ABSTRACT
Antimalarials, in particular artemisinin-based combination therapies (ACTs), are critical tools in reducing the global burden of malaria, which is concentrated in sub-Saharan Africa. Performing and reporting antimalarial efficacy studies in a transparent and standardized fashion permit comparison of efficacy outcomes across countries and time periods. This systematic review summarizes study compliance with WHO laboratory and reporting guidance pertaining to antimalarial therapeutic efficacy studies and evaluates how well studies from sub-Saharan Africa adhered to these guidelines. We included all published studies (January 2020 or before) performed in sub-Saharan Africa where ACT efficacy for treatment of uncomplicated Plasmodium falciparum infection was reported. The primary outcome was a composite indicator for study methodology consistent with WHO guidelines for statistical analysis of corrected efficacy, defined as an article presenting a Kaplan–Meier survival analysis of corrected efficacy or reporting a per-protocol analysis where new infections were excluded from the numerator and denominator. Of 581 articles screened, we identified 279 for the review. Molecular correction was used in 83% (232/279) to distinguish new infections from recrudescences in subjects experiencing recurrent parasitemia. Only 45% (99/221) of articles with therapeutic efficacy as a primary outcome and performing molecular correction reported corrected efficacy outcomes calculated in a way consistent with WHO recommendations. These results indicate a widespread lack of compliance with WHO-recommended methods of analysis, which may result in biases in how antimalarial effectiveness is being measured and reported from sub-Saharan Africa.
INTRODUCTION
With each introduction of a novel Plasmodium falciparum antimalarial comes a corresponding report of resistance shortly thereafter.1 Spread of resistance is a major challenge facing malaria control. The global malaria control community recommends regular programmatic monitoring of therapeutic efficacy of antimalarials to mitigate the effects of emerging resistance on malaria morbidity and mortality.
Since 1965, the WHO has advocated use of a standardized protocol for in vivo monitoring of antimalarial efficacy, with major revisions in 20032 and 2009.3 Adherence to a common protocol allows standardization of reporting, the ability to compare efficacy data across countries and over time, and coordination of effective response strategies.
In vivo therapeutic efficacy studies (TESs) administer antimalarials to individuals with laboratory-confirmed, symptomatic malaria infection and follow them over the course of several weeks to assess treatment outcome. These efficacy outcomes may serve as early warnings of antimalarial resistance emergence. Efficacy is measured as the proportion of patients “cured” with an adequate clinical and parasitological response (ACPR), defined as clearance of initial parasitemia and maintenance of blood slide negativity over the course of follow-up. Following treatment, a study subject is still likely to be exposed to infectious mosquito bites, especially in high-transmission settings where the likelihood of reinfection during the follow-up period is high, posing the risk of acquiring a new infection during the period. Consequently, recurrent parasitemia during follow-up can be attributed to either a new infection or persistence of a strain from the original infection (recrudescence), with only the latter considered evidence of true drug failure (for a glossary of commonly used terms in antimalarial efficacy monitoring, see Table 1). Determining whether a new infection or recrudescence has occurred is complicated by the fact that in areas of high transmission, the typical setting of TESs in sub-Saharan Africa, individuals are often infected with more than one strain both at treatment and/or recurrence.4,5 Differentiating between new infection and recrudescence is critical in assessing an antimalarial’s true efficacy—its ability to clear the original malaria infection(s) in the patient at time of treatment.
Table 1.
Glossary of commonly used terms related to antimalarial efficacy monitoring
| Term | Related terms | Definition |
|---|---|---|
| TES | In vivo study, therapeutic efficacy trial, and antimalarial efficacy trial | Clinical outcome trial to assess antimalarial efficacy in patients with malaria |
| Recurrent parasitemia | Late recurrence and late treatment failure* | Recurrence of microscopy-detectable parasitemia during follow-up despite initial clearance, typically defined as occurring 7 days or longer after initiation of therapy |
| New infection | Reinfection | Recurrent parasitemia due to a new infection with a parasite strain/clone different from the strain/clone from the original infection |
| Recrudescence | True treatment failure | Recurrent parasitemia due to inadequate clearance of a parasite strain/clone from the original infection |
| Locus | Marker and gene | A polymorphic site or region in the parasite genome that can be used to differentiate between different parasite strains |
| Allele | Band and haplotype | A distinct variant of a given locus; even though Plasmodium is haploid in the human host, multiple alleles can be observed in a given sample due to a multi-clone infection (concurrent infection by more than one strain) |
| Molecular correction | PCR correction | Process by which cases of recurrent parasitemia are classified as new infections or recrudescences by comparison of parasite genotypes |
| PCR-corrected efficacy | Corrected efficacy | Primary indicator from TESs, where only recrudescent infections are counted as treatment failures and reinfections are excluded or censored |
TESs = therapeutic efficacy studies.
“Recurrent parasitemia” and “late recurrence” are preferable terms to “late treatment failure” because they unambiguously encompass both recrudescences and new infections.
The WHO has published guidance on laboratory methods to classify a study subject’s recurrent parasitemia as either a new infection or recrudescence6 and also has provided guidance on incorporating these outcomes into a calculation of final antimalarial efficacy.3 Classifying recurrent parasitemia as a new infection or recrudescence is called molecular correction because it involves molecular genotyping to compare parasite genotype(s) present before treatment (day zero [D0]) with genotype(s) present on the day of recurrent parasitemia (DX). Because many approaches exist to genotype parasites and compare those genotypes to decide whether they “match,” the WHO issued standardized guidance on genotyping parasites in the context of antimalarial efficacy trials following a 2007 technical meeting.6 The guidance discussed microsatellite- and single-nucleotide polymorphism (SNP)-based genotyping strategies but primarily focused on the fragment length polymorphic loci msp1, msp2, and glurp, which had emerged as the most widespread genotyping loci at that time.
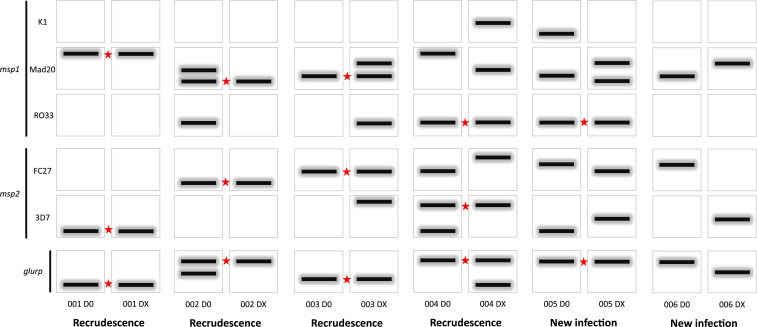
According to the WHO standard guidance, a recurrent parasitemia is only classified as a recrudescence if its DX sample matches at least one of the D0 sample’s alleles at every individual locus studied. This requirement for a match of at least one allele at each locus is decisive for calling the recurrent infection a recrudescence, irrespective of the loss or gain of additional alleles from D0 to DX. For the recurrent parasitemia to be classified as a new infection, all alleles for at least one locus at DX must be different from those present at D0. When using msp1, msp2, and/or glurp, a mixed infection (i.e., infection with more than one P. falciparum strain) may manifest as multiple bands on a gel or multiple peaks in capillary electrophoresis data. Furthermore, these multiple bands may represent different allelic families for msp1 and msp2, and a match at any allelic family for a locus is considered a match for that locus. For a recurrent parasitemia to be classified as a new infection, all alleles for at least one locus at DX must be different from those present at D0. These concepts are summarized in Figure 1 for the case of genotyping data with msp1, msp2, and glurp.
Figure 1.
Examples of msp1, msp2, and glurp electrophoretic band data from six pairs of day zero (D0) and day of failure (DX) samples. For clonal, single-strain infections, analysis is straightforward (participant 001 and 006). Analysis of multi-strain infections is more complicated and governed according to the 2007 WHO standard guidance. The presence of at least one shared allele (band) at any allelic family between D0 and DX samples at all genotyped loci is sufficient to provide evidence of recrudescence (participants 001–004, star denotes a shared allele). Notably, loss (participant 002), gain (participant 003), or both loss and gain (participant 004) of additional alleles is not evidence of new infection. Participant 005 would be classified as a new infection because there are no shared alleles at the msp2 locus at either allelic family. This figure appears in color at www.ajtmh.org.
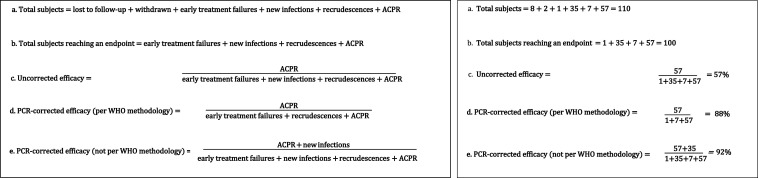
The second step of molecular correction is to calculate the corrected efficacy, often referred to as the PCR-corrected efficacy, to incorporate the molecular correction process which classifies recurrent parasitemias as either new infection or recrudescence. To calculate the PCR-corrected efficacy, which is the primary reportable outcome, the WHO recommends the use of the Kaplan–Meier survival analysis, which censors patients with new infections at the day of treatment failure. Although the Kaplan–Meier estimate is the preferred reportable indicator, the second best option is to report the per-protocol estimate of proportion ACPR at the final day of follow-up3,7 (Figure 2). Easier to estimate than the Kaplan–Meier estimator, and hence historically more widely used, this outcome is defined as ACPR/(ACPR + early treatment failure + recrudescence). Notably, WHO recommendations state that subjects with new infections should be removed entirely from the analysis (i.e., from both the numerator and denominator)3 and should not be grouped with subjects achieving ACPR (Supplemental Table S1).
Figure 2.
WHO classifications (left) for study participants in a therapeutic efficacy study, including the recommended approaches for calculating uncorrected and PCR-corrected efficacy (A–D). The final equation (E) is not a WHO-recommended approach but a common method observed in this systematic analysis. An example (right) using hypothetical data showing how WHO-endorsed calculation methods may yield an efficacy estimate with policy implications different from estimates obtained from calculations deviating from WHO-endorsed calculation methods (D vs. E; an efficacy threshold of < 90% is identified by the WHO as a trigger to reevaluate whether a drug should continue to be deployed as a first-line antimalarial). Hypothetical data used on the right: 110 subjects recruited, eight lost to follow-up, two withdrawn, one early treatment failure, 35 new infections, seven recrudescences, and 57 adequate clinical and parasitological response. ACPR = adequate clinical and parasitological response.
There has been no systematic evaluation of TES adherence to the WHO-recommended molecular correction laboratory methodology6 or the WHO-endorsed calculation methodology of the corrected efficacy.2,3 This systematic review will assess adherence to these two important methodological components of therapeutic efficacy monitoring in sub-Saharan African countries.
MATERIALS AND METHODS
We performed a systematic review of published studies from malaria-endemic countries in sub-Saharan Africa that reported artemisinin-based combination therapy (ACT) efficacy as an outcome. The purpose was to systematically characterize the methodologies used for 1) molecular genotyping of samples from participants with recurrent parasitemia, and 2) calculation of corrected efficacy.
Inclusion criteria.
We included all published studies (January 2020 or before) performed in sub-Saharan Africa where ACT efficacy for treatment of uncomplicated P. falciparum infection was a reported outcome. An ACT was defined as any artemisinin derivative plus partner drug taken orally, regardless of co-formulation or qualification status. Multicentric studies with any site(s) outside sub-Saharan Africa were excluded. Studies with non-efficacy primary outcomes (e.g., pharmacodynamics, safety, and gametocyte carriage) were included if they reported therapeutic efficacy as part of the report. Articles that compiled or reviewed results from already published studies were excluded, as were case reports, protocols, and nonhuman efficacy studies. There were no restrictions on language.
Search strategy and selection criteria.
We used PubMed to search for studies on antimalarial therapeutic efficacy, using keywords “antimalarial efficacy,” “ACT,” and “therapeutic efficacy.” This search was performed on May 17, 2020. In addition, the WorldWide Antimalarial Resistance Network publication library was searched for TESs conducted in sub-Saharan Africa.8 The U.S. President’s Malaria Initiative’s catalog of antimalarial TESs and bibliographies from the aforementioned articles were used to further identify studies.
Data collection.
A standardized data abstraction rubric was developed covering metadata, methods related to molecular correction of samples, and calculation of corrected efficacy. M. M. P. and E. S. H. identified possible studies to include in this review. M. M. P. and L. F. M. performed study abstractions, and discordances were resolved by consensus. Abstracted data were entered into an Excel spreadsheet. Study authors were not contacted.
Metadata.
Data on study country, year(s) of data collection, corresponding author contact, and journal were abstracted. Presence of a data availability statement was recorded, as was the inclusion of any genotyping data, either in the main text or supplemental information.
Methods for molecular genotyping of recurrent parasitemias.
Each included article was read to determine whether molecular correction had been performed to distinguish recrudescence from new infection in late recurrences. For articles attempting molecular correction, the methodology on how paired samples were genotyped and how these genotypes were compared was abstracted. Data on which loci were used, how fragment length was measured for fragment length polymorphic loci (e.g., msp1, msp2, glurp, or microsatellites), and whether a sequential algorithm approach was used or whether all loci were genotyped (for articles reporting the use of more than one locus) were abstracted.
Sequential approaches are sometimes applied because the 2007 WHO guidelines state that absence of a matching allele at any locus defines a new infection, and therefore there is no need to genotype any remaining loci. An example of a sequential genotyping approach is assaying all samples for the msp2 locus but only assaying samples for msp1 in those samples that showed evidence of recrudescence at msp2 (i.e., D0 and DX shared at least one allele at msp2). A systematic approach assays all of the loci for all samples.
For articles using fragment length polymorphic loci, the text was also analyzed to determine how D0 and DX genotypes were compared in the case of multiple alleles at a locus and categorized as consistent or inconsistent with the WHO methodology6 (a recrudescence requiring at least one allele match for all analyzed loci). Any indicator for which the text was ambiguous or missing was coded as unclear/not specified. The text and supplemental material were reviewed to determine whether genotyping data for recurrent parasitemia samples were reported, defined as fragment lengths for length polymorphic loci or bar codes for SNP-based genotyping methods.
Methods for statistical analysis of corrected efficacy.
The text and tables of each article were analyzed to characterize how the corrected efficacy outcome was calculated and reported. We determined whether a Kaplan–Meier survival analysis for the corrected efficacy was performed and reported. If a per-protocol analysis was reported, the methods section was analyzed to determine whether the original study authors had explicitly specified how new infections were categorized in the per-protocol analysis. The results table was then analyzed to verify whether new infections had been excluded from both the numerator and denominator (consistent with WHO guidelines2,3) or recoded as ACPR (inconsistent with WHO guidelines). This was performed by comparing the numerator and denominator of the uncorrected and corrected efficacy. If absolute numbers were not reported, the reported percentages were compared with back-calculated percentages under the two scenarios to ascertain how new infections had been categorized. If the numerator and denominator data were unavailable and the analysis of the percentages was ambiguous or not possible, the statistical methodology was coded as unclear.
A primary composite indicator for study methodology consistent with WHO guidelines for statistical analysis of corrected efficacy was created. This indicator was defined as an article presenting a Kaplan–Meier survival analysis of corrected efficacy or reporting a per-protocol analysis where new infections were excluded from both the numerator and denominator. For this indicator, studies that reported a Kaplan–Meier survival analysis of corrected efficacy but reported a per-protocol analysis where new infections were not excluded from both the numerator and denominator were classified as consistent with WHO guidelines.
Data analysis.
Frequencies for the abstracted parameters were calculated and reported. We used chi-square tests to quantify the association between a study methodology with the WHO guidelines2,3 regarding the per-protocol analysis. Studies were classified by the following criteria: study region; whether or not the methods section explicitly mentioned how new infections were categorized in the corrected efficacy calculations; whether or not the WHO protocol2,3 was cited or mentioned; and whether or not a Kaplan–Meier estimate was reported.
RESULTS
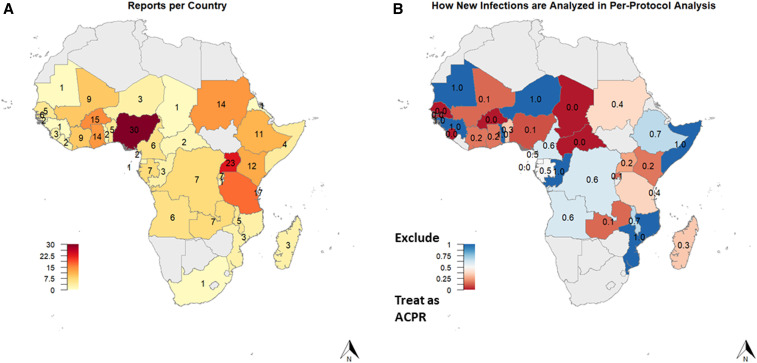
A total of 581 articles were screened, with 279 articles from 38 countries included in the analysis (Supplemental File S1). A PRISMA flow diagram detailing how articles were selected and excluded is provided in Supplemental Figure S1. Nearly all malaria-endemic sub-Saharan African countries were represented by at least one published report, but great variability in the number of reports per country existed across the continent (Figure 3A), ranging from 1 to 30. The median year of study end was 2008, ranging from 1996 (coinciding with the first published studies of ACTs in sub-Saharan Africa) to 2019.
Figure 3.
Number of therapeutic efficacy reports by country, excluding multicountry studies (A). Frequency of excluding new infections from the final analyses (rather than including them as adequate clinical and parasitological responses [ACPR]) in the per-protocol analysis by country (B).
A majority (232/279, 83%) of studies used molecular correction (Table 2). Nearly all (261/279, 94%) of the included articles listed therapeutic efficacy as a primary outcome. Four journals alone published 66% of all articles, with Malaria Journal accounting for 34% (96/279).
Table 2.
Summary characteristics of published therapeutic efficacy reports on artemisinin-based combination therapy from sub-Saharan Africa
| N | N | % | |
|---|---|---|---|
| Total reports included | 279 | – | – |
| Performed molecular correction | 232 | 279 | 83 |
| Main outcome | |||
| Therapeutic efficacy | 261 | 279 | 94 |
| Gametocyte carriage | 7 | 279 | 3 |
| Other | 11 | 279 | 4 |
| Single-country | 251 | 279 | 90 |
| Data availability statement | 22 | 279 | 8 |
| Journal | |||
| Malar J | 96 | 279 | 34 |
| Am J Trop Med Hyg | 27 | 279 | 10 |
| Trop Med Int Health | 18 | 279 | 6 |
| Trans R Soc Trop Med Hyg | 13 | 279 | 5 |
| Other | 125 | 279 | 45 |
N = number of articles assessed; n = number of articles with a finding.
Methods for molecular genotyping of recurrent parasitemia (Table 3).
Table 3.
Characteristics of molecular analysis reported by published therapeutic efficacy reports on artemisinin-based combination therapy from sub-Saharan Africa
| n | N | % | |
|---|---|---|---|
| Study shows genotyping data* | 6 | 232 | 3 |
| Study cites the 2007 WHO genotyping methodology† | 41 | 129 | 32 |
| Study explicitly states how samples with multiple alleles were analyzed | 97 | 232 | 42 |
| Consistent with the WHO methodology | 74 | 97 | 76 |
| Inconsistent with the WHO methodology | 14 | 97 | 14 |
| Unclear | 9 | 97 | 9 |
| Study specifies how fragment lengths were measured‡ | |||
| Capillary sequencing | 9 | 232 | 4 |
| Gel electrophoresis | 49 | 232 | 21 |
| Not specified | 174 | 232 | 75 |
| Study specifies whether all loci were analyzed or if sequential approach was used§ | |||
| All loci systematically assayed | 2 | 201 | 1 |
| Sequential | 23 | 201 | 11 |
| Not specified | 176 | 201 | 88 |
N = number of articles assessed; n = number of articles with a finding.
Does not include two additional studies with example, illustrative genotyping data.
Among articles conducted after 2009.
Among studies using fragment length polymorphic sites.
Among studies analyzing more than one locus.
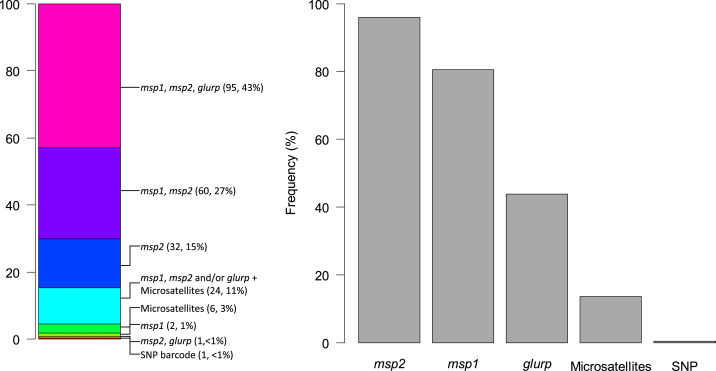
The most common set of loci, used in 43% (95/221) of evaluable studies, was msp1, msp2, and glurp (Figure 4A). msp2 was the single most frequently examined locus, reported to be used in 96% (212/221) of studies (Figure 4B). Only a handful of studies did not use any of the msp1, msp2, or glurp loci, all since 2013: six studies used only microsatellites and one used a SNP bar code.
Figure 4.
Distribution of molecular genotyping methodologies used by studies using molecular correction in sub-Saharan Africa (n = 221), by locus combination (A) and individual frequency for each locus (B). SNP = single-nucleotide polymorphism. This figure appears in color at www.ajtmh.org.
Fewer than half (97/232, 42%) of articles performing molecular correction explicitly described how cases of genotypes with multiple alleles were analyzed. Of these 97, 76% (74/97) described a protocol consistent with the WHO methodology,6 and 14% (14/97) performed the analysis of genotyping data in a way inconsistent with the WHO methodology, with the remainder lacking a clear description. Most of the 14 describing a manner inconsistent with the WHO methodology (11, 79%) were from studies conducted before 2009 (Supplemental Table S2).
Most studies using fragment length polymorphic loci (174/232, 75%) did not explicitly specify how fragment lengths were measured. An even larger proportion (176/201, 88%) did not specify whether loci were examined sequentially or whether all loci were genotyped.
Methods of calculation of corrected efficacy (Table 4).
Table 4.
Statistical methods used by therapeutic efficacy studies from sub-Saharan Africa for molecular correction
| n | N | % | |
|---|---|---|---|
| Study references WHO protocol for classification of treatment outcome | 233 | 279 | 84 |
| Explicitly cites 2003 protocol | 126 | 279 | 45 |
| Explicitly cites 2009 protocol | 80 | 279 | 29 |
| Study presents the Kaplan–Meier survival analysis for corrected efficacy* | 79 | 221 | 36 |
| Study presents per-protocol analysis for corrected efficacy* | 200 | 221 | 90 |
| Explicitly describes how new infections were considered in the per-protocol analysis† | 39 | 197 | 20 |
| How new infections were considered in per-protocol analysis, as verified retrospectively by abstractors† | |||
| Excludes new infections (consistent with WHO recommendations for per-protocol analysis) | 53 | 197 | 27 |
| New infections coded as adequate clinical and parasitological response | 118 | 197 | 60 |
| Does not provide clear enough description | 26 | 197 | 13 |
| Study presents analysis consistent with WHO recommendations (Kaplan–Meier or per-protocol excluding new infections) | 99 | 221 | 45 |
N = number of articles assessed; n = number of articles with a finding.
Among studies reporting therapeutic efficacy outcome with molecular correction.
Denominator excludes one study where definitions differed by site and two studies with no new infections.
Fewer than half (79/221, 36%) of studies with therapeutic efficacy as a primary outcome and performing molecular correction presented Kaplan–Meier estimates of corrected efficacy. By contrast, nearly all (200/221, 90%) of these studies reported a per-protocol analysis. Only a small proportion (39/197, 20%) explicitly described how new infections were categorized when calculating the per-protocol efficacy. When considering the verified retrospective review of the numerator/denominator/proportion data, approximately a quarter (53/197, 27%) could be verified as having per-protocol calculations consistent with the WHO guidelines (i.e., removing new infections from the calculations). More than half (118/197, 60%) were verified by abstractors as having categorized new infections as ACPR (i.e., treatment “successes”) in the calculation of the corrected efficacy (inconsistent with the WHO methodology), whereas the remaining studies (26/197, 13%) could not be assessed. Overall, only 45% (99/221) of studies with therapeutic efficacy as a primary outcome and performing molecular correction reported corrected efficacy results consistent with WHO recommendations.
There was heterogeneity in how new infections were categorized in the per-protocol analysis by country (Figure 3B), and regional differences reached statistical significance (P-value < 0.01) (Table 5). Studies that explicitly described how new infections were categorized in the per-protocol analysis were much more likely to exclude new infections rather than treat them as ACPR than studies which did not describe their calculation methods (P-value < 0.01). Similarly, studies performing a Kaplan–Meier analysis to calculate corrected efficacy were also more likely to exclude new infections rather than treat them as ACPR in the per-protocol analysis (34/57) than studies which did not perform a Kaplan–Meier analysis (19/140; P < 0.0001). Finally, studies that cited the WHO protocol were also more likely to exclude new infections rather than treat them as ACPR in the per-protocol analysis than studies which did not cite the WHO protocol. However, most of the studies citing the WHO protocol still categorized new infections as ACPR in the per-protocol analysis (Table 5).
Table 5.
Statistical methods for analysis of new infections in therapeutic efficacy studies from sub-Saharan Africa
| Exclude | Treat as ACPR | Unclear | ||||||
|---|---|---|---|---|---|---|---|---|
| N | n | % | n | % | n | % | P-value* | |
| Study region | ||||||||
| West Africa | 73 | 11 | 15 | 44 | 60 | 18 | 25 | 0.0003 |
| Central Africa | 32 | 16 | 50 | 14 | 44 | 2 | 6 | |
| East Africa | 53 | 19 | 36 | 31 | 58 | 3 | 6 | |
| Southern Africa | 17 | 7 | 41 | 10 | 59 | 0 | 0 | |
| Study explicitly describes statistical methods | ||||||||
| Yes | 38 | 20 | 53 | 17 | 45 | 1 | 3 | 0.0002 |
| No | 159 | 33 | 21 | 101 | 64 | 25 | 16 | |
| Study presents the Kaplan–Meier estimate | ||||||||
| Yes | 57 | 34 | 60 | 20 | 35 | 3 | 5 | < 0.0001 |
| No | 140 | 19 | 14 | 98 | 70 | 23 | 16 | |
| Study cites the WHO protocol | ||||||||
| Yes | 172 | 53 | 31 | 97 | 56 | 22 | 13 | 0.0049 |
| No | 25 | 0 | 0 | 21 | 84 | 4 | 16 | |
ACPR = adequate clinical and parasitological response; N = number of articles assessed; n = number of articles with a finding.
Difference between rates of studies excluding or reclassifying as ACPR, calculated using chi-squared test.
DISCUSSION
We have revealed notable variation in molecular correction practices in antimalarial TESs in sub-Saharan Africa, with studies often using approaches differing from WHO recommendations. Therapeutic efficacy studies for malaria drugs are sensitive to biases introduced during laboratory molecular correction and efficacy calculation. Alterations in methodology could lead to efficacious drugs being classified as failing or failing drugs being classified as efficacious. Of these two options, the latter is more probable and arguably more problematic. Classifying a new infection as a treatment “success” (i.e., ACPR), which was performed in 118 of 197 articles (with another 26 articles not providing enough information to determine), will always inflate a drug’s apparent efficacy. We found examples where an ACT’s apparent efficacy would fall below the WHO’s 90% threshold if it had been calculated according to WHO recommendations.9,10
We chose to focus our review solely on sub-Saharan Africa because, unlike Asia and Latin America, high levels of malaria transmission and endemicity are common across much of Africa, where 93% of the world’s cases occur.11 High transmission results in high rates of reinfection during study follow-up and a high multiplicity of infection. Therefore, molecular methods for genotyping recurrent parasitemia and calculation of corrected efficacy play an essential role in this region. The preponderance of P. falciparum cases occurs in Africa, so it is not surprising that well over half of the studies assessing ACT efficacy against P. falciparum in the last 5 years were conducted on the continent.8 Adhering to the highest standards of molecular correction and reporting remains particularly critical in sub-Saharan Africa, which is likely in the “calm before the storm” of impending antimalarial resistance.12
We assessed two main components of molecular correction: methods related to molecular correction in the laboratory and the calculation of the corrected efficacy outcome. For both, we observed two main findings: 1) inadequate description of methodology and data availability and 2) methodological approaches differing from WHO recommendations. Taken together, this combination makes comparison of efficacy results over time or between countries very challenging.
Most studies lacked adequate detail regarding the molecular genotyping methodologies; for example, only a small minority provided details on how length polymorphism was measured and whether a sequential genotyping of loci or systematic approach was used. Very few made genotyping data available, precluding readers from obtaining more than a superficial understanding of how genotypes were compared. The same lack of data availability precluded us from verifying how most of these studies analyzed their genotype data to obtain efficacy estimates. This problem in not unique to TESs. After the International Committee of Medical Journal Editors’ 2016 declaration for improved data availability and data sharing plans,13 most scientific journals still lacked an explicit data-sharing policy on their website.14 Of the 22 articles with data-sharing agreements in our analysis, all were published in 2016 or after, indicating the new policy may be working.
Notably, fewer than half of articles explicitly described how infections with multiple alleles were analyzed. Mixed strain infections are the norm rather than the exception in most of sub-Saharan Africa,15–19 and comparison of D0 and DX parasite genotypes in this context requires extra care and clear protocols. The current standard is to require the presence of at least one shared allele between D0 and DX at each molecular locus to classify a recurrent infection as recrudescence, yet we found multiple reports describing an approach that required all alleles to be identical in both samples to define recrudescence. Consequently, a sample would only be classified as recrudescence if 1) every single clone in the original infection survived therapy and recrudesced to detectable levels by recurrence, and 2) no new infections had become patent by recurrence. This stringent, and unrealistic, practice may lead to substantial underestimation of the true rate of recrudescence.
Our review also showed that many studies did not follow WHO recommendations in the description and calculation of the primary indicator, the PCR-corrected efficacy. The gold standard approach recommended by the WHO, the Kaplan–Meier estimator, is a largely unbiased estimator of the corrected efficacy, although more statistically complex “competing risk” methods can also be applied, especially in studies with high rates of missing genotyping data and high reinfection rates.7,20 Our analysis showed that only a minority of studies (36%) reported a Kaplan–Meier estimate of the corrected efficacy. Even when including studies that reported a per-protocol estimate where new infections were excluded from the numerator and denominator, in accordance with WHO guidelines, still fewer than half of the studies (45%) reported a primary outcome consistent with WHO recommendations. Worryingly, a majority of studies calculated the primary efficacy by treating new infections as treatment successes (i.e., ACPR). This inevitably results in underestimated treatment failure rates, potentially resulting in a failing drug being incorrectly classified as efficacious (Figure 2). Most of these studies did not adequately describe this important deviation from the WHO in vivo protocol, despite nominal adherence to the standard guidance. A reader would only be able to uncover the deviation through careful comparison of the numerator, denominator, and/or proportions data between the uncorrected and PCR-corrected efficacies.
Regardless of which loci are used, using molecular tools to classify recurrent parasitemias is challenging and can, depending on the setting, be biased either toward underestimation or overestimation of the true treatment efficacy.21 Allelic suppression, where minor-frequency alleles are not detected, is an important factor that can impact the sensitivity of detecting recrudescence, particularly in areas with high multiplicity of infection. With new approaches being proposed,22,23 including one attempting to quantify the uncertainty around the classification,24 and new laboratory techniques such as amplicon sequencing emerging, an update to the 2007 WHO genotyping guidance6 may be warranted. Such an update would also allow for clarification of commonly misapplied approaches, such as the calculation of PCR-corrected efficacy.
Although some of the issues we uncovered have been noted before,25 we believe that addressing them could be relatively straightforward. Investigators monitoring antimalarial efficacy are recommended to increase both transparency and adherence to global best practices. Investigators are urged to calculate the Kaplan–Meier estimate of corrected efficacy, for example by using the easy-to-use WHO spreadsheet tool (https://www.who.int/malaria/areas/drug_resistance/efficacy-monitoring-tools/en/). Investigators are encouraged to adequately describe the methodological approaches to molecular correction and to make available the data related to molecular correction either in the published report or publicly available supplements to the article, with the goal that a reader can reproduce the results. With the recent efforts to promote data availability, journals now routinely allow online-only Supplemental Material to be posted at no additional cost to authors. Making full genotyping data available, for example, as in the illustrative example shown in Table 6, will allow readers to understand how data were recorded, analyzed, and classified.
Table 6.
Sample genotyping data table for late recurrences from an antimalarial efficacy trial, showing band lengths, locus-by-locus comparisons, and final classification
| Sample ID | msp1 | msp2 | msp1 call | msp2 call | glurp call | Final classification | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day | K1 | Mad20 | RO33 | FC27 | 3D7 | Glurp | |||||
| 001 | 0 | – | 240 | – | – | 300 | 500 | R | R | R | R |
| 001 | 21 | – | 240 | – | – | 300 | 500 | ||||
| 002 | 0 | – | 210/190 | 215 | 200 | – | 900/1,000 | R | R | R | R |
| 002 | 21 | – | 190 | – | 200 | – | 1,000 | ||||
| 003 | 0 | – | 200 | – | 250 | – | 600 | R | R | R | R |
| 003 | 28 | – | 200/220 | 215 | 250 | 420 | 600 | ||||
| 004 | 0 | – | 240 | 215 | 250 | 300/400 | 1,000 | R | R | R | R |
| 004 | 21 | 210 | 210 | 215 | 350 | 400 | 500/1,000 | ||||
| 005 | 0 | 200 | 210 | 215 | 300 | 300 | 1,000 | R | NI | R | NI |
| 005 | 14 | – | 190/220 | 215 | 280 | 350 | 1,000 | ||||
| 006 | 0 | – | 210 | – | 300 | – | 1,000 | NI | NI | NI | NI |
| 006 | 28 | – | 220 | – | – | 350 | 900 | ||||
Samples correspond to band patterns shown in Figure 1. R = recrudescence; NI = new infection. Recrudescences were defined following the standard WHO protocol, where the presence of at least one shared allele (i.e., band) at all successfully genotyped loci was sufficient for evidence of recrudescence. Boldface indicates a match between the day 0 sample and the day of recurrent parasitemia sample.
The U.S. President’s Malaria Initiative funds a substantial proportion of TESs in sub-Saharan Africa and has pledged to adhere to a best-practice checklist for all future study reports published. This checklist appears in Table 7 and could be used with the more general Consolidated Standards of Reporting Trials (CONSORT) recommendations26 for reporting clinical trials to maximize transparency of published studies. One key item on this checklist is making all genotyping data publicly available. Journal editors and peer reviewers should similarly require greater data availability and better documentation of methods and results in antimalarial TES manuscripts. Investigators who had not previously adhered to WHO recommendations may change their analytical approach to be consistent with WHO methodology and recalculate PCR-corrected efficacy. This will invariably reduce their PCR-corrected efficacy results due purely to methodological reasons. In our view, such decreases do not warrant a revision of the WHO 90% threshold for reconsidering the continued use a first-line antimalarial. Many factors, clinical and operational, contributed to the choice of a 90% threshold. Rather, PCR-corrected efficacy trends should be interpreted in the context of updated analytical methods and, when possible, evaluated against this 90% benchmark. National malaria control programs, donors, and antimalarial resistance stakeholders are urged to interpret therapeutic efficacy data with a critical eye to the methods related to molecular correction and efficacy calculation, and to provide transparency in reporting, to ensure that future results are comparable, reliable, and informative.
Table 7.
A best-practice checklist identifying the minimum set of essential items to be included in articles reporting therapeutic efficacy of antimalarials for uncomplicated Plasmodium falciparum infection
| 1. Summary of study design and methods |
| 1.1 Summary |
| Dates and location(s) of study |
| Target sample size and rationale |
| 1.2 Antimalarial studied and dosing specifics |
| From which manufacturer |
| Whether the WHO supplied the medicine or if quality control was performed on the antimalarial(s) |
| Dosage details (e.g., in mg/kg) or given as tablet in age or weight bands; if the latter, details of bands |
| With or without food |
| Whether all doses were directly observed |
| How long subject was monitored after dosing |
| What was done in case of vomiting a dose |
| 1.3 Inclusion criteria |
| How malaria was diagnosed at the health facility |
| Age range |
| Fever specifics |
| How measured and defined (site: axillary, oral, etc; number, e.g., > 37.5°C) |
| Parasite density range required for study inclusion |
| Hemoglobin range |
| 1.4 Exclusion criteria |
| 1.5 Patient follow-up |
| Days participants followed up and in what approximate time windows |
| Treatment of patients in the case of early or late treatment failure |
| Laboratory tests performed at each follow-up visit |
| What else was done (e.g., clinical assessment) |
| 1.6 Definition of main outcome measures |
| Early and late failure definitions |
| Adequate clinical parasitological response definition |
| Whether new infections were eliminated from the per-protocol calculation |
| How loss to follow-up, protocol violation, indeterminate results, etc., were figured in per-protocol and Kaplan–Meier calculations |
| Kaplan–Meier methodology described |
| 1.7 Other |
| Data management: Which platform and any other data entry specifics such as double entry, data cleaning, and analysis |
| Human subjects review specifics |
| Consent/assent specifics |
| Funding source |
| Data availability statement |
| 2. Methods |
| 2.1 Microscopy |
| How were slides prepared (e.g., staining, thick, and thin) |
| How were slides read (e.g., how many readings, what defines a discrepancy, and what was done if there was a discrepancy) |
| How was parasite density calculated, including what parameter (e.g., white blood cells) was used and what assumption about density was assumed for that parameter |
| 2.2 Molecular correction (recrudescence vs new infection) description |
| Laboratory methods used to determine the presence of parasitemia in a late failure (e.g., microscopy and PCR) |
| Whether msp1/msp2/glurp, neutral microsatellites, or other markers were used |
| Which specific loci or microsatellites used |
| Whether background allele frequencies were obtained |
| Whether markers were assayed sequentially (e.g., msp1 and msp2 results used to determine whether glurp should be investigated) |
| Laboratory criteria/methods used to determine new infection vs recrudescence |
| Definition for new infection vs. recrudescence |
| Explicit mention of how recrudescence was defined when multiple alleles were present at day 0 or day of failure (i.e., explicitly state that presence of at least one shared allele was sufficient to define a match) |
| Range of fragment size that qualifies as a match for each marker |
| Cutoff settings for PCR artefacts and stutter peaks |
| Whether capillary electrophoresis by an automated sequencer or gels were used (e.g., to determine msp1 fragment length) |
| 3. Data and results |
| 3.1 Number enrolled, lost to follow-up, withdrawn, and protocol violations |
| Missing data and reason |
| 3.2 Participant composition by arm |
| Age, gender, initial parasite density, and initial hemoglobin |
| 3.3 Outcome by arm |
| Slide positivity on day 3 |
| Late treatment failures reported as clinical or parasitological |
| Late treatment failures reported as new infections or recrudescences |
| Adequate clinical and parasitological response |
| If day 42 results are provided (e.g., for dihydroartemisinin–piperaquine or artesunate–pyronaridine), then day 28 results are also reported |
| For per-protocol results, provide numerators and denominators, not just percentages |
| Kaplan–Meier estimates included |
| Two sets of results with confidence intervals, uncorrected, and PCR-corrected |
| Data collected at different sites reported by site and not just aggregated |
| Table or Supplemental Table of raw paired genetic data and classification for each late treatment failure |
Supplemental information, tables, and figure
ACKNOWLEDGMENTS
We thank Carol Sibley, Philippe Guerin, and Prabin Dahal at WWARN and Christian Nsanzabana at Swiss Tropical and Public Health Institute for sharing their curated lists of TESs and their review of this report.
Note: Supplemental information, tables, and figure appear at www.ajtmh.org.
REFERENCES
- 1.Blasco B, Leroy D, Fidock DA, 2017. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO , 2003. Assessment and Monitoring of Antimalarial Drug Efficacy for the Treatment of Uncomplicated Falciparum Malaria. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 3.WHO , 2009. Methods for Surveillance of Antimalarial Drug Efficacy. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 4.Abukari Z, Okonu R, Nyarko SB, Lo AC, Dieng CC, Salifu SP, Gyan BA, Lo E, Amoah LE, 2019. The diversity, multiplicity of infection and population structure of P. falciparum parasites circulating in asymptomatic carriers living in high and low malaria transmission settings of Ghana. Genes 10: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yakubu B, Longdet IY, Horsefield T, Davou DT, Obishakin E, 2019. High-complexity Plasmodium falciparum infections, north central Nigeria, 2015–2018. Emerg Infect Dis 25: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO , 2007. Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Genotyping to Identify Parasite Populations. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 7.Dahal P, et al. 2019. Competing risk events in antimalarial drug trials in uncomplicated Plasmodium falciparum malaria: a worldwide antimalarial resistance network individual participant data meta-analysis. Malar J 18: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takata J, Sondo P, Humphreys GS, Burrow R, Maguire B, Hossain MS, Das D, Commons RJ, Price RN, Guerin PJ, 2020. The worldwide antimalarial resistance network clinical trials publication library: a live, open-access database of Plasmodium treatment efficacy trials. Am J Trop Med Hyg 103: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siribié M, Diarra A, Tiono AB, Soulama I, Sirima SB, 2012. Efficacité de l’artéméther-luméfantrine dans le traitement du paludisme simple de l’enfant en milieu rural au Burkina Faso en 2009. Bull Soc Pathol Exot 105: 202–207. [DOI] [PubMed] [Google Scholar]
- 10.The PREGACT Study Group , 2016. Four artemisinin-based treatments in African pregnant women with malaria. N Engl J Med 374: 913–927. [DOI] [PubMed] [Google Scholar]
- 11.WHO . World Malaria Report 2019. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 12.Conrad MD, Rosenthal PJ, 2019. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis 19: e338–e351. [DOI] [PubMed] [Google Scholar]
- 13.Taichman DB, et al. 2016. Sharing clinical trial data — a proposal from the international committee of medical journal editors. N Engl J Med 374: 384–386. [DOI] [PubMed] [Google Scholar]
- 14.Siebert M, Gaba JF, Caquelin L, Gouraud H, Dupuy A, Moher D, Naudet F, 2020. Data-sharing recommendations in biomedical journals and randomised controlled trials: an audit of journals following the ICMJE recommendations. BMJ Open 10: e038887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Happi CT, Gbotosho GO, Sowunmi A, Falade CO, Akinboye DO, Gerena L, Kyle DE, Milhous W, Wirth DF, Oduola AMJ, 2004. Molecular analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. Am J Trop Med Hyg 70: 20–26. [PubMed] [Google Scholar]
- 16.Beck S, Mockenhaupt FP, Bienzle U, Eggelte TA, Thompson WN, Stark K, 2001. Multiplicity of Plasmodium falciparum infection in pregnancy. Am J Trop Med Hyg 65: 631–636. [DOI] [PubMed] [Google Scholar]
- 17.Mahdi Abdel Hamid M, Elamin AF, Albsheer MMA, Abdalla AAA, Mahgoub NS, Mustafa SO, Muneer MS, Amin M, 2016. Multiplicity of infection and genetic diversity of Plasmodium falciparum isolates from patients with uncomplicated and severe malaria in Gezira state, Sudan. Parasites Vectors 9: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiwuwa MS, Ribacke U, Moll K, Byarugaba J, Lundblom K, Färnert A, Fred K, Wahlgren M, 2013. Genetic diversity of Plasmodium falciparum infections in mild and severe malaria of children from Kampala, Uganda. Parasitol Res 112: 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldh M, et al. 2020. Multiplicity of asymptomatic Plasmodium falciparum infections and risk of clinical malaria: a systematic review and pooled analysis of individual participant data. J Infect Dis 221: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahal P, Simpson JA, Dorsey G, Guérin PJ, Price RN, Stepniewska K, 2017. Statistical methods to derive efficacy estimates of anti-malarials for uncomplicated Plasmodium falciparum malaria: pitfalls and challenges. Malar J 16: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juliano JJ, Gadalla N, Sutherland CJ, Meshnick SR, 2010. The perils of PCR: can we accurately ‘correct’ antimalarial trials? Trends Parasitol 26: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones S, Kay K, Hodel EM, Chy S, Mbituyumuremyi A, Uwimana A, Menard D, Felger I, Hastings I, 2019. Improving methods for analyzing antimalarial drug efficacy trials: molecular correction based on length-polymorphic markers msp-1, msp-2, and glurp. Antimicrob Agents Chemother 63: e00590–e00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felger I, Snounou G, Hastings I, Moehrle JJ, Beck HP, 2020. PCR correction strategies for malaria drug trials: updates and clarifications. Lancet Infect Dis 20: e20–e25. [DOI] [PubMed] [Google Scholar]
- 24.Plucinski MM, Morton L, Bushman M, Dimbu PR, Udhayakumar V, 2015. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob Agents Chemother 59: 6096–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins WJ, Greenhouse B, Rosenthal PJ, Dorsey G, 2006. The use of genotyping in antimalarial clinical trials: a systematic review of published studies from 1995–2005. Malar J 5: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anon . Consort - Downloads. Available at: http://www.consort-statement.org/downloads. Accessed October 17, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.