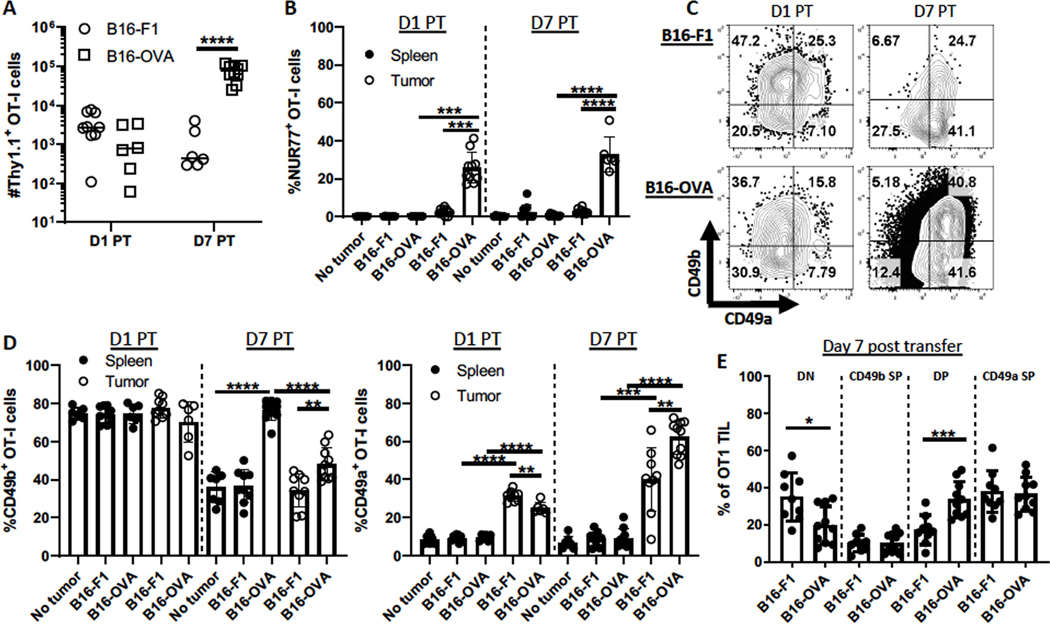
We next addressed the impact of TCR signaling on CD49a and CD49b expression, beyond the initial upregulation of CD49b. BRPKp110 expresses very low levels of MHC-I molecules, and BRPKp110 transfected with OVA is poorly recognized by OT-I cells. Activated CD49b SP OT-I cells from Nur77-GFP reporter mice were transferred into mice bearing B16-F1 melanomas, either non-transfected or transfected to express OVA (B16-OVA). The Nur77 reporter enabled measurement of TCR signaling. OT-I cells trafficked into both types of tumors, but their number increased substantially over 7 days when tumors expressed OVA (Fig. 4A). A significant fraction of B16-OVA OT-I TILs upregulated Nur77, indicating local antigen activation, whereas Nur77 was not expressed in OT-I cells in spleens of B16-OVA-bearing mice or in spleens and tumors of B16-F1-bearing mice (Fig. 4B). In B16-F1-bearing mice, OT-I cells lost CD49b in spleen and tumor comparably (Fig. 4C–D). Thus, in contrast to BRPKp110 (Fig. 2D), the B16-F1 TME did not support CD49b maintenance. In B16-OVA tumor-bearing mice, CD49b was maintained on OT-I cells in spleen and lost in the TME, although not to the same extent as in B16-F1 tumors (Fig. 4C–D). These results suggested that TCR stimulation not only upregulated CD49b, but also maintained its expression. However, other elements of the TME may diminish it. CD49a was upregulated on CD49b SP OT-I cells in B16 tumors but not spleens of tumor-bearing mice, independent of antigen (Fig. 4D–E), albeit to a lesser extent than in BRPKp110 tumors (Fig. 2D). However, the upregulation was significantly greater by day 7 in B16-OVA than B16-F1 tumors. Although it is possible that B16-OVA supported the selective proliferation of CD49a-expressing subpopulations, a more likely explanation is that antigen stimulation augmented the TME-dependent upregulation of CD49a expression.
Figure 4. Antigen engagement maintains CD49b and further upregulates CD49a.
Thy1.1+ OT-I Nur77-GFP splenocytes were transferred into C57BL/6 mice, which were subsequently immunized with OVA, polyIC, and anti-CD40. CD49b SP Thy1.1+ OT-I effectors were isolated and transferred into B16-F1 or B16-OVA tumor-bearing or tumor-free mice as in Figure 2A. Tumors and spleens were harvested 1 or 7 days post-transfer (PT). (A) The number of Thy1.1+ CD3+ CD8+ cells were quantitated, and (B) Nur77-GFP expression and (C-E) CD49b and CD49a expression was evaluated by flow cytometry. n=6–10 mice/group, 2 independent experiments. Groups were compared using Welch’s corrected T-tests. Bar graphs and error bars indicate mean +/− SD. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Expression of exhaustion markers is not linked to differential expression of CD49b and CD49a
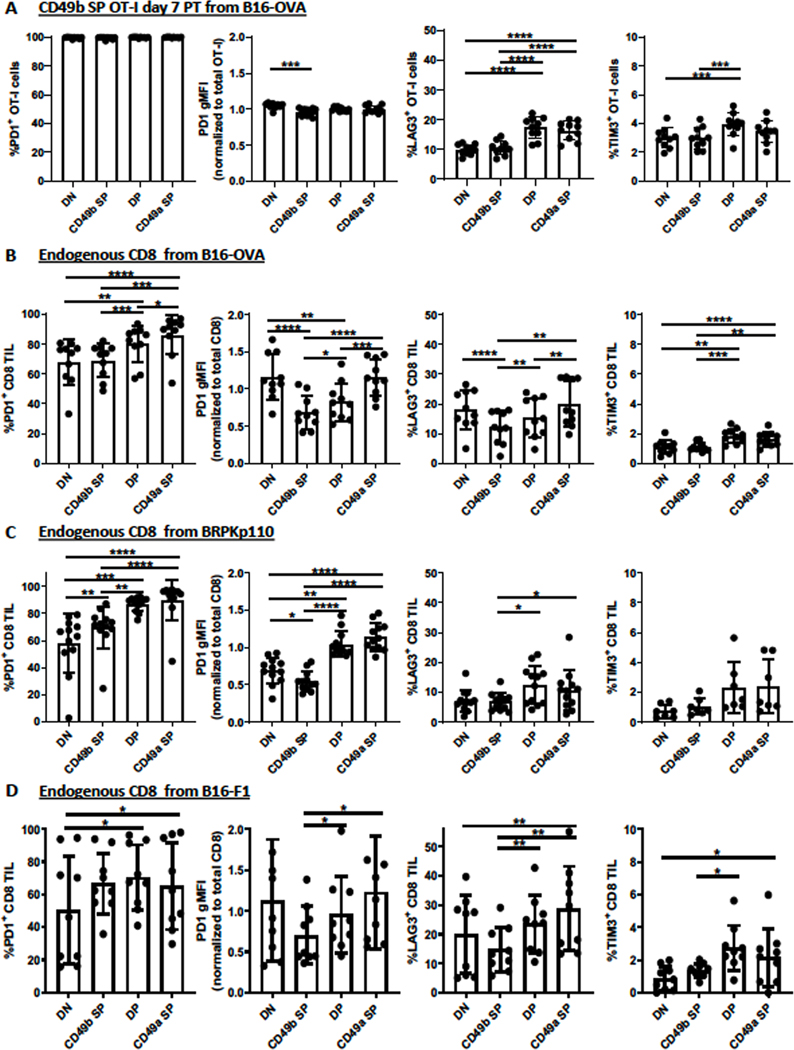
CD8+ TILs become exhausted over time due to chronic antigen stimulation (41). Because both the TME and antigen stimulation drove CD49a upregulation, we tested the hypothesis that CD49a expression and expression of exhaustion markers PD-1, LAG-3, and TIM-3 were associated. Due to the low numbers of transferred cells recoverable from BRPKp110 and B16-F1 tumors, it was not possible to evaluate this association in the absence of antigen. Thus, we evaluated integrin-expressing subpopulations in B16-OVA tumors after transfer of CD49b SP OT-I cells. At the time of transfer into tumor-free or B16-OVA-bearing mice, CD49b SP OT-I cells expressed PD-1 uniformly but had low expression of LAG-3 and TIM-3 (Supplementary Fig. S5). After 7 days, a fraction of OT-I cells in spleens of tumor-free mice had lost PD-1 expression, and the gMFI on the remaining PD-1+ cells was substantially lower (Supplementary Fig. S5A–B). Expression of LAG-3 and TIM-3 also decreased. Exhaustion markers on splenic OT-I cells after 7 days in B16-OVA tumor-bearing mice were similar (Supplementary Fig. S5B–C). In contrast, PD-1 expression was maintained on B16-OVA OT-I TILs, and the gMFI was substantially increased and LAG-3 and TIM-3 were also upregulated.
To test whether this exhaustion marker upregulation was associated with CD49a upregulation, we evaluated each integrin-expressing OT-I subpopulation 7 days post transfer. PD-1 was expressed on essentially all cells in all subpopulations, and the TME-induced upregulation of PD-1 gMFI was also consistent (Fig. 5A). The percentage of DP cells expressing LAG-3 and TIM-3 was low, but significantly higher than that of CD49b SP and DN cells, and no further increase in the CD49a SP population was seen (Fig. 5A). Similarly, when naïve OT-I cells were transferred prior to B16-OVA implantation and thus activated in vivo by tumor-derived OVA, these exhaustion markers were not differentially expressed among integrin-expressing subpopulations (Supplementary Fig. S6). Thus, upregulation of exhaustion markers on OT-I TILs in B16-OVA tumors was largely independent of CD49a expression.
Figure 5. Association of CD49a upregulation and gain of exhaustion markers on CD8+ TILs.
Experimental details were as in Figure 2A. (A) The indicated exhaustion markers were evaluated on either Thy1.1+ OT-I cells isolated from B16-OVA tumors seven days after transfer, or (B) endogenous Thy1.1neg CD3+ CD8+ cells isolated from the same B16-OVA (n=10, 2 independent experiments), (C) B16-F1 (n=9, 2 independent experiments), and (D) BRPKp110 (n=12, 4 independent experiments) tumors evaluated in Figure 4C–E and Figure 2D, F. Marker expression of subpopulations was compared with repeated-measures one-way ANOVA and Tukey’s multiple comparisons tests. Bar graphs and error bars indicate mean +/− SD. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
We also examined this association on endogenous CD8+ T cells from the same B16-OVA tumors, and endogenous CD8+ T cells from BRPKp110 and B16-F1 tumors. In contrast to the results above, the fractions of endogenous DP and CD49a SP subpopulations expressing PD-1, LAG-3, Tigit, and/or TIM-3 in B16-OVA and BRPKp110 tumors were significantly higher than those of the CD49b SP cells, as was the gMFI of PD-1 (Fig. 5B–C). These same trends were observed in B16-F1 tumors, although PD-1 expression did not differ (Fig. 5D). However, the CD49a SP subpopulation only significantly differed from the DP population in B16-OVA tumors, and only in relation to expression of PD-1 and LAG-3. Thus, there is an association between the upregulation of exhaustion markers and CD49a expression on endogenous CD8+ TILs. As described earlier (Fig. 1G), endogenous T cells were likely infiltrating into tumors continuously, in contrast to transferred OT-I cells. Endogenous CD49b SP cells were therefore “newer” on average, and for that reason, expressed fewer exhaustion markers. Thus, the association between integrin and exhaustion marker expression on endogenous CD8+ TILs was likely temporal but not mechanistically linked. This is consistent with the lack of a mechanistic linkage in adoptively transferred OT-I cells described above.
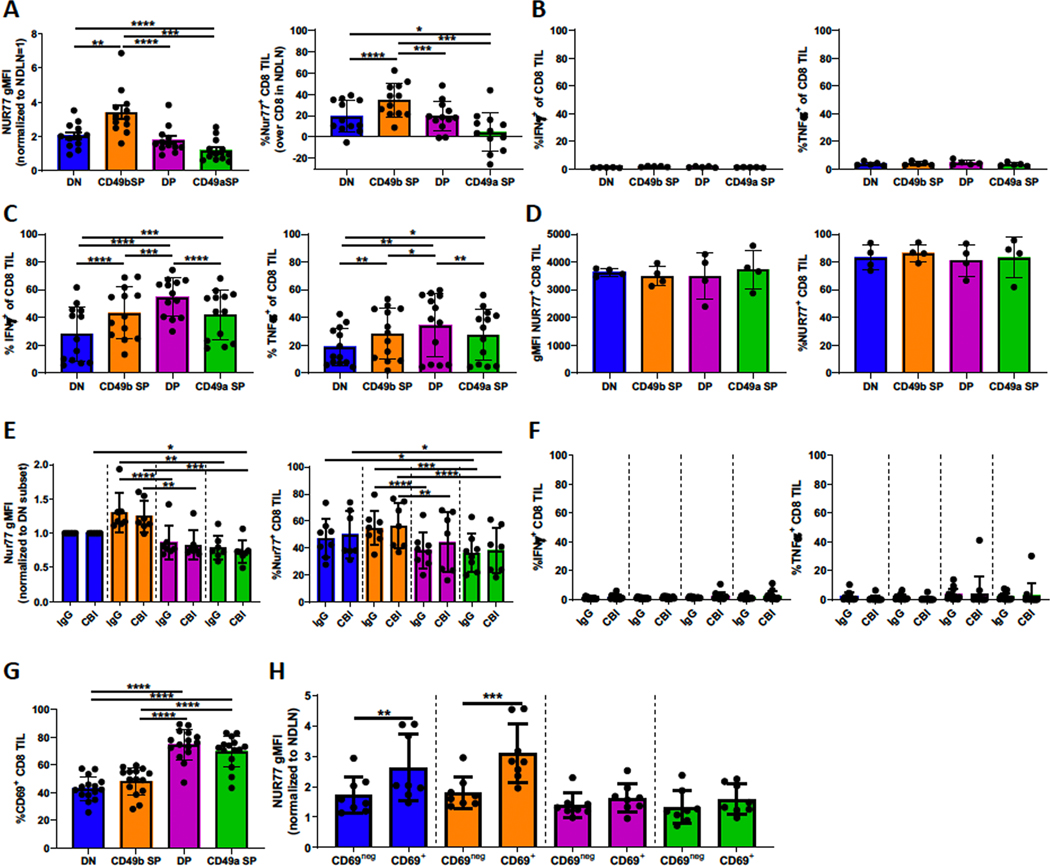
CD49b SP cells engage with antigen, but TCR signaling is low/absent in DP and CD49a SP cells
Despite the lack of a mechanistic linkage, we hypothesized that elevated expression of exhaustion markers on endogenous CD49a+ CD8+ T cells would render them less functional in vivo. To test this, BRPKp110 tumors were implanted into Nur77-GFP reporter mice. As hypothesized, DP and CD49a SP subpopulations showed significantly less TCR signaling than CD49b SP cells, based on Nur77 expression (Supplementary Fig. S7A). Nur77 expression in CD49a SP cells was not higher than baseline expression in CD8+ T cells from non-draining lymph nodes (Fig. 6A). Irrespective of CD49a or CD49b expression, no CD8+ TILs expressed IFNγ or TNFα directly ex vivo after in vivo brefeldin A treatment 4–6h prior to harvest (Fig. 6B). However, significant fractions expressed IFNγ, TNFα, and surface CD107a after in vitro restimulation for 4–6h with anti-CD3/CD28 (Fig. 6C, Supplementary Fig. S7B), demonstrating that they were active outside the TME. IFNγ, CD107a, and TNFα were expressed on larger fractions of DP cells than either SP subpopulation, and all subpopulations upregulated Nur77 equivalently upon re-stimulation for 12 hours with anti-CD3/CD28 in vitro (Fig. 6D). These data demonstrate that, although all subpopulations were suppressed downstream of Nur77 in vivo, CD49b SP cells in vivo responded to antigen based on Nur77 expression, whereas CD49a SP cells in vivo did not. This suggests either that these subpopulations were suppressed by different mechanisms or that CD49a SP T cells were not actively engaged with antigen in vivo.
Figure 6. CD49a-expressing subpopulations are not inhibited by PD-1, LAG-3, or TIM-3 and display a TRM-like phenotype.
(A, D, E, F, H) Nur77-GFP mice were implanted SC with BRPKp110. (A, D, H) Mice were either left untreated or (E, F) IP injected with a checkpoint blockade inhibitor cocktail 48 hours prior to harvest on day 14. (A, E, F, H, G) CD3+CD8+ cells were analyzed for Nur77-GFP, CD49a, CD49b and/or CD69 expression immediately or (D) after culture with CD3/CD28 beads for 12 hours. Nur77 gMFI was normalized to CD8+ T cells in non-draining lymph nodes (NDLN). %Nur77 on CD8+ T cells is after subtraction of values from NDLN in the same mice (Supplementary Fig. S7A). Intracellular IFNγ and TNFα expression was determined on (B, F) CD3+CD8+ cells immediately after harvest, 4–6 hours after IV injection with Brefeldin A, or (C) after culture with CD3/CD28 beads for 4–6 hours in presence of Brefeldin A. Marker expression of subpopulations was compared with repeated-measures one-way ANOVA and Tukey’s multiple comparisons tests (A-F). Differences between treatment groups within each subpopulation was tested with a Welch’s corrected T-test (E, F). CD69+ and CD69neg subsets within the integrin-expressing subpopulations were compared with a paired T-test (H). A, n=12, 3 independent experiments; B, n=5; C, n=13, 2 independent experiments; D, n=4; E/F, n=7/group, 2 independent experiments; G, n=15, 3 independent experiments; H, n=8/group, 2 independent experiments. Bar graphs and error bars indicate mean +/− SD. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
To test the hypothesis that inhibitory exhaustion pathways suppressed TCR signaling more in CD49a SP and DP TILs, we treated BRPKp110-bearing mice with a cocktail of checkpoint blockade inhibitors for 48 hours. At this timepoint, tumor weight, CD8+ T-cell infiltration, and CD49a and CD49b expression were unaltered (Supplementary Fig. S7C). Neither Nur77 (Fig. 6E) nor effector cytokine (Fig. 6F) expression was changed, indicating that diminished expression of Nur77 by CD49a SP and DP TILs was not due to inhibitory signaling via PD-1, LAG-3, or TIM-3. Extrinsic suppression mechanisms, such as myeloid-derived suppressor cells or regulatory T cells, could be responsible for this inhibition. However, the lack of inhibition of CD49b SP cells would demand that these integrin-expressing subpopulations be differentially localized in relation to these immunosuppressive cells or be differentially sensitive to their regulatory effectors.
CD49a-expressing subpopulations express elevated CD69 in an antigen-independent manner
CD49a has been described as a marker of TRM cells, together with PD-1 and CD69 (42,43). Consequently, we evaluated CD69 expression on subpopulations in conjunction with their expression of Nur77. All subpopulations expressed CD69, although the fraction was elevated in DP and CD49a SP subpopulations (Fig. 6G). In CD49b SP and DN subpopulations, CD69+ cells expressed elevated Nur77, tying CD69 expression to TCR stimulation (Fig. 6H). However, in DP and CD49a SP subpopulations, there was no difference in Nur77 expression between CD69+ and CD69neg cells. This is consistent with the antigen-independent expression of CD69 in TRM cells. It is also consistent with the hypothesis that CD49a SP and DP TIL do not upregulate Nur77 in vivo because they are not in contact with antigen-expressing cells.
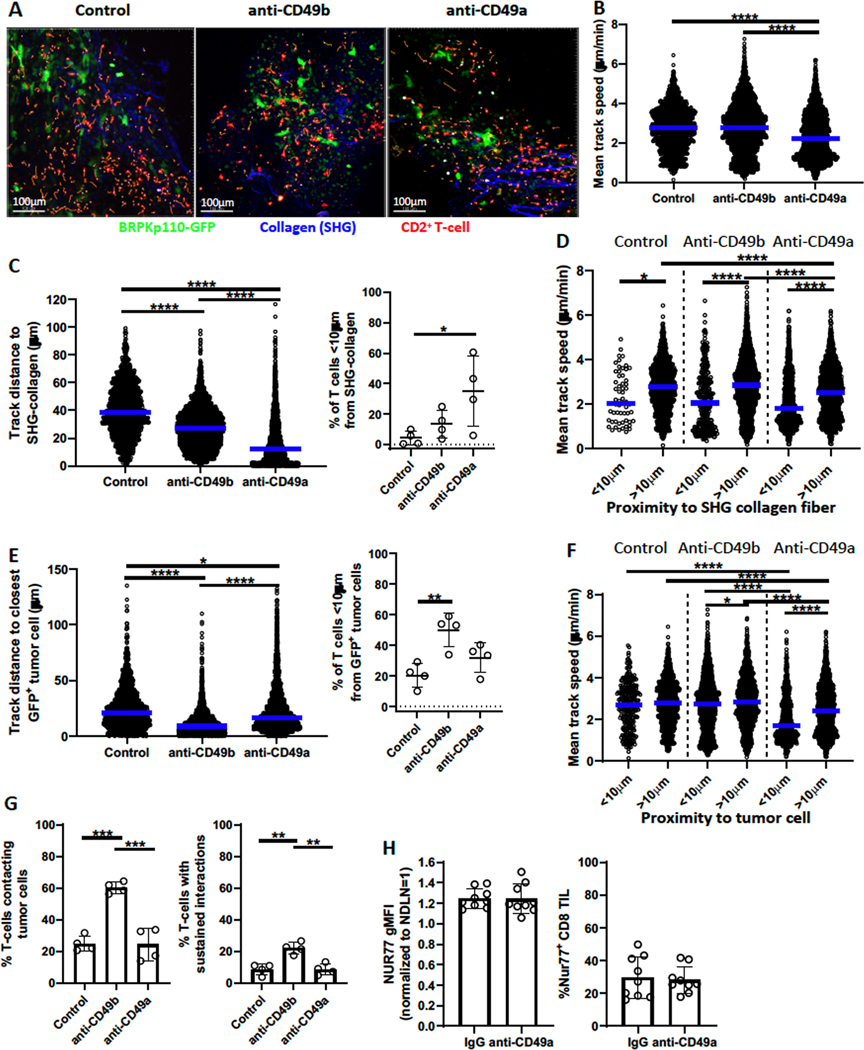
CD49a and CD49b ligation alters T-cell localization and interaction with tumor cells
Based on these results, we hypothesized that CD49a binding to collagen reduced the ability of CD8+ TILs to engage productively with antigen-expressing tumor cells, leading to diminished TCR stimulation. Conversely, CD49b interaction with collagen might enhance productive engagements, supporting TCR activation. Thus, we implanted GFP-expressing BRPKp110 cells into CD2-dsRed (labeling all T-cells) transgenic mice. Tumor cells, CD2+ T-cells, and collagen fibers (identified by second harmonic generation (SHG)) in 100–200μm thick live tumor slices were then imaged with 2-photon video microscopy (Fig. 7A; Supplementary Videos S1–S3). Separate slices from the same tumors were incubated with blocking antibodies against CD49a or CD49b, or control medium. For individual T cells, we quantitated motility (mean track speed) and time-averaged distances to collagen or tumor cells, as well as the number “contacting” (<1μm at 2 or more timepoints), or showing “sustained interactions” (<1μm for ≥10 minutes) with tumor cells.
Figure 7. CD49a affects localization and motility of T cells in tumors.
Day 21/22 BRPKp110-GFP tumors implanted SC into CD2-dsRed mice were cut into slices of 100–200 microns and incubated with blocking anti-CD49a or anti-CD49b for 2 hours or left untreated. Live video image acquisition and analysis of dsRed+ T cells, GFP+ BRPKp110 cells, and SHG collagen fibers is described in Methods and Results. (A) Composite image examples. Orange tracks represent dsRed+ T-cell movement for the preceding 10 minutes. (B) Mean track speed of each CD2+ T-cell was measured over 30 minutes. Numbers of CD2+ T-cells analyzed: 1311 (control), 2872 (anti-CD49b), and 2503 (anti-CD49a). (C) Left, Average distance of each T-cell track from panel B to the closest SHG collagen fiber over 30 minutes. Right, fraction of T-cells less than 10μm from the closest SHG collagen fiber. Each data point represents one of two analyzed fields in single tumor slices taken from one of two tumors. (D) Mean track speed of each T cell from (B) stratified by distance to the closest SHG collagen fiber. (E) Left, Average distance of each T-cell track from (B) to closest BRPKp110 cell over 30 minutes. Right, fraction of T-cells less than 10μm from closest BRPKp110 cell. Each data point represents one of two analyzed fields in single tumor slices taken from one of two tumors. (F) Mean track speed of each T-cell from (B), stratified by distance to BRPKp110 tumor cells. (G) Fraction of dsRed+ T cells localized <1μm from a BRPKp110 cell for 2 or more (left panel) and 10 or more (right panel) time points during a 30–32 time point observation period. Total number of CD2-dsRed T cells analyzed per group: 887 (control), 2868 (anti-CD49b), and 2714 (anti-CD49a). (H) Nur77-GFP reporter mice implanted SC with BRPKp110 (n=9, 2 independent experiments) were injected IV with anti-CD49a or control IgG 24 hours prior to tumor harvest on day 21. Nur77-GFP expression by CD3+CD8+ tumor-infiltrating T cells was analyzed. Groups were compared with an one-way ANOVA and Tukey’s multiple comparisons tests (A-G) or a Welch’s corrected T-test (H). Bar graphs and error bars indicate mean +/− SD. Blue lines indicate median. Factors of significance: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Under control conditions, T cells displayed a wide range of speeds, and relatively few were slow-moving (<1μm/min)(Fig. 7B; Supplementary Fig. S8A; Supplementary Videos S1–S3). Only 5% of T cells were in close proximity (<10μm) to a SHG-visualized collagen fiber (Fig. 7C). However, SHG enables visualization only of well-structured collagen fibers (44,45). Immunofluorescent staining of day 21 BRPKp110 sections confirmed a more pervasive presence of collagen type I and IV molecules (Supplementary Fig. S8B), suggesting that SHG underestimated the fraction of cells in close proximity to collagen. Nonetheless, cells in close proximity to visible collagen fibers moved significantly slower (Fig. 7D). About 22% of T cells were in close proximity (<10μm) to a tumor cell (Fig. 7E) but were not moving more slowly than T cells located further away (Fig. 7F). The fraction of T-cells “contacting” tumor cells was almost identical (Fig. 7G), indicating that most, if not all, T cells in close proximity to tumor cells also contacted them. However, the fraction of T cells that showed sustained interactions with tumor cells was lower (8.7%), consistent with low productive tumor cell engagement (46,47).
When CD49b was blocked, the fraction of cells in close proximity (<10μm) and contacting tumor cells increased by 150–200% compared to controls, while the fraction showing sustained interactions increased 240% (Fig. 7E, G). Despite this, the track speed of T cells in close proximity to tumor cells remained the same as that of T cells at a distance (Fig. 7F–G; Supplementary Fig. S8C). CD49b blockade also led to a significant reduction in the proximity of T cells to SHG-visualized collagen, although the fraction in close proximity (<10μm) only trended higher (Fig. 7C). Again, however, this was not associated with a significant decrease in mean track speed, overall or in relation to well-structured collagen fibers (Fig. 7B, D; Supplementary Fig. S8A). These results indicated that CD49b supports T cell localization away from tumor cells, and somewhat paradoxically, away from well-structured collagen fibers. It seems likely this reflects CD49b engagement with unstructured collagen.
When CD49a was blocked, a significantly larger fraction of T cells was located in close proximity (<10μm) to well-structured collagen fibers, and the average distance was significantly reduced (Fig. 7C). CD49a blockade also diminished the distance to tumor cells significantly but did not significantly increase the fraction of T cells contacting or showing sustained interactions (Fig. 7E, G). However, T cells in close proximity to tumor cells showed a significant reduction in motility relative to those at a distance, and also relative to T cells in close proximity to tumor cells in control tumors (Fig. 7F; Supplementary Fig. S8C). Although T cells in close proximity to well-structured collagen fibers also showed reduced motility relative to those at a distance, this was not significantly different from control or CD49b-blocked tumors (Fig. 7D). This decrease in motility was not due to a larger fraction of T cells in close proximity to collagen and tumor cells simultaneously (Supplementary Fig. S9A). Only T cells in close proximity to tumor cells alone, and not those in close proximity to both tumor cells and collagen fibers, showed significantly decreased speed when CD49a was blocked (Supplementary Fig. S9B). Together, these data demonstrate that CD49b and CD49a on T cells in tumors play distinct roles. Although both integrins promoted “distancing” of T cells from tumor cells, CD49b appeared to mediate a more pronounced re-localization of T cells, whereas CD49a promoted T-cell motility.
Because CD49a expression on TILs was associated with a lack of Nur77 upregulation, we tested the hypothesis that CD49a blockade would reverse this. We treated BRPKp110-bearing Nur77-GFP reporter mice with anti-CD49a 24 hours prior to tumor harvest. TILs from anti-CD49a-treated mice were not stained with fluorescent anti-CD49a, but the numbers of CD8+ T cells and the fraction expressing CD49b were unchanged, demonstrating that the in vivo blocking antibody was effective and did not kill CD49a+ cells (Supplementary Fig. S10). However, CD49a blockade did not lead to increased Nur77 expression (Fig. 7H). Thus, although CD49a blockade decreased the motility of T cells in close proximity to tumor cells, it did not increase sustained interactions, nor increase productive engagement of T cells with tumor antigen.
Discussion
The collagen-binding integrins CD49a and CD49b are considered to be involved in the retention of T cells in peripheral tissues, including tumors (29). Thus, their regulation and function on TILs may impact tumor control. Here, we found that CD8+ T cells expressed CD49b early after antigen activation and after tumor infiltration, gained CD49a, and lost CD49b. This was driven by antigen-independent elements in the TME, but antigen stimulation further enhanced CD49a expression. CD49a-expressing CD8+ TILs expressed higher levels of PD-1, LAG-3, TIM-3, and Tigit, but our work suggested that exhaustion markers and CD49a are associated temporally, not mechanistically. On the other hand, CD49a-expressing CD8+ TILs expressed CD69 in the absence of TCR signaling, whereas CD69 expression on CD49b populations was TCR signaling-associated. Co-expression of CD69 and CD49a is characteristic of TRM cells; thus, upregulation of CD49a may be associated with TILs that are not engaged with antigen. CD49a enhanced T-cell motility, especially in close proximity to tumor cells, suggesting that it may interfere with tumor cell recognition by distracting T cells from productive engagement, although we were not able to augment productive engagement by short-term CD49a blockade.CD49b also promoted relocalization of T cells at a greater distance from tumor cells. Thus, our results demonstrated that expression of these integrins impacts T-cell trafficking and localization in tumors via distinct mechanisms, and suggests a new way in which the TME, and likely collagen, could promote CD8+ TIL dysfunction.
CD49b is expressed on only a subset of specific effector CD8+ T cells in mouse models of arthritis and influenza or LCMV infection (17,19,25), and our work extends this to immunization with antigen in adjuvant. Thus, it is likely that a second signal or a specifically preprogrammed naïve T-cell subset is responsible for initial CD49b upregulation. We also showed that expression was maintained by elements in immunized mice and in the TME of BRPKp110, but not B16-F1, tumors. The B16-F1 TME may either lack these maintenance elements, or they may be antagonized by additional suppressive elements, such as TGFβ, which downregulated CD49b on activated OT-I cells in vitro. CD49b was maintained on OT-I cells in spleens of B16-OVA tumor bearing mice, and to a lesser extent on OT-I TILs, although this level was still higher than on TILs from B16-F1 tumors. We interpret this to suggest that TCR stimulation also promoted the expression of CD49b in both spleens and tumors of mice bearing B16-OVA, but continual stimulation or factors in the TME ultimately diminished expression.
We also established that CD49a was upregulated by TME factors that were elevated in BRPKp110 compared to B16 tumors, and present in lysates of BRPKp110 tumors but not cultured cells. It was also upregulated on activated OT-I cells in immunized mice, but not on activated CD49b+ OT-I cells transferred into naïve mice. Others have observed CD49a expression on influenza-specific T cells after localization in airways and lungs, and also on TRM T-cells but not circulating memory T-cells (17,27,28,48). In keeping with other work (27), we identified IL12 as a CD49a inducer in BRPKp110 tumor lysates, but could not confirm a role for TGFβ. IL12 expression by activated dendritic cells provides a potential explanation for CD49a upregulation on T cells in immunized mice, but not after adoptive transfer into naïve mice. However, it is important to identify sources of IL12 in peripheral tissues and the TME that promote CD49a expression, and whether other cytokines also play a role.
Adoptive transfer of CD49b SP cells demonstrated that CD49a upregulation and CD49b downregulation was a sequential process, but did not directly address the relationship between DP and CD49a SP cells. Although adoptive transfer of DP cells could provide more direct insight, the DP and CD49a SP populations obtainable after OT-I transfer and ovalbumin immunization in the absence of tumor are extremely small, with an unusually low expression of CD49a, and these experiments are not feasible. Our data is thus consistent with two models: one in which CD49b SP cells give rise to DP cells, which in turn give rise to CD49a SP cells; and one in which CD49b SP cells give rise to either DP or CD49a SP cells as independent lineages. We do not favor the latter model but cannot formally exclude it.
We observed that expression of CD69 on CD49b SP cells was strongly associated with TCR signaling, but its expression on CD49a+ cells was independent, and indeed, there was little evident TCR signaling in this population. Antigen-independent expression of CD69, together with PD-1, are cardinal markers of TRM cells (43), and others have shown that the environment plays a role in the generation of TRM cells in the skin, pointing to an antigen-independent regulation of CD49a (49,50). The overall upregulation of CD49a, CD69, and PD-1 could thus also be explained by a differentiation of CD49a+ CD8+ TILs towards a TRM-like phenotype.
We found a selective lack of TCR signaling in CD49a SP cells in the TME of BRPKp110 tumors, which was not restored by blockade of inhibitory pathways, and these cells were fully TCR responsive in vitro. This suggests that CD49a SP cells were not making productive contacts with antigen-expressing cells. Although we hypothesized that CD49a binding to collagens would trap T cells in areas well-separated from tumor cells, this instead appeared to be a property of CD49b. However, we found that CD49a increased motility, specifically of T cells in close proximity to tumor cells. CD49a binding to collagen also increased motility of T cells in vitro and in lung tissue (23,24). Blocking CD49a also caused substantial relocation of T cells into closer proximity to well-structured collagen fibers. This suggests that CD49a-driven motility was likely dependent on unorganized collagen molecules. Collagen alignment, length, width, density and straightness is frequently altered in tumors, although the level of disorganization is variable (51). Depending on the collagen state of the tumor, increased motility promoted by CD49a could distract T cells from engaging effectively with their antigen and thus point to a new mechanism of immune evasion.
Based on this model, we hypothesized that blocking CD49a in vivo would selectively enhance the functional activity of CD49a SP cells. However, short-term in vivo blockade of CD49a in our breast carcinoma model did not increase TCR signaling. An important consideration is that CD49a blockade promoted localization in close proximity to well-structured collagen fibers and did not lead to increases in T cell contacts or sustained interactions with tumor cells. It is possible that T cells may require longer term blockade of CD49a in vivo to regain function, or that their engagement with well-structured collagen may continue to interfere with productive tumor cell engagement. Our data suggest that further understanding the roles of CD49a and CD49b in tumors with various levels of collagen expression and organization may offer insights into the development of additional therapeutic strategies aimed at increasing the proximity and sustained interactions of T cells with tumor cells.
Supplementary Material
Synopsis:
A mechanism of CD8+ T-cell dysfunction is revealed. Collagen-binding CD49a and CD49b drive this dysfunction via modulation of CD8+ T-cell localization and motility, highlighting a new view on regulation of CD8+ T cells in the tumor microenvironment.
Acknowledgements
We thank the Rutkowski, Bullock, Slingluff and Engelhard laboratories for their constant input into this project, to Claire Rosean for sharing her knowledge regarding BRPKp110, and Andrew Dudley for help with tumor slicing.
Financial support: This work was supported by USPHS grants R01 CA197111 and R01 CA233716 (VHE); a Clinic and Laboratory Integration Program Award from the Cancer Research Institute (CLS); USPHS grant K22 NS104392 (UE); a Base Fund Award from the Oncode Institute (SvdB); and USPHS grant P30 CA044579 to the University of Virginia Cancer Center. RSL and ABR were supported by USPHS Training Grant T32 AI007496.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
References
- 1.Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 4.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–89. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson AR, Engelhard VH. CD8 T cells activated in distinct lymphoid organs differentially express adhesion proteins and coexpress multiple chemokine receptors. J Immunol. 2010;184:4079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods AN, Wilson AL, Srivinisan N, Zeng J, Dutta AB, Peske JD, et al. Differential Expression of Homing Receptor Ligands on Tumor-Associated Vasculature that Control CD8 Effector T-cell Entry. Cancer Immunol Res. 2017;5:1062–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debes GF, Arnold CN, Young AJ, Krautwald S, Lipp M, Hay JB, et al. Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues. Nat Immunol. 2005;6:889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown MN, Fintushel SR, Lee MH, Jennrich S, Geherin SA, Hay JB, et al. Chemoattractant receptors and lymphocyte egress from extralymphoid tissue: changing requirements during the course of inflammation. J Immunol. 2010;185:4873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matheu MP, Teijaro JR, Walsh KB, Greenberg ML, Marsolais D, Parker I, et al. Three phases of CD8 T cell response in the lung following H1N1 influenza infection and sphingosine 1 phosphate agonist therapy. PLoS ONE. 2013;8:e58033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda T, Egen JG, Lämmermann T, Kastenmüller W, Torabi-Parizi P, Germain RN. Tuning of Antigen Sensitivity by T Cell Receptor-Dependent Negative Feedback Controls T Cell Effector Function in Inflamed Tissues. Immunity. 2014;40:235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benechet AP, Menon M, Khanna KM. Visualizing T Cell Migration in situ. Front Immunol. 2014;5:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torcellan T, Hampton HR, Bailey J, Tomura M, Brink R, Chtanova T. In vivo photolabeling of tumor-infiltrating cells reveals highly regulated egress of T-cell subsets from tumors. Proc Natl Acad Sci U S A. 2017;114:5677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinosa-Carrasco G, Le Saout C, Fontanaud P, Michau A, Mollard P, Hernandez J, et al. Integrin β1 Optimizes Diabetogenic T Cell Migration and Function in the Pancreas. Front Immunol. 2018;9:1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steele MM, Churchill MJ, Breazeale AP, Lane RS, Nelson NA, Lund AW. Quantifying Leukocyte Egress via Lymphatic Vessels from Murine Skin and Tumors. J Vis Exp JoVE. 2019;143:e58704. [DOI] [PubMed] [Google Scholar]
- 15.Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean M-C, Validire P, Trautmann A, et al. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J Clin Invest. 2012;122:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bougherara H, Mansuet-Lupo A, Alifano M, Ngô C, Damotte D, Le Frère-Belda M-A, et al. Real-Time Imaging of Resident T Cells in Human Lung and Ovarian Carcinomas Reveals How Different Tumor Microenvironments Control T Lymphocyte Migration. Front Immunol. 2015;6:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter M, Ray SJ, Chapman TJ, Austin SJ, Rebhahn J, Mosmann TR, et al. Collagen distribution and expression of collagen-binding alpha1beta1 (VLA-1) and alpha2beta1 (VLA-2) integrins on CD4 and CD8 T cells during influenza infection. J Immunol. 2007;178:4506–16. [DOI] [PubMed] [Google Scholar]
- 18.Goldman R, Harvey J, Hogg N. VLA-2 is the integrin used as a collagen receptor by leukocytes. Eur J Immunol. 1992;22:1109–14. [DOI] [PubMed] [Google Scholar]
- 19.Fougerolles de AR, Sprague AG, Nickerson-Nutter CL, Chi-Rosso G, Rennert PD, Gardner H, et al. Regulation of inflammation by collagen-binding integrins α1β1 and α2β1 in models of hypersensitivity and arthritis. J Clin Invest. 2000;105:721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meharra EJ, Schön M, Hassett D, Parker C, Havran W, Gardner H. Reduced gut intraepithelial lymphocytes in VLA1 null mice. Cell Immunol. 2000;201:1–5. [DOI] [PubMed] [Google Scholar]
- 21.Sandoval F, Terme M, Nizard M, Badoual C, Bureau M-F, Freyburger L, et al. Mucosal Imprinting of Vaccine-Induced CD8+ T Cells Is Crucial to Inhibit the Growth of Mucosal Tumors. Sci Transl Med. 2013;5:172ra20–172ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter MV, Topham DJ. The α1β1 Integrin and TNF Receptor II Protect Airway CD8+ Effector T Cells from Apoptosis during Influenza Infection. J Immunol. 2007;179:5054–63. [DOI] [PubMed] [Google Scholar]
- 23.Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, et al. α1β1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13:836–42. [DOI] [PubMed] [Google Scholar]
- 24.Reilly EC, Emo KL, Buckley PM, Reilly NS, Smith I, Chaves FA, et al. TRM integrins CD103 and CD49a differentially support adherence and motility after resolution of influenza virus infection. Proc Natl Acad Sci U S A. 2020;117:12306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreasen SØ, Thomsen AR, Koteliansky VE, Novobrantseva TI, Sprague AG, Fougerolles de AR, et al. Expression and Functional Importance of Collagen-Binding Integrins, α1β1 and α2β1, on Virus-Activated T Cells. J Immunol. 2003;171:2804–11. [DOI] [PubMed] [Google Scholar]
- 26.Murray T, Fuertes Marraco SA, Baumgaertner P, Bordry N, Cagnon L, Donda A, et al. Very Late Antigen-1 Marks Functional Tumor-Resident CD8 T Cells and Correlates with Survival of Melanoma Patients. Front Immunol. 2016;7:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromley SK, Akbaba H, Mani V, Mora-Buch R, Chasse AY, Sama A, et al. CD49a Regulates Cutaneous Resident Memory CD8+ T Cell Persistence and Response. Cell Rep. 2020;32:108085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topham DJ, Reilly EC. Tissue-Resident Memory CD8+ T Cells: From Phenotype to Function. Front Immunol. 2018;9:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melssen MM, Olson W, Wages NA, Capaldo BJ, Mauldin IS, Mahmutovic A, et al. Formation and phenotypic characterization of CD49a, CD49b and CD103 expressing CD8 T cell populations in human metastatic melanoma. OncoImmunology. 2018;7:e1490855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang N, Bevan MJ. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, et al. Programs for the persistence, vigilance and control of human CD8 + lung-resident memory T cells. Nat Immunol. 2016;17:1467–78. [DOI] [PubMed] [Google Scholar]
- 32.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allegrezza MJ, Rutkowski MR, Stephen TL, Svoronos N, Perales-Puchalt A, Nguyen JM, et al. Trametinib Drives T-cell–Dependent Control of KRAS-Mutated Tumors by Inhibiting Pathological Myelopoiesis. Cancer Res. 2016;76:6253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allegrezza MJ, Rutkowski MR, Stephen TL, Svoronos N, Tesone AJ, Perales-Puchalt A, et al. IL15 Agonists Overcome the Immunosuppressive Effects of MEK Inhibitors. Cancer Res. 2016;76:2561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheen MR, Marotti JD, Allegrezza MJ, Rutkowski M, Conejo-Garcia JR, Fiering S. Constitutively activated PI3K accelerates tumor initiation and modifies histopathology of breast cancer. Oncogenesis. 2016;5:e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosean CB, Bostic RR, Ferey JCM, Feng T-Y, Azar FN, Tung KS, et al. Preexisting Commensal Dysbiosis Is a Host-Intrinsic Regulator of Tissue Inflammation and Tumor Cell Dissemination in Hormone Receptor–Positive Breast Cancer. Cancer Res. 2019;79:3662–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hargadon KM, Brinkman CC, Sheasley-O’Neill SL, Nichols LA, Bullock TNJ, Engelhard VH. Incomplete Differentiation of Antigen-Specific CD8 T Cells in Tumor-Draining Lymph Nodes. J Immunol. 2006;177:6081–90. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez AB, Peske JD, Engelhard VH. Identification and characterization of tertiary lymphoid structures in murine melanoma. Methods Mol Biol. 2018;1845:241–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullen A, Friedman RS, Krummel MF. Two-photon imaging of the immune system: a custom technology platform for high-speed, multicolor tissue imaging of immune responses. Curr Top Microbiol Immunol. 2009;334:1–29. [DOI] [PubMed] [Google Scholar]
- 40.Salerno EP, Olson WC, McSkimming C, Shea S, Slingluff CL Jr. T cells in the human metastatic melanoma microenvironment express site-specific homing receptors and retention integrins. Int J Cancer. 2014;134:563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, et al. CD8 T Cell Exhaustion in Chronic Infection and Cancer: Opportunities for Interventions. Annu Rev Med. 2018;69:301–18. [DOI] [PubMed] [Google Scholar]
- 42.Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Nadiarynkh O, Plotnikov S, Campagnola PJ. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat Protoc. 2012;7:654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostaço-Guidolin L, Rosin NL, Hackett T-L. Imaging Collagen in Scar Tissue: Developments in Second Harmonic Generation Microscopy for Biomedical Applications. Int J Mol Sci. 2017;18:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dustin ML, Bromley SK, Kan Z, Peterson DA, Unanue ER. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc Natl Acad Sci. 1997;94:3909–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman RS, Jacobelli J, Krummel MF. Mechanisms of T cell motility and arrest: Deciphering the relationship between intra- and extracellular determinants. Semin Immunol. 2005;17:387–99. [DOI] [PubMed] [Google Scholar]
- 48.Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, et al. CD49a Expression Defines Tissue-Resident CD8+ T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 2017;46:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon M-L, et al. The developmental pathway for CD103+ CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–301. [DOI] [PubMed] [Google Scholar]
- 50.Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med. 2015;21:1272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zunder SM, Gelderblom H, Tollenaar RA, Mesker WE. The significance of stromal collagen organization in cancer tissue: An in-depth discussion of literature. Crit Rev Oncol Hematol. 2020;151:102907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.